Abstract
There is an urgent need for a human immunodeficiency virus (HIV) vaccine that induces robust mucosal immunity. CD8+ cytotoxic T lymphocytes (CTLs) apply substantial antiviral pressure, but CTLs to individual epitopes select for immune escape variants in both HIV in humans and SIV in macaques. Inducing multiple simian immunodeficiency virus (SIV)-specific CTLs may assist in controlling viremia. We vaccinated 10 Mane-A1*08401+ female pigtail macaques with recombinant influenza viruses expressing three Mane-A1*08401-restricted SIV-specific CTL epitopes and subsequently challenged the animals, along with five controls, intravaginally with SIVmac251. Seroconversion to the influenza virus vector resulted and small, but detectable, SIV-specific CTL responses were induced. There was a boost in CTL responses after challenge but no protection from high-level viremia or CD4 depletion was observed. All three CTL epitopes underwent a coordinated pattern of immune escape during early SIV infection. CTL escape was more rapid in the vaccinees than in the controls at the more dominant CTL epitopes. Although CTL escape can incur a “fitness” cost to the virus, a putative compensatory mutation 20 amino acids upstream from an immunodominant Gag CTL epitope also evolved soon after the primary CTL escape mutation. We conclude that vaccines based only on CTL epitopes will likely be undermined by rapid evolution of both CTL escape and compensatory mutations. More potent and possibly broader immune responses may be required to protect pigtail macaques from SIV.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) pandemic continues to be a global health problem. Despite significant efforts, an effective vaccine has not yet been developed, and it is still unclear what a successful vaccination approach will comprise. Attempts to induce broadly reactive neutralizing antibodies have thus far been unsuccessful. Non-neutralizing antibodies appear to have afforded only modest protection in the recent Thai RV144 trial (1). Cytotoxic T lymphocyte (CTL) responses have been shown to assist in the control of HIV in humans and simian immunodeficiency virus (SIV) in macaques (2–9). Induction of CTL immunity by adenovirus vaccines was not protective in human trials (10), and a median of only one epitope-specific CTL response was induced by the regimen (11). A sieve analysis of breakthrough HIV sequence in the STEP adenovirus vaccine trial did, however, show some impact of the CTL responses in terms of selection for escape mutations (12). In the absence of complete control of viremia, selection of immune escape variants can reduce viral replication capacity (reduce “fitness”) and slow down disease progression. Mutations outside the CTL epitope can, however, partially compensate for the fitness cost of CTL escape mutations (13–17). The degree to which compensatory mutations negate fitness cost imputed by CTL escape mutations in the vaccination setting is not clear.
Broader CTL responses may impart more substantial control of viremia (18, 19), as suggested by the “heterozygous advantage” seen in subjects with a wider complement of HLA alleles (20, 21). Alternatively, eliciting CTL responses to a small number of critical epitopes pertinent for SIV control may be a more important factor for vaccine design (7).
We recently described an influenza vector SIV vaccine model whereby SIV CTL epitopes were inserted into the neuraminidase (NA) gene of mouse-adapted attenuated influenza viruses. Infection of pigtail macaques via the respiratory route induced SIV-specific CTL responses, but in pilot studies the induction of only one SIV-specific CTL response resulted in selection of immune escape variants and was not protective against SIVmac251 challenge (22). We expanded our vectors to include three SIV-specific CTL epitopes, all of which boosted CTL responses in small studies on SIV-infected animals. In the present study, we analyzed the immunogenicity, efficacy, and evolution of immune escape SIV variants after vaccination with recombinant influenza viruses expressing three SIV-specific CTL epitopes in pigtail macaques.
MATERIALS AND METHODS
Animals.
Fifteen naive female and two naive male colony-bred pigtail macaques (Macaca nemestrina) were studied, all expressing the common major histocompatibility complex (MHC) class I allele Mane-A1*08401 (previously named Mane-A*10) as confirmed using high-throughput sequencing (23). Animals were anesthetized intramuscularly with ketamine (10 mg/kg) prior to any procedures. All studies were approved by the University of Melbourne animal ethics committee.
Recombinant influenza virus-SIV constructs.
Six separate recombinant influenza A viruses used in the trial were generated using an eight-plasmid reverse genetics system as previously outlined and studied in macaques (22, 24–27). Briefly, the six DNA constructs encoded eight influenza virus segments, including a genetically manipulated neuraminidase (NA) segment containing one Mane-A*10-restricted CD8 T cell epitope sequence; either KP9 (SIV Gag164-172), KVA10 (SIV Tat114-123), or KSA10 (SIV Tat87-96). The CD8 T cell epitopes were inserted separately into the NA stalks of two mouse-adapted strains of influenza A virus using recombinant PCR techniques (25). These viruses share the same internal gene segments but differ in their surface glycoprotein hemagglutinin (HA) and NA genes—X31 (H3N2, A/HKx31) and PR8 (H1N1, A/Puerto Rico/8/1939). That is, the constructs consisted of X31-KP9, X31-KVA10, X31-KSA10, PR8-KP9, PR8-KVA10, and PR8-KSA10. The control influenza viruses consisted of X31 and PR8 without the inserted CD8 T cell epitopes. The stability of the inserted peptide epitopes within the expanded virions was confirmed by sequencing prior to vaccination.
Infection of pigtail macaques with recombinant influenza virus-SIV.
All three separate recombinant influenza virus-SIV constructs (either PR8 or X31) were administered to 10 vaccinated animals (Table 1). A total of 108 PFU of influenza virus was administered via the respiratory tract (intranasally [i.n.] and intratracheally) in phosphate-buffered saline as previously described (22). Corresponding wild-type influenza virus (108 PFU) was administered to “control” animals. All animals received the influenza viruses via the respiratory tract with the exception of four macaques (two vaccinees and two controls) that received the first two inoculations intravaginally (i.vag.), followed by two inoculations via the respiratory tract (Table 1). Vaccinations were given i.vag. based on the results from previous murine studies where i.vag. immunization with influenza A virus provided protective mucosal immunity against homosubtypic and heterosubtypic virus challenge via the respiratory tract (28). As a result, we hypothesized that i.vag. inoculation might result in sufficient infection to initiate an immune response in macaques.
Table 1.
Vaccine groups and regimens
| Test group | n | Animal no.a | Influenza virus-SIV vaccination (route, strain, dose [PFU]) at: |
SIVmac251 challenge (dose [TCID50], route) at wk 25f | |||
|---|---|---|---|---|---|---|---|
| Wk 0 | Wk 4 | Wk 9 | Wk 17 | ||||
| Vaccinees (n = 10) | 8 | 25377, 26359, B0527*, B0517, B0526, B0440, B0547, B0443* | i.n., H3N2-SIV, 108 | i.n., H1N1-SIV, 108 | i.n., H3N2-SIV, 108 | i.n., H1N1-SIV, 108 | 104, i.vag. |
| 2 | 45418†, C0933 | i.vag., H3N2-SIV, 108 | i.vag., H1N1-SIV, 108 | i.n., H3N2-SIV, 108 | i.n., H1N1-SIV, 108 | 104, i.vag. | |
| Controls (n = 7) | 3 | 19351, 19530†, B0508, | i.n.d, H3N2b, 108 | i.n., H1N1c, 108 | i.n., H3N2, 108 | i.n., H1N1, 108 | 104, i.vag. |
| 2 | 19341, C3754† | i.vag.e, H3N2, 108 | i.vag., H1N1, 108 | i.n., H3N2, 108 | i.n., H1N1, 108 | 104, i.vag. | |
| 2 | C3751‡, 5873‡ | 104, i.r. | |||||
*, Macaques euthanized at day 10 after SIV challenge; †, 3 macaques not infected after the SIVmac251 challenge at week 25 were rechallenged with a 10-fold-higher dose of SIVmac251 (105 TCID50) at week 32; ‡, male macaques infected with SIV mac251 at the same time used as controls for pyrosequencing studies.
X31 (H3N2, A/HKx31).
PR8 (H1N1, A/Puerto Rico/8/1939).
Macaques immunized intranasally (i.n.).
Macaques immunized intravaginally (i.vag.).
Note that female macaques were challenged i.vag., and male macaques were challenged intrarectally (i.r.).
The timing of influenza virus-SIV vaccination was as follows: week 0, X31 influenza virus-SIV; week 4, PR8 influenza virus-SIV; week 9, X31 influenza virus-SIV; and week 17, PR8 influenza virus-SIV (Table 1). Corresponding wild-type influenza virus (108 PFU) was administered to five “control” animals as follows: week 0, PR8 influenza virus; week 4, X31 influenza virus; week 9, X31 influenza virus; and week 17, PR8 influenza virus. Infection with influenza virus was monitored by quantitating influenza virus RNA levels in serials swabs of the upper respiratory tract postvaccination during infection (22). Briefly, swabs were analyzed using a SuperScript III One-Step reverse transcription-PCR (RT-PCR) system (Invitrogen) and primers and a probe targeting the matrix gene using the ABI 7500 real-time PCR system (Applied Biosciences). Seroconversion to influenza A virus in serial plasma samples was measured by a hemagglutination inhibition (HI) assay as previously described (29). Briefly, 25 μl of influenza virus was incubated at room temperature with 2-fold dilutions of sera treated with receptor-destroying enzyme (RDE) (Deka Seiken, Tokyo, Japan) for 1 h. Turkey red blood cells (25 μl [vol/vol]) were added, and hemagglutination was observed 30 min later. Titers are expressed as the reciprocal of the highest dilution of plasma where hemagglutination was prevented.
SIV challenge of pigtail macaques.
All female macaques (10 recombinant influenza virus-SIV-immunized animals and 5 influenza virus-immunized animals) were challenged i.vag. at week 25 with a 104 50% tissue culture infective dose (TCID50) of SIVmac251 (kindly provided by N. Miller, National Institutes of Health). In addition, two naive male macaques used solely for pyrosequencing studies were challenged intrarectally (i.r.) with a 104 TCID50 dose of SIVmac251 (Table 1). SIV infection was assessed by monitoring SIV plasma RNA levels as previously described (22). Three animals that did not become infected after the initial SIV challenge were rechallenged i.vag. 7 weeks later (at week 32) with a 10-fold-higher concentration of virus (105 TCID50 of SIVmac251). Two vaccinated animals (B0443 and B0527) were euthanized at day 10 to study tissue samples after SIVmac251 challenge, so the total number of vaccinated animals studied immunologically was 8. Animals were monitored at serial time points for total peripheral CD4 T cell depletion as previously described (30). β7high CD4+ T cells in the blood were also examined since it has previously been demonstrated that a reduction in β7high CD4+ T cells in the blood during SIV infection in rhesus macaques correlates with the loss of intestinal lamina propria CD4+ T cells, potentially providing a noninvasive method to measure disease progression (31).
CD8+ CTL responses by tetramer analysis.
SIV-specific CD8 T cell responses were measured using fresh whole heparinized blood by tetramer staining of CD3+ CD8+ lymphocytes using fluorescent Mane-A01*084 tetrameric protein folded around either the KVA10 (SIV Tat114-123), the KSA10 (SIV Tat87-96), or the KP9 (SIV Gag164-172) peptide epitope as previously described (22, 32). Expression of memory markers was assessed using a monoclonal antibody cocktail of anti-CD3-AF700 (clone SP34-2), anti-CD4-PE Cy7 (clone L200), anti-CD8-APC-H7 (clone SK1), anti-CD28 PerCP Cy5.5 (clone L293), and anti-CD95 fluorescein isothiocyanate (clone DX2). Expression of the mucosal homing marker α4β7 integrin on tetramer-positive cells was measured by counterstaining with rat anti-human integrin β7 APC (BD Pharmingen, catalog no. 551082, clone FIB504) (31).
Real-time PCR for CTL escape at KP9.
To quantify the viral levels of wild-type (WT) or escape mutant (EM) quasispecies at the KP9 epitope, we used a strain-specific real-time PCR assay developed in the laboratory (33, 34). This assay uses a forward primer specific for either the nucleotide mutation encoding the dominant K165R KP9 escape mutant or wild-type sequence. Briefly cDNA was made by reverse transcribing RNA extracted from EDTA-anticoagulated plasma and amplified by quantitative RT-PCR using both WT and EM primers specific for KP9 on an Eppendorf Realplex cycler. Analysis was performed using the Eppendorf Realplex software, where baselines were set two cycles earlier than real reported fluorescence.
Pyrosequencing for escape at KP9, KVA10, and KSA10.
To examine the kinetics of CD8+ T cell selection, we isolated and pyrosequenced plasma virus from 15 female animals in the vaccine trial and two additional male unvaccinated control animals at 10 serial time points from day 10 to week 20 after SIV infection as previously described (13). Briefly, viral RNA from EDTA anticoagulated plasma was extracted using the QIAamp MinElute virus spin kit or QIAamp Ultrasens RNA kit (Qiagen, Valencia, CA). Viral RNA was reverse transcribed and amplified using the SuperScript III One-Step RT-PCR system with Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) and MID-tagged primers (catalog no. 454; Sigma Aldrich) spanning Gag164-172 (KP9; KKFGAEVVP) and Tat114-123 (KVA10; KKETVEKAVA) and Tat87-96 (KSA10; KKAKANTSSA). The RT-PCR conditions were as follows: 50°C for 15 min; 94°C for 2 min; 40 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 50 s; and then 68°C for 5 min. DNA from PCR products was purified by cutting out appropriate bands from a 1% agarose gel and using a Qiagen gel extraction kit. Amplicons were pooled at equimolar ratios and sequenced by the Australian Genome Research Facility using a Roche/454 system. Amino acid variation within the three epitopes (Gag KP9, Tat KVA10, and Tat KSA10) was examined using a custom data analysis pipeline in Galaxy, the open source system for processing next-generation sequence data as described previously (18, 35–37). Sequence reads were translated and aligned into six reading frames and compared to a reference SIV sequence using BLAT (BLAST-Like Alignment Tool) (18, 38). Reads with low-quality bases within epitopes were discarded.
Calculation of escape rates.
We calculated the escape rate E between two time points ti and ti + 1 (see the bold line segments in Fig. 7A) assuming exponential growth/decay of viral strains during this time interval (39):
where fEM and fWT are the fractions of EM and WT in the total viral load.
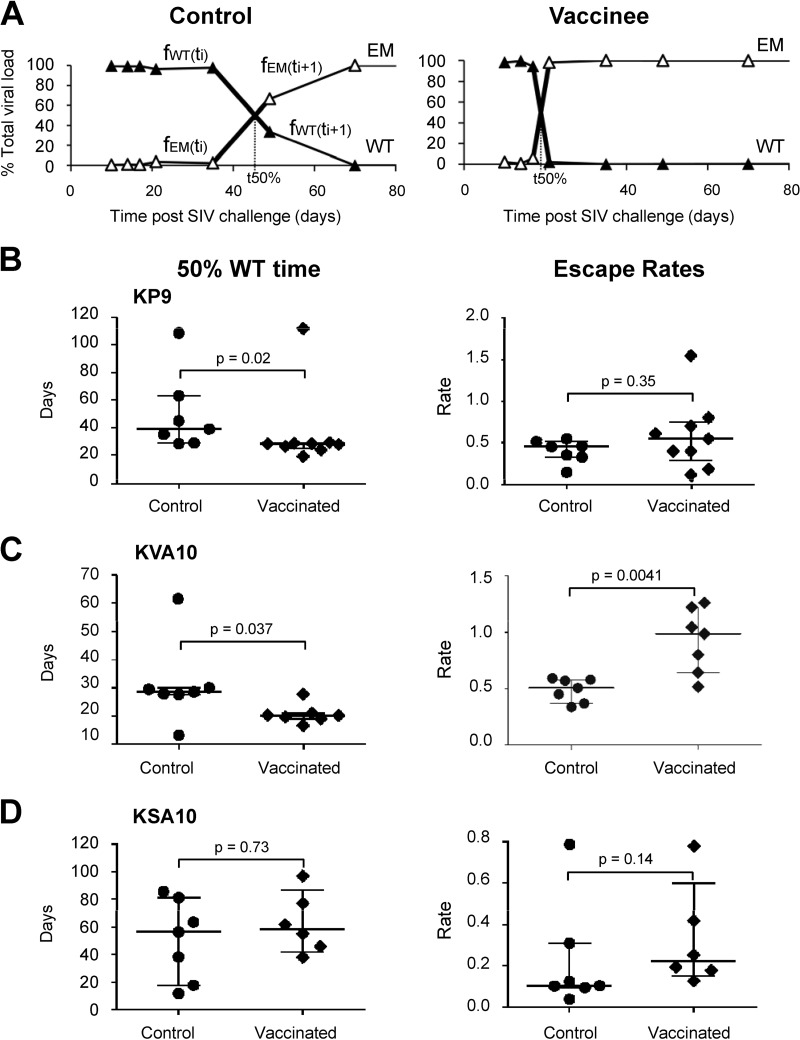
Fig 7.
Rates of CTL escape in vaccinees and controls. (A) Example of CTL escape at KP9 in a vaccinee and a control determined using pyrosequencing. The time taken to reach 50% EM (50:50 WT/EM, t50%) and maximal escape rates (bold lines, see Materials and Methods for the calculation) in a vaccinee and control animal is shown. (B to D) 50% WT time (t50%, first panel) and escape rates (second panel) for all of the animals at the three epitopes—KP9, KVA10, and KSA10—are shown. A Mann-Whitney test was used to determine whether differences between vaccinees and controls were statistically significant.
The time at which WT drops to 50% (t50%) was estimated using linear interpolation of fractions. For polymorphic escape, we defined the escape mutant as comprising all sequences different from the WT at the considered epitope (fEM = 1 − fWT). In this case, t50% lies at the intersection of fWT and fEM lines (see Fig. 7A).
RESULTS
Influenza virus-SIV inoculation of macaques.
Inducing broader CTL responses is an important goal of CTL-based HIV vaccines. We developed six separate influenza viruses expressing three SIV CTL epitopes that have previously been shown to induce SIV-specific CD8+ T cells responses in SIV-infected pigtail macaques when administered via the respiratory tract (22) and evaluated these vaccines in naive macaques. To assess the infectivity and immunogenicity of recombinant influenza viruses expressing three SIV Tat KVA10, Tat KSA10, and Gag KP9 epitopes, we immunized 10 Mane-A*10+ pigtail macaques over a period of 119 days. Influenza inoculation used two alternating doses of both H3N2 (X31) and H1N1 (PR8) strains (four vaccinations in total). Five macaques in the control group were inoculated with wild-type influenza virus strains at 108 PFU (n = 5; Table 1). The vaccine group was given a mixture of three recombinant influenza viruses (108 PFU of each) containing either KP9, KVA10 and KSA10 inserted into the neuraminidase stalk of H1N1 (PR8) and H3N2 (X31) (n = 10; Table 1) as previously described (22, 40).
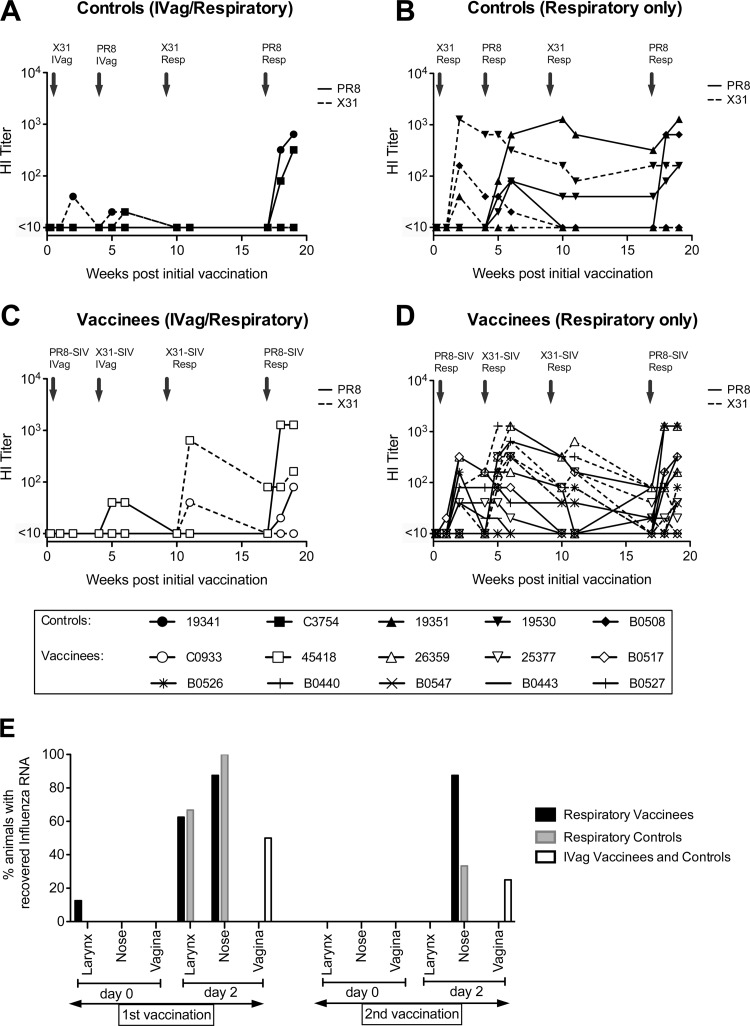
All animals received the influenza viruses via the respiratory tract with the exception of four macaques (two vaccinees and two controls) that received the first two inoculations i.vag. (Table 1) since we hypothesized that i.vag. inoculation might result in sufficient infection to initiate an immune response based on previous murine studies (28). To evaluate humoral recognition of influenza virus using the different routes of delivery of the influenza virus-SIV construct, we measured serum hemagglutination inhibition (HI) titers to both homologous parent viruses (H1N1 and H3N2) (Fig. 1). HI titers to PR8 (H1N1) and X31 (H3N2) for macaques given two i.vag. immunizations followed by two respiratory immunizations in the control group and the vaccinee group compared to those for controls and vaccinees given all immunizations via the respiratory tract are shown in Fig. 1. Influenza virus infection occurred in all macaques after two respiratory inoculations (Fig. 1B and D). The mean HI titers were significantly higher in both controls and vaccinees given two doses of influenza virus via the respiratory tract compared to intravaginally (controls: PR8, P = 0.02; X31, P = 0.03; vaccinees: PR8, P = 0.05; X31, P = 0.02 [Mann-Whitney]; Fig. 1). As a result, the four animals previously immunized via the vaginal tract received their third and fourth inoculations via the respiratory tract. These macaques subsequently seroconverted to influenza virus, with HI titers similar to macaques that had received all viruses via the respiratory tract (Fig. 1). Influenza virus RNA could be recovered from nose or larynx swabs 2 days after the first and second inoculations in most animals immunized by the respiratory route but was recovered less often in animals inoculated i.vag. (Fig. 1E).
Fig 1.
Vaccination of pigtail macaques with recombinant influenza virus-SIV. A comparison between routes of delivery of influenza virus (controls) and recombinant influenza virus-SIV vaccine (vaccinees) given intravaginally or via the respiratory tract. Hemagglutination inhibition (HI) titers in serial plasma samples to PR8 (H1N1, shown as solid lines) and X31 (H3N2, shown as dashed lines) are illustrated. Lines represent individual animals; HA titers to PR8 and X31 for the same animal are shown with the same symbol, and individual animals are identified by specific symbols as shown in the legend. HI titers of controls administered influenza virus via the vaginal tract and respiratory tract (A) and the respiratory tract only (B) are shown. HI titers of vaccinees administered recombinant influenza virus-SIV vaccine via the vaginal tract and respiratory tract (C) and the respiratory tract only are also shown (D). (E) Recovery of influenza virus RNA from nose, larynx, and vaginal swabs from vaccinees administered influenza virus-SIV via the respiratory tract (black), controls administered influenza virus via the respiratory tract (gray), and vaccinees and controls administered via the vaginal tract (white). RNA was recovered by PCR at day 0 and day 2 after the first and second influenza virus and influenza virus-SIV vaccinations.
T cell immunogenicity after vaccination and SIV challenge.
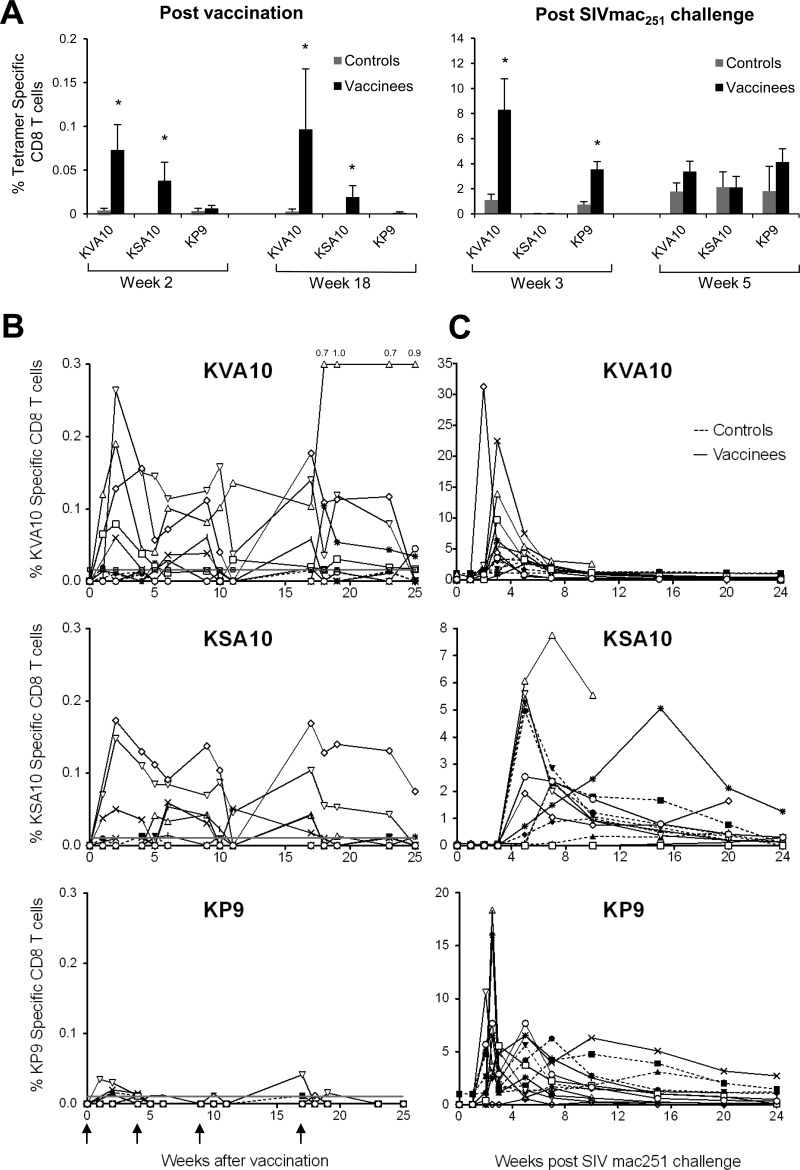
To assess the immunogenicity of the recombinant influenza-SIV vaccinations, we analyzed serial fresh blood samples for the presence of KVA10-, KSA10-, and KP9-specific CD8 T cells using specific tetramers. Modest, but significant KVA10 and KSA10 CD8 T cell responses in vaccinees compared to controls were observed 2 weeks after the first immunization and 1 week after the final vaccination at week 18 (Fig. 2A; *, P < 0.05). CTL responses to KVA10 and KSA10 in all vaccinees were above the threshold of detection (mean + 3× the standard deviation of the individual tetramer responses of all animals prior to vaccination) on at least three separate occasions postvaccination (Fig. 2B). In contrast, only one CTL response to KVA10 at one time point in the control group was above this threshold of detection.
Fig 2.
Detection of SIV Gag (KP9) and Tat (KVA10 and KSA10) specific CD8 T cells in peripheral blood. Pigtail macaques were given four vaccinations at weeks 0, 4, 9, and 17 with recombinant influenza virus-SIV (vaccinees) or influenza virus (controls) and challenged intravaginally at week 25 with pathogenic SIVmac251. (A) The means (±SE) of the proportion of CD8 T cells stained for KVA10/ManeA1*08401, KSA10/ManeA1*08401, and KP9/ManeA1*08401 tetramers at weeks 2 and 18 postvaccination and weeks 3 and 5 post SIVmac251 challenge were measured by using flow cytometry. Proportions of SIV-tetramer positive CD8+ T cells in vaccinees significantly higher than controls as determined by the Mann-Whitney test (P < 0.05) are highlighted with an asterisk (*). (B and C) Proportions of CD8 T cells stained for KVA10/ManeA1*08401, KSA10/ManeA1*08401, and KP9/ManeA1*08401 tetramers for individual macaques in vaccinees (open symbols, solid lines) and controls (solid symbols, dashed lines) after vaccination (B) and after SIVmac251 challenge (C). The horizontal line indicates the threshold of detection (mean + 3× the standard deviation of the individual tetramer responses of all animals prior to vaccination). Symbols for individual animals are shown in Fig. 1.
In contrast, the frequency of KP9-specific CD8 T cell responses in vaccinees postvaccination above the threshold of detection was lower (only 7 of 10 vaccinees had responses above the threshold). The magnitude of KP9-specific T cells responses postvaccination were also lower; however, these responses were of similar magnitude to those measured in a previous trial, where two macaques were also inoculated via the respiratory tract with the influenza virus-KP9 vaccine (22).
To address whether SIV-specific immune responses were primed by vaccination, we analyzed tetramer-specific CD8 T cell responses following SIVmac251 challenge. All female macaques were challenged intravaginally (i.vag.) and the two male macaques with challenged intrarectally (i.r.) with a 104 TCID50 of SIVmac251 8 weeks after the final vaccination (or 105 TCID50 of SIVmac251 if they did not become infected after the initial challenge). An analysis of the SIV-specific CD8 T cell responses after SIVmac251 challenge using tetramers showed that KVA10 and KP9 CD8 T cell responses in vaccinees were both earlier and significantly higher at 3 weeks postchallenge compared to the controls (Fig. 2A, KVA10, P = 0.011; KP9, P = 0.045 [Mann-Whitney test]), despite minimal KP9 CD8+ T cell responses after vaccination (Fig. 2B). It should also be noted that peripheral KSA10-specific CD8+ T cell responses were first detected at 5 weeks postchallenge, which is considerably later than for the KP9- and KVA10-specific CD8+ T cell responses that were first observed at 2 weeks postchallenge.
Induction of β7 integrin-expressing SIV-specific CD8 T cells after influenza virus-SIV vaccination and SIV challenge.
Studies in vitro have demonstrated that cells expressing high levels of α4β7 (α4+β7high) can bind to HIV and SIVsmm with high affinity, suggesting that these cells could be early targets for viral infection in vivo (41). The role of α4β7 has also been demonstrated in the mucosal homing of CD8 T cells in macaque models of SIV and other viral infections (42, 43), and it has been found that gp120 binds to and signals via an activated form of α4β7 on CD4+ T lymphocytes (41). In addition, intravenous administration of a recombinant rhesus monoclonal antibody against the α4β7 into rhesus macaques just prior to and during acute SIV infection resulted in a significant decrease in the plasma and gastrointestinal tissue viral load compared to control SIV-infected rhesus macaques (44). To assess the potential for the influenza virus-SIV inoculations to induce mucosal homing CD8 T cells, we examined SIV KVA10-, KSA10-, and KP9-specific CD8 T cells for high levels of β7 expression (β7high). Although the frequency of vaccine-induced tetramer-specific CD8 T cells was low, the proportion of high β7+ KVA10-, KSA10-, and KP9-specific CD8 T cells was greater after the fourth influenza virus-SIV boost (week 18) compared to 1 week after the first influenza virus-SIV boost (data not shown). This suggests that the mucosal vaccination may have induced SIV-specific CD8 T cells with mucosal homing capabilities.
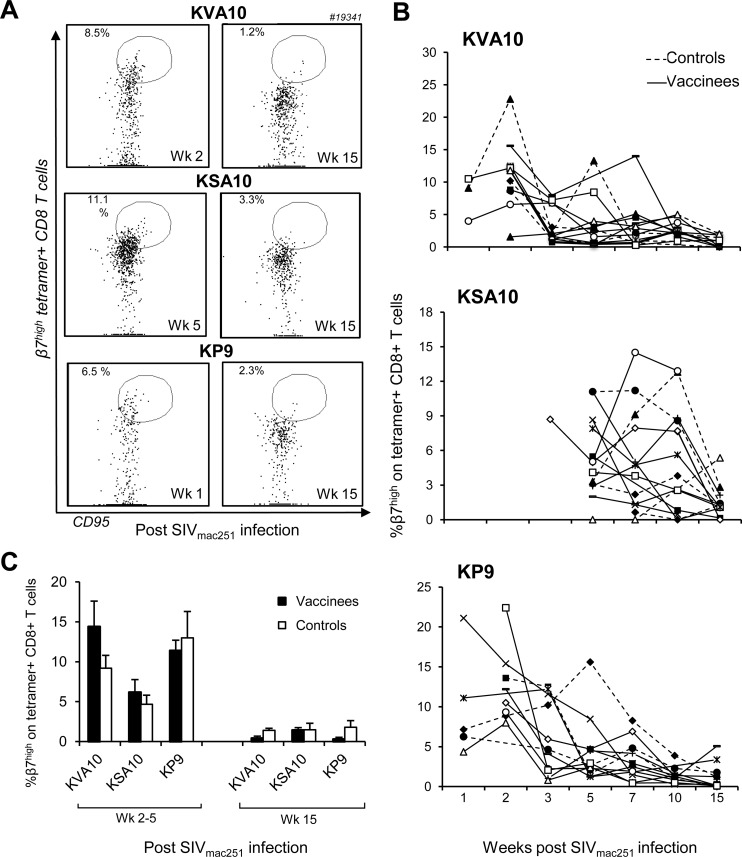
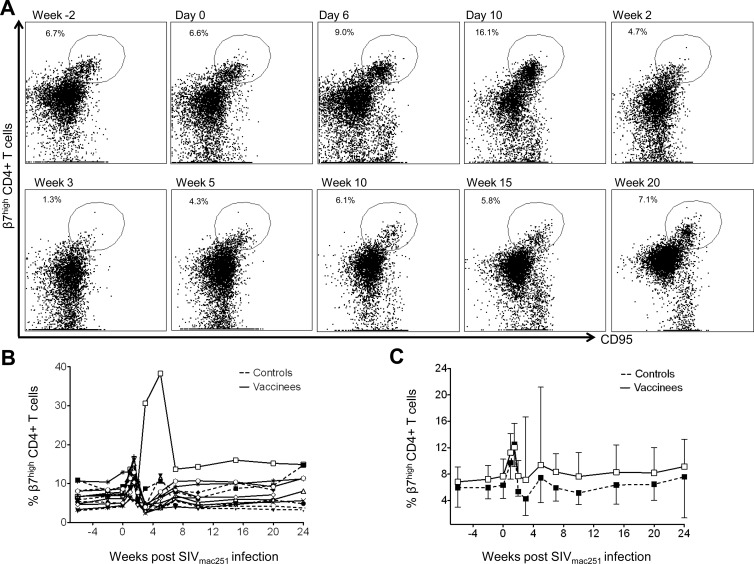
Since macaques were immunized with the influenza virus-SIV vaccine via the respiratory tract, we examined mucosal homing characteristics of SIV-specific CD8 T cells after SIV challenge (Fig. 3). A progressive decline in the proportion of peripheral β7high SIV KVA10-, KSA10-, and KP9-specific CD8 T cells was observed out to week 15 after SIV challenge. Note that the percentage of β7high tetramer-positive cells could only be measured once tetramer-specific CTLs appeared in the periphery, explaining the absences in time point values in the figures. For instance, peripheral KSA10-specific cells were not measured in the majority of animals until week 5, whereas KP9-specific and KVA10-specific CTL responses were measured at week 2. Flow cytometry dot plots from a representative vaccinated macaque showing the decline in CD95+ β7high CD8+ SIV tetramer-positive cells after the emergence of SIV tetramer positive cells in the periphery compared to at week 15 are shown in Fig. 3A. This decline is also evident when examining the proportions of β7high CD95+ CD8+ SIV tetramer-positive cells in individual animals at serial time points after SIV mac251 challenge (Fig. 3B). The decrease in the proportion of β7high CD95+ CD8+ SIV tetramer-positive cells shortly after challenge compared to during chronic infection was statistically significant (mean ± the standard error [SE] of controls [KVA10, P = 0.01; KSA10, P = 0.05; KP9, P = 0.002] and vaccinees [KVA10, P = 0.006; KSA10, P = 0.001; KP9, P = 0.01] calculated using the Wilcoxon test, Fig. 3C). The decline in β7high tetramer-positive cells in the periphery was observed in both controls and vaccinees and may be explained by their migration to SIV-infected mucosal tissues after SIV infection or death of this particular subset of cells.
Fig 3.
Expression of β7 integrin on SIV/Mane-A1*08401 tetramer-positive CD8 T cells. The expression of peripheral CD95+ β7high cells on CD3+ CD8+ SIV tetramer-positive cells after challenge with SIVmac251 was determined using flow cytometry. (A) Flow cytometry dot plots from a representative vaccinee macaque showing the proportion of CD95+ β7high cells when CD3+ CD8+ tetramer-positive cells first appear in the periphery: at week 2 postchallenge for KVA10- and KP9-positive CD8+ T cells and at week 5 for KSA10-positive CD8+ T cells and during chronic infection at 15 weeks after SIV challenge. (B) Individual vaccinees (open symbols, solid lines) and controls (solids symbols, dashed lines) macaques showing the proportion of CD95+ CD3+ CD8+ SIV tetramer-positive cells with β7high expression at serial time points after SIV mac251 challenge. Symbols for individual animals are as defined in Fig. 1. (C) Means ± the SE of the proportions of CD95+ β7high cells on CD3+ CD8+ SIV tetramer-positive cells on vaccinees (■) and controls (□) at week 2 after SIV challenge for KVA10 and KP9, at week 5 after SIV challenge for KSA10, and at week 15 after SIV challenge.
Outcome of SIV challenge.
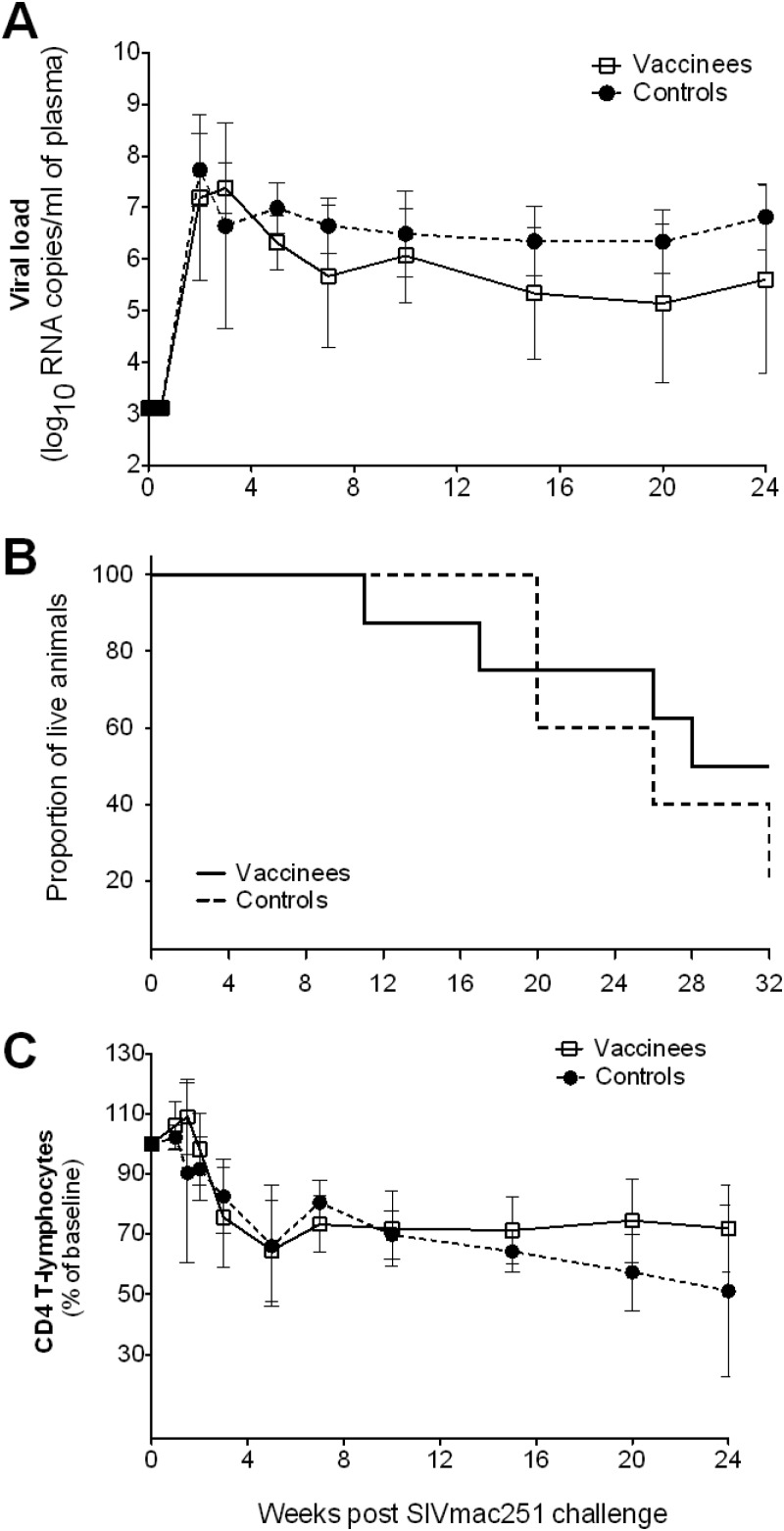
Eight weeks after the final vaccination, all 15 vaccinated female macaques were challenged i.vag. with a 104 TCID50 of SIVmac251. Two additional male macaques, added to the study to be used as controls for the pyrosequencing studies, were challenged i.r. with a 104 TCID50 dose of SIVmac251 (Table 1). Fourteen of the seventeen animals became infected after this initial challenge with SIVmac251; however, three macaques (one vaccinee and two controls) did not become infected. As a consequence, these three macaques were rechallenged with a 10-fold-higher dose of SIVmac251 (105 TCID50) at week 32, after which they became infected. There was no significant difference in mean viral loads (Fig. 4A) or survival (Fig. 4B) between influenza virus-SIV-vaccinated animals and influenza virus-vaccinated animals. The mean (±SE) peak viral loads 2 weeks after challenge between vaccinated and control groups were almost identical: 7.70 (±0.54) versus 7.73 (±0.71) log10 copies/ml, respectively. Although there was a subtle difference in the viral loads between controls and vaccinees during chronic infection, with the viral loads in the vaccinees being slightly lower than the controls (Fig. 4A), the difference was not found to be statistically significant when we compared the average chronic viral loads from week 5 and week 20 between controls and vaccinees using the Mann-Whitney test (P = 0.15).
Fig 4.
Outcome of SIVmac251 challenge of macaques vaccinated with influenza-SIV. Each macaque was challenged intravaginally with 104 TCID50 SIVmac251 at week 25 (8 weeks after the final immunization). Three macaques (one vaccinee and two controls) that did not become infected were rechallenged intravaginally with a 10-fold-higher dose of SIVmac251 (105 TCID50) at week 32, after which they came infected. Blood samples from vaccinees (□) and controls (●) were analyzed for SIV RNA by RT-PCR. (A) Mean (± the SE) of viral loads. (B) Kaplan-Meier survival graph for SIV-infected pigtail macaques. (C) CD4 T cell loss for each vaccine group. The mean values (± the SE) of the percentage of CD4 T cells at baseline are given. There were no significant differences between vaccinees and controls.
Loss of CD4+ T cells and β7high CD4+ T cells postchallenge.
A decrease in total peripheral CD4 T cells postchallenge was observed in both vaccinee and control groups, with no significant difference between the groups (Fig. 4C). It has previously been reported that memory CD4+ T cells in peripheral blood expressing high levels of the α4β7 receptor are preferentially infected and depleted during acute SIV infection, and this depletion correlated with depletion of gut CD4 T cells in rhesus macaques (31). We therefore measured β7high CD4 T cells in serial blood samples, and flow cytometry dot plots of CD95+ β7high CD4+ T cells of a representative vaccinated animal are shown in Fig. 5A. A decrease in the proportion of CD95+ β7high CD4 T cells in most animals was observed at week 2 to 3 during acute infection (31, 45) when examining individual macaques before and after challenge with SIVmac251 (Fig. 5B). Surprisingly, however, this decline was only transient, and by week 10 after SIV challenge, the proportions of peripheral CD95+ β7high CD4 T cells had returned to prechallenge levels. It is also interesting that the proportions of CD95+ β7high CD4 T cells in blood in all macaques, with the exception of one, increased at day 10 after SIV challenge prior to this decrease (Fig. 5B). There was no significant difference in the proportion of CD95+ β7high CD4 T cells in blood between the vaccinees and controls (Fig. 5C).
Fig 5.
Expression of β7high integrin on peripheral CD4 T cells. (A) Flow cytometry dot plot demonstrating transient loss of CD4+ CD95+ β7high T cells in blood 3 weeks after challenge with SIVmac251 in a representative vaccinated macaque. (B) Proportions of β7high CD4+ T cells of vaccinees (open symbols) and controls (filled symbols) in blood before and after SIVmac251 challenge for individual macaques. Symbols for individual animals are as defined in Fig. 1. (C) Means (± the SE) of the proportions of β7high CD4 T cells in blood in vaccinees and controls.
An increase, followed by a transient decline and then a return to baseline, of peripheral CD95+ β7high CD4 T cells may represent a redistribution of cells between the periphery and the mucosa. That is, CD95+ β7high CD4 T cells migrate from the mucosa to the periphery during acute infection, leading to an increase in CD95+ β7high CD4 T cells in the periphery. Shortly after, CD95+ β7high CD4 T cells are either killed or migrate back to the periphery and return to baseline levels as a result of an equilibration forming between redistribution and loss of cells between the mucosa and the periphery. Although these results are not consistent with the loss of β7high CD4 T cells observed after challenge with SIV in rhesus macaques (31), the physiological significance of α4β7 during SIV infection in pigtail macaques may be minimal.
Evolution of immune escape of SIV-specific CD8 T cells induced by influenza virus-SIV vaccines.
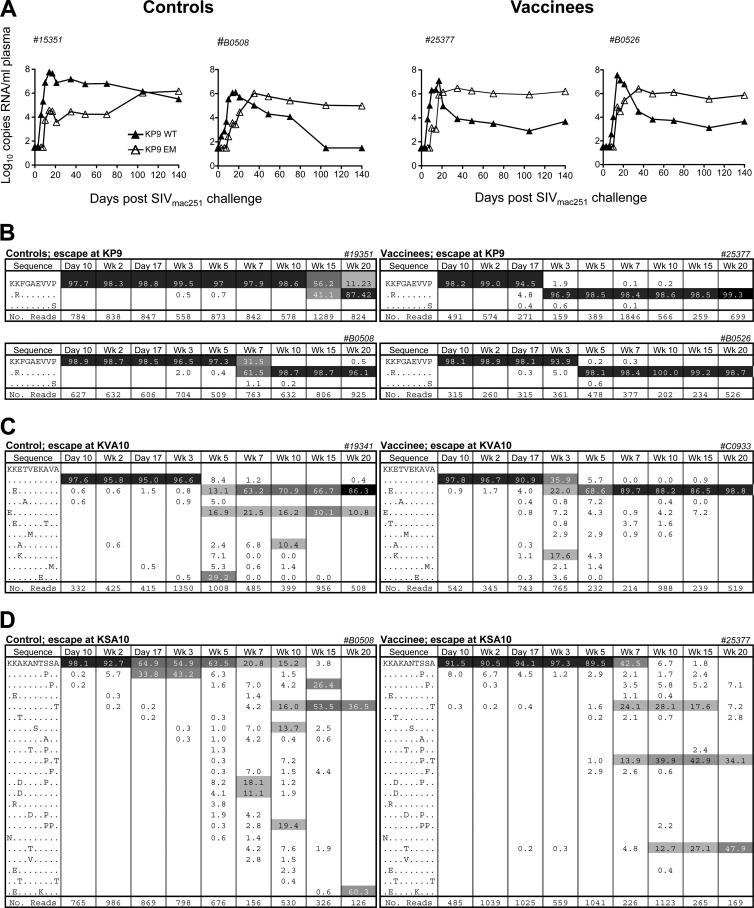
A possible explanation for the lack of control of SIV viremia despite the priming of multiple CTL responses was the rapid evolution of immune escape. We initially quantified CTL escape at the KP9 Gag epitope in plasma viral RNA at serial time points from day 10 to week 20 using a strain-specific PCR assay (33, 34) and found that vaccination with recombinant influenza virus-SIV generally resulted in an earlier generation of immune escape variants at the Gag KP9 CD8 T cell epitope compared to the control animals inoculated with influenza virus alone. The findings for KP9 escape in two representative vaccinees and controls are presented in Fig. 6A.
Fig 6.
Evolution of KP9, KVA10 and KSA10 CTL escape. (A) Examples of K165R CTL escape mutation kinetics by using a strain-specific real-time PCR assay in serial plasma RNA samples from control macaques (left panel) and vaccinees (right panel). (B to D) Examples of CTL escape by pyrosequencing in controls (left panels) and vaccinees (right panels) for the three CTL epitopes KP9 (B), KVA10 (C), and KSA10 (D). The CTL amino acid sequence is shown in the first column, with the percentage of the sequence in the subsequent columns and the time point after SIV challenge at the top of the column. The mutation identified is shown at each time point, with the total reads shown in the bottom row. Common variants at each time point are shaded. Rarer variants account for the remaining sequences.
Although the Gag KP9 CTL epitope usually escapes in a monomorphic way (K165R at position 2), the two previously observed Tat epitopes (KSA10, KVA10) escape in polymorphic ways (46) across animals, so a strain-specific real-time PCR assay is not suitable for these epitopes. As a result, to assess escape at the KSA10 and KVA10 epitopes, we performed pyrosequencing across these epitopes, as well as the KP9 epitope.
Analysis of immune escape from plasma RNA at the Gag KP9 epitope using pyrosequencing showed that immune escape not only occurred earlier in vaccinees, often occurring 14 days after inoculation, but also occurred very rapidly (i.e., at a high rate). The same two representative vaccinees and controls showing escape at KP9 by pyrosequencing are illustrated in Fig. 6B. Pyrosequencing was able to confirm results obtained using the KP9-specific PCR assay, as illustrated in Fig. 6A (33, 34). In addition, pyrosequencing revealed early and very rapid escape at the Tat KVA10 and KSA10 epitopes (Fig. 6C and D). As mentioned above, this escape was polymorphic in nature. The divergent nature of escape at the two Tat epitopes in control and vaccinee animals is represented in Fig. 6C and D.
Since we obtained an average of 500 reads for each sequencing time point using pyrosequencing, we could confidently detect the emergence of a range of viral variants with escape mutations with frequencies as low as 1% of circulating virus, which is consistent with the recently published pyrosequencing error rate (18).
The rate of CTL escape provides an estimate of the pressure applied by the CTL response (39, 47). We calculated both maximal escape rate assuming exponential growth/decay of WT and EM strains during this time interval (shown as bold lines in Fig. 7A) (39) and the time for the EM to reach 50% of the total variants (indicated as “t50%” in Fig. 7A). For polymorphic escape, we defined the escape mutant as comprising all sequences different from WT at the considered epitope. The analysis of the time taken for the virus to escape from 100 to 50% wild-type (WT) at the three CTL epitopes revealed that KP9 and KVA10 escape in the vaccinees emerged significantly earlier than in the control group. That is, 50% WT virus at the KP9 and KVA10 epitope occurred significantly earlier in vaccinees compared to controls (Mann-Whitney test; KP9, 50 ± 11 days versus 38 ± 12 days, P = 0.02; KVA10, 31 ± 6 days versus 21 ± 1 days, P = 0.037; Fig. 7B and C) but did not occur significantly faster for KSA10 (90 ± 14 days versus 68 ± 5 days, P = 0.73) (Fig. 7D). In addition to escape occurring earlier, the rate of escape was shown to be also significantly quicker only for KVA10 (Mann-Whitney test; P = 0.0041) (Fig. 7C). The rapid emergence of KP9 and KVA10 escape mutants may explain the failure to observe significantly lower viremia levels in vaccinated animals compared to controls. The emergence of both KSA10-specific CTL and KSA10 escape mutant virus was delayed in both vaccinated and control animals, and occurred significantly after the establishment of a “set-point” viral load. This suggests that this epitope may not be crucial to viral control in these animals. The failure of vaccination to significantly accelerate the development of KSA10 responses may suggest that the delayed response occurs as a result of late presentation of this epitope, which is not accelerated by vaccination.
Compensatory mutations.
Compensatory mutations outside an epitope can partially restore the replicative capacity of viruses. Certain patterns of linked mutations associated with CD8+ T cell epitope escape in highly conserved regions may also lead to variable levels of viral fitness (13). The K165R Gag CTL escape mutation has been shown to incur a significant fitness cost since the mutation rapidly reverts to WT following in vivo passage in MHC-mismatched animals (39, 48). This might suggest that, in the absence of compensatory mutations, the more rapid CTL escape observed in the vaccinees would incur a fitness cost and lead to reduced viremia. Since we did not in fact observe any virological efficacy, we hypothesized that rapid development of compensatory mutations may have evolved. We therefore examined the pyrosequencing data outside the immunodominant epitopes for evidence of mutations linked to the Gag K165R mutation.
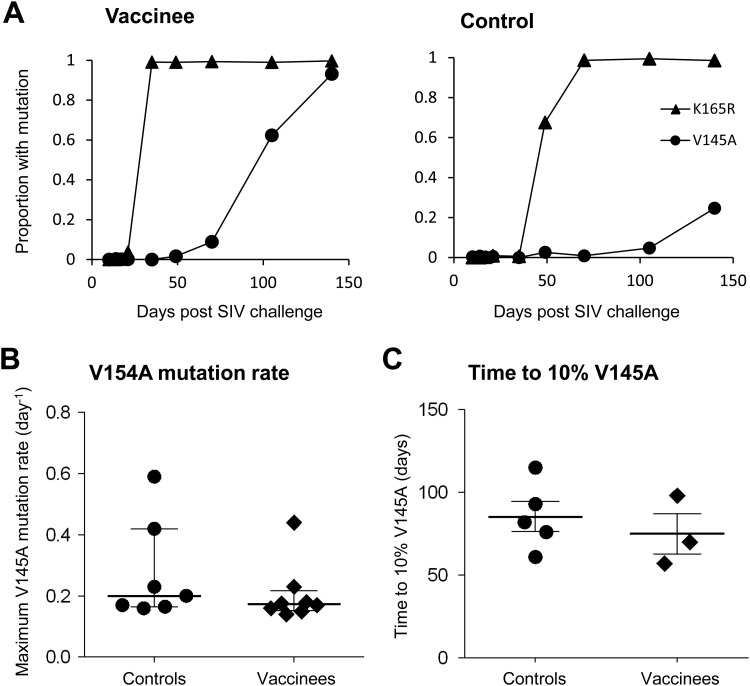
We found that a mutation 20 amino acids upstream of the KP9 epitope (V145A) that commonly followed the emergence of the K165R mutation and was detected in 9 of 15 animals. This mutation always emerged at a later time point relative to the KP9 (K165R) escape mutation. The V145A mutation could potentially be either a newly identified escape mutation or a compensatory mutation that occurs to overcome the substantial fitness cost of the K165R mutation. The likelihood that the V145A mutation is compensatory is strongly supported by (i) significant levels of the V145A mutation above background sequencing errors (>0.9%) only developed as the percentage of the K165R mutation approached 100% and not before and (ii) the fact that no CD4 or CD8 T cell responses were detected across the V145A site from peripheral blood mononuclear cell samples obtained from animals despite repeated efforts to detect them using peptide-stimulated intracellular cytokine staining assays (data not shown).
The potential V145A compensatory mutation only occurred relatively late in infection and the rate of selection of the V145A mutation was relatively slow compared to the earlier K165R mutations (Fig. 8A). Analysis of the maximum rate of development of this putative compensatory mutation showed no significant difference between controls and vaccines (Fig. 8B). Of the animals in which the V145A mutation was measured at 10% of total virus, there was a slight trend for the V145A mutation to reach 10% of total virus faster in vaccinated animals (76 ± 11 days versus 84 ± 9 days in controls, Fig. 8C), but this was not statistically significant. This slower selection of a compensatory mutation compared to the initial escape mutation might be expected, since the fitness cost of escape is always lower than the “immune benefit” of escape (39). Moreover, as previously shown, the fitness costs of mutation are directly proportional to total CD4+ T cell availability, so these fitness costs may reduce over time in chronic infection (49). In addition, it seems likely that the V145A mutation may only partly compensate for the fitness cost of K165R.
Fig 8.
Rate of emergence of putative V145A compensatory mutation. (A) Rate of selection of the V145A and K165R mutation in a representative vaccinee and control macaque as measured by the proportion of viruses containing the respective mutations using pyrosequencing data. (B) Maximum V154A mutation rates (per day) measured for controls and vaccinees determined using pyrosequencing data. (C) Time taken for the eight macaques (five controls, three vaccinees) displaying the putative compensatory mutation V145A to reach 10% V145A, as assessed using pyrosequencing data.
DISCUSSION
Developing a safe and effective HIV vaccine is a global health priority. CD8 T cell immunity is clearly important in controlling viremia in SIV-macaques (2, 5, 9, 50). Studying how CD8 T cells induce effective immunity and determining the optimal set of HIV-derived CD8+ T cell antigens and utilization of these antigens in appropriate vectors is a major goal of HIV vaccine design.
To understand the breadth of CD8 T cells required to facilitate control of SIV viremia by vaccination, we inserted three immunodominant SIV-specific CTL epitopes, Gag (KP9) and Tat (KSA10 and KVA10), into the influenza virus vector and analyzed the immunogenicity and protective efficacy of the vaccine. In addition, the evolution of immune escape variants of recombinant influenza viruses expressing the three SIV-specific CTL epitopes in pigtail macaques was examined. Despite priming SIV-specific CTL responses to these epitopes, there were no significant differences in the viral loads, loss of peripheral CD4+ T cells or survival between vaccinees and controls indicating that the vaccine did not provide any protection from SIV disease progression. We were unable to confirm that loss of β7high CD4 T cells in the periphery was an indicator of disease progression as previously reported (45). The physiological significance of α4β7 in SIV infection of pigtail macaques is unclear and may be minimal.
The inability of the influenza virus vaccine with the three SIV CTL epitopes to control viremia may illustrate the importance of a broader base (>3 epitopes) of CD8 T cell responses in facilitating control of SIV (50, 51). Alternatively, the three SIV CD8 T cell epitopes that we inserted into the influenza virus may not contain the critical epitopes necessary to control SIV replication in pigtail macaques. Recent studies have illustrated that a narrowly targeted vaccine containing only 3 SIV CTL epitopes inducing SIV-specific CD8+ T cell responses was sufficient to control SIV replication in Mamu-B*08+ Indian rhesus macaques (7). Future studies in pigtail macaques testing a range of immunodominant epitopes may assist in refining CTL-based control of primate lentiviruses. Furthermore, CD4+ T cell epitopes that generate functionally effective antibodies responses could be incorporated into vaccine vectors to further probe the minimal base of immunity required for protective efficacy.
The faster emergence of immune escape mutants at the Gag KP9 and Tat KVA10 epitopes in the vaccinated macaques identified by both real-time PCR and pyrosequencing (Fig. 6 and 7) suggests that the effectiveness of vaccine-induced CTL responses can easily be undermined by rapid viral evolution (52). This may be the consequence of only a small number of epitopes being targeted or the selection of epitopes that do not effectively control SIV viremia. Mudd et al. recently showed that vaccination with gene fragments containing SIV epitopes more highly associated with control of SIV viremia was more effective in controlling SIV viremia, although this was undermined by immune escape in a small subset of vaccinees (7). Recently, influenza virus A replicons have been engineered that enable the expression of recombinant proteins and green fluorescent protein (53, 54). If these vectors could be engineered to express whole HIV or SIV proteins or large gene fragments, they may be able to induce antibody or CD4+ and CD8+ T cell responses.
We initially reasoned that faster CTL escape might reduce virus fitness and lead to some reduction in viremia; however, we did not observe this. The extensive set of sequencing data from pyrosequencing from serial time points of viral RNA extracted from plasma day 10 to week 20 enabled us to identify a putative compensatory mutation (V145A) that was strongly associated with the Gag KP9 K165R escape mutation. This mutation may mitigate the fitness cost associated with the primary CTL escape mutation. Thus, the phenomenon of rapid viral escape and compensation may pose a significant impediment to CTL-based vaccines targeting a small number of epitopes (50, 51).
There are several limitations to our studies. First, the CTL responses induced by the vaccines were modest and more potent vaccines may reveal a greater impact on viral transmission, especially at the mucosae, as shown by Mudd et al. (7). We might also expect, however, that a higher magnitude of CTL response directed to a limited number or particular set of epitopes may drive faster viral escape and compensation as previously demonstrated when immunizing with a recombinant influenza virus construct containing only one immunodominant Gag CTL epitope, KP9 (55). Second, although there were no significant differences in viral loads between vaccinees and controls, we cannot exclude a modest effect of vaccination. Even compensatory mutations may not fully restore viral fitness. Third, although the putative V145A compensatory mutation was strongly associated with the K165R CTL escape mutation, detailed in vitro viral competition assays with molecular clones will be required to confirm this.
HIV infections remain a major global health threat, and an effective prophylactic vaccine is urgently needed. Our study demonstrated that a vaccine based on three immunodominant CTL epitopes in pigtail macaques was unable to protect animals from high levels of viremia and CD4 T cell loss after SIVmac251 challenge, despite elicitation of modest SIV-specific responses postvaccination. Rapid evolution of both CTL responses and compensatory mutations are likely to be the major causes of the failure of this vaccine. We conclude that CTL based vaccines need to induce stronger immune responses to specific CTL epitopes that are pertinent to the protection of pigtail macaques from SIV.
ACKNOWLEDGMENTS
We thank Jie Lin for supplying the tetramers used in the study and Ben Burwitz for his assistance with pyrosequencing. We also thank David O'Connor for helpful advice.
This study was supported by Australian NHMRC awards 628331, 1025567, and 510488, by the Australian Centre for HIV and Hepatitis Research, and by Gates Foundation award OPP1008294. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonaldo MC, Martins MA, Rudersdorf R, Mudd PA, Sacha JB, Piaskowski SM, Costa Neves PC, Veloso de Santana MG, Vojnov L, Capuano S, III, Rakasz EG, Wilson NA, Fulkerson J, Sadoff JC, Watkins DI, Galler R. 2010. Recombinant yellow fever vaccine virus 17D expressing simian immunodeficiency virus SIVmac239 gag induces SIV-specific CD8+ T-cell responses in rhesus macaques. J. Virol. 84:3699–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211 [DOI] [PubMed] [Google Scholar]
- 4. Evans DT, O'Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270–1276 [DOI] [PubMed] [Google Scholar]
- 5. Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Iii SC, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M, Jr, Haase AT, Lifson JD, Allen TM, Watkins DI. 2012. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 491:129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nixon DF, Townsend AR, Elvin JG, Rizza CR, Gallwey J, McMichael AJ. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484–487 [DOI] [PubMed] [Google Scholar]
- 9. Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860 [DOI] [PubMed] [Google Scholar]
- 10. Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, O'Sullivan A, Crossler J, Jones T, Nau M, Wong K, Zhao H, Raugi DN, Sorensen S, Stoddard JN, Maust BS, Deng W, Hural J, Dubey S, Michael NL, Shiver J, Corey L, Li F, Self SG, Kim J, Buchbinder S, Casimiro DR, Robertson MN, Duerr A, McElrath MJ, McCutchan FE, Mullins JI. 2011. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat. Med. 17:366–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burwitz BJ, Sacha JB, Reed JS, Newman LP, Norante FA, Bimber BN, Wilson NA, Watkins DI, O'Connor DH. 2011. Pyrosequencing reveals restricted patterns of CD8+ T cell escape-associated compensatory mutations in simian immunodeficiency virus. J. Virol. 85:13088–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dahirel V, Shekhar K, Pereyra F, Miura T, Artyomov M, Talsania S, Allen TM, Altfeld M, Carrington M, Irvine DJ, Walker BD, Chakraborty AK. 2011. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc. Natl. Acad. Sci. U. S. A. 108:11530–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedrich TC, Frye CA, Yant LJ, O'Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, Hughes AL, Wilson N, Watkins DI. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang KH, Goedhals D, Carlson JM, Brockman MA, Mishra S, Brumme ZL, Hickling S, Tang CS, Miura T, Seebregts C, Heckerman D, Ndung'u T, Walker B, Klenerman P, Steyn D, Goulder P, Phillips R, van Vuuren C, Frater J. 2011. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS One 6:e19018 doi:10.1371/journal.pone.0019018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Connor SL, Becker EA, Weinfurter JT, Chin EN, Budde ML, Gostick E, Correll M, Gleicher M, Hughes AL, Price DA, Friedrich TC, O'Connor DH. 2012. Conditional CD8+ T cell escape during acute simian immunodeficiency virus infection. J. Virol. 86:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103–2106 [DOI] [PubMed] [Google Scholar]
- 20. Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 21. O'Connor SL, Lhost JJ, Becker EA, Detmer AM, Johnson RC, Macnair CE, Wiseman RW, Karl JA, Greene JM, Burwitz BJ, Bimber BN, Lank SM, Tuscher JJ, Mee ET, Rose NJ, Desrosiers RC, Hughes AL, Friedrich TC, Carrington M, O'Connor DH. 2010. MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci. Transl. Med. 2:22ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sexton A, De Rose R, Reece JC, Alcantara S, Loh L, Moffat JM, Laurie K, Hurt A, Doherty PC, Turner SJ, Kent SJ, Stambas J. 2009. Evaluation of recombinant influenza virus-simian immunodeficiency virus vaccines in macaques. J. Virol. 83:7619–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fernandez CS, Reece JC, Saepuloh U, De Rose R, Ishkandriati D, O'Connor DH, Wiseman RW, Kent SJ. 2011. Screening and confirmatory testing of MHC class I alleles in pig-tailed macaques. Immunogenetics. 63:511–521 [DOI] [PubMed] [Google Scholar]
- 24. Andreansky SS, Stambas J, Thomas PG, Xie W, Webby RJ, Doherty PC. 2005. Consequences of immunodominant epitope deletion for minor influenza virus-specific CD8+-T-cell responses. J. Virol. 79:4329–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC. 2006. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc. Natl. Acad. Sci. U. S. A. 103:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Webby RJ, Andreansky S, Stambas J, Rehg JE, Webster RG, Doherty PC, Turner SJ. 2003. Protection and compensation in the influenza virus-specific CD8+ T cell response. Proc. Natl. Acad. Sci. U. S. A. 100:7235–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garulli B, Meola M, Stillitano MG, Kawaoka Y, Castrucci MR. 2007. Efficient vagina-to-lower respiratory tract immune trafficking in a murine model of influenza A virus infection. Virology 361:274–282 [DOI] [PubMed] [Google Scholar]
- 29. Mathews JD, Chesson JM, McCaw JM, McVernon J. 2009. Understanding influenza transmission, immunity and pandemic threats. Influenza Other Respir. Viruses 3:143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kent SJ, De Rose R, Mokhonov VV, Mokhonova EI, Fernandez CS, Alcantara S, Rollman E, Mason RD, Loh L, Peut V, Reece JC, Wang XJ, Wilson KM, Suhrbier A, Khromykh A. 2008. Evaluation of recombinant Kunjin replicon SIV vaccines for protective efficacy in macaques. Virology 374:528–534 [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Xu H, Gill AF, Pahar B, Kempf D, Rasmussen T, Lackner AA, Veazey RS. 2009. Monitoring α4β7 integrin expression on circulating CD4+ T cells as a surrogate marker for tracking intestinal CD4+ T-cell loss in SIV infection. Mucosal Immunol. 2:518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Rose R, Mason RD, Loh L, Peut V, Smith MZ, Fernandez CS, Alcantara S, Amarasena T, Reece J, Seddiki N, Kelleher AD, Zaunders J, Kent SJ. 2008. Safety, immunogenicity, and efficacy of peptide-pulsed cellular immunotherapy in macaques. J. Med. Primatol 37(Suppl 2):69–78 [DOI] [PubMed] [Google Scholar]
- 33. Loh L, Kent SJ. 2008. Quantification of simian immunodeficiency virus cytotoxic T lymphocyte escape mutant viruses. AIDS Res. Hum. Retrovir. 24:1067–1072 [DOI] [PubMed] [Google Scholar]
- 34. Loh L, Petravic J, Batten CJ, Davenport MP, Kent SJ. 2008. Vaccination and timing influence SIV immune escape viral dynamics in vivo. PLoS Pathog. 4:e12 doi:10.1371/journal.ppat.0040012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blankenberg D, Taylor J, Nekrutenko A. 2011. Making whole genome multiple alignments usable for biologists. Bioinformatics (Oxford, England) 27:2426–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19:Unit 19 10 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent WJ. 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez CS, Stratov I, De Rose R, Walsh K, Dale CJ, Smith MZ, Agy MB, Hu SL, Krebs K, Watkins DI, O'Connor HD, Davenport MP, Kent SJ. 2005. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J. Virol. 79:5721–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jegaskanda S, Reece JC, De Rose R, Stambas J, Sullivan L, Brooks AG, Kent SJ, Sexton A. 2012. Comparison of influenza and SIV specific CD8 T cell responses in macaques. PLoS One 7:e32431 doi:10.1371/journal.pone.0032431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9:301–309 [DOI] [PubMed] [Google Scholar]
- 42. Cromwell MA, Veazey RS, Altman JD, Mansfield KG, Glickman R, Allen TM, Watkins DI, Lackner AA, Johnson RP. 2000. Induction of mucosal homing virus-specific CD8+ T lymphocytes by attenuated simian immunodeficiency virus. J. Virol. 74:8762–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaufman DR, Liu J, Carville A, Mansfield KG, Havenga MJ, Goudsmit J, Barouch DH. 2008. Trafficking of antigen-specific CD8+ T lymphocytes to mucosal surfaces following intramuscular vaccination. J. Immunol. 181:4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, Wang X, Li J, Price AA, Little DM, Zaidi M, Lyles R, Villinger F. 2011. Blocking of α4β7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J. Immunol. 186:1044–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kader M, Wang X, Piatak M, Lifson J, Roederer M, Veazey R, Mattapallil JJ. 2009. α4+ β7hi CD4+ memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mason RD, Alcantara S, Peut V, Loh L, Lifson JD, De Rose R, Kent SJ. 2009. Inactivated simian immunodeficiency virus-pulsed autologous fresh blood cells as an immunotherapy strategy. J. Virol. 83:1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davenport MP, Loh L, Petravic J, Kent SJ. 2008. Rates of HIV immune escape and reversion: implications for vaccination. Trends Microbiol. 16:561–566 [DOI] [PubMed] [Google Scholar]
- 48. Fernandez CS, Smith MZ, Batten CJ, De Rose R, Reece JC, Rollman E, Venturi V, Davenport MP, Kent SJ. 2007. Vaccine-induced T cells control reversion of AIDS virus immune escape mutants. J. Virol. 81:4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petravic J, Loh L, Kent SJ, Davenport MP. 2008. CD4+ target cell availability determines the dynamics of immune escape and reversion in vivo. J. Virol. 82:4091–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu J, O'Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, Abbink P, Coffey RT, Grandpre LE, Seaman MS, Landucci G, Forthal DN, Montefiori DC, Carville A, Mansfield KG, Havenga MJ, Pau MG, Goudsmit J, Barouch DH. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hel Z, Tsai WP, Tryniszewska E, Nacsa J, Markham PD, Lewis MG, Pavlakis GN, Felber BK, Tartaglia J, Franchini G. 2006. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J. Immunol. 176:85–96 [DOI] [PubMed] [Google Scholar]
- 52. Rollman E, Mason RD, Lin J, Brooks AG, Kent SJ. 2008. Protection afforded by live attenuated SIV is associated with rapid killing kinetics of CTLs. J. Med. Primatol 37(Suppl 2):24–32 [DOI] [PubMed] [Google Scholar]
- 53. Krammer F, Pontiller J, Tauer C, Palmberger D, Maccani A, Baumann M, Grabherr R. 2010. Evaluation of the influenza A replicon for transient expression of recombinant proteins in mammalian cells. PLoS One 5:e13265 doi:10.1371/journal.pone.0013265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107:11531–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reece JC, Loh L, Alcantara S, Fernandez CS, Stambas J, Sexton A, De Rose R, Petravic J, Davenport MP, Kent SJ. 2010. Timing of immune escape linked to success or failure of vaccination. PLoS One 5:e12774 doi:10.1371/journal.pone.0012774 [DOI] [PMC free article] [PubMed] [Google Scholar]