Abstract
Flavivirus nonstructural protein 2A (NS2A) is a component of the viral replication complex that functions in virion assembly and antagonizes the host immune response. Although flavivirus NS2A is known to associate with the endoplasmic reticulum (ER) membrane, the detailed topology of this protein has not been determined. Here we report the first topology model of flavivirus NS2A on the ER membrane. Using dengue virus (DENV) NS2A as a model, we show that (i) the N-terminal 68 amino acids are located in the ER lumen, with one segment (amino acids 30 to 52) that interacts with ER membrane without traversing the lipid bilayer; (ii) amino acids 69 to 209 form five transmembrane segments, each of which integrally spans the ER membrane; and (iii) the C-terminal tail (amino acids 210 to 218) is located in the cytosol. Nuclear magnetic resonance (NMR) structural analysis showed that the first membrane-spanning segment (amino acids 69 to 93) consists of two helices separated by a “helix breaker.” The helix breaker is formed by amino acid P85 and one positively charged residue, R84. Functional analysis using replicon and genome-length RNAs of DENV-2 indicates that P85 is not important for viral replication. However, when R84 was replaced with E, the mutation attenuated both viral RNA synthesis and virus production. Remarkably, an R84A mutation did not affect viral RNA synthesis but blocked intracellular formation of infectious virions. Collectively, the mutagenesis results demonstrate that NS2A functions in both DENV RNA synthesis and virion assembly/maturation. The topology model of DENV NS2A provides a good starting point for studying how flavivirus NS2A modulates viral replication and evasion of host immune response.
INTRODUCTION
Dengue virus (DENV) is a mosquito-borne human pathogen that belongs to the genus Flavivirus, family Flaviviridae. Besides the four serotypes of DENV, the genus Flavivirus also includes other mosquito- and tick-borne viruses of public health importance, including West Nile virus (WNV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). DENV is prevalent in the tropical and subtropical regions of the world. DENV-infected patients develop dengue fever (DF); some infected individuals develop life-threating dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS). There are annually 50 to 100 million DENV infections, leading to 500,000 DHF cases and 22,000 deaths around the world (1). Due to the increase in urbanization and transportation, the global burden of DENV has grown dramatically, with over 2.5 billion people now at risk (http://www.who.int/csr/disease/dengue/en/). Unfortunately, neither a vaccine nor an antiviral is clinically available for prevention and treatment of DENV infection. It is therefore urgent to develop effective antivirals for DENV and other flaviviruses.
Flaviviruses are enveloped viruses containing a single-strand, plus-sense RNA genome 11 kb in length. The genomic RNA contains a 5′ untranslated region (UTR) with a type I cap (m7GpppAm) (2), a single open-reading frame (ORF), and a 3′ UTR. The ORF encodes a long polyprotein, which is co- and posttranslationally processed by a combination of viral and cellular proteases into three structural proteins (capsid [C], premembrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins are essential components of the virion and function in viral entry, fusion, and assembly. The nonstructural proteins are components of viral replication complexes (3). Among them, glycoprotein NS1 plays an essential role in viral RNA replication (4) and facilitates immune complex formation (5). NS3 contains serine protease (using NS2B as a cofactor), RNA helicase, and nucleotide triphosphatase activities (6, 7); in addition, NS3 functions in viral assembly (8, 9). The N-terminal one-third of NS5 contains a methyltransferase activity, responsible for viral RNA cap formation and internal RNA methylation (10–12). The methyltransferase domain was also proposed to have a weak guanylyltransferase activity (13). The C-terminal two-thirds of NS5 is an RNA-dependent RNA polymerase (RdRp) (14). NS5 also plays a role in evasion of innate immune response (15, 16). Limited information is known about the functions of hydrophobic membrane proteins NS2A, NS4A, and NS4B. These proteins serve as a scaffold for the replication complex (17). The integral membrane protein NS4A induces membrane rearrangement (18); NS4B colocates with double-stranded RNA (dsRNA) and plays a critical role in viral replication (19). NS4B also suppresses interferon α/β response (20, 21).
Flavivirus NS2A is a 22-kDa hydrophobic protein (22). Its N terminus is generated in the endoplasmic reticulum (ER) lumen by an unknown host protease (23); its C terminus is generated in cytoplasm by viral protease. NS2A was previously shown to be important for viral replication and pathogenesis. (i) NS2A functions in viral RNA synthesis. For example, Kunjin virus (KUNV) NS2A colocalizes with double-stranded viral RNA (dsRNA) and interacts with the 3′ UTR, NS3, and NS5 in the form of a replication complex (3). (ii) NS2A is important for viral assembly (see details in Discussion) (8, 24). (iii) Expression of DENV-2 NS2A alone inhibits interferon α/β response (21). An A30P substitution reduced the effect KUNV NS2A on antagonism of interferon response, leading to virulence attenuation in mice (25, 26). JEV NS2A was recently shown to block dsRNA-activated protein kinase PKR (27). (iv) A conserved slippery heptanucleotide motif, located at the beginning of the NS2A gene, contributes to the production of NS1′ in the JEV serogroup through a ribosomal frameshift mechanism (28). (v) NS2A participates in virus-induced membrane formation (24). Despite the above functions, the topology of flavivirus NS2A on the ER membrane has not been experimentally determined.
In this study, we used a number of biochemical approaches to establish the topology of DENV-2 NS2A on the ER membrane. Our results indicate that DENV-2 NS2A contains five integral transmembrane segments (amino acids 69 to 209) that span the lipid bilayer of the ER membrane; in addition, one membrane-associated segment (amino acids 32 to 51) interacts with the ER membrane without traversing the lipid bilayer. The nuclear magnetic resonance (NMR) structure of the first transmembrane segment (amino acids 69 to 93) showed two helices connected by a P85-mediated “helix breaker.” Functional analysis showed that amino acid P85 is not important for viral replication. Instead, the adjacent positively charged residue, R84, is critical for both viral RNA synthesis and intracellular virion assembly/maturation.
MATERIALS AND METHODS
Bioinformatics.
The amino acid sequence of DENV-2 NS2A (strain NGC) was used for membrane topology prediction using the following webservers: HMMTOP (http://www.enzim.hu/hmmtop/index.php), TMHMM2 (http://www.cbs.dtu.dk/services/TMHMM/), DAS (http://www.sbc.su.se/∼miklos/DAS/maindas.html), TOPCONS (http://topcons.cbr.su.se/), Split (http://split.pmfst.hr/split/4), and MEMSAT3 (http://bioinf.cs.ucl.ac.uk/psipred/). The amphipathic helix prediction was performed using the HeliQuest program (http://heliquest.ipmc.cnrs.fr/).
Cell culture, viruses, and antibodies.
Baby hamster kidney (BHK-21) cells were maintained in high-glucose Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 4 mM l-glutamine, 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). HEK293T (human embryo kidney 293T) cells were grown in low-glucose DMEM (Invitrogen) with 10% FBS and 1% penicillin/streptomycin. All cells were incubated at 37°C with 5% CO2. Recombinant DENV-2 (NGC strain) was produced from plasmid pACYC NGC FL, a full-length (FL) infectious cDNA clone of DENV-2 (see details below). A mouse monoclonal antibody against enhanced green fluorescence protein (eGFP) was purchased from Roche. The mouse monoclonal antibody 4G2 against DENV envelope protein was prepared from hybridoma cell lines obtained from the American Type Culture Collection (ATCC). Goat anti-mouse antibodies conjugated to horseradish peroxidase (HRP) were purchased from Sigma-Aldrich. Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes) was bought from Life Technologies.
Plasmid construction.
Standard molecular biology methods were used in all plasmid constructions. Our experience suggests that the low-copy-number plasmid pACYC177 (Thermo Fisher Scientific Inc.) is an ideal vector for construction of stable infectious cDNA clone of flavivirus (29–31). Therefore, we swapped the full-length (FL) cDNA of DENV-2 (NGC strain) from a previous infectious clone plasmid (32) into vector pACYC177. The resulting plasmid, pACYC-NGC FL, contained a long DNA fragment covering a T7 promoter, the DENV-2 NGC genome, and the hepatitis delta virus ribozyme sequence (HDVr). The DNA fragment was engineered into pACYC177 via two unique restriction sites: SacII (upstream of T7 promoter) and ClaI (downstream of HDVr).
Replicon pACYC-NGC was constructed by replacing the viral structural genes (C-prM-E) with a Renilla luciferase gene followed by a foot-and-mouth disease virus (FMDV) 2A peptide. In the replicon clone, the first 38 amino acids of C protein and the last 31 amino acids of E protein were retained. To facilitate the construction, a SacI restriction site was introduced into both pACYC NGC FL and the pACYC-NGC replicon clone by silent mutation (A4338G, A4342T, and G4343C) using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Inc.) with the primer pair NGC_SacI_F and NGC_SacI_R (Table 1). For site-directed mutagenesis, subclone TA-D was constructed by amplifying viral genome from nucleotide position 2,544 to 4,344 using the primer pair NGC_NheI_F and NGC_SacI_R (Table 1), the PCR DNA was directly cloned into vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA). NS2A mutations (R84A, R84E, R84S, and P85A) were individually introduced into the subclone TA-D by site-directed mutagenesis using the corresponding primers (Table 1); the DNA containing each mutation was inserted into pACYC NGC FL or the pACYC NGC replicon clone at the NheI and SacI restriction sites.
Table 1.
Sequences of DNA primers used in this study
| Primer name | Sequence (5′ to 3′) |
|---|---|
| NGC_SacI_F | CAGGCAGAGATATCAGGGAGCTCTCCAATCCTGTCAATAAC |
| NGC_SacI_R | GTTATTGACAGGATTGGAGAGCTCCCTGATATCTCTGCCTG |
| NGC_NheI_F | CCTTCAAAGCTAGCTTCAGCT |
| N2A_R84A_F | CTAGCAGCCTTCAAAGTCGCACCAACTTTTGCAGCTGG |
| N2A_R84A_R | CCAGCTGCAAAAGTTGGTGCGACTTTGAAGGCTGCTAG |
| N2A_R84E_F | CTAGCAGCCTTCAAAGTCGAACCAACTTTTGCAGCTGG |
| N2A_R84E_R | CCAGCTGCAAAAGTTGGTTCGACTTTGAAGGCTGCTAG |
| N2A_R84S_F | CTAGCAGCCTTCAAAGTCTCCCCAACTTTTGCAGCTGGAC |
| N2A_R84S_R | GTCCAGCTGCAAAAGTTGGGGAGACTTTGAAGGCTGCTAG |
| N2A_P85A_F | CAGCCTTCAAAGTCAGAGCAACTTTTGCAGCTGG |
| N2A_P85A_R | CCAGCTGCAAAAGTTGCTCTGACTTTGAAGGCTG |
| 2A-p1-F | CGCGGATCCACCATGGTGAGCAAGGGCGAGGAGC |
| 2A-p1-R | ACCCTCGAGTTACTTGTACAGCTCGTCCATGCCG |
| 2A-p3-R | ACCCTCGAGTTAGGCGGAGGTGGAGTTCTTGTACAGCTCGTCCATGCCGAGAG |
| 2A-p4-F1 | GGCGAATTCACCATGAGCACCTCACTGTCTGTGT |
| 2A-p4-R1 | TCTGTTATGAGTTTTCCAGAGGCGGCCGCTTTCTGGATAGCTG |
| 2A-p4-F2 | CAGCTATCCAGAAAGCGGCCGCCTCTGGAAAACTCATAACAGA |
| 2A-p4-R2 | GGTGGATCCGGCTGTGACCAAGGAGTTGAC |
| NGC 7764V | CGTCGAGAGAAATATGGTCACACC |
| NGC 7844C | CCACAATAGTATGACCAGCCT |
| M_GAPDH-F | AGGTCGGTGTGAACGGATTTG |
| M_GAPDH-R | TGTAGACCATGTAGTTGAGGTCA |
A mammalian expression vector, pXJ, driven by a cytomegalovirus (CMV) promoter was used to express various NS2A fusion proteins for topology study. Multiple restriction enzyme sites (EcoRI, NotI, BamHI, XhoI, and KpnI) of vector pXJ were used for cloning different NS2A fragments; each NS2A fragment was C-terminally fused in frame with enhanced green fluorescence protein (eGFP). pACYC NGC FL was used as a template for amplification of different NS2A fragments. The eGFP was amplified from pEGFPluc vector (Clontech). For construction of pXJ-TM-eGFP (TM represents various transmembrane fragments of NS2A), different NS2A fragments were fused with eGFP and inserted into vector pXJ at the BamHI and XhoI sites. For construction of pXJ-leader-TM-eGFP-Glyc, an N-glycosylation (Glyc) acceptor site (Asn-Ser-Thr-Ser-Ala) (18) was fused to the C terminus of eGFP by PCR using the primer pair 2A-p1-F and 2A-p3-R (Table 1); the eGFP-Glyc fragment was inserted into vector pXJ at BamHI and XhoI sites, resulting in plasmid pXJ-eGFP-Glyc. To construct the “leader” (containing viral proteins upstream of NS2A to form the right topology of NS2A; see below), we performed an overlap PCR using two primer pairs (2A-p4-F1/2A-p4-R1 and 2A-p4-F2/2A-p4-R2) to fuse a signal peptide comprising the last 24 amino acids of E protein (E24) with a fragment containing the first 50 amino acids of NS1 protein (N50), an NotI restriction site, and the last 50 amino acids of NS1 (C50). The leader was inserted into plasmid pXJ-eGFP-Glyc (described above) at the EcoRI and BamHI sites, resulting in construct pXJ-leader-eGFP-Glyc. Plasmid pXJ-leader-eGFP-Glyc was used for insertion of various NS2A fragments using the NotI and BamHI sites, resulting in various pXJ-leader-TM-eGFP-Glyc constructs.
For constructs used for in situ fluorescence protease protection assay, the eGFP sequence, amplified from the pEGFPluc vector with the primer pair 2A-p1-F1 and 2A-p1-R (Table 1), was engineered into vector pXJ at BamHI and XhoI restriction sites, resulting in plasmid pXJ-eGFP. A modified pACYC-B plasmid (using pACYC177 as a vector and containing a T7 promoter followed by nucleotides 1 to 5434 of the DENV-2 NGC genome) was initially constructed by silent mutation (T2874C and T2343C) to knock out two internal EcoRI sites. PCR fragments containing E24, full-length NS1, and different truncated versions of NS2A were amplified from the modified pACYC-B plasmid, digested with EcoRI and BamHI, and inserted into plasmid pXJ-eGFP, resulting in constructs pXJ-E24-NS1-TM-eGFP. All constructs were validated by restriction enzymes digestion and DNA sequencing.
Membrane flotation assay.
A membrane flotation assay was performed according to a previously described procedure with some modifications (19). Briefly, 293T cells were seeded in a 10-cm dish (Nunc) and transfected with 10 μg plasmids encoding different TM-eGFP fusion proteins using X-tremeGENE 9 DNA transfection reagent (Roche) following the manufacturer's instructions. At 10 to 16 h posttransfection (p.t.), the cells were washed once with phosphate-buffered saline (PBS), detached into PBS by gentle pipetting, and centrifuged at 1,000 × g for 5 min. Cell pellets were resuspended in prechilled hypotonic lysis buffer (10 mM Tris-HCl [pH 7.5], 2 mM MgCl2), incubated on ice for 10 min, and disrupted with 15 strokes of a Dounce homogenizer (Sigma). Cell lysates were centrifuged at 1,000 × g at 4°C for 5 min to remove nuclei, unlysed cells, and debris. Postnuclear lysates were mixed with an equal volume of 75% (wt/vol) Nycodenz (Axis Shield, Oslo, Norway) solution in PBS. The lysates (0.96 ml) were loaded at the bottom of a 3.5-ml thick-walled polycarbonate ultracentrifugation tube (Beckman). The tube was sequentially overlaid with 2.16 ml of 35% Nycodenz in PBS (first later) and 0.24 ml of 5% Nycodenz in PBS (second layer). Equilibrium centrifugation was performed in a Beckman SW 55 Ti rotor at 30,000 rpm for 20 h. After centrifugation, an equal volume (30 μl) of sample from each fraction (0.42 ml per fraction) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were detected by Western blotting (described below).
In situ fluorescence protease protection assay.
The in situ fluorescence protease protection assay was performed using a previously described protocol (33). BHK-21 cells were seeded in a Lab-Tek chamber (1.2 × 104 cells/chamber). After 16 to 24 h of incubation, cells were transfected with 1 μg plasmids encoding different NS2A-eGFP fusion proteins. At 24 h p.t., the cells were washed twice in KHM buffer (110 mM potassium acetate [pH 7.2], 20 mM HEPES, and 2 mM MgCl2). Plasma membrane was selectively permeabilized in KHM buffer supplemented with 50 μM digitonin (Merck) at room temperature for 5 min. After being washed once with KHM buffer, the cells were treated with 50 μg/ml protease K (New England BioLabs) in KHM buffer and immediately quantified for fluorescence intensities at 8-s intervals using a Zeiss LSM 5 DUO laser scanning confocal microscope. Signals from 5 to 7 fields (each field contained about 4 to 6 positive cells) were averaged and normalized. Immunofluorescence staining images were captured using Zeiss LSM 5 META system and processed using Adobe Photoshop CS3 (Adobe Systems, San Jose, CA). The mean value of fluorescence intensities of the whole time series (5 min) is presented. Figures were plotted using Adobe illustrator CS3 software. The following formulae were used to calculate the relative fluorescence intensity and the initial rate of fluorescence degradation: relative fluorescence intensity = florescence intensity collected at 300 s/florescence intensity collected at time zero × 100; initial rate of fluorescence degradation (%/min) = (100% − retained relative fluorescence intensity at 8 s) × 60/8.
In vitro deglycosylation assay.
Deglycosylation assay was performed by following a previously reported method (34). HEK293T cells were transfected in a 6-well plate (4 × 105 cells/well and 4 μg DNA) using X-tremeGENE 9 DNA transfection reagent (Roche). At 24 h p.t., the cells expressing GFP-tagged NS2A were washed twice in PBS and detached by gently pipetting. The cells were pelleted by centrifuging at 500 × g for 5 min and resuspended in a solubilization buffer comprising 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM magnesium acetate, 1% Triton X-100 (Sigma), and an EDTA-free protease inhibitor cocktail (Roche). After the cells had been solubilized by rotating the tube at 4°C for 2 h, the cell lysates were centrifuged at 15,000 rpm and 4°C for 1 h. The clarified supernatants were treated with 50,000 units/ml PNGase F (New England Biolabs) or PBS at 37°C for 2 h according to the manufacturer's instructions, except that the denaturation procedure was omitted. The samples were heated at 65°C for 15 min and analyzed by 10% SDS-PAGE followed by Western blotting.
In vitro protease K protection assay.
HEK293T cells were transfected with various NS2A expression constructs, as described in the preceding section. At 20 h p.t., cell lysates were prepared according to a previously described protocol (19), with some modifications. Briefly, the transfected cells were washed twice with PBS, detached in cold PBS, and centrifuged at 1,000 × g for 5 min. Cells were resuspended in a hypotonic lysis buffer (10 mM Tris-HCl, pH 7.5, and 2 mM MgCl2). After incubation on ice for 10 min, the cells were disrupted with 15 strokes of a homogenizer (Sigma). The lysates were clarified by centrifugation at 1,000 × g for 5 min to pellet nuclei, unlysed cells, and debris. The supernatant (postnuclear lysates) were centrifuged at 15,000 rpm for 30 min to pellet membranes and their associated proteins. Pellets were resuspended in PBS and treated with 50 μg/ml protease K (New England BioLabs) for 1 h on ice in the absence or presence of 0.5% Triton X-100. The digestion reaction was stopped by adding 20 mM phenylmethylsulfonyl fluoride (PMSF; Sigma), and the reaction mixture was further incubated on ice for 15 min. The samples were then mixed with a preheated (95°C) 4× lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) supplemented with 100 mM dithiothreitol (DTT) (Sigma), heated at 95°C for 15 min, and analyzed by Western blotting.
SDS-PAGE and Western blotting.
Samples were analyzed on a 10% polyacrylamide–SDS gel. Proteins were transferred onto a nitrocellulose or polyvinylidene difluoride (PVDF) membrane using a Trans-Blot apparatus (Bio-Rad), followed by incubation in blocking buffer (comprising 5% skim milk, 20 mM Tris-HCl [pH 7.5], 137 mM NaCl, and 0.1% Tween 20) for 1 h. The blots were incubated in blocking buffer containing a mouse monoclonal antibody against eGFP (1:1,000 dilution) overnight at 4°C. After being washed three times (10 min each time) with TBST (Tris-buffered saline–Tween) buffer, the blots were further incubated with a goat anti-mouse antibody conjugated to horseradish peroxidase (1:5,000 dilution) in blocking buffer for 1 h, followed by three washes with TBST buffer. The antibody-protein complexes were detected using the ECL Western blotting system (GE Healthcare).
RNA transcription, electroporation, and replicon transient-transfection assay.
FL DENV-2 and replicon RNAs were in vitro transcribed using a T7 mMessage mMachine kit (Ambion, Austin, TX) from cDNA plasmids prelinearized by XbaI. The RNA transcripts (10 μg) were electroporated into BHK-21 cells following a protocol described previously (35). The cells transfected with replicon RNA were seeded in a 12-well plate (2 × 105 cells/well). At various time points, the cells were washed once with phosphate-buffered saline (PBS) and lysed using lysis buffer (Promega). The plates containing the lysis buffer were sealed with Parafilm and stored at −80°C. Once samples for all time points had been collected, luciferase signals were measured in a Clarity luminescence microplate reader (BioTek) according to the manufacturer's protocol.
Immunofluorescence assay (IFA).
BHK-21 cells transfected with viral RNA were grown in an 8-well Lab-Tek chamber slide (Thermo Fisher Scientific). At various time points, the cells were fixed in PBS supplemented with 4% paraformaldehyde at room temperature for 20 to 30 min and permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min. After 1 h of incubation in a blocking buffer containing 1% FBS in PBS, the cells were treated with mouse monoclonal antibody 4G2 for 1 h and washed three times for 5 min with PBS. The cells were then incubated with Alexa Fluor 488 goat anti-mouse IgG for 1 h in blocking buffer, after which the cells were washed as described above. The cells were mounted in a mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Inc.). Fluorescence images were taken with a Leica DM4000 B system. Images were merged using Adobe Photoshop CS3 software.
Virus production and reverse transcriptase PCR (RT-PCR) assay.
BHK-21 cells were transfected with genome-length RNA as descried above. The transfected cells were seeded in one T-75 flask (8 × 106 cells in 15 ml DMEM supplemented with 10% FBS). After incubation at 37°C for 24 h, the cells were cultured in DMEM with 2% FBS at 30°C for another 4 days. At every 24 h p.t., 200 μl culture fluids was collected and stored at −80°C. On day 5 p.t., all culture fluids were centrifuged at 4°C and 500 × g for 5 min, aliquoted, and stored at −80°C. For each sample, viral titer was quantified by plaque assay using BHK-21 cells (30).
Quantification of extra- and intracellular viral RNA and infectious virions.
After electroporation, BHK-21 cells were seeded in a 6-well plate (about 4.5 × 105 cells per well) in DMEM with 10% FBS and incubated at 37% with 5% CO2. At 5 h p.t., the transfected cells were washed once with DMEM to remove residual in vitro-transcribed RNA in the culture fluids and maintained in DMEM containing 2% FBS. At 12, 24, and 48 p.t., culture fluids were harvested, clarified by centrifugation at 500 × g at 4°C for 5 min, and stored at −80°C. Cell samples were harvested by using a previously described method, with some modifications (36). Briefly, at various time points, cells were washed once with cold PBS and incubated for 3 min at 4°C in a cold alkaline–high-salt solution (1 M NaCl and 50 mM sodium bicarbonate, pH 9.5) to remove cell surface-associated virus. The cells were then washed twice with cold PBS, treated with 0.25% Trypsin-EDTA (Gibco) to detach them from the plate surface, and collected by centrifugation at 1,000 × g for 5 min. The cell pellets were resuspended in 250 μl RPMI 1640 medium with 2% FBS. One hundred microliters of the cell suspensions was centrifuged to pellet the cells; the pelleted cells were then quantified for intracellular viral RNA (see below). The remaining 150 μl of cell suspensions was lysed using a single freeze-thaw cycle (frozen at −80°C and thawed at 37°C), cellular debris was removed by centrifugation at 3,200 × g for 5 min at 4°C, and the supernatant was quantified for intracellular infectious virus using a plaque assay.
Viral RNAs in culture fluids were extracted using a QIAamp viral RNA minikit (Qiagen), and intracellular total RNAs were isolated using an RNeasy minikit (Qiagen). Extracted RNAs were eluted in 50 μl RNase-free water. Two microliters of RNA elution was used for real-time RT-PCR. Viral RNAs were measured using an iScript one-Step RT-PCR kit with SYBR green (Bio-Rad) and the primer pair NGC 7764V/7844C (Table 1). The mRNA level of the housekeeping gene glyceraldehyde-3-phophate dehydrogenase (GAPDH) was measured in parallel using the primer pair M_GAPDH-F/M_GAPDH-R (Table 1). The intracellular viral RNAs were normalized according to the mRNA level of GAPDH.
Peptide synthesis, purification, and NMR spectroscopy.
A peptide representing the first transmembrane segment, corresponding to residues D67 to R94 of DENV-2 NS2A (acetyl-DIGMGVTYLALLAAFKVRPTFAAGLLLR), was synthesized and purified by reverse-phase high-pressure liquid chromatography (RP-HPLC) to a purity of ≥95% (GL Biochem Ltd., Shanghai, China). The sequence of the peptide was confirmed by mass spectroscopy. The purified peptide was dissolved to 3 mg/ml in 250 μl of the deuterated solvent (Cambridge isotope) containing 4:4:1 of CDCl3, CD3OH, and H2O (pH = 7.0) (37). The NMR sample was transferred to a 3-mm NMR tube for data collection. All NMR data were collected at 25°C on a Bruker Avance II700 MHz spectrometer equipped with a 5-mm z-gradient TXI cryoprobe. A total correlation spectroscopy (TOCSY) experiment was recorded with a mixing time of 80 ms. Two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY) experiments were recorded using mixing times of 200 ms and 300 ms. The proton chemical shifts were directly referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). The nitrogen and carbon chemical shifts were referenced indirectly to DSS. The 1H-13C and 1H-15N heteronuclear single quantum correlation spectroscopy (HSQC) in 13C and 15N natural abundance were also acquired to facilitate resonance assignment. All NMR spectra were processed using Topspin (Bruker, version 2.1) and analyzed using Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Peptide structure determination.
The peptide assignment in solvent was accomplished using the previously reported procedures, in which spin systems were identified by a TOCSY spectrum and sequential connectivity was determined using a NOESY spectrum (37, 38). Cross-peaks from the NOESY spectrum with a mixing time of 200 ms were selected, assigned, and integrated in Sparky. The NOEs were calibrated with CYANA and transferred to distance constrains for structural determination (39). The structure was determined using torsion angle dynamics-simulated annealing as implemented in CYANA using NOE restraints. One hundred structures were calculated using CYANA. Fifteen structures with the lowest energy function were selected and viewed using PyMOL (www.pymol.org).
RESULTS
Bioinformatic analysis of DENV NS2A membrane topology.
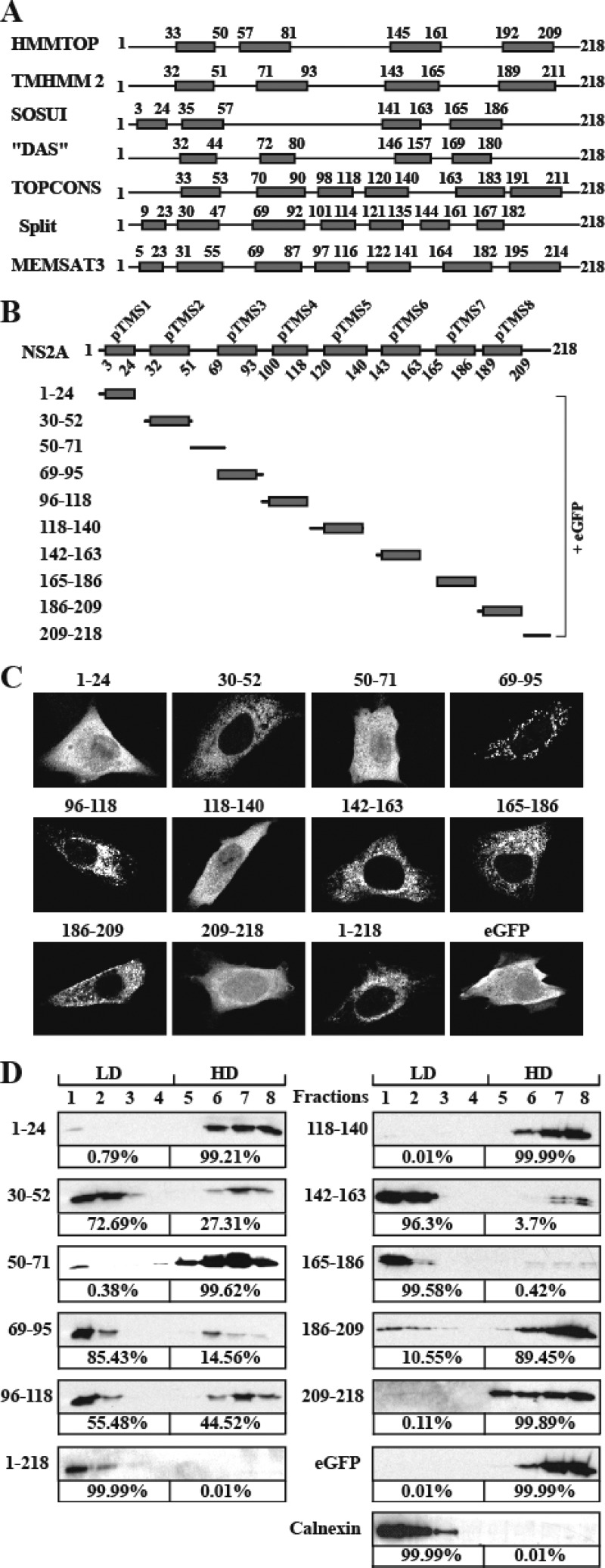
Approximately 42% of DENV NS2A amino acids are hydrophobic residues. Flavivirus NS2A was previously shown to associate with ER membrane in the replication complex (3). To examine which regions of NS2A are responsible for membrane association, we initially analyzed the membrane topology using several bioinformatics algorisms. As shown in Fig. 1A, different algorithms predicted different numbers and positions of transmembrane segments (pTMS). To avoid discounting any pTMS, we combined all the predicted topologies into a reference model (Fig. 1B). Based on this reference model, we experimentally analyzed individual segments as either membrane associated (integrally or peripherally membrane associated) or non-membrane associated (see below). Each NS2A segment was C-terminally fused with an eGFP reporter (Fig. 1B). The length of each selected segment was slightly different from that of the predicted pTMS to balance the intact flexibility between pTMS and eGFP according to charged residues around the C terminus of each pTMS.
Fig 1.
Prediction of membrane topology and analysis of membrane-associated activity of DENV-2 NS2A. (A) Schematic representation of DENV-2 NS2A transmembrane segments predicted by HMMTOP, TMHMM2, SOSUI, DAS, TOPCONS, Split, and MEMSAT3. The gray boxes indicate predicted transmembrane segments (pTMS). The positions of the first and last amino acid of pTMS are indicated. (B) A reference model of DENV-2 NS2A topology. Different fragments covering the entire NS2A were C-terminally fused with eGFP. The amino acid positions of each NS2A fragment are indicated on the left. (C) IFA analysis of BHK-21 cells transfected with various NS2A-eGFP constructs. At 24 h p.t., the expression of eGFP was monitored by a mouse monoclonal antibody against eGFP and a goat anti-mouse IgG conjugated with Alexa Fluor 488. The eGFP signal is in white. (D) Membrane flotation analysis of 293T cells transfected with plasmids expressing NS2A fragment-eGFP fusion proteins. NS2A fragment-eGFP proteins in each fraction were detected using an antibody against eGFP. Calnexin, probed with rabbit IgG against calnexin (Sigma), was used as an integral membrane protein control. The percentages of signals detected in the low-density (LD) fractions (1 to 4) and high-density (HD) fractions (5 to 8) were calculated by ImageJ software and are indicated below the panels.
Cellular localization of NS2A fragments.
To define the regions of NS2A that has membrane-associated activity, we transfected BHK-21 cells with plasmids that express various NS2A fragment-eGFP fusion proteins (Fig. 1B). At 20 h p.t., the expression and intracellular localization of each fusion protein were monitored by IFA. A fusion protein containing a real membrane-targeting segment would present an ER staining pattern, whereas a protein without any membrane-targeting segment would display an even distribution of intracellular staining pattern. As shown in Fig. 1C, expression of eGFP alone (without any NS2A fragment) exhibited an even distribution of fluorescence throughout the cell, whereas expression of the full-length NS2A-eGFP (construct 1–218) displayed fluorescence in the perinuclear region, confirming that NS2A is a membrane-associated protein. Expression of NS2A fragments 1–24, 50–71, 118–140, and 209–218 fused with eGFP showed fluorescence patterns similar to that of expression of eGFP alone, suggesting that these NS2A fragments do not have membrane-associated activity. In contrast to the above result, expression of NS2A fragments 30–52, 69–95, 96–118, 142–163, 165–186, and 186–209 fused with eGFP exhibited a reticular fluorescence pattern, indicating that these NS2A fragments have membrane-associated activity. It should be noted that these regions that showed ER membrane-interacting activity do not necessarily span the entire lipid bilayer of ER membrane; some of these fragments could peripherally interact with the membrane without traversing the complete lipid bilayer.
Membrane flotation analysis.
To validate the above membrane-associated activities of NS2A fragments, we performed a membrane flotation assay (Fig. 1D). As controls, we found that soluble eGFP alone was exclusively distributed in the high-density (HD) fractions, whereas calnexin, a well-known ER integral membrane protein, completely floated to low-density (LD) fractions. For the NS2A fragment-eGFP fusion proteins, more than 99% of the expressed proteins containing NS2A fragments 1–24, 50–71, 118–140, and 209–218 were found in the HD fractions, indicating that these segments are dissociated with membrane. In contrast, more than 55% of the proteins containing fragments 30–52, 69–95, 96–118, 142–163, 165–186, and 1–218 (full-length NS2A) were detected in the LD fractions, indicating that these fragments have membrane-associated activity. The above membrane flotation results are in good agreement with the cellular localization results (Fig. 1C).
For fragment 186–209, the correlation between the membrane flotation result and the cellular localization data is not strong: only 10.6% of this fragment-eGFP fusion protein was detected in the LD fraction (Fig. 1D), whereas the same protein exhibited a reticular fluorescent pattern inside the cell (Fig. 1C). However, it should be noted that, in the membrane flotation assay, all non-membrane-associated fragments (1–24, 50–71, 118–140, and 209–218) exhibited <0.8% protein distribution in the LD fractions. The 10.6% distribution of fragment 186–209 in the LD fractions suggests that this fragment has a weak membrane-associated activity.
Next, we used three additional methods to further investigate the topology of NS2A which have been well established for the study of the topology of membrane proteins: (i) the in situ fluorescence protease protection assay (34), (ii) the in vitro deglycosylation assay (18, 19), and (iii) the in vitro protease protection assay (19).
In situ fluorescence protease protection analysis.
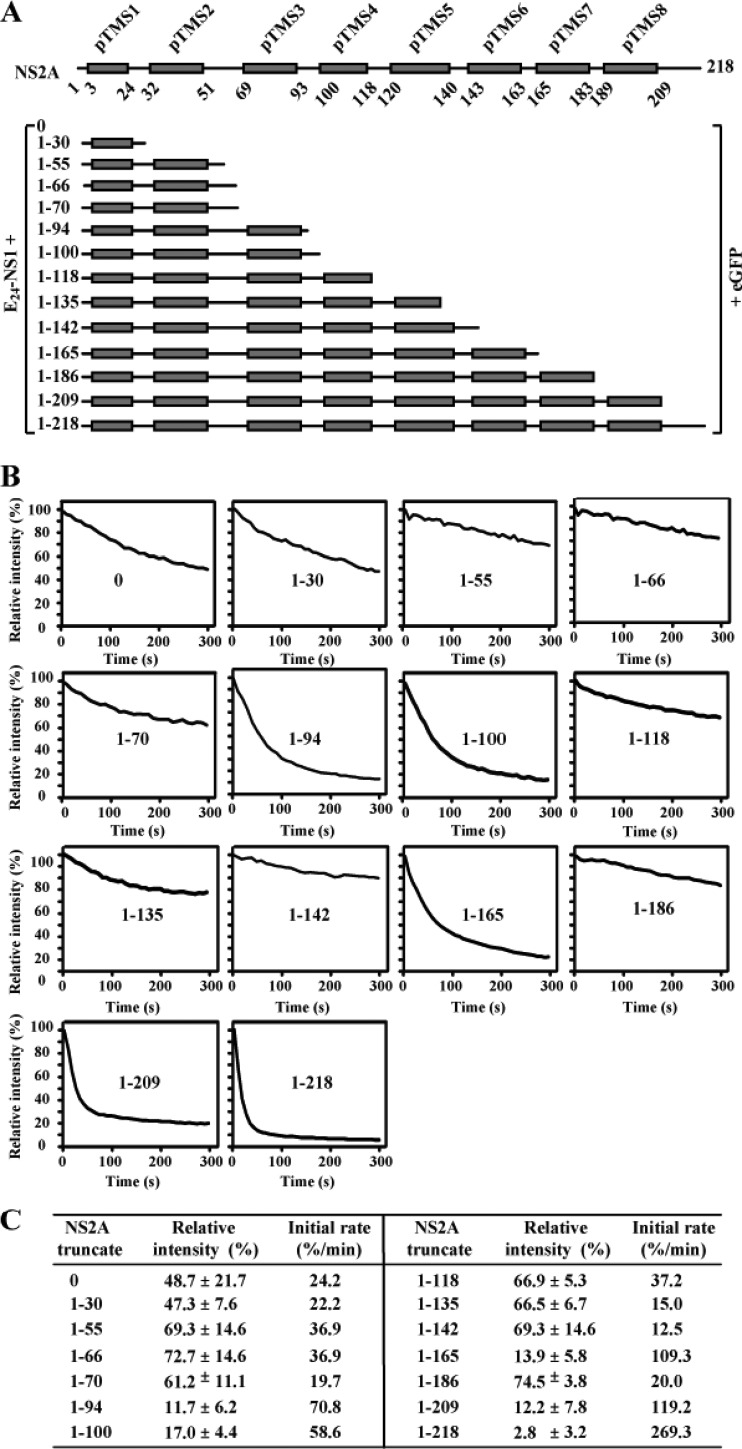
We probed the membrane topology of NS2A in the presence of full-length NS1 using an in situ fluorescence protease protection assay. To ensure the correct topology of NS2A on the ER membrane, we constructed a panel of plasmids in which E24 (the last 24 amino acids of E protein, which serves as a leader signal for NS1 localization into the ER lumen) and the full-length NS1 were fused to various C-terminally truncated NS2A-eGFP proteins (Fig. 2A). Individual plasmids were transfected into BHK-21 cells. At 24 h p.t., the cells were treated with digitonin, which selectively permeabilizes the plasma membrane without affecting the ER membrane. After the digitonin treatment, the cells were incubated with protease K, and the intensity of eGFP fluorescence was quantified. If the construct expresses eGFP on the cytosolic side of ER, eGFP will be efficiently digested by protease K. If the construct expresses eGFP on the lumen side of ER, eGFP will be less accessible to protease K digestion. The initial rate of fluorescence degradation and the relative intensity of fluorescence at the end of protease digestion (5 min) could be used to differentiate the luminal or cytosolic localization of eGFP. We first determined the optimal concentration of digitonin to be 50 μM (for selective permeabilization of plasma membrane). Under this condition, the cytosolic fluorescence in cells expressing eGFP alone disappeared within 5 min after addition of protease K (data not shown).
Fig 2.
In situ fluorescence protease protection assay. (A) E24-NS1-NS2A (truncated)-eGFP fusion constructs. Each NS2A fragment was N-terminally fused with E24-NS1 (representing a signal peptide derived from the last 24 residues of E protein [E24] and NS1 protein), and C-terminally fused with eGFP. pTMS are shown as gray boxes. (B) In situ fluorescence protease protection assay. BHK-21 cells were transfected with the indicated NS2A constructs. At 24 h p.t., the cells were permeabilized with digitonin followed by protease K treatment. Once the protease K was added, the fluorescence intensities were continuously quantified for 300 s (see Materials and Methods). Relative intensities were calculated using the fluorescence intensity at every 8 s divided by that at time zero when protease K was added. The means of relative intensities derived from 5 to 7 fields (each field containing 4 to 6 positive cells) are presented. The positions of NS2A truncates are indicated in the corresponding panels. (C) Summary of relative fluorescence intensity and initial rate of fluorescence degradation. Average results from at least two independent experiments are presented. See Materials and Methods for calculations.
Next, we used the above conditions to analyze various constructs consisting of E24, NS1, and truncated NS2A proteins conjugated to eGFP. As expected, in cells expressing E24-NS1-NS2A (complete)-eGFP (construct 1–218 in Fig. 2A and B), the fluorescence intensity reduced rapidly upon protease K addition with an initial rate of 269.3% per minute (see Materials and Methods for calculation); only 2.8% of the fluorescence intensity was retained at 5 min postdigestion, indicating that eGFP is located on the cytosolic side. In contrast, in cells expressing E24-NS1-eGFP (i.e., without NS2A; construct 0 in Fig. 2), the fluorescence signal decreased slowly at an initial rate of 24.2% per minute; about 50% of fluorescence intensity was retained at 5 min after protease digestion, indicating that eGFP resides in the ER lumen. These results are in agreement with the current understanding that the N and C termini of NS2A are generated by a lumen-resident cellular protease and the cytosolic viral protease, respectively (23). The results also validate the in situ fluorescence protease assay for membrane protein analysis.
Figure 2B shows the in situ fluorescence protease digestion of various E24-NS1-NS2A fragment-eGFP constructs. The results revealed two distinct patterns of fluorescence degradation. Pattern I consisted of a high initial rate of fluorescence degradation (>50% per minute) and a low fluorescence intensity (<20%) at 5 min postdigestion. Pattern I was observed for constructs 1–94, 1–100, 1–165, 1–209, and 1–218 (Fig. 2B and C), indicating that the C termini of these NS2A constructs are located in cytosol. Pattern II consisted of a lower initial rate of fluorescence degradation and a higher fluorescence upon protease K digestion. Pattern II was detected for constructs 0, 1–30, 1–55, 1–66, 1–70, 1–118, 1–135, 1–142, and 1–186 (Fig. 2B and C), indicating that the C termini of these NS2A truncates reside in the ER lumen. Statistical analysis showed that the differences in relative fluorescence intensity between pattern I and pattern II constructs are statistically significant (two-way ANOVA, P < 0.0001). Notably, among the constructs exhibiting pattern II, the fluorescence intensities of cells expressing E24-NS1-NS2A(0)-eGFP and E24-NS1-NS2A(1–30)-eGFP decreased slightly more than those of other constructs. This is likely because constructs 0 and 1–30, respectively, generated the proteins NS1-eGFP and NS1-NS2A(1–30)-eGFP, which have low affinity for the ER membrane; these proteins were partially secreted (like NS1) and became accessible for protease K digestion. Overall, the switch of fluorescence behavior between patterns I and II among various NS2A constructs (Fig. 2B) allowed us to estimate the topology of NS2A on the ER membrane (see below).
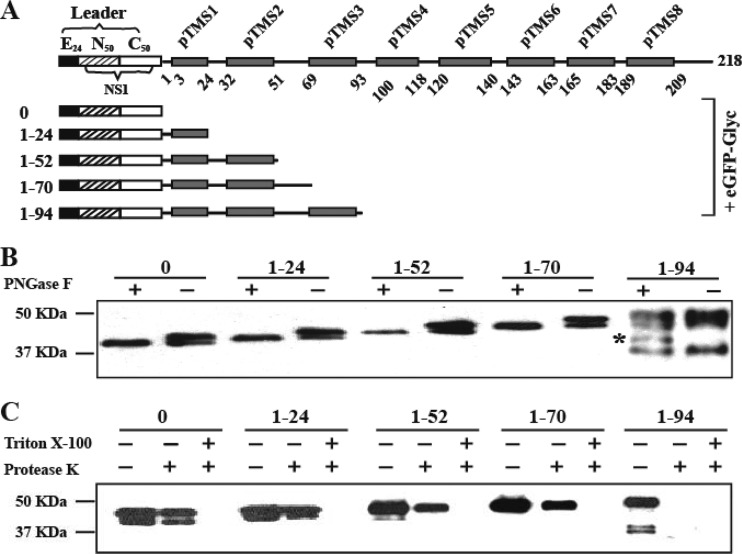
In vitro deglycosylation analysis.
We performed an in vitro deglycosylation assay to further evaluate the membrane topology of NS2A. Figure 3A shows the constructs used for the deglycosylation analysis. For correct ER localization, a leader was generated by in-frame fusion of three components: the last 24 amino acids of E protein (E24, serving as a signal sequence for NS1 localization) (40), the first 50 amino acids of NS1, and the last 50 amino acids of NS1 (containing the octapeptide sequence essential for cleavage of NS2A) (40, 41). It should be noted that the internal NS1 N-glycosylation sites (N130 and N201) were excluded in these constructs. The leader was then fused with various NS2A truncates, an eGFP, and an exogenous glycosylation acceptor sequence (Glyc) (Fig. 3A). When expressed in cells, eGFP-Glyc is glycosylated when the C terminus of NS2A truncate is located in the ER lumen, where the N-glycosylation enzymes reside. The glycosylation status of each NS2A-eGFP-Glyc construct could be detected by the digestion of endoglycosidase PNGase F. For construct 0 (without NS2A), two distinct bands of eGFP-Glyc derivatives were observed without PNGase F treatment; upon PNGase F digestion, only the faster-migrating band was observed (Fig. 3B). As a control, expression of eGFP-Glyc alone without the leader sequence generated only the faster-migrating band (data not shown). Similar glycosylation patterns were observed for NS2A constructs 1–24, 1–52, and 1–70 (Fig. 3A). These results again indicate that the first 70 amino acids of DENV-2 NS2A are located in the ER lumen.
Fig 3.
Deglycosylation and in vitro protease protection assays. (A) Leader-NS2A truncate-eGFP-Glyc constructs. The leader sequence contains a signal peptide derived from the last 24 residues of E protein (E24; black box), the first 50 residues of NS1 (striped box), and the last 50 residues of NS1 (white box). pTMS are shown as gray boxes. Each NS2A fragment was N-terminally fused with the leader sequence and C-terminally fused with eGFP-Glyc (a glycosylation sequence). The amino acid positions of each NS2A constructs are indicated on the left. (B) In vitro deglycosylation assay. HEK293T cells were transfected with various constructs depicted in (A). An extra band of unknown identity is indicated with an asterisk. At 24 h p.t., the expressed fusion proteins were extracted and treated with PNGase F (+) or PBS (−). A mouse monoclonal antibody against GFP was used to probe eGFP-tagged proteins (see Materials and Methods). Molecular mass markers are shown on the left. (C) In vitro protease protection assay. Cell membranes of the transfected cells as described for panel B were extracted and digested with protease K in the presence or absence of 0.5% Trion X-100. The samples were analyzed by Western blotting as described for panel B.
In contrast, when NS2A construct 1–94 was expressed, treatment with of PNGase F did not eliminate the top band representing NS2A-eGFP-Glyc (Fig. 3A). However, an extra band of unknown identity (Fig. 3B) was observed upon PNGase F treatment. This result suggests that amino acid 94 of NS2A is located in the cytosol, and amino acids 70 to 94 (corresponding to pTMS3) span the ER membrane from the lumen to the cytosol. Our attempts to examine the glycosylation status of fusion proteins carrying longer NS2A fragments failed: many bands of smaller sizes were observed, and the majority of the expressed proteins were trapped in stacking gels (data not shown).
In vitro protease protection assay.
We performed an in vitro protease protection assay (18, 19) to further confirm the NS2A topology. HEK293T cells were transfected with the same plasmids used in the deglycosylation assay (Fig. 3A). At 24 h p.t., ER membrane fractions were isolated, treated with protease K in the presence or absence of the detergent Triton X-100, and analyzed by Western blotting using an eGFP antibody. In this experiment, the N-terminal leader–NS2A fragment served as a signal for the translocation of the C-terminal eGFP on the ER membrane. eGFP located in the ER lumen is protected from protease K cleavage and is digested upon detergent treatment. In contrast, eGFP located in the cytoplasm will be readily digested by protease K without detergent treatment. As shown in Fig. 3C, eGFP was partially protected from protease K digestion when expressed from constructs 0, 1–24, 1–52, and 1–70. Once the membrane was disrupted by Triton X-100, the protein was completely degraded by protease K. The results further indicate that the first 70 amino acids of NS2A are located in the ER lumen. Conversely, eGFP protein expressed from construct 1–94 was completely digested by protease K without any detergent, again indicating that amino acid 94 of NS2A is located in the cytosol.
Our attempt to use the in vitro protease assay to analyze the constructs carrying longer NS2A fragments failed (data not shown) for the reason described above. It should be noted that in our deglycosylation and in vitro protease protection assays, the N-terminal leader was not cleaved from the NS2A fragments (Fig. 3B and C). The result demonstrates that the N-terminal 90 residues of NS2A is not sufficient for the cleavage at the NS1-NS2A junction. This observation agrees with a previous report that most of the N-terminal NS2A (>70%) is required for the cleavage at the NS1-NS2A junction (23).
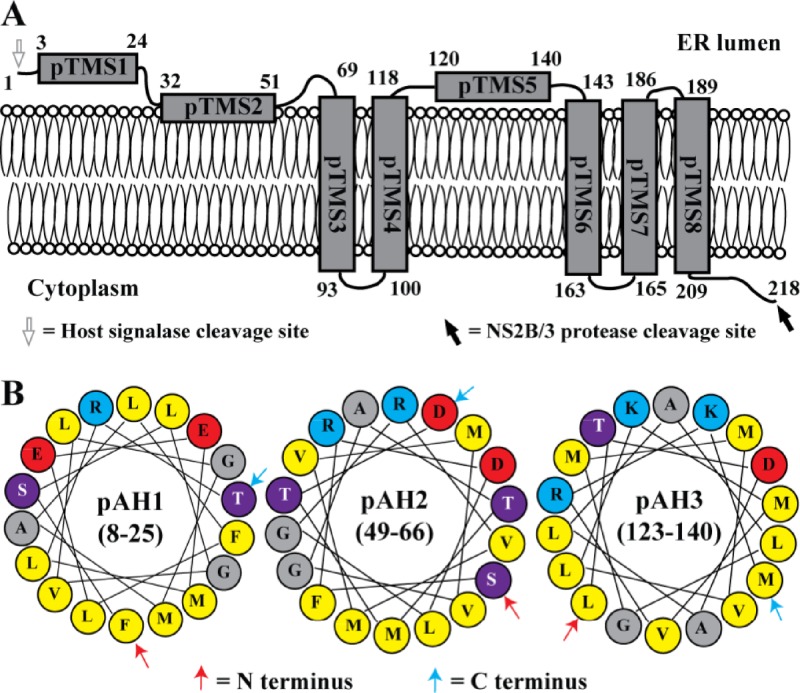
A topology model of DENV NS2A on the ER membrane.
To consolidate the above results, we propose a topology model of DENV-2 NS2A on the ER membrane (Fig. 4A). The N-terminal 68 amino acids are located in the ER lumen; within this region, pTMS1 does not have membrane-associated activity, whereas pTMS2 is likely to peripherally associate with ER membrane without spanning the lipid bilayer. Amino acids 69 to 209 form five real TMS, each of which integrally spans the ER membrane. pTMS3, pTMS6, and pTMS8 span the membrane from lumen to cytosol; pTMS4 and pTMS7 transverse the membrane from cytosol to lumen. pTMS5 does not have membrane-associated activity. The C-terminal amino acids 210 to 218 are located in cytosol.
Fig 4.
Membrane topology of DENV NS2A. (A) A topology model of DENV-2 NS2A on the ER membrane. The N-terminal 68 residues are located in the ER lumen; pTMS1 does not associate with the membrane, whereas pTMS2 is likely to peripherally associate with ER membrane. pTMS3, pTMS6, and pTMS8 span the membrane from lumen to cytosol, whereas pTMS4 and pTMS7 span the membrane from cytosol to lumen. The C-terminal 9 residues are located in cytosol. It should be noted that the topology of pTMS4-8 is supported only by results from Fig. 1 and 2; due to technical difficulties (see details in the text), we were not able to validate the topology of pTMS4-8 by the deglycosylation and in vitro protease protection assays (Fig. 3). (B) Three amphipathic helices (pAH1, pAH2, and pAH3) predicted by HeliQuest. Each pAH construct is an α-helix composed of 18 amino acids. The numbers represent amino acid positions of NS2A for each pAH. Red, blue, purple, gray, and yellow circles represent negatively charged, positively charged, polar, glycine/alanine, and hydrophobic residues, respectively.
Sequence analysis of DENV-2 NS2A using the HeliQuest program suggests that amino acids 8 to 25 (overlapping pTMS1), 49 to 66 (overlapping three residues with pTMS2), and 123 to 140 (within pTMS5) could form amphipathic helices (pAH1-3) (Fig. 4B). The charged residues on one side of the helices could potentially form transient interactions with the negatively charged phosphates on the lipid membrane.
NMR structure of pTMS3.
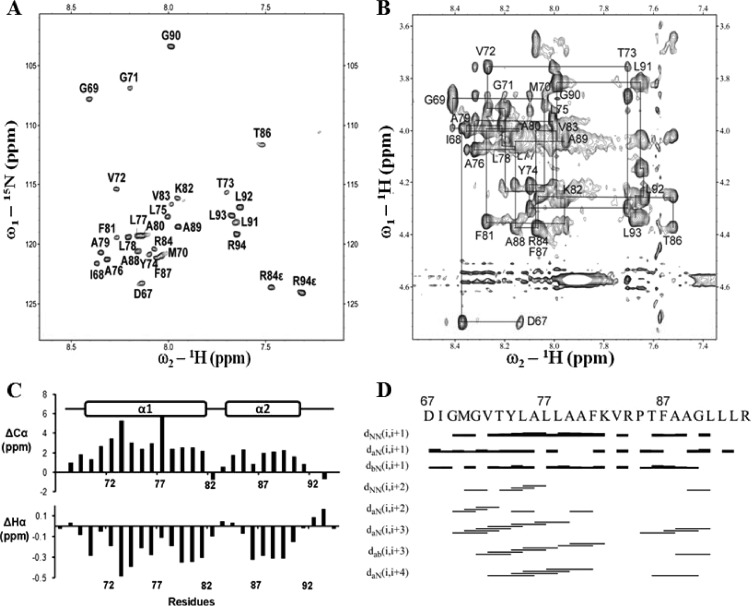
The data above clearly showed that pTMS3 is a real transmembrane domain. This result prompted us to solve the NMR structure of pTMS3 (Protein Data Bank [PDB] identification number 2M0S). A peptide corresponding to amino acids D67-R94 was synthesized and purified. Due to its hydrophobic nature, we solubilized the peptide in deuterated detergents, including SDS and dodecylphosphocholine (DPC) (42). However, poorly resolved NMR spectra were obtained under these conditions (data not shown). Interestingly, when solubilized in an organic solvent (see Materials and Methods), the peptide yielded a well-resolved spectrum that allowed us to complete the resonance assignment for the HSQC and NOESY spectra (Fig. 5A and B). Next, chemical shift index (CSI) analysis (43) was performed using the deviation of 1Hα and 13Cα chemical shifts from the random coil values (Fig. 5C). The negative variation of 1Hα chemical shifts and the positive variation of the 13Cα chemical shifts were observed for residues G69-K82 and T86-L91, indicating that these amino acids are folded as α helices. There was a “break” present between the two helices due to the presence of P85. The NOE assignment for this peptide further supported the CSI analysis (Fig. 5C). The NOE connectivity of a typical α-helix was observed for these two helical fragments containing residues G69 to K82 and T86 to L91, in which strong dNN (i, i + 1) (NOESY connection observed between amide protons) and weak dαN (i, i + 4) (NOESY connections observed between alpha and amide protons) NOE connections were observed (Fig. 5D).
Fig 5.
Resonance assignment of pTMS3. (A) 1H-15N-HSQC of pTMS3 in 15N natural abundance. The assigned cross-peaks are labeled with amino acid and residue position. (B) Assignment of the 2D NOESY spectrum of pTMS3 in the Hα region. Connections between residues are shown with lines. (C) Secondary structure analysis of pTMS3. Deviations of Cα (top) and Hα (bottom) from random coil values are shown. (D) Summary of NOE connectivity. The NOE plot was made from CYANA.
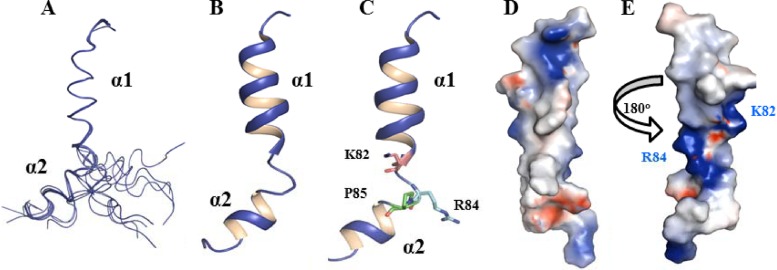
The NMR data suggest that pTMS3 consists of two α-helices separated by a helix “breaker” (Fig. 5). The three-dimensional (3D) structure was determined with 239 distance constraints, including 86 intraresidue, 85 sequential, and 68 medium-range NOEs (Table 2). No hydrogen bond constraints were applied during the structural determination. Figure 6A shows the set of 15 structures with the lowest energy function out of 100 calculated structures with Cα traces. The two α-helical structures were well defined, with root mean square deviations (RMSD) of 0.05 Å for the backbone and 0.78 Å for the heavy atoms (Fig. 6A and B). The structures of the N- and C-terminal residues are flexible due to a lack of distance restraints; this result is consistent with the CSI analysis (Fig. 5). Figure 6C is a ribbon representation of pTMS3 showing two positively charged residues (K82 and R84) and P85; these three residues make the two helices flexible. A surface representation of the pTMS3 displays a hydrophobic surface at one side of this domain (Fig. 6D) and two positively charged patches on the other side of this domain, which arise from the presence of K82 and R84 (Fig. 6E).
Table 2.
Statistics for the structural models of pTMS3
| Parameter | Value |
|---|---|
| No. of: | |
| Restraints | 239 |
| Intraresidue NOEs | 86 |
| Sequential NOEs (i to i + 1) | 85 |
| Medium range NOEs (i to i + 2,3,4) | 68 |
| Dihedral angle restraints | 0 |
| NOE violations > 0.5 Å | 0 |
| Ramachandran plot (%) | |
| Residues in most favored regions | 82 |
| Residues in additional allowed regions | 17.5 |
| Residues in generously allowed regions | 0.2 |
| Residues in disallowed regions | 0.2 |
| RMSD (Å) for: | |
| Backbone atoms (4–16) | 0.05 |
| Heavy atoms (4–16) | 0.78 |
| Backbone atoms (20–24) | 0.05 |
| Heavy atoms (4–16) | 0.74 |
Fig 6.
Structures of NS2A pTMS3. (A) Stereo view of the ensemble of 15 NMR structures from CYANA. Only the Cα trace is shown with PyMOL. (B) Cartoon representation of the pTMS3 structure. Two α-helices are shown with labels. (C) Positions of K82, R84, and P85 in pTMS3. In the cartoon representation, the side chains of K82, R84, and P85 are shown in stick mode. (D) Surface representation of pTMS3. The charge analysis was conducted in Pymol. The positively charged residues, negatively charged residues, and hydrophobic residues are shown in blue, red, and white, respectively. (E) Surface representation of pTMS3 with a 180° rotation from panel D. All images were prepared using Pymol (www.pymol.org).
Functional analysis of NS2A R84 and P85.
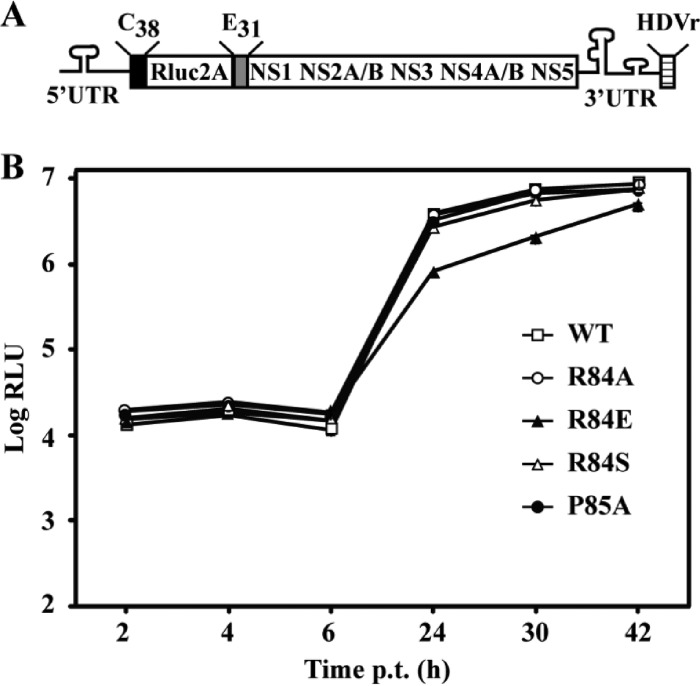
To examine the biological relevance of the helix breaker identified in pTMS3, we performed mutagenesis of R84 and P85 in the context of replicon and genome-length RNA of DENV-2. R84 was replaced by the amino acid A, E, or S. P85 was mutated to A to eliminate the helix breaker. In the transient transfection assay using a luciferase replicon (Fig. 7), none of the mutations affected luciferase signals at 2, 4, and 6 h p.t. (representing input RNA translation). Only the mutation R84E reduced luciferase activity by 20 to 59% at 24, 30, and 42 h p.t. (indicating RNA synthesis); all other mutations did not change the luciferase signals compared with the wild-type (WT) replicon. The replicon results indicate that, unlike the mutation R84E (which attenuates viral RNA synthesis), the substitutions R84S and P85A do not affect viral translation or RNA synthesis.
Fig 7.
Replicon analysis of NS2A R84 and P85. (A) Schematic diagram of a luciferase replicon of DENV-2. Rluc2A, Renilla luciferase gene followed by the foot-and-mouth disease virus 2A peptide; C38, nucleotides encoding the first 38 amino acids of C protein; E31, nucleotides encoding the last 31 amino acids of E protein; HDVr, hepatitis delta virus ribozyme sequence. (B) Transient replicon assay. Equal amounts of replicon RNA (WT or mutant R84A, R84E, R84S, or P85A) were electroporated into BHK-21 cells. At the indicated time points, the transfected cells were lysed and assayed for luciferase activities. The y axis shows the log10 value of Renilla luciferase activity (RLU). Each data point is the average for three replicates, and error bars show the standard deviations.
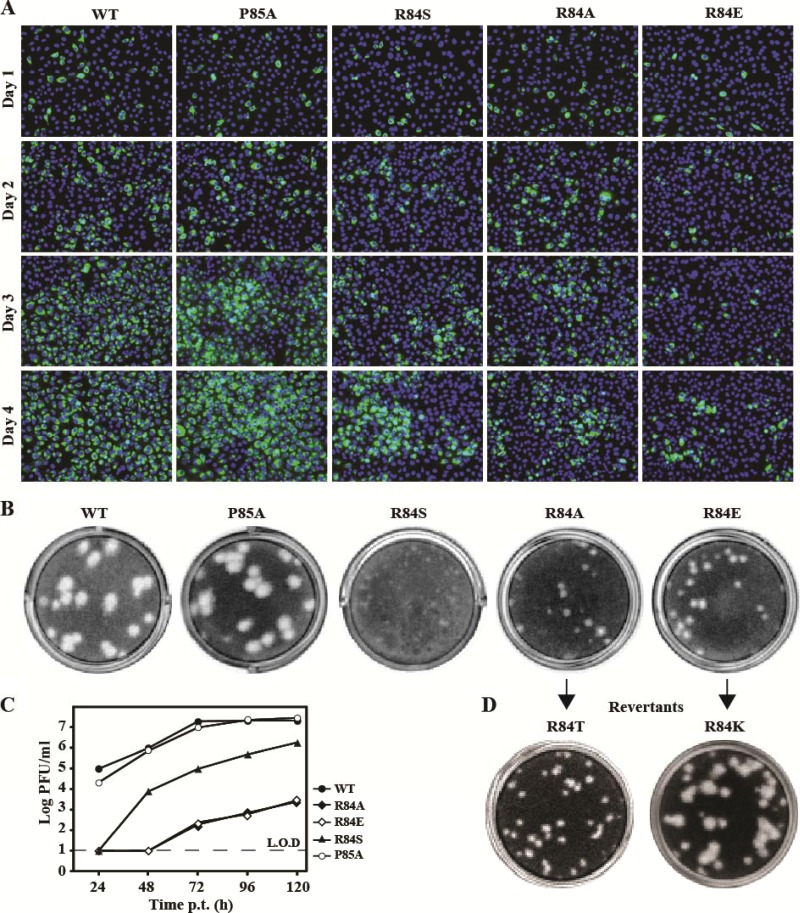
Using an infectious cDNA clone of DENV-2, we examined the above mutations (R84A, R84E, R84S, and P85A) in a complete viral replication cycle. Equal amounts of WT and mutant genome-length RNAs were transfected into BHK-21 cells. The transfected cells were monitored for viral E protein expression using an IFA assay (Fig. 8A). Similar levels of E-positive cells were observed for the WT and P85A mutant from day 1 to 4 p.t.; fewer E-positive cells were detected for mutants R84A and R84S; almost no increase in E-positive cells was observed for mutant R84E. Since the replicon results showed that, except for the R84E substitution, other mutations had no effect on viral RNA synthesis (Fig. 7), the decrease in the number of E-positive cells suggests that R84A and R84S mutants were defective in virus assembly/release. In agreement with the IFA result, mutant virus P85A displayed plaque morphology similar to that of the WT virus, whereas the R84A, R84E, and R84S mutants exhibited plaques smaller than those of the WT virus (Fig. 8B). In addition, cells transfected with mutant R84S, R84A, or R84E RNAs did not yield any infectious virus at 24 and 48 h p.t., and generated much less virus than those transfected with the WT RNA at 72 to 120 h posttransfection. In contrast, mutant P85A and WT RNAs generated similar amounts of infectious viruses (Fig. 8C). Sequencing of the recombinant viruses confirmed that the engineered mutations were retained in the recovered viruses without any other changes (data not shown).
Fig 8.
Analysis of NS2A R84 and P85 in DENV-2 genome-length RNA. (A) IFA analysis. Equal amounts of WT or mutant genome-length RNA were electroporated into BHK-21 cells. The transfected cells were monitored for E protein expression at indicated time points. An anti-E monoclonal antibody 4G2 and a goat anti-mouse IgG conjugated with Alexa Fluor 488 were used as primary and secondary antibodies, respectively. Green and blue indicate E protein and nucleus staining, respectively. (B) Plaque morphology of WT and mutant viruses. The viruses were derived from the media collected on day 5 posttransfection. (C) Virus production. Culture media from cells transfected with genome-length RNAs from (A) were collected at the indicated time points; viral titers were quantified by plaque assay. Average results with standard deviations are presented. The dashed line indicates the limit of detection (L.O.D; 10 PFU/ml). (D) Plaque morphology of revertant viruses after 5 rounds of passaging on Vero cells. Amino acid substitutions identified at position 84 of NS2A are indicated.
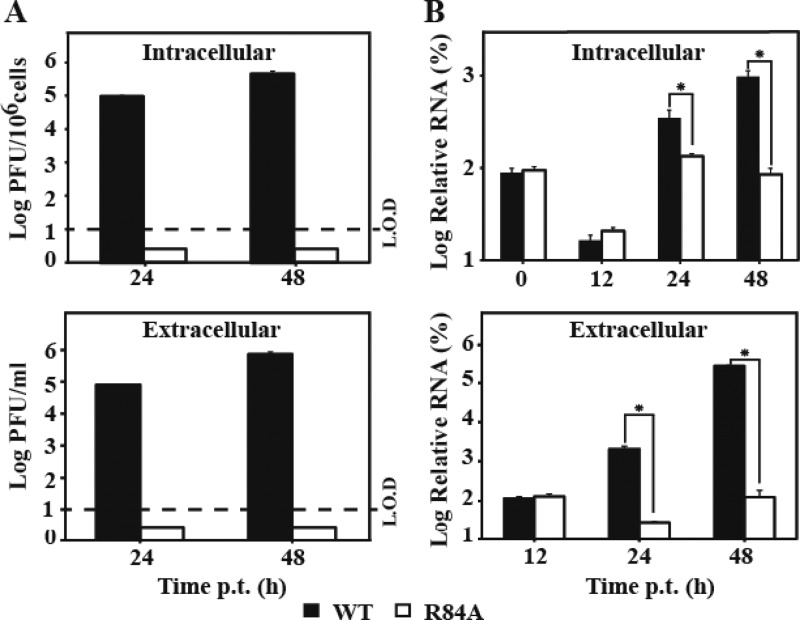
The NS2A R84A mutation abolishes the formation of intracellular infectious virus.
To further examine the role of R84 in virion assembly/release, we measured intra- and extracellular amounts of viral RNA as well as infectious viruses after BHK-21 cells were transfected with the WT or R84A genomic RNA. As shown in Fig. 9A, increasing amounts of intra- and extracellular infectious viruses were observed for the WT RNA at 24 and 48 h p.t., whereas neither intra- nor extracellular infectious viruses were detected for the R84A RNA. In contrast, at 0 and 12 h p.t., no statistical significant difference in intra- or extracellular levels of viral RNA was observed between the WT and the R84A mutant (Fig. 9B), indicating that the two RNAs were transfected into cells with similar efficiencies. At 24 and 48 h p.t., the WT RNA generated significantly more intra- and extracellular viral RNA than the R84A RNA (Fig. 9B). Since the replicon results showed that the R84A mutation does not affect viral RNA synthesis (Fig. 7), the higher intracellular levels of WT viral RNA at 24 and 48 h p.t. were most likely due to the reinfection of the newly produced virus. Collectively, the results indicate that the NS2A R84A mutation abolishes the formation of intracellular infectious virus.
Fig 9.
Comparison of intra- and extracellular infectious viral particles and viral RNA between the WT and the R84A mutant. BHK-21 cells were transfected with equal amounts of WT and mutant genome-length RNA. At the indicated time points, intra- and extracellular infectious viral particles were quantified by plaque assay (A); viral RNAs were measured by quantitative RT-PCR (B) (for details, see Materials and Methods). The intracellular viral particles were normalized to the cell number and presented as PFU per 106 cells. The relative RNA was calculated using formula 100% × 2(CTi − CTo), where CTi is the CT value for the individual virus sample and CTo is the CT value derived from the intracellular WT viral RNA collected at 0 h p.t. for intracellular RNA calculation or at 12 h p.t. for extracellular RNA calculation. The intracellular viral RNAs were normalized to the CT values derived from GAPDH mRNA. An asterisk indicates statistical significance based on Student's t test (P < 0.05). L.O.D, limit of detection. Each data point is an average; error bars indicate the standard deviations derived from three independent experiments (n = 3).
Revertant analysis.
Since R84A and R84E exhibited the most defective phenotype among the analyzed mutants (Fig. 8A to C), we chose these two mutants for revertant analysis. Both mutant viruses were continuously cultured on Vero cells and monitored for improved viral replication. After five rounds of passaging (4 days per round), the R84E virus produced larger plaques than the unpassaged virus (Fig. 8D) and generated viral titers equivalent to those of the WT virus (2.2 × 106 PFU/ml); sequencing of the resulting virus showed that the R84E mutation had been changed to R84K. Similarly, after five rounds of passaging, the R84A virus was able to generate higher viral titers (1.7 × 105 PFU/ml). However, the plaque morphology of the resulting virus remained small (Fig. 8D), and sequencing of the resulting virus revealed that the R84A mutation had been changed to R84T. For both revertant viruses, no other mutation was found throughout the genomic RNAs.
DISCUSSION
Flavivirus NS2A colocalizes with dsRNA, NS1, NS3, and NS5 within the replication complex (3). Besides viral RNA synthesis, NS2A also functions in flavivirus assembly/release (24). The goal of this study was to determine the topology of flavivirus NS2A on the ER membrane. Using a panel of biochemical methods, we established DENV-2 NS2A as an ER membrane protein with five membrane-spanning segments. NMR structure showed that the first transmembrane segment (pTMS3) of NS2A consists of two helices separated by a helix breaker mediated by amino acids R84 and P85. Functional analysis of the breaker residues showed that only R84, but not P85, is critical for viral RNA synthesis as well as virion assembly/release.
We initially used multiple bioinformatics algorithms to predict the topology of NS2A that could be experimentally tested. Various algorithms predicted different topology models with only one common TMS from amino acids 30 to 57 (Fig. 1A). To avoid discounting any potential TMS, we consolidated all eight predicted TMS into a test model (Fig. 1B). When each predicted TMS was individually fused to GFP, six of the eight predicted pTMS (pTMS2, -3, -4, -6, -7, and -8) could anchor GFP to the ER membrane, whereas the other two predicted pTMS (pTMS1 and -5) could not (Fig. 1C). The IFA-based cellular localization results (Fig. 1C) are in general agreement with the membrane flotation data (Fig. 1D), with the exception that the correlation of the results between the two methods is weak for pTMS8 (equivalent to fragment 186–209). The membrane-associated activity of pTMS8 was strengthened by the in situ fluorescence protease protection result (Fig. 2B), which clearly showed that pTMS8 spans from the ER lumen to the cytosol. In the in situ fluorescence protease protection assay, the membrane-associated activity of pTMS8 could be enhanced through its interaction with upstream NS2A sequence; no such interaction occurred in the cellular localization and membrane flotation assays when only individual NS2A fragments were examined in the absence of upstream NS2A sequence.
It should be noted that pTMS that showed ER membrane-interacting activity do not necessarily span the entire lipid bilayer of ER membrane; some of the pTMS (such as pTMS2) could peripherally interact with the membrane without traversing the complete lipid bilayer. Sequence analysis of DENV-2 NS2A using HeliQuest program suggests that amino acids 8 to 25 (overlapping pTMS1), 49 to 66 (overlapping three residues with pTMS2), and 123 to 140 (within pTMS5) could form amphipathic helices (pAH1-3) (Fig. 4B). The hydrophilic sides of these amphipathic helices may peripherally or transiently interact with ER membrane on the lumen side. However, the results presented in Fig. 1C and D demonstrate that none of these three fragments are associated with ER membrane when expressed individually, thus arguing against the possibility of their membrane-associated activity.
We used an in situ fluorescence protease protection assay to examine a panel of C-terminally truncated forms of NS2A fused with a GFP reporter (Fig. 2). This assay allowed us to differentiate between the cytosolic and noncytosolic residues in cells expressing the various NS2A-GFP fusion proteins. The results indicate that, among the six pTMS that showed membrane-interacting activities (Fig. 1C and D), pTMS2 is located in the ER lumen, whereas pTMS3, pTMS4, pTMS6, pTMS7, and pTMS8 span the ER membrane (Fig. 2B). Based on these results, we propose a topology model of DENV-2 NS2A on the ER membrane (Fig. 4A). In this model, the first 69 amino acids of NS2A are located in the ER lumen. This conclusion is supported by three lines of evidence. (i) Both the N and C termini of pTMS2 were only weakly accessible to protease digestion in situ (Fig. 2B). (ii) Fragments representing various portions of the first 70 amino acids of NS2A were protected from protease cleavage in vitro (Fig. 3C). (iii) When these fragments were C-terminally fused with a glycosylation signal, the resulting peptides were all glycosylated (Fig. 3B). As discussed above, when pTMS2 was individually fused with GFP, it targeted GFP to the ER membrane (Fig. 1C and D). To consolidate all these results, we surmise that pTMS2 may peripherally interact with the ER membrane on the lumen side (Fig. 4A).
Our model contradicts a previous report that YFV NS2A residues 189 to 191 (corresponding to DENV-2 NS2A residues 186 to 188) are cytosolic. These residues form an internal cleavage site for YFV protease NS2B/NS3, leading to the production of NS2Aα (8, 44). This discrepancy may imply a variation in the NS2A topology among flaviviruses.
We solved the NMR structure of pTMS3 (Fig. 6). pTMS3 consists of two helices separated by a helix breaker mediated by residue P85. Two charged residues, K82 and R84, are located upstream of the breaker within this transmembrane region. We explored the function of this unique breaker structure by mutagenesis analysis. In both replicon (Fig. 7) and genome-length (Fig. 8) RNA, substitution of P85A did not show any defects in viral infection cycle, indicating that the breaker structure itself is not important for DENV-2 replication. However, the R84E mutation reduced viral RNA synthesis and the production of infectious virus. The decrease in virus production could be caused by the reduction of viral RNA synthesis; alternatively, this mutation could directly reduce the efficiency of virion assembly/release. Interestingly, when the same residue was changed to amino acid A or S (R84A or R84S), the mutation showed no effect on viral RNA synthesis, as indicated by the replicon results (Fig. 7); remarkably, both substitutions generated fewer virus particles (Fig. 8 and 9). Further analysis showed that no infectious virion was produced inside the cells transfected with the mutant R84A RNA, whereas high levels of intracellular infectious virion were generated in the WT RNA-transfected cells (Fig. 9). Collectively, these results demonstrate that residue R84 functions in both RNA synthesis (as demonstrated by using the R84E mutation) and virion assembly/maturation (as demonstrated by using R84A).
In agreement with our results, the Rice group and the Khromykh group previously showed that NS2A plays a role in virion assembly/release in YFV (8) and KUNV (24), respectively. In the case of KUNV, the compensatory mutation T149P in NS2A (corresponding to amino acid 154 of DENV-2 NS2A) was able to rescue the defects in virus assembly caused by the I59N mutation (corresponding to residue 58 of DENV-2 NS2A) (24). If the current topology model of DENV-2 NS2A is applicable to KUNV, the genetic data imply that I59 with in the predicted pAH2 (Fig. 4B) may interact with pTMS6 to regulate virus assembly. In addition to the intramolecular interactions reported in KUNV NS2A, an intermolecular interaction between YFV NS2A residue K190S (corresponding to residue 188 in DENV-2 NS2A) and NS3 D343 (within helicase domain) was reported to be important for YFV assembly/release (8). It remains to be determined how these nonstructural protein interactions modulate flavivirus assembly. In the present study, we also performed revertant analysis using R84A and R84E mutants. Reversions of R84A to R84K and R84E to R84T were recovered; both revertants were found to enhance viral replication (Fig. 8D); unfortunately, no intramolecular or intermolecular interaction was obtained in our study.
In summary, the present study identified the DENV-2 NS2A as a protein with five membrane-spanning segments. The proposed topology model could serve as a starting point to investigate the mechanism of flavivirus NS2A in viral replication and its modulation of host immune response. Structural and functional analysis of the first ER membrane-spanning region (pTMS3) of DENV-2 NS2A showed its function in both viral RNA synthesis and virion assembly. Studies are ongoing to understand how flavivirus NS2A modulates the two events of viral RNA synthesis and virion assembly.
ACKNOWLEDGMENTS
We thank Jing Zou and other colleagues at Novartis Institute for Tropical Diseases for technical support and helpful discussions during the course of this study.
This work was partially supported by the TCR flagship “STOP Dengue” program of the National Medical Research Council in Singapore. C.K. is supported by an A*STAR Investigatorship. Z.Y.'s group is supported by Important National Science & Technology Specific Project (2009ZX10004-504) and National Natural Scientific Fund of China (81072675).
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Lindenbach BD, Thiel H-J, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott William & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Cleaves GR, Dubin DT. 1979. Methylation status of intracellular dengue type 2 40 S RNA. Virology 96:159–165 [DOI] [PubMed] [Google Scholar]
- 3. Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215 [DOI] [PubMed] [Google Scholar]
- 4. Lindenbach BD, Rice CM. 1997. Trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, et al. 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J. Infect. Dis. 193:1078–1088 [DOI] [PubMed] [Google Scholar]
- 6. Warrener P, Tamura JK, Collett MS. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wengler G, Czaya G, Farber PM, Hegemann JH. 1991. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 72:851–858 [DOI] [PubMed] [Google Scholar]
- 8. Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 8:e1002642 doi:10.1371/journal.ppat.1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. 2009. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15:2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926–39937 [DOI] [PubMed] [Google Scholar]
- 15. Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882 [DOI] [PubMed] [Google Scholar]
- 19. Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the Dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854–8863 [DOI] [PubMed] [Google Scholar]
- 20. Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chambers TJ, McCourt DW, Rice CM. 1989. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169:100–109 [DOI] [PubMed] [Google Scholar]
- 23. Falgout B, Markoff L. 1995. Evidence that flavivirus NS1-NS2A cleavage is mediated by a membrane-bound host protease in the endoplasmic reticulum. J. Virol. 69:7232–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 82:4731–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tu YC, Yu CY, Liang JJ, Lin E, Liao CL, Lin YL. 2012. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J. Virol. 86:10347–10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, Hall R, Khromykh AA. 2010. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 84:1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou G, Chen YL, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi PY. 2011. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 286:14362–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zou G, Xu HY, Qing M, Wang QY, Shi PY. 2011. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res. 91:11–19 [DOI] [PubMed] [Google Scholar]
- 32. Gualano RC, Pryor MJ, Cauchi MR, Wright PJ, Davidson AD. 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 79:437–446 [DOI] [PubMed] [Google Scholar]
- 33. Lorenz H, Hailey DW, Wunder C, Lippincott-Schwartz J. 2006. The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat. Protoc. 1:276–279 [DOI] [PubMed] [Google Scholar]
- 34. Hooper R, Churamani D, Brailoiu E, Taylor CW, Patel S. 2011. Membrane topology of NAADP-sensitive two-pore channels and their regulation by N-linked glycosylation. J. Biol. Chem. 286:9141–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, Shi PY. 2011. Inhibition of dengue virus by targeting viral NS4B protein. J. Virol. 85:11183–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. 2012. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology 432:511–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gayen S, Kang C. 2011. Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex. Biochem. Biophys. Res. Commun. 409:572–576 [DOI] [PubMed] [Google Scholar]
- 38. Wuthrich K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY [Google Scholar]
- 39. Guntert P. 2004. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278:353–378 [DOI] [PubMed] [Google Scholar]
- 40. Somnuke P, Hauhart RE, Atkinson JP, Diamond MS, Avirutnan P. 2011. N-linked glycosylation of dengue virus NS1 protein modulates secretion, cell-surface expression, hexamer stability, and interactions with human complement. Virology 413:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hori H, Lai CJ. 1990. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J. Virol. 64:4573–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kang C, Li Q. 2011. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 15:560–569 [DOI] [PubMed] [Google Scholar]
- 43. Mielke SP, Krishnan VV. 2004. An evaluation of chemical shift index-based secondary structure determination in proteins: influence of random coil chemical shifts. J. Biomol. NMR 30:143–153 [DOI] [PubMed] [Google Scholar]
- 44. Nestorowicz A, Chambers TJ, Rice CM. 1994. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication, and evidence for alternative processing of the NS2A protein. Virology 199:114–123 [DOI] [PubMed] [Google Scholar]