Abstract
Dengue virus (DENV) is an important human pathogen, especially in the tropical and subtropical parts of the world, causing considerable morbidity and mortality. DENV replication occurs in the cytoplasm; however, a high proportion of nonstructural protein 5 (NS5), containing methyltransferase (MTase) and RNA-dependent RNA polymerase (RdRp) activities, accumulates in the nuclei of infected cells. The present study investigates the impact of nuclear localization of NS5 on its known functions, including viral RNA replication and subversion of the type I interferon response. By using a mutation analysis approach, we identified the most critical residues within the αβ nuclear localization signal (αβNLS), which are essential for the nuclear accumulation of this protein. Although we observed an overall correlation between reduced nuclear accumulation of NS5 and impaired RNA replication, we identified one mutant with drastically reduced amounts of nuclear NS5 and virtually unaffected RNA replication, arguing that nuclear localization of NS5 does not correlate strictly with DENV replication, at least in cell culture. Because NS5 plays an important role in blocking interferon signaling via STAT-2 (signal transducer and activator of transcription 2) degradation, the abilities of the NLS mutants to block this pathway were investigated. All mutants were able to degrade STAT-2, with accordingly similar type I interferon resistance phenotypes. Since the NLS is contained within the RdRp domain, the MTase and RdRp activities of the mutants were determined by using recombinant full-length NS5. We found that the C-terminal region of the αβNLS is a critical functional element of the RdRp domain required for polymerase activity. These results indicate that efficient DENV RNA replication requires only minimal, if any, nuclear NS5, and they identify the αβNLS as a structural element required for proper RdRp activity.
INTRODUCTION
Dengue fever is the most common mosquito-borne viral disease affecting humans (1). An estimated 40% of the world's population lives in areas where dengue is endemic, and ∼230 million infections occur annually worldwide (2, 3). With nearly 500,000 patients developing dengue hemorrhagic fever annually, leading to more than 20,000 deaths, dengue has emerged as a major public health problem with significant economic, political, and social impacts (1). However, neither antiviral drugs nor an approved vaccine is currently available (4, 5). The lack of a vaccine is due mainly to the existence of 4 different dengue virus (DENV) serotypes that can cause severe disease symptoms, especially upon secondary infection with a heterologous serotype.
DENV is an enveloped, single-stranded RNA virus belonging to the genus Flavivirus in the family Flaviviridae (2). The DENV genome has a length of ∼10,700 nucleotides and possesses a type I cap at its 5′ end. The genome encodes a single polyprotein, which is cotranslationally and posttranslationally cleaved by host and viral proteases into 3 structural proteins (capsid [C], premembrane protein [prM], envelope protein [E]) and 7 nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (6). The structural proteins, together with the RNA genome, are the main constituents of the infectious virion, whereas the NS proteins are part of cytoplasmic replication complexes (7). In addition, some NS proteins are involved in counteracting cellular antiviral defenses (8, 9).
NS5 is the largest DENV protein, with a molecular size of ∼105 kDa, and comprises two domains. It contains methyltransferase (MTase) and guanylyltransferase activities (10) in the N-terminal domain and RNA-dependent RNA polymerase (RdRp) activity in the C-terminal domain (11). Thus, NS5 is involved in both RNA capping and RNA replication. In addition, NS5 is responsible for blocking the type I interferon (IFN) response by binding to STAT-2 (signal transducer and activator of transcription 2) and inducing its proteasomal degradation (9, 10, 12, 13). It is assumed that most, if not all, of these functions of NS5 are exerted in the cytoplasm. However, in DENV-infected cells, a high proportion of NS5 resides in the nucleus (14, 15), a pattern similar to that found for yellow fever virus-infected cells (16). Two functional nuclear localization signals (NLSs), designated the βNLS and the αβNLS, have been identified in DENV NS5. The latter plays the major role in nuclear translocation and viral RNA replication (17–20). However, the crystal structure of the DENV NS5 polymerase domain (12) suggests that the two NLSs might be integral parts of the NS5 polymerase domain, raising the possibility that the reduced replication of viruses containing mutations in the αβNLS might be due to impairment of their RdRp activity. Moreover, nuclear localization and RdRp activity might be linked, as deduced from the observation that nuclear NS5 is hyperphosphorylated and has only weak interaction with NS3 (21).
DENV NS5 is known to play a crucial role in the modulation of the innate immune response. Earlier studies identified NS5 as a key factor for increased production of the immunomodulatory cytokine interleukin-8 (IL-8) (22). Secretion of IL-8 appears to be influenced by NS5 localized in the nucleus (18, 23). However, it is not clear whether increased IL-8 secretion is linked to impaired replication of NS5 mutants with reduced nuclear accumulation. Much better established is the role of NS5 in blocking the JAK-STAT signal pathway that is activated by type I IFN. Infection of cells with DENV activates double-stranded RNA sensors such as RIG-I, MDA-5, or TLR-3, thus triggering IFN synthesis and secretion (24). NS5 abrogates IFN signaling by interacting with STAT-2, resulting in the proteasomal degradation of STAT-2 and the IFN resistance of DENV-infected cells (9, 25).
While these studies establish NS5 as a main IFN antagonist, the role of the nuclear localization of NS5 in DENV replication is unclear. In this study, we compared a set of mutants in which the nuclear localization of NS5 was impaired to different extents. We determined the virological parameters of the mutants and their properties with respect to the activation and inhibition of the type I IFN system, as well as the biochemical and structural features of the NLS in the context of full-length NS5.
MATERIALS AND METHODS
Cell lines and culture conditions.
The Huh7, BHK-21, HEK 293T, Vero E6, and A549 cell lines were grown in Dulbecco's modified minimal essential medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum in a 37°C incubator with 5% CO2. C6/36 cells derived from Aedes albopictus were maintained in minimal essential medium (MEM; Invitrogen, Karlsruhe, Germany) supplemented with 10 mM HEPES, 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum at 28°C.
Plasmid construction.
Plasmid pDVWS601, containing the full-length cDNA of DENV type 2 (DENV-2) strain New Guinea C (NGC) (26, 27), was modified by the insertion of a firefly luciferase reporter gene or a hygromycin phosphotransferase gene, replacing the structural genes, to yield a DENV luciferase reporter replicon or a selectable subgenomic DENV replicon, respectively (28). To introduce NLS mutations into NS5, restriction sites for AgeI and SacI were introduced as silent mutations at positions flanking the βNLS and αβNLS. Mutagenesis was carried out by two-step overlap PCR using forward primer 5′GAATGTAAGAGAAGTCAAAGGCCTGACAAAAGGA3′ and reverse primer 5′TGGAACTACAAGTACGCGTCCGTCTTTCATGATTAACTCA3′, carrying the unique StuI and MluI sites, respectively, and the following primers for overlap PCR: the AgeI forward primer (5′ACCATGGCAGCTATGAAACAAAACAAACCGGTTCAGCATCATCCATGGTG3′), the AgeI reverse primer (5′CACCATGGATGATGCTGAACCGGTTTGTTTTGTTTCATAGCTGCCATGGT3′), the SacI forward primer (5′TGTTGAAGATAGTAGGTTTTGGGAGCTCGTTGACAAGGAAAGGA3′), and the SacI reverse primer (5′TCCTTTCCTTGTCAACGAGCTCCCAAAACCTACTATCTTCAACA3′) (the restriction sites used for cloning and the mutations introduced are indicated by underlining). Amplicons were inserted via StuI and MluI restriction sites into pDVWS601or pDVWS601-Fluc. The αβNLS mutations were also generated by overlap PCR using the AgeI forward primer and the SacI reverse primer, and the amplicons were inserted via AgeI and SacI into pDVWS601 or pDVWS601-Fluc. The following overlap primers were used for the generation of NLS mutants: KK371-372 forward (5′ACCCAAGAACCGAAAGAAGGCACAGCGGCACTAATGAAAATCACGGCA3′) and KK371-372 reverse (5′TGCCGTGATTTTCATTAGTGCCGCTGTGCCTTCTTTCGGTTCTTGGGT3′); KK387-388 forward (5′GAGTGGCTTTGGAAAGAACTAGGGGCCGCAAAGACACCTAGGATGTGCAC3′) and KK387-388 reverse (5′GTGCACATCCTAGGTGTCTTTGCGGCCCCTAGTTCTTTCCAAAGCCACTC3′); KK388-389 forward (5′GAGTGGCTTTGGAAAGAACTAGGGAAGGCAGCGACACCTAGGATGTGCAC3′) and KK388-389 reverse (5′GTGCACATCCTAGGTGTCGCTGCCTTCCCTAGTTCTTTCCAAAGCCACTC3′); KKK387-389 forward (5′GAGTGGCTTTGGAAAGAACTAGGGGCCGCAGCGACACCTAGGATGTGCAC3′) and KKK387-389 reverse (5′GTGCACATCCTAGGTGTCGCTGCGGCCCCTAGTTCTTTCCAAAGCCACTC3′); RE396-397 forward (5′AAGACACCTAGGATGTGCACTGCAGCCGAATTCACAAGAAAGGTGAGAAG3′) and RE396-397 reverse (5′CTTCTCACCTTTCTTGTGAATTCGGCTGCAGTGCACATCCTAGGTGTCTT3′); EE397-398 forward (5′AAGACACCTAGGATGTGCACTAGAGCCGCATTCACAAGAAAGGTGAGAAG3′) and EE397-398 reverse (5′CTTCTCACCTTTCTTGTGAATGCGGCTCTAGTGCACATCCTAGGTGTCTT3′); REE396-398 forward (5′AAGACACCTAGGATGTGCACTGCAGCCGCATTCACAAGAAAGGTGAGAAG3′) and REE396-398 reverse (5′CTTCTCACCTTTCTTGTGAATGCGGCTGCAGTGCACATCCTAGGTGTCTT3′); and RK401-402 forward (5′AGGATGTGCACTAGAGAAGAATTCACAGCAGCCGTGAGAAGCAATGCAGCC3′) and RK401-402 reverse (5′GGCTGCATTGCTTCTCACGGCTGCTGTGAATTCTTCTCTAGTGCACATCCT3′). The pcDNA-based NS5 expression construct was generated by PCR using pDVWS601 as the template with the following primers, bearing unique BamHI and XbaI restriction sites, respectively: forward, 5′TACCGAGCTCGGATCCATGGGAACTGGCAACATA3′; reverse, 5′ATAATTCTAGACTACCACAGGACTCCTGCCTCTTCCTCTTC3′. Amplicons were treated with BamHI and XbaI and were inserted into pcDNA 3.1+ treated with the same enzymes. DNA fragments containing the NLS mutations were transferred from pDVWS601 via StuI and PmlI restriction sites into pcDNA 3.1-NS5. Deletion mutations affecting the NS5 NLS were generated by PCR, and the mutants were inserted via AgeI and PmlI into pcDNA-NS5. The following primers were used: 5′CAAAACAAACCGGTTCAGCATCATCCAAGAAACTAATGAAAATCACGGCAGAGTGGCTTTGG3′ (forward) and 5′TTCAACAGCCTCACGTGCCGACTTCCACTT3′ (reverse) for deletion of the βNLS; 5′TATGAAACAAAACAAACCGGTTCAGCA3′ (forward) and 5′CAGCCTCACGTGCCGACTTCCACTTGTTCTCATCAGTGAATATGGCCCCCAAGGCTGCATTTGTGCCTTCTTTCGGTTCTTGGGTTCTCGTGTCCACTTT3′ (reverse) for deletion of the αβNLS; and 5′GCTATGAAACAAAACAAACCGGTTCAGCATCATCCAATGCAGCCTTGGGGGCCATATTCACTGATGAGAACAAGTGGAAGTCGGCACGTGAGGCTGTTGAA3′ (forward) and 5′TTCAACAGCCTCACGTGCCGACTTCCACTTGTTCTCATCAGTGAATATGGCCCCCAAGGCTGCATTGGATGATGCTGAACCGGTTTGTTTTGTTTCATAG3′ (reverse) for deletion of both NLSs. The pET21b-based vector used for the expression of full-length NS5 carrying an N-terminal hemagglutinin (HA) tag and a C-terminal hexahistidine tag in Escherichia coli was generated by PCR using forward primer 5′TTCCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGTACCCATACGACGTCCCAGACTACGCTGGAACTGGCAACATAGGAGA3′ and reverse primer 5′AGAGGATCCCTACTAGTGATGGTGATGGTGATGCCACAGGACTCCTGCCTCTT3′. After restriction digestion of PCR products with XbaI and BamHI, they were inserted into pET21b. αβNLS mutations were inserted into pET21b-NS5 by fragment exchange using the corresponding DENV mutants and the StuI and PmlI restriction sites.
Indirect immunofluorescence.
Huh7 or BHK-21 cells electroporated with DENV RNA were seeded onto glass coverslips in 24-well plates at a density of 1 × 105 cells per well. Cells were processed for immunofluorescence as explained below. For the transfection of plasmid DNA, Huh7 cells were seeded onto glass coverslips in 24-well plates at a density of 1 × 105 cells per well. Approximately 18 h later, cells were transfected with pcDNA-based expression constructs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol and were incubated for ∼16 h. Cells were fixed with 2% paraformaldehyde (Applichem GmbH, Darmstadt, Germany) and were permeabilized with 0.5% (vol/vol) Triton X-100 in phosphate-buffered saline (PBS). Primary staining was carried out by a 45-min incubation with either an NS5-specific rabbit polyclonal antiserum (dilution, 1:200) (7), a DENV E-specific monoclonal antibody (1:300) (ATCC), or a STAT-2-specific rabbit polyclonal antiserum (1:100) (Santa Cruz Biotechnology), each diluted in PBS containing 3% goat serum. After extensive washes with PBS, secondary staining was carried out by a 45-min incubation with an Alexa 488- or Alexa 546-conjugated secondary antibody, each diluted 1:1,000 in PBS containing 3% goat serum. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Karlsruhe, Germany). Samples were mounted on glass slides with Fluoromount G (Southern Biotechnology Associates, Birmingham, AL), and images were acquired using a Leica CTR MIC fluorescence microscope or a Nikon Eclipse Ti spinning-disc confocal laser microscope. Images were processed with Adobe Photoshop software, and immunofluorescence signals were quantified by using the ImageJ software package (National Institutes of Health, Bethesda, MD).
In vitro transcription.
pDVWS601-derived constructs were linearized by restriction digestion with XbaI. Ten micrograms of a phenol-chloroform-purified DNA template was used in an in vitro transcription reaction mixture containing 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM of each ATP, UTP, and CTP, 1.56 mM GTP, 1 mM m7GpppG cap analogue (New England Biolabs), 1 U RNasin (Promega, Mannheim, Germany) per μl, 0.1 μg plasmid DNA per μl, and 0.6 U T7 RNA polymerase (Promega) per μl. After incubation for 2.5 h at 37°C, 0.3 U T7 RNA polymerase per μl reaction mixture was added, and the mixture was incubated for an additional 2.5 h at 37°C. Transcription was terminated by the addition of 1.2 U RNase-free DNase (Promega) per μg of plasmid DNA, followed by a 60-min incubation at 37°C. RNA was extracted with acidic phenol and chloroform, precipitated with isopropanol, and dissolved in RNase-free water. Denaturing agarose gel electrophoresis was used to check RNA integrity, and the concentration was determined by measurement of the optical density at 260 nm.
Electroporation of DENV RNA.
BHK-21 or Huh7 cells were detached from the plate by trypsinization, and PBS-washed cells were suspended in Cytomix (29) containing 2 mM ATP and 5 mM glutathione at a density of 1.5 × 107 or 1.0 × 107 cells per ml, respectively. Ten micrograms of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension, and the cells were electroporated using a Gene Pulser system (Bio-Rad, Munich, Germany) in a cuvette with a gap width of 0.4 cm (Bio-Rad) at 960 μF and 270 V. The cells were immediately transferred to 12 ml of complete DMEM, and 2.5 × 105 cells were seeded into each well of a 6-well plate.
Virus replication assays.
Cells plated in duplicate wells in 6-well plates were lysed at different time points in 350 μl lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT [pH 7.8]) using repetitive freeze-thaw cycles. For each well, two 100-μl portions of the lysate were each mixed with 360 μl assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, and 15 mM K2PO4 [pH 7.8]), and after the addition of 200 μl of a luciferin solution (200 μM luciferin, 25 mM glycylglycine [pH 8.0]), luminescence was measured for 20 s in a luminometer (Lumat LB 9507; Berthold, Freiburg, Germany). The kinetics of replication was determined by normalizing the relative light units (RLU) determined at different time points after transfection to the 4-h RLU value, which reflects translation from the input RNA and thus transfection efficiency.
Protein expression.
Escherichia coli cells [strain Rosetta(DE3); kindly provided by Stéphane Bressanelli, Gif-sur-Yvette, France] were transformed with pET21b-NS5 and were plated onto LB agar plates containing 100 μg/ml ampicillin and 1% glucose. Starter cultures were prepared by inoculating single colonies into 5 ml of LB medium containing 100 μg/ml ampicillin and 1% glucose and incubating overnight. The starter culture was diluted 1:100 with fresh medium (final volume, 500 ml) containing 100 μg/ml ampicillin and was grown at 37°C until an optical density at 600 nm of 0.8 to 1.0 was reached. Protein expression was induced by cold shock at 4°C for 30 min prior to the addition of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and 2% ethanol. After overnight incubation at 18°C, cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, and the cell pellet was stored at −80°C until it was required for further processing.
Protein purification.
Cell pellets were resuspended in lysis buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 20% glycerol, 1% octyl glucoside, 50 mM imidazole, 1 mM DTT, and a protease inhibitor mix [Roche]) containing 1 mg/ml lysozyme and 5 U/ml Benzonase, incubated at 4°C for 30 min, and lysed by sonication. Lysates were clarified by centrifugation at 12,000 × g for 15 min at 4°C. Soluble NS5 was purified by metal affinity chromatography using a nickel-nitrilotriacetic acid (Ni-NTA) column equilibrated with lysis buffer. Unbound proteins were removed by washing the gel bed with 5 column volumes of wash buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 20% glycerol, 0.1% octyl glucoside, 50 mM imidazole, 1 mM DTT, and a protease inhibitor mix [Roche]). Bound protein was eluted by incubating the gel bed with elution buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 20% glycerol, 1% octyl glucoside, 400 mM imidazole, 1 mM DTT, and a protease inhibitor mix [Roche]). NS5-containing fractions were quantified by using the Bradford method, and purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Peak fractions were pooled and stored at −80°C.
RNA-dependent RNA polymerase assay.
The RdRp assay was carried out using 400 ng of DENV genomic RNA generated by in vitro transcription and 200 ng of purified NS5. The reaction was performed in a total volume of 50 μl, and the reaction mixture contained 50 mM HEPES (pH 8.0), 10 mM KCl, 5 mM MgCl2, 3 mM MnCl2, 1 mM DTT, 1 U/μl RNasin, 0.5 mM (each) ATP, CTP, and UTP, 10 μM GTP, and 10 μCi [32P]GTP (3,000 Ci/mmol; Perkin-Elmer). After 2 h of incubation at 30°C, RNA was precipitated with 10% trichloroacetic acid on GF-C microfilters (GE Healthcare), and the incorporation of radioactivity was measured in a liquid scintillation counter. All measurements were carried out in duplicate.
Methyltransferase assay.
The 2′O-methyltransferase assay was carried out in a total volume of 50 μl containing 1 μg of m7-capped DENV subgenomic RNA (corresponding to nucleotides 1 to 175 of the DENV-2 NGC genome) generated by in vitro transcription as the template and 100 ng of purified NS5. The assay buffer contained 50 mM Tris (pH 7.0), 10 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 0.05% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2 mM DTT, and 2 μCi 3H-labeled S-adenosylmethionine (Perkin-Elmer). After 2 h of incubation at 22°C, the RNA was precipitated with ethanol, and the incorporation of radioactivity was measured by liquid scintillation counting.
RNA quantification by qRT-PCR.
Total cellular RNA from ∼5 × 105 virus-infected cells was isolated by using the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) as recommended by the manufacturer. The cDNA was prepared using a High Capacity cDNA reverse transcription (RT) kit (Applied Biosystems, Life Technologies). Quantitative real-time PCR (qRT-PCR) was carried out using an ABI Prism 7000 sequence detector system (Applied Biosystems, Foster City, CA). For each primer set, reactions were conducted in triplicate by using the Green Dye RT-PCR master mix (PJK Gmbh, Hilden, Germany) according to the manufacturer's instructions with the following primers (with their specificities given in parentheses): 5′TTGAGTAAACTGTGCAGCCTGTAGCTC3′ (forward) and 5′GGGTCTCCTCTAACCTCTAGTCCT3′ (reverse) (DENV); 5′GAAGGTGAAGGTCGGAGT3′ (forward) and 5′GGGTCTCCTCTAACCTCTAGTCCT3′ (reverse) (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). The total volume of the reaction mixture was 15 μl, and reactions were performed in two stages: stage 1, 15 min at 95°C; stage 2, 40 cycles of 15 s at 95°C and 60 s at 60°C. Quantities of DENV RNA were calculated by using serial dilutions of known amounts of DENV in vitro transcripts that were processed in parallel. For cellular genes, the ΔΔCT method (30) was used to calculate relative expression levels.
RESULTS
Mutations within αβNLS abrogate the nuclear accumulation of NS5.
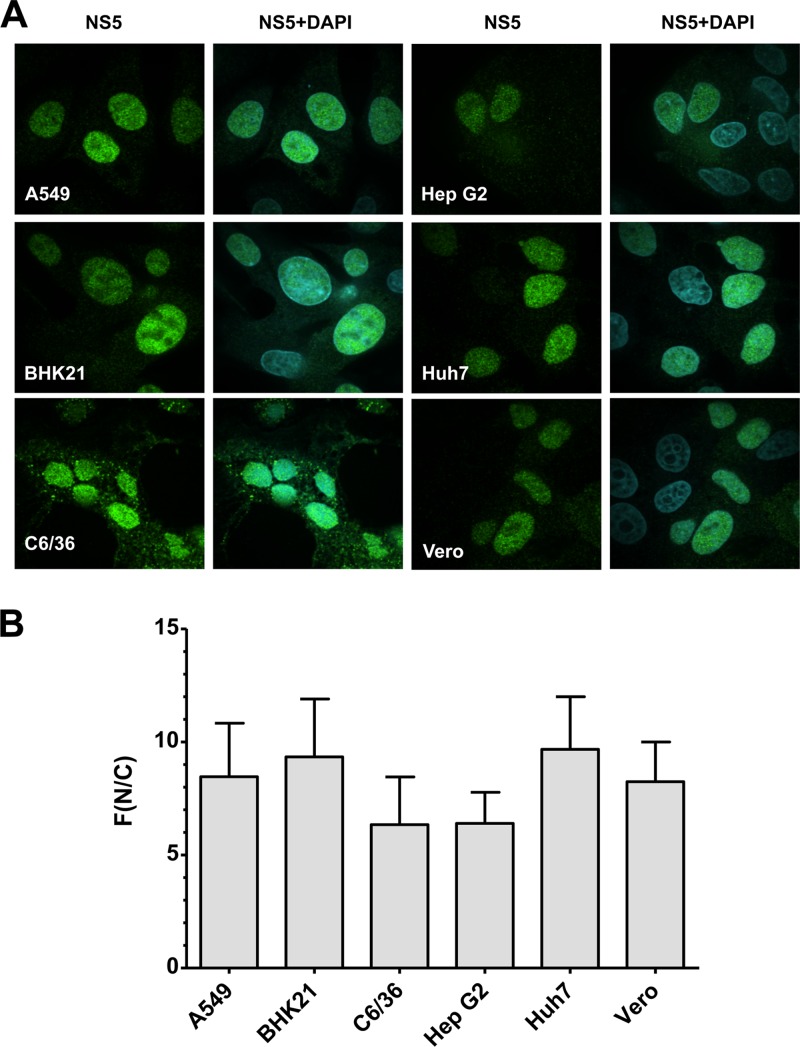
In the initial set of experiments, we determined whether the subcellular localization of DENV NS5 depends on the host cell type (Fig. 1). We used human hepatoma cells (Huh7, HepG2), human lung carcinoma cells (A549), African green monkey kidney cells (Vero), baby hamster kidney cells (BHK-21), and mosquito cells (Aedes albopictus; C6/36). Quantitation of the NS5-specific immunofluorescence signal revealed that ∼80 to 90% of NS5 detectable in this assay localized in the nucleus 24 h after infection with DENV strain NGC, which was used throughout this study, and this pattern was independent of the cell line used (Fig. 1A and B). We therefore concluded that nuclear accumulation of NS5 is cell type independent and is a conserved feature in human and nonhuman cells.
Fig 1.
Nuclear localization of NS5 is independent of the cell line used. (A) The indicated cell lines were infected with DENV-2 (strain NGC; multiplicity of infection, 1), and cells were fixed 24 h after infection. Subcellular localization of NS5 was determined by immunofluorescence microscopy using an NS5-specific antibody. Images were captured by confocal laser scanning microscopy using a 100× objective. (B) Quantification of NS5-specific immunofluorescence by use of the ImageJ software package. The extent of nuclear localization of NS5 was determined by quantifying the NS5 signals detected in the nucleus (N) and the cytoplasm (C) and calculating the ratio of mean fluorescence (F) in these two compartments. Results are means ± standard deviations (n, ≥30).
To determine the relative contributions of the βNLS and the αβNLS in NS5 to nuclear accumulation, we generated NS5 deletion mutants in which either the βNLS, the αβNLS, or both were deleted (Fig. 2). Deletion of the βNLS reduced the nuclear accumulation of NS5 about 3-fold, whereas deletion of the αβNLS completely abrogated nuclear localization (Fig. 3A). Thus, the αβNLS plays a major role in the nuclear localization of NS5.
Fig 2.
Overview of constructs and mutants used in this study. (A) (Top) Diagram of the full-length DENV genome. The 5′ and 3′ UTRs are shown with their putative secondary structures (9). Polyprotein cleavage products are separated by vertical lines and are labeled as specified in the introduction. (Center) Genomic organization of the selectable subgenomic replicon. The gene encoding hygromycin resistance is fused via the ubiquitin coding sequence (Ubi) to NS1. (Bottom) Structure of the DENV reporter genome. The gene encoding firefly luciferase (Ffluc) is fused via Ubi to the N terminus of the polyprotein. All constructs are derived from the DENV-2 (DV2) NGC isolate (26). (B) (Top) Schematic diagrams of wild-type (WT) and mutant DENV-2 NS5. Wild-type NS5 is composed of an N-terminal methyltransferase domain and a C-terminal RNA-dependent RNA polymerase domain (shaded). The latter contains the βNLS (amino acids 321 to 368) and the αβNLS (amino acids 369 to 405). The positions of the amino acids deleted are given above each NLS deletion mutant. (Bottom) Amino acid sequences of the wild-type αβNLS and mutants derived from it. The minimal αβNLS according to reference 17 (amino acids 369 to 393) is shaded in the wild-type sequence. For each mutant, the double or triple alanine substitutions are indicated by AA or AAA, and they are given below the corresponding positions of the wild type.
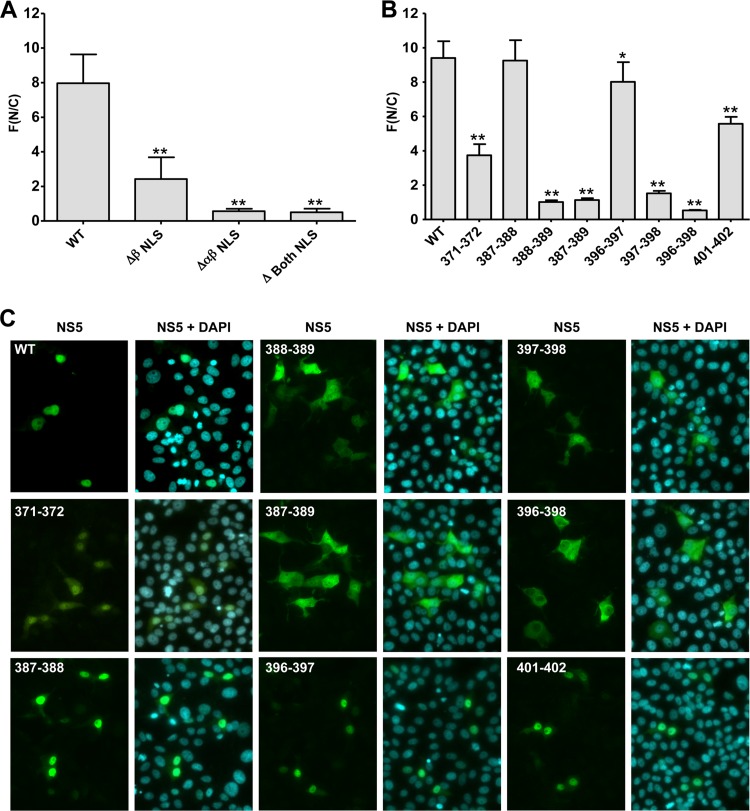
Fig 3.
Subcellular localization of NS5 NLS mutants. Huh7 cells were transfected with plasmids directing the expression of NS5 bearing NLS deletions or the indicated amino acid substitutions under the control of the cytomegalovirus promoter, and cells were fixed 16 h later. The subcellular localization of NS5 was determined by immunofluorescence using an NS5-specific antibody. Images were captured with a 20× objective using a confocal laser scanning microscope. NS5-specific immunofluorescence was quantified by using the ImageJ software package. (A and B) The extent of the nuclear localization of NLS mutants was determined by quantifying the NS5 signals inside the nucleus (N) and the cytoplasm (C) and calculating the ratio of the mean fluorescence (F) in the nucleus to that in the cytoplasm. Results are means ± standard deviations (n, ≥50) for two independent experiments. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.001) from the wild type. (A) Nuclear accumulation of NLS deletion mutants lacking amino acids 321 to 368 (ΔβNLS), amino acids 369 to 405 (ΔαβNLS), or amino acids 321 to 405 (Δ Both NLS). (B) Nuclear accumulation of αβNLS mutants. (C) Representative immunofluorescence images of NS5 mutants.
We next conducted mapping experiments in order to identify amino acid residues in the αβNLS that are crucial for the nuclear localization of NS5. A panel of mutants was generated in which clusters of charged amino acid residues were replaced by alanine residues (Fig. 2B). These mutants were transiently expressed in Huh7 cells, and the subcellular localizations of these NS5 variants were determined (Fig. 3B and C). Two regions that were most sensitive to mutations were found: amino acid residues 388 and 389, corresponding to the second lysine cluster in the αβNLS, and the two acidic amino acid residues at positions 397 and 398, residing in the C terminus of the αβNLS (Fig. 3B and C). Mutations affecting residues 387 and 388 or residues 396 and 397 had only minor effects on the nuclear localization of NS5, whereas mutations affecting residues 401 and 402 or residues 371 and 372 had an intermediate phenotype. The mutation affecting amino acid residues 387 to 389 strongly reduced nuclear localization, to levels similar to those for the mutation affecting residues 388 and 389, whereas alanine substitutions of amino acid residues 396 to 398 had the most drastic effect, reducing the nuclear accumulation of NS5 to levels comparable to those obtained with the αβNLS deletion mutant. These results suggested that not only the bipartite NLS within the αβNLS but also residues C-terminal to this NLS play crucial roles in the nuclear localization of NS5.
Nuclear localization of NS5 is not strictly required for viral RNA replication.
We next determined whether nuclear localization of NS5 is required for DENV replication. The αβNLS mutations were introduced into a genomic DENV luciferase reporter virus (Fig. 2A), and replication competence was determined in BHK-21 cells that had been transfected with capped in vitro transcripts corresponding to the mutant genomes. Time course experiments were conducted, and values obtained 24, 48, 72, and 96 h after transfection were normalized to the 4-h value, which reflects transfection efficiency (Fig. 4A). The wild-type DENV reporter virus genome served as a positive control and the RdRp active-site mutant (GND) as a negative control. Overall, the replication competence of the mutant viruses (mutants 371-372, 387-388, 388-389, 387-389, 396-397, and 396-398), as well as virus production, correlated with the degree of nuclear localization of NS5 (compare Fig. 4A and B with 3B). However, there were two remarkable exceptions. First, mutant 401-402 replicated to levels only about 2-fold above the background, even though the nuclear localization of NS5 was only moderately affected. Second, mutant 397-398 replicated to wild-type levels, even though the nuclear localization of NS5 was highly reduced (compare Fig. 4A and 3B).
Fig 4.
Replication competence of NLS mutants in BHK-21 cells. (A) Capped RNAs generated by in vitro transcription of cloned DENV reporter genomes were transfected by electroporation into BHK-21 cells. Lysates of cells prepared at the time points given above the graph were used to determine luciferase activity. Values were normalized to the 4-h value, which reflects transfection efficiency, and are expressed as means ± standard deviations for three independent experiments. (B) Titers of infectious virus released into culture supernatants of cells that had been transfected with the indicated DENV reporter genomes. Supernatants of the cells used for the experiment for which results are shown in panel A were harvested 96 h postelectroporation and were used to infect naïve BHK-21 cells. Seventy-two hours later, DENV replication was determined by a luciferase reporter assay reflecting the titers of infectious virus released from transfected cells. Values are means ± standard deviations for three independent experiments and are expressed relative to wild-type (WT) titers, which were set at 100%. Asterisks indicate significant differences (**, P < 0.001) from the wild type. (C) Replication of nonreporter DENV genomes carrying NLS mutations. BHK-21 cells were electroporated with the indicated DENV genomes. RNA replication was analyzed 72 h postelectroporation by determining the number of viral RNA copies by use of quantitative real-time RT-PCR. Values are means ± standard deviations for two independent experiments with three replicates each. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.001) from the wild type. (D) Quantification of nuclear NS5 as described in the legend to Fig. 1B. Results are means ± standard errors of the means (n, ≥50) from two independent experiments. (E) Localization of NS5 in nonreporter DENV genomes carrying NLS mutations. The subcellular localization of selected mutants was determined by immunofluorescence using an NS5-specific polyclonal antibody 72 h postelectroporation. Nuclear DNA was visualized by DAPI staining. Images were captured with a 60× objective using a confocal laser scanning microscope.
Although these results were corroborated by measuring titers of infectious virus in the culture supernatants of transfected cells (Fig. 4B), we could not rule out the possibility that the discrepancy between replication and NS5 nuclear localization observed for the two mutants was due to the use of a reporter virus genome. Therefore, we introduced the αβNLS mutations into a nonreporter DENV genome (Fig. 2A). In vitro transcripts derived from these genomes were transfected into BHK-21 cells, and RNA replication was monitored by qRT-PCR. Although the nonreporter genomes replicated more efficiently than the reporter virus constructs, the relative differences between the wild type and the various mutants were comparable (Fig. 4C). Importantly, in this setting also, mutant 397-398 was fully viable. Moreover, the replication of mutant 401-402 was reduced ca. 50,000-fold from that of the wild-type virus, even though nuclear localization was reduced only ∼2-fold (Fig. 3B). In comparison, the nuclear localization of NS5 in mutant 371-372 was reduced more, whereas replication was hardly affected (compare Fig. 3B with Fig. 4C).
We also determined the subcellular localization of NS5 in the context of infected BHK-21 cells by using the full-length nonreporter virus (Fig. 4D and E). The nuclear accumulation of the mutant NS5 proteins in infected cells was comparable to that observed for cells transiently expressing only NS5. Importantly, in mutant 396-398, virtually no nuclear NS5 was detected, yet this virus replicated ∼500-fold above background, and mutant 397-398 replicated to the wild-type level even though the amount of nuclear NS5 was severely reduced. The possibility of high replication due to reversion of mutation 397-398 to wild type was ruled out by two complementary approaches. First, to avoid the accumulation of possible revertants in the virus stock used for the infection experiments, we monitored nuclear localization after electroporation and obtained identical results (data not shown). Second, we generated a modified 397-398 construct with two nucleotide exchanges at each affected codon. The replication of this construct was identical to that of the original 397-398 construct, thus excluding the possibility that the observed phenotype was caused by revertants (data not shown). Finally, analogous results were obtained when these mutants were tested in Huh7 cells (data not shown), arguing against a cell line-specific effect. We therefore concluded that nuclear localization of DENV NS5 is not a strict requirement for efficient RNA replication, at least in cell culture.
The replication deficiency of αβNLS mutants cannot be rescued by trans-complementation.
Earlier studies suggested that nuclear localization of NS5 is required in order to suppress the induction of antiviral cytokines, most notably interleukin-8 (IL-8) (18). Therefore, we investigated the effects of the NLS mutations on IL-8 induction in Huh7 cells transfected with the various mutants. Although wild-type NS5 induced IL-8 ca. 12-fold as measured by qRT-PCR, the level of induction was lower with two selected NLS mutants (397–398 and 396–398) and correlated with their replication competence rather than with the nuclear abundance of NS5 (data not shown). However, these results did not exclude a role for nuclear NS5 in the induction of other antiviral cytokines, and therefore, we conducted trans-complementation experiments. Assuming that nuclear NS5 would block the activation of such cytokines, the expression of nuclear NS5 should be sufficient to rescue replication-defective NLS mutants. To test this assumption, we determined the replication competence of the αβNLS mutant viruses in the presence of wild-type NS5 provided in trans by a subgenomic replicon (Fig. 5A). Reporter virus genomes containing αβNLS mutations were transfected into a cell line containing a stably replicating selectable DENV replicon, and the replication of the transfected genomes was measured by a luciferase assay. A reporter virus carrying a deletion within NS1 was used as a positive control, because earlier reports had described the rescue of NS1 mutations by trans-complementation (31, 32). As shown in Fig. 5B, none of the αβNLS mutants could be rescued in trans by the helper replicon, even though the replicon provided large amounts of nuclear NS5 (Fig. 5C). In contrast, efficient rescue of the NS1 mutant, analyzed in parallel, was achieved, whereas this mutant was unable to replicate in the absence of the helper replicon (Fig. 5B). This result suggested that the replication defect of the αβNLS mutants could not be attributed directly to the reduced amount of NS5 in the nucleus. Moreover, the data imply that the lack of inhibition of antiviral cytokine induction, due to the small amount of nuclear NS5, did not account for the low level of replication of the αβNLS mutants.
Fig 5.
Mutations in the αβNLS affecting viral replication cannot be rescued by trans-complementation. (A) Schematic of the experimental strategy. A Huh7-derived cell line containing a stably replicating subgenomic NGC replicon (as shown in Fig. 2A) was transfected with full-length genomes containing mutations in the αβNLS. Cells were seeded into multiwell culture dishes and were harvested 4, 24, 48, 72, and 96 h after transfection. The replication of the transfected reporter virus genome was determined by a luciferase assay. Note that the subgenomic helper replicon contains the selectable marker but lacks a luciferase reporter gene. p.e., postelectroporation. (B) The αβNLS mutants (x axis) were transfected into Huh7 replicon cells. The replication of the NLS mutants was quantified by a luciferase assay using cell lysates prepared at the indicated time points. The NS1 deletion mutant (ΔNS1) was used as a positive control. The replication of the wild-type DENV reporter genome and of the ΔNS1 mutant, each transfected into naïve Huh7 cells that lack the subgenomic helper replicon, served as references. (C) NS5 expression in DENV replicon cells was determined by immunofluorescence using an NS5-specific polyclonal antibody. Nuclear DNA was visualized by DAPI staining. Images were captured with a 100× objective using a confocal laser scanning microscope.
Mutations in the αβNLS do not affect interferon resistance.
Apart from its role in viral RNA replication, NS5 plays an important role in subverting the antiviral defense, most notably IFN signaling, by binding to STAT-2 and triggering its degradation (9, 25). We therefore investigated the possible effects of αβNLS mutations on IFN-induced antiviral defense. In the first set of experiments, we determined the IFN-α or IFN-γ sensitivities of three αβNLS mutants with severe (397–398), moderate (371–372), or minor (396–397) decreases in NS5 nuclear localization. Addition of IFN-α at 6 h, 12 h, and 24 h after electroporation caused time-of-addition- and dose-dependent reductions in the levels of DENV replication in Huh7 cells (Table 1). However, at 24 h postelectroporation, even with the highest IFN-α concentration (10,000 U/ml), viral replication was reduced only ∼10-fold. In contrast, the replication of a hepatitis C virus (HCV) luciferase-based reporter replicon that we used in parallel as a positive control was reduced ∼15-fold by the lowest IFN-α concentration (100 U/ml) at the same time point. Importantly, none of the DENV αβNLS mutants showed altered sensitivity to the antiviral activity of IFN-α, even though the replication competence of the wild type differed from those of the mutants (Fig. 4).
Table 1.
IFN sensitivities of αβNLS mutantsa
| Time of IFN addition (h post-epo) | IFN added | Concn (U/ml) | Virus replication (%)b |
||||
|---|---|---|---|---|---|---|---|
| WT | 371–372 | 396–397 | 397–398 | HCV | |||
| 6 | IFN-α | 1,000 | 3.7 ± 0.7 | ND | 5.7 ± 2.8 | 2.8 ± 0.2 | 0.04 ± 0.02 |
| IFN-γ | 100 | 1.9 ± 0.2 | ND | 4.1 ± 2.6 | 1.2 ± 0.1 | 0.13 ± 0.1 | |
| 12 | IFN-α | 1,000 | 2.9 ± 0.6 | ND | 3.6 ± 0.4 | 2.6 ± 0.2 | 0.08 ± 0.04 |
| IFN-γ | 100 | 2.0 ± 0.2 | ND | 4.5 ± 2.7 | 1.9 ± 0.2 | 0.19 ± 0.13 | |
| 24 | IFN-α | 100 | 44.3 ± 13.0 | 29.5 ± 5.1 | 48.1 ± 13.7 | 46.2 ± 9.8 | 6.8 ± 7.7 |
| 1,000 | 17.3 ± 2.8 | 20.1 ± 3.9 | 17.8 ± 8.6 | 16.8 ± 2.6 | 2.9 ± 3.4 | ||
| 10,000 | 8.6 ± 2.3 | ND | 7.8 ± 3.7 | 4.8 ± 0.8 | 2.1 ± 2.5 | ||
| IFN-γ | 10 | 6.9 ± 4.4 | 13.2 ± 1.8 | 8.0 ± 3.5 | 5.0 ± 1.2 | 11.6 ± 6.7 | |
| 100 | 1.9 ± 0.9 | ND | 2.3 ± 1.3 | 1.3 ± 0.2 | 5.5 ± 5.8 | ||
| 1,000 | 1.1 ± 0.3 | ND | 1.4 ± 0.7 | 0.9 ± 0.2 | 5.0 ± 5.9 | ||
Huh7 cells transfected with DENV-2 αβNLS mutants were treated 6 h, 12 h, or 24 h after electroporation (epo) with IFN-α or IFN-γ at different concentrations. Three days later, cells were harvested, and virus replication was measured by a luciferase assay. As a positive control, an HCV subgenome (isolate JFH-1) containing a luciferase gene was used (40). Values were normalized to those for cells electroporated with the corresponding constructs but not treated with IFN, which were set at 100%. Results are means ± standard deviations for three independent experiments.
WT, wild type; ND, below the detection limit.
To corroborate these results, we assessed the abilities of selected αβNLS mutants to reduce STAT-2 abundance. The genome of wild-type DENV or the αβNLS mutant 371–372, 388–389, 397–398, or 396–398 was transfected into Huh7 cells, which were analyzed by immunofluorescence for E protein and STAT-2. We used this single-cell analysis because of the various replication capacities of the mutants, which precluded Western blot analysis of total-cell lysates. As shown in Fig. 6A and B, the extents of STAT-2 degradation by the wild type and the mutants were comparable, suggesting that the mutations affecting the αβNLS do not impair efficient STAT-2 degradation.
Fig 6.
STAT-2 degradation is not affected by mutations in the αβNLS. (A) Huh7 cells were transfected either with the wild-type NGC genome (WT) or with the indicated αβNLS mutants. Seventy-two hours later, the cells were treated with 100 U of IFN-α/ml. After 2 h, the cells were fixed, and E protein and STAT-2 were detected by indirect immunofluorescence using E- and STAT-2-specific antisera. The images were acquired with a 100× objective using a confocal laser scanning microscope. Note that in cells expressing DENV E-protein, STAT-2 is absent, as manifested by the “empty” nucleus. (B) The amount of STAT-2 was determined by measuring the average STAT-2 signal inside the nuclei of infected cells. The values, expressed as percentages of the amount of STAT-2 in uninfected control cells, are means ± standard deviations (n, ≥30) for two independent experiments.
We also determined whether αβNLS mutations affected the antiviral response against type II IFN (Table 1). Although IFN-γ treatment had a stronger antiviral effect than IFN-α on DENV replication, the replication of all αβNLS mutants was reduced to the same extent, and their inhibition was comparable to that observed with the wild type. These results show that reduced nuclear accumulation of NS5 does not affect sensitivity to IFN-α or IFN-γ.
Mutations affecting the NS5 αβNLS have minimal effects on RdRp and MTase activities.
Structural studies of the NS5 RdRp domain suggest that the αβNLS forms an integral part of the polymerase domain (33). Moreover, genetic studies point to cross talk between the RdRp domain and the MTase domain (34), arguing that at least some of the mutations in the αβNLS might affect either of these enzymatic activities and thus impair replication competence. To analyze RdRp and MTase activities, we purified bacterially expressed full-length NS5 containing the αβNLS mutations. In the initial set of experiments, we used an NS5 expression construct containing a hexahistidine affinity tag at the C terminus only. However, this protein was highly insoluble and massively degraded, as seen in the Coomassie blue stain and NS5-specific Western blot analysis (Fig. 7A and B, lanes 1 and 2). In contrast, when we also fused a hemagglutinin (HA) affinity tag to the N terminus of NS5, the fusion protein was stabilized, and a large proportion of expressed NS5 was full length (Fig. 7B, lanes 5 and 6). Although most of the full-length NS5 protein expressed was in the insoluble fraction of the cell lysate, sufficient amounts were soluble and could be purified with high efficiency under nondenaturing conditions by hexahistidine-specific affinity chromatography. Yields were in the range of 2 to 5 mg of purified protein per liter of E. coli culture, representing 1 to 2% of total expressed full-length NS5 protein. By using this strategy, we were able to generate NS5 proteins containing the αβNLS mutations (Fig. 8A). Inactive-MTase (S56A) and inactive-RdRp (D663N) mutants served as negative controls; wild-type NS5 was used as a positive control.
Fig 7.
Expression of singly or doubly tagged full-length NS5 in E. coli. (A) Schematic representation of the expression constructs encoding full-length NS5 fused either to a C-terminal hexahistidine tag alone (construct 5His) or to an N-terminal HA tag and a C-terminal hexahistidine tag (construct 5HAHis). Mutations abrogating the methyltransferase (MTase) activity (S56A) or the RdRp activity (D663N), used as controls in the biochemical assays, are indicated. (B) Analysis of full-length NS5 proteins expressed in E. coli. Protein samples were analyzed by SDS-PAGE and Coomassie blue staining of the gel (top) or by Western blotting (WB) using an NS5-specific polyclonal antiserum (bottom). For each lane, 1 μg protein, as determined by the Bradford assay, was loaded onto the gel. SM, size marker; UI, uninduced; I, induced; S, soluble fraction; IS, insoluble fraction; FT, flowthrough; E, eluate fraction. Because the amount of protein in the wash fraction (W) was below the detection limit, a 1:700 dilution of the total wash fraction was loaded onto the gel.
Fig 8.
Biochemical characterization of MTase and RdRp activities of full-length NS5 containing mutations in the αβNLS. (A) The tagged full-length NS5 proteins given above the gel were expressed in E. coli as described in Materials and Methods and were extracted from cell lysates by affinity purification using Ni-NTA affinity chromatography. Proteins were eluted with imidazole, and 5 μg protein was analyzed for purity and integrity by SDS-PAGE and Coomassie blue staining. Numbers on the left are the sizes of molecular mass standards (Marker). (B) A 2′O-methyltransferase assay of NLS mutants was carried out using a capped DENV-2 genomic RNA template (corresponding to nucleotides 1 to 175 of the genome), 100 ng purified NS5, and 2 μCi of 3H-radiolabeled S-adenosylmethionine. 3H incorporation was determined by liquid scintillation counting. (C) RdRp assay of NLS mutants using in vitro-transcribed DENV-2 genomic RNA as the template and 100 ng purified NS5. The amount of 32P-radiolabeled GMP incorporated was measured by liquid scintillation counting. The 2′O-MTase and RdRp activities are expressed as percentages of activities for wild-type (WT) NS5, with means and standard deviations estimated from three independent measurements.
We first determined the impact of mutations affecting the αβNLS on 2′O-MTase activity by measuring the transfer of a 3H-radiolabeled methyl group contained in S-adenosylmethionine to a 5′-terminal fragment (corresponding to nucleotides 1 to 172) of the DENV-2 RNA genome (35). As shown in Fig. 8B, the 2′O-MTase activities of all NLS mutants were comparable to that of the wild type, arguing that the αβNLS does not contribute to in vitro MTase activity.
The RdRp activities of the NS5 proteins were determined by using noncapped in vitro transcripts corresponding to DENV RNA genomes as the template and measuring the incorporation of [32P]GMP into newly synthesized RNA. As shown in Fig. 8C, except for one mutant, the RdRp activities of all the NS5 proteins were not affected by the αβNLS mutations and were comparable to that of wild-type NS5. The exception was the NS5 protein containing alanine substitutions at positions 401 and 402, which showed ∼20-fold-reduced RdRp activity. This impairment of RdRp enzymatic activity explains the low level of replication of this αβNLS mutant, even though these mutations affected NS5 nuclear localization only to a very minor extent.
Structure modeling of αβNLS mutations in the context of full-length NS5.
To gain insight into possible roles of residues R401 and K402 in RdRp activity, we used a structure model of the RdRp domain of DENV-2 that was derived from the crystal structure of the DENV-3 RdRp domain (36) in order to localize the positions of the αβNLS as well as residues R401 and K402. Figure 9 shows that most of the mutated residues in the αβNLS, including R401 at the C terminus of the second helix, are exposed on the surface of the finger subdomain. In contrast, residue K402 is buried and is located in the RNA template tunnel next to the active site. A model of a de novo initiation complex of the DENV-2 polymerase domain (36) containing the 3′ end of the positive-strand RNA genome, the two nucleotides forming the 5′ end of the negative-strand RNA (ATP and GTP), and the catalytic ions shows that K402 is positioned near the phosphate and ribose of the 3′ end of the template, potentially maintaining interactions with the partially negatively charged O atoms. K402 may thus be involved in the binding of the 3′ end of the template and/or in threading the template through the active site. In the model based on the DENV-3 RdRp domain structure without a template, K402 is engaged in a salt bridge with residue E491. Breaking and rebuilding of this salt bridge might be involved in template transport, and therefore, mutations affecting K402 might interfere with this process. Alternatively, the observed loss of polymerase activity in the R401-K402 mutant might be due to altered domain structure or stability of the NS5 RdRp resulting from the loss of this salt bridge.
Fig 9.
Structural model of the DENV-2 RNA polymerase domain and localization of the NLS mutations. (A) Model of the DENV-2 RdRp domain derived from the structure of the DENV-3 polymerase domain (Protein Data Bank entry 2J7W). Shown is the classical closed “right-hand” structure of the RdRp domain, with the active site provided mainly by elements from the palm subdomain, situated in the center. The finger subdomain contributes the RNA template tunnel, and the thumb subdomain contributes the priming loop, which is essential for de novo initiation. The αβNLS consists of two helices, which reside on the surface of the finger subdomain. The N-terminal helix harbors residues K371-K372 at its N terminus and residues K387-K388-K389 at its C terminus. The C-terminal helix bears residues R396-E397-E398 at its N terminus. The C terminus of this helix contributes to the RNA template tunnel. Whereas residue R401 is at the surface of the domain, K402 is orientated toward the tunnel. (B) Model of the DENV-2 RdRp domain in complex with the 3′ end of the genome (CU-3′, represented as sticks color coded according to the atom type: white, C; red, O; blue, N; orange, P), as well as the first and second nucleotides used to initiate synthesis of the negative-strand RNA: ATP (represented as sticks and color coded according to the atom type: yellow, C; red, O; blue, N; orange, P) and GTP (represented as sticks color coded according to the atom type: light blue, C; red, O; blue, N; orange, P). Green spheres represent the two catalytic Mg2+ ions. R401 resides on the surface of the domain and is not engaged in an interaction. In contrast, K402 is engaged in a salt bridge to residue E491 (red dotted lines) and is situated in close proximity to partially negatively charged groups of the RNA template (O atoms of the phosphodiester and ribose backbone) to allow possible interactions (black dotted line). The models were generated as described elsewhere (36). PyMol was used to create the images.
In conclusion, our in vitro studies demonstrate that except for mutant 401-402, decreased replication of the αβNLS mutant viruses was not due to impaired enzymatic activities of NS5. Mutations inside the minimal αβNLS barely affected polymerase activity, in contrast to the 401-402 mutation at the very C-terminal end of the αβNLS, supporting the notion that this region is a critical element of the RdRp domain.
DISCUSSION
DENV NS5 is a multifunctional enzyme that plays an important role in viral RNA replication and the inhibition of the innate immune response. Although all these processes take place in the cytoplasm, a high proportion of NS5 accumulates in the nuclei of DENV-2-infected cells. While similar results have been reported for yellow fever virus (16), nuclear localization of NS5 is not a conserved feature of all flaviviruses; West Nile virus (WNV) NS5 does not localize to the nucleus (34, 37).
We observed nuclear accumulation of DENV NS5 in a wide range of mammalian and insect cell lines, indicating that this phenomenon is highly conserved among various DENV hosts. Moreover, nuclear localization of NS5 did not depend on the expression system and was virtually identical for cells expressing this protein alone and DENV-infected cells (18, 23). However, this does not rule out a possible modulation of this process by NS5 interaction partners, such as viral RNA or other viral proteins.
The results obtained with deletion mutants identified the αβNLS as the primary determinant for nuclear localization, since deletion of the βNLS reduced, but did not abolish, the nuclear accumulation of NS5. Nevertheless, several other regions in NS5 might influence nuclear transport, probably due to their role in proper NS5 topology or their influence on importin binding. In contrast to earlier studies that targeted the minimal bipartite NLS motif (KK-X14-KKK) (18, 23), we systematically mutated all charged amino acid clusters within the αβNLS to identify the residues within this region that are most essential for NS5 nuclear transport. Although mutations within the bipartite NLS reduced nuclear transport, mutations affecting a charged amino acid cluster (396REE398) C-terminal of the bipartite NLS completely abrogated nuclear transport of NS5. This observation was remarkable, because this mutation (396–398) blocked the activities of both NLS motifs without altering them directly. It is possible that this charged amino acid cluster contributes to NS5 topology required for interaction with importins. Importantly, alanine substitutions affecting these amino acid residues impaired nuclear translocation tremendously yet did not affect, or only moderately affected, viral RNA replication. In fact, mutant 397–398 replicated to wild-type levels in spite of severely reduced amounts of nuclear NS5. Moreover, mutant 396–398, which did not express nuclear NS5 to a detectable level, was viable (Fig. 4C and D). We therefore conclude that nuclear NS5 is not a strict requirement for DENV replication.
Structural studies on DENV-3 NS5 suggest that both NLS regions form integral parts of the polymerase domain and that mutations affecting the NLSs might affect the enzymatic activities of the protein (12). By using recombinant full-length NS5, we found that, with one exception, all αβNLS mutants had RdRp activity comparable to that of the wild type, and none of the mutations affected MTase activity. These results indicate that the reduction in the level of replication of the NLS mutants was due not to disruption of NS5 enzymatic activities but possibly to an effect on the interaction of NS5 with other viral or cellular proteins or with viral RNA in the replication complex. Alternatively, the mutations might have altered the overall structure of NS5 while retaining the subdomain structures and thus their enzymatic activities. The structure of full-length NS5 has not yet been elucidated, but it can be assumed that NS5 adopts several conformations according to the particular step of genome replication (RNA synthesis and capping) and nuclear import. Indeed, small-angle X-ray scattering (SAXS) studies with soluble full-length DENV-2 NS5 suggest multiple conformations (38). In this respect, we note that K402, which was altered in mutant 401-402 (the only mutant with impaired RdRp activity), might be involved in interactions between the MTase and the RdRp domain. Such a crucial function would be underscored by the fact that K402 is strictly conserved across the Flavivirus genus (18). Moreover, our results obtained with mutant 401-402 are consistent with a previous study (39) showing that mutations of the corresponding NS5 residues (RK 400-401) of DENV-2 strain 16681 abrogate RdRp activity.
NS5 has been identified as a major determinant for IL-8 induction upon DENV infection (22), and impaired nuclear localization of NS5 has been reported to enhance IL-8 secretion (18). However, we observed that the replication competence of the virus, rather than the extent of NS5 nuclear accumulation, was the major determinant for IL-8 induction. This was found by measuring the induction of endogenous IL-8 and by using an IL-8-specific reporter assay (data not shown), arguing that DENV-induced IL-8 production is influenced by multiple factors, including replication competence and the amount of nuclear NS5.
NS5 plays a crucial role in subverting the innate immune response by blocking IFN signaling, which is mediated by interaction with STAT-2, and targeting STAT-2 for degradation (9, 25). We found that none of the αβNLS mutations affected this property, suggesting that nuclear transport of NS5 is dispensable for NS5-mediated STAT-2 degradation. Likewise, the IFN-α and IFN-γ sensitivities of the mutant viruses were not different from those of the wild type, showing that reduced nuclear accumulation of NS5 does not sensitize DENV to the antiviral effects of type I and type II interferons. We therefore conclude that the various replication efficiencies of the αβNLS mutants are independent of their abilities to counteract the IFN response.
In agreement with this conclusion, we found that αβNLS mutants with impaired RNA replication could not be rescued in trans by wild-type NS5. Rescue of replication would, however, be expected if nuclear NS5 exerted an interferon-antagonizing function outside the viral replication complex, and the same should apply to other antiviral cytokines, such as IL-8. To ensure the proper expression and processing of NS5 in the context of a polyprotein, which appears to be critical at least for the IFN-antagonistic function (9), we used a subgenomic replicon as a helper. Although a previous study (18) directly correlated reduced replication of NLS mutants with a reduction in nuclear accumulation, the effects of NLS mutations on viral replication appear to be more complex. It is possible that residues essential for nuclear accumulation at the same time exert additional functions, such as proper NS5 structure or interaction with viral and cellular proteins, that are directly related to replication. In this case, mutant 397-398 would allow the genetic uncoupling of these different functions, thus providing an important tool for future studies. Moreover, a very recent independent study found that nuclear accumulation of NS5 appears to be limited to distinct DENV serotypes. While nuclear accumulation was found for DENV-2 and DENV-3, it was much lower or undetectable for DENV-1 or DENV-4, respectively (H. Hannemann, P. Sung, J. Bird, S. P. Lim, and A. Davidson, unpublished data). Finally, other flaviviruses, such as WNV, also do not express nuclear NS5 (37). These results reveal striking differences not only between the various flaviviruses, but also between different DENV serotypes. It remains to be determined whether these differences reflect alternative strategies used by these viruses to achieve efficient replication and whether these differences correlate with pathogenesis in vivo.
ACKNOWLEDGMENTS
We thank Ulrike Engel, Christian Ackermann, and the Nikon Imaging Center (Heidelberg University) for providing assistance with confocal microscopy. We are grateful to U. Herian and F. Huschmand for excellent technical support and to Volker Lohmann for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB638, TP5) and the European Commission for Research and Innovation (FP7 HEALTH-2010 Collaborative Project SILVER; contract 260644).
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Kyle JL, Harris E. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 62:71–92 [DOI] [PubMed] [Google Scholar]
- 2. Halstead SB.2007. Dengue. Lancet 370:1644–1652 [DOI] [PubMed] [Google Scholar]
- 3. Gubler DJ. 2012. The economic burden of dengue. Am. J. Trop. Med. Hyg. 86:743–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guy B, Almond JW. 2008. Towards a dengue vaccine: progress to date and remaining challenges. Comp. Immunol. Microbiol. Infect. Dis. 31:239–252 [DOI] [PubMed] [Google Scholar]
- 5. Guy B, Guirakhoo F, Barban V, Higgs S, Monath TP, Lang J. 2010. Preclinical and clinical development of YFV 17D-based chimeric vaccines against dengue, West Nile and Japanese encephalitis viruses. Vaccine 28:632–649 [DOI] [PubMed] [Google Scholar]
- 6. Bartenschlager R, Miller S. 2008. Molecular aspects of Dengue virus replication. Future Microbiol. 3:155–165 [DOI] [PubMed] [Google Scholar]
- 7. Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. 2009. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15:2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidson AD. 2009. Chapter 2. New insights into flavivirus nonstructural protein 5. Adv. Virus Res. 74:41–101 [DOI] [PubMed] [Google Scholar]
- 12. Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317–325 [DOI] [PubMed] [Google Scholar]
- 14. Kapoor M, Zhang L, Mohan PM, Padmanabhan R. 1995. Synthesis and characterization of an infectious dengue virus type-2 RNA genome (New Guinea C strain). Gene 162:175–180 [DOI] [PubMed] [Google Scholar]
- 15. Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the Dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854–8863 [DOI] [PubMed] [Google Scholar]
- 16. Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. 1992. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 73(Part 5):1125–1130 [DOI] [PubMed] [Google Scholar]
- 17. Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. 2002. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin β1 and importin α/β-recognized nuclear localization signals. J. Biol. Chem. 277:36399–36407 [DOI] [PubMed] [Google Scholar]
- 18. Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. 2007. Nuclear localization of dengue virus nonstructural protein 5 through its importin α/β-recognized nuclear localization sequences is integral to viral infection. Traffic 8:795–807 [DOI] [PubMed] [Google Scholar]
- 19. Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. 1999. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 257:731–737 [DOI] [PubMed] [Google Scholar]
- 20. Johansson M, Brooks AJ, Jans DA, Vasudevan SG. 2001. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-β and the viral helicase, NS3. J. Gen. Virol. 82:735–745 [DOI] [PubMed] [Google Scholar]
- 21. Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100–19106 [DOI] [PubMed] [Google Scholar]
- 22. Medin CL, Fitzgerald KA, Rothman AL. 2005. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J. Virol. 79:11053–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. 2009. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J. Biol. Chem. 284:15589–15597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. 2011. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl. Trop. Dis. 5:e926 doi:10.1371/journal.pntd.0000926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200:1261–1270 [DOI] [PubMed] [Google Scholar]
- 26. Gualano RC, Pryor MJ, Cauchi MR, Wright PJ, Davidson AD. 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 79(Part 3):437–446 [DOI] [PubMed] [Google Scholar]
- 27. Pryor MJ, Carr JM, Hocking H, Davidson AD, Li P, Wright PJ. 2001. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 65:427–434 [DOI] [PubMed] [Google Scholar]
- 28. Miller S. 2007. Untersuchungen zur Topologie, Funktion und Lokalisation der Proteine NS4A und NS4B des Dengue Virus. Ph.D. thesis University of Heidelberg, Heidelberg, Germany [Google Scholar]
- 29. van den Hoff MJ, Christoffels VM, Labruyere WT, Moorman AF, Lamers WH. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48:185–197 [DOI] [PubMed] [Google Scholar]
- 30. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 31. Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khromykh AA, Sedlak PL, Westaway EG. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yap TL, Chen YL, Xu T, Wen D, Vasudevan SG, Lescar J. 2007. A multi-step strategy to obtain crystals of the dengue virus RNA-dependent RNA polymerase that diffract to high resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B. 2007. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282:10678–10689 [DOI] [PubMed] [Google Scholar]
- 35. Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E. 2010. Biochemical characterization of the (nucleoside-2′O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides 7MeGpppACn and GpppACn. J. Gen. Virol. 91:112–121 [DOI] [PubMed] [Google Scholar]
- 36. Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur J-J, Canard B. 2012. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 8:e1002912 doi:10.1371/journal.ppat.1002912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mackenzie JM, Kenney MT, Westaway EG. 2007. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J. Gen. Virol. 88:1163–1168 [DOI] [PubMed] [Google Scholar]
- 38. Bussetta C, Choi KH. 2012. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry 51:5921–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iglesias NG, Filomatori CV, Gamarnik AV. 2011. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J. Virol. 85:5745–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]