Abstract
The human kinome comprises over 800 individual kinases. These contribute in multiple ways to regulation of cellular metabolism and may have direct and indirect effects on virus replication. Kinases are tempting therapeutic targets for drug development, but achieving sufficient specificity is often a challenge for chemical inhibitors. While using inhibitors to assess whether c-Jun N-terminal (JNK) kinases regulate hepatitis C virus (HCV) replication, we encountered unexpected off-target effects that led us to discover a role for a mitogen-activated protein kinase (MAPK)-related kinase, MAPK interacting serine/threonine kinase 1 (MKNK1), in viral entry. Two JNK inhibitors, AS601245 and SP600125, as well as RNA interference (RNAi)-mediated knockdown of JNK1 and JNK2, enhanced replication of HCV replicon RNAs as well as infectious genome-length RNA transfected into Huh-7 cells. JNK knockdown also enhanced replication following infection with cell-free virus, suggesting that JNK actively restricts HCV replication. Despite this, AS601245 and SP600125 both inhibited viral entry. Screening of a panel of inhibitors targeting kinases that may be modulated by off-target effects of AS601245 and SP600125 led us to identify MKNK1 as a host factor involved in HCV entry. Chemical inhibition or siRNA knockdown of MKNK1 significantly impaired entry of genotype 1a HCV and HCV-pseudotyped lentiviral particles (HCVpp) in Huh-7 cells but had only minimal impact on viral RNA replication or cell proliferation and viability. We propose a model by which MKNK1 acts to facilitate viral entry downstream of the epidermal growth factor receptor (EGFR) and extracellular signal-regulated kinase (ERK), both of which have been implicated in the entry process.
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) is a major cause of liver disease worldwide. Most infected persons fail to eliminate the virus following acute infection, placing them at risk for chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (for a review, see reference 1). Classified within the genus Hepacivirus of the family Flaviviridae, HCV is an enveloped virus with a positive-sense, single-stranded RNA genome approximately 9.6 kb in length. A single open reading frame encodes a large precursor polyprotein, which is processed into 10 proteins: core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (2). Core, E1, and E2 proteins are structural proteins that are present within the virion, while the processed polyprotein segment extending from NS3 to NS5B assembles into a replicase complex that directs synthesis of the viral RNA (3). Because of its importance as a human pathogen, both HCV replication mechanisms and host responses that restrict HCV infection have been intensively studied. Such knowledge has led to the development of new direct-acting antiviral drugs that offer increasing opportunities for eliminating HCV infections and either arresting or reversing the progression of liver disease. Despite this, major gaps remain in our understanding of the biology of HCV.
One unanswered question that has relevance for both vaccine development and antiviral therapy is why host responses are able to successfully eliminate HCV in a minority of infected individuals while infections become persistent in others. Innate host responses to HCV seem likely to play a primary role in the early elimination of virus, as this correlates strongly with polymorphisms in the interleukin-28B (IL-28B) (gamma 3 interferon [IFN-λ3]) gene (4). IFN-mediated Janus kinase signal transducer and activator of transcription (JAK-STAT) signaling results in strong anti-HCV activity and likely contributes to viral control by stimulating the expression of numerous antiviral proteins, including protein kinase R, 2′,5′-oligoadenylate synthetase, MxA, viperin, interferon-stimulated gene 21, and others (5). Other cellular signaling pathways also have been shown to suppress HCV replication, including those triggered by protein kinase C (6), phosphoinositide-3 (PI3) kinase (7, 8), SMAD (9), extracellular signal-regulated kinase (ERK) (7, 10, 11), and p38 kinase (12).
Although there is limited understanding of how intracellular signaling restricts HCV replication, mitogen-activated protein kinases (MAPKs) appear to play an essential role. These are versatile serine/threonine kinases involved in signal transduction that modulate gene transcription in response to changes in extracellular stimuli (13). They control numerous fundamental cellular processes, including proliferation, differentiation, migration, and apoptosis. Three MAPK cascades have been defined, including those centered on ERK, c-Jun N-terminal kinase (JNK), and p38. ERK and p38 signaling are both antagonistic to HCV replication (7), while the impact of JNK activation has not been studied. JNKs (comprised of JNK1, JNK2, and JNK3) are activated by a variety of cytotoxic stresses, such as UV irradiation, heat/osmotic shock, and oxidative stress, and proinflammatory cytokines, including tumor necrosis factor-α (TNF-α), interleukin-1 (IL-1), and transforming growth factor-β (TGF-β) (14). Moreover, innate immune responses induced by Toll-like receptor (TLR) recognition of invading pathogens activate JNK signaling (15), suggesting that JNKs act as intermediates in antiviral signaling. Activated JNKs are phosphorylated and, in turn, phosphorylate a variety of substrates, including transcription factors, Bcl2 family members, and cytoskeleton molecules.
While such cell signaling may limit infection, signaling via other pathways appears to be essential for entry of HCV into the host cell. Several host proteins are involved in this process, including the tetraspanin CD81 (16, 17), scavenger receptor class B type I (SR-BI) (18, 19), and the tight junction proteins claudin-1 (CLDN1) (20) and occludin (OCLN) (21), as well as the recently identified Niemann-Pick C1-like 1 (NPC1L1) protein (22). Viral entry is initiated by the binding of virus to glycosaminoglycans (23, 24) and/or the low-density lipoprotein (LDL) receptor (25) on the cell surface, followed by subsequent steps involving scavenger receptor B1 (SR-BI), CD81, CLDN1, and OCLN, ultimately leading to clathrin-mediated endocytosis of the virus (26, 27; for a review, see reference 28). While mechanistic details of this process remain to be elucidated, several lines of evidence suggest that host kinases play critical roles. Inhibition of protein kinase A (PKA) limits HCV entry and alters the pattern of CLDN1 expression (29), while a comprehensive RNA interference (RNAi) screen identified epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) as additional host factors (30). EGFR activation enhanced HCV entry by regulating CD81 interactions with CLDN1 and membrane fusion, although the downstream signaling pathways were not elucidated. ERK activation has also been linked to CD81 engagement (31).
Here, we describe experiments aimed at determining whether JNK signaling regulates HCV infection. We found that chemical inhibitors of JNK (AS601245 and SP600125) or siRNA knockdown of JNK expression enhanced replication of viral RNA, indicating a negative regulatory role for JNK kinases. Nonetheless, an off-target effect of these JNK inhibitors blocked infection with cell-free virus, leading to recognition of the involvement of a MAPK-related kinase, MAPK interacting serine/threonine kinase 1 (MKNK1), in viral entry.
MATERIALS AND METHODS
Cells and viruses.
The human hepatocellular carcinoma cell lines Huh-7 and Huh-7.5 were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 1× penicillin-streptomycin at 37°C in a 5% CO2 environment. NNeo/3-5B/RG (RG) and NNeo/C-5B/2-3 (2, 3), Huh-7 clonal cell lines that harbor HCV-N (genotype 1b) subgenomic replicon RNA and genome-length replicon RNA, respectively (32), were maintained in complete DMEM with 250 μg/ml of G418. HJ3-5 and H77S.2 viruses have been described previously (33, 34). Plasmids encoding infectious chimeric genotype 1a (structural) and 2a (nonstructural proteins) HCV genomes, HJ3-5, H-NS2/NS3-J (34, 35), and HJ3-5/GLuc2A (that expresses Gaussia princeps luciferase [GLuc]) (36) have been described previously. Retroviral particles pseudotyped with the H77c envelope (HCVpp) and vesicular stomatitis envelope protein (VSVpp) were prepared as described previously (37).
Reagents and antibodies.
Antibodies to JNK, phospho-JNK, MKNK1, NPC1L1, and Myc-Tag were purchased from Cell Signaling Technology. Antibodies to β-actin (A-5441) and FLAG (F-3165) were from Sigma-Aldrich. Antibodies to claudin-1 (clone 2H10D10) and occludin (OC-3F10) were from Invitrogen. Antibodies to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AM4300) and SR-BI (NB400-104) were purchased from Ambion and Novus Biologicals, respectively. A mouse monoclonal antibody to the HCV core protein (C7-50) and rabbit polyclonal antibody to CD81 (PA5-13582) were obtained from Thermo Scientific. Rabbit antibody to HCV NS5A was the generous gift of Craig Cameron. JNK inhibitors SP600125 and AS601245 were purchased from Calbiochem.
Plasmids and DNA transfection.
pRLHL, a dicistronic, dual luciferase reporter plasmid containing the HCV internal ribosome entry site (IRES) within its intercistronic space, has been described (38). pCMV-GLuc was purchased from New England BioLabs, Inc. An expression vector for JNK2 (pcDNA-FLAG-JNK2) was constructed by amplifying specific cDNA using conventional reverse transcription-PCR (RT-PCR) methods and cloning the amplified sequences into the HindIII-EcoRI site of pcDNA6/V5-HisB with FLAG sequence at the N terminus of the gene. pcDNA-Myc-MKK73E, an expression vector for constitutively active MKK7, was constructed by inserting the cDNA of MKK7 into the KpnI-XbaI site of pcDNA6/V5-HisB with a myc sequence at the N terminus of the gene, followed by site-specific mutagenesis of S271E, T275E, and S277E using a QuikChange site-directed mutagenesis kit (Stratagene). Transfection of plasmid DNAs was accomplished with Fugene 6 (Roche) according to the manufacturer's recommended procedures.
RNA transcription and transfection.
In vitro transcription of HCV RNA and transfection were performed as previously described (39).
Kinase inhibitor screen.
Naïve Huh-7.5 cells (2 × 105 cells/well) were plated in 6-well culture dishes, and 24 h later they were treated with various chemical inhibitors of selected kinases (see Results) at 10 μM for 1 h prior to inoculation with virus (HJ3-5) at a multiplicity of infection (MOI) of 1 in the presence of the inhibitor. Inhibitors were diluted in dimethyl sulfoxide (DMSO) (final concentration, <0.1%). The cells were then washed twice with PBS and refed with fresh culture medium with no inhibitor. Cells were harvested 3 days later for immunoblotting with antibodies to HCV core protein. Dose-ranging experiments to determine the effective inhibitory concentrations (ICs) of kinase inhibitors were carried out similarly.
Inhibition of FFU formation and HCVpp entry.
Huh-7.5 cells (1 × 105 cells/well) were plated in an 8-well chamber slide (Lab-Tek). Inhibitors were added to the medium 1 h prior to inoculation with HJ3-5 virus, H77c HCVpp, or VSVpp. VSVpp was diluted 1:20 prior to inoculation, so that it produced luciferase activity approximating that obtained with HCVpp in untreated cells. Virus was allowed to adsorb for 6 h in the presence of the inhibitor, following which the cells were washed twice with PBS and refed with fresh medium containing no inhibitor. The cells were processed 2 days later for detection of foci of infected cells by immunostaining with antibody to core protein, as described previously (34) (HJ3-5-infected cells), or lysed for luciferase assay (HCVpp and VSVpp). In related experiments, cells were treated with the inhibitor for 6 h prior to virus inoculation or after virus adsorption and then similarly processed.
Luciferase assay.
Cell lysates were prepared with Passive Lysis buffer (Promega), and luciferase activity was measured by Luciferase Assay systems (Promega) by following the manufacturer's protocol. For the measurement of GLuc activity, we followed the method described by Shimakami et al. (33)
RNAi.
Short interfering RNAs (siRNAs) for JNK1 (J-003514-17), JNK2 (J-003505-18), MKNK1 (L-004879-00-005), and MKNK2 (L-004908-00-0005) were purchased from Thermo Scientific. Transfection of siRNAs was accomplished by either Oligofectamine (Invitrogen) or TransIT-TKO transfection reagent (Mirus) by following the manufacturer's recommended procedures. For the infection experiment, 2 days after transfection, the cells were trypsinized and seeded in a 48-well culture dish (1 × 105 cells/well). One day after seeding, the cells were incubated with viruses (HJ3-5 or HCVpp) for 6 h. The cells were washed with phosphate-buffered saline (PBS) and refed with fresh medium. The cells were fixed 48 h later for counting the number of infectious foci or lysed for luciferase assay.
Cell proliferation assay.
Cell proliferation was assessed by WST-1 (2-[2-methoxy-4-nitrophenyl]-3-[4-nitrophenyl]-5-[2,4-disulpho-phenyl]-2H-tetrazolium, monosodium salt) assay according to the manufacturer's suggested protocol (Millipore).
Northern blotting.
Total cellular RNA was extracted using TRIzol reagent (Invitrogen). Six μg of total RNA was electrophoresed and transferred to a nylon membrane (Hybond-N; GE Healthcare). The membrane was probed with in vitro-transcribed negative-strand HCV RNA labeled with digoxigenin (DIG), using reagents provided with the DIG Northern Starter kit (Roche Applied Science) and by following the manufacturer's recommended procedures. After extensive washing and blocking, the membrane was hybridized with anti-digoxigenin-AP, incubated with disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD), and exposed to a BioMax MR X-ray film (Eastman Kodak).
Immunoblotting.
Standard immunoblotting procedures were followed (40). Proteins transferred to a polyvinylidene difluoride (PVDF) membrane were probed with specific primary antibodies (as described above), followed by further incubation with a secondary antibody conjugated with horseradish peroxidase (GE Healthcare) or IRDye 800CW goat anti-mouse IgG or IRDye 680 goat anti-rabbit IgG (LI-COR Biosciences). Proteins were visualized by ECL Western blotting detection reagents (GE Healthcare) or an Odyssey infrared imaging system (LI-COR Biosciences).
qRT-PCR.
For quantitative RT-PCR (qRT-PCR), total cellular RNA was prepared with an RNeasy Minikit (Qiagen). mRNAs for MKNK1, MKNK2, and GAPDH were quantified by a CFX96 real-time system (Bio-Rad) using an iScript One-Step RT-PCR kit with SYBR green (Bio-Rad). Primer sequences were 5′ CCAGGCAGGCCGTGGTGAAG 3′ and 5′ AGCGGTGAGAGCAGGCTGGA 3′ for MKNK1 mRNA and 5′ AGGAAGATGTGCTGGGGGAG 3′ and 5′ CCTGGCACTGGTACAGCATC 3′ for MKNK2 mRNA.
RESULTS
JNK activation restricts replication of HCV replicon RNA.
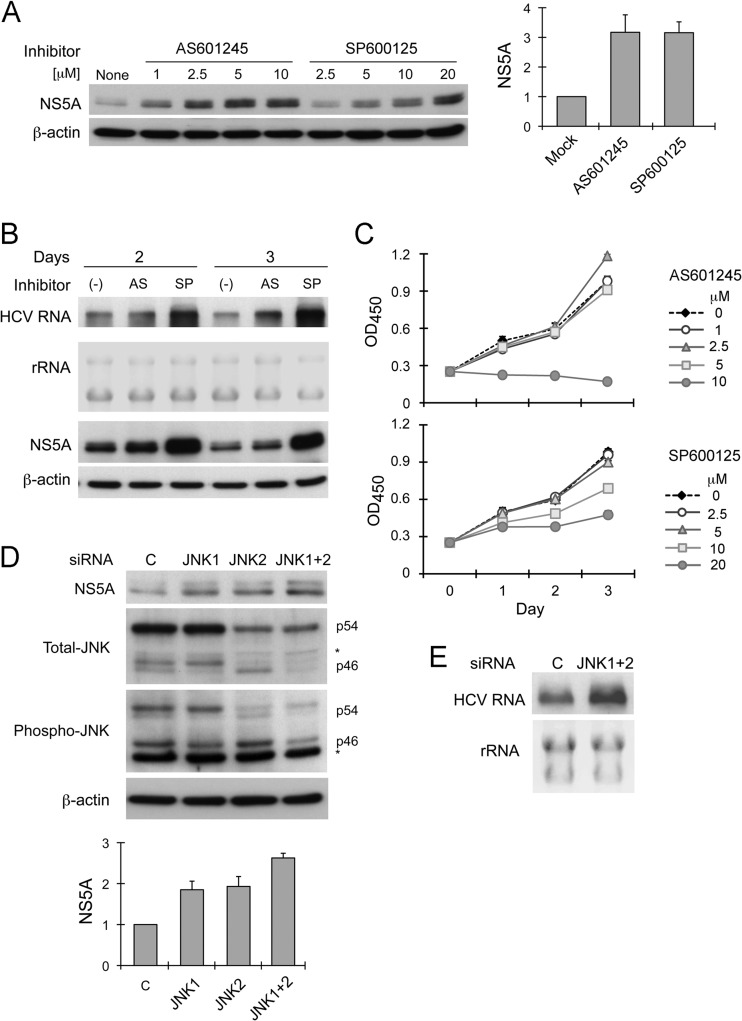
To determine whether JNK signaling regulates replication of HCV RNA, we treated a cell line containing a subgenomic HCV RNA replicon, RG, with two chemical inhibitors of JNK, AS601245 (1 to 10 μM) and SP600125 (2.5 to 20 μM). Both inhibitors increased NS5A abundance in a dose-dependent fashion (Fig. 1A). Both JNK inhibitors also increased the abundance of the HCV replicon RNA, although SP600125 had a greater effect on this than AS601245 (Fig. 1B). Significantly, cellular proliferation was reduced at the higher concentrations of these compounds, as evidenced in WST-1 assays (Fig. 1C). Thus, the increase in viral RNA and viral protein abundance could not be attributed to a reversal of JNK suppression of cell growth (41). Moreover, since the reduced cellular proliferation observed at these concentrations could be expected to have a negative effect on HCV replication (42), the increase in HCV RNA abundance that we observed when RG cells were treated with these high concentrations of the inhibitors may underrepresent the degree to which JNK-mediated control of replication is derepressed. Similar effects were observed with a genome-length replicon cell line (data not shown).
Fig 1.
JNK inhibition enhances HCV RNA abundance in replicon cells. (A) Effect of JNK inhibitors AS601245 and SP600125 on HCV replicon abundance. RG cells were treated with AS601245 (1 to 10 μM) or SP600125 (2.5 to 20 μM) for 24 h prior to harvesting and immunoblot assay for NS5A expression. (Left) Immunoblots of NS5A and β-actin, a loading control. (Right) Mean relative NS5A abundance ± standard deviations (SD) at 2.5 μM AS601245 or 10 μM SP600125 in 3 independent experiments. Protein abundance was quantified by densitometry and calculated as the amount relative to that in lysates of untreated cells (considered 1). (B) Northern blot analysis showing abundance of HCV replicon RNA in RG cells treated with 2.5 μM AS601245 (AS) or 10 μM SP600125 (SP) for 2 or 3 days. 18S and 28S ribosomal RNAs are shown as a loading control. At the bottom are immunoblots of NS5A and β-actin in cell lysates. (C) Impact of AS601245 (0 to 10 μM) or SP600125 (0 to 20 μM) on cellular proliferation. WST-1 assay was carried out on cells incubated for 1, 2, or 3 days in media containing inhibitors. Mean values at an optical density of 540 nm (OD450) were calculated from triplicate experiments. (D) RNAi knockdown of JNK1 and JNK2 enhances replicon RNA abundance. RG cells were transfected with JNK1-specific, JNK2-specific, a combination of JNK1- and JNK2-specific siRNAs (JNK1+2), or negative-control siRNAs 3 days prior to being harvested for immunoblotting. (Upper) Immunoblots of NS5A, total JNK, phosphorylated JNK, and β-actin. (Lower) Relative NS5A abundance (means ± SD) in 3 independent transfection experiments. Protein bands were quantified as described for panel A; an asterisk signifies a nonspecific protein band. (E) Northern blot showing replicon RNA abundance in RG cells transfected 3 days previously with JNK1- and JNK2-specific or control siRNAs.
To confirm the results obtained with these chemical inhibitors, we assessed the effect of siRNA-mediated knockdown of JNK on the replicon. There are three JNK isoforms (JNK1, JNK2, and JNK3), each of which is expressed as a short (46 kDa) and long species (54 kDa) (43). RT-PCR assays detected only JNK1 and JNK2 transcripts in RG cells (data not shown). This was as expected, since RG cells are derived from Huh-7 hepatoma cells, and JNK3 expression is restricted to heart, brain, testis, and pancreatic β cells (43). Thus, we used only JNK1- and JNK2-specific siRNA pools to knock down JNK expression. Immunoblots using anti-JNK antibody revealed one major 54-kDa band and three bands migrating with a mass close to 46 kDa in these cells (Fig. 1D). The most rapidly migrating 46-kDa band was reduced by JNK1-specific siRNA, while the 54-kDa and middle 46-kDa bands were decreased with JNK2-specific siRNA. The most slowly migrating 46-kDa band was unchanged with either siRNA and is likely to be nonspecific. Antibodies to phospho-JNK (Thr183/Tyr185) reacted with both the 54-kDa and most rapidly migrating 46-kDa species (corresponding to JNK2 and JNK1, respectively). Thus, both JNK1 and JNK2 are activated and likely to contribute to JNK signaling in these Huh-7-derived cells, although the underlying mechanism responsible for JNK activation is uncertain. As we anticipated from the studies with chemical inhibitors, knockdown of either JNK1 or JNK2 resulted in an increase in NS5A abundance, while combined knockdown of both JNK1 and JNK2 had an additive effect on NS5A abundance (Fig. 1D) and resulted in a clear increase in replicon RNA abundance (Fig. 1E). We conclude from these results that JNK activation restricts the replication of HCV replicon RNAs.
JNK suppresses replication of infectious virus.
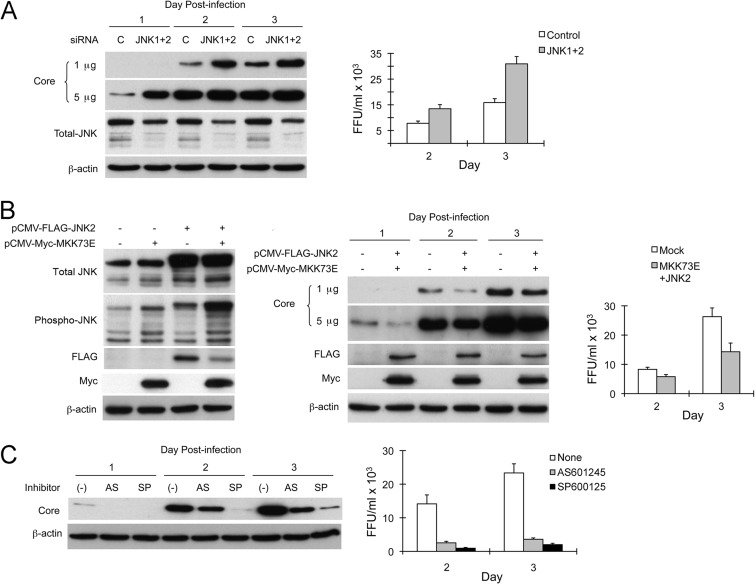
To confirm that JNK has similar effects on replication of infectious virus, we transfected Huh-7 cells with JNK1- and JNK2-specific siRNAs prior to inoculation with HJ3-5, a genotype 1a/2a chimeric virus (35). Cells were infected at an MOI of 2 and then monitored for viral protein abundance and yields of infectious virus released into the media. As shown in Fig. 2A, knockdown of JNK1 and JNK2 expression led to a significant increase in core protein expression and a doubling of the infectious virus yield at both 48 and 72 h postinfection. Conversely, JNK activation suppressed HCV replication. To activate JNK, we cotransfected expression vectors for JNK2 and MKK73E, a constitutively active form of mitogen-activated protein kinase kinase 7 (MAP2K7, or MKK7) that acts as an upstream activator of JNK (44). While independent expression of either JNK2 or MKK73E had little effect on the abundance of phosphorylated JNK, their coexpression resulted in substantial JNK activation (Fig. 2B, left). This was reflected in reduced core protein expression (Fig. 2B, center) and infectious virus yields (Fig. 2B, right). These results indicate that JNK activation not only suppresses amplification of replicon RNAs but also restricts replication of infectious virus.
Fig 2.
Contrasting effects of genetic manipulation versus chemical inhibition of JNK on infection with cell-free HCV. (A) JNK knockdown enhances the replication of HCV in Huh-7 cells. (Left) Cells were transfected with negative-control or JNK1- and JNK2-specific siRNAs prior to inoculation with HJ3-5 virus at an MOI of 2. Cell lysates prepared 1, 2, and 3 days postinfection were assayed for core, JNK, and β-actin proteins. Immunoblottings for core protein were carried out using 1 or 5 μg of total protein resolved in each lane. (Right) HJ3-5 virus titers in supernatant media collected 2 and 3 days postinfection. Error bars indicate SD in replicate experiments. (B) JNK activation suppresses HCV replication in Huh-7 cells. (Left) Cells were transfected with expression vectors for JNK2 (pCMV-FLAG-JNK2) and constitutively active MKK7 (pCMV-Myc-MKK73E). After incubation for 2 days, cells were harvested, followed by immunoblot analyses for total and phosphorylated JNK, ectopically expressed JNK2 (using anti-FLAG antibody), ectopically expressed MKK73E (using anti-Myc antibody), and β-actin. (Middle) Cells were cotransfected with pCMV-FLAG-JNK2 and pCMV-Myc-MKK73E or were transfected only with an equal amount of empty vector (pcDNA6/V5-HisB) prior to inoculation with HJ3-5 virus. Cells were harvested 1, 2, and 3 days later, and lysates were assayed for core, FLAG (tagged to JNK2), Myc (tagged to MKK73E), and β-actin proteins. (Right) HJ3-5 virus titers in supernatant fluids were determined 2 and 3 days postinfection. (C) JNK inhibitors AS601245 and SP600125 suppress infection by cell-free virus. (Left) Cells were pretreated with 2.5 μM AS601245, 10 μM SP600125, or DMSO 1 h prior to inoculation with HJ3-5 virus at an MOI of 2. Cells were then cultured in media containing the inhibitors for 1, 2, and 3 days prior to preparation of lysates for assay of core and β-actin proteins. (Right) HJ3-5 virus titers in supernatant fluids 2 and 3 days postinfection.
Off-target effects of AS601245 and SP600125 inhibit HCV entry.
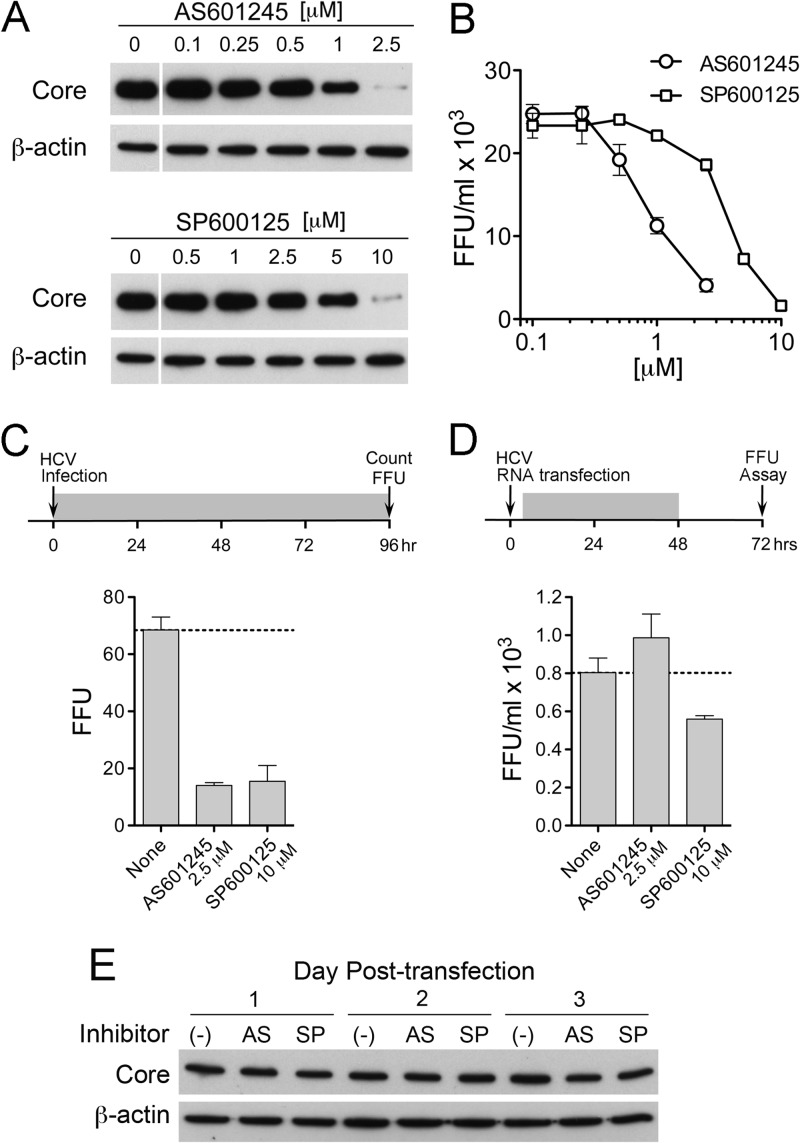
Given the results described above, we anticipated that the chemical inhibitors of JNK would also enhance replication of infectious virus. To confirm this, we pretreated Huh-7 cells with 2.5 μM AS601245 or 10 μM SP600125 for 1 h prior to inoculation with HJ3-5 virus at a multiplicity of 2. After 6 h of incubation at 37°C for virus adsorption, the cells were washed with PBS and refed with medium containing the inhibitors. Surprisingly, both core protein expression levels and infectious virus yields were sharply reduced by both inhibitors compared to levels for nontreated cells (Fig. 2C). Subsequent studies demonstrated that AS601245 possessed a 50% effective antiviral concentration (EC50) of 0.92 μM, and SP600125 had an EC50 of 3.8 μM (Fig. 3A and B). These concentrations are within the range commonly used for JNK inhibition and do not negatively impact proliferation of Huh-7 cells in WST-1 assays (Fig. 1C).
Fig 3.
Suppression of HCV infection of Huh-7 cells by increasing concentrations of AS601245 or SP600125. (A) Cells were pretreated with AS601245 (0 to 2.5 μM) or SP600125 (0 to 10 μM) for 1 h prior to inoculation with HJ3-5 virus and then were maintained in media containing the inhibitors. Lysates prepared 3 days postinoculation were assayed for core and β-actin protein abundance by immunoblotting. (B) Titer of infectious virus released from HJ3-5-infected cells treated as described for panel A. Infectious virus yields were measured in supernatant fluids collected on day 3 postinoculation using an FFU assay (45). (C) Inhibition of infectious focus formation by 2.5 μM AS601245 or 10 μM SP600125 versus DMSO control (None). (Top) Huh-7 cells were inoculated with genotype 1a H77S.2 virus (33) in the presence of compounds at time point 0. Medium was replaced at 24-h intervals, and the cells were fixed for detection of replication foci by indirect immunofluorescence for core protein at 96 h. The shaded bar represents the period of exposure to compounds. (Bottom) FFU enumerated at 96 h. (D) Lack of suppression of HCV RNA replication and infectious virus production by equivalent concentrations of the JNK inhibitors tested in panel C. (Top) Huh-7 cells were electroporated with in vitro-transcribed H77S.2 RNA and then fed 4.5 h later, when they were adherent to the vessel surface, with medium containing 2.5 μM AS601245, 10 μM SP600125, or DMSO only. Cells were refed at 24 h with media containing the compounds (shaded segment of timeline) and then at 48 h with media containing no inhibitors. Supernatant fluids harvested from the cells at 72 h were assayed for the presence of infectious virus in an FFU assay. (Bottom) Infectious virus yields from RNA-transfected cells. Data shown in panels C and D represent means ± SD from triplicate or duplicate cultures. (E) Cells were electroporated with H-NS2/NS3-J RNA (35) and then treated with AS601245 (2.5 μM) or SP600125 (10 μM). Cells were harvested at 24-h intervals to monitor expression of HCV core and β-actin (loading control) proteins by immunoblotting.
Since both AS601245 and SP600125 inhibit replication of infectious virus (Fig. 2C and 3) yet stimulate replication of replicon RNAs (Fig. 1A and B), we suspected that they block a step in viral entry, or possibly assembly and release. This would be an off-target effect of these kinase inhibitors, since specific RNAi-mediated knockdown of JNK enhances replication of infectious virus (Fig. 2A). To better characterize this off-target effect, we measured the number of infectious foci formed in cells inoculated at a low multiplicity of infection with genotype 1a H77S.2 virus (33, 45) in the presence or absence of AS601245 or SP600125. Both inhibitors resulted in an approximately 80% reduction in the number of infectious foci (Fig. 3C). This was not due to inhibition of H77S RNA replication, as there was only a minimal (SP600125) or no (AS101245) effect on yields of infectious virus released from cells that were treated with the inhibitors for 48 h after transfection with H77S.2 RNA (Fig. 3D). Similarly, there was little effect on core protein expression, a measure of viral replication, in cells transfected with a replication-competent HCV RNA (H-NS2/NS3-J) that is incapable of directing assembly of infectious virus particles (35) (Fig. 3E). These results point strongly toward an off-target effect of these JNK inhibitors that hits a key step in HCV entry.
Identification of MKNK as a host factor in HCV entry.
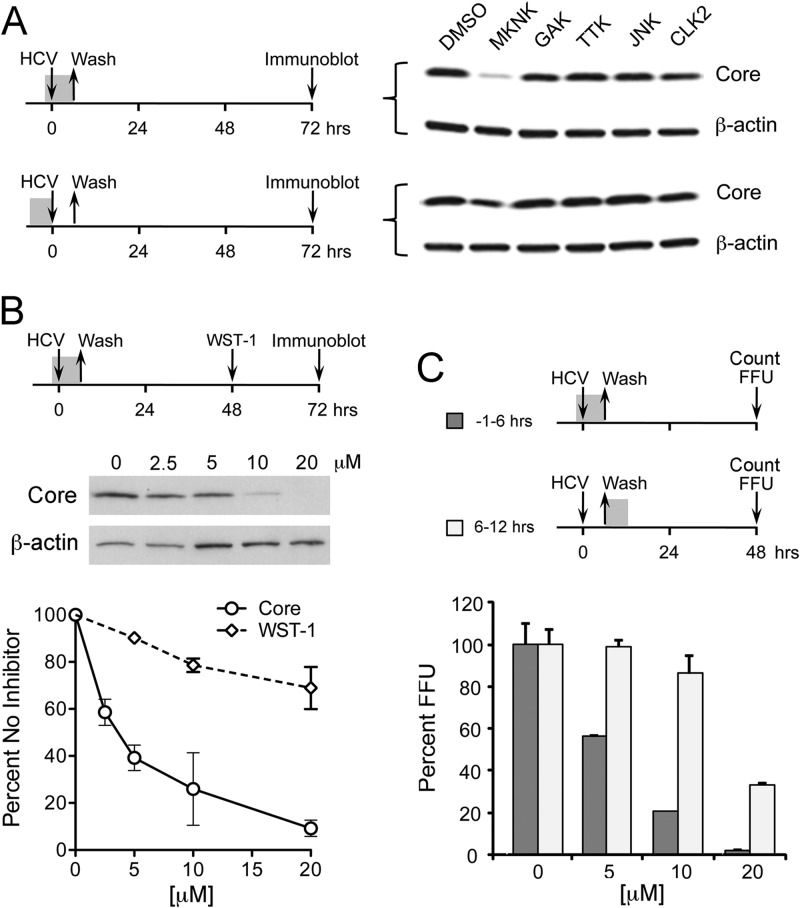
The specificity of chemical inhibitors of kinases is notoriously difficult to control, as exemplified by these results. Several kinases have been implicated previously in HCV entry, as described above, and the JNK inhibitors are likely to have nonspecific activity against an additional, unidentified kinase involved in this process. To identify this kinase, we screened a panel of chemical inhibitors targeting four candidate kinases, each of which was considered to be subject to off-target inhibition by the JNK inhibitors studied above (unpublished data from Roche): RO5096129, targeting cyclin G-associated kinase (GAK); RO4475417, targeting MKNK; RO4567182, targeting CDC-like kinase 2 (CLK2); and RO5071631, targeting TTK protein kinase (TTK). We also tested RO5108581, a novel inhibitor with high specificity for JNK. Each of these compounds inhibits its target kinase by more than 90% at 10 μM (data not shown), the concentration used for initial screening experiments. Cells were treated with compound for 1 h prior to inoculation with HJ3-5 virus (MOI of 1), following which the virus was allowed to adsorb to cells in the presence of the compound for 6 h. This was followed by extensive washing of the cells and refeeding with normal media (no compound). Under these conditions, the MKNK inhibitor (RO4475417) caused a significant decrease in HCV core protein expression, as measured by immunoblots of cell lysates prepared 72 h postinfection (Fig. 4A, upper). Importantly, the compound had a much more limited effect on core protein expression when it was used to treat the cells for 6 h prior to infection and washed out prior to virus inoculation (Fig. 4A, lower), while the other inhibitors had no effect under either set of conditions. These results are consistent with the MKNK inhibitor blocking an early step in the virus life cycle.
Fig 4.
Screening of kinases that may be targeted nonspecifically by JNK inhibitors. (A) Huh-7.5 cells were treated with selected kinase inhibitors (10 μM concentration) for either (top) 1 h prior to infection by HJ3-5 virus and during a 6-h period of viral adsorption thereafter, or (bottom) 6 h prior to virus inoculation. Kinases included RO4475417, which inhibits MKNK; RO5096129, an inhibitor of GAK; RO5071631, a TTK protein kinase inhibitor; RO5108581, a JNK inhibitor; and RO4567182, a CLK2 inhibitor. Following virus adsorption, cells were cultured in the absence of inhibitors. Cell lysates were prepared on day 3 for immunoblotting for HCV core protein. (B) Comparison of HCV inhibitory and cytotoxic effects of increasing concentrations of RO4475417 (MKNK inhibitor). Huh-7.5 cells were pretreated with the inhibitor for 1 h prior to a 6-h HJ3-5 virus adsorption period, then they were washed prior to refeeding with medium containing no inhibitors. HCV inhibitory activity was assessed by quantitative immunoblotting for core protein at 72 h, while cytotoxicity was determined by WST-1 cellular proliferation assay 48 h after inhibitor treatment (the midpoint in the virus growth period). Means ± standard errors (SE) were calculated from triplicate experiments. (C) RO4475417 inhibition of infectious focus formation. Huh-7.5 cells were treated with increasing concentrations of compound for either 1 h prior to and then during a 6-h period of HJ3-5 virus adsorption (−1 to 6 h) or a 6-h period following virus adsorption (6 to 12 h), as shown schematically. Cells were then washed and refed with media containing no compound. Cells were fixed at 48 h, and FFU were enumerated as described in the legend to Fig. 3C. Means ± SE were calculated from two independent experiments.
To more fully characterize this effect, we treated cells with a range of concentrations of RO4475417 for 1 h prior to and during a 6-h period of virus adsorption (Fig. 4B). We then compared core protein expression (quantitative immunoblotting) at 72 h to effects on cellular metabolism (WST-1 assay) at 48 h. The reduction in core protein expression was dose related and virtually complete at 20 μM. At this concentration, RO4475417 had only a modest negative effect in the WST-1 assay, consistent with a role for MKNK in cell death and survival (46). We also infected cells at a low multiplicity in the presence or absence of RO4475417 and counted the number of foci of infected cells formed at 48 h (Fig. 4C). Increasing concentrations of the MKNK inhibitor, added 1 h prior to infection and continued through a 6-h virus adsorption period, resulted in a progressive decrease in the number of foci, with very few foci present at the highest concentration tested, 20 μM. In contrast, the addition of RO4475417 to the cultures for a 6-h period beginning at the end of the virus adsorption period had much less effect on the focus count. This difference was dramatic at a 10 μM concentration of RO4475417 (Fig. 4C).
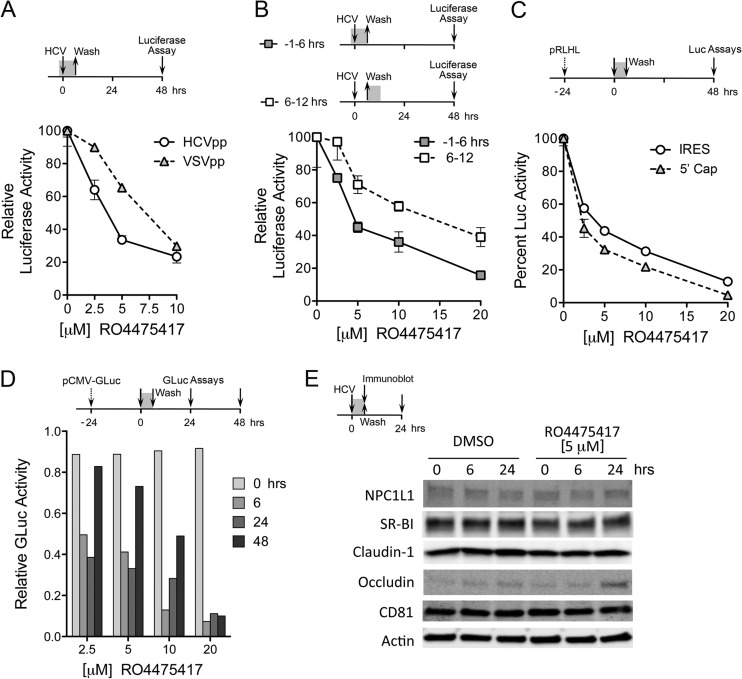
To confirm that the MKNK inhibitor RO4475417 inhibits a step in viral entry, we assessed its ability to block entry of pseudotyped HCV particles (HCVpp) (37). These experiments are complicated by the fact that MKNK contributes to the regulation of eukaryotic translation through its phosphorylation of eIF4E, a rate-limiting component of the eukaryotic 7-methylguanosine cap-binding 4F complex (47). Inhibition of MKNK thus reduces the efficiency of cap-dependent translation and therefore would be expected to nonspecifically reduce the expression of luciferase from pseudotyped lentivirus particles. Nonetheless, we observed differences in the ability of RO4475417 to suppress luciferase expression mediated by infection with HCVpp compared to that of control particles pseudotyped with the vesicular stomatitis virus envelope (VSVpp) when cells were treated with 2.5 to 5.0 μM from 1 h before to 6 h after inoculation (Fig. 5A). In replicate independent experiments, the mean 50% infectious concentration (IC50) of RO4475417 for inhibition of HCVpp entry was 3.35 ± 0.05 μM, and that for VSVpp entry was 6.95 ± 1.05 μM (mean P value of <0.01 by the extra-sum-of-squares F test). Higher concentrations of the inhibitor led to nearly complete suppression of luciferase expression induced by both HCVpp and VSVpp, consistent with an effect on cap-dependent translation (Fig. 5A). To control for nonspecific suppression of luciferase translation, we compared the effects of RO4475417 on HCVpp-mediated luciferase expression when cells were treated with the compound during infection (from 1 h before to 6 h after inoculation) versus the 6-h period immediately following (from 6 to 12 h postinoculation) (Fig. 5B). There was significantly less reduction in luciferase under the latter conditions, indicating that the effect of RO4475417 is greatest when it is present during HCVpp entry (Fig. 5B). In contrast, the JNK inhibitor RO5108581 demonstrated no dose-related effect on entry of HCVpp (data not shown).
Fig 5.
MKNK inhibition and cellular entry of HCVpp. (A) Impact of increasing concentrations of RO4475417 (MKNK inhibitor) on luciferase expressed by HCVpp versus VSVpp. Cells were treated with the inhibitor for 1 h prior to and for 6 h during pseudotyped particle infection. Luciferase activity was assayed at 48 h, as shown at the top. Results shown represent mean ± SD relative luciferase activity in triplicate cultures and are representative of two independent experiments. (B) Inhibition of HCVpp-mediated luciferase expression by RO4475417 when added to cell culture media 1 h prior to and during HCVpp infection (−1 to 6 h, as described for panel A) or following a 6-h incubation with HCVpp (6 to 12 h). Results shown represent mean ± SE relative luciferase activity. (C) Inhibition of cap-dependent and HCV IRES-dependent translation following a 6-h exposure to increasing concentrations of RO4475417 (MKNK inhibitor). Cells were transfected with pRLHL, which expresses a dicistronic transcript containing the HCV IRES within the intercistronic space. Twenty-four h later (time point 0), cells were treated with RO4475417 for 6 h. Cells were lysed and luciferase activities measured at 48 h. Results shown represent RLuc (cap-dependent translation initiation) and FLuc (IRES-dependent) activities relative to those in the absence of the compound (100%) and are the means ± SD from triplicate cultures. (D) Kinetic analysis of recovery from RO4475417-mediated inhibition of cap-dependent translation. Cells were transfected with pCMV-GLuc, which expresses secreted GLuc, 24 h prior to drug exposure, as described for panel C. Supernatant fluids were harvested for GLuc assay and replaced with fresh medium at 6, 24, and 48 h. (E) Lack of an effect of a 6-h exposure to RO4475417 (5 μM) on cellular abundance of the HCV coreceptors CD81, SR-B1, claudin-1, occludin, and NPC1L1. Cells were treated with compound for 6 h starting at time point 0 and harvested for immunoblot assays at 6 and 24 h.
Given the impressive effect of RO4475417 on luciferase expressed by VSVpp, we further characterized its effects on cellular translation. For this, we transfected cells with pRLHL (38), a dicistronic reporter plasmid in which translation of the upstream cistron (Renilla luciferase [RLuc]) is initiated in a cap-dependent fashion, while translation of the downstream cistron (firefly luciferase [FLuc]) is controlled by the HCV IRES placed within the intercistronic space. Increasing concentrations of RO4475417 strongly inhibited cap-dependent translation (Fig. 5C). Unexpectedly, the compound also inhibited HCV IRES-dependent translation under these conditions, albeit to a lesser extent. To examine the kinetics of RO4475417-mediated suppression of translation, we similarly treated cells transfected with pCMV-GLuc, a reporter plasmid that expresses secreted Gaussia princeps luciferase (GLuc) in a cap-dependent fashion. Cell culture supernatant fluids were collected and replaced at the end of a 6-h exposure to increasing concentrations of R04475417 and at 24 and 48 h after translation. At each interval, secreted GLuc activity was compared to that from untreated cells, allowing a real-time determination of the rate of recovery from suppression of translation. These results revealed a dose-dependent length of translation suppression (Fig. 5D), with 75% recovery by 48 h in cells treated with 5 μM the compound.
To assess whether suppression of cap-dependent translation by R04475417 results in reduced expression of known HCV coreceptors, we determined the abundance of CD81, SR-B1, claudin-1, occludin, and NPC1L1 proteins in lysates of cells exposed to a 5 μM concentration of the compound for 6 h (Fig. 5E). Remarkably, this revealed no loss of expression of any of these receptor molecules either during the period of drug exposure or over the ensuing 24 h. Thus, despite the strong effects of the compound on translation, its ability to inhibit HCV entry cannot be attributed to impaired expression of known cellular receptors.
siRNA-mediated depletion confirms a role for MKNK1 in HCV entry.
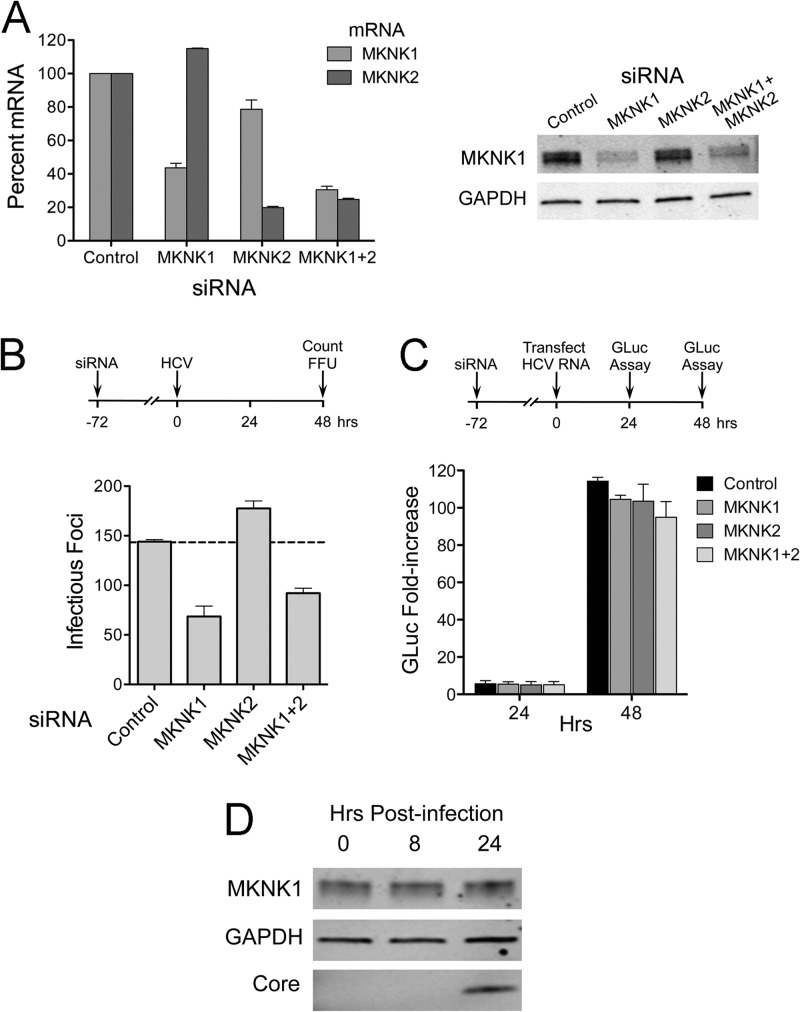
There are two genetically distinct isoforms of MKNK, MKNK1 and MKNK2 (48). To confirm that entry inhibition by RO4475417 was indeed related to inhibition of MKNK and to identify the specific isoform involved, we transfected cells with siRNAs targeting either or both isoforms. Three days after transfection, the cells were infected with HJ3-5 virus, and infectious foci were enumerated 2 days later. These knockdowns proved to be difficult, but ∼60% knockdown of MKNK expression was confirmed by qRT-PCR (MKNK1 and MKNK2) or immunoblotting (MKNK1 only, due to a lack of antibodies specific for MKNK2) (Fig. 6A, left and right). MKNK1 knockdown reproducibly caused a >50% reduction in the number of infectious foci in cells inoculated with HJ3-5 virus, while MKNK2 knockdown caused a slight but reproducible increase (Fig. 6B). The decreased numbers of infectious foci following MKNK1 knockdown could not be attributed to impaired viral RNA replication, as depletion of neither MKNK1 nor MKNK2 significantly reduced replication of a transfected viral RNA (HJ3-5/GLuc2A) in which the GLuc reporter was inserted between p7 and NS2 of the polyprotein (Fig. 6C).
Fig 6.
RNAi-mediated knockdown of MKNK1 inhibits HCV cell entry. (A) (Left) RNAi-mediated decreases in MKNK mRNAs. Total cellular RNA was extracted from cells following transfection with MKNK1- and MKNK2-specific siRNAs and assayed for MKNK1 and MKNK2 mRNAs by qRT-PCR. Means ± SD were calculated from replicate experiments. (Right) Immunoblots of MKNK1 in lysates from siRNA-transfected cells. MKNK2-specific antibody was not available. (B) Number of infectious foci in HJ3-5 virus-inoculated cells following knockdown of MKNK1 and MKNK2. Huh-7.5 cells were infected by HJ3-5 virus 3 days after siRNA transfection. Two days later, foci of infected cells were visualized by indirect immunofluorescence and enumerated. Results shown represent means ± SE from two independent experiments. (C) Effect of MKNK knockdown on HCV RNA replication. Cells were transfected with HJ3-5/GLuc2A RNA 72 h following transfection with the indicated siRNAs. GLuc activities secreted by the transfected cells were measured and plotted as fold change relative to GLuc present 6 h after HCV RNA transfection (33). Means ± SE were calculated from two independent experiments. (D) Immunoblot showing MKNK1 expression in Huh-7.5 cells following infection with HJ3-5 virus (MOI of ∼5). Lysates were prepared at 0 h (before infection) and 8 and 24 h after infection and were subjected to immunoblotting for MKNK1, GAPDH, and HCV core protein.
Sustained suppression of cellular translation following siRNA knockdown of MKNK (as opposed to short-term chemical inhibition, as shown in Fig. 4C) is likely to interfere with the interpretation of experiments using pseudotyped lentivirus particles, since their entry is monitored by measuring cap-dependent expression of a luciferase reporter protein. Indeed, while we observed a 39% ± 10% (means ± standard errors of the means [SEM]) decrease in luciferase activity expressed by HCVpp following MKNK1 knockdown in 3 independent experiments, similar reductions were observed with control VSVpp particles (data not shown). These results are consistent with general inhibition of translation. In contrast, knockdown of MKNK2 resulted in a modest increase (17% ± 15%) in luciferase expressed by HCVpp.
Despite the confounding effects of MKNK1 inhibition or depletion on cellular translation, the data presented in Fig. 5 and 6 collectively provide strong evidence for a role for MKNK1 in HCV cell entry. However, we observed no significant change in the expression level of MKNK1 at 8 or 24 h after infection at a high MOI (MOI = 5) (Fig. 6D). We did not assess changes in its phosphorylation status of MKNK1 under these conditions due to a lack of phospho-MKNK1-specific antibodies.
DISCUSSION
JNKs are an evolutionarily conserved subgroup of the MAPK family that exert a broad influence on a variety of cellular functions, such as proliferation, apoptosis, differentiation, and development. Previous studies suggested that both ERK and p38, other MAPKs, act to restrict HCV replication. We have demonstrated that ERK activates mTOR in cooperation with the phosphoinositide-3 kinase pathway, in turn activating p70 S6 kinase and eIF4E binding protein and suppressing HCV replication (7). Zhu and Liu (11) also reported that IL-1β acts to inhibit replication in part through activation of ERK. Similarly, p38 activation may contribute to interferon-α-mediated anti-HCV activity in association with activation of MAPK-activated protein kinase 2 (MAPKAPK2) (12). Although these observations indicate that MAPKs are able to negatively regulate HCV replication, the JNKs have not been investigated previously. We show here, using a cell line harboring a subgenomic HCV RNA replicon, that both chemical inhibition and RNAi-mediated knockdown of JNK expression enhances HCV protein and RNA abundance (Fig. 1 and 2A), while JNK activation induced by overexpression of JNK2 and MKK73E suppresses replicon amplification as well as replication of infectious virus in cell culture (Fig. 2B). These data lead us to conclude that JNK activation suppresses HCV replication, similar to what has been observed previously with ERK and p38, other members of the MAPK family.
JNK is known to modulate the replication of certain viruses. Herpes simplex virus (49), Kaposi's sarcoma-associated herpesvirus (50), rhesus rotavirus (51), and human immunodeficiency virus 1 (52) are all suppressed by JNK inhibition, while inhibitors of JNK enhance replication of influenza virus (53) and varicella-zoster virus (54). In contrast, coxsackievirus B3 yields are not altered by JNK inhibition with SP600125 (55). Hence, the effects of JNK appear to vary with different viruses, possibly reflecting differences in the host cells and culture conditions as much as in the virus itself. Ludwig et al. (53) demonstrated that JNK inhibition suppressed AP-1-dependent IFN-β promoter activity, resulting in increased yields of influenza virus. We did not detect any change in basal IFN-β promoter activity in RG cells following siRNA knockdown of JNK (data not shown). This may reflect an absence of IFN-β promoter activation in the replicon cells, because the HCV NS3/4A protease severely restrains IRF3 activation by directing the proteolysis of MAVS and TRIF, key adaptor proteins in RIG-I and TLR-3 signaling, respectively (56). Thus, IRF3-activated genes, including IFN-β, are unlikely to play a role in the suppression of HCV replication by JNK, and the mechanism by which this occurs remains to be completely elucidated.
In sharp contrast to RNAi-mediated knockdown of JNK (Fig. 2A), the JNK inhibitors AS601245 and SP600125 demonstrated an inhibitory effect on infection with cell-free HCV (Fig. 2C). There is little doubt about the specificity of the knockdown, as we observed similar results with 4 different siRNAs targeting distinct JNK sequences (data not shown). Thus, we attribute this difference and the inhibition of de novo HCV infection (Fig. 2C) to an off-target effect of the chemical compounds. This is not surprising, as Bain et al. (57) reported that SP600125 inhibited 13 of 28 kinases tested with potency at least as great as its activity against JNK. Although these results were derived from in vitro assays, the compound is likely to have similar broad effects on kinases in vivo. The specificity of AS601245 has not been similarly examined, but it would be no surprise were it to similarly target a variety of related kinases. Based on the potential for such off-target effects, we selected 4 kinases as candidates for further study, GAK, CLK2, TTK, and MKNK, and assessed the ability of chemical compounds targeting these kinases to interfere with HCV infection. Of these, RO4475417, an inhibitor targeting MKNK, was uniquely capable of blocking both an early step in HCV infection (Fig. 4) and HCVpp entry (Fig. 5). While this compound has strong, negative effects on both cap-dependent and HCV IRES-dependent translation (Fig. 5C and D), it did not reduce the abundance of known HCV receptor molecules under the conditions in which it was used (Fig. 5E).
As with the JNK inhibitors we tested, it is possible that the effects we observed with RO4475417 treatment could in part reflect off-target inhibition of a kinase other than MKNK. Although the compound has demonstrated a high degree of specificity against an expanded panel of kinases, it is designed to compete with ATP in the binding site and thus could block the activity of another kinase. However, unlike the results we obtained with RNAi-mediated knockdown of JNK, MKNK1 knockdown provided strong support for a specific role for MKNK1 in HCV entry, as it significantly reduced the number of cells successfully infected by HCV in a fluorescent focus formation assay (Fig. 6B) while having no effect on replication of transfected viral RNA (Fig. 6C). These results could not be confirmed using pseudotyped lentivirus particles due to suppression of cellular translation following MKNK1 knockdown. Taken collectively, however, the results we obtained with authentic infectious HCV following chemical (Fig. 4) or genetic (Fig. 6) knockdown of MKNK1 strongly support a specific role for this kinase in HCV entry, or possibly another very early step in replication of infectious virus, such as uncoating of the viral genome.
The MKNKs were first discovered as ERK substrates (58, 59) and comprise four proteins, with two isotypes produced from each of two genes (MKNK1 and MKNK2) (48, 60). These differ in their cellular localization and degree of regulation by ERK, with MKNK1a being predominantly cytoplasmic and highly regulated by ERK and p38 MAP kinases. MKNK1b differs from MKNK1a in lacking a C-terminal MAPK-binding domain and nuclear export signal, and it is at least partly nuclear in its distribution and minimally regulated by ERK/p38 kinases (48, 60). MKNK1 is involved in stress responses, cytokine expression, and control of translation. It positively regulates cap-dependent cellular translation via phosphorylation of the cap-binding protein, eIF4E, a rate-limiting component of the cap-binding complex eIF4F, and its promotion of translation is important for replication of herpesviruses in quiescent cells (61, 62) and reactivation of herpesviruses from latency (63). MKNK1-mediated phosphorylation of eIF4E also stimulates picornaviral cap-independent translation, and its induction by oncogenic Ras has been associated with the growth of oncolytic polioviruses in glioblastoma cells (64). While MKNK1 is expressed in liver tissue, it has not been recognized previously to be a host factor in the HCV life cycle.
Substantial prior evidence supports an important role for kinases in HCV entry. Lupberger et al. (30) recently reported the results of a genome-wide siRNA screen of kinases involved in HCV entry. Their primary screen identified a total of 106 kinases potentially involved in HCVpp entry. Included among these was MKNK2 but, interestingly, not MKNK1. Further screening with infectious virus and individual siRNAs reduced this list to 58 kinases involved in HCV entry, and apparently it excluded any role for MKNK2 (30). These authors subsequently focused on kinases for which specific inhibitors have been approved for clinical use and demonstrated that the epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2), both receptor tyrosine kinases, act as host factors in HCV entry. EGFR activation appeared to enhance a late step in HCV entry by facilitating interactions between the virus coreceptors CD81 and CLDN1. Exactly how this happens, and what events occur downstream of the receptor tyrosine kinases, is not clear. Brazzoli et al. (31) demonstrated that CD81 engagement activates several Rho GTPases, and that this mediates actin-dependent relocalization of CD81-E2 complexes toward the tight junction. This was shown to occur in association with Raf/MEK(MAP2K)/ERK signaling, which could lead to MKNK1 activation. U0126, a chemical inhibitor of MEK-1 and MEK-2, inhibited HCV infection, and MEK signaling appeared to be required for a very late entry or early postentry step in replication (31).
Based on these prior studies, it is possible that MKNK1 is activated by ERK downstream of Ras GTPase signaling induced by the receptor tyrosine kinases during HCV entry. While MKNK1 is likely to be part of a complex network of kinases that are activated during viral entry (30), the sizable impact of chemical and genetic inhibition of MKNK1 activity on HCV infection suggests that it plays a significant role in this process, at least in the Huh-7 hepatoma cells we studied. At the same time, it is clear that each of the 3 MAPKs, ERK, p38, and JNK (as shown here), exert strong negative effects on HCV RNA replication. These contrasting effects on viral entry and viral RNA replication are surprising and suggest that there must be exquisite dynamic regulation of their activities both temporally and spatially during the infection process.
ACKNOWLEDGMENTS
We thank Charles Rice for Huh-7.5 cells, Craig Cameron for NS5A antibody, Takaji Wakita for reagents related to pseudotyped viruses, and Lemon laboratory members for helpful comments and suggestions.
This study was supported in part by grants from the National Institutes of Health (RO1-DA024565, RO1-AI095690, and P20-CA150343).
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Seeff LB. 1997. The natural history of chronic hepatitis C virus infection. Clin. Liver Dis. 1:587–602 [DOI] [PubMed] [Google Scholar]
- 2. Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 4. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fimia GM, Evangelisti C, Alonzi T, Romani M, Fratini F, Paonessa G, Ippolito G, Tripodi M, Piacentini M. 2004. Conventional protein kinase C inhibition prevents alpha interferon-mediated hepatitis C virus replicon clearance by impairing STAT activation. J. Virol. 78:12809–12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishida H, Li K, Yi M, Lemon SM. 2007. p21-activated kinase 1 is activated through the mammalian target of rapamycin/p70 s6 kinase pathway and regulates the replication of hepatitis C virus in human hepatoma cells. J. Biol. Chem. 282:11836–11848 [DOI] [PubMed] [Google Scholar]
- 8. Mannova P, Beretta L. 2005. Activation of the N-Ras-PI3K-Akt-mTOR pathway by hepatitis C virus: control of cell survival and viral replication. J. Virol. 79:8742–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murata T, Ohshima T, Yamaji M, Hosaka M, Miyanari Y, Hijikata M, Shimotohno K. 2005. Suppression of hepatitis C virus replicon by TGF-beta. Virology 331:407–417 [DOI] [PubMed] [Google Scholar]
- 10. Murata T, Hijikata M, Shimotohno K. 2005. Enhancement of internal ribosome entry site-mediated translation and replication of hepatitis C virus by PD98059. Virology 340:105–115 [DOI] [PubMed] [Google Scholar]
- 11. Zhu H, Liu C. 2003. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J. Virol. 77:5493–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishida H, Ohkawa K, Hosui A, Hiramatsu N, Kanto T, Ueda K, Takehara T, Hayashi N. 2004. Involvement of p38 signaling pathway in interferon-alpha-mediated antiviral activity toward hepatitis C virus. Biochem. Biophys. Res. Commun. 321:722–727 [DOI] [PubMed] [Google Scholar]
- 13. Turjanski AG, Vaque JP, Gutkind JS. 2007. MAP kinases and the control of nuclear events. Oncogene 26:3240–3253 [DOI] [PubMed] [Google Scholar]
- 14. Weston CR, Davis RJ. 2007. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19:142–149 [DOI] [PubMed] [Google Scholar]
- 15. Takeuchi O, Akira S. 2001. Toll-like receptors; their physiological role and signal transduction system. Int. Immunopharmacol. 1:625–635 [DOI] [PubMed] [Google Scholar]
- 16. Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 17. Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C, Soulier E, Schvoerer E, Schuster C, Stoll-Keller F, Bartenschlager R, Pietschmann T, Barth H, Baumert TF. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731 [DOI] [PubMed] [Google Scholar]
- 20. Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 21. Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 18:281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003–41012 [DOI] [PubMed] [Google Scholar]
- 24. Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. 2007. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46:411–419 [DOI] [PubMed] [Google Scholar]
- 26. Meertens L, Bertaux C, Dragic T. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. 2009. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5:e1000702 doi:10.1371/journal.ppat.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ploss A, Evans MJ. 2012. Hepatitis C virus host cell entry. Curr. Opin. Virol. 2:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farquhar MJ, Harris HJ, Diskar M, Jones S, Mee CJ, Nielsen SU, Brimacombe CL, Molina S, Toms GL, Maurel P, Howl J, Herberg FW, van Ijzendoorn SC, Balfe P, McKeating JA. 2008. Protein kinase A-dependent step (s) in hepatitis C virus entry and infectivity. J. Virol. 82:8797–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ikeda M, Yi M, Li K, Lemon SM. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shimakami T, Yamane D, Welsch C, Hensley L, Jangra RK, Lemon SM. 2012. Base pairing between hepatitis C Virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 86:7372–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell GA, Lemon SM. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152–162 [DOI] [PubMed] [Google Scholar]
- 39. Kim S, Welsch C, Yi M, Lemon SM. 2011. Regulation of the production of infectious genotype 1a hepatitis C virus by NS5A domain III. J. Virol. 85:6645–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY [Google Scholar]
- 41. Wolter S, Mushinski JF, Saboori AM, Resch K, Kracht M. 2002. Inducible expression of a constitutively active mutant of mitogen-activated protein kinase kinase 7 specifically activates c-JUN NH2-terminal protein kinase, alters expression of at least nine genes, and inhibits cell proliferation. J. Biol. Chem. 277:3576–3584 [DOI] [PubMed] [Google Scholar]
- 42. Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waetzig V, Herdegen T. 2005. Context-specific inhibition of JNKs: overcoming the dilemma of protection and damage. Trends Pharmacol. Sci. 26:455–461 [DOI] [PubMed] [Google Scholar]
- 44. Holtmann H, Winzen R, Holland P, Eickemeier S, Hoffmann E, Wallach D, Malinin NL, Cooper JA, Resch K, Kracht M. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chrestensen CA, Eschenroeder A, Ross WG, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Sturgill TW. 2007. Loss of MNK function sensitizes fibroblasts to serum-withdrawal induced apoptosis. Genes Cells 12:1133–1140 [DOI] [PubMed] [Google Scholar]
- 47. Shveygert M, Kaiser C, Bradrick SS, Gromeier M. 2010. Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol. Cell. Biol. 30:5160–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buxade M, Parra-Palau JL, Proud CG. 2008. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front. Biosci. 13:5359–5373 [DOI] [PubMed] [Google Scholar]
- 49. McLean TI, Bachenheimer SL. 1999. Activation of cJUN N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J. Virol. 73:8415–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan H, Xie J, Ye F, Gao SJ. 2006. Modulation of Kaposi's sarcoma-associated herpesvirus infection and replication by MEK/ERK, JNK, and p38 multiple mitogen-activated protein kinase pathways during primary infection. J. Virol. 80:5371–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Holloway G, Coulson BS. 2006. Rotavirus activates JNK and p38 signaling pathways in intestinal cells, leading to AP-1-driven transcriptional responses and enhanced virus replication. J. Virol. 80:10624–10633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muthumani K, Wadsworth SA, Dayes NS, Hwang DS, Choo AY, Abeysinghe HR, Siekierka JJ, Weiner DB. 2004. Suppression of HIV-1 viral replication and cellular pathogenesis by a novel p38/JNK kinase inhibitor. AIDS 18:739–748 [DOI] [PubMed] [Google Scholar]
- 53. Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S. 2001. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J. Biol. Chem. 276:10990–10998 [PubMed] [Google Scholar]
- 54. Rahaus M, Desloges N, Wolff MH. 2004. Replication of varicella-zoster virus is influenced by the levels of JNK/SAPK and p38/MAPK activation. J. Gen. Virol. 85:3529–3540 [DOI] [PubMed] [Google Scholar]
- 55. Si X, Luo H, Morgan A, Zhang J, Wong J, Yuan J, Esfandiarei M, Gao G, Cheung C, McManus BM. 2005. Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J. Virol. 79:13875–13881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lemon SM. 2010. Induction and evasion of innate antiviral responses by hepatitis C virus. J. Biol. Chem. 285:22741–22747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bain J, McLauchlan H, Elliott M, Cohen P. 2003. The specificities of protein kinase inhibitors: an update. Biochem. J. 371:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fukunaga R, Hunter T. 1997. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16:1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goto S, Yao Z, Proud CG. 2009. The C-terminal domain of Mnk1a plays a dual role in tightly regulating its activity. Biochem. J. 423:279–290 [DOI] [PubMed] [Google Scholar]
- 61. Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 5:e1000334 doi:10.1371/journal.ppat.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goetz C, Everson RG, Zhang LC, Gromeier M. 2010. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol. Ther. 18:1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]