Abstract
Drug resistance occurs through a series of subtle changes that maintain substrate recognition but no longer permit inhibitor binding. In HIV-1 protease, mutations at I50 are associated with such subtle changes that confer differential resistance to specific inhibitors. Residue I50 is located at the protease flap tips, closing the active site upon ligand binding. Under selective drug pressure, I50V/L substitutions emerge in patients, compromising drug susceptibility and leading to treatment failure. The I50V substitution is often associated with amprenavir (APV) and darunavir (DRV) resistance, while the I50L substitution is observed in patients failing atazanavir (ATV) therapy. To explain how APV, DRV, and ATV susceptibility are influenced by mutations at residue 50 in HIV-1 protease, structural and binding thermodynamics studies were carried out on I50V/L-substituted protease variants in the compensatory mutation A71V background. Reduced affinity to both I50V/A71V and I50L/A71V double mutants is largely due to decreased binding entropy, which is compensated for by enhanced enthalpy for ATV binding to I50V variants and APV binding to I50L variants, leading to hypersusceptibility in these two cases. Analysis of the crystal structures showed that the substitutions at residue 50 affect how APV, DRV, and ATV bind the protease with altered van der Waals interactions and that the selection of I50V versus I50L is greatly influenced by the chemical moieties at the P1 position for APV/DRV and the P2 position for ATV. Thus, the varied inhibitor susceptibilities of I50V/L protease variants are largely a direct consequence of the interdependent changes in protease inhibitor interactions.
INTRODUCTION
With no cure available for the treatment of human immunodeficiency virus (HIV) infection at present, slowing down the progression of the infection to AIDS has been a major focus in anti-HIV therapy development. In this effort, the aspartyl protease of HIV-1 has been an important drug target. Presently, there are nine FDA-approved protease inhibitors (PIs), and some of these are an important part of highly active antiretroviral therapy (HAART) (1). However, the selection of HIV-1 variants with inhibitor resistance mutations in the protease gene impairs the ability of PIs to effectively block protease activity (2–4).
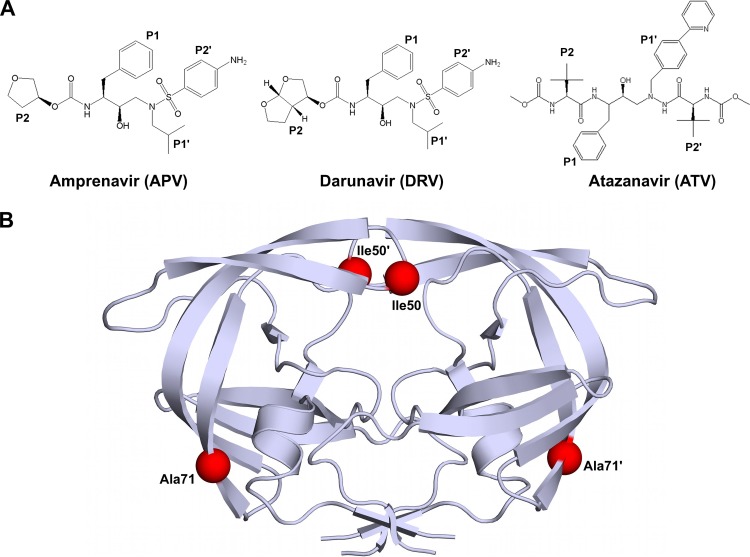
The HIV-1 protease is a homodimeric protein with 99 amino acid residues in each monomer. At least 10 nonhomologous and asymmetric substrate sites within the Gag and Gag-Pro-Pol polyproteins are cleaved by the HIV-1 protease to release viral enzymes and structural proteins essential for virion maturation (5–8). Drug resistance emerges under the selective pressure of inhibitor therapy when the protease mutates to no longer efficiently bind PIs but continue to cleave substrates. Many major primary drug resistance mutations observed in the clinic occur at the flap region of protease, which is critical in controlling ligand (substrate and inhibitor) access to the active site. In particular, the substitutions accumulating at the active-site residue position 50, located at the flap tip (Fig. 1B), are commonly associated with resistance to amprenavir (APV), darunavir (DRV), and atazanavir (ATV), three potent FDA-approved PIs (Fig. 1A) (8–10).
Fig 1.
(A) Chemical structures of APV, DRV, and ATV. Chemical groups that correspond to substrate residues when bound within the active site are indicated P1, P2, P1′, and P2′. (B) Homodimeric HIV-1 protease with mutation sites I50 and A71 indicated by red spheres in each monomer.
The Ile-to-Val substitution at residue 50 (I50V) is the signature resistance mutation in patients failing APV and DRV therapy (11–14). On the other hand, mutation to Leu at this position (I50L) is observed in patients failing ATV therapy (15, 16). However, patients with the I50L substitution in protease respond significantly better to PIs other than ATV, indicating that the I50L substitution renders the protease hypersusceptible to other PIs (16). The substitutions at residue 50 are often observed together with a secondary A71V mutation that is outside the active site (Fig. 1B). More than 60 and 50% of patient sequences in the HIV drug resistance database (17) with the I50L and I50V mutations, respectively, have the A71V comutation. The A71V substitution compensates for the loss of viral fitness resulting from primary drug resistance mutations (18). Due to their high clinical significance, the I50L/A71V and I50V/A71V double mutations have been studied for their effect on binding a few PIs, mostly by modeling and computation (19). However, a detailed comparative thermodynamic and X-ray structural analysis on binding of the three clinically significant PIs to these two double mutants is missing.
In the present study, structural and biophysical methods were used to determine the impact of substitutions at residue 50 on APV, DRV, and ATV susceptibility. Binding thermodynamics and X-ray crystal structures were obtained for protease with I50V/L and A71V mutations. The in vitro binding affinities agree well with clinical observations in confirming that the I50V and I50L substitutions differentially affect protease susceptibility to APV, DRV, and ATV. Both double mutants display reduced binding entropy compared to wild-type (WT) protease, and the extent of enthalpic compensation of this reduction determines the changes in inhibitor susceptibility. The crystal structures of protease inhibitor complexes reveal that the I50(V,L) and A71V mutations cause significant changes in van der Waals (vdW) contacts between the inhibitor and protease and, hence, provide insights into the molecular basis for varied inhibitor susceptibility.
MATERIALS AND METHODS
Nomenclature.
The following nomenclature is followed to refer to each inhibitor complex: inhibitorprotease variant. For example, APVWT, APVI50V/A71V, and APVI50L/A71V refer to the WT, I50V, and I50L variants in complex with APV. Prime notation is used to distinguish the two monomers in the protease dimer according to the binding orientation of the ligand in the dimer active site. For example, residue 30 from the first monomer is referred to as D30 if it interacts with the N terminus of the ligand. The same residue from the second monomer is referred to as D30′.
Protease gene construction.
The WT protease gene was generated as previously described (20), with the Q7K substitution introduced to prevent autoproteolysis (21). I50V/A71V and I50L/A71V variants were generated by introducing the appropriate mutations into the wild-type gene by site-directed mutagenesis using a Stratagene QuikChange site-directed mutagenesis kit (Agilent Technologies, La Jolla, CA). Mutagenesis was confirmed by DNA sequencing.
Protein expression and purification.
Each variant was subcloned into the heat-inducible pXC35 expression vector (American Type Culture Collection [ATCC], Manassas, VA) and transformed into Escherichia coli TAP-106 cells. Protein overexpression, purification, and refolding were carried out as previously described (22). Protein used for crystallographic studies was further purified with a Pharmacia Superdex 75 fast-performance liquid chromatography column (GE Healthcare, Chalfont St. Giles, United Kingdom) equilibrated with refolding buffer (50 mM sodium acetate, pH 5.5, 10% glycerol, 5% ethylene glycol, 5 mM dithiothreitol).
ITC.
Binding affinities and thermodynamic parameters of APV, DRV, and ATV binding to the WT, I50V/A71V, and I50L/A71V protease variants were determined by isothermal titration calorimetry (ITC) on a VP isothermal titration calorimeter (MicroCal, LLC, Northampton, MA). The buffer used for all protease and inhibitor solutions consisted of 10 mM sodium acetate (pH 5.0), 2% dimethyl sulfoxide, and 2 mM tris(2-carboxyethyl)phosphine. Binding affinities for all protease variants were obtained by competitive-displacement titration using acetyl-pepstatin as the weaker binder. A solution of 6 to 40 μM protease was titrated with 200 to 400 μM acetyl-pepstatin to saturation. The pepstatin was then displaced by titrating 200 to 330 μM APV, DRV, or ATV. Heats of dilution were subtracted from the corresponding heats of reaction to obtain the heat resulting solely from the binding of the ligand to the enzyme. All experiments were carried out at 20°C. Data were processed and analyzed with the ITC data analysis module (Microcal) for Origin 7 data analysis and graphing software (OriginLab, Northampton, MA). Final results represent the average of at least two measurements.
Crystallization and structure determination.
Protease solutions of WT, I50V/A71V, and I50L/A71V at concentrations between 1.0 and 2.0 mg ml−1 were equilibrated with a 3- to 5-fold molar excess of APV, DRV, and ATV for 1 h on ice. Crystals were grown over a reservoir solution consisting of 126 mM phosphate buffer (pH 6.2), 63 mM sodium citrate, and 18% to 40% ammonium sulfate by the hanging-drop vapor-diffusion method. X-ray diffraction data for I50V/A71V in complex with DRV were collected on BioCARS beamline 14-BMC at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) at a wavelength tuned to 0.9 Å with a Quantum 315 charge-coupled-device (CCD) X-ray detector (Area Detector Systems Corporation, Poway, CA). Diffraction data for I50V/A71V in complex with APV and ATV were collected on BioCARS beamline 14-IDB at the Advanced Photon Source (Argonne National Laboratory) at a wavelength tuned to 1.03 Å with a MarCCD 165 X-ray detector (Rayonix, LLC, Evanston, IL). Data for all other complexes were collected in-house on an R-Axis IV imaging-plate system (Rigaku Corporation, Tokyo, Japan) mounted on a rotating-anode X-ray source (Rigaku Corporation) at a wavelength of 1.54 Å. All data were collected under cryo-cooled conditions.
The data were indexed, integrated, and scaled using HKL/HKL-2000 software (HKL Research, Charlottesville, VA) (23). Structure determination and refinement were carried out using the CCP4 program suite (24), as previously described (25). The tensor (T), libration (L), and screw (S) parameter files used in TLS refinement of the I50V variant complexes were generated using the TLS Motion Determination server (26). Model building was carried out, followed by real-space refinement, with either the O molecular graphics software (27) or the COOT molecular graphics software (28). Structural comparisons were made by superimposing structures using the Cα atoms of the terminal regions (residues 1 to 9 and 86 to 99) from both monomers. Structures were visualized using PyMol molecular graphics software (29). Crystallographic and refinement statistics are given in Table 1. The DRVI50V, ATVI50V/A71V, and ATVI50L/A71V protease complexes crystallized with two protease dimers in the asymmetric unit. The APVI50V/A71V protease had a second inhibitor molecule bound outside the active site of one of the dimers and is likely a crystallographic artifact.
Table 1.
Crystallographic statistics for WT, I50V/A71V, and I50L/A71V variants in complex with APV, DRV, and ATV
| Parametera | Value for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT |
I50V/A71V |
I50L/A71V |
|||||||
| APV | DRVb | ATV | APV | DRV | ATV | APV | DRV | ATV | |
| Resolution (Å) | 1.8 | 1.2 | 1.8 | 1.6 | 1.9 | 1.6 | 2.2 | 2.1 | 2.1 |
| Space group | P212121 | P212121 | P212121 | P21 | P21 | P21 | P212121 | P212121 | P21 |
| Cell dimensions | |||||||||
| a (Å) | 50.7 | 54.9 | 50.9 | 50.6 | 50.62 | 51.2 | 50.9 | 51.4 | 51.2 |
| b (Å) | 57.9 | 57.8 | 58.1 | 63.3 | 63.34 | 58.4 | 58.1 | 57.9 | 59.5 |
| c (Å) | 61.7 | 62.0 | 62.0 | 58.6 | 58.64 | 62.1 | 61.2 | 61.4 | 59.9 |
| β (°) | 96.6 | 97.3 | 95.8 | 82.2 | |||||
| No. of reflections | |||||||||
| Total | 49,469 | 302,022 | 114,163 | 148,290 | 118,410 | 146,999 | 32,037 | 33,870 | 56,701 |
| Unique | 14,987 | 50,056 | 16,905 | 36283 | 26,674 | 42,520 | 9,311 | 10,762 | 20,006 |
| Rmerge (%) | 2.9 | 3.8 | 7.1 | 7.0 | 7.0 | 4.9 | 8.6 | 6.2 | 4.7 |
| Completeness (%) | 79.7 | 95.5 | 95.7 | 98.0 | 99.6 | 96.2 | 95.7 | 96.6 | 95.8 |
| I/σI | 19.0 | 25.0 | 11.3 | 8.7 | 8.4 | 11.6 | 6.4 | 7.9 | 9.8 |
| Rwork (%) | 19.6 | 14.1 | 17.7 | 16.9 | 16.3 | 17.3 | 19.6 | 18.0 | 19.0 |
| Rfree (%) | 22.6 | 17.9 | 20.9 | 20.5 | 21.2 | 21.5 | 25.8 | 24.0 | 24.8 |
| RMSD | |||||||||
| Bond length (Å) | 0.007 | 0.004 | 0.008 | 0.008 | 0.010 | 0.009 | 0.008 | 0.004 | 0.008 |
| Bond angle (°) | 1.4 | 1.5 | 1.2 | 1.4 | 1.4 | 1.4 | 1.4 | 1.2 | 1.4 |
| PDB accession no. | 3EKV | 1T3R | 3EKY | 3OXV | 3OXW | 3OXX | 3EM3 | 3EM6 | 3EM4 |
Rmerge = Σ|I − 〈I〉|/ΣI × 100, where I is the intensity of a reflection and 〈I〉 is the average intensity; I/σI is signal-to-noise ratio, Rwork = Σ|Fo–Fc|/Σ|Fo| × 100, where Fo is observed electron density and Fc is calculated electron density; Rfree was calculated from 5% of reflections, chosen randomly, which were omitted from the refinement process; RMSD, root mean square deviation.
Adapted from King et al. (9).
Distance-difference matrices were generated for each WT-inhibitor and mutant-inhibitor protease structure pair to reveal structural differences between the WT-inhibitor and mutant-inhibitor complexes, as previously described (25). Briefly, distances between all Cα atoms within the dimer were calculated for each complex. A distance-difference matrix was then computed for each atom for a given pair of complexes. The average deviation for each Cα atomic distance was calculated from the distance-difference matrix (average of the absolute values for each row or column), and the backbone structure was colored for increasing average deviation from blue to red using PyMol (29).
The inhibitor-protease vdW contacts were estimated for each of the nine complexes, DRVWT, DRVI50V/A71V, DRVI50L/A71V, APVWT, APVI50V/A71V, APVI50L/A71V, ATVWT, ATVI50V/A71V, and ATVI50L/A71V, using a simplified Lennard-Jones potential as previously described (30). The values for the WT were subtracted from those for the corresponding mutant-inhibitor complexes, and the differences were plotted as bar graphs.
RESULTS
Binding thermodynamics.
Inhibitor binding thermodynamics of double mutant I50V/A71V and I50L/A71V proteases were compared to those of WT protease by ITC (Table 2). In agreement with clinical observations and previous reports (31), the I50V variant had 2.2-fold and 16.7-fold weaker affinities for APV and DRV, respectively, than WT protease. Interestingly, we found that this variant had a 7.7-fold enhanced binding affinity for ATV (Kd = 0.03 nM versus 0.23 nM for WT). This is in contrast to the reduced affinity reported for the mutant with the I50V single mutation (32). Thus, the I50V/A71Vdouble mutation causes a reduced affinity for APV and DRV, while rendering the protease more susceptible to ATV.
Table 2.
Thermodynamic parameters for APV, DRV, and ATV binding to WT, I50V/A71V, and I50L/A71V HIV-1 proteases
| Inhibitor and protease variant | Kd (nM) | Kd ratioa | ΔH (kcal mol−1) | −TΔS (kcal mol−1) | ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| APV | |||||
| WT | 0.39 ± 0.20 | 1.0 | −7.3 ± 0.9 | −5.3 | −12.6 |
| I50V/A71V | 0.88 ± 0.02 | 2.2 | −11.8 ± 0.3 | −0.3 | −12.1 |
| I50L/A71V | 0.14 ± 0.02 | 0.4 | −9.5 ± 0.2 | −3.8 | −13.2 |
| DRV | |||||
| WT | 0.004 ± 0.002 | 1.0 | −12.1 ± 0.8 | −3.1 | −15.2 |
| I50V/A71V | 0.067 ± 0.021 | 16.7 | −12.0 ± 0.6 | −1.6 | −13.6 |
| I50L/A71V | 0.016 ± 0.001 | 4.0 | −13.4 ± 0.9 | −1.1 | −14.5 |
| ATV | |||||
| WT | 0.23 ± 0.12 | 1.0 | −1.1 ± 0.2 | −11.8 | −12.9 |
| I50V/A71V | 0.03 ± 0.01 | 0.1 | −7.3 ± 1.1 | −6.6 | −13.9 |
| I50L/A71V | 0.65 ± 0.05 | 2.8 | −3.6 ± 0.4 | −8.7 | −12.3 |
Fold change with respect to WT protease.
I50L is a signature mutation for ATV resistance, and as expected, the I50L/A71V variant exhibited a 2.8-fold weaker affinity for ATV. It is well established that the I50L mutation is associated with hypersusceptibility to PIs other than ATV (31), but its effect on the binding of the newest and most potent PI, DRV, is less known. We found that I50L/A71V variant had a 4.0-fold decreased affinity to DRV. Despite this weakening in affinity, the highly potent DRV nonetheless bound 40.6-fold more strongly (Kd = 0.016 nM) to the I50L/A71V variant than ATV (Kd = 0.65 nM). In contrast to the cases of ATV and DRV, APV had a 2.8-fold higher affinity to the I50L/A71V variant than the WT protease, indicating that I50L/A71V mutations render protease more susceptible to APV, as previously described for seven PIs, including APV (33).
In addition to the binding affinities, the enthalpic and entropic contributions to the free energy (ΔG) of inhibitor binding were compared between the double mutant and WT proteases (Fig. 2). The drug resistance mutations I50 (V,L)/A71V resulted in compensatory changes in the enthalpy (ΔH) and entropy of binding (−ΤΔS) with respect to those for the WT protease, with enhanced enthalpy but a decreased entropy of binding being observed. The compensation was the largest for ATV binding, with significantly reduced entropy (−6.6 kcal/mol for the I50V/A71V variant and −8.7 kcal/mol for the I50L/A71V variant compared to −11.8 kcal/mol for the WT protease) largely compensated for by an increase in the binding enthalpy. This entropy-enthalpy compensation was observed for all complexes except DRVI50V/A71V. Binding of DRV to all protease variants is largely enthalpy driven, as previously described (34). Similar to the other complexes, I50V/A71V mutations rendered the entropy of DRV binding unfavorable (by 1.5 kcal/mol); however, this entropic loss was not compensated for by an enhanced enthalpy, leading to the largest fold decrease in binding affinity among all complexes examined here. Nevertheless, the highly potent DRV still binds with a higher affinity to the I50V/A71V variant than the other two PIs.
Fig 2.

Changes in inhibitor binding thermodynamics of double mutant protease variants I50V/A71V and I50L/A71V with respect to WT. ΔΔG, ΔΔH, and Δ(−ΤΔS) are differences in the free energy, enthalpy, and entropy of binding compared to those for WT protease binding to the same inhibitor, respectively. (ΔΔG = ΔG for the mutant − ΔG for the WT; ΔG, ΔH, and −TΔS for mutant and WT proteases are presented in Table 2).
Crystal structures.
Six new crystal structures of the I50(V,L)/A71V protease variants in complex with DRV, APV, and ATV were determined and compared with those of the previously determined respective WT protease-inhibitor complexes (Protein Data Bank [PDB] accession numbers 3EKV, 1T3R, and 3EKY). The crystallographic statistics are in Table 1. All structures have resolutions of 2.2 Å or better, enabling a detailed analysis of the changes induced by the mutations.
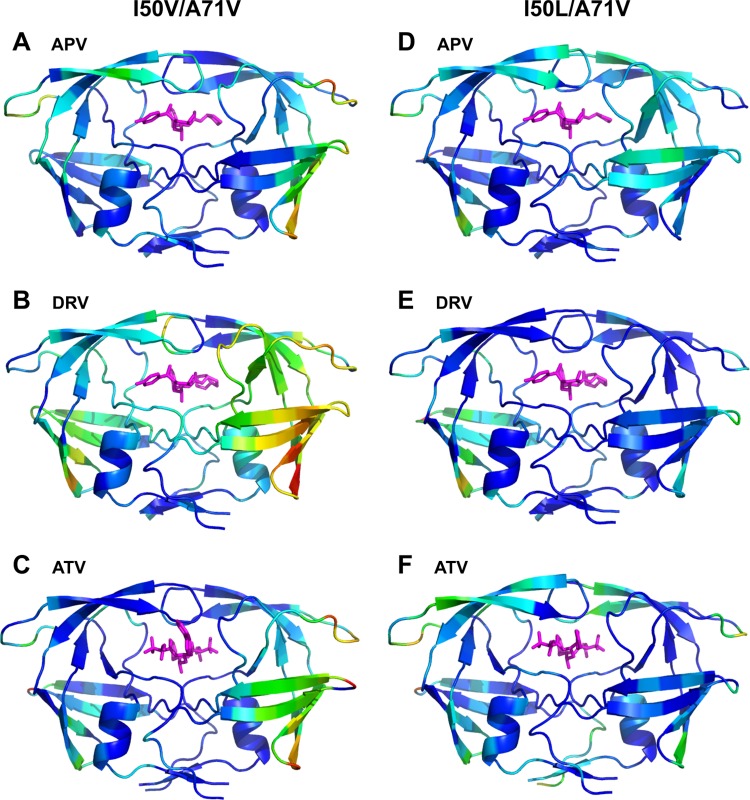
The crystal structures of all six mutant complexes are displayed in Fig. 3, where the protease backbone is colored from blue to red to indicate an increase in the average deviation compared with that for the corresponding WT structure. These deviations were computed from distance-difference matrices of Cα atoms, to reveal global changes in the overall structure without any superposition bias (see Materials and Methods).
Fig 3.
Crystal structures of I50V/A71V and I50L/A71V complexes with residues colored according to the average displacement of Cα atoms with respect to the corresponding WT structure, increasing from blue to red (0 to 1.66 Å). For structures with two dimers in an asymmetric unit, the structure for the dimer with the higher deviations from the WT structure is displayed. (A) APVI50V/A71V; (B) DRVI50V/A71V; (C) ATVI50V/A71V; (D) APVI50L/A71V; (E) DRVI50L/A71V; (F) ATVI50L/A71V.
In general, the deviations from the WT structure were more asymmetric and widespread in the I50V/A71V complex structures than in the I50L/A71V complex structures, notably, in the beta-sheet region, including the G68 to G71 loop, where the A71V mutation is located, and the elbow of the flap in the same monomer. Other notable changes for both variants were in the flap region (G40 to R57), where the I50V mutation is located. The rearrangement of flaps was minimal for ATVI50V/A71V but the most pronounced for ATVI50L/A71V, in accordance with the preferential resistance of the I50L/A71V variant to ATV. The complex that displayed the most widely spread backbone shifts upon the mutations was DRVI50V/A71V, which also displayed the highest fold change loss in affinity with respect to that for the WT protease.
The possible effect of mutations at residues 50 and 71 on the hydrogen bonds between the inhibitors and protease variants was investigated for the crystal structures. Considering that the estimated error in atomic positioning is 0.10 Å to 0.15 Å in structures with 1.5- to 1.8-Å resolutions (35), there were no significant changes in the hydrogen bonding pattern in the mutant complexes. All the structurally important hydrogen bonds in the APVWT structure were conserved in the APVI50V/A71V mutant, similarly to the single APVI50V mutant (35). In the APVI50L/A71V structure, one hydrogen bond between the protease D29 N and O6 of APV was lost, while two new hydrogen bonds formed between N3 of APV and D30 OD2 and with O1 of APV and a water molecule. The hydrogen bond interactions of DRV with the protease in the WT, I50V/A71V, and I50L/A71V complexes were almost identical, with the exception of a direct interaction with the Asp30′ side chain in the mutant complexes instead of the water mediated in the DRVWT structure. No differences in hydrogen bonding between the inhibitor and protease were found for the ATV complexes. Thus, contrary to modeling and simulation predictions with I50V/A71V-DRV complex (19), the varied inhibitor susceptibilities were not due to any changes in protease-inhibitor hydrogen bonding.
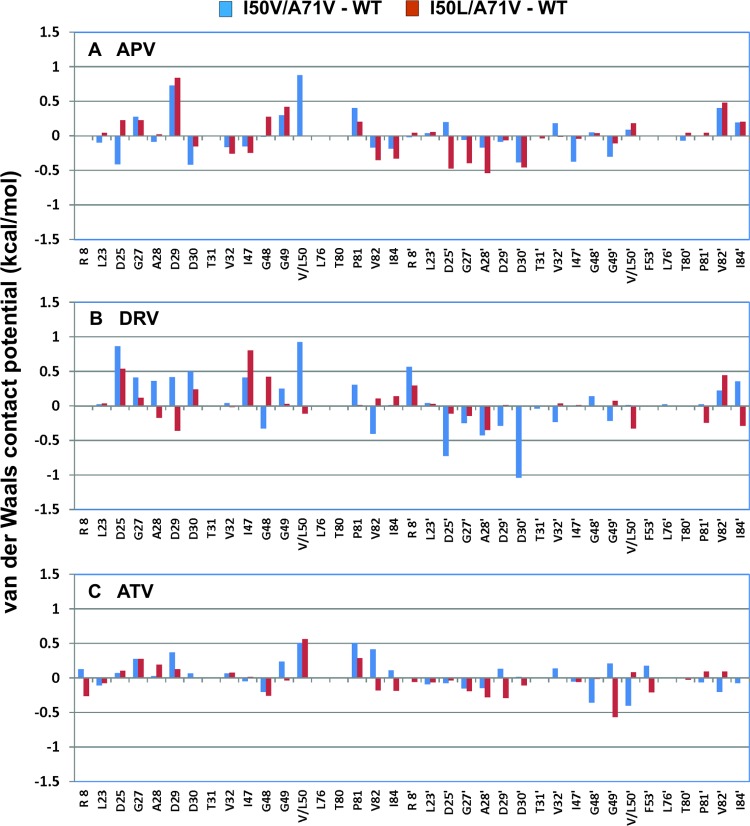
Next, the distributions of vdW interactions between the protease and inhibitors were analyzed for each complex (Fig. 4 and 5). Positive and negative values in Fig. 5 indicate loss and enhancement, respectively, of favorable inhibitor packing in the I50(V,L)/A71V complex compared to that in the WT. In the WT protease, the I50 side chain interacted with the sulfonyl moiety at the P1′ position in the APV and DRV complexes but packed against the t-butyl group at the P2′ position in the ATV complex (Fig. 4A, D, and G). While DRV and APV share the same (hydroxyethylamine)sulfonamide core, the ATV structure is very different and contains an aza-hydroxyethylamine core (Fig. 1A). These differences in the chemical moieties between APV/DRV and ATV resulted in different subsite (P1′ versus P2′) packing interactions with protease. Accordingly, APV and DRV lost favorable packing interactions in the mutant with the I50V mutation (Fig. 4B and E), unlike ATV (Fig. 4H). This is reflected in the high positive (unfavorable) change at residue 50 in the vdW interaction potentials of APV and DRV for the I50V variant (blue bars in Fig. 5A and B). In contrast, ATV lost a similar degree of favorable packing with residue 50 in both double mutants, which was compensated for by more favorable packing with residue 50′ of the other monomer in the case of I50V/A71V (Fig. 5C). The DRVI50V/A71V structure displayed repacking around the bound inhibitor relative to the structure in the WT complex, such that the loss of vdW energy for one monomer was compensated for by the gain of vdW contacts by the second monomer (Fig. 5B). This asymmetric inhibitor repacking was accompanied by an overall asymmetric rearrangement of protease structure (Fig. 3B). Curiously, ATVI50(V,L)/A71V structures exhibited minimal changes in the vdW contact potentials (<±0.5 kcal/mol) compared to that in the ATVWT structure (Fig. 5C). Although residue 50 lost favorable packing in both variant structures, this loss was compensated for by enhanced packing around residue 50′ of the other monomer in ATVI50V/A71V but not in ATVI50L/A71V.
Fig 4.
van der Waals interactions of residue 50 with APV, DRV, and ATV bound to the protease variants. Protease variants are colored gray for WT, blue for I50V/A71V, and green for I50L/A71V. The inhibitors are colored pink. (A) APVWT; (B) APVI50V/A71V; (C) APVI50L/A71V; (D) DRVWT; (E) DRVI50V/A71V; (F) DRVI50L; (G) ATVWT; (H) ATVI50V/A71V; (I) ATVI50L/A71V.
Fig 5.
Differences of van der Waals interaction potentials between the mutant and WT protease inhibitor complexes. (A) APV complex; (B) DRV complex; (C) ATV complex. The vdW interaction values are averaged per residue for the inhibitor complexes when there are two dimers in the asymmetric unit. Protease variants are colored blue for I50V/A71V and red for I50L/A71V.
For the I50V/A71V variant, the loss of vdW interactions of residue 50 with the three inhibitors (APV, 0.9 kcal/mol; DRV, 0.9 kcal/mol; ATV, 0.5 kcal/mol) (Fig. 5A to C) correlated with the weaker binding affinities of DRV and APV (Table 2). The minimal disturbance around the active site (Fig. 2C) and enhanced packing around residue 50′ (Fig. 5C) correlated with tighter binding of ATV to I50V/A71V, contributing to hypersusceptibility. On the other hand, ATV had reduced vdW interactions with both residues 50 and 50′ in I50L/A71V (Fig. 4I and 5C), in agreement with the reduced affinity of binding of this variant to ATV. The change in the vdW interactions of residue 50 for the I50L/A71V variant with APV and DRV was negligible (Fig. 5A and B). Hence, the packing interactions of residue 50 were not significantly altered in APV- and DRV-I50L/A71V variant complexes, but there was a rearrangement of vdW packing around these inhibitors.
Overall, changes in the vdW interactions with the inhibitors are in agreement with, although they do not completely explain, the differential inhibitor susceptibilities of the two double mutants. Specifically, the changes in packing at residue 50 with Val or Leu substitutions correlated with differential susceptibilities to the DRV, APV, and ATV inhibitors.
DISCUSSION
The flaps are the most dynamic region in the unliganded form of HIV-1 protease and control access of substrates and inhibitors to the active site. Upon ligand binding, residue 50 of one monomer contacts I54 of the other monomer, stabilizing the bound conformation. Mutations at residue 50 at the flap tips of HIV-1 protease can severely impact inhibitor susceptibility and viral fitness by altering interactions with ligands and/or flap dynamics. We have investigated the impact of I50V and I50L mutations on binding to three PIs in the context of A71V comutation. The compensatory mutation A71V partially recovers viral fitness (18), possibly by reversing the negative impact of these primary mutations on ligand binding and dynamics. The effect of the A71V mutation propagates asymmetrically through the β-sheet region in one of the monomers, especially in the I50V/A71V double mutant complex structures (Fig. 3). This structural readjustment may be responsible for the contrasting inhibitor binding behavior of the I50V/A71V double mutant studied here and the I50V single mutant (32).
The double mutant I50V/A71V protease exhibits a reduced affinity to both APV and DRV, which have closely related chemical structures (Table 1 and Fig. 1A). This reduced affinity is in agreement with the accumulation of the I50V/A71V mutations in patients who fail APV and DRV therapy. Conversely, we found that this variant is more susceptible to ATV. While there have been reports of increased or no change in susceptibility to ATV in patients whose isolates carry the I50V mutation in the protease gene (15, 31, 36, 37), the idea of hypersusceptibility of the I50V/A71V variant to ATV has not been established. In one instance, the hypersusceptibility was observed in the presence of Gag cleavage site mutations (37). Our binding thermodynamics data, in combination with clinical observations, suggest that the I50V substitution has the potential to make the protease more susceptible to ATV. However, secondary mutations or cleavage site mutations might be required in order to enhance the binding of ATV to I50V protease in vivo.
The other double mutation, I50L/A71V, is more extensively studied and known to cause ATV resistance and hypersusceptibility to APV (33, 38). On the other hand, binding of DRV to the I50L/A71V variant has been addressed only by Molecular Dynamics simulation studies, starting from the complex crystal structure determined here, and by computational binding free energy estimations (19). We have experimentally established the impact of this double mutation on binding to DRV. In contrast to computational estimations, not hypersusceptibility but a 4-fold loss of affinity was observed for DRV binding to the I50L/A71V variant. The thermodynamics data indicate that the loss in binding enthalpy relative to that for the WT is not compensated for by the entropy gain, unlike in the case of APV, resulting in a net effect of loss in binding free energy (Table 2).
Both double mutant proteases displayed an unfavorable change in inhibitor binding entropy relative to that of the WT protease for all three inhibitors (Fig. 2). The entropic effects in the double mutants indicate changes in protease flexibility and/or solvation. More flexibility in the unliganded form or rigidity in the inhibitor-bound form of the protease would cause unfavorable binding entropy relative to that for the WT enzyme. The contrasting entropic changes between the I50V single mutant (32) and the I50V/A71V variant indicate that the A71V mutation is responsible for altered protein flexibility, causing unfavorable effects on binding entropy. While entropic changes are the main driving force behind drug resistance, overcompensation by a favorable enthalpy gain leads to hypersusceptibility in the cases of the ATVI50V/A71V and APVI50L/A71V variants. We found that the favorable changes in enthalpy are not due to hydrogen bonding but rather are due to enhanced packing around the inhibitor.
Despite not being the only determinant, packing around the residue 50 mutation site correlates with the altered affinity of inhibitors. With the I50V substitution, as a result of the loss of a methyl group, vdW interactions with the sulfonyl moiety in APV/DRV are weakened compared to those in the WT complexes (Fig. 4 and 5). On the other hand, the valine substitution did not cause such a loss of packing around the bulky t-butyl group of ATV at the P2′ position. Thus, V50 can pack well against the t-butyl group of ATV and allow the inhibitor to bind with minimal structural perturbations, in agreement with the hypersusceptibility of this double mutant to ATV.
Conversely, in the I50L/A71V complex, the bulky P2′ moiety of ATV is unfavorable, as it potentially results in steric clashes with the longer L50 side chain. Therefore, to accommodate the inhibitor, residue L50 is slightly shifted relative to I50 in the WT complex (Fig. 4G and I). This shift causes both a loss of favorable vdW contacts with the inhibitor and an overall rearrangement of the whole flap region (Fig. 3). These crystal structures confirm similar predictions by modeling studies on ATV with I50L/A71V protease (33). Conversely, with the I50L substitution, the additional methyl group in Leu allows residue 50 to maintain its interactions with the smaller sulfonyl group at P2, in agreement with the increased affinity observed for APV and DRV.
The importance of P1/P1′ and P2/P2′ moieties in targeting multidrug-resistant protease variants has recently become evident (39–41). Considering the effect of both longer and shorter side chain substitutions at residue 50 in the two double mutants analyzed here (I50V,L), we propose that these moieties in PIs should optimally accommodate S1/S1′ and S2/S2′ sites in the protease, consistent with the substrate envelope hypothesis (30, 42). Binding of such PIs would not be substantially affected by substitutions at these sites due to either loss of favorable packing interactions or potential steric clashes causing major rearrangement of the protease to accommodate the PI.
In conclusion, the thermodynamics of binding and high-resolution crystal structures presented here largely explain the differential susceptibility to PIs of two double mutants with V and L substitutions at residue 50 and set the stage for further investigations to fully decipher the molecular basis of drug resistance. Our results demonstrate that relatively subtle packing rearrangements around the inhibitors and the flap region correlate with the distinct pattern of resistance mutations observed in the clinic in response to APV, DRV, and ATV treatment. Other possible effects on inhibitor binding affinities, such as solvation and an altered conformational flexibility of the protease/inhibitor, which cannot be inferred from static structural analysis alone, will shed further light into molecular mechanisms of PI resistance and hypersusceptibility.
ACKNOWLEDGMENTS
This work was supported by a National Institutes of Health grant (P01-GM66524) and Tibotec Inc. Use of the Advanced Photon Source for X-ray data collection was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Use of BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR007707.
The protease inhibitors used in this study were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The initial cell pellets from which the recombinant I50L/A71V protease was purified were kindly provided by Bristol-Myers Squibb Research and Development.
We also thank William Royer for helpful discussions.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Debouck C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retroviruses 8:153–164 [DOI] [PubMed] [Google Scholar]
- 2. Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires KE, Deutsch PJ, Emini E. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569–571 [DOI] [PubMed] [Google Scholar]
- 3. Emini EA, Schleif WA, Deutsch P, Condra JH. 1996. In vivo selection of HIV-1 variants with reduced susceptibility to the protease inhibitor L-735,524 and related compounds. Adv. Exp. Med. Biol. 394:327–331 [DOI] [PubMed] [Google Scholar]
- 4. Roberts NA. 1995. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS 9(Suppl 2):S27–S32 [PubMed] [Google Scholar]
- 5. Chou K-C, Tomasselli AG, Reardon IM, Heinrikson RL. 1996. Predicting human immunodeficiency virus protease cleavage sites in proteins by a discriminant function method. Proteins 24:51–72 [DOI] [PubMed] [Google Scholar]
- 6. Henderson LE, Copeland TD, Sowder RC, Schultz AM, Oraszlan S. 1988. Human retroviruses, cancer and AIDS: approaches to prevention and therapy. Liss, New York, NY [Google Scholar]
- 7. Pettit SC, Sheng N, Tritch R, Erickson-Vitanen S, Swanstrom R. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15–25 [DOI] [PubMed] [Google Scholar]
- 8. Sadler BM, Stein DS. 2002. Clinical pharmacology and pharmacokinetics of amprenavir. Ann. Pharmacother. 36:102–118 [DOI] [PubMed] [Google Scholar]
- 9. King NM, Prabu-Jeyabalan M, Wigerinck P, De Bethune M-P, Schiffer CA. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012–12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson BS, Riccardi KA, Gong YF, Guo Q, Stock DA, Blair WS, Terry BJ, Deminie CA, Djang F, Colonno RJ, Lin PF. 2000. BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents. Antimicrob. Agents Chemother. 44:2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Partaledis JA, Yamaguchi K, Tisdale M, Blair EE, Falcione C, Maschera B, Myers RE, Pazhanisamy S, Futer O, Cullinan AB, Stuver CM, Byrn RA, Livingston DJ. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tisdale M, Myers RE, Ait-Khaled M, Snowden WA. 1999. HIV drug resistance analysis during clinical studies with the protease inhibitor amprenavir, abstr. 118. Abstr. Sixth Conf. Retroviruses Opportunistic Infect., Chicago, IL [Google Scholar]
- 13. Van Marck H, Dierynck I, Kraus G, Hallenberger S, Pattery T, Muyldermans G, Van Vijmen H, Hertogs K, Bethune M. 2007. Unraveling the complex resistance pathways of darunavir using the bioinformatics resistance determination (BIRD). Antivir. Ther. 12:S141 [Google Scholar]
- 14. Vermeiren H, Van Craenenbroeck E, Alen P, Bacheler L, Picchio G, Lecocq P. 2007. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods 145:47–55 [DOI] [PubMed] [Google Scholar]
- 15. Colonno R, Parkin N, McLaren C, Seekins D, Hodder S, Schnittman S, Kelleher T. 2004. Pathways to atazanavir resistance in treatment-experienced patients and impact of residue 50 substitutions, abstr. 656. Abstr. Conf. Retroviruses Opportunistic Infect., San Francisco, CA [Google Scholar]
- 16. Colonno R, Rose R, McLaren C, Thiry A, Parkin N, Friborg J. 2004. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J. Infect. Dis. 189:1802–1810 [DOI] [PubMed] [Google Scholar]
- 17. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher CA. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349–2359 [DOI] [PubMed] [Google Scholar]
- 19. Meher BR, Wang Y. 2012. Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): molecular dynamics simulation and binding free energy studies. J. Phys. Chem. B 116:1884–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prabu-Jeyabalan M, Nalivaika E, King NM, Schiffer CA. 2004. Structual basis for coevolution of the human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 78:12446–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rose JR, Salto R, Craik CS. 1993. Regulation of HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J. Biol. Chem. 268:11939–11945 [PubMed] [Google Scholar]
- 22. King NM, Melnick L, Prabu-Jeyabalan M, Nalivaika EA, Yang S-S, Gao Y, Nie X, Zepp C, Heefner DL, Schiffer CA. 2002. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 11:418–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 25. Prabu-Jeyabalan M, Nalivaika EA, Romano K, Schiffer CA. 2006. Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate. J. Virol. 80:3607–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 27. Jones TA, Bergdoll M, Kjeldgaard M. 1990. O: a macromolecular modeling environment, p 189–195 In Bugg C, Ealick S. (ed), Crystallographic and modeling methods in molecular design. Springer-Verlag Press, Berlin, Germany [Google Scholar]
- 28. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. DeLano WL. 2002. The PyMol molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 30. Nalam MN, Ali A, Altman MD, Reddy GS, Chellappan S, Kairys V, Ozen A, Cao H, Gilson MK, Tidor B, Rana TM, Schiffer CA. 2010. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 84:5368–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 54:4253–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, Amzel LM, Freire E. 2007. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J. Virol. 81:5144–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yanchunas J, Jr, Langley DR, Tao L, Rose RE, Friborg J, Colonno RJ, Doyle ML. 2005. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob. Agents Chemother. 49:3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Surleraux DL, Tahri A, Verschueren WG, Pille GM, de Kock HA, Jonckers TH, Peeters A, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. 2005. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 48:1813–1822 [DOI] [PubMed] [Google Scholar]
- 35. Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. 2010. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. FEBS J. 277:3699–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elston R, Scherer J, Schapiro J, Bethell R, Kohlbrenner V, Mayers D. 2006. De-selection for the I50V mutation occurs in clinical isolates during Aptivus/r (tipranavir/ritonavir) based therapy. Antivir. Ther. 11:S102 [Google Scholar]
- 37. Wainberg MA, Martinez-Cajas JL, Brenner BG. 2007. Strategies for the optimal sequencing of antiretroviral drugs toward overcoming and preventing drug resistance. Future HIV Ther. 1:291–313 [Google Scholar]
- 38. Brower ET, Bacha UM, Kawasaki Y, Freire E. 2008. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem. Biol. Drug Des. 71:298–305 [DOI] [PubMed] [Google Scholar]
- 39. Agniswamy J, Shen CH, Aniana A, Sayer JM, Louis JM, Weber IT. 2012. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry 51:2819–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cai Y, Schiffer CA. 2010. Decomposing the energetic impact of drug resistant mutations in HIV-1 protease on binding DRV. J. Chem. Theory Comput. 6:1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ide K, Aoki M, Amano M, Koh Y, Yedidi RS, Das D, Leschenko S, Chapsal B, Ghosh AK, Mitsuya H. 2011. Novel HIV-1 protease inhibitors (PIs) containing a bicyclic P2 functional moiety, tetrahydropyrano-tetrahydrofuran, that are potent against multi-PI-resistant HIV-1 variants. Antimicrob. Agents Chemother. 55:1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369–381 [DOI] [PubMed] [Google Scholar]