Abstract
Tetherin/BST-2 (here called tetherin) is an antiviral protein that restricts release of diverse enveloped viruses from infected cells through physically tethering virus envelope and host plasma membrane. For HIV-1, specific recruitment of tetherin to assembly sites has been observed as its colocalization with the viral structural protein Gag or its accumulation in virus particles. Because of its broad range of targets, we hypothesized that tetherin is recruited through conserved features shared among various enveloped viruses, such as lipid raft association, membrane curvature, or ESCRT dependence. We observed that reduction of cellular cholesterol does not block tetherin anti-HIV-1 function, excluding an essential role for lipid rafts. In contrast, mutations in the capsid domain of Gag, which inhibit induction of membrane curvature, prevented tetherin-Gag colocalization detectable by confocal microscopy. Disruption of Gag-ESCRT interactions also inhibited tetherin-Gag colocalization when disruption was accomplished via amino acid substitutions in late domain motifs, expression of a dominant-negative Tsg101 derivative, or small interfering RNA (siRNA)-mediated depletion of Tsg101 or Alix. However, further analyses of these conditions by quantitative superresolution localization microscopy revealed that Gag-tetherin coclustering is significantly reduced but persists at intermediate levels. Notably, this residual tetherin recruitment was still sufficient for the full restriction of HIV-1 release. Unlike the late domain mutants, the capsid mutants defective in inducing membrane curvature showed little or no coclustering with tetherin in superresolution analyses. These results support a model in which both Gag-induced membrane curvature and Gag-ESCRT interactions promote tetherin recruitment, but the recruitment level achieved by the former is sufficient for full restriction.
INTRODUCTION
Tetherin (BST-2/CD317/HM1.24/PDCA-1) is an interferon-inducible protein, which restricts the release of HIV-1 virions and causes their accumulation at the cell surface (1, 2). Evidence obtained by fluorescence and transmission electron microscopy studies shows that tetherin accumulates at sites of virus assembly (3–10). This is consistent with a model where tetherin inhibits virus release by directly linking cell and viral membranes (3, 5, 6, 9). This restrictive function is most likely a result of the unique topology of tetherin, which contains both an N-terminal transmembrane domain and a predicted C-terminal glycosylphosphatidylinositol (GPI) anchor. The ability of tetherin to form dimers or possibly higher-order multimers has also been shown to contribute to its antiviral function (11–15). As with other GPI-anchored proteins, tetherin is thought to reside in lipid rafts, based on its resistance to detergent solubilization and cholesterol dependence of this detergent resistance (16–18). Tetherin is able to inhibit a broad range of unrelated enveloped viruses, ruling out a direct interaction with any particular viral protein as a requirement for restriction of virus release (19, 20).
HIV-1 is able to overcome tetherin-mediated restriction through the action of its accessory protein Vpu (1, 2). Recent studies have determined the nature of the interaction between Vpu and tetherin and revealed that Vpu promotes degradation of tetherin and sequestration of tetherin away from assembly sites (for reviews, see references 19 to 21). In contrast, the mechanism by which tetherin interacts with assembling viruses remains unknown.
The assembly process of HIV-1 likely proceeds in a stepwise fashion (22, 23). Early steps include binding of Gag to the plasma membrane, mediated by the matrix (MA) domain (24). Dimerization of Gag is mediated by the capsid (CA) domain, and higher-order multimerization is mediated by interactions between the nucleocapsid (NC) domain and RNA (23). Finally, as multimerization progresses, Gag is able to induce membrane curvature and, via the late domain (in p6) and NC regions, recruit cellular ESCRT machinery to ultimately facilitate scission of host and viral membranes (25). During these steps, assembling Gag also associates with cholesterol-rich lipid rafts, as well as other membrane microdomains, such as tetraspanin-enriched microdomains (26, 27). Similar to dependence on cellular ESCRT machinery for efficient virus release (28, 29), association with lipid rafts is a common aspect shared among the assembly of many enveloped viruses (30). Because of the association between Gag and lipid rafts, many have hypothesized that lipid rafts play an important role in the recruitment of tetherin to virus assembly sites (7, 31).
To better understand the mechanisms by which tetherin restricts a broad spectrum of enveloped viruses, we sought to determine how tetherin is recruited to sites of HIV-1 assembly in the absence of Vpu. In this study, we examined the contributions of lipid rafts, membrane curvature, and interactions with cellular ESCRT machinery to the process of tetherin recruitment to viral assembly sites.
MATERIALS AND METHODS
Plasmids and siRNA.
pNL4-3 (32), pNL4-3/Udel (a kind gift from Klaus Strebel) (33), pNL4-3/Gag-Venus, pNL4-3/Fyn(10)ΔMA/Gag, and pNL4-3/Fyn(10)fullMA/Gag (34), pNL4-3/PHPLCδ1ΔMA/Gag (26), pCMV/Tsg-5′ (a kind gift from Stanley Cohen) (35), and expression vectors encoding NFP-GPI and NFP-HATM (26) were described previously. Udel versions of previously described pNL4-3-based plasmids were created by replacing the SalI/BamHI fragment of pNL4-3 with the corresponding region of pNL4-3/Udel, which does not express Vpu (33). A plasmid encoding the full-length cDNA of human BST-2/tetherin under the control of a cytomegalovirus (CMV) promoter, pCMV6XL-5 hBST-2, was obtained from Origene (Rockville, MD). Plasmids used to make pseudotyped virus stocks, pCMVNLGagPolRRE (38) and pHCMV-G (a kind gift from Jane Burns) (39), were described previously. Capsid mutations previously shown to disrupt membrane curvature and efficient particle formation, P99A (40) and EE75,76AA (41), were introduced into pNL4-3/Gag-Venus/Udel by PCR mutagenesis to give pNL4-3/P99A/Gag-Venus/Udel, pNL4-3/EE75,76AA/Gag-Venus/Udel. pNL4-3/PTAP−/Gag-Venus/Udel (p6 residues 7 to 10 were changed from PTAP to LIRL [42]), pNL4-3/CCY−/Gag-Venus/Udel (bearing the NC C28S, NC C49S, and p6 Y36S mutations [43]), and pNL4-3/Y36S/Gag-Venus/Udel (p6 Y36S mutation [44]) were generated by standard PCR mutagenesis techniques. The gene encoding the photoswitchable fluorescent protein mEos2 was obtained from the pRSETa mEos2 plasmid from Addgene (Cambridge, MA). The bright, monomeric version of the Eos fluorescent protein, mEos3.2, was generated by PCR mutagenesis of the parental mEos2 protein (bearing the I102N, H158E, and Y189A mutations) as described previously (45). pNL4-3/Gag-mEos3.2/Udel and its derivatives were constructed by replacing the Venus coding sequence with mEos3.2. The CA/NC mutant used in this study, pNL4-3/20LK/WM/14A1G/Gag-mEos3.2/Udel, bears the MA mutation 20LK (46) to facilitate efficient membrane binding in the absence of normal multimerization, which is disrupted by the capsid C-terminal mutations W184A and M185A (WM) and mutations of the 15 basic amino acids of the NC domain to alanine or glycine (14A1G) (47). Small interfering RNA (siRNA) sequences directed against Tsg101 (CCUCCAGUCUUCUCUCGUC) and Alix (GAAGGAUGCUUUCGAUAAA) were described previously (48, 49). A control siRNA, which does not target any known human mRNA sequences (CUCCGAACGUGUCAC), was also used (50). Double-stranded RNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Antibodies.
Anti-BST-2/tetherin polyclonal rabbit serum, generated by Klaus Strebel (51), and anti-HIV-Ig polyclonal human serum were obtained from the NIH AIDS Research and Reference Reagent Program (Germantown, MD). Monoclonal mouse antibodies against CD46, CD55, CD59, and Tsg101 were obtained from BD Biosciences (San Diego, CA). Polyclonal rabbit anti-green fluorescent protein (GFP) was obtained from Clontech (Mountain View, CA). Mouse polyclonal anti-PDC6I (Alix) was obtained from Abcam (Cambridge, MA). Monoclonal mouse anti-α-tubulin was obtained from Sigma (St. Louis, MO). Species- and/or isotype-specific Alexa Fluor 488-, 594-, and 647-conjugated secondary antibodies were obtained from Invitrogen (Carlsbad, CA). Alexa Fluor 647-conjugated monoclonal anti-human CD317 (tetherin) was obtained from BioLegend (San Diego, CA).
Cells.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS). HT-1080 cells (ATCC CCL-121) were maintained in Eagle's minimal essential medium (EMEM) supplemented with 10% FBS. For virus release assays, cells were plated at a density of 5.6 × 105 per well in 6-well culture plates and incubated overnight. Cells were then transfected with 2 μg of pNL4-3-based plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. In HT-1080 cells, 2 μg pNL4-3-based plasmids were used with or without 50 ng pCMV6XL-5 hBST2. Transfection for microscopy was performed as previously described (26). For T-cell experiments, A3.01 and a derivative cell line, P2 (52), were maintained in RPMI 1640 with 10% FBS.
Cholesterol depletion and virus release assays.
Cholesterol depletion (38) and virus release assays (34) were performed as previously described. Virus-like particle (VLP) release assays of PR− Gag, Gag-Venus, and Gag-mEos3.2 were performed in a similar manner with the exception that cell lysates were subjected to boiling with a low concentration of SDS prior to immunoprecipitation and VLP lysates were loaded directly, without immunoprecipitation, to minimize epitope masking resulting from the lack of functional protease in these molecular clones (53).
Antibody copatching assay, confocal microscopy, and image analysis.
HeLa cells were plated and transfected as described above, followed by 16-h of incubation at 37°C. For antibody copatching assay, cells were stained by primary antibodies at 1:100 in DMEM–5% FBS for 10 min at room temperature, washed 3 times with phosphate-buffered saline (PBS), and stained by secondary antibodies at 1:200 in DMEM–5% FBS for 10 min at room temperature. Cells were then washed 3 times and fixed with 4% paraformaldehyde (PFA) in PBS for 30 min at room temperature. For standard immunofluorescence microscopy, cells were fixed with 4% PFA in PBS for 30 min at room temperature and stained by primary and secondary antibodies for 1 h each. Samples were mounted in Fluoromount-G medium (Southern Biotech, Birmingham, AL) and imaged on a Leica SP5X microscope (Leica, Wetzlar, Germany). Images were acquired with a 100× PL APO objective (numerical aperture [NA] = 1.40) with 5× scanning zoom at a resolution of 1,024 × 1,024 (30 nm per pixel). Excitation was done with 488-nm, 514-nm, and 590-nm laser lines. Acquisition bandwidths were 500 to 550 nm, 525 to 575 nm, and 600 to 650 nm, respectively.
To calculate the degree of colocalization between markers, at least 10 regions of interest, each from different fields, were analyzed per condition. Regions of interest were randomly selected from cells where two proteins of interest were present. For Gag-tetherin colocalization measurements, we excluded cells that do not show distinct Gag puncta from analyses. Pearson's correlation coefficient was calculated using the ImageJ 1.43u software program (NIH; http://rsb.info.nih.gov/ij/) with the JACoP plugin (54). For confocal images of tetherin, a smoothing filter was applied in ImageJ after images were analyzed to remove excess background signal. This filter sets the intensity value at each pixel equal to the average of its nearest neighbors in a 3 × 3-pixel square area.
T-cell infection and microscopy.
Generation of pseudotyped virus stocks, infection of A3.01 and P2 T cells, and immunofluorescence microscopy were performed as described previously (52). Images were analyzed as described above for HeLa cells, with the exception that a median filter was applied in ImageJ prior to analysis to remove out-of-focus signals from epifluorescence images. For the virus release assay, 4 × 105 P2 T cells were spinoculated with 200 µl of vesicular stomatitis virus (VSV)-G-pseudotyped virus stock in the presence of 0.8 μg Polybrene and incubated for 2 days prior to metabolic labeling.
Transmission electron microscopy analysis of Gag-Venus mutants.
HeLa cells were plated and transfected as described above. Cells were fixed 16 h posttransfection with 2% glutaraldehyde in PBS. Cells were analyzed on a Hitachi H7600 transmission electron microscope as previously described (55).
Superresolution localization microscopy and cross-correlation analysis.
HeLa cells were plated and transfected as described above. Cells were fixed at 16 h posttransfection in 4% PFA with 0.1% glutaraldehyde for 10 min at room temperature. Cells were then stained by Alexa Fluor 647-conjugated monoclonal antitetherin antibody for 2 h at room temperature. Cells were washed extensively with PBS and placed in imaging buffer composed of 50 mM Tris-HCl (pH 8.0), 10 mM NaCl, 1% β-mercaptoethanol (β-ME), 10% glucose, and an enzymatic oxygen scavenging system as described previously (56). Samples were imaged in total internal reflection using an inverted IX81-ZDC microscope with a cellTIRF module (Olympus America, Center Valley, PA) and a 100× UAPO total internal reflection fluorescence (TIRF) objective (NA = 1.49). Images were acquired on an electron-multiplying charge-coupled-device (EMCCD) camera (iXon-897; Andor, Belfast, Ireland). Red and far-red emissions were separated onto two halves of the EMCCD camera using a DV2 dual-view imaging system equipped with 650-nm long-pass dichroic and emission filters (Photometrics, Tucson, AZ). Alexa-647 fluorophores were activated and imaged with 640-nm laser excitation (Cube 640-75FP; Coherent, Santa Clara, CA), while Gag-mEos3.2 fluorophores were activated at 405 nm (Cube 405-50FP; Coherent, Santa Clara, CA) and imaged using laser excitation at 561 nm (Sapphire 561-150 CW; Coherent, Santa Clara, CA). In both cases, laser intensities were adjusted such that single fluorophores could be distinguished in individual images. Superresolution images were reconstructed from 7,500 individual diffraction-limited images according to previously described methods (56) implemented in custom software written in the Matlab programming language (Mathworks, Natick, MA). Briefly, single-molecule peaks were identified and fit to a two-dimensional Gaussian shape. The ensemble of peaks was then culled to remove outliers in brightness, size, aspect ratio, and localization error. The culling algorithm is designed to remove likely contributions from signals that do not originate from single activated fluorophores so that they do not contribute to the final image. Culled events are not correlated in space. Since both fluorophores are known to reversibly photoswitch under our imaging conditions (57), and therefore a single fluorophore could emit signals more than once during acquisition of an image series, we interpret the smallest/dimmest puncta in final images, those containing one to several activation events, as likely representing single molecules. In our quantification of coclustering (see below), we circumvent possible artifacts from over- (or under)counting single proteins by quantifying coclustering averaged over large areas of the cell surface with cross-correlation functions. Fiduciary markers were used to transform Alexa 647 and mEos3.2 images, following previously published methods (58), and stage drift was corrected by aligning single-color superresolution images generated from signals acquired every 100 to 500 frames by localizing the maxima of cross-correlation functions. Superresolution images were reconstructed by incrementally increasing the intensity of pixels at positions corresponding to localized single molecules after correcting for stage drift. Image resolution is estimated by comparing the autocorrelation of images reconstructed from all identified single-molecule centers to those of images reconstructed from data grouped to account for localized single molecules that remain activated in sequential frames as described previously (59). Cross-correlation functions, c(r), as a function of radius, r, for cells were evaluated from ungrouped reconstructed images, computed using fast Fourier transforms as described previously (60), and normalized to 1 at large radius.
To facilitate the comparison of the levels of cross-correlations between conditions, we reported the integrated intensity under cross-correlation curves, out to a radius of 1 μm. To accomplish this, curves tabulated from individual cells were first fit to the sum of Gaussian and exponential functions in order to average noise at large radii. Best-fit curves were then integrated according to the equation , where Δr is the distance between adjacent points in the tabulated correlation functions (25 nm). The number of Gag (tetherin) proteins correlated with the average tetherin (Gag) protein out to a radius of 1 μm is given by ρI, where ρ is the average surface density of Gag (tetherin). The values presented in Fig. 5F and 7C (integrated cross-correlation) are integrated intensity (I) divided by integrated area (). The resulting value is equal to the increased surface density of Gag (tetherin) coclustered with tetherin (Gag).
Fig 5.
siRNA-mediated depletion of Tsg101 or Alix decreases tetherin recruitment in HeLa cells. (A) Western blotting of Tsg101 and Alix in cells transfected with indicated siRNA and lysed at 64 h posttransfection. Alpha-tubulin is shown as a loading control. (B and C) HeLa cells were transfected sequentially with the indicated siRNA and a molecular clone encoding Gag-YFP without Vpu. At 64 h post-transfection with siRNA (16 h post-transfection with Gag-YFP expression plasmids), cells were fixed by 4% PFA for 30 min and immunostained for endogenous tetherin. (B) Representative images of the dorsal surface of cells are shown. Scale bar = 5.0 μm. (C) Degree of colocalization was calculated in the same manner as for Fig. 1. The result is shown for one of two independent experiments and displayed as mean R values ± SEM. *, P < 0.01. (D) HeLa cells were transfected with indicated siRNA, followed by transfection with a Vpu-deficient molecular clone expressing Gag-mEos3.2. Cells were then fixed, stained with Alexa Fluor 647 antitetherin antibody, and imaged by TIRF. Superresolution images were reconstructed as described in Materials and Methods. Representative images are shown. Magnified images for boxed areas in the left panels are shown on the right. Scale bars are 2 μm for the left panels and 500 nm for the right panels. (E) Images of five cells per condition were acquired in each of two independent experiments (total, 10 cells) and used for measurement of Gag-tetherin cross-correlation. Cross-correlation values are shown as means ± SEM. The dashed line indicates random distribution (cross correlation = 1). (F) Integrated cross-correlation was calculated as described in Materials and Methods and is shown as mean integrated cross-correlation values ± SEM. *, P < 0.05.
Fig 7.
Both membrane curvature and Gag-ESCRT interactions enhance tetherin recruitment in HeLa cells. (A) Cells expressing molecular clones encoding either WT Gag-mEos3.2 or CA/NC mutant Gag-mEos3.2 and lacking the vpu gene were fixed and imaged by conventional TIRF microscopy. (B) Cells expressing the indicated Gag-mEos3.2 constructs in the absence of Vpu were stained for tetherin and imaged by TIRF as for Fig. 5, and superresolution microscopy images were reconstructed as described in Materials and Methods. Representative images are shown for WT, PTAP−, and CA/NC Gag-mEos3.2. Magnified images for boxed areas in the left panels are shown on the right. Scale bars are 2 μm for the left panels and 500 nm for the right panels. (C) Images of five cells per condition were acquired in each of two independent experiments (total, 10 cells) and used for measurement of Gag-tetherin cross-correlation. Integrated cross-correlation was calculated as described in Materials and Methods and is shown as means ± SEM. *, significantly less than that for the WT (P < 0.05); †, significantly greater than that for CA/NC (P < 0.05).
The Pearson's correlation coefficient used to quantify colocalization in confocal microscopy images and the integrated cross-correlation analysis method used for superresolution localization microscopy images are not directly comparable. The Pearson's coefficient is the covariance (cross-correlation at r = 0) between the two images divided by the variance (square root of the autocorrelation function at r = 0) of each individual image (60), whereas the integrated cross-correlation values are for r values of 0 to 1 μm and are not affected by the autocorrelation functions. Unfortunately, it is not possible to evaluate reliable Pearson's coefficients from superresolution images due to overcounting artifacts in autocorrelation functions (details are described in reference 59).
siRNA knockdowns.
HeLa cells were plated at a density of 105 cells per well in 24-well culture plates (Corning, Fairport, NY) and incubated overnight. Cells were then transfected with 20 pM siRNA using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After 24 h, cells were trypsinized, transferred into Lab-Tek 8-well chamber slides (Thermo Fisher, Waltham, MA; for confocal microscopy), Lab-Tek 4-well chamber coverslips (for superresolution microscopy), or 6-well culture plates (Corning, Fairport, NY; for Western blot analysis and virus release assays) and further incubated for 24 h. For microscopy, cells were then transfected with 0.6 μg pNL4-3-based plasmids and incubated for an additional 16 h prior to fixation and staining as described above for confocal or superresolution localization microscopy (a total of 64 h after siRNA transfection). For Western blot analysis, cells were lysed at 64 h after siRNA transfection, and lysates were subjected to SDS-PAGE and immunoblotting. For virus release assays, cells were transfected with siRNA as described above, transfected with pNL4-3-based plasmids at 48 h after siRNA transfection, and incubated for an additional 16 h (a total of 64 h after siRNA transfection) prior to metabolic labeling.
RESULTS
Tetherin does not copatch significantly with lipid raft markers in HeLa cells.
Given the known involvement of lipid rafts in the assembly of many tetherin-susceptible enveloped viruses, it is possible that lipid rafts may direct tetherin to sites of HIV-1 assembly. To determine whether tetherin is associated with lipid rafts, we used an antibody copatching assay to examine the distribution of endogenous tetherin and lipid raft markers on the surface of HeLa cells. By the same assay, Gag has been previously shown to strongly associate with lipid raft markers in HeLa cells (26). In the present study, we observed substantial copatching of tetherin with Gag-Venus yellow fluorescent protein (YFP) but not with a nonraft marker, CD46 (Fig. 1). We also observed a moderate level of copatching between two lipid raft markers, NFP-GPI (a GPI-anchored nonfluorescent GFP variant) and CD59, and between NFP-HATM (a fusion between the transmembrane domain of influenza virus hemagglutinin [HA] and NFP) and CD55 in the absence of Gag (Fig. 1). However, no significant copatching was observed between tetherin and four different lipid raft markers, CD55, CD59, NFP-GPI, and NFP-HATM, in the absence of Gag (Fig. 1). These results suggest that although tetherin can be recovered from detergent-resistant membrane fractions, it does not appear to have a strong affinity for lipid rafts detected in this experiment at the plasma membrane of HeLa cells.
Fig 1.
Tetherin copatches with Gag-YFP but not with CD46 or lipid raft markers. HeLa cells were treated with primary antibodies against indicated proteins expressed on the cell surface for 10 min, followed by treatment with species- and/or isotype-specific fluorescent secondary antibodies for 10 min and fixation with 4% PFA for 30 min. Cells expressing Gag-YFP were treated with antibodies to patch tetherin alone prior to fixation. (A) Representative images of the dorsal surface of cells are shown. (B) Pearson's correlation coefficients were calculated for fluorescence intensities of green and red signals at each pixel. Data shown are from 10 regions of interest, each from different cells, from one representative experiment out of two performed, displayed as mean Pearson's correlation (R) values ± SEM. P values were determined using Student's t test. *, P < 0.05. Scale bar = 5.0 μm.
Tetherin antiviral function is insensitive to cholesterol depletion.
While a recent report showed that treatment of cells by antitetherin antibody does not apparently alter cell surface distribution of tetherin (61), we cannot rule out the possibility that antibody treatment may alter the native microdomain partitioning of tetherin. To assess tetherin-raft association and its significance by an alternative approach, we sought to determine whether lipid rafts play a role in tetherin antiviral function. To this end, we depleted cellular cholesterol that is essential for lipid raft integrity by methyl-β-cyclodextrin (MβCD) and examined the effect of this treatment on tetherin-mediated inhibition of virus release. We found that under both mild (30 min with 5 mM MβCD) and more stringent (30 min with 10 mM MβCD) cholesterol-depleting conditions, tetherin remains a potent inhibitor of wild-type (WT) Gag release in HeLa cells as assessed by Vpu dependence of virus release (Fig. 2A to C). While association of Vpu with lipid rafts is debated (17, 62, 63), such association could be a confounding factor in the experiments using HeLa cells. To directly examine the effect of tetherin, we used HT-1080 cells that do not express endogenous tetherin. We observed that in these cells, exogenously expressed tetherin inhibits virus release regardless of cholesterol depletion treatments (Fig. 2D to F). However, cholesterol depletion is also known to significantly inhibit membrane binding and release of WT Gag (38, 64). To separate the effects of cholesterol depletion on virus particle production and tetherin function, we conducted similar experiments using Fyn(10)fullMA Gag, which contains the first 10 amino acids of Fyn kinase at the N terminus of Gag. Virus particle production of this Gag derivative is less sensitive to cholesterol depletion than that of WT Gag (64). We found that tetherin also potently inhibits release of Fyn(10)fullMA Gag under cholesterol-depleting conditions in HeLa and HT-1080 cells (Fig. 2). These results indicate that tetherin antiviral function is insensitive to cholesterol depletion. Altogether, cholesterol depletion and microscopy data suggest that lipid rafts do not play an essential role in tetherin association with assembling Gag and tetherin-mediated inhibition of HIV-1 release.
Fig 2.
Tetherin antiviral function is insensitive to cholesterol depletion. Virus release assays were performed in HeLa cells transfected with HIV-1 molecular clones that express Vpu or not (A, B, and C) or in HT-1080 cells transfected with Vpu-deficient molecular clones with or without exogenously expressed tetherin (D, E, and F). These molecular clones encode either WT Gag or Fyn(10)fullMA Gag. Cells were cultured in Met/Cys-deficient medium containing FBS or cholesterol-depleted serum (CDS) and 5 mM or 10 mM MβCD prior to metabolic labeling with [35S]Met/Cys for 2 h. Virus release efficiency was determined as virus-associated Gag/total Gag and normalized to virus release efficiency in cells cultured in the medium containing FBS under Vpu (+) or tetherin (−) conditions. Data shown are from three independent experiments, displayed as means ± 1 standard deviation. *, P < 0.05; **, P < 0.01.
CA mutations that impair induction of membrane curvature block tetherin recruitment.
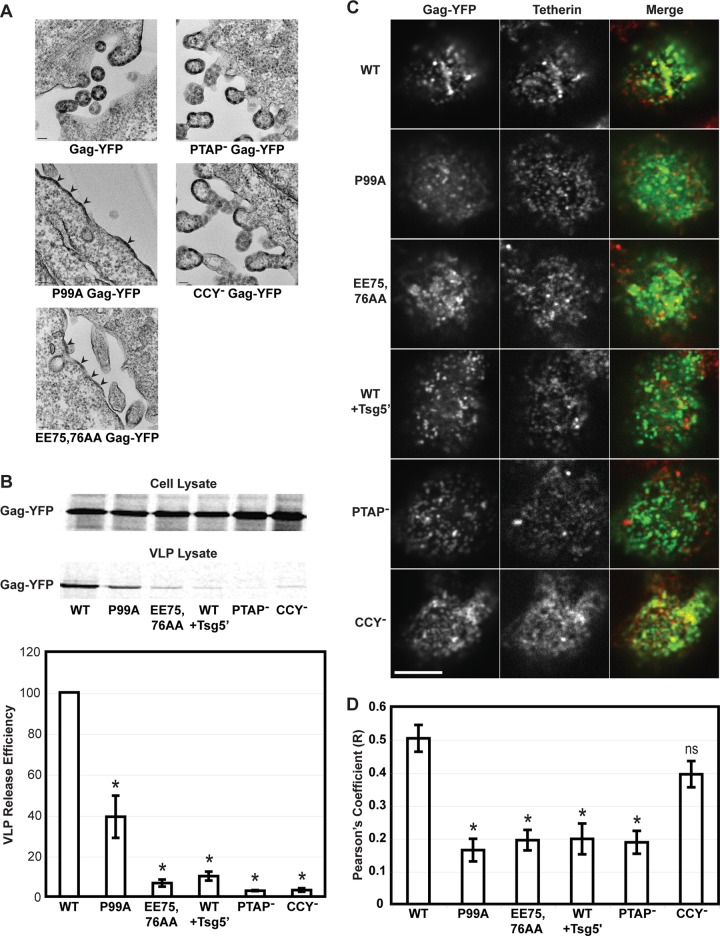
Next, we focused on whether membrane curvature and recruitment of cellular ESCRT machinery, which are common events among assembly of many unrelated enveloped viruses, are required for tetherin recruitment to assembly sites. To test the role of membrane curvature in tetherin recruitment, we utilized two different Gag mutants that are able to bind the plasma membrane and multimerize but are unable to induce the typical membrane curvature seen with WT Gag (26). These two mutations, CA P99A (40) and CA EE75,76AA (41), were introduced into an HIV-1 molecular clone encoding a Gag-YFP fusion. To validate whether these Gag mutations have the expected impacts on membrane curvature in the context of Gag-YFP fusions, we first performed transmission electron microscopy analyses and VLP release assays using HeLa cells transfected with Vpu+ molecular clones expressing WT, P99A, and EE75,76AA Gag-YFP (Fig. 3A and B). We found that consistent with the membrane curvature defect observed for non-YFP-tagged versions (unpublished data) (26, 41), both P99A and EE75,76AA produced electron-dense patches beneath the plasma membrane and were reduced in VLP production, although the defect was much more severe with the EE75,76AA mutant. Residual release of Gag by the P99A mutant is likely due to assembly, budding, and release of the spherical VLPs that are still observed to occur in an apparently WT-like manner, albeit at the low levels (unpublished data) (26). We next introduced the same CA mutations into a molecular clone lacking Vpu expression (pNL4-3/Gag-YFP/Udel) and examined the distributions of tetherin and Gag in the absence of Vpu-mediated downmodulation of tetherin by immunostaining. Cells were fixed prior to immunostaining to prevent antibody-driven copatching. Both CA mutants showed significantly lower colocalization with tetherin than WT Gag-YFP (Fig. 3C and D). These results identify the CA amino acid residues replaced in these mutants as molecular determinants in tetherin recruitment to HIV-1 assembly sites and suggest a possible role for membrane curvature in tetherin recruitment. Alternatively, these mutants may be defective at a stage of assembly prior to when tetherin recruitment occurs.
Fig 3.
Membrane curvature and Tsg101 binding correlate with tetherin recruitment in HeLa cells. (A) HeLa cells expressing Vpu+ molecular clones encoding indicated mutants of Gag-YFP were analyzed by transmission electron microscopy. Arrowheads indicate electron-dense patches that are likely to represent membrane-associated Gag-YFP multimers. Scale bar = 100 nm. (B) A VLP release assay of Vpu+ Gag-YFP constructs was performed for indicated Gag-YFP constructs in HeLa cells as for Fig. 2. A representative autoradiogram is shown, along with quantitation of VLP release efficiency. Data shown are from 2 independent experiments, displayed as mean VLP release efficiencies. (C and D) HeLa cells expressing molecular clones encoding indicated Gag-YFP constructs and lacking the vpu gene were fixed at 16 h posttransfection with 4% PFA for 30 min and immunostained for endogenous tetherin expressed on the cell surface. (C) Representative images of the dorsal surface of cells are shown. Scale bar = 5.0 μm. (D) Degree of colocalization between Gag-YFP and tetherin was calculated in the same manner as for Fig. 1. Results are shown for one of two independent experiments and displayed as mean R values ± SEM. *, P < 0.01; ns, not significant.
The interaction between Gag and Tsg101 enhances tetherin recruitment in HeLa cells.
Following the induction of membrane curvature and the formation of viral buds, ESCRT-mediated scission of viral and host membranes occurs. An ESCRT-I protein, Tsg101, as well as an ESCRT-related protein, Alix (AIP1/PDCD6IP), serve as an interface of the ESCRT machinery with Gag and facilitate efficient virus release (65–69). To determine whether the ESCRT complex contributes to tetherin recruitment, we examined the impact of a dominant-negative form of Tsg101, called Tsg-5′, on tetherin-Gag colocalization. Tsg-5′ corresponds to the N-terminal half of Tsg101, which binds HIV-1 p6 (65, 70), and lacks the C-terminal half of the protein, responsible for recruiting other ESCRT proteins (69, 71). Expression of this protein has been shown to inhibit virus release of HIV-1 (67). We observed a significant reduction in tetherin-Gag colocalization when Tsg-5′ was expressed (Fig. 3C and D), suggesting that the interaction between ESCRT and Gag is responsible for specific recruitment of tetherin.
Tsg101 binds to Pro-(Thr/Ser)-Ala-Pro (PT/SAP) motifs, including the PTAP motif in the Gag p6 domain (65, 70, 72). To abrogate this binding, we exchanged the PTAP motif sequence of the p6 domain for Leu-Ile-Arg-Leu, referred to as PTAP− Gag-YFP. This change is known to inhibit virus release (42, 73). Binding of Alix occurs through two different regions of Gag. The first known motif, LYPxnL, is within p6, and its binding to Alix can be disrupted by the p6 Y36S mutation (43, 44). The second binding site lies within the NC domain (43, 74), and its interaction with Alix can be disrupted by the double mutant NC C28S/C49S (43). To remove both Alix binding sites from Gag, we constructed a triple mutant (NC C28S/C49S and p6 Y36S), referred to as CCY− Gag-YFP. In the vpu-positive context, both PTAP− and CCY− mutants exhibited a budding-arrested phenotype characteristic of late domain/ESCRT disruption and failed to release VLPs (Fig. 3A and B). In the case of the CCY− mutant, the failure of VLP release may also be partially due to an additional defect in Gag assembly caused by mutations in the zinc fingers of the NC domain (75, 76). When expressed in HeLa cells, in the vpu-negative context, PTAP− Gag-YFP showed significantly less colocalization with tetherin, while CCY− Gag-YFP showed levels of colocalization which were not significantly less than those with WT Gag-YFP (Fig. 3C and D). We also observed similar levels of colocalization with the single mutant p6 Y36S (data not shown). These results suggest that in HeLa cells, the interaction between Tsg101 and the PTAP motif of the Gag p6 domain promotes recruitment of tetherin.
The interaction between Gag and ESCRT enhances tetherin recruitment in T cells.
To test whether members of the ESCRT complex participate in tetherin recruitment in T cells, which are a natural host cell type for HIV-1 in vivo, we infected A3.01 T cells with stocks of VSV-G-pseudotyped HIV-1 encoding WT Gag-YFP, PTAP− Gag-YFP and Y36S Gag-YFP and lacking vpu. After 48 h, cells were fixed and immunostained for surface tetherin. In contrast to the observation that only the Tsg101 binding site of Gag is required for tetherin recruitment in HeLa cells, both PTAP− and Y36S mutations decreased colocalization of Gag-YFP with tetherin in A3.01 T cells (Fig. 4). However, we noted that the impact of the Y36S change was modest (Fig. 4). Similar results were also observed in primary human CD4+ T cells (data not shown). These data show that both Tsg101 and Alix binding sites within the Gag p6 domain enhance tetherin recruitment to Gag assembly sites in T cells. In A3.01 cells, the CCY− triple mutant failed to form distinct puncta at the plasma membrane, unlike in HeLa cells, and therefore was not assessed for tetherin recruitment (data not shown).
Fig 4.
Both Tsg101- and Alix-binding sites in Gag are required for maximal tetherin recruitment in T cells. A3.01 T cells were infected with VSV-G-pseudotyped viruses expressing indicated Gag-YFP derivatives and lacking the vpu gene. After 48 h, cells were fixed with 4% PFA for 30 min and immunostained for endogenous tetherin expressed on the cell surface. (A) Representative images of the dorsal surface of cells are shown. Scale bar = 5.0 μm. (B) Quantitation of colocalization was performed as for Fig. 1 after application of the median filter. Results are shown for one of two independent experiments and are displayed as mean R values ± SEM. *, P < 0.01.
siRNA knockdown of Tsg101 or Alix decreases tetherin recruitment in HeLa cells.
To further assess the importance of Tsg101 and Alix in tetherin recruitment, we analyzed the effect of siRNA-mediated depletion of endogenous Tsg101 and Alix proteins in HeLa cells (Fig. 5). Using previously reported siRNA duplex sequences (48, 49), substantial depletion was achieved for both proteins (Fig. 5A). We found that depletion of either Tsg101 or Alix caused a marked decrease in tetherin colocalization with Gag-YFP. In contrast, significant colocalization was observed in cells transfected with nontarget control siRNA (Fig. 5B and C). These data indicate that both Tsg101 and Alix enhance tetherin recruitment to assembling Gag-YFP in HeLa cells. In addition to the observed loss of colocalization between tetherin and Gag after Tsg101 or Alix depletion, it is of note that both conditions also appear to show a more diffuse staining pattern for tetherin (Fig. 5B). Such a diffuse signal was also observed for control-siRNA-treated cells that do not express Gag (data not shown). These observations are consistent with the possibility that recruitment of tetherin to assembly sites results in an enhanced punctate staining pattern of tetherin, as seen with WT Gag-YFP.
To analyze the contribution of ESCRT-Gag interactions to tetherin recruitment at the level of single virus particles, we utilized superresolution localization microscopy. To achieve high-resolution information by this technique, proteins of interest are either tagged with photoswitchable or photoactivatable fluorescent proteins or immunolabeled by certain organic fluorophores, which are capable of reversible photoswitching in a reducing buffer (56, 77). For our experiments, we fused Gag to the recently developed, green-to-red photoswitchable fluorescent protein mEos3.2 (45). Unlike its parent protein, mEos2 (78), which is able to form dimers and tetramers, mEos3.2 is truly monomeric and presumably does not affect the multimerization state of fusion proteins. mEos3.2 has also been shown to exhibit superior spectral qualities and is capable of reversible photoswitching in reducing buffer, making it an ideal fusion protein for superresolution localization microscopy (45). For our experiments, we depleted Tsg101 and Alix in HeLa cells by siRNA as in previous experiments. Cells were then transfected with a molecular clone expressing a Gag-mEos3.2 fusion protein and lacking vpu. Cells were then fixed, labeled with an Alexa Fluor 647-conjugated monoclonal antibody directed to tetherin, and placed in a reducing buffer (57) before imaging by TIRF microscopy (56). Given the high resolution that was achieved by this technique (25 nm for Gag-mEos3.2 and 15 nm for tetherin), we chose to quantify images using cross-correlation analysis, which reflects the degree of coclustering of two proteins as a function of distance, rather than Pearson's correlation, which reflects whether or not two proteins are located within the same image pixel. To determine the degree of coclustering between tetherin and Gag-mEos3.2, we employed a statistical cross-correlation analysis, which measures the increased probability of finding a detected tetherin protein at a given distance from a detected Gag protein or vice versa (59). A normalized cross-correlation of 1 is observed when the distribution of detected molecules in one color channel is independent of the distribution of detected molecules in the other color channel. In contrast, a correlation greater than 1 is observed when signals from the two channels are not independent and show coclustering. When we quantified images of WT Gag-mEos3.2 and tetherin for cross-correlation up to radii of 1 μm, we observed a high degree of cross-correlation, particularly within 100 nm, when cells were treated with a nontarget siRNA (Fig. 5D and E). In contrast, when Tsg101 or Alix was depleted by siRNA, we observed intermediate levels of tetherin-Gag coclustering at the same range of radii (Fig. 5D and E). To facilitate the comparison between conditions, we used the integrated cross-correlation between r = 0 and r = 1,000 nm (see Materials and Methods) (Fig. 5F). By this method, both Tsg101- and Alix-depleted cells showed significantly less coclustering between tetherin and Gag-mEos3.2 than cells treated with control siRNA. The reduction in Gag-tetherin colocalization caused by the same siRNA treatment (Fig. 5C) appears more severe than that in coclustering (Fig. 5F). This is likely due to differences between the methods used to quantify colocalization and coclustering in the two experiments. In particular, diffuse tetherin signals observed in Tsg101- or Alix-depleted cells, which reduce the contrast in tetherin localization inside versus outside Gag patches, are likely to contribute to reduction of Pearson's correlation coefficients, whereas they would not necessarily affect cross-correlations. Regardless, both confocal and superresolution microscopy data indicate that both Tsg101 and Alix promote tetherin recruitment to HIV-1 assembly sites and yet intermediate levels of recruitment still occur in Tsg101- or Alix-depleted HeLa cells.
Intermediate levels of tetherin recruitment are sufficient for inhibition of virus release.
To determine whether the maximal colocalization/coclustering between tetherin and Gag, which was observed to be ESCRT dependent (Fig. 5), is required for tetherin function, we sought to assess the ability of tetherin to inhibit release of WT HIV-1 upon depletion of Alix. Previous studies showed that unlike depletion of other ESCRT proteins, such as Tsg101, depletion of Alix has little effect on ESCRT-mediated release of HIV-1 (79, 80). Therefore, the effect of Alix depletion on tetherin function can be examined specifically without a confounding effect on particle scission. We treated HeLa cells with siRNAs in the same transfection procedure as for Fig. 5 and measured tetherin activity by comparing Vpu+ and Vpu− molecular clones in these HeLa cells. We found that virus release of Vpu− HIV-1 was inhibited compared with that of Vpu+ HIV-1 in Alix-depleted HeLa cells as potently as, or even slightly more potently than, that in HeLa cells treated with control siRNA (∼25-fold versus ∼21-fold; Fig. 6A to C). These results suggest that while Alix is required for maximal colocalization and coclustering between tetherin and Gag, this high level of clustering is not necessary for the full antiviral activity of tetherin.
Fig 6.
Disruption of Gag-ESCRT interactions does not affect tetherin function. (A to C) HeLa cells were transfected with either nontarget or Alix-directed siRNA. At 48 h after transfection, cells were further transfected with either Vpu+ or Vpu− HIV-1 molecular clones. Cells were incubated for 16 h (64 h post-transfection with siRNA), followed by metabolic labeling and immunoprecipitation with HIV-Ig as for Fig. 1. Representative autoradiograms of cell- and virus-associated proteins are shown for experiments using HeLa cells (A). (B) The cellular amounts of Alix and α-tubulin in cells treated as in panel A were examined by immunoblotting. Note that the same transfection procedure was used to deplete Tsg101 and Alix for Fig. 5. (C) Quantitation of virus release efficiency from 3 independent experiments is shown as means ± 1 standard deviation. (D and E) P2 T cells were spinoculated with the indicated viruses with or without Vpu and incubated for 48 h, followed by metabolic labeling and immunoprecipitation. One of three autoradiograms is shown (D). Quantitation of 3 independent experiments is shown (E). *, P < 0.05.
Enhancement of virus release by Vpu is well documented for a T cell line, A3.01 (33), which has been shown to express a detectable level of tetherin on the cell surface (6, 51) (Fig. 4). In contrast to HeLa cells, late domain motifs and hence ESCRT recruitment appear to play a less-critical role in virus release in T cells, including an A3.01 cell clone (73). Taking advantage of this difference, we sought to assess the contribution of both Tsg101 and Alix interactions in tetherin function in an A3.01-derived cell clone, P2. For these experiments, P2 cells were spinoculated with VSV-G-pseudotyped viruses expressing either WT, PTAP−, or Y36S Gag proteins either with or without vpu. In these experiments, the p6 mutants produced virus particles at a markedly impaired but still detectable level in the presence of Vpu (Fig. 6D and E). Notably, deletion of Vpu reduced virus release to similar extents for WT and p6 mutant Gag proteins. In other words, we did not observe any reversal of virus release restriction when either the Tsg101- or primary Alix-binding sites were mutated in Gag. These results corroborate the results obtained with siRNA experiments performed using HeLa cells (Fig. 6A to C) and suggest that Gag-ESCRT interactions are not required for the antiviral activity of tetherin.
Altogether, these results indicate that although Gag-ESCRT interactions cause a detectable and significant increase in tetherin recruitment, the intermediate levels of tetherin recruitment observed when Tsg101 or Alix are depleted are sufficient for full inhibition of virus particle release.
Intermediate levels of tetherin recruitment, which are sufficient for inhibition of virus release, are dependent on intact CA sequences required for Gag-induced membrane curvature.
To identify molecular determinants for the intermediate levels of tetherin recruitment observed in previous experiments, which are sufficient for full inhibition of virus release, we analyzed coclustering between endogenous tetherin and several Gag mutants in HeLa cells by superresolution localization microscopy. As in the work shown in Fig. 3, we utilized two CA mutants that form electron-dense Gag patches at the plasma membrane but fail to induce membrane curvature efficiently (P99A Gag-mEos3.2 and EE75,76AA Gag-mEos3.2). We also examined the effect of Tsg5′ expression and mutations of Tsg101- and Alix-binding motifs in Gag (PTAP− Gag-mEos3.2 and CCY− Gag-mEos3.2) on tetherin-Gag coclustering. Virus release assays showed that replacement of YFP with mEos3.2 does not grossly affect efficiencies of VLP release by Gag mutants except that the virus release defect of P99A Gag was more severe in the mEos3.2 context than in the YFP context (data not shown). To establish a baseline for analysis, we also used a Gag derivative that contains mutations at both CA dimer interface and NC basic residues (20LK/WM/14A1G/Gag-mEos3.2; here called CA/NC Gag-mEos3.2). Due to these mutations, this Gag derivative was expected to be highly defective in multimerization and therefore was expected to not cocluster with tetherin. As expected, by standard TIRF microscopy, which is inherently diffraction limited, CA/NC Gag displays a largely uniform, diffuse membrane-lining phenotype with some patches (Fig. 7A) that may represent membrane topology (81). Reconstructed superresolution images show various sizes of small puncta of CA/NC Gag-mEos3.2 at the plasma membrane, which likely represent single molecules and perhaps small clusters of Gag molecules (Fig. 7B). WT and other mutants appeared to show more bright and extended puncta (Fig. 7B; data not shown). After analyzing the Gag mutants described above in the same manner as described for Fig. 5, we found that there are three distinct categories of tetherin-Gag coclustering (Fig. 7B and C). First, WT Gag-mEos3.2 showed a high level of tetherin-Gag coclustering. Second, when Gag-ESCRT interactions are disrupted (WT plus Tsg5′, PTAP−, and CCY−), significantly less tetherin coclustered with Gag (Fig. 7B and C). Notably, as was observed in the siRNA experiments (Fig. 5F), intermediate levels of tetherin-Gag coclustering were detected under these conditions, in which Gag interactions with Tsg101 or Alix were blocked genetically rather than by reducing the expression levels of these host factors. Third, we observed negligible levels of tetherin-Gag coclustering with P99A and EE75,76AA Gag-mEos3.2, which are not significantly more than that seen with CA/NC Gag-mEos3.2. The intermediate levels of tetherin recruitment observed upon disruption of Gag-ESCRT interactions (second category) were significantly greater than that seen for CA mutants that fail to induce membrane curvature (P99A and EE75,76AA Gag-mEos3.2) and CA/NC Gag-mEos3.2 (third category) (Fig. 7C).
These results indicate that intermediate levels of tetherin recruitment, which are sufficient for virus release inhibition (Fig. 6), are observed when Tsg101 or Alix is depleted (Fig. 5) or when ESCRT-interacting motifs of Gag are disrupted (Fig. 7). In addition, these intermediate levels of tetherin recruitment are abolished by mutations that disrupt either Gag multimerization or virus-induced membrane curvature (Fig. 7). Given that the two CA N-terminal domain mutants used in this study (P99A and EE75,76AA mutants) still contain the intact major Gag multimerization domains (the CA C-terminal domain and the NC domain) and show dense patches of multimerized Gag at the plasma membrane by electron microscopy (Fig. 3), it is likely that membrane curvature but not Gag multimerization per se is required for tetherin recruitment to assembly sites.
DISCUSSION
HIV-1 encodes four accessory genes that have been implicated in combating host defense responses (Vif, Vpr, Vpu, and Nef) (82), exemplifying the necessity for immune evasion by successful viral pathogens. Such evolutionary interplay between virus and host is epitomized by the relationship between HIV-1 Vpu and human tetherin. Tetherin appears to have arisen early in mammalian evolution and remains a potent inhibitor of many enveloped viruses (20). Considering the broad range of susceptible viruses, it is unlikely that tetherin recruitment is mediated by a direct interaction between tetherin and the Gag polyprotein. In support of this notion, we and others have observed that tetherin potently inhibited release of Gag derivatives lacking the juxta-membrane MA domain, which could potentially interact with the short cytoplasmic tail of tetherin (83) (unpublished data). Instead of the MA sequence, this study identified amino acid residues in the CA N-terminal domain and late domain motifs as molecular determinants for recruitment of tetherin to HIV-1 assembly sites at the plasma membrane. Importantly, mutations in the former impair Gag-induced membrane curvature, whereas those in the latter disrupt Gag-ESCRT interactions. Therefore, these results support a model in which both membrane curvature and the presence of ESCRT promote tetherin accumulation to the assembly sites of HIV-1.
We and others have hypothesized that tetherin recruitment may be mediated by lipid rafts on the plasma membrane. This seemed an attractive model because lipid rafts are incorporated into the membranes of many enveloped viruses. Another line of evidence in support of this hypothesis is the presence of tetherin in Triton X-insoluble membrane fractions (16, 18). Even though a study of tetherin chimeras in which the GPI anchor is replaced with a heterologous transmembrane domain showed that lipid raft association measured by detergent resistance is not sufficient (84), it was possible that association of tetherin with lipid rafts may play an important role. In the present study, however, using an antibody copatching assay, we found that tetherin itself does not appear to associate strongly with multiple lipid raft markers (CD55, CD59, NFP-GPI, and NFP-HATM) at the plasma membrane, all of which were previously shown to copatch strongly with WT Gag (26). Another recent study also failed to detect significant colocalization between tetherin and a lipid raft marker, GM1, by superresolution microscopy techniques (10). It is a valid concern that tetherin localization in the copatching assay may not reflect its native partitioning to microdomains unlike other raft proteins, since antibody treatment alters tetherin function (61). However, we also found that cholesterol depletion that disrupts the integrity of lipid rafts has little effect on tetherin function. These results indicate that although tetherin may still have an affinity for a specific subset of lipid rafts before its recruitment to virus assembly sites, such prior association with cholesterol-dependent lipid rafts is not required for tetherin antiviral function.
Conventional and superresolution microscopy of Gag late domain mutants, however, did reveal a potential role for Gag-ESCRT interaction in enhancing tetherin recruitment to virus assembly sites in HeLa and T cells. Moreover, siRNA knockdown experiments confirmed that expression of both Tsg101 and Alix is required for the high levels of tetherin recruitment seen with WT Gag in HeLa cells. A previous study that analyzed nascent particles formed by PTAP− Gag using immuno-scanning electron microscopy (SEM) showed that WT tetherin does not appear to specifically accumulate at HIV-1 assembly sites (9). Although WT Gag particles were not examined in that study, this observation is consistent with our findings supporting a role for ESCRT in enhancing tetherin recruitment. Notably, and somewhat unexpectedly, the high levels of tetherin recruitment, which is manifested as tetherin-Gag colocalization detected by confocal microscopy, were dispensable for the full activity of virus release restriction.
Conventional and superresolution microscopy showed largely consistent results for most conditions. However, a triple Gag mutant defective in Alix interaction (CCY−) did not show a significant decrease in tetherin recruitment as quantified by Pearson's coefficients of confocal images in HeLa cells but did show significantly less coclustering with tetherin in HeLa cells by superresolution microscopy analysis. While the origin of this difference remains unclear, it is possible that tetherin accumulates at or near the relatively large protrusive tubular membrane structures containing CCY− Gag patches (Fig. 3A) and thereby leads to the WT-level Pearson's correlation coefficient values without actually associating with Gag clusters. It is also possible that these structures may form more frequently on the top surface of cells than on the bottom surface. Indeed, we noticed that the Gag-positive tubular structures were detected on the bottom surface of cells imaged by superresolution microscopy but only in a minority of cells (unpublished observation). Superresolution analyses also revealed a difference in the levels of tetherin association with Gag mutants defective in ESCRT interactions versus those defective in inducing membrane curvature (Fig. 7), which were indistinguishable in confocal microscopy analyses (Fig. 3). Analysis of a multimerization-defective Gag (CA/NC Gag) mutant by superresolution techniques also suggests the presence of Gag clusters on the cell surface (Fig. 7). These observations, together with findings of recent studies (10, 85, 86), highlight the utility of using superresolution microscopy to analyze virus-host interactions at the level of single virus assembly sites and to study early as well as late events in virus assembly.
While the exact mechanism by which Tsg101 and Alix enhance tetherin recruitment to HIV-1 assembly sites remains to be elucidated, this activity likely occurs later in the assembly process, since tetherin is not recruited to the same extent by Gag mutants defective in inducing membrane curvature (Fig. 4) despite the presence of an intact late domain in these mutants. Although HIV-1 Gag may associate with Alix early during assembly, as seen with equine infectious anemia virus (EIAV) Gag, recruitment of downstream ESCRT components occurs later during the process (87, 88). It is possible that ESCRT proteins may form a barrier, which limits diffusion away from budding viruses. Notably, while recruitment of Tsg101 and Alix and hence the ESCRT machinery to assembly sites appears to be important for enhanced tetherin recruitment, formation of the pinching-off machinery composed of ESCRT-III is not sufficient. In Alix-depleted cells, the ESCRT-III machinery is still functional (79, 80) (Fig. 6), but the high levels of tetherin recruitment were not observed (Fig. 5), indicating that this recruitment relies not only on the presence of ESCRT but on the presence of Alix or an Alix-dependent process.
Although the additional level of tetherin recruitment promoted by Gag-ESCRT interactions appears unnecessary for tetherin-mediated inhibition of HIV-1, which forms relatively small, spherical virus particles, it is possible that larger enveloped viruses, such as filamentous orthomyxoviruses or filoviruses, may require greater amounts of tetherin to prevent their escape from infected cells. It would be interesting to examine the effects of ESCRT disruption on tetherin-mediated inhibition of these viruses.
Our findings, however, did provide evidence that Gag-induced membrane curvature results in intermediate levels of tetherin recruitment, which are sufficient to inhibit virus release. This result suggests that tetherin itself may be attracted to membrane curvature. Unfortunately, the contribution of membrane curvature to the antiviral effect of tetherin cannot be examined using virus release assays. This is because, in contrast to Gag-ESCRT interactions, membrane curvature is inseparable from virus particle release. Most membrane-curvature-defective Gag constructs fail to release VLPs regardless of Vpu expression (e.g., EE75,76AA Gag-YFP). Some curvature mutants (e.g., P99A Gag-YFP) release a low level of VLPs, but this is through assembly, budding, and release that proceed in an apparently WT-like manner, albeit less frequently (unpublished data) (26). As such, the low but detectable VLP release of P99A Gag-YFP was dependent on Vpu (unpublished data). Thus, in either case, it is not feasible to examine using Gag mutants to what extent membrane curvature promotes tetherin-mediated inhibition of virus release. However, it may be possible to address this point using tetherin mutants. There are several examples of cellular proteins that are able to both sense and manipulate membrane curvature via protein domains that form curved structures (89, 90). Intriguingly, based on crystallography analyses, it was hypothesized that a tetherin tetramer can form curved assemblies that may sense membrane curvature (14). In this regard, it will be interesting to examine requirements for recruitment of tetherin mutants that can dimerize but are predicted to fail to oligomerize (13, 15, 86). All enveloped viruses that assemble at the plasma membrane, whether dependent upon ESCRT machinery or not, must deform this membrane during budding, and therefore the membrane-curvature-dependent tetherin recruitment can explain the broad range of tetherin-susceptible viruses. It remains to be seen whether tetherin distinguishes virus-induced and non-virus-induced membrane curvature and whether higher levels of tetherin recruitment, which are dependent on ESCRT engagement, are relevant in the context of other enveloped viruses.
ACKNOWLEDGMENTS
We thank Kathleen Collins, Eric Freed, Alice Telesnitsky, and members of our laboratory for helpful discussions and critical review of the manuscript. We also thank Matthew Stone for help with image analysis and Jingga Inlora and Madeline Nye for technical assistance. We thank Jane Burns, Stanley Cohen, Eric Freed, Heinrich Göttlinger, and Klaus Strebel for reagents. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-Ig from NABI and NHLBI and anti-BST-2 from Klaus Strebel.
This work was supported by NIH grants, R00 GM087810 (to S.L.V.), R56 AI089282 (to A.O.), and R21 AI095022 (to A.O.). J.R.G. is supported by NIH training grant T32 AI007527-13.
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 2. Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. 2010. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog. 6:e1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Krausslich HG, Fackler OT, Keppler OT. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297 [DOI] [PubMed] [Google Scholar]
- 5. Habermann A, Krijnse-Locker J, Oberwinkler H, Eckhardt M, Homann S, Andrew A, Strebel K, Krausslich HG. 2010. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J. Virol. 84:4646–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hammonds J, Wang JJ, Yi H, Spearman P. 2010. Immunoelectron microscopic evidence for Tetherin/BST2 as the physical bridge between HIV-1 virions and the plasma membrane. PLoS Pathog. 6:e1000749 doi:10.1371/journal.ppat.1000749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83:1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5:e1000450 doi:10.1371/journal.ppat.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehmann M, Rocha S, Mangeat B, Blanchet F, Uji IH, Hofkens J, Piguet V. 2011. Quantitative Multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog. 7:e1002456 doi:10.1371/journal.ppat.1002456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80 doi:10.1186/1742-4690-6-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinz A, Miguet N, Natrajan G, Usami Y, Yamanaka H, Renesto P, Hartlieb B, McCarthy AA, Simorre JP, Gottlinger H, Weissenhorn W. 2010. Structural basis of HIV-1 tethering to membranes by the BST-2/tetherin ectodomain. Cell Host Microbe 7:314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang H, Wang J, Jia X, McNatt MW, Zang T, Pan B, Meng W, Wang HW, Bieniasz PD, Xiong Y. 2010. Structural insight into the mechanisms of enveloped virus tethering by tetherin. Proc. Natl. Acad. Sci. U. S. A. 107:18428–18432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swiecki M, Scheaffer SM, Allaire M, Fremont DH, Colonna M, Brett TJ. 2011. Structural and biophysical analysis of BST-2/tetherin ectodomains reveals an evolutionary conserved design to inhibit virus release. J. Biol. Chem. 286:2987–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schubert HL, Zhai Q, Sandrin V, Eckert DM, Garcia-Maya M, Saul L, Sundquist WI, Steiner RA, Hill CP. 2010. Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations. Proc. Natl. Acad. Sci. U. S. A. 107:17951–17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709 [DOI] [PubMed] [Google Scholar]
- 17. Lopez LA, Yang SJ, Exline CM, Rengarajan S, Haworth KG, Cannon PM. 2012. Anti-tetherin activities of HIV-1 Vpu and Ebola virus glycoprotein do not involve tetherin removal from lipid rafts. J. Virol. 86:5467–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masuyama N, Kuronita T, Tanaka R, Muto T, Hirota Y, Takigawa A, Fujita H, Aso Y, Amano J, Tanaka Y. 2009. HM1.24 is internalized from lipid rafts by clathrin-mediated endocytosis through interaction with alpha-adaptin. J. Biol. Chem. 284:15927–15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Fruh K. 2010. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6:e1000913 doi:10.1371/journal.ppat.1000913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dube M, Bego MG, Paquay C, Cohen EA. 2010. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology 7:114 doi:10.1186/1742-4690-7-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology (Bethesda) 26:236–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 410:512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weiss ER, Gottlinger H. 2011. The role of cellular factors in promoting HIV budding. J. Mol. Biol. 410:525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. 2011. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 85:9749–9766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krementsov DN, Rassam P, Margeat E, Roy NH, Schneider-Schaulies J, Milhiet PE, Thali M. 2010. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hurley JH, Hanson PI. 2010. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11:556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ono A. 2010. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell 102:335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammonds J, Spearman P. 2010. An imperfect rule for the particle roost. Cell Host Microbe 7:261–263 [DOI] [PubMed] [Google Scholar]
- 32. Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. 1990. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 64:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 82:2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun Z, Pan J, Hope WX, Cohen SN, Balk SP. 1999. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer 86:689–696 [DOI] [PubMed] [Google Scholar]
- 36. Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. 1999. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 9:1489–1492 [DOI] [PubMed] [Google Scholar]
- 37. Ahmad N, Maitra RK, Venkatesan S. 1989. Rev-induced modulation of Nef protein underlies temporal regulation of human immunodeficiency virus replication. Proc. Natl. Acad. Sci. U. S. A. 86:6111–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U. S. A. 98:13925–13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yee JK, Friedmann T, Burns JC. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Part A):99–112 [DOI] [PubMed] [Google Scholar]
- 40. Kong LB, An D, Ackerson B, Canon J, Rey O, Chen IS, Krogstad P, Stewart PL. 1998. Cryoelectron microscopic examination of human immunodeficiency virus type 1 virions with mutations in the cyclophilin A binding loop. J. Virol. 72:4403–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popov S, Popova E, Inoue M, Gottlinger HG. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9:727–729 [DOI] [PubMed] [Google Scholar]
- 46. Kiernan RE, Ono A, Freed EO. 1999. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J. Virol. 73:4728–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J. Virol. 83:7322–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Janvier K, Pelchen-Matthews A, Renaud JB, Caillet M, Marsh M, Berlioz-Torrent C. 2011. The ESCRT-0 component HRS is required for HIV-1 Vpu-mediated BST-2/tetherin down-regulation. PLoS Pathog. 7:e1001265 doi:10.1371/journal.ppat.1001265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. 2006. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J. Biol. Chem. 281:34640–34650 [DOI] [PubMed] [Google Scholar]
- 50. Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. 2010. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. U. S. A. 107:12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miyagi E, Andrew AJ, Kao S, Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog. 6:e1001167 doi:10.1371/journal.ppat.1001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ono A, Waheed AA, Joshi A, Freed EO. 2005. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 [DOI] [PubMed] [Google Scholar]
- 55. Gonda MA, Aaronson SA, Ellmore N, Zeve VH, Nagashima K. 1976. Ultrastructural studies of surface features of human normal and tumor cells in tissue culture by scanning and transmission electron microscopy. J. Natl. Cancer Inst. 56:245–263 [DOI] [PubMed] [Google Scholar]
- 56. Rust MJ, Bates M, Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3:793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Endesfelder U, Malkusch S, Flottmann B, Mondry J, Liguzinski P, Verveer PJ, Heilemann M. 2011. Chemically induced photoswitching of fluorescent probes—a general concept for super-resolution microscopy. Molecules 16:3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. 2005. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. U. S. A. 102:1419–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, Baird BA. 2012. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7:e31457 doi:10.1371/journal.pone.0031457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hammond S, Wagenknecht-Wiesner A, Veatch SL, Holowka D, Baird B. 2009. Roles for SH2 and SH3 domains in Lyn kinase association with activated FcεRI in RBL mast cells revealed by patterned surface analysis. J. Struct. Biol. 168:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miyagi E, Andrew A, Kao S, Yoshida T, Strebel K. 2011. Antibody-mediated enhancement of HIV-1 and HIV-2 production from BST-2/tetherin-positive cells. J. Virol. 85:11981–11994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fritz JV, Tibroni N, Keppler OT, Fackler OT. 2012. HIV-1 Vpu's lipid raft association is dispensable for counteraction of the particle release restriction imposed by CD317/Tetherin. Virology 424:33–44 [DOI] [PubMed] [Google Scholar]
- 63. Ruiz A, Hill MS, Schmitt K, Stephens EB. 2010. Membrane raft association of the Vpu protein of human immunodeficiency virus type 1 correlates with enhanced virus release. Virology 408:89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ono A, Waheed AA, Freed EO. 2007. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology 360:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 66. Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 [DOI] [PubMed] [Google Scholar]
- 67. Demirov DG, Ono A, Orenstein JM, Freed EO. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699 [DOI] [PubMed] [Google Scholar]
- 69. von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. 2003. The protein network of HIV budding. Cell 114:701–713 [DOI] [PubMed] [Google Scholar]
- 70. Pornillos O, Alam SL, Davis DR, Sundquist WI. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812–817 [DOI] [PubMed] [Google Scholar]
- 71. Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059–36071 [DOI] [PubMed] [Google Scholar]
- 72. VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Demirov DG, Orenstein JM, Freed EO. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. 2011. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J. Virol. 85:2304–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375:592–610 [DOI] [PubMed] [Google Scholar]
- 76. Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. 2007. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 4:54 doi:10.1186/1742-4690-4-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 [DOI] [PubMed] [Google Scholar]
- 78. McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. 2009. A bright and photostable photoconvertible fluorescent protein. Nat. Methods 6:131–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Rheenen J, Jalink K. 2002. Agonist-induced PIP(2) hydrolysis inhibits cortical actin dynamics: regulation at a global but not at a micrometer scale. Mol. Biol. Cell 13:3257–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 83. Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Andrew AJ, Kao S, Strebel K. 2011. C-terminal hydrophobic region in human bone marrow stromal cell antigen 2 (BST-2)/tetherin protein functions as second transmembrane motif. J. Biol. Chem. 286:39967–39981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528 [DOI] [PubMed] [Google Scholar]
- 86. Hammonds J, Ding L, Chu H, Geller K, Robbins A, Wang JJ, Yi H, Spearman P. 2012. The tetherin/BST-2 coiled-coil ectodomain mediates plasma membrane microdomain localization and restriction of particle release. J. Virol. 86:2259–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13:469–474 [DOI] [PubMed] [Google Scholar]
- 88. Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bhatia VK, Hatzakis NS, Stamou D. 2010. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin. Cell Dev. Biol. 21:381–390 [DOI] [PubMed] [Google Scholar]
- 90. Antonny B. 2011. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 80:101–123 [DOI] [PubMed] [Google Scholar]