Abstract
Myxoma virus (MYXV) provides an important model for investigating host-pathogen interactions. Recent studies have also highlighted how mutations in transformed human cells can expand the host range of this rabbit virus. Although virus growth depends upon interactions between virus and host proteins, the nature of these interactions is poorly understood. To address this matter, we performed small interfering RNA (siRNA) screens for genes affecting MYXV growth in human MDA-MB-231 cells. By using siRNAs targeting the whole human genome (21,585 genes), a subset of human phosphatases and kinases (986 genes), and also a custom siRNA library targeting selected statistically significant genes (“hits”) and nonsignificant genes (“nonhits”) of the whole human genome screens (88 genes), we identified 711 siRNA pools that promoted MYXV growth and 333 that were inhibitory. Another 32 siRNA pools (mostly targeting the proteasome) were toxic. The overall overlap in the results was about 25% for the hits and 75% for the nonhits. These pro- and antiviral genes can be clustered into pathways and related groups, including well-established inflammatory and mitogen-activated protein kinase pathways, as well as clusters relating to β-catenin and the Wnt signaling cascade, the cell cycle, and cellular metabolism. The validity of a subset of these hits was independently confirmed. For example, treating cells with siRNAs that might stabilize cells in G1, or inhibit passage into S phase, stimulated MYXV growth, and these effects were reproduced by trapping cells at the G1/S boundary with an inhibitor of cyclin-dependent kinases 4/6. By using 2-deoxy-d-glucose and plasmids carrying the gene for phosphofructokinase, we also confirmed that infection is favored by aerobic glycolytic metabolism. These studies provide insights into how the growth state and structure of cells affect MYXV growth and how these factors might be manipulated to advantage in oncolytic virus therapy.
INTRODUCTION
Myxoma virus (MYXV) is the prototypic member of the Leporipoxvirus genus of chordopoxviruses and causes the disease myxomatosis in European (Oryctolagus spp.) rabbits. The virus was introduced into Australia in the 1940s in an attempt to control feral rabbit populations, and subsequent field and laboratory investigations have provided the foundations of our understanding of host-pathogen coevolution in the natural environment (1–3). Myxomatosis has also provided an important model for investigating molecular mechanisms of viral pathogenesis, and its study has provided key insights into how large DNA viruses can manipulate the host to avoid immune surveillance. MYXV is now known to encode many proteins that interfere in processes broadly related to innate and adaptive immune defenses and which, when deleted or mutated, dramatically reduce virus virulence. Examples include proteins that bind to cytokines and chemokines, proteins that inhibit apoptotic and inflammatory signaling networks, and proteins that perturb antigen presentation. Other mechanisms have also been identified wherein MYXV uses gene products like M005/M-T5 (4) and M010/MGF (5) to create a more favorable cellular growth environment. Many of these virus proteins exhibit a narrow species specificity, and thus MYXV naturally infects only rabbits and hares. However, it can replicate in some human and mouse cells if key defenses, such as those regulated by Akt/protein kinase B (PKB) (4) or type I interferons (6), are disrupted. This has led to the suggestion that MYXV may have value as a safe and selective oncolytic agent, since these systems are often impacted by cell transformation (7, 8). A more detailed description of these genes and processes can be found in several reviews (9–11).
Although these and other studies have provided important insights into the mechanisms of viral pathogenesis, well-characterized virulence factors comprise only a small fraction of the 159 unique gene products of MYXV (strain Lausanne) (12). Most of these MYXV genes are widely conserved between different poxviruses, and this homology can be used to assign one or more biological roles to core processes, like entry, gene transcription, DNA replication, assembly, and exit. To accomplish this complex and coordinated developmental program, MYXV depends (like all viruses) upon cellular anabolic and catabolic processes to provide supplies of energy and biosynthetic precursors, as well as the macromolecular components (cytoskeleton, ribosomes, tRNAs, organelles, etc.) that are needed to productively complete an infectious cycle (13).
While it is broadly understood that poxviruses, like MYXV, rely upon the cell to provide an environment conducive to growth, it seems likely that the 109 hits detected in a 2-hybrid screen of vaccinia virus versus human proteins (14) represent only a small fraction of the possible interactions in a poxvirus-infected cell that contains >20,000 genes. In fact, new evidence continues to emerge showing that poxviruses depend more upon the host for specific cellular resources than previously anticipated. For example, it was long thought that poxviruses were generally self-sufficient when it came to viral DNA replication. However, our studies have shown that vaccinia virus (and by extension, viruses like MYXV) specifically recruits cellular DNA topoisomerase II to sites of virus replication and assembly, through an interaction with the virus-encoded DNA ligase (15). It has also been shown that poxviruses exhibit different degrees of dependence upon the cell for a supply of metabolites, like deoxynucleoside triphosphates (dNTPs). Leporipoxviruses encode only the smaller (R2) subunit of the heterodimeric (R12·R22) ribonucleotide reductase (16). Thus, in order to produce a functional enzyme and a secure supply of dNTPs, the MYXV R2 protein must form hybrid molecules with the cellular R1 protein (17). This situation reflects virus adaption to how the cell cycle determines the levels of dNTPs and cellular R2 protein, which are induced in G1 and become most abundant in S phase (18), and it also illustrates the role that cell growth state plays in determining host tropism.
Unfortunately, single-gene studies can provide only limited insights into the many possible cellular functions that affect the growth of viruses. As an alternative, one can use RNA interference (RNAi) screens to explore this problem on a much larger, and even genomic, scale. These methods were pioneered in Drosophila melanogaster cells, which have been used to study factors affecting Drosophila C (19), influenza virus (20), and dengue virus (21) replication. Subsequently, a number of whole-genome studies have been conducted to survey what mammalian cell functions modulate the growth of HIV (22, 23), West Nile virus (24), hepatitis C virus (25, 26), influenza virus (27), and vesicular stomatitis virus (28). Poxviruses have been less studied by these methods, although a study in Drosophila cells supported an abortive early infection by vaccinia virus, and a kinome screen has identified several proteins supporting virus endocytosis (29).
That said, the results obtained through use of small interfering RNA (siRNA) screens need to be interpreted with caution and are generally best interpreted as only a rough guide to cell and viral biology. This is illustrated by a meta-analysis of the human host genes that have been linked to the replication of HIV in three independent siRNA screens. Those studies noted that the pairwise degree of overlap of hits between the independent screens was <7% (22, 23, 30–32). A similar low-level reproducibility of only 25% was also seen in Drosophila screens for JAK/STAT signaling pathways (33–35). This situation reflects a high rate of false-positive hits and false-negative nonhits, with the former being more experimentally problematic. The occurrence of false-positive hits is mostly due to the siRNA reagents targeting mRNAs other than those they were designed to deplete (36–41), although the risk of these “off-target” effects can be reduced by retesting using siRNAs designed to target different sites in the same putative gene transcripts.
Bearing these concerns in in mind, but also recognizing the substantial gap in our understanding of poxvirus-host interactions, we decided to conduct a series of high-throughput RNAi screens by using siRNA libraries targeting both the whole human genome and also a subset of genes comprising the human kinome. These screens have identified >1,000 genes that appear to modulate the growth of MYXV in MDA-MB-231 cells, a human breast cancer cell line. Screening of a custom siRNA library targeting the hits and nonhits of the whole-genome screens showed that about 25% of the hits were very likely to be true positives. These genes modulate a variety of pathways, some of which have been previously identified as being important for poxvirus replication (e.g., the cytokine/chemokine-mediated inflammatory pathways and the mitogen-activated protein kinase [MAPK] pathways), whereas others (e.g., the cell cycle and glycolysis) are more novel discoveries. The validity of a subset of these hits was confirmed by independent methods. These methods included toxicity screens, which identified 32 siRNA pools (mostly targeting the proteasome) that are toxic in MDA-MB-231 cells. The results provide insights into both basic poxvirus biology as well as the host factors that affect MYXV growth in transformed human cells.
MATERIALS AND METHODS
Cells and viruses.
MDA-MB-231 cells were obtained from M. Hitt (University of Alberta) and maintained in Dulbecco's modified Eagle's medium–nutrient F-12 Ham medium (DMEM/F12; Sigma, St. Louis, MO) with 10% fetal bovine serum (FBS; Sigma) and 1% l-glutamine (Invitrogen, Carlsbad, CA). Buffalo green monkey kidney (BGMK) cells were cultured in modified Eagle's medium (MEM; Invitrogen) with 10% FBS. PANC-1 (human pancreatic epithelioid carcinoma), MCF-7 (human breast cancer), and U118-MG (human glioma) cell lines were maintained in DMEM (Invitrogen) with 10% FBS and 1% l-glutamine. U87 cells (human glioblastoma) were cultured in MEM containing 10% FBS, 1% MEM nonessential amino acids (Invitrogen), and 1% l-glutamine. T47D (human breast cancer) and CAPAN-2 (human pancreatic adenocarcinoma) cell lines were maintained in RPMI 1640 medium (Sigma) containing 10% FBS and 1% l-glutamine. All of the cells, with the exception of MDA-MB-231 cells, were obtained from the American Type Culture Collection, and all were tested and shown to be negative for mycoplasma. A recombinant MYXV (strain Lausanne), vMYX-LacZ, in which the lacZ gene is inserted into an intergenic locus and driven by a late poxvirus promoter (42), was a gift from G. McFadden (University of Florida). The virus was cultured on BGMK cells and purified by sucrose gradient centrifugation.
Plaque assays.
Infected cells were lysed in three rounds of freeze-thawing, serial dilutions were prepared, and the diluted viruses were used to infect BGMK cells. The viruses were cultured for 4 days, fixed with 2% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS), stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Invitrogen) solution containing 0.5 M K3Fe(CN)6, 0.5 M K4Fe(CN)6, 1 M MgCl2, and 100 mg/ml of X-Gal and counted to determine the number of LacZ+ focus-forming units (FFU).
siRNA libraries and chemical inhibitors.
The Silencer phosphatase and kinase (“kinome”) library was purchased from Ambion (Austin, TX). The library comprises three different siRNAs targeting each of 267 human phosphatases and 719 human kinases, and these were each resuspended in 96-well plates at a concentration of 5 μM. We pooled the three siRNAs that target each gene prior to performing the screens (43). The Silencer Select whole-genome siRNA library was also purchased from Ambion. The library comprises 21,585 pooled siRNAs, targeting all the known genes in the human genome, and was resuspended at a concentration of 2 μM in 96-well plates. Each pool contained three different siRNAs directed against a given gene. The Dharmacon siGENOME Human SMARTpool custom siRNA library was purchased from Dharmacon/Thermo Scientific (Waltham, MA). This library contains 88 siRNAs that were selected from the hits and nonhits of our whole-genome siRNA screens. The library contains four siRNAs targeting a specific gene pooled and resuspended at a concentration of 2 μM in 96-well plates. The separate and individual siRNAs that were used to validate the hits identified in these screens were purchased from Qiagen (Chatsworth, CA). Etoposide was purchased from the university hospital pharmacy, and 2-deoxy-d-glucose (2DG) and PD-0332991 were obtained from Sigma and Selleckchem (Houston, TX), respectively.
Plasmids and plasmid transfection assays.
Plasmids carrying the genes for the human phosphofructokinase muscle isoform (PFKM) or rat 6-phosphofructose-2-kinase/fructose-2,6-bisphosphatase isoform 3 (PFKFB3) were a gift from J. Bolaños (Universidad de Salamanca, Spain). Plasmid carrying Myc-tagged PRIM2 was purchased from Origene (Rockville, MD). To perform transfections, MDA-MB-231 cells were seeded overnight in 24-well plates, at a density of ∼200,000 cells per well, and then 0.5 μg to 2 μg of plasmid DNA was mixed with Lipofectamine 2000 transfection reagent (Invitrogen) at a 3:1 ratio and applied to the cells as directed by the manufacturer.
siRNA transfection and vMYX-LacZ infection of cells.
A reverse transfection protocol was used to perform all of the siRNA screens, using final siRNA concentrations of 10 nM for the kinome screens, 5 nM for the whole-genome screens, and 20 nM for the Dharmacon custom library screens. Briefly, aliquots of the pooled siRNAs were added into 96-well plates and resuspended to a volume of 20 μl/well in a solution containing 0.1 μl/well of DharmaFECT 4 transfection reagent (Thermo Scientific, Rockford, IL) and Opti-MEM I medium. The mixture was incubated for 30 to 45 min at room temperature and then overlaid with 5,000 cells per well with freshly trypsinized MDA-MB-231 cells in DMEM/F12 medium supplemented with 10% FBS and 1% l-glutamine. The cells were incubated at 37°C in a 5% CO2 atmosphere for 3 days, and then the medium was replaced with 20 μl of PBS containing vMYX-LacZ. After incubation of the cells in the infectious inoculum for 1 h, 80 μl of fresh medium was added. The virus was applied at a multiplicity of infection (MOI) of 0.1 for the kinome, Dharmacon custom library, and whole-genome primary screens or an MOI of 1 for the whole-genome validation screen. The cells were cultured for 48 h, lysed using a single freeze-thaw cycle in reporter lysis buffer (Promega, Madison, WI), and then assayed for β-galactosidase activity as described below.
Other siRNA transfection assays used a forward method of transfection in which 5,000, 200,000, or 500,000 cells were seeded per well in 96-, 24-, or 6-well plates overnight, respectively. The cells were treated for 3 h with a mix of siRNA and DharmaFECT 4 transfection reagent, appropriate medium was added, and the cells were cultured for another 2 to 3 days before applying MYXV as described above.
Enzyme and cell viability assays.
β-Galactosidase assays were conducted using a commercial kit (Promega). The lysed cells were mixed with one volume of 2× assay buffer (200 mM sodium phosphate buffer [pH 7.3], 2 mM MgCl2, 100 mM β-mercaptoethanol, and 1.33 mg/ml o-nitrophenyl-β-d-galactopyranoside) and incubated for 1 h at 37°C, and the reaction was stopped with three volumes of 1 M Na2CO3 (ACROS Organics, NJ). The absorbance was measured at 420 nm using a PerkinElmer EnVision plate reader. Additional spectrophotometric assays were used to measure lactate (with the method described in reference 44), ATP (with an ATP kit [Invitrogen]), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; with a KDalert GAPDH kit [Ambion]), again as per the manufacturers' instructions.
An alamarBlue dye assay was used to measure cell viability (45). For example, to identify toxic siRNAs, MDA-MB-231 cells were reverse transfected as described above, cultured for 3 days, and then incubated with 50 μM resazurin (Sigma) in phenol red-free DMEM/F12 medium for 2 to 3 h at 37°C. The fluorescence was also measured using an EnVision plate reader, with 560-nm excitation and 590-nm emission filter settings.
RT-PCR assays.
Cells were lysed with TRIzol reagent (Invitrogen), mixed with chloroform, and centrifuged. The aqueous phase was collected, and the RNA was precipitated with isopropyl alcohol. The pellet was then washed with 70% ethanol and dissolved in water, and the amount of RNA was determined by spectrophotometry. Approximately 0.5 μg of RNA was processed into cDNA by using SuperScript III reverse transcriptase (RT) as directed by the supplier (Invitrogen). Quantitative PCRs (qPCRs) used the primers described in Table S1 of the supplemental material and Taq DNA polymerase as directed by the supplier (Thermo Scientific, Rockford, IL).
Western blotting.
Cells were lysed in buffer (150 mM NaCl, 20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5% NP-40) containing a protease inhibitor tablet (Roche, Mannheim, Germany) for 1 h on ice. The proteins were separated by SDS-PAGE, transferred to nitrocellulose (Thermo Fisher Scientific), and analyzed in a Western blot assay using a protocol optimized for an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The primary antibodies were goat polyclonal antibodies directed against the R1 subunit of ribonucleotide reductase and vaccinia-related kinase 1 (VRK-1; both from Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit polyclonal antibody for enhanced green fluorescent protein (EGFP; BD BioSciences, San Jose, CA), a rat monoclonal antibody for PRIM2 (Cell Signaling Technology, Danvers, MA), a mouse monoclonal antibody for Myc (Cell Signaling Technology), and a mouse monoclonal antibody for β-actin (Sigma). The membranes were probed with IRDye 680CW or IRDye 800CW secondary antibodies (LI-COR Biosciences) and imaged with an Odyssey scanner.
Immunofluorescence.
Cells were seeded on coverslips in 24-well plates, washed twice with PBS, fixed with 4% paraformaldehyde for 30 min, quenched with 0.1 M glycine for 10 min, and then incubated in a blocking buffer containing 3% bovine serum albumin (Sigma) in 1% Triton X-100 (MP Biochemicals) in PBS for 1 h at 20°C. Then, the cells were incubated overnight at 4°C in primary antibody, washed three times with PBS, and incubated in secondary antibody for 3 h at 20°C. Finally, the cells were washed three times and incubated in 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen) and rhodamine phalloidin (Invitrogen) for 30 min at room temperature, and the coverslips were mounted on the slides. An Applied Precision Deltavision microscope was used to obtain images of the specimens.
Flow cytometry.
Cells were treated with drugs for 24 to 72 h, or transfected with siRNAs for 48 to 72 h, and then detached with trypsin. The cells were recovered by centrifugation, washed twice with PBS, centrifuged, resuspended in 70% ethanol, and incubated on ice for 1 h. The cells were then treated with 1 μg/ml of RNase for 30 min, stained for 1 h in 1 μg/ml propidium iodide, and counted using an LSR Fortessa cell analyzer (BD Biosciences, San Jose, CA). FlowJo software was used for cell cycle analysis.
Statistics and bioinformatics.
Interplate variability for all the screens was normalized by subtracting the median of each plate from the raw results of each well on a specific plate. Following this normalization step, the mean of the replicates was calculated. Data quality analysis was conducted using frequency distribution and correlation plots. We used a strictly standardized mean difference (SSMD) method to analyze the kinome, whole-genome validation, and Dharmacon custom library screens and the Z-score method to analyze the primary whole-genome and the siRNA toxicity screens (46–48). The online tools PANTHER (49), DAVID (50, 51), and PINdb (52) were used to search for gene associations and relationships. In other studies, Student's t test was used to compare the data, and the results were considered to be significant if P was ≤0.05.
RESULTS AND DISCUSSION
Optimization of siRNA screening methods.
Before embarking upon the large-scale siRNA screen, we performed experiments to optimize the choice of cells and the transfection, infection, and assay methods. We also tested these technologies by using a screen of the kinome (∼1,000 genes for siRNA pools targeting human phosphatases and kinases) prior to screening the whole human genome.
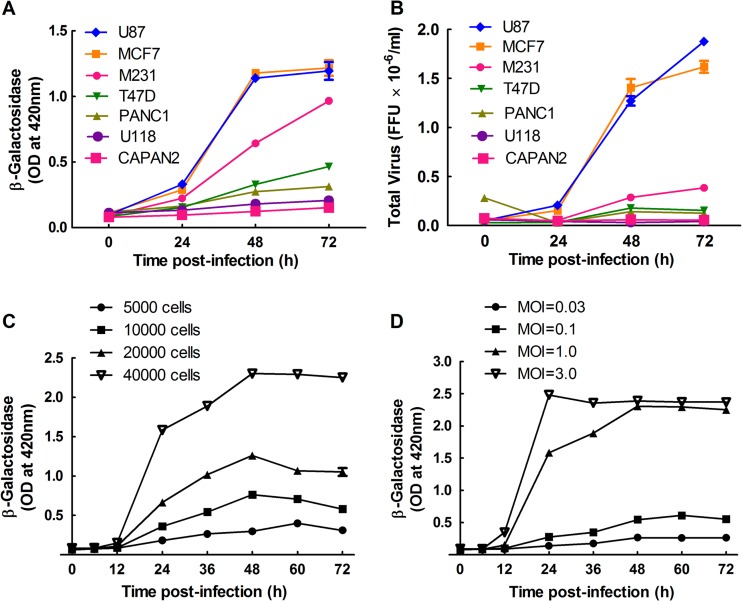
For the optimization experiments, as well as the final screens, we used an MDA-MB-231 human breast cancer cell line (53) and a recombinant MYXV encoding an Escherichia coli lacZ gene inserted between the M10L and M11L genes and regulated by a late virus promoter (42). The lacZ gene permits more accurate titration of the virus and, because poxvirus late genes are not expressed in the absence of replication, the β-galactosidase produced by vMYX-LacZ provides a measure of the level of virus replication. It also produced a better signal-to-noise ratio than did fluorescent protein reporter constructs (data not shown). We used MDA-MB-231 cells because they showed an intermediate level of permissiveness to MYXV infection compared to other common human cell lines (Fig. 1A), and thus they were expected to facilitate detection of both stimulatory and inhibitory siRNAs. These cells have also been studied intensively over many years, providing a wealth of literature on specific signaling pathways. We also examined how well the β-galactosidase assay correlated with virus yield (i.e., FFU). Although there was not a linear relationship between the two assays, the same relative relationship was seen between the levels of β-galactosidase and the number of FFU, when different cell types were infected with vMYX-LacZ at an MOI of 0.1 (Fig. 1B). This showed that the LacZ assay is a good surrogate that should be predictive of MYXV yields.
Fig 1.
MYXV growth in MDA-MB-231 cells. (A and B) MDA-MB-231 cells support an intermediate level of virus growth compared to other human cell lines. U87, MCF7, MDA-MB-231 (M231), T47D, PANC1, U118, and CAPAN2 cells were seeded into 96-well plates, cultured for 24 h, and then infected with MYXV at an MOI of 0.1. The cells were lysed at the indicated time points and assayed for β-galactosidase activity (A) and FFU (B). (C and D) MYXV growth kinetics in MDA-MB-231 cells. MDA-MB-231 cells were cultured for 24 h in 96-well plates at densities ranging from 5,000 to 40,000 cells per well and then infected with MYXV at an MOI of 1 (C). Alternatively, 40,000 cells were plated per well and infected with MYXV at the indicated multiplicities of infection. β-Galactosidase activity was measured at the indicated time points and is reported as the optical density (OD). These data are representative of two independent experiments.
We also tested what effects the cell density and multiplicity of infection have on MYXV growth kinetics when assayed using the β-galactosidase reporter (Fig. 1C and D). Based upon these experiments, we identified an MOI between 0.1 and 1 and a 48-h infection period as the infection conditions that were least likely to result in signal saturation while still generating a good signal-to-noise ratio.
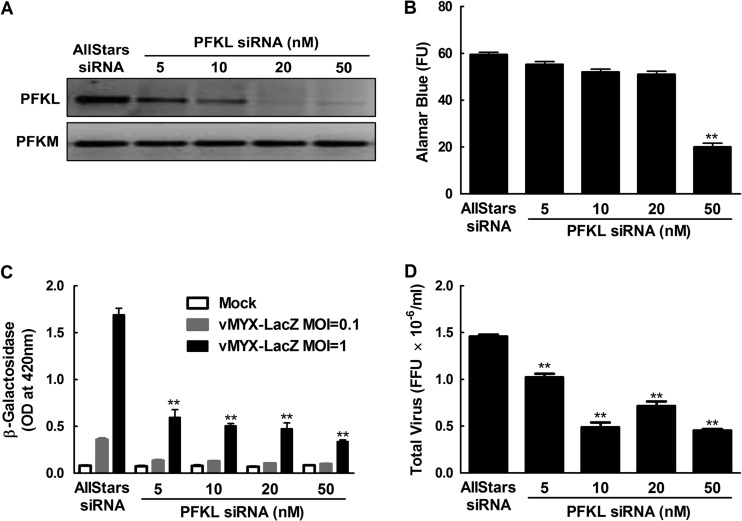
We also optimized the efficiency of siRNA delivery and silencing by transfecting the MDA-MB-231 cells with different amounts of GAPDH siRNAs and measuring the silencing in a GAPDH activity assay. We observed that siRNA concentrations of ≥1 nM reduced the GAPDH activity by ∼70% (see Fig. S1 in the supplemental material). We also determined that the DharmaFECT 4 transfection reagent produced no cytotoxicity under these conditions, as judged by alamarBlue cell viability assays (data not shown). We subsequently used siRNAs targeting the mRNAs for the large subunit of cellular ribonucleotide reductase (R1) and the phosphofructokinase liver isoform (PFKL) as positive controls in the larger screens. As noted above, MYXV encodes an R2 subunit of ribonucleotide diphosphate reductase, but during infection the virus R2 protein must complex with the cellular R1 subunit to form an enzyme that catalyzes the rate-limiting step in dNTP biogenesis (17, 18). As predicted, siRNA knockdown of cellular R1 produced a 55-to-60% reduction in virus replication (see Fig. S2 in the supplemental material). PFKL was initially identified as a strong and reproducible hit in the kinome screen, which is described in more detail below (see Fig. 5). In all of the screens, an “AllStars” siRNA was used as a negative control, as it produced no detectable effects on virus growth or cell viability.
Fig 5.
siRNA silencing of PFKL inhibits MYXV replication. (A) MDA-MB-231 cells were transfected with siRNA against PFKL for 3 days and then lysed, and RT-PCR was performed to measure specific knockdown of the liver (PFKL) rather than the muscle (PFKM) isozyme. (B to D) The transfected cells were also screened for viability in an alamarBlue assay (FU, fluorescence units) (B) or infected with MYXV for 48 h at MOIs of 0.1 and 1 for the β-galactosidase assay (C) and an MOI of 1 for virus yield determination (D). The RT-PCRs showed that 10 to 50 nM siRNA was needed to knock down the PFKL transcript, with toxicity only being observed at ≥50 nM PFKL siRNA (B). Virus growth was significantly reduced (**, P ≤ 0.001) even with the lowest doses of siRNAs, relative to cells treated with the negative-control siRNA. These data are representative of two independent experiments.
Based on the experiments, we designed a screening method in which we seeded MDA-MB-231 cells into 96-well plates at a density of 5,000 cells per well, reverse transfected the cells with 5 to 10 nM siRNA for 3 days, and then infected the cells with vMYX-LacZ at an MOI of 0.1 to 1. Two days later, the cells were lysed and assayed for β-galactosidase activity.
Kinome test screen.
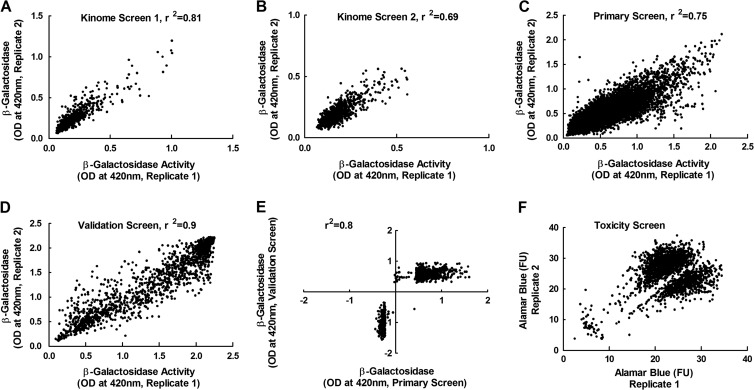
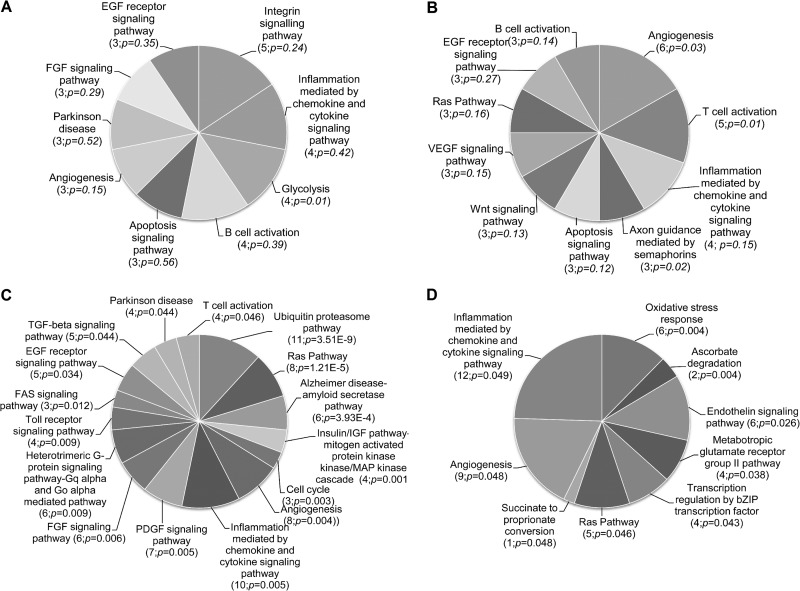
We next tested these methods by performing two duplicate RNAi screens with an siRNA library targeting ∼1,000 human kinases and phosphatases. Each gene in the screen was targeted with a pool of three different siRNAs, based upon our previous study that had shown that pooled siRNAs produced better penetrance than single siRNAs without causing an unmanageable increase in false positives (43). Analysis of the degree of correlation between the duplicates within each screen showed that the methods were generally robust, with the coefficient of determination (R2) varying from 0.7 to 0.8 in the second and first screens, respectively (Fig. 2A and B). The siRNAs that affected virus replication were identified by using the SSMD method, which minimizes the rates of false discovery and false nondiscovery in siRNA screens (46–48). Out of the 986 genes targeted by this library, we identified 70 siRNA pools that inhibited (SSMD, ≤−3) and 26 that increased (SSMD, ≥3) virus growth (see Table S2 in the supplemental material). These hits could be assigned to many different pathways, with genes related to apoptosis, inflammation, and cell growth regulation comprising the most prominent targets (Fig. 3A and B). Perhaps most notable were the several strongly inhibitory hits within the ERB-B signaling pathway. Although the ErbB-2 receptor is not highly expressed in MDA-MB-231 cells (54), MYXV growth factor could still potentially signal through this route (55) and would affect the Akt/PKB pathway, which is important for MYXV growth (56). PIK3CA (phosphoinositide-3-kinase alpha) regulates this pathway, and siRNAs targeting PIK3CA were highly inhibitory. Many inhibitory siRNA pools also targeted enzymes linked to glycolysis (Fig. 3A), in a logically consistent manner. For example, silencing PFKL reduced MYXV replication, whereas silencing a phosphatase that opposes PFK-1 activity, FPB1 (fructose-1,6-bisphosphatase 1), increased virus replication (see Table S2).
Fig 2.
Experimental reproducibility. Each experiment was performed in duplicate, and β-galactosidase assays were conducted to measure virus growth. (A to D) The reproducibility of paired replicates from the first (A) and the second (B) kinome screens, from the primary whole-genome screen (C), and from the validation screen (D). (E) Comparison of data obtained in the validation screen (a rescreen of the proviral and antiviral hits identified by the whole-genome screen, along with a small number of neutral control hits) versus the signals detected in the original whole-genome primary screen. (F) How a cluster of toxic siRNAs were identified, using alamarBlue assays. Shown are the correlation coefficients (r2) for the different experiments.
Fig 3.
Pathway analysis of gene hits. PANTHER (Protein ANalysis THrough Evolutionary Relationships) was used to look for relationships between the genes identified by the kinome (A and B) or the whole-genome (C and D) screens, and which genes either decreased (A and C) or increased (B and D) virus replication. The values in parentheses indicate the number of gene hits in each pathway and the estimated P value.
We also retested four of the hits by using different siRNA silencing reagents and virus yields, to gain some insights into the reliability of the screen. Three of the four hits (PFKL, pyruvate dehydrogenase kinase isozyme 3 [PDK3], and vaccinia virus-related kinase 2 [VRK2]) could be independently verified, whereas the fourth (vaccinia virus-related kinase 1 [VRK1]) could not. We did not detect hits involving the AMPK gene, as has been reported for Drosophila (29), but humans encode multiple isoforms of AMPK and its subunits (29), and this redundancy may have obscured any hits. This small-scale survey indicated that the methods we were using were sufficiently robust to identify practical numbers of true-positive hits.
Whole-genome primary siRNA screen.
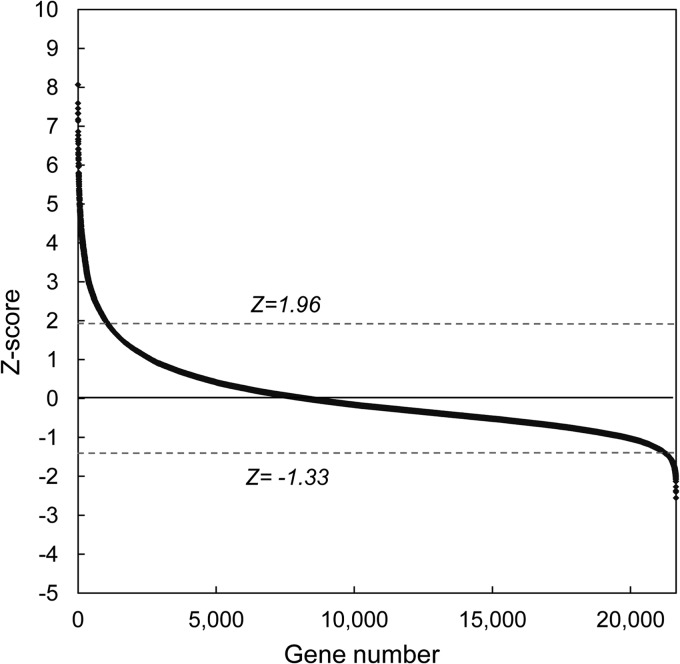
We next screened 21,585 pooled siRNAs, which target all the known genes in the human genome. The screens were conducted in duplicate, using the same methods described above. The data quality analysis showed that both replicates produced positively skewed frequency distribution curves (see Fig. S3A and B in the supplemental material), suggesting that more siRNAs increased virus replication than decreased it. The duplicates showed a good degree of correlation (R2, 0.75) (Fig. 2C). These data were analyzed by applying Z-score statistics (46, 57) to the normalized results of the screen; this method was used in preference to SSMD because the number of siRNAs was large enough to produce a frequency distribution approximating a Gaussian curve, albeit with a slight positive skew (see Fig. S3A and B). Like the frequency distribution curves (see Fig. S3A and B), the Z-score method also detected more siRNAs that increased virus replication than decreased it (Fig. 4), and this asymmetry complicated how one should select the genes identified in this screen. We thus applied two approaches to initially define these hits. First, we included all the siRNAs that produced Z-scores of ≥1.96 or ≤−1.96 and which comprised 5% of the genes in the screen. However, this included relatively few siRNAs that inhibited MYXV growth, and we wanted to retest to determine if this effect was real. Therefore, we also included a further subset of siRNAs that comprised 5% of the most-inhibitory siRNAs based on the Z-score curve. This led to the further inclusion of siRNAs with Z-scores between −1.96 and −1.33 (Fig. 4). In the end, we identified 1,588 siRNAs that fit these criteria, 1,048 with a Z-score of ≥1.96 and 540 with a Z-score of ≤−1.33. These siRNAs were picked and replated and used in another round of screening.
Fig 4.
Z-score analysis of data from the whole-genome screen. A Z-score was calculated for each gene, using the means of the replicates after prior normalization using the plate means. Genes with Z-scores of ≥1.96 or ≤−1.33 were identified as potential hits and were included in the validation screen. See the text for further discussion of the selection criteria.
Validation of the whole-genome primary siRNA screen.
We rescreened 1,721 siRNAs in duplicate in a validation screen. These siRNAs comprised 1,588 siRNA hits from the whole-genome primary screen, 69 additional high-scoring hits from the kinome screens (hits not detected by the whole-genome screen), and 64 randomly selected “nonhit” controls (i.e., Z-score of ≈0) from the whole-genome screen. We also incorporated onto all of the plates the positive-control siRNAs, which targeted PFKL as well as the R1 subunit of the cellular ribonucleotide reductase.
The validation screen was conducted in duplicate and showed a high degree of correlation between the duplicates with R2 value of 0.9 (Fig. 2D). We also determined the degree of correlation between the normalized means of the whole-genome screen and of the validation screens and again noted a high degree of agreement between the screens, (R2 = 0.8) (Fig. 2E). As one would expect, these data also reclustered into the three preselected groups representing inhibitory, stimulatory, and neutral siRNA pools (Fig. 2E). The controls also clustered into predicted groups, with the mock-infected wells producing the lowest reading for β-galactosidase activity while the level of activity detected in infected cells treated with the two inhibitory positive controls (R1 and PFKL siRNAs) clustered between the levels detected in mock-infected cells and in the MYXV-infected cells treated with the Allstars control siRNA (see Fig. S4A in the supplemental material). The validation screen thus provided some confidence that these methods can reproducibly detect siRNA pools that modulate the growth of MYXV in MDA-MB-231 cells.
Determined reproducibility based on the Dharmacon custom siRNA library screen.
Some insights into the reproducibility and reliability of the methods were obtained by comparing the results of the kinome and whole-genome screens. By definition, the kinome siRNAs comprise a subset of the whole-genome set, but the two libraries were designed separately and employ different chemical modifications. Of the 96 genes detected in the two initial kinome screens, 28% were subsequently detected again in the whole-genome primary screen. We included all 96 hits in the whole-genome validation screen, and again we detected 37.5% as hits.
To further analyze the degree of reproducibility of our screens, we purchased a custom siRNA library from another source. That library contains a pool of four siRNAs targeting each of 88 genes (59 hits and 29 nonhits of the whole-genome screens) in 96-well plates. The hits were selected randomly from statistically significant pathway hits. After the screens were performed in triplicate and normalized, we again used cutoffs of SSMD ≥3 and ≤−3 for hit selection. This independent screen detected 25.4% of the hits of the whole-genome screen as high-probability true positives and 75.9% of the nonhits as high-probability true negatives (Table 1; see also Table S4 in the supplemental material). Thus, taking the two comparisons together, 25 to 30% of the genes identified by our methods could be verified using independent libraries and screens. This is higher than the aforementioned meta-analysis of genes affecting HIV replication, but we had the advantage of using similar transfection, infection, and assay protocols.
Table 1.
Comparison of results of the whole-genome validation and custom siRNA library screens
| Whole-genome validation screen | Dharmacon custom siRNA library screen |
Total | ||
|---|---|---|---|---|
| No. (%) of hits associated with: |
No. (%) of nonhits | |||
| Increased MYXV replication | Decreased MYXV replication | |||
| Hit, increased MYXV replication | 8 (27.6) | 3 (10.3) | 18 (62.1) | 29 |
| Hit, decreased MYXV replication | 4 (13.3) | 7 (23.3) | 19 (63.3) | 30 |
| Nonhit | 5 (17.2) | 2 (6.9) | 22 (75.9) | 29 |
| Total | 22 | 21 | 45 | 88 |
Screen for siRNA toxicity.
We were concerned that these methods do not exclude cytotoxic siRNAs. To identify which of the 1,721 siRNAs might be toxic, we measured cell viability at the end of the 3-day transfection period, using the same transfection protocol and an alamarBlue cell viability assay (45). The data clustered into three groups (Fig. 2F). Most of the siRNAs in the replicate assays clustered into two groupings in the upper right quadrant, which exhibited no signs of toxicity as judged by the strong fluorescence signals. (Why we saw two clusters of signals is unclear, although this was considered unrelated to the siRNAs or to transfection because untreated cells, and also cells transfected with the Allstars control, produced the same two data clusters [see Fig. S4B in the supplemental material].) In contrast, a small group of siRNAs produced very low viability scores in both replicates. These scores are typically produced by toxic agents, such as etoposide, which was added to wells in each plate as an internal control in the screen (see Fig. S4B and S5). These findings showed that the alamarBlue screen readily detects toxic siRNAs.
The data obtained from the siRNA toxicity screen were normalized, and the frequency distributions (see Fig. S3C and D in the supplemental material) permitted analysis based on Z-score statistics. Toxic siRNAs were defined as those with Z-scores of ≤−1.96. The screen identified 32 toxic siRNAs, of which many belong to the ubiquitin-proteasome pathway, including the UBC gene that encodes ubiquitin C (Table 2). Many of these proteasomal genes play some role in regulating or supporting the cell cycle, as do other toxic gene targets, like the human WEE1 homolog, PLK1, and RAN. In hindsight, this was not too surprising. Bortezomib (Velcade) is a recognized chemotherapeutic agent that inhibits the 26S proteasome and induces apoptosis in transformed cells (58). Our screen clearly identified MDA-MB-231 cell sensitivity to siRNAs targeting different components of the 26S proteasome, and our results further validate the method used to discover siRNAs that “inhibit” MYXV replication. Recent papers have suggested that proteasome inhibitors might find some utility as antipoxvirus agents (59, 60). However, our data do highlight the critical importance of examining the effects of these drugs on cell viability, especially over longer times of exposure and with considerations for cell type.
Table 2.
Toxic siRNAs
| Gene symbol | Gene name | Z score |
|---|---|---|
| LOC729093 | Hypothetical protein LOC729093 | −1.97 |
| PSMD11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | −2.01 |
| CYP51A1 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | −2.01 |
| PSMC4 | Proteasome (prosome, macropain) 26S subunit, ATPase, 4 | −2.02 |
| RAN | RAN, member RAS oncogene family | −2.02 |
| LOC728242 | X antigen family, member 2-like | −2.02 |
| IFITM1 | Interferon-induced transmembrane protein 1 (9–27) | −2.04 |
| PSMD3 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | −2.07 |
| TFAP4 | Transcription factor AP-4 (activating enhancer binding protein 4) | −2.10 |
| SNRPEL1 | Small nuclear ribonucleoprotein polypeptide E-like 1 | −2.14 |
| RGS3 | Regulator of G-protein signaling 3 | −2.15 |
| LOC119358 | Similar to hCG2040270 | −2.18 |
| ZFP3 | Zinc finger protein 3 homolog (mouse) | −2.21 |
| LOC728991 | Hypothetical LOC728991 | −2.23 |
| FAM55B | Family with sequence similarity 55, member B | −2.43 |
| SLC38A10 | Solute carrier family 38, member 10 | −2.45 |
| KIAA1604 | KIAA1604 protein | −2.52 |
| PLK1 | Polo-like kinase 1 (Drosophila) | −2.53 |
| FLJ22675 | Hypothetical gene supported by AK026328 | −2.63 |
| PSMD1 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | −2.84 |
| C9orf135 | Chromosome 9 open reading frame 135 | −2.85 |
| PSMD8 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | −2.93 |
| WEE1 | WEE1 homolog (S. pombe) | −2.98 |
| TTLL8 | Tubulin tyrosine ligase-like family, member 8 | −3.06 |
| ALB | Albumin | −3.08 |
| LOC644828 | Hypothetical LOC644828 | −3.30 |
| LOC728689 | Similar to eukaryotic translation initiation factor 3, subunit 8, 110 kDa | −3.35 |
| RPL21 | Ribosomal protein L21 | −3.47 |
| LOC728962 | Similar to 60S ribosomal protein L7 | −3.47 |
| UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | −3.49 |
| SF3B1 | Splicing factor 3b, subunit 1, 155 kDa | −3.52 |
| UBC | Ubiquitin C | −5.45 |
Analysis of hits.
We used the SSMD method to analyze the results of the validation screen. Those siRNAs that exhibited SSMD scores of ≤−3 or ≥3 were defined as the final hits in the screen, although these conservative criteria likely overlook weaker but still biologically real hits (see below). Overall, we identified 711 siRNAs that promoted MYXV growth in MDA-MB-231 cells and 333 siRNAs that inhibited virus replication. These represent 4.8% of all the siRNAs/genes we screened (see Table S3 in the supplemental material). Those siRNAs that increased or decreased MYXV replication were defined as proviral and antiviral, respectively. Consequently, those genes that increased MYXV replication when silenced were called “antiviral” genes, and vice versa.
These hits were categorized using the PANTHER classification system (49). The method compares the list of hits (and, by assumption, genes) affecting MYXV growth against a reference list that contains all the genes in the human genome, categorized as to putative or known function. Those pathways that were statistically overrepresented, with a P value of ≤0.05, were considered biologically significant. Among the pathways involving genes that decreased the replication of MYXV when silenced were the previously noted ubiquitin-proteasome system, the Ras and mitogen-activated protein kinase (MAPK) pathways, chemokine and cytokine inflammatory pathways, and the cell cycle (Fig. 3C). Among the pathways that significantly increased virus growth, when genes in these pathways were silenced, were the oxidative stress response, ascorbate degradation, and endothelin signaling pathways (Fig. 3D). A few pathways are represented in both categories of hits (the Ras and chemokine/cytokine signaling pathways), although the statistics are more solid for the MYXV growth-inhibiting siRNA category.
We also examined the relationship between the hits using the PIN database (52) of interacting nuclear proteins. We noted that several genes encoding proteins that are part of transcription complexes appeared to have an antiviral activity, that is, knocking down these genes enhanced MYXV growth. For example, we detected hits in proteins that are components of the TFIID transcription initiation factor (TAF4 and TAF9) and a TFIID-related complex (TRRAP, TAF4, and TAF9). Several (3 of 10) proteins comprising the TFIIH initiation/elongation complex (ERCC2, GTF2H1, and CCNH) were also detected in our screen.
Although 1,044 gene hits are still too many to investigate in detail, we elected to explore a subset of these hits and pathways to gain some insights into whether the screen had reliably identified new cell factors that affect MYXV replication. Based upon the data summarized in Table S3 in the supplemental material and also illustrated in Fig. 3, we chose to study a subset of genes that play important roles in glycolysis, MAPK signaling, DNA replication, and the cell cycle.
Role of aerobic glycolysis in MYXV growth.
Glycolysis converts glucose to pyruvate and in the process makes intermediary metabolites that are critical substrates for nucleotide biogenesis and other anabolic pathways (61). The glycolytic enzymes and substrates also regulate cell proliferation and apoptosis downstream of phosphatidylinositol 3-kinase and Akt (62–65). Thus, it is of critical importance to rapidly dividing cells, especially cancer cells, where the shift to glycolysis is called the Warburg effect. Over 60 years ago, Kun and Smith noted that MYXV infection of chicken embryo chorioallantoic membranes caused increased conversion of glucose to lactic acid, and this finding led them to suggest that infection was associated with increased glycolysis (66). Our screen clearly identified the important role glycolysis plays in supporting MYXV growth in MDA-MB-231 cells.
After uptake, glucose is converted to glucose 6-phosphate by hexokinases (HK) and then converted into pyruvate in eight additional steps. The rate-limiting step in glycolysis is catalyzed by PFK-1 (67), and three PFK-1 isoforms have been identified in liver (PFKL), muscle (PFKM), and platelets (PFKP). PFK-1 is allosterically regulated by fructose-2,6-bisphosphate, which is produced from fructose 6-phosphate via 6-phosphofructose-2-kinase/fructose-2,6-bisphosphatase (PFKFB) (68–70). PFKFB is a homodimeric enzyme, and there are four different isoforms, each encoded by different genes (PFKB1 to -4). Many of these enzymes in (or regulating) the glycolytic pathway were major hits in the kinome screens (Fig. 3), with siRNAs targeting PFKL, PFKM, HKIII, and PFKFB1 all inhibiting MYXV replication and siRNAs targeting glucose-6-phosphate isomerase (GPI) and fructose-1,6-bisphosphatase 1 (FBP1) stimulating virus growth. Most of these hits (except for HKIII and GPI) were also detected in the whole-genome screens, although the effects on LacZ expression were obscured by other higher-scoring siRNAs, and thus the glycolytic pathways did not appear among the final hits from the whole-genome screen (Fig. 3). The facts that the enzymes that promote glycolysis (e.g., PFKL) also promote MYXV replication and enzymes that negatively regulate glycolysis (e.g., FBP1) reduce MYXV growth led us to hypothesize that MYXV infection is highly favored in cells utilizing glycolytic metabolism. The following experiments were conducted to confirm this.
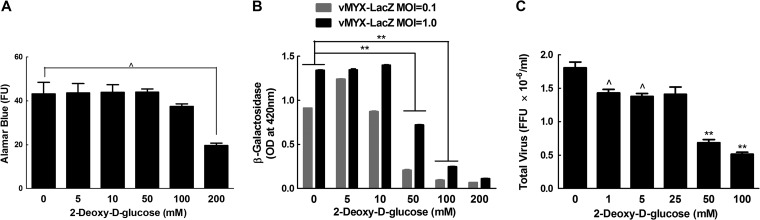
We elected to focus on PFK-1 (PFKL and PFKM) and PFKFB. We first used RT-PCR to verify the knockdown of PFKL mRNA in MDA-MB-231 cells. Cells were transfected with different concentrations of PFKL siRNAs for 3 days, and then RT-PCR, with PFKL-specific primers, was used to measure the residual levels of PFKL transcripts. As little as 5 nM siRNA was needed to achieve some knockdown of PFKL over a 3-day period, although better silencing was seen with 20 to 50 nM siRNA and without off-target effects on PFKM (Fig. 5A). However, the highest concentrations of PFKL siRNA (e.g., 50 nM) produced significant toxicity (Fig. 5B). These concentrations of PFKL siRNA were also retested to see what effects they had on β-galactosidase expression and virus yields over a 48-h infection cycle. Both assays detected a 2- to 3-fold reduction in virus gene expression and yields (FFU) when we used 10 to 20 nM siRNA. These were conditions under which one detects little or no toxicity (Fig. 5C and D). We also examined the growth kinetics of MYXV in PFKL siRNA-transfected cells and determined that silencing of PFKL resulted in a reduction in virus replication at all time points, starting from 24 h postinfection (see Fig. S6A in the supplemental material). Similar results were obtained with 2DG, a chemical inhibitor of glycolysis. In these experiments, 2- to 3-fold reductions in gene expression (Fig. 6B) and virus yields (Fig. 6C) were again detected in the presence of 50 to 100 nM 2DG, concentrations causing little or no toxicity (Fig. 6A). To prove that the 2DG was working, we also measured the levels of lactate and ATP. Both compounds were significantly reduced in cells treated with higher (100 nM) concentrations of 2DG, showing that glycolysis was inhibited under these conditions (see Fig. S7 in the supplemental material).
Fig 6.
A glycolytic inhibitor, 2DG, also inhibits MYXV growth. MDA-MB-231 cells were pretreated with 2DG for a day and then infected with MYXV for 48 h in the continued presence of 2DG. An alamarBlue assay was then used to measure cell viability (A), and virus growth was measured using β-galactosidase (B) and plaque (C) assays. The plaque assays used an MOI of 1. A significant reduction (^, P ≤ 0.05; **, P ≤ 0.001) in virus growth was seen in cells treated with ≥50 mM 2DG in the β-galactosidase assay or with doses as low as 1 mM in the plaque assay. No significant toxicity was detected at 2DG concentrations of <100 mM. These data are representative of three independent experiments.
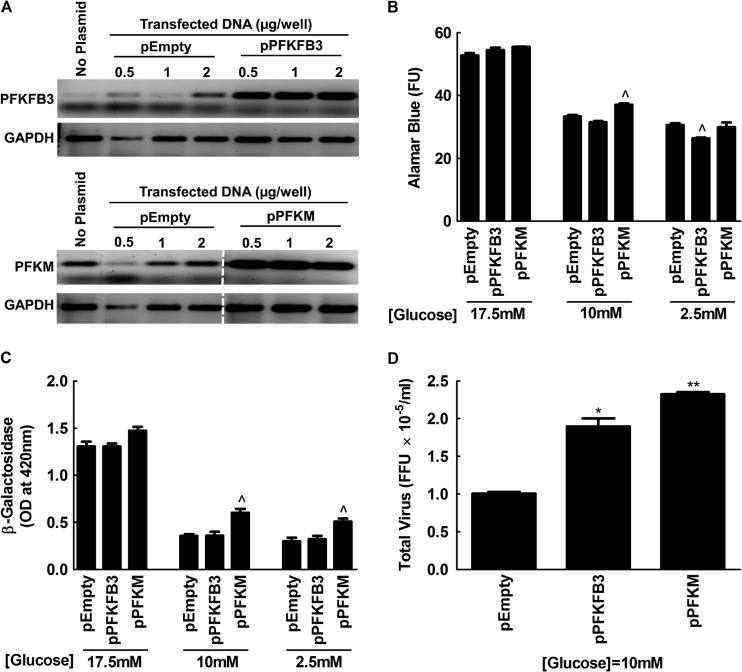
We also tested whether a reciprocal increase in virus yield could be obtained by increasing the amount of glycolysis in MYXV-infected cells. For this purpose, we used pIRES-EGFP plasmids carrying genes for PFKM or PFKFB3 that have been shown to increase glycolytic activity in transfected COS-7 cells (71). RT-PCR showed one can also obtain higher levels of expression of these genes in MDA-MB-231 cells (Fig. 7A), although this did not have any effect on MYXV growth under standard cell culture conditions (Fig. 7B and C). However, we noted that the culture medium used in all of these studies contained high levels of glucose (17.5 mM), and we were concerned that this might have saturated the glucose flux through the glycolytic pathways. Therefore, we repeated the experiments in media containing 10 or 2.5 mM glucose. Although these concentration somewhat reduced the cell viability, relative to cells grown in DMEM/F-12 (Fig. 7B), we saw increased virus growth in PFKM-transfected cells whether measured using β-galactosidase (∼1.5-fold [Fig. 7C]) or using plaque assays (∼2.5-fold [Fig. 7D]). We also detected an ∼2-fold increase in virus yield in cells transfected with PFKB3 (Fig. 7D), although this was not correlated with an increase of β-galactosidase expression (Fig. 7C). Collectively, these data support the observations from the original siRNA screens and show that MYXV growth is positively correlated with glycolytic activity in a transformed human cell.
Fig 7.
MYXV growth is enhanced by phosphofructokinase in low-glucose media. MDA-MB-231 cells were transfected with either an empty plasmid (pEmpty) or with plasmids carrying genes for PFKM or PFKFB3 and cultured for 3 days in standard DMEM/F12 medium. (A) RT-PCR was used to detect expression of the transfected genes. (B) In a parallel study, cell survival was measured after transfecting MDA-MB-231 cells using 1 μM plasmid and then culturing the cells for 3 days in DMEM/F-12 medium containing 17.5 (normal), 10, and 2.5 mM glucose. (C and D) Other cells were also infected with MYXV at an MOI of 0.1 for 2 days, and virus replication was measured in β-galactosidase (C) and plaque (D) assays. Although dropping the glucose concentration reduced cell viability and thus virus yields, the effect on virus yields was partly reversed by enhancing the expression of PFKM (and to a lesser extent, PFKFB3 expression). The data are representative of two independent experiments. Significance is indicated relative to the pEmpty vector controls, as follows: ^, P ≤ 0.05; *, P ≤ 0.01; **, P ≤ 0.001.
MAPK pathways.
The MAPK signaling pathways mediate a number of cellular responses induced by extracellular signals and/or cellular stress. There are three major MAPK signaling pathways. The extracellular signal-regulated kinases ERK1/2 mediate mitogen-activated proliferative and survival responses, while the Jun N-terminal kinase (JNK) and p38 MAPK signaling pathways regulate cellular responses to stress (72, 73). The importance of these pathways for poxviruses was first suggested by the discovery of growth factor homologs encoded by poxviruses like vaccinia virus (VACV) (74, 75) and Shope fibroma virus and MYXV (76, 77) and the subsequent demonstration that these signal through the Raf/MEK/ERK pathway (78, 79). However, the role Erk1/2 plays in determining MYXV tropism is complex, because although Erk1/2 activation might be advantageous from the perspective of promoting cell proliferation, this activation can also promote Rig-I-dependent expression of a TNF- and type I interferon-dependent antiviral response (6, 80). This is further complicated by the fact that MDA-MB-231 cells encode an activated KRAS gene and express high levels of basal phosphorylated Erk1/2 (81), and such oncogenic RAS mutations can suppress this RNA-induced and Rig-I-mediated IFN-β response (82).
We noted that knocking down many genes upstream of MEK/ERK enhanced MYXV growth (Table 3), consistent with the hypothesis that such interventions suppress activation of an Erk1/2-linked Rig-I-dependent antiviral response. This included genes for brain-derived neurotrophic factor (BDNF), which is expressed by and exerts mitogenic effects on MDA-MB-231 cells (83). We also detected a proviral siRNA targeting the neurotrophic tyrosine kinase receptor NTRK3 (see Table S3 in the supplemental material). However, we noted that knocking down genes in the distal portions of this pathway generally inhibited virus replication. Genes in this region included that for ERK1 (MAPK3) itself, as well as a number of ribosomal protein S6 kinases of uncertain specificity that are phosphorylated and activated by Erk1/2 (RPS6KA6, -A3, and -L1). A proviral role for Erk1/2 itself was also suggested by the fact that knocking down the dual-specificity phosphatases DUSP5 and DUSP6 favored viral growth. DUSP5 and -6 bear some distant similarity to MYXV m069L and VACV H1L, and these human phosphatases inhibit Erk1/2 (84, 85). One of the ribosomal protein S6 kinases, RPS6KA1 (also called RSK1), increased virus replication when targeted with an siRNA. Given the many postulated, overlapping, and observed targets of the ribosomal protein S6 kinases, it is difficult to rationalize the mix of stimulatory and inhibitory effects that result from reducing the expression of this family of ribosomal protein S6 kinases.
Table 3.
Hits detected in MAPK pathways by using the DAVID algorithm
| Gene symbol | Gene name | SSMD |
|---|---|---|
| RPS6KA1 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 1 | 7.06 |
| MAP3K6 | Mitogen-activated protein kinase kinase kinase 6 | 6.89 |
| PRKACG | Protein kinase, cAMP dependent, catalytic, gamma | 6.86 |
| MAP3K7IP2 | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 | 6.83 |
| MOS | v-mos Moloney murine sarcoma viral oncogene homolog | 6.76 |
| PDGFRA | Platelet-derived growth factor receptor, alpha polypeptide | 6.41 |
| MAP2K7 | Mitogen-activated protein kinase kinase 7 | 6.14 |
| PLA2G2A | Phospholipase A2, group IIA (platelets, synovial fluid) | 5.83 |
| MAPKAPK2 | Mitogen-activated protein kinase-activated protein kinase 2 | 5.65 |
| PPP3CC | Protein phosphatase 3 (formerly 2B), catalytic subunit, gamma isoform | 5.62 |
| DUSP6 | Dual-specificity phosphatase 6 | 5.56 |
| MAP4K1 | Mitogen-activated protein kinase kinase kinase kinase 1 | 5.36 |
| BDNF | Brain-derived neurotrophic factor | 5.26 |
| CACNG3 | Calcium channel, voltage dependent, gamma subunit 3 | 5.12 |
| PLA2G1B | Phospholipase A2, group IB (pancreas) | 5.03 |
| MAP3K3 | Mitogen-activated protein kinase kinase kinase 3 | 4.99 |
| TAOK3 | TAO kinase 3 | 4.48 |
| SOS1 | Son of sevenless homolog 1 (Drosophila) | 4.37 |
| DUSP5 | Dual-specificity phosphatase 5 | 4.23 |
| CACNB1 | Calcium channel, voltage dependent, beta 1 subunit | 4.08 |
| RPS6KA6 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 6 | −3.41 |
| CDC25B | Cell division cycle 25 homolog B (S. pombe) | −5.26 |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | −5.77 |
| MAPK7 | Mitogen-activated protein kinase 7 | −6.29 |
| RPS6KA3 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | −6.31 |
| PAK1 | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | −6.92 |
| MAPK3 | Mitogen-activated protein kinase 3 (ERK1) | −7.39 |
| PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | −7.81 |
| MAP2K4 | Mitogen-activated protein kinase kinase 4 | −7.96 |
| MAPK10 | Mitogen-activated protein kinase 10 | −11.32 |
What roles the JNK and p38 signaling pathways might play under these culture conditions are less clear, although relatively more hits, both stimulatory and inhibitory, were detected in the JNK than in the p38 pathway (Table 3; see also Table S3 in the supplemental material). For example, one of the strongest stimulatory hits was in RIPK4 (see Table S3 in the supplemental material), a receptor-interacting stress-activated kinase that promotes JNK and NF-κB activation (86). There was confusion when we saw that although silencing of the kinases in one of the JNK-activating arms (MAP4K1/HPK1 ≫ MAP2K7) also increased MYXV replication, knocking down the kinases in the other arm of the JNK pathway (MAP3K8 > MKK4 > MAPK10/JNK3) paradoxically decreased MYVX replication. It has been noted that a basal level of Jnk1/2 activity is required to avoid G2/M arrest in MDA-MB-231 cells (87, 88), which could provide a rationale for the inhibitory effects linked to knocking down MAP3K8 and MKK4; however, judging from different microarray studies, MDA-MB-231 cells probably express too little of the Jnk3 neuronal isoform to attribute any significance to a hit in the JNK3 gene.
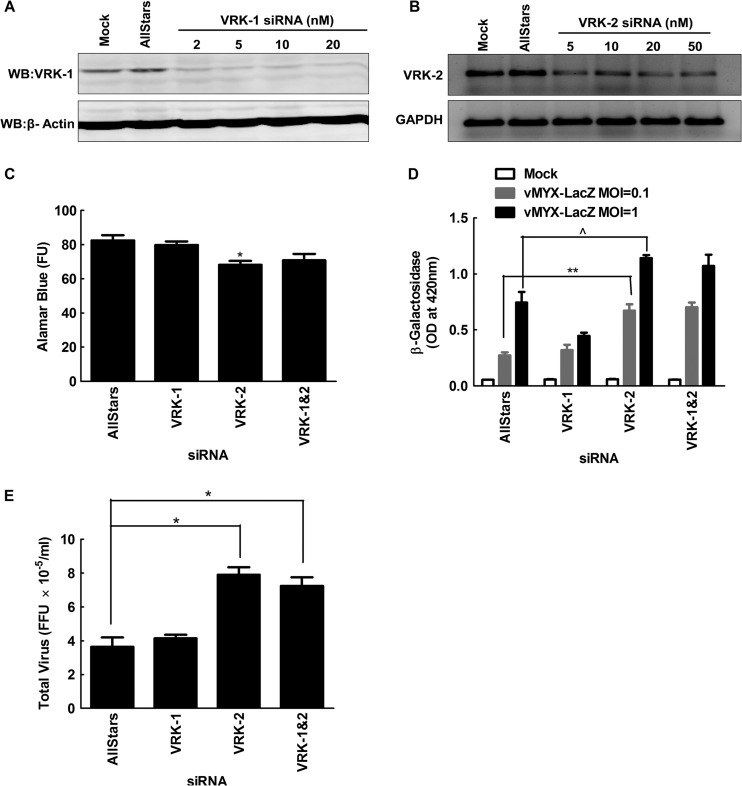
In the course of the initial kinome screen, we also detected hits in the VRK1 and VRK2 vaccinia virus-related kinases, human homologs of the VACV B1R (89) and MYXV m142R genes. VRK1 and VRK2 phosphorylate cellular BAF protein in vitro (90), the antiviral protein also targeted by the B1 kinase. The VRK1 hit subsequently proved to be a false lead, but the stimulation of MYXV growth by siRNAs directed against VRK2 was very reproducible. Its effect seemed to be reduced or eliminated at 72 h and 96 h after infection, suggesting a delay in virus growth kinetics rather than a persistent inhibition of virus replication (Fig. 8; see also Fig. S6B and C in the supplemental material). Previous studies had shown that VRK2 can modulate the activity of the Erb signaling pathway in breast cancer cells by inhibiting Erk phosphorylation (91), as well as the stress response to IL-1β/hypoxia through inhibition of Jnk phosphorylation (92). This effect is probably due to VRK2 interfering with the activity of the scaffolding proteins Jip1 (92) and Ksr1 (91), which promote phosphorylation of Jnk (by Map2k7) and Erk (by Mek1/2), respectively. Further study showed that these siRNAs do knock down VRK2 mRNA (Fig. 8B) and also showed that siRNAs targeting VRK2 had only a minor effect on cell viability (Fig. 8C), while clearly enhancing MYXV replication in these cells (Fig. 8D and E). This is consistent with the observation that knocking down Map2k7 also enhances MYXV growth and that some level of pErk is needed to promote MYXV growth (Table 3). Given the complexity of these interactions, it is difficult to guess exactly what role VRK2 plays in modifying MYXV growth. However, it is striking that poxviruses encode homologs of proteins directly affecting Jnk and/or Erk phosphorylation and that these gene homologs (DUSP5/6 and VRK2) turned up in the screen.
Fig 8.
Silencing VRK-2 enhances MYXV replication in MDA-MB-231 cells. (A and B) MDA-MB-231 cells were transfected with the indicated concentrations of siRNAs for 3 days and then assayed for the effects on VRK-1 (A) or VRK-2 (B) expression by Western blotting or RT-PCR, respectively. (C) Subsequent studies used 10 nM AllStars and VRK-1 siRNAs and 50 nM VRK-2 siRNA, although we detected a slight but still significant (P ≤ 0.01) reduction in the viability of cells transfected with 50 nM VRK-2 siRNA. (D and E) Cells were also treated with siRNAs for 3 days, infected with MYXV for 48 h, and assayed for virus growth in β-galactosidase (D) or plaque (E) assays. Knockdown of VRK-2 significantly enhanced MYXV growth, but the same effect was not seen in cells treated with siRNAs against VRK-1. These data are representative of three independent experiments, and significance is indicated as described for Fig. 7.
Other signal transduction pathways.
Few hits were detected with high statistical likelihood in other cell growth-regulating pathways. Constitutive upregulation of the Akt pathway has been highlighted as playing an important role in MYXV tropism (93), and we did detect a strong proviral siRNA affecting the PTEN locus (see Table S3 in the supplemental material). The PTEN gene product antagonizes the Akt/PKB pathway by dephosphorylating phosphatidylinositol-3,4,5-trisphosphate. It is expressed in MDA-MB-231 cells, and these cells normally exhibit low levels of activated phospho-Akt (94), creating a situation where PTEN knockdown could create an environment more favorable for MYXV growth. We also noted several hits centered on β-catenin (CTNNB1) and the Wnt signaling cascade, which is active with many component genes upregulated in triple-negative breast cancer cell lines like MDA-MB-231 (95). Proviral siRNAs targeted β-catenin itself, the transcription factor TCF/LEF (TCF7L2), which is activated when β-catenin relocates to the nucleus, one of the protein phosphatase regulatory subunits that regulate β-catenin stability (PPP2R4), Axin (AXIN2), a scaffolding protein involved in β-catenin proteolysis, a γ-subunit of protein kinase A (PRKACG) (96), and the Frizzled receptor, although we detected FZD6, and it is FZD7 that is reportedly expressed at the highest levels among the eight FZD genes in MDA-MB-231 cells (95). While the correlation was not perfect (another proviral siRNA was detected affecting DKK4, which may or may not be an inhibitor of Wnt signaling [97]), the impression is that destabilizing or decreasing the levels of β-catenin favors virus growth. It is unclear whether this relates to Wnt signaling or to the role β-catenin plays in stabilizing intercellular adherens junctions and the actin cytoskeleton (98). However, it is notable that knocking down LIMK2, which would be expected to destabilize the actin cytoskeleton (99), and DIAPH1, which would disrupt adherens junctions (100), also promotes MYXV growth. We recently showed that MYXV has difficulties in disrupting the cortical actin layer during exit, and this can be partially ameliorated by incorporating the VACV F11L gene (101). This provides some rationale in support for the later structural hypothesis, although the two biological effects are not mutually exclusive.
The cell cycle.
The cell cycle is known to play an important role in modulating MYXV replication, as illustrated by studies showing that the MYXV M-T5 protein promotes p27 degradation and exit from the G0/G1 checkpoint (102). It is thus perhaps not surprising that a striking pattern of siRNA hits was observed among genes linked to, or regulating, the cell cycle (Table 4). Interventions that might promote the transition into G1, or inhibit passage from G1 into S phase, in uninfected cells consistently stimulated MYXV growth, whereas knockdown of genes that regulate passage throughout the rest of the cell cycle consistently decreased virus growth (Table 4). For example E2F5 appears to suppress expression of genes required for entry into G1 (103), and its knockdown stimulates MYXV growth. Similarly, CDK6 regulates progression out of G1 in MDA-MB-231 cells through hyperphosphorylation of retinoblastoma protein and release of activator E2Fs (104), and also by catalyzing the assembly of the prereplicative complex during G1 (105). Our screen detected proviral siRNAs targeting both CDK6 and its regulatory binding partner cyclin D2 (CCND2) (106). The process of “DNA licensing” in G1 also requires that the Mcm proteins be incorporated into a hexameric MCM2-7 helicase (105), and siRNAs targeting one of these genes (MCM2) also stimulated MYXV growth. Finally, another proviral siRNA was also detected targeting cyclin H (CCNH), a protein linked to many cellular activities, including the aforementioned role as part of TFIIH (107).
Table 4.
Hits detected in the cell cycle by using the DAVID algorithm
| Gene symbol | Gene name | SSMD |
|---|---|---|
| E2F5 | E2F transcription factor 5, p130 binding | 6.67 |
| MCM2 | Minichromosome maintenance complex component 2 | 5.95 |
| CDK6 | Cyclin-dependent kinase 6 | 5.89 |
| LOC646096 | Protein kinase CHK2-like; CHK2 checkpoint homolog (S. pombe); similar to hCG1983233 | 5.70 |
| CCNH | Cyclin H | 5.39 |
| CCND2 | Cyclin D2 | 4.68 |
| ANAPC13 | Anaphase-promoting complex subunit 13 | 3.86 |
| BUB1B | Budding uninhibited by benzimidazoles 1 homolog beta (yeast) | −4.42 |
| PRKDC | Similar to protein kinase, DNA activated, catalytic polypeptide | −4.85 |
| CDC25B | Cell division cycle 25 homolog B (S. pombe) | −5.26 |
| PLK1 | Polo-like kinase 1 (Drosophila); toxic | −6.45 |
| TTK | TTK protein kinase | −7.88 |
| CHEK1 | CHK1 checkpoint homolog (S. pombe) | −8.21 |
| STAG2 | Stromal antigen 2 | −8.89 |
| WEE1 | WEE1 homolog (S. pombe); toxic | −9.57 |
In contrast, MYXV growth was inhibited by siRNAs targeting genes affecting or regulating subsequent events in the cell cycle. For example, the NPAT gene is required for S-phase histone expression and cell cycle progression (108), and siRNAs targeting NPAT were highly inhibitory. Two of the siRNA pools were toxic for MDA-MB-231 cells (WEE1 and PLK1). Wee1 and Plk1 comprise a regulatory triad with Cdc2 (also called Cdk1), and inhibiting the expression of either Wee1 or Plk1 within this negative feedback loop might be expected to improperly promote the G2/M transition by releasing Cdc2/Cdk1 from Wee1 inhibition (109). We also noted that knocking down genes involved in regulating the DNA damage checkpoints had deleterious effects on virus growth (although an important caveat is that MDA-MB-231 cells encode a mutated p53 gene, which complicates the interpretation of signaling patterns [110, 111]). For example, DNA protein kinase (PRKDC) normally phosphorylates RPA2 in the presence of DNA damage and delays S-phase progression and mitotic entry (112), and interfering with this system by knocking down PRKDC inhibited MYXV growth. Additional antiviral siRNAs targeting CHEK1 (also called CHK1) and a downstream effector, CDC25B, were discovered. When activated by DNA damage signals, Chk1 phosphorylates (and inhibits) Cdc25B (113), stabilizing the phosphorylated form of Cdc2/Cdk1 and favoring G2 arrest. (Use of siRNAs to inhibit Wee1 and CHEK1 during S phase also causes unscheduled DNA replication and chromosome breaks and could deplete cells of dNTPs [114].) Adding further to the complexity, Cdc25B, Cdc2/Cdk1, and Plk1 form another regulatory triad (115, 116) that when perturbed can promote aberrant mitosis (113). These data suggest that defective regulation of events outside of G1 creates a situation deleterious to MYXV growth. This hypothesis can perhaps be extended by noting that knockdown of two genes required by the spindle assembly checkpoint TTK (also called Mps1) (117) and BUB1B (118) also inhibits MYXV growth, although siRNAs directed against TPR (a component of the nuclear pore that is specifically required to assemble the mitotic spindle checkpoint complex [119]) seemed to stimulate virus growth. Some of these effects may be related to the fact that cap-dependent translation is inhibited in mitotic cells (120), or to regulation of the APC complex (118, 121). However, further studies are needed to clarify this situation.
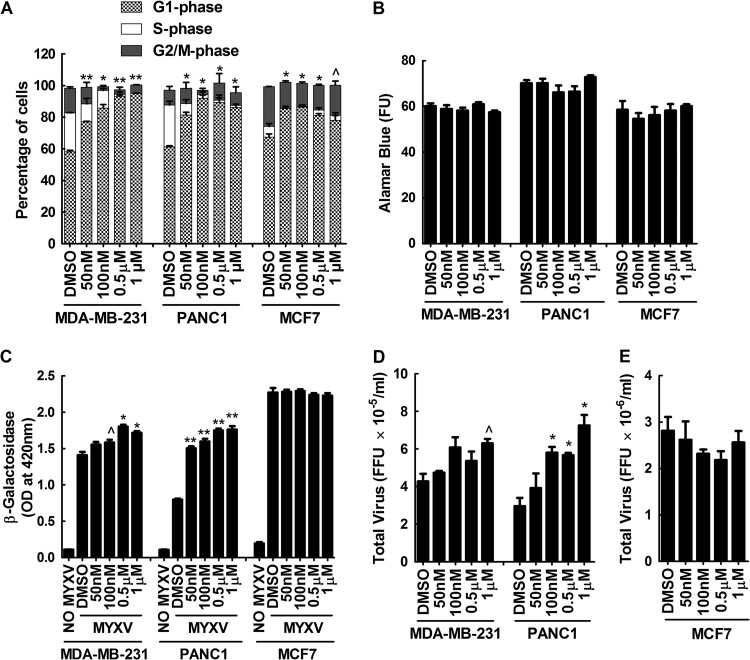
These observations led us to test the hypothesis that MYVX replication is favored in cells when they are initially encountered at the G1 (or the G1/S boundary) portion of the cell cycle. First we tested whether treating cells with the Cdk4/6 inhibitor PD-0332991 (122) would produce the same proviral effects seen with siRNAs targeting CDK6. Such treatments should stabilize cells in G1, and this was tested by flow cytometric measurement of the DNA content in three different kinds of PD-0332991-treated cells: MDA-MB-231, PANC1, and MCF7 (Fig. 9A). These cells were also then tested to measure the effects of PD-0332991 on MYXV growth, and the effects of the siRNA targeting CDK6 were duplicated in MDA-MB-231 and PANC1 cells. Applying 1 μM PD-0332991 to MDA-MB-231 cells (twice the 50% inhibitory concentration [122]) increased the proportion of cells in G1 from 58% to 95% and enhanced MYXV growth by about 30% (Fig. 9C and D). A similar effect was seen in PANC1 cells (Fig. 9C and D). The drug did not enhance MYXV growth in MCF7 cells, although PD-0332991 also had a lesser effect upon the proportion of G1 cells (Fig. 9A, C, and E). MCF7 cells are also far more supportive of MYXV growth than are MDA-MB-231 or PANC1 cells, suggesting that the cell cycle may not be a factor that limits virus growth in these cells. If we assume that PD-0332991 is a specific inhibitor of Cdk4/6, these studies support the hypothesis that the G1 phase of the cell cycle is more advantageous for virus growth.
Fig 9.
Inhibition of cell cycling with the CDK4/6 inhibitor PD-0332991 enhances MYXV growth in some cancer cell lines. (A and B) MDA-MB-231, PANC1, and MCF7 cells were treated with the indicated doses of PD-0332991 for 24 h, trypsinized, and stained with propidium iodide, and flow cytometry was used to determine the distribution of cells across the cell cycle. Low (50 nM) doses of the drug significantly increased the proportion of G1-phase MDA-MB-231 and PANC1 cells and (to a lesser extent) also increased the proportion of G1-phase MCF7 cells (A), without significant toxicity (B). (C to E) The drug-treated cells were also infected with MYXV for 48 h and assayed for virus-encoded β-galactosidase and FFU. PD-0332991 significantly enhanced MYXV growth in PANC1 and MDA-MB-231 cells relative to the solvent controls but had no effect on MCF7 cells. These data are representative of three independent experiments, and significance is indicated as described for Fig. 7.
DNA primase.
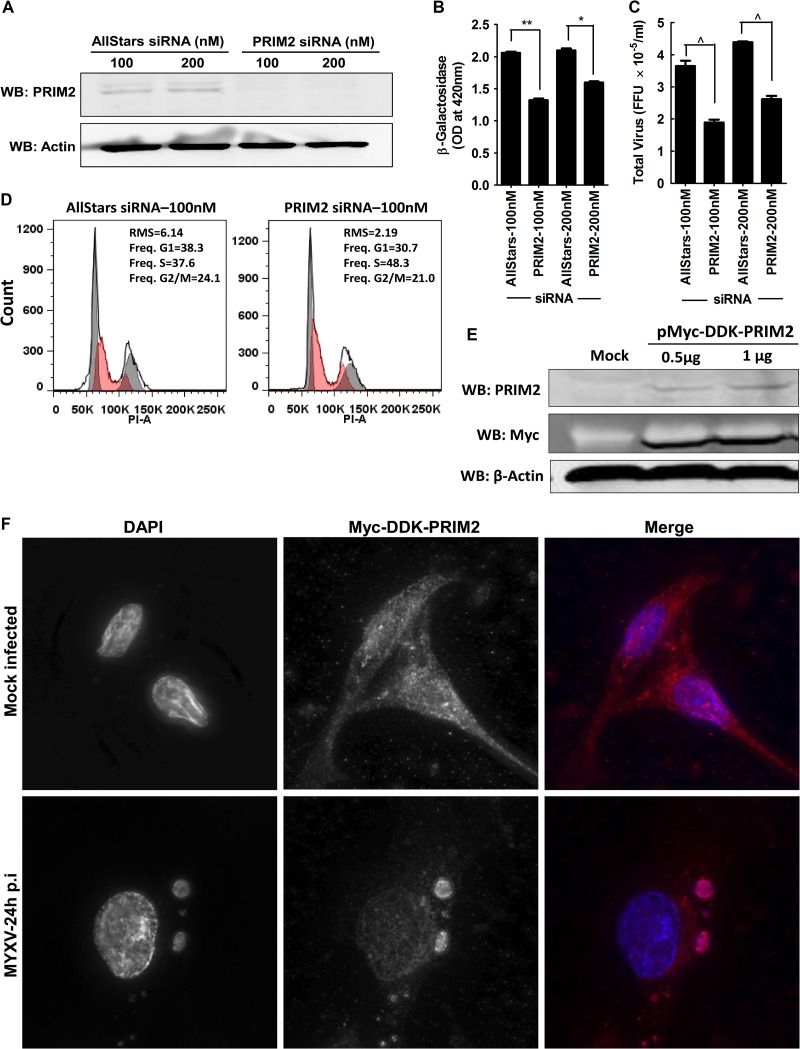
We also wanted to test how well MYXV grows when cells are stabilized in S phase, but this issue is complicated by the fact that most S-phase inhibitors poison DNA replication and thus also directly inhibit virus replication. However, we had noted that siRNA silencing of PRIM2 expression also inhibited MYXV growth (see Table S3 in the supplemental material), and this provided another way of exploring how progression through the cell cycle affects MYXV growth. The mammalian DNA primase is a heterodimer composed of a catalytic subunit (PRIM1) and an accessory subunit (PRIM2) (123–125). In Schizosaccharomyces pombe, disruption of primase function activates the Chk1 checkpoint response (126), and we suspected that this effect might also be observed in human cells. Independent experiments confirmed that siRNA silencing of PRIM2 (Fig. 10A) caused a 25 to 40% decrease in MYXV growth as demonstrated in both β-galactosidase (Fig. 10B) and plaque (Fig. 10C) assays, although the degree of reduction in virus replication was reduced as time of infection advanced beyond 48 h (see Fig. S6D in the supplemental material). We also observed that inhibition of PRIM2 function had no negative effects on cell survival at doses up to 200 nM (data not shown) while it increased the proportion of cells in S phase (Fig. 10D). This is consistent with knockdown of PRIM2 causing a delay in S-phase progression and supports the hypothesis that MYXV grows best when cells are in the G1 or the G1/S part of the cell cycle.
Fig 10.
PRIM2 modulates MYXV growth in MDA-MB-231 cells and relocates to virus factories. (A) MDA-MB-231 cells were transfected with control or PRIM2 siRNA for 72 h, and then a Western blot assay was performed to show PRIM2 knockdown. (B and C) A 3-day treatment with PRIM2 siRNA also caused a significant reduction in MYXV growth (MOI, 0.1; 48 h) as judged with β-galactosidase (B) and plaque (C) assays. (D) Propidium iodide staining and flow cytometry showed that siRNA silencing of PRIM2 produced a 10% increase in the proportion of S-phase cells at the expense of G1- and G2/M-phase cells. The curve fit for S-phase cells is shaded in red. (E and F) To gain further understanding of the behavior of Prim2 under these circumstances, a myc- and DDK-tagged form of the gene was also transfected into MDA-MB-231 cells, where it was overexpressed, as judged by Western blotting (E) and appeared to relocate to MYXV factories (F). These data are representative of three independent experiments, and significance is indicated as described above for Fig. 7.
These conclusions are complicated by one further observation. We had assumed that since poxviruses encode their own DNA primase (the VACV D5 protein exhibits a helicase-primase activity [127]), there would be no reason to expect that cellular PRIM2 would play any direct role in virus DNA replication. However, as a check on this assumption, we transfected cells with a plasmid carrying a myc-tagged version of PRIM2 (none of the available antibodies were suitable for use in imaging applications) and then monitored the distribution of the protein in mock- and MYXV-infected cells. In the absence of virus, PRIM2 was mostly distributed throughout the cytoplasm, with some small amount of nuclear staining, but curiously, it appeared to be recruited to factories in MYXV-infected cells (Fig. 10F). This may not have any significance, as many DNA-binding proteins are recruited to virus factories (summarized in reference 15), but we cannot exclude the possibility that PRIM2 also serves a more direct role in supporting MYXV growth.
Enhancement of oncolysis.
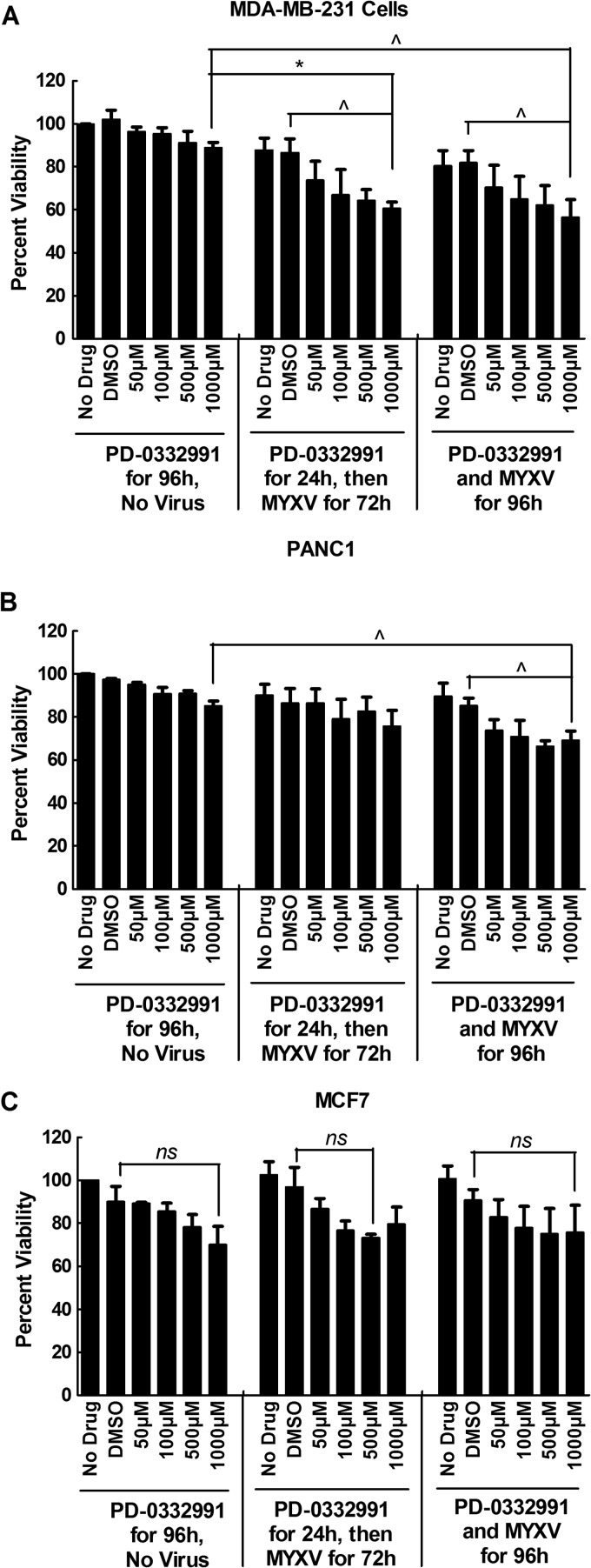
Finally, we also examined whether these studies might produce methods for improving virus killing of tumor cells. We hypothesized that combining a drug causing G1-phase growth arrest, PD-0332991, with MYXV infection would cause enhanced oncolysis. By itself, treating MDA-MB-231 cells with up to 1 mM PD-0332991 for 4 days caused only a small (but not significant) reduction in cell viability, while infecting these cells with MYXV (at an MOI of 1) for 3 or 4 days killed 15 to 20% of the cells relative to uninfected and nontreated controls (Fig. 11). The amount of cell killing was further enhanced ∼20% either by simultaneous exposure of the cells to PD-0332991 and MYXV for 4 days or by pretreating the cells with PD-0332991 for a day followed by 3 days of MYXV exposure (Fig. 11A). A similar effect was seen with PANC1 cells, albeit statistical significance was obtained only if the cells were exposed to virus for 4 days (Fig. 11B). In contrast, these effects were not seen with PD-0332991-treated MCF7 cells, which as we previously noted do not show the same drug-induced changes in the cell cycle that are seen in MDA-MB-231 or PANC1 cells (Fig. 9). Overall, these results suggest that MYXV oncolytic activity could be modestly enhanced by cotreatment with a class of chemotherapeutic drugs that promote accumulation of cells in G1 phase, although this effect would likely be seen only in select cancer cells.
Fig 11.
Treating cells with the CDK4/6 inhibitor PD-0332991 enhances the oncolytic activity of MYXV. MDA-MB-231 (A), PANC1 (B), and MCF7 (C) cells were treated with the indicated concentrations of PD-0332991 alone for 4 days or pretreated with PD-0332991 for 24 h and then infected with MYXV for 3 days (MOI, 1), or they were simultaneously infected and treated with drug and virus for 4 days. Drug treatment appeared to reduce cell viability, but not enough to achieve significance. However, the effects of drug and virus appeared to be additive in MDA-MB-231 cells, with the combination treatment killing ∼40% of the cells, compared to the 10 to 15% killing caused by virus alone. The effect was also seen in PANC1 cells, but not in MCF7 cells. These data combine the results of three independent experiments, each employing three replicates.
Conclusions.
Our study has provided a broad survey of the role individual genes play in regulating the growth of myxoma virus in human cells. The screens encompassed ∼22,000 genes and returned ∼1,000 possible hits, with ∼25% probability of true positivity for any one gene hit, absent independent confirmation. Of the hits, twice as many genes appeared to serve an antiviral rather than a proviral role. That is, virus growth was stimulated by siRNAs targeting 711 genes and was inhibited by 333 siRNA pools. This is not the outcome that one would expect if many siRNAs simply decreased cell fitness and thus rendered cells less supportive of virus growth, and it suggests that organisms might encode a great many minor systems that collectively create innate antiviral defenses. Although concerns have been raised about the reliability of siRNA screening methods and the effects on virus replication are sometimes quite modest, our approach is reproducible (Fig. 2; see also Fig. S4 in the supplemental material) and did identify several cellular pathways that have been shown to affect poxvirus growth (e.g., the MAPK and ubiquitin-regulated proteolytic pathways) as well as toxic siRNAs that would, of course, be expected to “inhibit” virus replication. Many new hits were mapped to the glycolytic pathway, an important downstream target of the PTEN/Akt axis, and these provide further insights into why Akt mutations affect MYXV tropism (56). The more limited complement of nucleotide biosynthetic genes of leporipoxviruses (16), relative to VACV, may also account for why MYXV favors initiation of growth in cells occupying the G1 or G1/S boundary of the cell cycle, and a similar deficiency in genes mediating MYXV exit (101) may also partly explain several hits in the Wnt/Rho signaling cascade. Although much more effort will be needed to fully document these and other hits detected in the screen, our approach has provided a first-pass filter for genes affecting poxvirus growth that are encoded in the human genome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ulrike Strunk for her assistance, M. Hitt and M. Shmulevitz for their comments on the manuscript, and the Faculty of Medicine and Dentistry for support of the siRNA screening core.
W.M.T. is a recipient of an Alberta Cancer Foundation graduate scholarship. This work was funded by grants to D.H.E. from the Natural Sciences and Engineering Research Council of Canada and from the Canada Foundation for Innovation.
Footnotes
Published ahead of print 13 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02617-12.
REFERENCES
- 1. Fenner F. 2000. Adventures with poxviruses of vertebrates. FEMS Microbiol. Rev. 24:123–133 [DOI] [PubMed] [Google Scholar]
- 2. Fenner F, Fantini B. 1999. Biological control of vertebrate pests: the history of myxomatosis—an experiment in evolution. CABI Publishing, New York, NY [Google Scholar]
- 3. Kerr PJ. 2012. Myxomatosis in Australia and Europe: a model for emerging infectious diseases. Antiviral Res. 93:387–415 [DOI] [PubMed] [Google Scholar]
- 4. Werden SJ, Barrett JW, Wang G, Stanford MM, McFadden G. 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J. Virol. 81:2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Opgenorth A, Nation N, Graham K, McFadden G. 1993. Transforming growth factor alpha, Shope fibroma growth factor, and vaccinia growth factor can replace myxoma growth factor in the induction of myxomatosis in rabbits. Virology 192:701–709 [DOI] [PubMed] [Google Scholar]
- 6. Wang F, Ma Y, Barrett JW, Gao X, Loh J, Barton E, Virgin HW, McFadden G. 2004. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 5:1266–1274 [DOI] [PubMed] [Google Scholar]
- 7. Kim M, Williamson CT, Prudhomme J, Bebb DG, Riabowol K, Lee PW, Lees-Miller SP, Mori Y, Rahman MM, McFadden G, Johnston RN. 2010. The viral tropism of two distinct oncolytic viruses, reovirus and myxoma virus, is modulated by cellular tumor suppressor gene status. Oncogene 29:3990–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang F, Barrett JW, Ma Y, Dekaban GA, McFadden G. 2009. Induction of alpha/beta interferon by myxoma virus is selectively abrogated when primary mouse embryo fibroblasts become immortalized. J. Virol. 83:5928–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377–423 [DOI] [PubMed] [Google Scholar]
- 10. Spiesschaert B, McFadden G, Hermans K, Nauwynck H, Van de Walle GR. 2011. The current status and future directions of myxoma virus, a master in immune evasion. Vet. Res. 42:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Werden SJ, McFadden G. 2008. The role of cell signaling in poxvirus tropism: the case of the M-T5 host range protein of myxoma virus. Biochim. Biophys. Acta 1784:228–237 [DOI] [PubMed] [Google Scholar]
- 12. Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao JX, Macaulay C, Willer D, Evans D, McFadden G. 1999. The complete DNA sequence of myxoma virus. Virology 264:298–318 [DOI] [PubMed] [Google Scholar]
- 13. Duteyrat JL, Gelfi J, Bertagnoli S. 2006. Ultrastructural study of myxoma virus morphogenesis. Arch. Virol. 151:2161–2180 [DOI] [PubMed] [Google Scholar]
- 14. Zhang L, Villa NY, Rahman MM, Smallwood S, Shattuck D, Neff C, Dufford M, Lanchbury JS, Labaer J, McFadden G. 2009. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J. Proteome Res. 8:4311–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin YC, Li J, Irwin CR, Jenkins H, DeLange L, Evans DH. 2008. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J. Virol. 82:5922–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willer DO, McFadden G, Evans DH. 1999. The complete genome sequence of Shope (rabbit) fibroma virus. Virology 264:319–343 [DOI] [PubMed] [Google Scholar]
- 17. Gammon DB, Gowrishankar B, Duraffour S, Andrei G, Upton C, Evans DH. 2010. Vaccinia virus-encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis. PLoS Pathog. 6:e1000984 doi:10.1371/journal.ppat.1000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reichard P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349–374 [DOI] [PubMed] [Google Scholar]
- 19. Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 19:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]
- 23. Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504 [DOI] [PubMed] [Google Scholar]
- 24. Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 106:16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822 [DOI] [PubMed] [Google Scholar]
- 28. Panda D, Das A, Dinh PX, Subramaniam S, Nayak D, Barrows NJ, Pearson JL, Thompson J, Kelly DL, Ladunga I, Pattnaik AK. 2011. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:19036–19041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. 2010. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 6:e1000954 doi:10.1371/journal.ppat.1000954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goff SP. 2008. Knockdown screens to knockout HIV-1. Cell 135:417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeung ML, Houzet L, Yedavalli VS, Jeang KT. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284:19463–19473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6:e292 doi:10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohr S, Bakal C, Perrimon N. 2010. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 79:37–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199–204 [DOI] [PubMed] [Google Scholar]
- 37. Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635–637 [DOI] [PubMed] [Google Scholar]
- 38. Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qiu S, Adema CM, Lane T. 2005. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 33:1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. 2012. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods 9:363–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagenblatt A, Barrionuevo LS, Lichter P, Mertens D. 2008. Off-target effects of siRNA specific for GFP. BMC Mol. Biol. 9:60 doi:10.1186/1471-2199-9-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Opgenorth A, Graham K, Nation N, Strayer D, McFadden G. 1992. Deletion analysis of two tandemly arranged virulence genes in myxoma virus, M11L and myxoma growth factor. J. Virol. 66:4720–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parsons BD, Schindler A, Evans DH, Foley E. 2009. A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One 4:e8471 doi:10.1371/journal.pone.0008471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lundholm L, Mohme-Lundholm E, Vamos N. 1963. Lactic acid assay with L(+)lactic acid dehydrogenase from rabbit muscle. Acta Physiol. Scand. 58:243–249 [DOI] [PubMed] [Google Scholar]
- 45. Hamid R, Rotshteyn Y, Rabadi L, Parikh R, Bullock P. 2004. Comparison of alamar blue and MTT assays for high throughput screening. Toxicol. In Vitro 18:703–710 [DOI] [PubMed] [Google Scholar]
- 46. Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE. 2009. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang XD, Ferrer M, Espeseth AS, Marine SD, Stec EM, Crackower MA, Holder DJ, Heyse JF, Strulovici B. 2007. The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen. 12:497–509 [DOI] [PubMed] [Google Scholar]
- 48. Zhang XD, Lacson R, Yang R, Marine SD, McCampbell A, Toolan DM, Hare TR, Kajdas J, Berger JP, Holder DJ, Heyse JF, Ferrer M. 2010. The use of SSMD-based false discovery and false nondiscovery rates in genome-scale RNAi screens. J. Biomol. Screen. 15:1123–1131 [DOI] [PubMed] [Google Scholar]
- 49. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang da W, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 52. Luc PV, Tempst P. 2004. PINdb: a database of nuclear protein complexes from human and yeast. Bioinformatics 20:1413–1415 [DOI] [PubMed] [Google Scholar]
- 53. Cailleau R, Young R, Olive M, Reeves WJ., Jr 1974. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 53:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wright M, Grim J, Deshane J, Kim M, Strong TV, Siegal GP, Curiel DT. 1997. An intracellular anti-erbB-2 single-chain antibody is specifically cytotoxic to human breast carcinoma cells overexpressing erbB-2. Gene Ther. 4:317–322 [DOI] [PubMed] [Google Scholar]
- 55. Tzahar E, Moyer JD, Waterman H, Barbacci EG, Bao J, Levkowitz G, Shelly M, Strano S, Pinkas-Kramarski R, Pierce JH, Andrews GC, Yarden Y. 1998. Pathogenic poxviruses reveal viral strategies to exploit the ErbB signaling network. EMBO J. 17:5948–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang G, Barrett JW, Stanford M, Werden SJ, Johnston JB, Gao X, Sun M, Cheng JQ, McFadden G. 2006. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc. Natl. Acad. Sci. U. S. A. 103:4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boutros M, Bras LP, Huber W. 2006. Analysis of cell-based RNAi screens. Genome Biol. 7:R66 doi:10.1186/gb-2006-7-7-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bonvini P, Zorzi E, Basso G, Rosolen A. 2007. Bortezomib-mediated 26S proteasome inhibition causes cell-cycle arrest and induces apoptosis in CD-30+ anaplastic large cell lymphoma. Leukemia 21:838–842 [DOI] [PubMed] [Google Scholar]
- 59. Satheshkumar PS, Anton LC, Sanz P, Moss B. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 83:2469–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Teale A, Campbell S, Van Buuren N, Magee WC, Watmough K, Couturier B, Shipclark R, Barry M. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J. Virol. 83:2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Levine AJ, Puzio-Kuter AM. 2010. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330:1340–1344 [DOI] [PubMed] [Google Scholar]
- 62. Gatenby RA, Gillies RJ. 2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4:891–899 [DOI] [PubMed] [Google Scholar]
- 63. Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. 2001. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 15:1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH. 2002. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim. Biophys. Acta 1555:14–20 [DOI] [PubMed] [Google Scholar]
- 65. Wallace DC. 2005. Mitochondria and cancer: Warburg addressed. Cold Spring Harbor Symp. Quant Biol. 70:363–374 [DOI] [PubMed] [Google Scholar]
- 66. Kun E, Smith MH. 1950. Effect of infectious myxoma virus on glycolysis of chorioallantoic membrane of chick embryo. Proc. Soc. Exp. Biol. Med. 73:628–631 [DOI] [PubMed] [Google Scholar]
- 67. Weber G. 1977. Enzymology of cancer cells (second of two parts). N. Engl. J. Med. 296:541–551 [DOI] [PubMed] [Google Scholar]
- 68. Van Schaftingen E, Hers HG. 1981. Phosphofructokinase 2: the enzyme that forms fructose 2,6-bisphosphate from fructose 6-phosphate and ATP. Biochem. Biophys. Res. Commun. 101:1078–1084 [DOI] [PubMed] [Google Scholar]
- 69. Van Schaftingen E, Hue L, Hers HG. 1980. Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem. J. 192:897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Van Schaftingen E, Jett MF, Hue L, Hers HG. 1981. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc. Natl. Acad. Sci. U. S. A. 78:3483–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bando H, Atsumi T, Nishio T, Niwa H, Mishima S, Shimizu C, Yoshioka N, Bucala R, Koike T. 2005. Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin. Cancer Res. 11:5784–5792 [DOI] [PubMed] [Google Scholar]
- 72. Kyriakis JM, Avruch J. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807–869 [DOI] [PubMed] [Google Scholar]
- 73. Sears RC, Nevins JR. 2002. Signaling networks that link cell proliferation and cell fate. J. Biol. Chem. 277:11617–11620 [DOI] [PubMed] [Google Scholar]
- 74. Brown JP, Twardzik DR, Marquardt H, Todaro GJ. 1985. Vaccinia virus encodes a polypeptide homologous to epidermal growth factor and transforming growth factor. Nature 313:491–492 [DOI] [PubMed] [Google Scholar]
- 75. Twardzik DR, Brown JP, Ranchalis JE, Todaro GJ, Moss B. 1985. Vaccinia virus-infected cells release a novel polypeptide functionally related to transforming and epidermal growth factors. Proc. Natl. Acad. Sci. U. S. A. 82:5300–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chang W, Upton C, Hu SL, Purchio AF, McFadden G. 1987. The genome of Shope fibroma virus, a tumorigenic poxvirus, contains a growth factor gene with sequence similarity to those encoding epidermal growth factor and transforming growth factor alpha. Mol. Cell. Biol. 7:535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Upton C, Macen JL, McFadden G. 1987. Mapping and sequencing of a gene from myxoma virus that is related to those encoding epidermal growth factor and transforming growth factor alpha. J. Virol. 61:1271–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Andrade AA, Silva PN, Pereira AC, De Sousa LP, Ferreira PC, Gazzinelli RT, Kroon EG, Ropert C, Bonjardim CA. 2004. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 381:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schweneker M, Lukassen S, Spath M, Wolferstatter M, Babel E, Brinkmann K, Wielert U, Chaplin P, Suter M, Hausmann J. 2012. The vaccinia virus O1 protein is required for sustained activation of extracellular signal-regulated kinase 1/2 and promotes viral virulence. J. Virol. 86:2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang F, Gao X, Barrett JW, Shao Q, Bartee E, Mohamed MR, Rahman M, Werden S, Irvine T, Cao J, Dekaban GA, McFadden G. 2008. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 4:e1000099 doi:10.1371/journal.ppat.1000099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lonne GK, Masoumi KC, Lennartsson J, Larsson C. 2009. Protein kinase Cδ supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J. Biol. Chem. 284:33456–33465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shmulevitz M, Pan LZ, Garant K, Pan D, Lee PW. 2010. Oncogenic Ras promotes reovirus spread by suppressing IFN-beta production through negative regulation of RIG-I signaling. Cancer Res. 70:4912–4921 [DOI] [PubMed] [Google Scholar]
- 83. Adriaenssens E, Vanhecke E, Saule P, Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X, Hondermarck H. 2008. Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Res. 68:346–351 [DOI] [PubMed] [Google Scholar]
- 84. Arkell RS, Dickinson RJ, Squires M, Hayat S, Keyse SM, Cook SJ. 2008. DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5. Cell. Signal. 20:836–843 [DOI] [PubMed] [Google Scholar]
- 85. Mandl M, Slack DN, Keyse SM. 2005. Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase DUSP5. Mol. Cell. Biol. 25:1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J. 2002. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 3:1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mingo-Sion AM, Marietta PM, Koller E, Wolf DM, Van Den Berg CL. 2004. Inhibition of JNK reduces G2/M transit independent of p53, leading to endoreduplication, decreased proliferation, and apoptosis in breast cancer cells. Oncogene 23:596–604 [DOI] [PubMed] [Google Scholar]
- 88. Wang L, Pan Y, Dai JL. 2004. Evidence of MKK4 pro-oncogenic activity in breast and pancreatic tumors. Oncogene 23:5978–5985 [DOI] [PubMed] [Google Scholar]
- 89. Nezu J, Oku A, Jones MH, Shimane M. 1997. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45:327–331 [DOI] [PubMed] [Google Scholar]
- 90. Nichols RJ, Wiebe MS, Traktman P. 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17:2451–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fernandez IF, Blanco S, Lozano J, Lazo PA. 2010. VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol. Cell. Biol. 30:4687–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Blanco S, Sanz-Garcia M, Santos CR, Lazo PA. 2008. Modulation of interleukin-1 transcriptional response by the interaction between VRK2 and the JIP1 scaffold protein. PLoS One 3:e1660 doi:10.1371/journal.pone.0001660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stanford MM, Barrett JW, Nazarian SH, Werden S, McFadden G. 2007. Oncolytic virotherapy synergism with signaling inhibitors: Rapamycin increases myxoma virus tropism for human tumor cells. J. Virol. 81:1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yu K, Toral-Barza L, Discafani C, Zhang WG, Skotnicki J, Frost P, Gibbons JJ. 2001. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr. Relat. Cancer 8:249–258 [DOI] [PubMed] [Google Scholar]
- 95. Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, Deng X, Chen L, Kim CC, Lau S, Somlo G, Yen Y. 2011. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene 30:4437–4446 [DOI] [PubMed] [Google Scholar]
- 96. Ke J, Zhang C, Harikumar KG, Zylstra-Diegel CR, Wang L, Mowry LE, Miller LJ, Williams BO, Xu HE. 2012. Modulation of beta-catenin signaling by glucagon receptor activation. PLoS One 7:e33676 doi:10.1371/journal.pone.0033676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pendas-Franco N, Garcia JM, Pena C, Valle N, Palmer HG, Heinaniemi M, Carlberg C, Jimenez B, Bonilla F, Munoz A, Gonzalez-Sancho JM. 2008. DICKKOPF-4 is induced by TCF/beta-catenin and upregulated in human colon cancer, promotes tumour cell invasion and angiogenesis and is repressed by 1α,25-dihydroxyvitamin D3. Oncogene 27:4467–4477 [DOI] [PubMed] [Google Scholar]
- 98. Shapiro L, Weis WI. 2009. Structure and biochemistry of cadherins and catenins. Cold Spring Harb. Perspect. Biol. 1:a003053 doi:10.1101/cshperspect.a003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sumi T, Matsumoto K, Takai Y, Nakamura T. 1999. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147:1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Carramusa L, Ballestrem C, Zilberman Y, Bershadsky AD. 2007. Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J. Cell Sci. 120:3870–3882 [DOI] [PubMed] [Google Scholar]
- 101. Irwin CR, Evans DH. 2012. Modulation of the myxoma virus plaque phenotype by vaccinia virus F11 protein. J. Virol. 86:7167–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Johnston JB, Wang G, Barrett JW, Nazarian SH, Colwill K, Moran M, McFadden G. 2005. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J. Virol. 79:10750–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen HZ, Tsai SY, Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cover CM, Hsieh SJ, Tran SH, Hallden G, Kim GS, Bjeldanes LF, Firestone GL. 1998. Indole-3-carbinol inhibits the expression of cyclin-dependent kinase-6 and induces a G1 cell cycle arrest of human breast cancer cells independent of estrogen receptor signaling. J. Biol. Chem. 273:3838–3847 [DOI] [PubMed] [Google Scholar]
- 105. Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333–374 [DOI] [PubMed] [Google Scholar]
- 106. Sherr CJ. 1995. D-type cyclins. Trends Biochem. Sci. 20:187–190 [DOI] [PubMed] [Google Scholar]
- 107. Nigg EA. 1996. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr. Opin. Cell Biol. 8:312–317 [DOI] [PubMed] [Google Scholar]
- 108. Gao G, Bracken AP, Burkard K, Pasini D, Classon M, Attwooll C, Sagara M, Imai T, Helin K, Zhao J. 2003. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 23:2821–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc. Natl. Acad. Sci. U. S. A. 101:4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Negrini M, Sabbioni S, Haldar S, Possati L, Castagnoli A, Corallini A, Barbanti-Brodano G, Croce CM. 1994. Tumor and growth suppression of breast cancer cells by chromosome 17-associated functions. Cancer Res. 54:1818–1824 [PubMed] [Google Scholar]
- 111. Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. 2007. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11:175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liaw H, Lee D, Myung K. 2011. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One 6:e21424 doi:10.1371/journal.pone.0021424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Loffler H, Rebacz B, Ho AD, Lukas J, Bartek J, Kramer A. 2006. Chk1-dependent regulation of Cdc25B functions to coordinate mitotic events. Cell Cycle 5:2543–2547 [DOI] [PubMed] [Google Scholar]
- 114. Sorensen CS, Syljuasen RG. 2012. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lee HJ, Hwang HI, Jang YJ. 2010. Mitotic DNA damage response: Polo-like kinase-1 is dephosphorylated through ATM-Chk1 pathway. Cell Cycle 9:2389–2398 [DOI] [PubMed] [Google Scholar]
- 116. Lobjois V, Froment C, Braud E, Grimal F, Burlet-Schiltz O, Ducommun B, Bouche JP. 2011. Study of the docking-dependent PLK1 phosphorylation of the CDC25B phosphatase. Biochem. Biophys. Res. Commun. 410:87–90 [DOI] [PubMed] [Google Scholar]
- 117. Lan W, Cleveland DW. 2010. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J. Cell Biol. 190:21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. 2011. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 124:4332–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lee SH, Sterling H, Burlingame A, McCormick F. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pyronnet S, Dostie J, Sonenberg N. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Barford D. 2011. Structural insights into anaphase-promoting complex function and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3605–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ. 2009. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11:R77 doi:10.1186/bcr2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Frick DN, Richardson CC. 2001. DNA primases. Annu. Rev. Biochem. 70:39–80 [DOI] [PubMed] [Google Scholar]
- 124. Tseng BY, Ahlem CN. 1982. DNA primase activity from human lymphocytes. Synthesis of oligoribonucleotides that prime DNA synthesis. J. Biol. Chem. 257:7280–7283 [PubMed] [Google Scholar]
- 125. Yagura T, Kozu T, Seno T, Saneyoshi M, Hiraga S, Nagano H. 1983. Novel form of DNA polymerase alpha associated with DNA primase activity of vertebrates. Detection with mouse stimulating factor. J. Biol. Chem. 258:13070–13075 [PubMed] [Google Scholar]
- 126. Griffiths DJ, Liu VF, Nurse P, Wang TS. 2001. Role of fission yeast primase catalytic subunit in the replication checkpoint. Mol. Biol. Cell 12:115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B. 2007. Poxvirus DNA primase. Proc. Natl. Acad. Sci. U. S. A. 104:18724–18729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.