Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) infections of endothelial and B cells are etiologically linked with Kaposi's sarcoma (KS) and primary effusion B-cell lymphoma (PEL), respectively. KS endothelial and PEL B cells carry multiple copies of the nuclear episomal latent KSHV genome and secrete a variety of inflammatory cytokines, including interleukin-1β (IL-1β) and IL-18. The maturation of IL-1β and IL-18 depends upon active caspase-1, which is regulated by a multiprotein inflammasome complex induced by sensing of danger signals. During primary KSHV infection of endothelial cells, acting as a nuclear pattern recognition receptor, gamma interferon-inducible protein 16 (IFI16) colocalized with the KSHV genome in the nuclei and interacted with ASC and procaspase-1 to form a functional inflammasome (Kerur N et al., Cell Host Microbe 9:363-375, 2011). Here, we demonstrate that endothelial telomerase-immortalized human umbilical cells (TIVE) supporting KSHV stable latency (TIVE-LTC cells) and PEL (cavity-based B-cell lymphoma 1 [BCBL-1]) cells show evidence of inflammasome activation, such as the activation of caspase-1 and cleavage of pro-IL-1β and pro-IL-18. Interaction of ASC with IFI16 but not with AIM2 or NOD-like receptor P3 (NLRP3) was detected. The KSHV latency-associated viral FLIP (vFLIP) gene induced the expression of IL-1β, IL-18, and caspase-1 mRNAs in an NF-κB-dependent manner. IFI16 and cleaved IL-1β were detected in the exosomes released from BCBL-1 cells. Exosomal release could be a KSHV-mediated strategy to subvert IL-1β functions. In fluorescent in situ hybridization analyses, IFI16 colocalized with multiple copies of the KSHV genome in BCBL-1 cells. IFI16 colocalization with ASC was also detected in lung PEL sections from patients. Taken together, these findings demonstrated the constant sensing of the latent KSHV genome by IFI16-mediated innate defense and unraveled a potential mechanism of inflammation induction associated with KS and PEL lesions.
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is etiologically associated with KS, an angioproliferative malignancy of human skin, as well as with two angiolymphoproliferative disorders: body cavity-based B-cell lymphoma (BCBL) (or primary effusion lymphoma [PEL]) and some forms of polyclonal B-cell proliferative multicentric Castleman's disease (MCD) (1). In vivo, viral DNA and transcripts have been detected in human B cells, macrophages, keratinocytes, endothelial cells, and epithelial cells (2). Similar to other herpesviruses, KSHV displays latent and lytic cycles in the infected cells. The spindle-shaped proliferating endothelial cells, considered a hallmark of KS lesions, harbor the KSHV genome in a latent state. A lytic cycle is also detected in a low percentage of infiltrating inflammatory monocytes of KS lesions (2).
Human B-cell lines from PEL, such as BCBL-1 and BC-3, carry more than 80 copies of the KSHV genome per cell in a latent form (2). The KSHV lytic cycle and virus production induced from these cells by chemicals or by the lytic cycle switch open reading frame 50 (ORF50) gene product serve as the source of virus for in vitro studies. The KSHV latency-associated ORF73 (LANA-1), ORF72 (vCyclin), ORF71 (vFLIP), K12 (Kaposin), and ORF10.5 (LANA-2) gene products as well as 12 microRNAs are expressed in PEL cells. These KSHV gene products ensure tethering of the viral genome as an episome to host cell chromatin, control the KSHV lytic ORF50 gene, and evade host responses, including apoptosis, autophagy, interferons (IFNs), etc., which are essential for the maintenance of latent infection and cell survival (1).
KSHV infects a variety of in vitro target cells, such as human dermal microvascular endothelial (HMVEC-d) cells, human foreskin fibroblasts (HFFs), embryonic kidney epithelial cells (293 cells), monocytic cells (THP-1), and B cells. KSHV entry into target cells is mediated by endocytosis, followed by rapid transit of the viral genome containing capsid along the microtubule network to nuclear pores and subsequent delivery of the viral genome into the nucleus (3). Unlike primary infection with alpha- and betaherpesviruses, primary infection of adherent target cells and THP-1 cells with γ2-KSHV in vitro does not result in a productive lytic cycle and progeny viral particle formation. Instead, the virus enters into latency with limited viral gene expression. The angioproliferative KS lesion microenvironment is enriched with proangiogenic inflammatory cytokines (interleukin-1β [IL-1β], IL-6, gamma IFN [IFN-γ], tumor necrosis factor [TNF], granulocyte-macrophage colony-stimulating factor, prostaglandin E2), angiogenic factors (angiogenin, basic fibroblast growth factor, vascular epidermal growth factor, platelet-derived growth factor), and chemokines (monocyte chemoattractant protein 1, IL-8) (4), which are critical factors contributing to the growth, survival, and spread of KSHV-infected cells in both KS and PEL (5, 6). Elucidating the pathways regulating the secretion of these cytokines and growth factors is critical in designing therapeutic strategies.
During virus and other pathogen infection, induction of inflammatory cytokines depends on recognition of viral components by host pattern recognition receptors (PRRs). Three different classes of PRRs, including several Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and multiple NOD-like receptors (NLRs), are known to recognize various viral pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs). Signaling through these pathways results in type I interferon induction or maturation of powerful proinflammatory cytokines, such as IL-1β and IL-18 (7). The PRRs are localized in different cellular compartments for efficient detection of the invading virus and other pathogens. These distinct classes of PRRs participate in synergistic and cooperative signaling to elicit an efficient and well-coordinated innate immune response to pathogen infection.
Emerging lines of evidence indicate that the innate immune response plays a central role in regulating the latency program of gammaherpesviruses, including KSHV. Studies have shown that activation of TLR7/8 and the NLRP1 inflammasome regulate viral reactivation and progeny production in KSHV latently infected PEL cells (8, 9). Inflammasomes are cytoplasmic sensors of foreign molecules, including pathogens, and function to induce caspase-1 activation, which in turn regulates the cleavage (maturation) of cytokines such as IL-1β and IL-18.
We have recently shown that gamma interferon-inducible protein 16 (IFI16) recognizes the KSHV genome during primary infection in endothelial HMVEC-d cells (10). IFI16 is a PYD-containing HIN-200 protein with predominant nuclear expression in several cells, such as endothelial, epithelial, and hematopoietic cells as well as keratinocytes (11). Our studies demonstrated that during de novo KSHV infection of HMVEC-d cells, IFI16 interacted with the adaptor molecule ASC and procaspase-1 to form a functional inflammasome complex. Initially, IFI16 colocalized with ASC in the nucleus and subsequently in the perinuclear area of infected endothelial cells. Caspase-1 activation by KSHV was reduced by IFI16 and ASC silencing. Studies with UV-inactivated KSHV suggested that KSHV gene expression and/or the viral genome is crucial for inducing the inflammasome. Transduction of endothelial cells with lentivirus vectors carrying latent ORF71, ORF72, ORF73, and K12 genes, lytic K8 and ORF74 genes, and the green fluorescent protein (GFP) gene as a control did not induce caspase-1 activation. This suggested that viral genes individually do not play a role in KSHV-induced inflammasome activation and IFI16 does not recognize linear integrated foreign DNA (10). In addition, IFI16 colocalized with the KSHV genome in the infected endothelial cell nucleus, suggesting that the KSHV genome is probably required for IFI16 inflammasome activation (10).
It is widely believed that the intracellular innate response to viral DNA is largely restricted to the cytoplasm. However, our recent seminal discovery suggested that IFI16 has uniquely evolved as a sensor of nuclear pathogens (10). All herpesviruses, including KSHV, establish lifelong latent infection characterized by the persistence of multiple viral episomal genomes in their target cell nuclei. It is not known whether such latency-associated nuclear viral genomes activate an innate immune response. Since KS spindle cells and PEL cells contain KSHV in the episomal latent state, we set out to determine (i) whether innate sensors detect the presence of the latent KSHV genome, (ii) the identity of the sensor(s), and (iii) whether this sensing leads to inflammasome activation. Our comprehensive studies presented here show activation of the IFI16-dependent inflammasome and cleavage of IL-1β and IL-18 in latently infected endothelial (telomerase-immortalized human umbilical vein [TIVE] cells supporting KSHV stable latency [TIVE-LTC cells]) and B (PEL) cells and detection of the IFI16 inflammasome in in vivo PEL samples. These studies demonstrate the existence of a constant innate immune response against the KSHV genome during latency which could be one of the driving forces for the inflammatory and angiogenic responses detected in KSHV-associated KS and PEL malignancies.
MATERIALS AND METHODS
Cells.
TIVE cells and TIVE-LTC cells carrying KSHV in a latent state (12) (a generous gift from Rolfe Renne, University of Florida) were grown in endothelial basal medium 2 (EBM-2) with growth factors (Clonetics). Epstein-Barr virus (EBV)-negative and KSHV-negative (EBV− KSHV−) lymphoma BJAB cells, KSHV-positive BCBL-1 PEL cells, as well as BJAB-KSHV cells (13) (a generous gift from Blossom Damaina, University of North Carolina) were cultured in RPMI 1640 medium (Gibco BRL) with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 2 mM l-glutamine, and antibiotics (14). HMVEC-d cells (CC-2543; Lonza, Walkersville, MD) were cultured in EBM-2 with growth factors (Lonza, Walkersville, MD).
Virus.
Procedures for the induction of the KSHV lytic cycle in BCBL-1 cells, supernatant collection, and virus purification were described previously (15–17). KSHV DNA was extracted from the purified virus, and copy numbers were quantitated by real-time DNA PCR using primers amplifying the KSHV ORF73 gene (15–17).
Antibodies and reagents.
Rabbit polyclonal anti-human IL-1β and caspase-1 antibodies were from Cell Signaling Technology, Beverly, MA, and Invitrogen Corporation, Carlsbad, CA, respectively. Goat polyclonal antibodies against human ASC/TMS1 were from Ray Biotech, Norcross, GA. Mouse monoclonal antibodies against ASC and human IL-18 were from MBL International, Woburn, MA, and mouse anti-Alix antibody was from Cell Signaling Technology, Beverly, MA. Rabbit and mouse antibodies against human IFI16 were from Santa Cruz Biotechnology Inc., Santa Cruz, CA. Rabbit and mouse anti-AIM2, anti-NLRP3, anti-Rab27a, and anti-Tsg101 and mouse anti-TATA binding protein (TBP) antibodies were from Abcam Inc., Cambridge, MA. Anti-rabbit, anti-goat, and anti-mouse antibodies linked to horseradish peroxidase, Alexa Fluor 488, and Alexa Fluor 594 were from KPL Inc., Gaithersburg, MD, or Molecular Probes, Eugene, OR. Protein G-Sepharose 4 Fast Flow beads were from GE Healthcare Bio-Sciences Corp., Piscataway, NJ.
Quantitative real-time RT-PCR.
Expression of genes for various inflammasome proteins (IL-1β, IL-18, and caspase-1) was examined by real-time reverse transcription-PCR (RT-PCR) using a SYBR green detection system (10). The expression levels of these genes were normalized to the level of 18S rRNA gene expression. The final mRNA levels of the genes studied were normalized using the comparative cycle threshold method.
Western blot analysis.
The cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer containing a protease inhibitor cocktail. Equal amounts of protein samples were resolved by 10 to 20% SDS-PAGE and subjected to Western blotting. Processing of caspase-1 was examined by specific antibodies detecting the proform and mature form of each protein. Processing of IL-1β was examined by specific antibodies detecting only the mature form of the IL-1β protein. To examine the intracellular distribution of ASC, caspase-1, and IFI16, nuclear and cytoplasmic extracts were prepared from BJAB, BCBL-1, TIVE, and TIVE-LTC cells as per methods described previously (10). The nuclear and cytoplasmic fractions were subjected to immunoblotting with anti-ASC, anti-caspase-1, and anti-IFI16 antibodies. These membranes were stripped and immunoblotted with anti-tubulin and anti-TBP antibodies to confirm equal loading of cytoplasmic and nuclear lysates, respectively. The immunoreactive bands were developed by enhanced chemiluminescence reaction (NEN Life Sciences Products, Boston, MA) and quantified by following standard protocols (18).
Coimmunoprecipitation (co-IP).
KSHV infection-induced protein-protein interactions were examined by co-IP experiments. BJAB, BCBL-1, BJAB-KSHV, TIVE, and TIVE-LTC cells were washed, lysed in RIPA lysis buffer, and clarified by centrifugation for 15 min at 4°C. Equal amounts of total protein lysates were used for co-IP assays. The lysates were incubated for 2 h with immunoprecipitating antibody (anti-ASC or anti-caspase-1 antibody) at 4°C, and immune complexes were captured using 10 μl of protein G-Sepharose. They were washed three times with phosphate-buffered saline, boiled with SDS-PAGE sample buffer, resolved by 10% SDS-PAGE, and subjected to Western blotting.
Laser-scanning confocal immunofluorescence.
BJAB and BCBL-1 cells were fixed and permeabilized with acetone, washed, and blocked with Image-iT FX signal enhancer (Invitrogen) for 20 min, and TIVE-LTC cells were fixed for 10 min with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100 for 5 min, washed, and blocked with Image-iT FX signal enhancer (Invitrogen) for 20 min. The cells were incubated with goat anti-ASC, rabbit anti-caspase-1, or mouse anti-IFI16 antibodies, followed by anti-goat Alexa Fluor 488-, anti-rabbit Alexa Fluor 594-, or anti-mouse-Alexa 594-labeled antibodies, respectively. An Olympus Fluoview 300 fluorescence confocal microscope was used for imaging, and analysis was performed using Fluoview software (Olympus, Melville, NY).
Immunofluorescence (IFA) microscopy.
TIVE, TIVE-LTC, and HMVEC-d cells infected with KSHV (20 to 30 DNA copies/cell) were fixed for 10 min with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100 for 5 min, washed, and blocked with Image-iT FX signal enhancer (Invitrogen) for 20 min. BJAB and BCBL-1 cells were fixed and permeabilized with acetone, washed, and blocked with Image-iT FX signal enhancer (Invitrogen) for 20 min. Mock- and KSHV-infected cells were incubated with goat anti-ASC, rabbit anti-caspase-1, or mouse anti-IFI16 antibodies for 2 h at 37°C, followed by anti-goat Alexa Fluor 488-, anti-rabbit Alexa Fluor 594-, or anti-mouse Alexa Fluor 594-labeled antibodies, respectively, for 1 h at 37°C. A Nikon Eclipse 80i fluorescence microscope was used for imaging, and analysis was performed using Metamorph imaging software.
Immunofluorescence and FISH.
Uninfected and KSHV-infected HMVEC-d cells were fixed as described above and incubated with mouse anti-IFI16 antibodies, followed by donkey anti-mouse Alexa Fluor 594 secondary antibodies. These cells were fixed with 2% paraformaldehyde for 10 min, permeabilized in 0.2% Triton X-100 for 5 min, and treated with 0.1 M Tris-HCl (pH 7.0) for 2 min and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) twice for 2 min. The cells were treated with denaturing solution (70% formamide in 2× SSC) at 70°C for 2 min, washed, dried, and subjected to in situ hybridization. A KSHV fluorescent in situ hybridization (FISH) probe was developed and validated by the Anatomic Pathology FISH Laboratory, Medical College of Virginia, Richmond, VA. A Spectrum green-labeled BAC36 KSHV entire genome was used as a probe. The KSHV probe was diluted (1:10) in DenHyb hybridization solution (Insitus Biotechnologies, Albuquerque, NM) and incubated with cells for hybridization in a humidified chamber at 37°C overnight. The slides were sequentially washed in 2× SSC (10 min) and deionized water (1 min), and nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole) and examined.
Immunohistochemistry.
Sections of lung from healthy individuals and solid PEL lesions of lung from PEL patients were obtained from the AIDS and Cancer Specimen Resource (ACSR). The immunohistochemistry staining procedure was carried out at the Pathology Core Facility, Northwestern University, Feinberg School of Medicine, Chicago, IL. The sections were stained with mouse anti-IFI16 monoclonal and goat anti-ASC polyclonal antibodies and visualized by anti-goat Alexa Fluor 488 and anti-mouse Alexa Fluor 594, respectively. The Olympus Fluoview 300 fluorescence confocal microscope was used for imaging, and analysis was performed using Fluoview software (Olympus, Melville, NY).
Production of lentivirus vector for expressing KSHV genes in HMVEC-d cells.
The production and infection of lentivirus vectors were carried out as described by Vart et al. (19). Vesicular stomatitis virus G envelope-pseudotyped lentivirus was produced using a four-plasmid transfection system as previously described (20, 21). Lentivirus vector particles for overexpressing ORF71, ORF72, ORF73, ORF74, and K12 were prepared as described previously (22). Lentivirus vector plasmids for vFLIP-HA-wt and vFLIP-HA-A57L were generously provided by Thomas F. Schulz (Hannover Medical School, Hannover, Germany) (23). The lentiviral preparation was directly added to HMVEC-d cells to transduce the indicated KSHV genes. The cells were split at 24 h after lentivirus transduction and subsequently examined.
Exosome preparation.
BJAB and BCBL-1 cells grown in RMPI 1640 medium plus 10% FBS were harvested, washed with Hanks balanced salt solution, and resuspended at the same cell density in exosome-depleted RMPI 1640 medium plus 10% FBS. These cultures were incubated for 3 days, and released exosomes were harvested by stepwise high-speed and ultracentrifugation methods (24). Exosome-depleted cell culture medium was prepared by ultracentrifugation of the medium at 100,000 × g for 16 h to remove naturally occurring exosomes from fetal bovine serum (24). The purity of exosomes obtained was assessed by Western blot analysis for the presence of the multivesicular body proteins Alix and Tsg101 and the absence of the endoplasmic reticulum (ER) protein calnexin (24).
RESULTS
The inflammasome is activated in TIVE-LTC and BCBL-1 cells latently infected with KSHV.
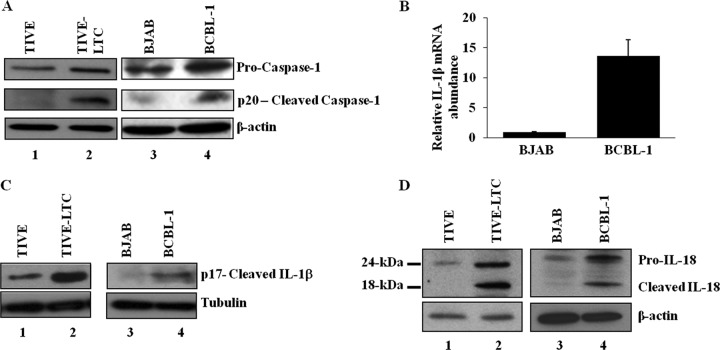
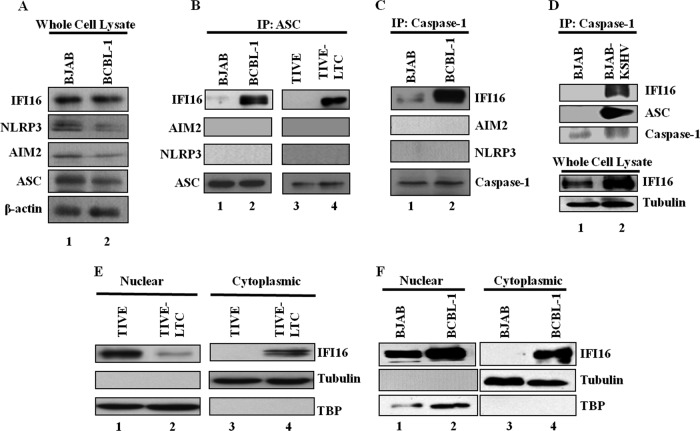
The endothelial TIVE-LTC cells with latent KSHV gene expression support long-term episomal maintenance, which is similar to viral gene expression in KSHV-infected KS endothelial cells (12), and the PEL cell line BCBL-1 contains >80 copies of the KSHV genome in a latent episomal form (1). Since these cells secrete a variety of cytokines and growth factors, including IL-1β and IL-18 (5, 6), we first determined whether the inflammasome that is activated in these cells mediates the cleavage of procaspase-1 into active caspase-1, which in turn cleaves pro-IL-1β (31 kDa) and pro-IL-18 (24 kDa) into active IL-1β (17 kDa) and IL-18 (18 kDa) (25). Compared to KSHV-negative control TIVE and BJAB cells, increased processed caspase-1 (p20) protein levels were observed in KSHV-positive TIVE-LTC and BCBL-1 cells (Fig. 1A, lanes 1 to 4). Analysis of gene expression demonstrated a higher IL-1β mRNA abundance in BCBL-1 cells than BJAB cells (Fig. 1B). We observed only about a 1.2- to 1.4-fold increase in IL-1β mRNA expression in TIVE-LTC cells over the level in control TIVE cells (data not shown), which could have been due to the limited (10 to 20%) number of LANA-1-positive cells in the TIVE-LTC cell population. Despite this, immunoblotting showed higher levels of processed IL-1β (p17) protein in KSHV-positive TIVE-LTC and BCBL-1 cells than the control TIVE and BJAB cells (Fig. 1C, lanes 1 to 4). Processing of IL-1β was examined by specific antibodies detecting only the mature form of IL-1β protein. Similarly, higher levels of processed IL-18 (18-kDa) protein were detected in TIVE-LTC and BCBL-1 cells than in control TIVE and BJAB cells (Fig. 1D, lanes 1 to 4). Collectively, these results demonstrated constitutive inflammasome activation in cells harboring a latent KSHV genome.
Fig 1.
Activation of the inflammasome in cells latently infected with KSHV. (A) Lysates of endothelial cells latently infected with KSHV (TIVE-LTC), KSHV-negative endothelial (TIVE) cells, latent KSHV-positive B cells (BCBL-1), and KSHV-negative B cells (BJAB) were analyzed for procaspase-1 and activated caspase-1 (p20) proteins by immunoblot analysis. (B) BJAB and BCBL-1 cells were analyzed for IL-1β gene expression by real-time RT-PCR. Each bar represents the fold increase in gene expression ± SD of three independent experiments. Fold changes were calculated after normalization with expression of the 18S rRNA gene. (C and D) TIVE, TIVE-LTC, BJAB, and BCBL-1 cell lysates were analyzed for cleaved IL-1β (p17) protein (C) and for pro-IL-18 and cleaved IL-18 proteins (D) by immunoblot analysis. Tubulin and β-actin were used as loading controls.
The KSHV latency-associated vFLIP gene induces the expression of IL-1β, IL-18, and caspase-1.
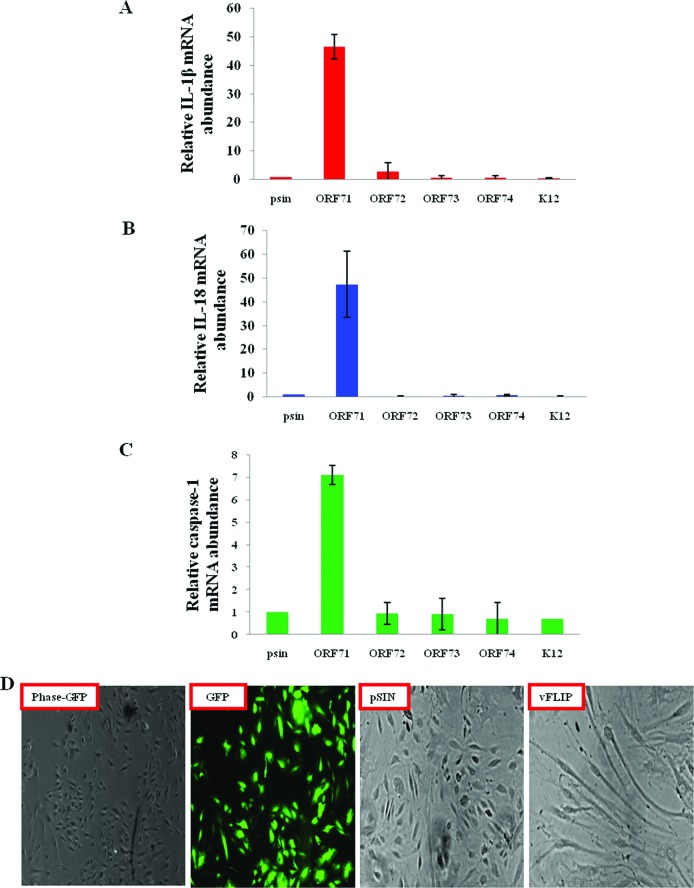
Induction of the inflammasome requires two signals: (i) initial signal 1, which induces NF-κB-mediated transcriptional activation and synthesis of increased proforms of the IL-1β and IL-18 proteins (25), and (ii) signal 2, which is initiated by a sensor protein such as NLRP3, AIM-2, or IFI16 that recognizes the danger signal and induces the formation of a multiprotein complex for processing of procaspase-1 (26). Since we observed elevated IL-1β gene expression and processing of caspase-1, IL-1β, and IL-18 in KSHV latently infected BCBL-1 and TIVE-LTC cells, we investigated whether any of the KSHV latency program-associated gene products activates signal 1 and induces the transcription of inflammasome-associated genes. For this, we expressed the various KSHV latent ORF71, ORF72, ORF73, and K12 genes as well as the lytic ORF74 gene individually in HMVEC-d cells via a lentiviral vector system and analyzed IL-1β, IL-18, and caspase-1 gene expression by quantitative RT-PCR. Compared to the vector control (pSIN), only vFLIP (ORF71), a known inducer of NF-κB (2), induced the expression of the IL-1β, IL-18, and caspase-1 genes by about 47-, 47-, and 7-fold, respectively (Fig. 2A to C). In contrast, other KSHV genes induced a modest to no effect on IL-1β, IL-18, and caspase-1 gene expression (Fig. 2A to C). Similar to earlier reports, expression of vFLIP induced a typical spindle cell shape in HMVEC-d cells (Fig. 2D) (27).
Fig 2.
KSHV latent protein vFLIP induces expression of genes associated with inflammasome activation. (A to C) Primary HMVEC-d cells were transduced with control (pSIN)-, ORF71-, ORF72-, ORF73-, ORF74-, or K12-expressing lentiviral vectors. At 72 h postransduction, expression of inflammasome-associated IL-1β, IL-18, and caspase-1 genes was analyzed by real-time RT-PCR. Each bar represents the fold increase in gene expression ± SD of three independent experiments. Fold changes were calculated by considering the levels of control transduced cells to be 1 after normalization with expression of the 18S rRNA gene. (D) Primary HMVEC-d cells transduced with control (pSIN), GFP-expressing, or vFLIP-expressing lentiviral vectors were examined after 72 h by phase-contrast and fluorescence microscopy. Magnification, ×40.
KSHV latency-associated vFLIP induces inflammasome-associated gene expression in an NF-κB-dependent manner.
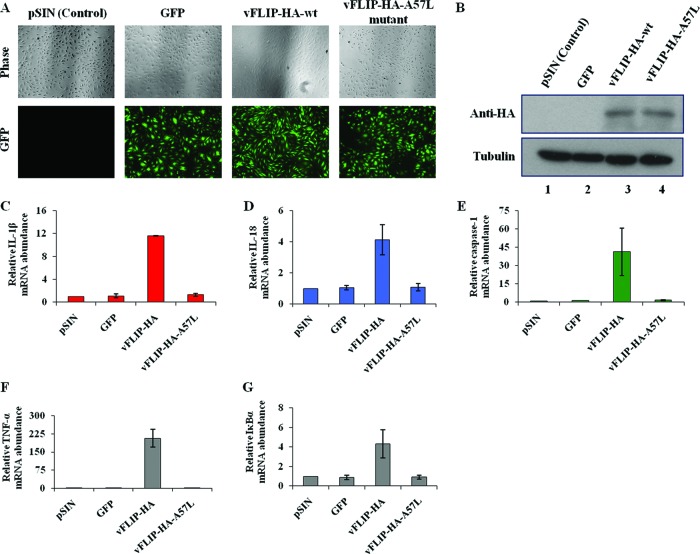
To determine whether vFLIP induces the inflammasome-associated genes in an NF-κB-dependent manner, we employed lentiviral vectors expressing hemagglutinin (HA)-tagged vFLIP wild type (wt) and its mutant with the A57L point mutation, which is incapable of interacting with IKKγ and activating NF-κB signaling (23). As shown before (23), expression of vFLIP wt but not its IKKγ-binding-deficient A57L mutant induced the spindle cell shape phenotype in HMVEC-d cells (Fig. 3A). Furthermore, compared to the vector control (pSIN), only vFLIP-wt induced the expression of IL-1β, IL-18, and caspase-1 genes by about 11-, 4-, and 41-fold, respectively (Fig. 3C to E). In contrast, GFP expression or the vFLIP A57L point mutation had no appreciable effect on IL-1β, IL-18, and caspase-1 gene expression (Fig. 3C to E). Real-time mRNA expression analysis of NF-κB-induced TNF alpha (TNF-α) and IκBα genes confirmed the vFLIP-mediated activation of NF-κB signaling in transduced HMVEC-d cells (Fig. 3F and G). Transduction efficiency and equal expression were confirmed using anti-HA antibodies by immunoblotting (Fig. 3B, lanes 3 and 4).
Fig 3.
vFLIP-induced expression of inflammasome-associated genes is NF-κB dependent. (A) Primary HMVEC-d cells transduced with control (pSIN)-, GFP-, vFLIP-HA-wt-, and vFLIP-HA-A57L (mutant for NF-κB activation)-expressing lentiviral vectors were examined after 72 h by phase-contrast and fluorescence microscopy. Magnification, ×40. (B) Expression of HA-tagged vFLIP and its mutant protein in transduced HMVEC-d cells was confirmed by immunoblot analysis. Tubulin was used as a loading control. (C to G) Primary HMVEC-d cells were transduced with control (pSIN)-, GFP-, vFLIP-HA-wt-, or vFLIP-HA-A57L-expressing lentiviral vectors. At 72 h postransduction, expression of inflammasome-associated IL-1β-, IL-18-, caspase-1-, and NF-κB-induced TNF-α and IκBα genes was analyzed by real-time RT-PCR. Each bar represents the fold increase in gene expression ± SEM of three independent experiments. Fold changes were calculated by considering the levels of control transduced cells to be 1 after normalization with expression of the 18S rRNA gene.
Collectively, results in Fig. 2 and 3 demonstrated that the latent vFLIP gene (gene product) provides a constitutive signal 1 in cells latently infected with KSHV for the constant expression of inflammasome-associated genes.
ASC and caspase-1 colocalize in TIVE-LTC and BCBL-1 cells latently infected with KSHV.
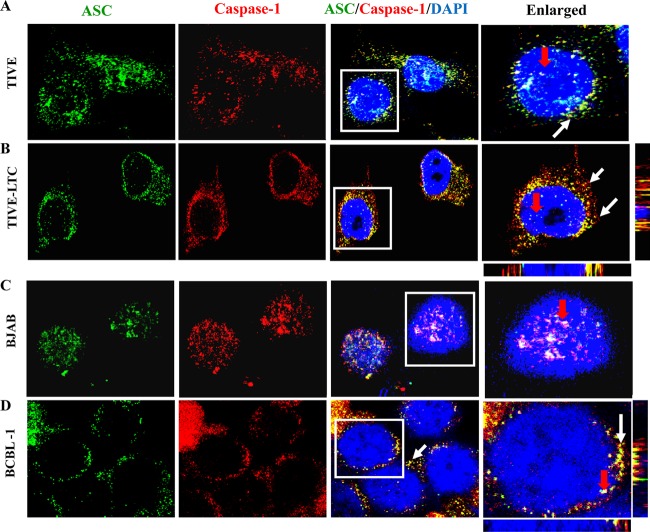
Activation of the inflammasome results in homotypic interactions between proteins of the inflammasome complex that can be visualized by fluorescence microscopy (10, 28, 29). Since the adaptor ASC molecule interacts with procaspase-1 upon inflammasome activation, we visualized KSHV-positive (TIVE-LTC and BCBL-1) and control KSHV-negative (TIVE and BJAB) cells with ASC- and caspase-1-specific antibodies. In TIVE cells, we observed moderately speckled nuclear staining of ASC and caspase-1, with a relatively low number of colocalizing spots in the nucleus and in the cytoplasm (Fig. 4A, white and red arrows, respectively). Similarly, more nuclear ASC and caspase-1 staining was detected in the BJAB cells, with a low number of colocalizing spots detected only in the nucleus (Fig. 4C, red arrow). In contrast, in KSHV-positive cells, ASC and caspase-1 colocalization was readily observed in the perinuclear area, with some colocalization detected in the nuclei (Fig. 4B and D, white and red arrows, respectively). These observations demonstrate that the ASC–caspase-1 interaction is induced by KSHV in these cells. The marked differences in the pattern of ASC and caspase-1 staining between KSHV-positive and -negative cells also suggested that the presence of latent KSHV infection induces the redistribution of ASC and caspase-1 from the nuclei to the perinuclear area.
Fig 4.
ASC and caspase-1 interact and redistribute in the cytoplasm of cells latently infected with KSHV. TIVE and TIVE-LTC cells fixed with 2% paraformaldehyde (A and B) or BJAB and BCBL-1 cells fixed with ice-cold acetone (C and D) were subjected to immunofluorescence analysis. Cells were stained with anti-ASC and anti-caspase-1 antibodies and visualized by incubation with Alexa Fluor 488 (green) and Alexa Fluor 594 (red) secondary antibodies, respectively. Cell nuclei were stained with DAPI (blue). The boxed areas were enlarged and are shown in the rightmost panels. White and red arrows, colocalization of ASC and caspase-1 in the cytoplasm and nucleus, respectively. Image results are depicted from a representative field taken after three independent experiments were performed. Magnification, ×60.
IFI16 forms an inflammasome complex with ASC and caspase-1 in TIVE-LTC and BCBL-1 cells latently infected with KSHV.
ASC functions as an adaptor molecule in this multiprotein complex, recruiting and interacting with sensor proteins such as NLRP3, AIM2, or IFI16. Hence, we next sought to identify the ASC-interacting sensor protein involved in inflammasome activation during latent KSHV infection. Examination of whole-cell lysates revealed that the adaptor ASC protein, the sensor HIN-200 IFI16 and AIM2 proteins, as well as the NLR protein NLRP3 were expressed at similar levels in BJAB and BCBL-1 cells (Fig. 5A, lanes 1 and 2). Equal quantities of protein lysates were immunoprecipitated with anti-ASC antibodies and Western blotted for IFI16, AIM2, and NLRP3. IFI16 was observed only in the ASC immunoprecipitates from KSHV-positive BCBL-1 and TIVE-LTC cells and not those from control BJAB and TIVE cells (Fig. 5B, lanes 1 to 4). NLRP3 and AIM2 were not detected in any of these immunoprecipitates (Fig. 5B). In addition, co-IP studies also demonstrated the presence of only IFI16 and not AIM2 and NLRP3 in caspase-1 immunoprecipitates in KSHV-positive BCBL-1 cells (Fig. 5C, lanes 1 and 2). These results suggested that IFI16, ASC, and caspase-1 are probably in the same complex.
Fig 5.
IFI16 interacts with ASC and caspase-1 and translocates to the cytoplasm in cells latently infected with KSHV. (A) Whole-cell lysates of BJAB and BCBL-1 cells were examined for protein expression with IFI16, AIM2, NLRP3, ASC, and β-actin antibodies. (B) TIVE, TIVE-LTC, BJAB, and BCBL-1 cell lysates were immunoprecipitated with anti-ASC antibody and Western blotted for IFI16, AIM2, and NLRP3 proteins. (C) BJAB and BCBL-1 cell lysates were immunoprecipitated with anti-caspase-1 antibody and Western blotted for IFI16, AIM2, and NLRP3 proteins. ASC and caspase-1 IP blots are shown in Fig. 5. (B and C) (Bottom) IP efficiency. (D) BJAB and BJAB-KSHV cell lysates were immunoprecipitated with anti-caspase-1 antibody and Western blotted for IFI16, ASC, and caspase-1 proteins. Whole-cell lysates were also examined for protein expression with IFI16 and tubulin antibodies. (E and F) Expression and subcellular distribution of IFI16 were examined in nuclear and cytoplasmic fractions of TIVE and TIVE-LTC cells (E) and BJAB and BCBL-1 cells (F) by immunoblot analysis. TBP-1 and tubulin were used to demonstrate the purity and equal loading of nuclear and cytoplasmic fractions, respectively.
To confirm that the presence of the KSHV genome is responsible for the induction of IFI16 association with ASC and caspase-1, we employed a stably selected recombinant KSHV-infected BJAB-KSHV cell line (13). We observed IFI16 and ASC in the caspase-1 immunoprecipitates only in KSHV-positive BJAB cells and not in KSHV-negative parental BJAB cells (Fig. 5D, lanes 1 and 2). These results clearly demonstrated that IFI16 is the sensor protein that could be detecting the presence of the nuclear KSHV genome, leading to the multiprotein IFI16, ASC, and caspase-1 inflammasome complex.
Latent KSHV in TIVE-LTC and BCBL-1 cells induces subcellular redistribution of IFI16 and ASC.
We have previously shown that during de novo KSHV infection of HMVEC-d cells, IFI16 and the adaptor protein ASC undergo subcellular redistribution and form colocalizing aggregates in the perinuclear area (10). In contrast, upon infection with vaccinia virus (a poxvirus), which replicates its DNA genome in the cytoplasm, IFI16 remained in the nucleus (10). Here, we used biochemical and immunofluorescent methods to examine the IFI16 distribution in cells with latent KSHV. Nuclear and cytoplasmic fractions of cells were immunoblotted for IFI16. In TIVE cells, IFI16 was predominantly detected in the nucleus, with a low signal detected in the cytoplasm (Fig. 5E, lanes 1 and 3). In contrast, in KSHV-positive TIVE-LTC cells, only a low level of IFI16 was detected in the nucleus, with higher levels detected in the cytoplasm (Fig. 5E, lanes 2 and 4). Similarly, IFI16 was predominantly detected in the nucleus of BJAB cells, with a small amount detected in the cytoplasm (Fig. 5F, lanes 1 and 3). In contrast, in BCBL-1 cells, higher levels of IFI16 were detected in the nucleus as well as in the cytoplasm (Fig. 5F, lanes 2 and 4).
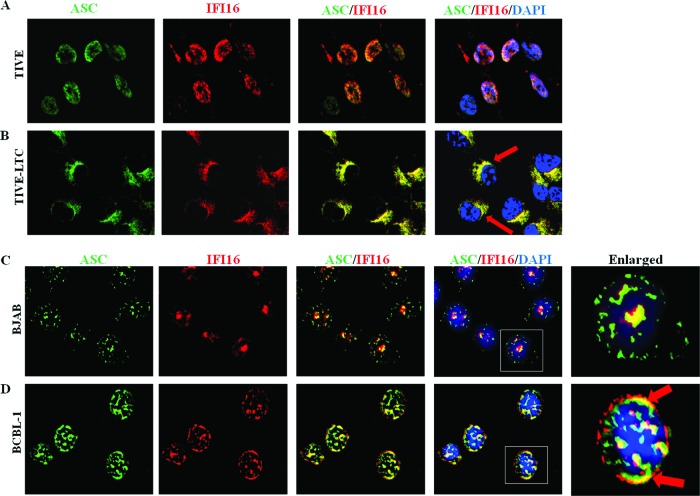
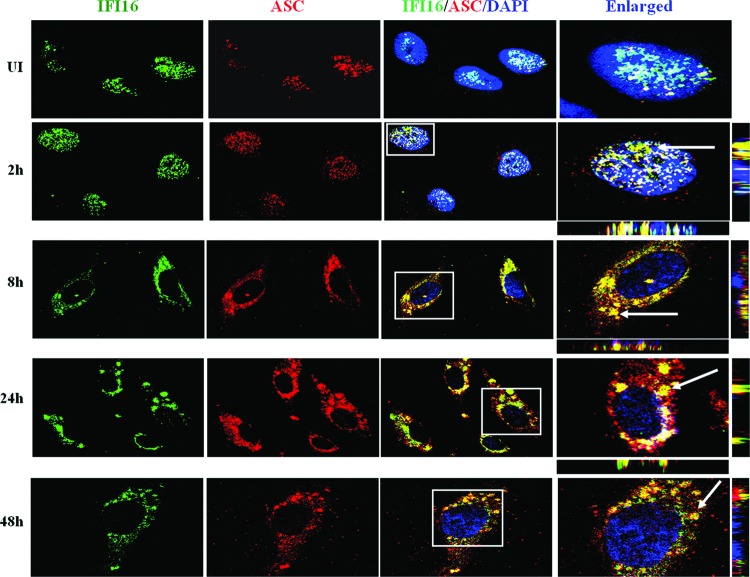
Corroborating these findings, immunofluorescence imaging also showed a nuclear to cytosolic redistribution of IFI16 and a strong cytosolic and nuclear ASC-IFI16 colocalization in TIVE-LTC and BCBL-1 cells (Fig. 6B and D, red arrows) compared to that in TIVE and BJAB cells (Fig. 6A and C). IFI16 also colocalized with ASC and redistributed in the cytoplasm of the KSHV-positive PEL cell line BC-3 (data not shown). In addition, redistribution of IFI16-ASC in the cytoplasm at the later time points (24 and 48 h postinfection [p.i.]), representing latency during de novo KSHV infection of primary HMVEC-d cells, was also observed (Fig. 7). Taken together, these results demonstrated that latent KSHV infection induces the formation of an inflammasome protein complex involving IFI16, ASC, and procaspase-1.
Fig 6.
IFI16 colocalizes with ASC and redistributes in the cytoplasm of latently infected cells. (A and B) TIVE and TIVE-LTC cells fixed with 2% paraformaldehyde (A) or BJAB and BCBL-1 cells fixed with ice-cold acetone (B) were subjected to immunofluorescence analysis. Cells were reacted with anti-ASC and anti-IFI16 antibodies, washed, and incubated with Alexa Fluor 488 (green) and Alexa Fluor 594 (red) secondary antibodies, respectively. Cell nuclei were visualized by DAPI (blue). The boxed areas were enlarged, and the enlargements are shown in the far right panels. Red arrows, colocalization of ASC and IFI16. Image results are depicted from a representative field taken after three independent experiments were performed. Magnification, ×60.
Fig 7.
Induction of the IFI16-ASC inflammasome and subcellular redistribution of IFI16-ASC during early and late stages of KSHV infection of endothelial cells. HMVEC-d cells were infected with KSHV (30 DNA copies per cell) for 2 h, uninternalized virus was removed by washing, and infected and uninfected cells were further incubated at 37°C for the indicated times. The cells were washed, fixed, permeabilized, and blocked with Image-iT FX signal enhancer. Cells were stained with anti-IFI16 and anti-ASC antibodies and visualized by incubation with Alexa Fluor 488 (green) and Alexa Fluor 594 (red) secondary antibodies, respectively. The image was merged with DAPI-stained nuclei. The boxed areas were enlarged, and the enlargements are presented in the rightmost panels. White arrows, colocalization. Image results are depicted from a representative field taken after three independent experiments were performed. UI, uninfected. Magnification, ×60.
IFI16 and cleaved IL-1β are released in the exosomes of BCBL-1 cells latently infected with KSHV.
Our studies show cleaved IL-1β in the total lysates of cells latently infected with KSHV (Fig. 1C). To detect mature IL-1β secreted into the cellular supernatants of KSHV-infected TIVE-LTC and BCBL-1 cells, we performed an enzyme-linked immunosorbent assay (ELISA). In multiple experiments, we observed only very low levels of cleaved IL-1β in the supernatants (data not shown). One plausible explanation could be that the sensitivity of the assay is too low. Recently, however, it has been shown that cells do secrete immunomodulatory cytokines in the microvascular bodies called exosomes (30). Exosomes are 40- to 100-nm-diameter small membrane vesicles that correspond to the internal vesicles present in multivesicular endosomes (MVEs) (30). They are released from many different cell types upon fusion of MVEs with the cell surface. They have also been shown to transfer oncogenic signaling receptors and β-amyloid peptides from one cell to another, as well as a specific subset of mRNAs and microRNAs (31). Exosomes contain endosome-associated proteins (Rab GTPases, annexins, flotillin, Alix, and TSG101), integrins, and tetraspanins (CD63, CD9, CD81, and CD82) and are enriched in raft lipids, such as cholesterol, ceramide, and sphingolipids (32). Rab GTPases control trafficking in the endocytic and secretory pathways by recruiting specific effector proteins onto membrane surfaces to drive either cargo collection, organelle motility, or vesicle docking at target membranes (33). Specifically, Rab27a has been implicated in exosome release (34).
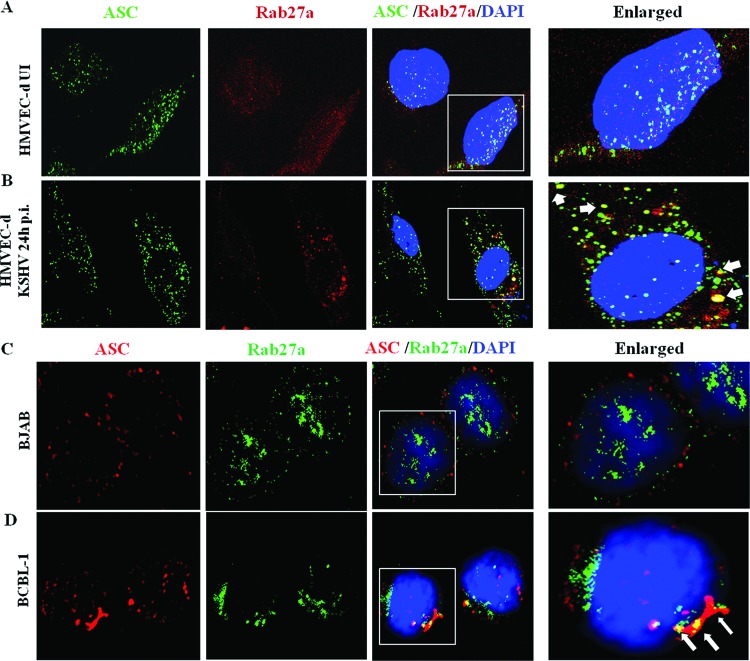
To investigate whether KSHV-induced inflammasome components are part of the exosome compartment, we first analyzed for the association of Rab27a and ASC in KSHV-infected cells. Compared to uninfected cells, at 24 h after de novo infection of HMVEC-d cells, a time when latency has been established, we observed very prominent colocalization of all ASC spots with Rab27a (Fig. 8A and B, white arrows). Similarly, we observed colocalization of ASC and Rab27a (Fig. 8D; white arrows) as well as IFI16 and Rab27a (data not shown) in BCBL-1 cells but not in BJAB cells (Fig. 8C).
Fig 8.
Subcellular localization of ASC with Rab27a during latency establishment during primary KSHV infection and in latently infected PEL cells. Primary HMVEC-d cells either uninfected (A) or KSHV infected for 24 h (B) were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100 for 5 min, blocked with Image-iT FX signal enhancer, reacted with anti-ASC and anti-Rab27a antibodies, washed, and incubated with Alexa Fluor 488 (green) and Alexa Fluor 594 (red) secondary antibodies, respectively. (C and D) BJAB and BCBL-1 cells were fixed with ice-cold acetone, blocked with Image-iT FX signal enhancer, reacted with anti-ASC and anti-Rab27a antibodies, washed, and incubated with Alexa Fluor 994 (red) and Alexa Fluor 488 (green) secondary antibodies, respectively. Cell nuclei were visualized by DAPI (blue). The boxed areas were enlarged, and the enlargements are shown in the far right panels. White arrows, colocalization of ASC and Rab27a. Image results are depicted from a representative field taken after three independent experiments were performed. Magnification, ×60.
Next, we analyzed the exosomes secreted from BJAB and BCBL-1 cells. The purity of the exosomes was shown by the presence of multivesicular body proteins Alix and Tsg101 and the absence of the ER protein calnexin (Fig. 9A, lanes 1 and 2). Very interestingly, cleaved IL-1β, IFI16, and Rab27a were prominently detected in lysates of exosomes from BCBL-1 cells but not in those from BJAB cells (Fig. 9A, lanes 1 and 2). Corroborating these findings, an increased association of IFI16 with the exosomal marker Tsg101 was also observed in BCBL-1 cells (Fig. 9B, bottom, white arrows) compared to that in BJAB cells (Fig. 9B, top). We also detected increased levels of Tsg101 in BCBL-1 cells over BJAB cells (Fig. 9B). Consistent with IFA data, immunoblot analysis of whole-cell lysates of BJAB and BCBL-1 cells also demonstrated an increase in Tsg101 expression in BCBL-1 cells (Fig. 9C, lane 2). Since IL-1β in the exosomes is not detected in the capture ELISA, as IL-1β is washed away after the initial period of incubation of the supernatants, these results suggested a possible reason for the low detection of IL-1β in the supernatants of KSHV-infected BCBL-1 cells. Taken together, these studies suggested that KSHV-induced inflammasome activation leads to release of mature IL-1β and IFI16 in the exosomes of latently infected cells.
Fig 9.
Detection of IFI16 and cleaved IL-1β in the exosomes released from cells latently infected with KSHV. (A) Exosomes released by BJAB and BCBL-1 cells in culture for 3 days were harvested and analyzed for the presence of IFI16, Rab27a, and cleaved IL-1β by Western blot analysis. The purity of exosome fractions was determined by the presence of multivesicular body-derived proteins Alix and Tsg101 and the absence of the ER protein calnexin. Ten micrograms of proteins was loaded per lane for Western blot analysis. Representative blots of 3 independent experiments are presented. (B) BJAB and BCBL-1 cells were immunostained for colocalization of the multivesicular body marker Tsg101 (green) and IFI16 (red). Cells were fixed, permeabilized, and blocked prior to immunostaining as described in the legend to Fig. 8. Alexa Fluor 594 (red) and Alexa Fluor 488 (green) secondary antibodies were used to detect IFI16 and Tsg101, respectively. Cell nuclei were visualized by DAPI (blue). White arrows, colocalization of IFI16 and Tsg101. Image results are depicted from a representative field taken after three independent experiments were performed. Magnification, ×60. (C) Whole-cell lysates of BJAB and BCBL-1 cells were examined for Tsg101 protein expression by immunoblot analysis. Tubulin was used as a loading control.
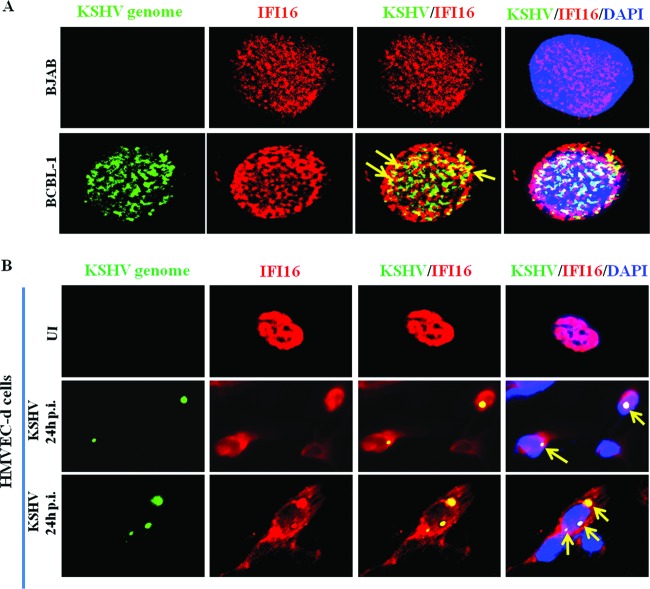
IFI16 colocalizes with the KSHV genome in latently infected cell nuclei.
We have previously demonstrated that IFI16 colocalizes with the KSHV genome in the nucleus during primary infection, an event that initiates the formation of the inflammasome with ASC and caspase-1 (10). Using a whole-viral-genome FISH probe (10), we explored whether IFI16 interacts with the KSHV genome in BCBL-1 cell nuclei. We observed that almost all the KSHV genome in the BCBL-1 cell nucleus colocalized with nuclear IFI16 (Fig. 10A, yellow arrows). As demonstrated in Fig. 6D, IFI16 was also detected in the cytoplasm of BCBL-1 cells without any colocalization. This finding indicated the specificity of the FISH analyses as well as the absence of free viral genome in the cytoplasm. IFI16 was not detected at appreciable levels in the cytoplasm of KSHV-negative BJAB cells, and as expected, the KSHV genome was not detected in these cells (Fig. 10A). Since the BCBL-1 cell nucleus carries >80 episomal DNA copies, the multiple IFI16 colocalization spots must be due to IFI16 interaction with all of the viral genome. This is further substantiated by IFI16 colocalization with the KSHV genome during de novo infection of HMVEC-d cells (Fig. 10B). In cells containing one copy of KSHV DNA, as shown by a single FISH spot, only one IFI16 immunofluorescent colocalization spot was observed (Fig. 10B, middle, yellow arrows). In contrast, in cells carrying multiple KSHV genomes, multiple IFI16-colocalizing spots, representing the sensing of individual genomes by IFI16, were observed (Fig. 10B, bottom, yellow arrows). These findings clearly indicate that IFI16 must be sensing the KSHV genome continuously in the latently infected cells and thereby contributing to constitutive inflammasome activation.
Fig 10.
Colocalization of IFI16 with the KSHV genome in infected cell nuclei. (A) BJAB and BCBL-1 cells were fixed and immunostained with mouse anti-IFI16 antibody followed by anti-mouse Alexa Fluor 594 (red) secondary antibody. (B) Primary HMVEC-d cells were infected with KSHV (30 DNA copies/cell) for 2 h, washed, and incubated for the indicated time points. The cells in panels A and B were then subjected to in situ hybridization with a Spectrum green-labeled whole-KSHV-genome probe and subjected to immunofluorescence image analysis. Cell nuclei were visualized by DAPI (blue). Yellow arrows, colocalization of the KSHV genome and IFI16. Magnification, ×60. (B) (Middle) Cells containing one copy of KSHV DNA shown by a single FISH spot with only one IFI16 colocalization spot; (bottom) a cell carrying multiple KSHV genomes with multiple IFI16 colocalization spots.
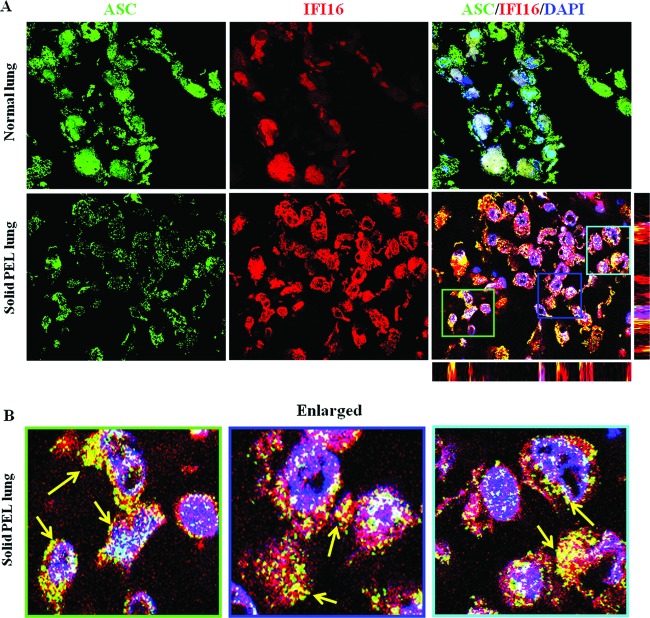
KSHV infection-associated human lesions exhibit IFI16 and ASC interaction.
We next assessed whether the ASC-IFI16 interaction occurs in an in vivo system such as a KSHV-infected solid PEL lesion. The KSHV-associated solid PEL lesions in lungs and healthy subject lungs were examined by immunostaining with anti-ASC and IFI16 antibodies. A predominantly nuclear localization for ASC and IFI16 with no prominent colocalization was observed in the normal healthy lung tissue sections (Fig. 11A). In contrast, perinuclear cytoplasmic colocalization of ASC and IFI16 was observed in the PEL lesion sections (Fig. 11A, bottom, and B, enlarged). Detection of such a distinct staining pattern in vivo not only corroborated in vitro KSHV-infected cell data but also indicated the potential in vivo involvement of an IFI16-mediated inflammasome in KSHV pathogenesis.
Fig 11.
In vivo interaction of IFI16 and ASC in KSHV-infected solid PEL lesions. (A) Sections of lung from healthy individuals and solid PEL lesions from PEL patients were stained with mouse anti-IFI16 monoclonal and goat anti-ASC polyclonal antibodies and visualized by anti-goat Alexa Fluor 488 (green) and anti-mouse Alexa Fluor 594 (red) antibodies, respectively. Cell nuclei were visualized by DAPI (blue). The boxed areas were enlarged, and the enlargements are shown in the bottom panels (B). Yellow arrows point to the colocalization of ASC and IFI16. Image results are taken from a representative field (n = 3 healthy subject and patient tissue section samples). Magnification, ×60.
DISCUSSION
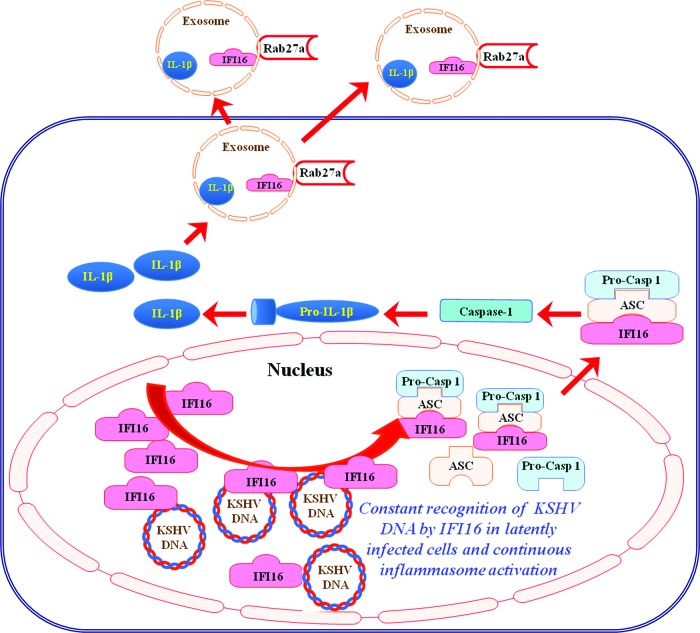
Vertebrate cells express many different PRRs that detect a variety of viral pathogen-associated molecular and danger patterns (PAMPs/DAMPs) and elicit antiviral interferon and proinflammatory responses. During herpesvirus infection, several virus-associated molecules, including viral DNA, as well as the consequences of viral infection are probably the main PAMPs and DAMPs detected by host innate immune receptors, which include TLRs, RLRs, and inflammasomes. All herpesviruses maintain a latent life cycle during which they must evade immune detection. During latency, the herpesvirus persists in the host cell nuclei without lytic replication and maintains an episomal form of the viral genome with minimal gene expression. The cellular innate response to different human and animal herpesviruses during the process of primary viral infection has been studied for many years. However, the identification of such a cellular innate response to a herpesvirus that has already established a latent life cycle has been elusive. Our findings here demonstrate that the KSHV genome in the latently infected cell nuclei triggers an intrinsic response via nuclear IFI16, and sensing of the viral genome by IFI16 culminates in the formation of an inflammasome complex with ASC and caspase-1 with the concomitant maturation of IL-1β and IL-18 (Fig. 12).
Fig 12.
Schematic model depicting the sensing of the KSHV genome by IFI16 and inflammasome complex formation during latency. Results presented here demonstrate that KSHV episomal DNA is sensed in the infected cell during latency by innate DNA sensor IFI16 in the nucleus, leading to recruitment of adaptor protein ASC and pro-caspase-1 (Pro-Casp-1) to form an inflammasome complex. Inflammasome complex formation is followed by its translocation to the cytoplasm and activation of caspase-1, leading to cleavage of pro-IL-1β and pro-IL-18 to their mature forms. Both IFI16 and cleaved IL-1β are sorted and released into exosomal compartments, which could be a KSHV-mediated strategy to regulate their cognate and potential antiviral functions. A single IFI16-colocalizing spot in cells carrying a single copy of viral genome (Fig. 10B) and multiple IFI16-colocalizing spots in cells carrying multiple KSHV genomes (Fig. 10B), representing the sensing of individual genomes by IFI16, suggest that IFI16 must be sensing the KSHV genome continuously in the latently infected cells and thereby contributing to constitutive inflammasome activation. IFI16 interaction with the KSHV genome probably triggers changes that allow it to interact with ASC, leading to the formation of the inflammasome complex with ASC and caspase-1 and translocation to the cytoplasm, while a new IFI16 molecule probably comes into contact with the viral genome and thus results in a continuum of the above-described processes.
Establishment of an inflammatory milieu with elevated levels of proinflammatory cytokines, chemokines, and growth factors has been shown to be important for the development of KS (4, 6, 18). Other inflammatory mediators like IL-6, IL-10, Cox-2, leukotrienes, and various others are associated with PEL (35, 36). Similarly, latent herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV) are also associated with the chronic inflammatory response (37, 38). van de Berg et al. (37) and Theil et al. (38) observed that patients with latent CMV infection had elevated levels of IL-18 compared to CMV-seronegative patients. The trigeminal ganglia of patients with latent HSV-1 infection were characterized by lymphocytic cell infiltration and elevated cytokine/chemokine expression, suggesting the presence of chronic inflammation (37, 38). However, how these latent viruses induce secretion of elevated inflammatory mediators is not clearly understood. On the basis of our finding that the latent KSHV genome activates the IFI16 inflammasome, it is reasonable to speculate that HSV-1 and HCMV genomes might also be eliciting an IFI16 inflammasome-activating signal. Further studies are ongoing to determine the role of IFI16 in sensing other viruses replicating their genome in the host cell nucleus.
The presence of abundant inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, is known to promote the pathogenesis of KSHV-associated diseases (35, 36, 39). In addition, the inflammatory cytokines and activation of innate response pathways of TLRs and inflammasomes control KSHV reactivation from latency. Activation of TLR7/8 promotes KSHV reactivation from latency and enhances viral progeny production (8). On the other hand, activation of the NALP1/3 inflammasome prevents KSHV reactivation from latency (8), suggesting that inflammasome activation promotes KSHV latency. Taken together, these findings indicate that inflammatory and innate immune signaling pathways greatly regulate the KSHV latency/reactivation program.
Release of IFI16 in the exosomes derived from BCBL-1 cells further suggests a higher-order complexity of viral regulation of host-mediated responses. Furthermore, detection of cleaved IL-1β in the exosomes of KSHV-infected cells also suggests a potential mechanism by which KSHV subverts the host innate response (Fig. 9 and 12). For example, to mediate its function, mature IL-1β released from the cells has to bind its cognitive cell surface IL-1 receptor that initiates downstream signaling. However, the IL-1β released in the exosomes of cells latently infected with KSHV could be functionally inactive, as it cannot engage the IL-1 receptors. Hence, exosomal release of IL-1β could be a virus-mediated strategy to subvert constitutive secretion of proapoptotic mature IL-1β, which may otherwise prove detrimental in the establishment of viral latency. Further studies are required to comprehend the role of exosome-mediated modulation of immune regulation during KSHV latency.
A recent report described how IFI16 acts as a restriction factor for HCMV replication (40). The authors demonstrated that small interfering RNA-mediated silencing of IFI16 in human embryo lung fibroblasts (HELFs) resulted in enhanced replication of HSV-1, HSV-2, and HCMV. Consequently, IFI16 overexpression in HELFs strongly inhibited HCMV replication by inhibiting early and late viral gene expression (40). This implicates IFI16 as a novel restriction factor in the lytic replicative cycles of herpesviruses. Further studies are essential to determine the effect of knocking down IFI16 on the establishment of KSHV latency during primary infection as well in PEL cells with established latency. Studies are under way to investigate the role of IFI16 in maintenance of KSHV latency and its potential effect on lytic reactivation.
Unterholzner et al. (41) showed that IFI16 is essential for STING- and TBK1-mediated IFN-β induction in response to transfected DNA and HSV-1 infection. Furthermore, we earlier demonstrated that during de novo KSHV infection, IFI16 colocalizes with the KSHV genome in the nucleus and activates the inflammasome (10). More recently, Li et al. have shown that the overexpression of IFI16 senses HSV-1 DNA primarily in the nucleus of U2OS cell lines and that the viral DNA-sensing ability is influenced by the subcellular localization of IFI16 (42). The authors demonstrated that the acetylation status of IFI16 was a major determinant of IFI16 subcellular distribution and that acetylation within the nuclear localization sequence (NLS) of IFI16 promoted cytoplasmic localization and inhibited its nuclear importation (42). In addition, inhibition of deacetylase activity by broad-spectrum deacetylase inhibitors was demonstrated to prevent nuclear accumulation of IFI16, suggesting the role of histone deacetylases (HDACs) in regulating IFI16 localization (42). It is plausible that the modulation of HDACs/histone acetyltransferase (HAT) by KSHV during its latency could be promoting the subcellular redistribution of IFI16, and further comprehensive studies are ongoing to shed more light on these aspects.
Recently, the crystal structures of the HIN domains of AIM2 and IFI16 in complex with double-stranded DNA have been described (43). It was demonstrated that IFI16 and AIM2 recognize DNA in a non-sequence-specific manner, which is accomplished through electrostatic attraction between the positively charged HIN domain residues and the sugar phosphate backbone of the double-stranded DNA. Furthermore, the Pyrin and HIN domains of AIM2 are shown to be in an autoinhibited intramolecular complex state which is liberated by DNA binding, thus facilitating the assembly of inflammasomes along the DNA staircase (43). It is reasonable to speculate that acting as a true pattern recognition receptor, IFI16 may be binding similarly to KSHV episomal DNA in a non-sequence-specific way to induce assembly of the inflammasome during latency (Fig. 12).
IFI16 is known to associate with proteins of DNA damage response signaling pathway molecules and has been shown to accumulate at the genomic site of DNA damage along with BRCA1, Mre/Rad50/NBS1, and ATM (44). Interestingly, HATs and HDACs, which regulate IFI16 localization, also mediate the DNA damage response through acetylation of important DNA repair and checkpoint proteins (45, 46). In addition, UV-B irradiation of human keratinocytes was reported to lead to redistribution of IFI16 from the nucleus to the cytoplasm and further into the supernatants of the cells (47). Similar nucleus-to-cytoplasm translocation of IFI16 was also observed in diseased skin sections from patients with systemic lupus erythematosus (SLE), which was implied to contribute to the detection of autoantibodies against IFI16 in the sera of patients with SLE (47). From these findings, it is conceivable that other forms of DNA damage might also be able to activate IFI16-mediated innate immunity downstream of DNA damage response (DDR). It is possible that the presence of several copies of KSHV episomal genomes in the host cell nucleus is sensed as DNA damage, fomenting IFI16-mediated inflammasome activation. It is also interesting to note that IFI16 colocalization with ASC as well as evidence of inflammasome activation, albeit weak, was also observed in TIVE and BJAB cells and could be due to DDR in these cells. Further studies are in progress to define the role of IFI16 in DDR and its connection to inflammasome activation.
Overall, from our present and earlier findings, we not only demonstrated that IFI16 acts as a nuclear pathogen sensor and detects nuclear KSHV episomal DNA but we also discovered the existence of a constant cellular innate immune response against the KSHV genome during latency (Fig. 12). Since this could be one of the factors associated with the chronic inflammation seen in KSHV-associated KS and PEL malignancies, if further studies unravel a role of IFI16 in KSHV latency, then IFI16 could serve as an attractive target to reduce inflammation associated with KSHV as well eradicate KSHV latent infection and associated diseases.
ACKNOWLEDGMENTS
This study was supported in part by Public Health Service grants CA 075911 and CA 168472 to B.C. and RFUMS-H. M. Bligh Cancer Research Fund to B.C.
We thank Keith Philibert and Alice Gilman-Sachs for critically reading the manuscript. We thank Thomas F. Schulz for providing us with the plasmids required for the production of the vFLIP-HA-wt and vFLIP-HA-A57L lentivirus vectors.
Footnotes
Published ahead of print 6 February 2013
REFERENCES
- 1. Ganem D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273–296 [DOI] [PubMed] [Google Scholar]
- 2. Ganem D. 2007. Kaposi's sarcoma-associated herpesvirus, p 2847–2888 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Chandran B. 2010. Early events in Kaposi's sarcoma-associated herpesvirus infection of target cells. J. Virol. 84:2188–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ensoli B, Sturzl M. 1998. Kaposi's sarcoma: a result of the interplay among inflammatory cytokines, angiogenic factors and viral agents. Cytokine Growth Factor Rev. 9:63–83 [DOI] [PubMed] [Google Scholar]
- 5. Gasperini P, Sakakibara S, Tosato G. 2008. Contribution of viral and cellular cytokines to Kaposi's sarcoma-associated herpesvirus pathogenesis. J. Leukoc. Biol. 84:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma-Walia N, Paul AG, Bottero V, Sadagopan S, Veettil MV, Kerur N, Chandran B. 2010. Kaposi's sarcoma associated herpes virus (KSHV) induced COX-2: a key factor in latency, inflammation, angiogenesis, cell survival and invasion. PLoS Pathog. 6:e1000777 doi:10.1371/journal.ppat.1000777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. 2011. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 11:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. 2009. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc. Natl. Acad. Sci. U. S. A. 106:11725–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. 2011. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Veeranki S, Choubey D. 2012. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol. Immunol. 49:567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. An FQ, Folarin HM, Compitello N, Roth J, Gerson SL, McCrae KR, Fakhari FD, Dittmer DP, Renne R. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 80:4833–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nun TK, Kroll DJ, Oberlies NH, Soejarto DD, Case RJ, Piskaut P, Matainaho T, Hilscher C, Wang L, Dittmer DP, Gao SJ, Damania B. 2007. Development of a fluorescence-based assay to screen antiviral drugs against Kaposi's sarcoma associated herpesvirus. Mol. Cancer Ther. 6:2360–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paul AG, Sharma-Walia N, Chandran B. 2011. Targeting KSHV/HHV-8 latency with COX-2 selective inhibitor nimesulide: a potential chemotherapeutic modality for primary effusion lymphoma. PLoS One 6:e24379 doi:10.1371/journal.pone.0024379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma-Walia N, Krishnan HH, Naranatt PP, Zeng L, Smith MS, Chandran B. 2005. ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79:10308–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naranatt PP, Krishnan HH, Svojanovsky SR, Bloomer C, Mathur S, Chandran B. 2004. Host gene induction and transcriptional reprogramming in Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8)-infected endothelial, fibroblast, and B cells: insights into modulation events early during infection. Cancer Res. 64:72–84 [DOI] [PubMed] [Google Scholar]
- 17. Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma-Walia N, Raghu H, Sadagopan S, Sivakumar R, Veettil MV, Naranatt PP, Smith MM, Chandran B. 2006. Cyclooxygenase 2 induced by Kaposi's sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J. Virol. 80:6534–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, Gratrix F, Takeuchi Y, Boshoff C. 2007. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res. 67:4042–4051 [DOI] [PubMed] [Google Scholar]
- 20. Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sadagopan S, Sharma-Walia N, Veettil MV, Raghu H, Sivakumar R, Bottero V, Chandran B. 2007. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 81:3949–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alkharsah KR, Singh VV, Bosco R, Santag S, Grundhoff A, Konrad A, Sturzl M, Wirth D, Dittrich-Breiholz O, Kracht M, Schulz TF. 2011. Deletion of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein, vFLIP, from the viral genome compromises the activation of STAT1-responsive cellular genes and spindle cell formation in endothelial cells. J. Virol. 85:10375–10388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 25. Stutz A, Golenbock DT, Latz E. 2009. Inflammasomes: too big to miss. J. Clin. Invest. 119:3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832 [DOI] [PubMed] [Google Scholar]
- 27. Grossmann C, Podgrabinska S, Skobe M, Ganem D. 2006. Activation of NF-kappaB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J. Virol. 80:7179–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bryan NB, Dorfleutner A, Rojanasakul Y, Stehlik C. 2009. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 182:3173–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thery C, Ostrowski M, Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9:581–593 [DOI] [PubMed] [Google Scholar]
- 31. Zhu M, Li Y, Shi J, Feng W, Nie G, Zhao Y. 2012. Exosomes as extrapulmonary signaling conveyors for nanoparticle-induced systemic immune activation. Small 8:404–412 [DOI] [PubMed] [Google Scholar]
- 32. Simons M, Raposo G. 2009. Exosomes—vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21:575–581 [DOI] [PubMed] [Google Scholar]
- 33. Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 34. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12:19–30 [DOI] [PubMed] [Google Scholar]
- 35. Jones KD, Aoki Y, Chang Y, Moore PS, Yarchoan R, Tosato G. 1999. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi's sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood 94:2871–2879 [PubMed] [Google Scholar]
- 36. Arguello M, Paz S, Hernandez E, Corriveau-Bourque C, Fawaz LM, Hiscott J, Lin R. 2006. Leukotriene A4 hydrolase expression in PEL cells is regulated at the transcriptional level and leads to increased leukotriene B4 production. J. Immunol. 176:7051–7061 [DOI] [PubMed] [Google Scholar]
- 37. van de Berg PJ, Heutinck KM, Raabe R, Minnee RC, Young SL, van Donselaar-van der Pant KA, Bemelman FJ, van Lier RA, ten Berge IJ. 2010. Human cytomegalovirus induces systemic immune activation characterized by a type 1 cytokine signature. J. Infect. Dis. 202:690–699 [DOI] [PubMed] [Google Scholar]
- 38. Theil D, Derfuss T, Paripovic I, Herberger S, Meinl E, Schueler O, Strupp M, Arbusow V, Brandt T. 2003. Latent herpesvirus infection in human trigeminal ganglia causes chronic immune response. Am. J. Pathol. 163:2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ensoli B, Nakamura S, Salahuddin SZ, Biberfeld P, Larsson L, Beaver B, Wong-Staal F, Gallo RC. 1989. AIDS-Kaposi's sarcoma-derived cells express cytokines with autocrine and paracrine growth effects. Science 243:223–226 [DOI] [PubMed] [Google Scholar]
- 40. Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8:e1002498 doi:10.1371/journal.ppat.1002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U. S. A. 109:10558–10563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, Wang Y, Johnstone RW, Deng C, Qin J, Ouchi T. 2003. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene 22:8931–8938 [DOI] [PubMed] [Google Scholar]
- 45. Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M. 2011. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471:74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17:1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Costa S, Borgogna C, Mondini M, De Andrea M, Meroni PL, Berti E, Gariglio M, Landolfo S. 2011. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. Br. J. Dermatol. 164:282–290 [DOI] [PubMed] [Google Scholar]