Abstract
We report a highly reproducible method to crystallize the RNA-dependent RNA polymerase (RdRp) domain of dengue virus serotype 3 (DENV-3), allowing structure refinement to a 1.79-Å resolution and revealing amino acids not seen previously. We also present a DENV-3 polymerase/inhibitor cocrystal structure at a 2.1-Å resolution. The inhibitor binds to the RdRp as a dimer and causes conformational changes in the protein. The improved crystallization conditions and new structural information should accelerate structure-based drug discovery.
TEXT
Many flaviviruses are significant human pathogens. However, no antiviral therapy is currently available for the treatment of flavivirus infections. The flavivirus RNA-dependent RNA polymerase (RdRp), located at the C-terminal two-thirds of nonstructural protein 5 (NS5), is an attractive target for antiviral development (1–3). Like other polymerases, the flavivirus RdRp adopts a right-hand configuration composed of the fingers, palm, and thumb subdomains (4–6). Unfortunately, because of their flexibility, several segments of the protein are missing in the current flavivirus RdRp unliganded crystal structures, including several loops at the interface between the fingers and thumb subdomains (5, 6). Moreover, crystals of the dengue virus serotype 3 (DENV-3) RdRp were obtained at 4°C following a tedious dehydration procedure (7), making high-throughput structure-based drug discovery very inconvenient. It is therefore critical to develop a robust crystallization protocol to allow structural determination of DENV RdRp in complex with small-molecule inhibitors. A simplified crystallization protocol will be invaluable for structure-based rational design of inhibitors of DENV RdRp.
Here we report such a procedure to grow crystals of DENV-3 RdRp at the temperature of 18°C that routinely diffract to a resolution higher than 2.0 Å. This allowed an improved refinement of the original RdRp structure (PDB code 2J7U [6]), revealing several amino acids hitherto not seen. Using these improved conditions for crystallization, we present the first cocrystal structure of a flavivirus RdRp with an inhibitor. This cocrystal structure shows that the inhibitor induces major conformational changes in the DENV-3 RdRp.
An improved protein purification protocol for the reproducible crystallization of DENV-3 RdRp.
The previous conditions used to grow DENV-3 RdRp crystals required a complicated dehydration protocol through gradual transfer of the crystals into increasing concentrations of polyethylene glycol (PEG) (7). This method was difficult to routinely reproduce because crystals were obtained only after a few weeks at 4°C. To improve crystallization, we established a new purification protocol of the DENV-3 RdRp that avoided the use of 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), EDTA, or β-mercaptoethanol. Instead, we included Tris (2-carboxyethyl)-phosphine (TCEP; Thermo Scientific) in the final RdRp solution to prevent protein oxidation. The addition of TCEP minimizes protein precipitation, leading to reproducible crystallization of various protein batches. Specifically, the DENV-3 RdRp domain, spanning residues 272 to 900 of NS5 (GenBank accession number AY662691), was cloned into pET15b and expressed as described previously (6, 7). The cell pellet was lysed by sonication in buffer A (20 mM Na HEPES at pH 7.0, 300 mM NaCl, 5 mM imidazole, and EDTA-free complete protease inhibitors [Roche]). The lysate was clarified by centrifugation at 100,000 × g for 1 h at 4°C. The supernatant was purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography by washing unbound protein with buffer A supplemented with 40 mM imidazole. The RdRp was eluted in a linear imidazole gradient ranging from 40 to 500 mM. For removing the N-terminal His tag, 500 U of thrombin (Sigma) was added to the pooled fractions containing the RdRp; the mixture was dialyzed overnight against buffer A supplemented with 5 mM TCEP. The RdRp was further purified by size exclusion chromatography using the same buffer. SDS-PAGE analysis of the resulting RdRp indicated a purity of >95% (Fig. 1A).
Fig 1.
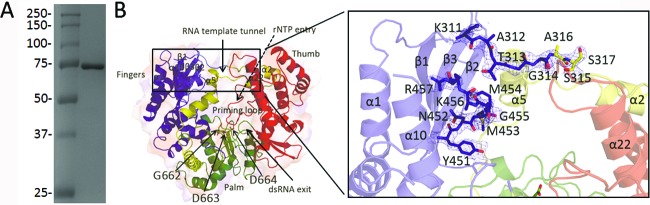
The free DENV-3 RdRp structure. (A) SDS-PAGE analysis of purified recombinant DENV-3 RdRp. Molecular mass standards are labeled. (B) “Front view” of the overall structure of the DENV-3 RdRp (PDB code 4HHJ; this work), showing tunnels for single-stranded RNA (ssRNA) template entry, rNTP entry (dashed arrow at the back of the RdRp), and exit of the dsRNA product (arrow at the front of the RdRp). The palm domain (green) contains the active-site residues (Asp663 and Asp664 from the GDD motif, represented as sticks), and the fingers and thumb domains are colored blue and red, respectively. The priming loop is colored red and is labeled, while the NLS region is colored yellow. The inset shows a magnified view of residues 311 to 317 and 451 to 457 that are now visible in the refined DENV-3 RdRp structure using the improved crystallization protocol. The 2Fo-Fc electron density map calculated with phases from the refined model is displayed at 1σ level.
Highly reproducible method to obtain RdRp crystals.
Crystallization was set up manually at 18°C using hanging-drop vapor diffusion. DENV-3 RdRp was concentrated to 7 to 10 mg/ml in 20 mM HEPES at pH 7.0, 300 mM NaCl, and 5 mM TCEP. One microliter of RdRp solution was mixed with 1 μl of precipitant (20 to 25% PEG 550 monomethyl ether and 0.1 M Tris-HCl, pH 8.0). Crystals of size 100 by 100 by 300 μm3 grew at 18°C over 2 to 4 days. For cryoprotection, crystals were transferred to the crystallization solution supplemented with 10% glycerol. Diffraction data, collected on beam-line X10SA (PXII) at the Swiss Light Source, were integrated using MOSFLM (8) and scaled using SCALA from the CCP4 suite (9). The structure was refined using REFMAC5 (9) starting from the deposited DENV-3 RdRp structure (PDB code 2J7U) (6). Data collection showed that the space group and diffraction quality of the crystals obtained at 18°C (Table 1) were comparable (isomorphous) to those of the crystals previously obtained at 4°C following the dehydration procedure (7).
Table 1.
Data collection and refinement statistics
| Statistic | Values for: |
|
|---|---|---|
| Free RdRp | RdRp-NITD107 complex | |
| Space group | C2221 | C2221 |
| Cell parameters (a, b, c) (Å) | 161.32, 177.61, 57.82 | 161.46, 177.73, 57.93 |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution range (Å) | 59.7–1.79 | 48.5–2.1 |
| No. of observed reflections | 440,985 | 250,191 |
| No. of unique reflectionsa | 78,264 (11,270) | 49,099 (7,072) |
| Completeness (%) | 99.9 (99.9) | 99.9 (99.9) |
| Multiplicity | 5.6 (5.1) | 5.1 (5.1) |
| Rmergeb | 0.063 (0.493) | 0.081 (0.661) |
| I/σ(I) | 14.5 (3.0) | 11.6 (2.4) |
| Solvent content (%) | 54.8 | 55.4 |
| No. of reflections: | ||
| Used for refinement | 78,166 (5,149) | 49,017 (3,430) |
| Used for Rfree calculation | 3,935 (294) | 2,482 |
| No. of nonhydrogen atoms | 5,528 | 5,342 |
| No. of water molecules | 665 | 432 |
| Average B factorc (Å2) (NITD107-1, NITD107-2) | 38.5 | 47.6 (77.1, 83.6) |
| Rfactord (%) | 17.7 (24.1) | 18.1 (27.3) |
| Rfreee (%) | 20.9 (25.4) | 22.4 (30.0) |
| Rms deviations from ideality | ||
| Bond length (Å) | 0.010 | 0.010 |
| Bond angle (°) | 1.00 | 1.06 |
| Residues in Ramanchandran plot | ||
| Residues in favored regions (%) | 97.9 | 97.9 |
| Ramachandran outliers (%) | 0.0 | 0.2 |
| PDB code | 4HHJ | 3VWS |
The numbers in parentheses refer to the last (highest) resolution shell.
Rmerge = ∑h∑i|Ihi − <Ih>|/∑h,i Ihi, where Ihi is the ith observation of the reflection h and <Ih> is its mean intensity.
The average temperature factors are given for each of the two bound NITD107 molecules. Their occupancy was set to 1.
Rfactor = ∑||Fobs| − |Fcalc||/∑ |Fobs|. Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated with 5% of reflections excluded from the refinement.
Refinement of the free RdRp structure reveals amino acids hitherto not seen.
Several segments that were hitherto disordered in the deposited flavivirus RdRp structure (6) are now visible in the electron density map of the free RdRp (Fig. 1B, inset). Loop 1 (residues 311 to 316) adopts an extended conformation that connects strand β1 from the finger subdomain to helix α2 attached to the thumb subdomain. Residues 451 to 455 are now well ordered: residues 451 to 453 form a short β-strand (named β1′), completing the β2-β3-β1 sheet in the fingers subdomain (Fig. 1B, inset). Nonetheless, three regions of the protein comprising a total of 41 residues remain poorly ordered in the refined model: residues 407 to 419 that form loop L3, residues 458 to 468 from the finger subdomain, and the C-terminal end spanning residues 884 to 900. Of note, the same amino acid segments are also disordered in an unrelated crystal form of a longer DENV-3 free RdRp construct with two molecules per asymmetric unit obtained in the space group P21212 (S. P. Lim, C. C. Seh, C. W. Liew, P.-Y. Shi, and J. Lescar, unpublished data), suggesting their intrinsic mobility in the absence of ligand regardless of detailed crystal-packing forces.
NITD107 inhibits DENV-4 RdRp.
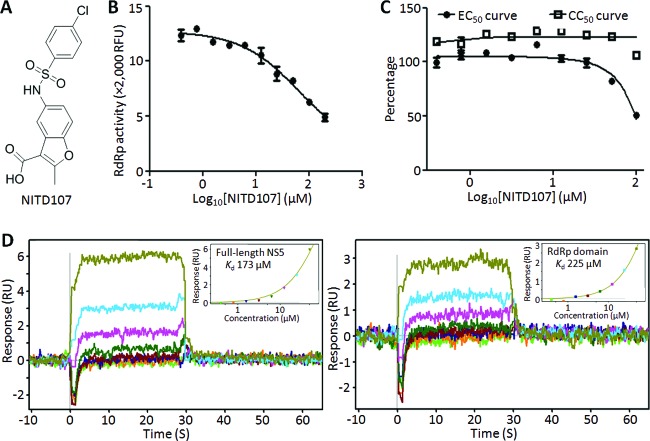
NITD107 (Fig. 2A) was identified through a high-throughput screen using a previously reported RNA elongation assay of DENV-4 RdRp (10). NITD107 was originally purchased from SPECS (catalog no. AQ-390/42861805). We used the RdRp from DENV-4 rather than the RdRps from other serotypes in the screening assay, because DENV-4 RdRp exhibits the highest enzymatic activity (data not shown). NITD107 inhibits the RdRp activity in a dose-responsive manner, with a 50% inhibitory concentration (IC50) of 113 μM (Fig. 2B). The compound also inhibits DENV-2 replication using a replicon assay (11), with a 50% effective concentration (EC50) of about 100 μM (Fig. 2C). Cell viability assays (11) showed that the antiviral activity was not due to compound-mediated cytotoxicity (Fig. 2C).
Fig 2.
NITD107 inhibits DENV RdRp. (A) Structure of NITD107. (B) Inhibition of RdRp activity. Two-fold serially diluted concentrations of NITD107 were incubated using a previously reported fluorescence-based RdRp assay (10). The inhibitory effect of NITD107 on RdRp activity was measured by the decrease in relative fluorescent units (RFU). An average result of three independent experiments is shown, with errors bars indicating standard deviations. (C) Antiviral activity in cell culture. The antiviral activity of NITD107 was assayed in a DENV-2 luciferase-reporting replicon assay (11). The cytotoxicity of the compound was measured using a cell proliferation-based Cell Counting Kit-8 by following the manufacturer's instructions (Dojindo Molecular Technologies, Rockville, MD). Both replicon activity and cell viability are presented as percentages; activities derived from samples with no compound treatment are set at 100%. Error bars indicate standard deviations (n = 4). (D) SPR measurements of the interaction between NITD107 and full-length NS5 or the RdRp domain of DENV-4. Biotinylated full-length NS5 or the RdRp domain was immobilized on the surface of a streptavidin chip. Three-fold serially diluted NITD107 (0.2 to 50 μM) was injected across the immobilized NS5/RdRp for 30 s (to allow compound-protein association), followed by injections of running buffer without compound for 1 min (to allow compound-protein dissociation). Insets show the plots of steady-state response (RU) against analyte concentration (μM). Binding affinities were calculated by fitting the data to the 1:1 steady-state model using BIAevaluation 4.1. The experiments were performed in triplicate; only one of the triplicates is shown for clarity.
NITD107 weakly but selectively binds to DENV RdRp.
We performed a surface plasmon resonance (SPR) analysis to demonstrate the binding of NITD107 to DENV RdRp. SPR measurements were done on a Biacore 3000 (Uppsala, Sweden). Streptavidin was immobilized on a CM5 sensor chip surface using amine coupling in 10 mM Na HEPES and 150 mM NaCl (pH 7.4). The surface was activated by a 15-min injection of N-hydroxysuccinimide (NHS)/ethyl(dimethylaminopropyl) carbodiimide (EDC), followed by injection of streptavidin in 10 mM acetate (pH 4.5) until the required density (8,000 resonance units [RU]) was achieved; the chip was then blocked by a 4-min ethanolamine injection at a flow rate of 10 μl/min at 25°C. The surface was primed with running buffer (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 0.05% Tween 20, 2 mM dithiothreitol [DTT], and 3% dimethyl sulfoxide [DMSO]), and the temperature was decreased to 4°C. The N-terminally biotinylated DENV-4 full-length NS5 protein (10) was captured at a density of ∼12,000 RU. The DENV-4 RdRp domain (residues 274 to 900 of NS5) was minimally biotinylated and captured at a density of ∼6,000 RU. Raw sensorgrams were reduced and solvent-corrected using a DMSO calibration curve and the Scubber software package (BioLogic Software, Campbell, Australia). Compound NITD107 was injected in running buffer using series of 3-fold dilutions starting at 50 μM (highest concentration) and at a flow rate of 30 μl/min. Association between DENV-3 NS5/RdRp and NITD107 was measured for 30 s, and dissociation was measured for 1 min. Binding affinities were evaluated by fitting the data to the 1:1 steady-state model using BIAevaluation 4.1. The SPR results showed that the compound directly bound to the full-length NS5 protein and to the NS5 RdRp domain with Kd (dissociation constant) estimates of 173 μM and 225 μM, respectively (Fig. 2D). As a negative control, using SPR analysis, NITD107 did not bind to the DENV-4 methyltransferase domain, encompassing the N-terminal 272 amino acids of the NS5 protein (Kd > 500 μM). Collectively, these results demonstrate that NITD107 weakly but selectively binds to the DENV RdRp domain.
Complex between DENV-3 RdRp and NITD107.
Since the molecular mass (366 Da) of NITD107 is relatively low, the compound may serve as a starting point for improvement using a fragment-based approach aided by X-ray crystallography. We therefore cocrystallized the DENV-3 RdRp with this inhibitor. To this end, we mixed at 4°C the DENV-3 RdRp in 20 mM HEPES at pH 7.0, 300 mM NaCl, and 5 mM TCEP with NITD107 (from a stock solution; 10 mM in 100% DMSO) to reach final concentrations of ∼130 μM (RdRp) and 1 mM NITD107, respectively. The DENV-3 RdRp-NITD107 mixture was immediately used for cocrystallization using the optimized conditions described above. Crystals were obtained on days 2 to 4. Diffraction data were collected at SLS, and the structure of the NITD107-RdRp complex was refined to a 2.1-Å resolution (Table 1).
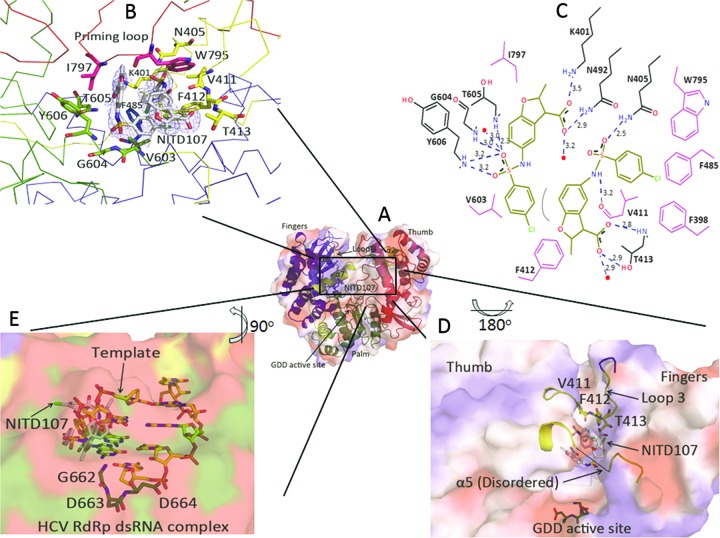
Figure 3A to C depicts the interactions between NITD107 and the DENV-3 RdRp. Two NITD107 molecules adjacent to each other are located in the RNA binding groove: a stacking interaction is established between the phenyl group of one NITD107 molecule and the indole ring of the other bound NITD107 molecule (Fig. 3B and C). NITD107 makes extensive contacts with loop 3 (residues 410 to 419), which becomes ordered upon compound binding (Fig. 3D). The sulfonamide and carboxylic groups of both molecules are engaged in several hydrogen bonds and salt bridges with residues Lys401, Thr605 (molecule NITD107-1), Asn405, and Thr413 (molecule NITD107-2) (Fig. 3C). The relatively low number of specific interactions observed in the cocrystal structure is consistent with the weak inhibitory and binding activities of this inhibitor toward RdRp outlined above. A superposition of the NITD107-RdRp complex with the RdRp from hepatitis C virus (HCV) bound to double-stranded RNA (dsRNA) (PDB code 4E7A) (12) shows that NITD107 would provoke steric hindrance with the template strand of a RNA duplex (Fig. 3E), suggesting a mode of inhibition via competition with the RNA substrate.
Fig 3.
Interactions between DENV-3 RdRp and the compound NITD107. (A) Overall view of the DENV-3 RdRp with the same orientation and color coding as in Fig. 1. The active site where compound NITD107 binds is boxed. (B) Detailed view of the NITD107 binding site, with residues from DENV-3 RdRp participating in the interaction, with the NITD107 dimer represented by sticks and colored using the same color coding as in Fig. 1. A difference electron density map at a level of 3σ with the coefficients Fo-Fc, in which atoms from the ligands were omitted from the phase calculation, is overlaid. (C) Schematic view of the atomic interactions between the DENV-3 RdRp and the NITD107 dimer. Interatomic distances between 2.5 to 3.2 Å are depicted. (D) The main conformational changes in the RdRp (in surface representation) induced by NITD107 binding are (i) the stabilization of loop 3 and a helical turn (yellow sticks and ribbon), which come in contact with the compound (represented by sticks and labeled by an arrow) and (ii) that helix α5 becomes disordered (represented by a dashed line). Residues from the active site (GDD motif) are represented by green sticks. (E) Superposition of the NITD107-RdRp complex (this work) with the RdRp from HCV bound to dsRNA (PDB code 4E7A), showing that NITD107 (gray sticks, arrow) would provoke steric hindrance with the template strand of the RNA duplex, shown by yellow sticks.
Conformational changes in RdRp induced by NITD107 binding.
NITD107 binding to RdRp causes several distinct conformational changes in the protein (Fig. 3D). Binding of the compound stabilizes an extra turn of helix α7, and loop 3, which is disordered in the free protein, becomes ordered upon the formation of several interactions with the ligand via residues Val411, Phe412, and Thr413 (Fig. 3C and D). In addition, compound binding is accompanied by an order to disorder transition of helix α5 that is more than 10 Å away from the ligand, near the ribonucleotide triphosphate (rNTP) entry tunnel. These results indicate that loop 3 and helix α5 are two hotspots of conformational flexibility of the flavivirus RdRp. Overall, the conformation of the protein bound to NITD107 remains closed. Concerted movements of the thumb, the priming loop, and loop 3 (which obstructs the path of the primer and template strands) would be required to create enough space to accommodate an RNA duplex.
Implications for drug discovery.
Viral polymerases are a proven target for effective antiviral therapy. The lack of a robust crystallization method for DENV RdRp has so far hampered efforts to develop antivirals against this important human pathogen. In this short communication, we have addressed this issue by achieving two specific aims. The first aim was to develop a highly reproducible crystallization method that enables determination of the high-resolution structure of DENV RdRp. The previous protocol requires crystallization at 4°C for a few weeks and a complicated dehydration procedure (7). In contrast, the new protocol yielded crystals at 18°C in 2 to 4 days without the tedious dehydration procedure. The second aim was to demonstrate that this improved protocol can be used to determine the cocrystal structure of the RdRp bound to a small-molecule inhibitor. As a proof of concept, we determined the cocrystal structure of the NITD107-RdRp complex at a 2.1-Å resolution. This strategy can now be used to analyze other inhibitors of DENV RdRp and also for fragment screening using X-ray crystallography in order to identify chemical starting points for drug discovery.
NITD107 exhibited weak antiviral activities against DENV RdRp enzyme and replicon. The compound also selectively binds to the full-length NS5 from DENV and its RdRp domain but not to the MTase domain of NS5. The cocrystal structure shows that two molecules of NITD107 bind to the RNA binding groove of the polymerase. This mode of action is similar to that of an N-sulfonyl-anthranilic acid derivative that was reported to inhibit DENV RdRp (IC50 of 0.7 μM) through binding to the RNA template tunnel of the polymerase (13). Since the two NITD107 molecules are located adjacent to each other, linking the two molecules to form an inhibitor of higher affinity is a strategy worth pursuing. As mentioned above, during the initiation of RNA polymerization, the polymerase needs to switch from a closed conformation to an open conformation in order to accommodate the viral RNA template. The binding of inhibitors, such as NITD107 or its derivatives, may potentially lock the polymerase in the closed conformation and, thus, prevent viral RNA synthesis.
Protein structure accession numbers.
The coordinates have been deposited in the Protein Data Bank with access codes 4HHJ and 3VWS.
ACKNOWLEDGMENTS
We thank colleagues at Novartis Institute for Tropical Diseases for technical help and scientific discussions during the course of this work.
This research was supported by BMRC grant 0912219/599 to the J.L. lab.
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926–39937 [DOI] [PubMed] [Google Scholar]
- 2. Davidson AD. 2009. New insights into flavivirus nonstructural protein 5. Adv. Virus Res. 74:41–101 [DOI] [PubMed] [Google Scholar]
- 3. Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff MP, Canard B. 2006. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351:145–158 [DOI] [PubMed] [Google Scholar]
- 4. Lescar J, Canard B. 2009. RNA-dependent RNA polymerases from flaviviruses and picornaviridae. Curr. Opin. Struct. Biol. 19:759–767 [DOI] [PubMed] [Google Scholar]
- 5. Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B. 2007. Crystal structure of the RNA polymerase domain of the West Nile virus nonstructural protein 5. J. Biol. Chem. 282:10678–10689 [DOI] [PubMed] [Google Scholar]
- 6. Yap TL, Xu T, Chen YL, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. 2007. The crystal structure of the dengue virus RNA-dependent RNA polymerase at 1.85-angstrom resolution. J. Virol. 81:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yap TL, Chen YL, Xu T, Wen D, Vasudevan SG, Lescar J. 2007. A multi-step strategy to obtain crystals of the Dengue virus RNA-dependent RNA polymerase that diffract to high resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leslie AGW. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4-ESF-EAMCB newsletter on protein crystallography, no. 26. [Google Scholar]
- 9. Collaborative Computational Project, number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50(Part 5):760–763 [DOI] [PubMed] [Google Scholar]
- 10. Niyomrattanakit P, Abas SN, Lim CC, Beer D, Shi PY, Chen YL. 2011. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 16(2):201–210 [DOI] [PubMed] [Google Scholar]
- 11. Ng CY, Gu F, Phong WY, Chen YL, Lim SP, Davidson A, Vasudevan SG. 2007. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res. 76(3):222–231 [DOI] [PubMed] [Google Scholar]
- 12. Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. 2012. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 86:6503–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niyomrattanakit P, Chen Y-L, Dong H, Yin Z, Qing M, Glickman JF, Lin K, Mueller D, Voshol H, Lim JYH, Nilar S, Keller TH, Shi P-Y. 2010. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84:5678–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]