Abstract
Alphaviruses establish a persistent infection in arthropod vectors which is essential for the effective transmission of the virus to vertebrate hosts. The development of persistence in insects is not well understood, although it is thought to involve the innate immune response. Using a transgenic fly system expressing a self-replicating viral RNA genome analog, we have previously demonstrated antiviral roles of the Drosophila Imd (immune deficiency) and Jak-STAT innate immunity pathways in response to alphavirus replication. In the present study, comparative microarray analysis of flies harboring an alphavirus replicon and control green fluorescent protein flies identified 95 SINrep-sensitive genes. Furthermore, a subset of these genes is regulated by Rel or STAT transcription factors of the Imd and Jak-STAT pathways, respectively. We identified two antimicrobial peptide genes, attC and dptB, which are SINrep sensitive and regulated by STAT and Rel, respectively. SINrep flies heterozygous for attC had an increased viral RNA level, while knocking down dptB in SINrep flies resulted in impaired development. When injected with whole virus, the double-stranded RNA knockdowns of either attC or dptB showed a significant increase in virus titers. Our data demonstrate an antiviral response involving the Imd and Jak-STAT mediated expression of dptB and attC.
INTRODUCTION
Alphaviruses are arthropod-born RNA viruses. Through enzootic transmission, alphaviruses circulate between an amplifying host, such as birds, and an arthropod vector, typically mosquitoes. Transmission to humans often leads to disease and death in severe cases. In recent years, the resurgence of Chikungunya virus (CHIKV) in Europe, Asia and the Americas has brought alphaviruses to the attention of public health organizations as emergent pathogens (1, 2). In humans, symptoms of alphavirus infection range from rash and arthritis to encephalitis in severe cases (3, 4). Currently, no effective human vaccination is available for the prevention and treatment of alphavirus infection.
Alphaviruses can replicate efficiently in both vertebrate and invertebrate hosts; however, the pattern of infection differs in a host-dependent manner. Vertebrates infected with alphaviruses develop an acute infection that is cleared by the host immune system, while arthropods maintain a persistent infection, which is crucial for efficient transmission (5, 6). How arthropod hosts respond to alphaviruses and influence their amplification leading to a persistent infection is still not well understood. Here, we examine aspects of the innate immune response of arthropods to Sindbis virus (SINV), the type species of the Alphavirus genus.
Innate immunity pathways play a vital role in combating microbial infections in arthropods. Pathways, such as Toll, Imd, and Jak-STAT, are known to respond to infection and lead to the transcription of genes involved in fighting the pathogen (7–9). Common downstream processes include the production of humoral factors, such as antimicrobial peptides secreted from the fat body and phagocytosis, encapsulation and melanization by hemolymph (7, 8, 10). In terms of antiviral defense, it is known that arthropods employ the RNA interference (RNAi) pathway, the innate immunity pathways mentioned above, and processes such as autophagy and the phenoloxidase cascade to fight virus infection (11, 12). Studies have shown that in Drosophila, virus replication initiates and becomes the target of RNAi silencing (13–18). Depleting Dicer-2 of the RNAi machinery in Drosophila results in higher susceptibility to infection by flock house virus (Nodaviridae), Drosophila C virus (DCV; Dicistroviridae), Sindbis virus (SINV; Togaviridae), and Drosophila X virus (DXV; Birnaviridae) (13, 19). This mechanism is supported by the discovery of viral proteins that serve as RNAi suppressors (17, 20). Recently, Sabin et al. reported that Ars2 was important in the biogenesis of both small interfering RNA (siRNA) and microRNA (miRNA), suggesting the involvement of miRNA-mediated silencing as well (21).
Viral infection also triggers innate immunity signaling cascades, resulting in changes in transcription. The Toll, Imd, and Jak-STAT pathways have been shown to be activated by viral infection and responsible for an antiviral response. The Jak-STAT pathway is activated by DCV, West Nile virus, SINV, and dengue virus among other viruses (22–24). In both Drosophila and mosquitoes, knockdowns and mutations in the Jak-kinase led to an increase in viral infection, indicating an antiviral response involving the Jak-STAT pathway, presumably as a result of STAT-dependent expression of an antiviral effector molecule (23, 24). When considering a specific response to virus infection, certain downstream molecules, vir-1, tepII, and DVRF1 and DVRF2, have been shown to be upregulated by various viruses (23–25).
NF-κB-like signaling pathways also play an antiviral role. The Toll pathway is activated in DXV-injected flies and the mutants of its transcriptional regulator, Dif, were more sensitive to viral injection (26). Similar effects were also observed in Aedes aegypti mosquitoes upon DENV infection (27). We have previously reported the involvement of the Imd pathway (which activates Relish an NF-κB ortholog) in Drosophila antiviral defense to SINV (28). Similar effects were demonstrated in cricket paralysis virus-injected flies (29). While it is clear that these signaling pathways play a role in antiviral immunity, the complete profile of downstream effectors and their antiviral mechanisms are yet to be determined.
Our study focuses on the antiviral response of Drosophila melanogaster innate immune system induced by RNA replication of SINV. Mosquitoes, the natural vector of SINV, share conserved immune system pathways with fruit flies (30–32). By using Drosophila, we are able to exploit the fully sequenced genome and powerful genetic tools to investigate the arthropod response to alphaviruses.
SINV has a positive-sense, single-stranded RNA (ssRNA) genome with a 5′ cap and a 3′ poly(A) tail. The virus genome consists of two open reading frames; the first encodes the nonstructural proteins (nsPs) essential for viral genome replication, and the second encodes the structural proteins and is translated from a subgenomic mRNA synthesized by the nsPs (33). Previously, our lab generated a transgenic fly line encoding a SINV replicon RNA capable of expressing green fluorescent protein (GFP) in place of the structural proteins (UAS-SINrep:GFP) (28). Crossing this fly line to tissue-specific GAL4 driver lines results in the launch of the transcription of the SINrep:GFP RNA from the fly genome. This RNA is capable of autonomous replication and expression of GFP, which serves as visual demonstration of viral genome replication in these flies (28).
Using this transgenic fly system with the SINV replicon, Avadhanula et al. demonstrated both the Imd and the JAK-STAT pathways played an antiviral role in response to SINV replicon-mediated RNA synthesis. Reduced expression of Relish or STAT, the terminal transcription factors of the Imd and Jak-STAT pathways, respectively, resulted in increased viral genome replication. Moreover, injecting SINV into Relish mutant flies led to higher virus titers. These results suggested an overall antiviral mechanism for both pathways (28).
To further characterize the antiviral responses mediated by Relish and STAT, we performed microarray analyses in search for SINrep-sensitive genes. Using the SINV replicon system allowed us to focus solely on the host response to viral genome replication, a process requiring the production of viral nsPs, the formation of the RNA synthetic complex, and the production of viral RNAs. We report genes found to be SINrep sensitive and responsive to Relish and/or STAT. We identified 95 SINrep-sensitive genes with a ≥2-fold change in expression; 80 were upregulated and 15 downregulated compared to the control. Of these genes, two encode antimicrobial peptides, attC and dptB. Knocking down these genes led to either an increase in viral RNA synthesis or defects in development in the presence of SINrep. Furthermore, intrathoracic injection of infectious virus into the attC or dptB knockdown flies resulted in a higher viral load. These findings demonstrate that the antimicrobial peptides, AttC and DptB, are involved in Drosophila antiviral response to SINV.
MATERIALS AND METHODS
Fly stocks.
Fly stocks (listed in Table 1) were obtained from the Bloomington Stock Center and the Vienna Drosophila RNAi Center. Fly stocks were raised on standard cornmeal-agar medium at 25°C. Penicillin-streptomycin was added to the agar in fly crosses to prevent bacterial contamination. Fly lines used for microarray analysis, and all fly crosses tested as Wolbachia-free. The FlyBase (www.flybase.org) accession numbers for the genes used in the text included actin5C (CG4027), relish (CG11992), stat92E (CG4257), dptB (CG10794), and attC (CG4740).
Table 1.
Fly stocksa
| Stock no. | Gene | Genotype |
|---|---|---|
| 3954 | act5c | y1 w*; P{Act5C-GAL4}17bFO1/TM6B, Tb1 |
| 9457 | relish | w1118; RelE20 es |
| 11681 | stat92E | ry506 P{PZ}Stat92E06346/TM3, ryRK, Sb1, Ser1 |
| 31417 | rfp | W1118; P{UAS-RFP.W}3/TM3, Sb1 |
| 28975 | dptB | y1v1; P{TRiP.HM05186}attP2 |
| 25598 | attC | w1118; Mi{ET1}AttCMB05438 |
| v101213 | attC | P{KK107193}VIE-260B |
All of the stocks were obtained from the Bloomington Stock Center, except for v101213, obtained from the Vienna Drosophila RNAi Center.
Microarray analysis.
Four replicate pools of adult flies from both Act5C-GAL4, UAS-GFP and Act5C-GAL4, UAS-SINrep:GFP lines were collected 3 days after eclosion. Flies were snap-frozen in liquid nitrogen, and RNA was extracted using TRIzol reagent (Life Technologies, Grand Island, NY). A whole-genome microarray was conducted using the Drosophila Genome 2.0 Array (Affymetrix, Santa Clara, CA) to compare the transcriptome between Act5C-GAL4, UAS-GFP, and Act5C-GAL4, UAS-SINrep:GFP. Sample preparation, hybridization, and analysis of variance were conducted by Indiana University-Purdue University Indianapolis Center for Medical Genomics. Annotations on the cellular compartments and biological processes of the identified genes were based on information from Flybase. Analysis parameters were set to a fold change of ≥2 and a P value of ≤0.01.
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (34) and are accessible through GEO series accession number GSE42726 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42726).
Real-time quantitative RT-PCR analysis.
Adult flies were snap-frozen in liquid nitrogen 3 days after eclosion and stored at −80°C. Flies were homogenized in TRIzol reagent, followed by RNA extraction. cDNA was synthesized used AffinityScript QPCR cDNA synthesis kit (Agilent, Santa Clara, CA) with random hexamer primers. Negative (no reverse transcriptase [RT]) controls were performed for each sample. Quantitative RT-PCRs (qRT-PCRs) were prepared using Brilliant III SYBR green QPCR master mix (Agilent) with gene-specific primers according to the manufacturer's protocol and with the Applied Bioscience StepOnePlus qRT-PCR machine (Life Technologies). The expression levels were normalized to the 18S rRNA expression using the delta delta comparative threshold method (ΔΔCT). Fold changes were determined using the comparative threshold cycle (CT) method. The primer sequences are available upon request.
Fly crosses. (i) Knocking down attC in SINrep.
We first crossed attC−/−; +/+(25598) to the balancer Sco/Cyo; +/+ (stock was a gift from Justin Kumar). The resulting progeny, attC−/Cyo; +/+, was then crossed to +/+; SINrep/TM3, yielding siblings of attC−/+; SINrep/TM3 and the control Cyo/+; SINrep/+ (SIN WT) (see Fig. 4A).
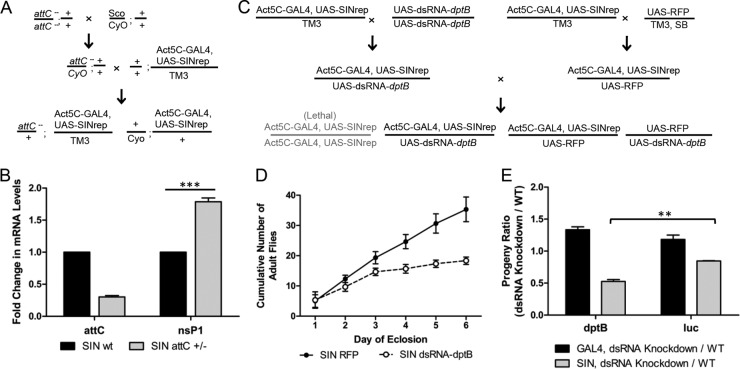
Fig 4.
Characterization of the effect of antimicrobial peptides on SIN replication. (A) Crossing scheme to generate +/CyO; Act5C-GAL4, UAS-SINrep/+ (SINrep WT) and attC−/+; Act5C-GAL4, UAS-SINrep/+ (SINrep attC+/−) flies (see Materials and Methods). (B) qRT-PCR analysis of attC and nsP1 transcripts in SINrep WT and SINrep attC+/− flies. (C) Crossing scheme to generate Act5C-GAL4, UAS-SINrep/UAS-RFP (SIN RFP), Act5C-GAL4, UAS-SINrep/UAS-dsRNA-dptB (SIN dsRNA-dptB), and UAS-RFP/UAS-dsRNA-dptB (see Materials and Methods). (D) Cumulative numbers of SIN RFP and SIN dsRNA-dptB progeny eclosing from an individual cross. (E) Progeny composition ratio of dsRNA-dptB knockdown flies in the presence (gray) or absence (black) of SINrep. dsRNA-luc serves as a nontargeting control. Ratios represent SIN-dsRNA-gene of interest/SIN-RFP, where both are siblings of the same cross. Error bars represent the SEM. **, P < 0.01; ***, P < 0.005 (as calculated by the Student t test).
(ii) Knocking down dptB in SINrep.
We first crossed SINrep to UAS-dsRNA-dptB or UAS-RFP (red fluorescent protein). The UAS-RFP line served as a selection marker, as well as a promoter control to ensure a similar level of promoter usage, i.e., basal UAS-SINrep:GFP transcription level. The F1 progeny Act5C-GAL4, UAS-SINrep:GFP/UAS-dsRNA was crossed to the F1 Act5C-GAL4, UAS-SINrep:GFP/UAS-RFP. In this way, we were able to yield both the F2 Act5C-GAL4, UAS-SINrep:GFP/UAS-RFP (SIN WT) control and the F2 SIN-dsRNA-dptB knockdown mutant (Act5C-GAL4; UAS-SINrep:GFP/UAS-dsRNA-dptB) as siblings within the same cross.
Cells and viruses.
BHK-21 cells (American Type Culture Collection, Rockville, MD) were cultured in Eagle minimum essential medium with Earle's balanced salt solution, supplemented with 10% fetal bovine serum and 0.1% penicillin-streptomycin, at 37°C and 5% CO2.
TE3′2J-mcs Sindbis virus construct was a gift from Zachary Adelman and Kevin Myles (Virginia Tech) and modified to insert the eGFP sequence in the multiple cloning site. Virus was generated through the transfection of the in vitro-transcribed TE3′2J-GFP RNA into BHK-21 cells. Virus was harvested after 48 h, purified through a 27% sucrose cushion, and resuspended in 1× phosphate-buffered saline (PBS). The concentration was determined by titering on BHK-21 cells.
Virus injections.
Adult flies were injected with 106 PFU of TE3′2J-GFP 3 days after eclosion. A Nanoject II automatic nanoliter injector (Drummond, Broomall, PA) was used for intrathoracic injection. Flies were snap-frozen 2 or 4 days postinjection. Single fly lysates were made using 1× PBS with protease inhibitor cocktail (EMD; Millipore, Billerica, MA) and filtered through a 0.22-μm-pore-size polyvinylidene difluoride membrane (Sigma, St. Louis, MO). Lysates were titered on BHK cells and scanned with a Typhoon 9210 imager to detect GFP fluorescence (GE Healthcare, Pittsburgh, PA). Plaque images were analyzed using ImageJ software (35).
Statistical analyses.
All P values were calculated using the Student t test. In the injection assays, outliers were determined using the Grubb's test. The average fold changes in titer (see Fig. 5C) were calculated using the variable bootstrapping method, measuring the fold change between each potential pair of flies to determine the variability of the mean (36).
Fig 5.
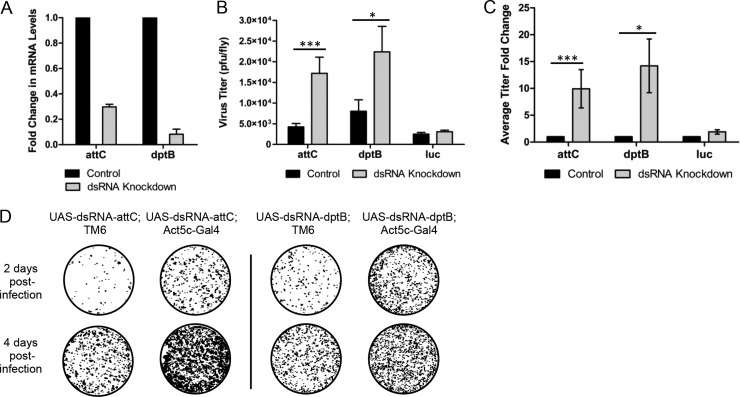
Characterization of the effect of antimicrobial peptides on virus production. (A) dsRNA knockdown of attC or dptB in undriven UAS-dsRNA/TM6 flies (control) and driven Act5C-GAL4; UAS-dsRNA flies. The data represent three independent biological replicates. The expression is normalized to 18S rRNA. (B) Average of individual fly titers of undriven UAS-dsRNA/TM6 (control) and driven Act5C-GAL4; UAS-dsRNA knockdown flies injected with 106 PFU of TE3′2J-GFP Sindbis virus (n = 20 for each sample.) (C) The average titer fold change was determined by variable bootstrapping of fly titers shown in panel B. (D) Plaques of individual TE3′2J-GFP injected flies harvested 2 or 4 days postinjection and titered on BHK cells. Plaques were imaged by GFP signal (black). All error bars represent the SEM. *, P < 0.05; ***, P < 0.005 (as calculated by the Student t test).
RESULTS
Identification of transcripts altered by SINV replication.
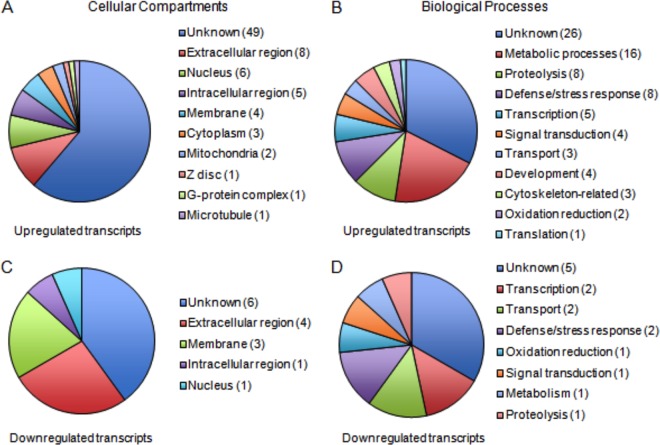
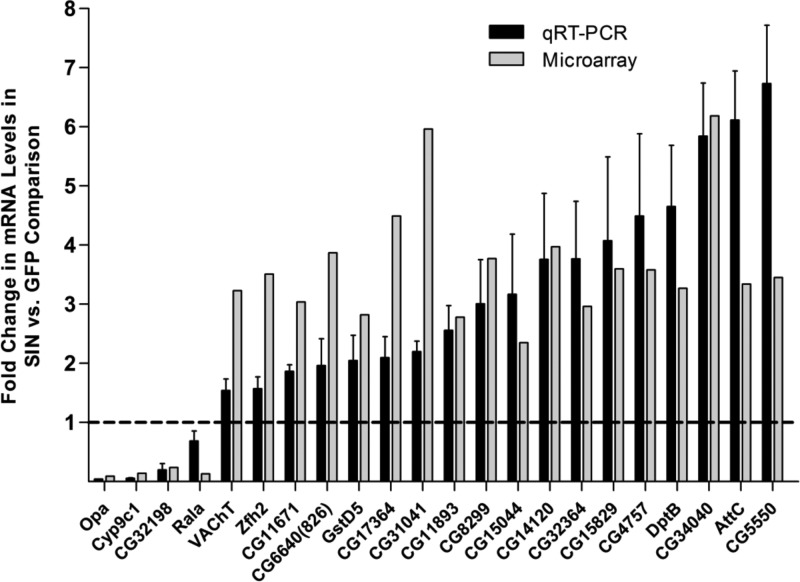
Our lab and others previously demonstrated the importance of the Imd and Jak-STAT pathways in inducing an inhibitory response to viral infection (23, 24, 28). These pathways result in the translocation of a transcription factor (Rel or STAT) to the nucleus, activating the transcription of downstream effector genes. To identify downstream effectors that are involved in the host response to SINV replication, we performed a microarray analysis. Act5C-GAL4, UAS-SINrep:GFP (hereafter referred to as SINrep) and Act5C-GAL4, UAS-GFP (GFP) flies were collected 3 days after eclosion. Total RNA was isolated from whole flies and submitted for microarray analysis in search for SINrep-sensitive transcripts. We set our analysis parameters at a fold change of ≥2 with P value of ≤0.01. A total of 95 genes were differentially transcribed in the SINrep flies compared to the control GFP flies (see Dataset S1 in the supplemental material). The most abundant categories of biological processes were metabolic processes, proteolysis, defense responses, transcription, and signal transduction (Fig. 1). The protein products of these transcripts are found in compartments throughout the cell: extracellular regions, intracellular regions, the nucleus, and membranes. We validated these results by qRT-PCR using gene-specific primers. Twenty-two genes that showed alterations in expression levels in the microarray analysis were chosen for validation. These genes were chosen to represent the range and distribution of expression changes seen in the microarray results. Genes that showed a fold change of ≥2 compared to the control in the microarray analysis exhibited similar changes by qRT-PCR analysis, confirming the differential transcription profiles demonstrated in the microarray analysis (Fig. 2).
Fig 1.
Functional annotations of SIN-sensitive genes. Genes were identified through a microarray of Act5-Gal4, UAS-SINrep:GFP (SINrep) flies versus Act5c-Gal4, UAS-GFP (GFP) flies. Analysis parameters were set to a fold change of ≥2 and a P value of ≤0.01. The 95 genes that are differentially expressed in SINrep versus GFP are categorized according to their cellular compartments (A and C) and biological processes (B and D). Panels A and B categorize the transcripts that are upregulated; panels C and D categorize the genes that are downregulated.
Fig 2.
Validation of SIN-sensitive genes. The levels of SIN-sensitive transcripts were tested by qRT-PCR of RNA samples from SINrep flies and compared to GFP flies with GFP fly levels set to 1. The results were normalized to 18S rRNA. The data represent four independent biological replicates. Error bars represent the standard errors of the mean (SEM).
Identification of SINrep-sensitive genes regulated by the Imd and Jak-Stat pathways.
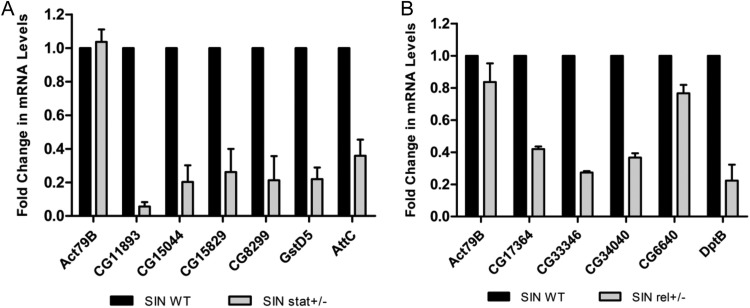
It has been previously shown that mutations in the Imd and Jak-STAT innate immunity pathways lead to an increase in SINV replication. We examined the regulation of transcriptional changes found in the microarray by the Imd and Jak-STAT pathways. We analyzed the 2-kb-upstream sequences of all 95 SINrep-sensitive genes for Rel and STAT binding sites. Thirty-eight genes had at least one predicted Rel binding site (GGGGATTTTT/CCCNN, where GGGGA are the critical residues) (37). Sixty-one genes had at least one canonical STAT binding site (TTCNNNGAA, where the +3C and +7G are critical residues) in combination with at least one canonical STAT binding site with one nucleotide change (38). Thirty-seven genes had a least one STAT binding site with one nucleotide change. To examine the dependence of these genes on Imd and JAK-STAT signaling in responding to SINV replication, candidate genes with Rel or STAT binding sites were chosen for validation (Table 2). SINrep flies were crossed with Relish or STAT mutant flies. SINrep flies which were heterozygous for the rel or stat mutation were compared to sibling flies containing only SINrep (SIN WT). RNA was extracted from progeny of these crosses and used for qRT-PCR analysis with gene-specific primers to examine the transcription of genes of interest. Transcription of genes examined in the SINrep/stat+/− flies decreased by at least 50% compared to the sibling SINrep WT control (Fig. 3A). SINrep/rel+/− mutant flies also showed a decrease in the transcription of genes with Rel binding sites compared to the SINrep WT flies (Fig. 3B). These changes in SINrep/stat+/− and SINrep/rel+/− mutants indicate that the transcription of these genes is under the control of STAT or Rel. In contrast, levels of a housekeeping gene, actin79B, did not significantly change with either mutation. Among these SINrep-sensitive and STAT- or Rel-regulated genes, we chose to characterize the effects of attC and dptB on virus replication. These genes have been previously shown to code for antimicrobial peptides involved in insect innate immunity (39–41).
Table 2.
Predicted STAT-/REL-binding sites within 2 kb upstream of the tested genes
| Site and gene symbol | Sequence(s)a |
|---|---|
| STAT-binding sites | |
| CG11893 | TTCNNNGAT (2), GTCNNNGAA, TTCNNNGAC, TTCNNNGTA (2), ATCNNNGAA (2), TTCNNNGAG, TTCNNNGCA |
| CG15044 | ATCNNNGAA (2), TTCNNNGAT, TGCNNNGAA (2), TTCNNNGGA (3), TCCNNNGAA, TACNNNGAA |
| CG15829 | CTCNNNGAA, TTCNNNGAG, TCCNNNGAA, TTCNNNGTA, TTCNNNGAT |
| CG8299 | TTCNNNGAG, GTCNNNGAA, TACNNNGAA |
| GstD5 | TACNNNGAA, ATCNNNGAA, TCCNNNGAA (2), TGCNNNGAA (2), TTCNNNGAC (2), TTCNNNGAG |
| AttC | TCCNNNGAA (2), TTCNNNGAC (2), TTCNNNGAG, TACNNNGAA (3), TTCNNNGAT (2), TTCNNNGTA, TGCNNNGAA, TTCNNNGGA |
| REL-binding sites | |
| CG17364 | GGGGAATTTA |
| CG33346 | GGGGATTCGCCC |
| CG34040 | GGGGCATTT |
| CG6640 | GGGGAAGACCC |
| DptB | GGGGATCCAC, GGGGAATCTC, GGGGATTACC |
Numbers in parentheses indicate the numbers of the same sequence appearing within 2 kb upstream.
Fig 3.
SIN-responsive gene transcription in STAT and Relish mutants. Transcript levels of SIN-responsive genes with STAT (A) or Rel (B) binding sites were measured in SINrep WT and SINrep/stat+/− or SINrep/rel+/− flies. The levels in SINrep WT flies were set to 1. Expression was normalized to 18S rRNA. The data shown represent three independent biological replicates of five flies each. Error bars represent the SEM.
Effect of antimicrobial peptides on virus replication.
To examine the effect of attC and dptB on viral genome replication, we effectively reduced expression of both genes using mutant stocks according to the crossing schemes in Fig. 4A and 4C (for details, see Materials and Methods). The heterozygous attC mutant fly exhibited a 70% knockdown in attC mRNA levels (Fig. 4B). The heterozygous mutant progeny, attC−/+; SINrep/TM3, demonstrated a modest 1.79-fold increase in viral nsP1 levels in comparison to the sibling SINrep WT control fly (P < 0.005; Fig. 4B). This indicates an antiviral effect of AttC on viral replication.
We also efficiently knocked down dptB transcription (92% knockdown) using a UAS-dsRNA-dptB line (28975) driven by Act5c-Gal4. The crossing scheme is described in Fig. 4C (see also Materials and Methods). The viral nsP1 levels in dptB knockdown mutants were not significantly changed compared to their sibling controls (data not shown). However, during progeny collection, contrary to the theoretical 1:1 ratio, we noticed a very low number of SIN-dsRNA-dptB progeny compared to their SIN-RFP siblings, indicating that many of the SINrep-mutant progeny were dying, leaving those least affected to be assayed (Fig. 4D). We observed that the survival issues of SINrep-dsRNA-dptB knockdown mutants seemed to occur during early pupation (data not shown). This biased progeny composition was gene specific since, when we examined UAS-dsRNA-luc as a nonspecific targeting control, the ratio of (SINrep dsRNA)/(SINrep wt) was 0.85, significantly higher than that of the dptB knockdown mutants (0.53, P < 0.01; Fig. 4E, gray bars). In another set of control crosses where the double-stranded RNA (dsRNA) lines were crossed to the wild-type Act5C-GAL4/TM6 (WT) driver line in the absence of SINrep, the ratio of (WT dsRNA)/(WT wt) were very close to the predicted 1:1 (Fig. 4E, black bars). Therefore, knocking down dptB in the presence of viral genome replication appeared to have a detrimental effect on fly development and survival. Due to this problem, examination of RNA synthesis in dptB mutant flies was not continued. Analysis of the effect of dptB on virus replication was continued using infection with whole virus (see below).
Effect of antimicrobial peptides on virus production.
We continued to characterize the putative antiviral role of AttC and DptB by performing injections in dsRNA-attC or dsRNA-dptB knockdown flies using whole SINV (TE3′2J-GFP). UAS-dsRNA-attC flies or UAS-dsRNA-dptB flies were crossed with Act5C-GAL4/TM6 to drive expression of the dsRNA-attC and the dsRNA-dptB constructs. Undriven UAS-dsRNA-attC; TM6 progeny were used as controls for their sibling knockdown progeny UAS-dsRNA-attC; Act5C-GAL4. The dsRNA-attC knockdown flies showed a 70% decrease in attC transcript levels in comparison to the control flies as determined by qRT-PCR (Fig. 5A). Virgin female flies with reduced (UAS-dsRNA-attC; Act5C-GAL4) and normal (UAS-dsRNA-attC;TM6) levels of attC were infected by intrathoracic injection with of TE3′2J-GFP Sindbis virus and then collected and snap-frozen 2 or 4 days postinjection. Single fly lysates were titered on BHK cells. At 2 days postinjection, the flies with reduced attC (UAS-dsRNA-attC; Act5C-GAL4) showed an average virus titer of 1.72 × 104 PFU/fly, a 4.05-fold increase over the control flies (4.24 × 103 PFU/fly, P < 0.005) (Fig. 5B). In contrast, flies expressing a nonviral, non-Drosophila dsRNA to luciferase (UAS-dsRNA-luc/Act5C-GAL4) did not show a significant difference in virus replication compared to control UAS-dsRNA-luc/TM6 flies. This showed that the increase in virus replication was due to the knockdown of attC and not due simply to the expression of a dsRNA.
When determining differences between dsRNA-attC and control fly titers, we also performed variable bootstrapping to determine the variability of the mean (36). This method allows us to examine every potential pair of dsRNA-attC and control flies as individual experiments to determine a more accurate average fold change. Using this method, the dsRNA-attC flies showed an average fold change in virus titer of 9.93 ± 3.56 (P < 0.005; Fig. 5C). In contrast, the dsRNA-luc flies do not show a significant change from the control flies. When examining the flies over time, the individual fly titers in the dsRNA-attC flies continue to increase over time, maintaining the 4-fold increase over the control flies at 4 days postinjection (Fig. 5D). Taken together, the SINrep, attC− crosses and injection experiments demonstrate that knocking down attC leads to an increase in virus replication and an even greater increase in whole virus production, indicating an antiviral role for AttC.
We also continued the analysis of dptB using whole virus injections. UAS-dsRNA-dptB flies were crossed with Act5C-Gal4 flies to generate the driven RNAi knockdown progeny (UAS-dsRNA-dptB; Act5c-GAL4) and its sibling control (UAS-dsRNA-dptB; TM6) Reduction of dptB transcript levels in the dsRNA knockdown compared to the control flies was 92% (Fig. 5A). Flies were injected with TE3′2J-GFP Sindbis virus and harvested at 2 or 4 days postinjection. The UAS-dsRNA-dptB; Act5c-GAL4 flies had an average virus titer of 2.24 × 104 PFU/fly, a 2.78-fold increase over the control flies (8.04 × 103 PFU/fly, P < 0.05) (Fig. 5B.) To further analyze the fold change, we used variable bootstrapping to determine the variability of the mean. When taking every potential dsRNA-dptB and control comparison into account, the virus titers in the dsRNA-dptB flies demonstrated a fold change of 14.21 ± 5.83 (P < 0.05) (Fig. 5C). Virus titers in UAS-dsRNA-luc/Act5c-GAL4 flies showed no significant difference from those in control UAS-dsRNA-luc/TM6 flies, again indicating that the proviral effect of knocking down dptB was not simply a consequence of generalized activation of the RNAi pathway. The individual fly titers continue to increase over time as shown by the titers at 4 days postinjection (Fig. 5D). These data indicate an antiviral role for DptB during SINV infection.
DISCUSSION
Arthropods infected with alphaviruses develop a persistent infection that is controlled but not cleared by the immune system. The arthropod immune system consists of the RNAi pathway, as well as the Toll, Imd, and Jak-STAT signaling pathways, which result in changes in the transcriptome leading to the production of proviral and antiviral factors. Previously published microarray studies have reported many genes that are regulated in response to alpha- and flavivirus infections in Drosophila S2 cells, Aedes aegypti midguts, and whole Aedes aegypti mosquitoes (25, 42, 43). In the present study we identified 95 genes with significantly altered expression as a result of SINV replication in whole Drosophila. Although genes found in our whole Drosophila microarray did not specifically match those previously published in microarrays, there are categories of genes that are found in all, such as genes involved in proteolysis, the defense response, and regulation of transcription. The differences between studies clearly show that particular aspects of the response to virus infection vary depending on the level at which the infection is studied, i.e., cellular, tissue, or organismal. Likewise, various responses to different pathogens indicate the ability of the arthropod innate immune system to distinguish between pathogens and mount a specific response. In the current microarray study using the transgenic SINrep flies, we were able to examine the responses specific to viral RNA replication at the organismal level. Through this method we were able to determine changes that are a result of the components and processes of viral genome replication in the absence of the structural proteins and entry/release processes.
To examine the response of Drosophila to SINV replication, we focused on the Imd and Jak-STAT pathways that have been implicated in the antiviral response. Among the 95 altered transcripts, several genes have been previously identified as part of the Imd-mediated defense response. PGRP-SD plays a role in detecting bacterial infections by binding peptidoglycan (44). A proline-rich antimicrobial peptide, Mtk (Metchnikowin), and a catalytic PGRP (PGRP-SB1) are both expressed upon Imd activation and showed significant upregulation in SINrep flies (45, 46). We did not see any upregulation of signaling molecules responsible for the antibacterial response of the Imd pathway, including relish, imd, dfadd, dredd, ird5, tab2, and key, though we have previously shown that SINrep flies heterozygous for these molecules had higher viral RNA load (28).
To identify other Imd-regulated genes, we examined the 2-kb-upstream sequences of all 95 SINrep-sensitive genes for Rel binding sites. Using the Rel-binding sequence parameters determined by Busse et al. (37), we identified 38 genes with Rel binding sites and validated a subset of these as Rel responsive. Variations in κB binding sequence and numbers of κB sites determine the differential regulation by the three NF-κB homologues involved in fly innate immunity: Relish, Dif, and Dorsal (37). This may explain the fact that certain genes with a single κB site (e.g., CG6640) only showed slight downregulation in a Rel heterozygous background (Fig. 3B).
Studies of the Jak-STAT pathway have identified transcripts expressed through activation of the Jak-STAT pathway by different stimuli. Previously, totA and other tot family members were shown to be upregulated in response to cellular stress (47, 48). Other identified genes such as tepI, tepII, and raf code for proteins involved in phagocytosis, opsonization, and hemocyte proliferation in response to Gram-negative bacterial infection (25, 49, 50). During DCV or dengue virus infections, the Jak-STAT pathway was shown to be activated and induce the transcription of vir-1 or DVRFs, respectively (23, 24). In the present study, we report that of the 95 SINrep-sensitive genes 61 of these genes have STAT binding sites. Upon validating a subset of these genes, we demonstrated that expression is inhibited with the removal of STAT. We did not find significant changes in the transcription of tepI, tepII, raf, vir-1, or DVRF1 and DVRF2 in response to SINrep; however, this is not surprising since different stimuli have been shown to invoke different responses from the Jak-STAT pathway (7, 23). Surprisingly, totA and totC show decreased mRNA levels in response to SINrep, contrary to previous reports of increased expression during septic injury. The difference could be explained by different inputs to the Jak-STAT pathway, leading to different outcomes. A recent study identified Vago as an alternative input to the Jak-STAT pathway during virus infection through a yet-to-be-identified receptor (51). We observed increased expression of vago in Drosophila as a result of SINV replication (Table 1). An alternative input to the Jak-STAT pathway, such as Vago, could be responsible for differential regulation of downstream effector genes. The tot genes have been shown to be expressed through the activation of Jak-STAT through the canonical Upd-Domeless ligand-receptor pair (47). The decreased levels of the tot transcripts in response to SINrep may be indicative of competition for Hop-STAT machinery between Upd-Domeless and the unidentified Vago receptor. The use of an alternative receptor-ligand pair, as well as the potential downstream effects on STAT modification and activation, may provide information about arthropods' ability to distinguish between pathogens and mount a specific response. More research is needed to further confirm alternative activation of Jak-STAT and its role in differential gene expression.
Through our analysis of SINrep-responsive genes in whole Drosophila, we identified two antimicrobial genes, attC and dptB, which were upregulated in response to SINV replication. Levels of attC or dptB mRNA decreases significantly in stat+/− or rel+/− flies, indicating regulation by the Jak-STAT and Imd pathways, respectively. In addition, when attC or dptB is knocked down, SINV levels increase, suggesting an antiviral mechanism. attC and dptB are both found on chromosome 2. dptB is 0.6 kb downstream of another Imd-responsive antimicrobial peptide, dpt, on chromosome 2, and they share a 43% overall gene sequence identity (52). Despite its close proximity and potentially similar regulation pattern, dpt was not identified as SINrep sensitive. attC is one of four attacin-encoding genes; attA and attB are located on chromosome 2, while attD is on chromosome 3. The AttA and AttB proteins share 98% identity with each other, and AttC is 73% identical to AttA and AttB (52). The expression of attA, attB, and attD was not found to be SINrep sensitive.
In regard to protein structure, DptB is very similar to the attacins (52) (Fig. 6). Both DptB and AttC have a signal sequence that is thought to be required for secretion from the cell. The proteins are further processed by the removal of the proline-rich propeptide domain that is similar to the proline-rich domain found in drosocins (52). DptB shares the canonical furin cleavage site (RVRR) with AttA and AttB, while the AttC cleavage site differs by one amino acid (RARR). The AttC propeptide, also known as Maturated Prodomain of Attacin-C (MPAC), has been shown to have antibacterial activity, independent of the rest of the protein (53). AttC and DptB also include glycine-rich regions. AttC has an additional glycine-rich region and an N-terminal attacin domain that is similar to that of AttA, AttB, and AttD (52). Previously, AttC has been characterized as an antimicrobial peptide in response to Gram-negative bacteria in Hyalophora cecropia (39, 54). Carlsson et al. determined that AttC was able to interact with the lipopolysaccharide of Escherichia coli and disturb the structure of the outer membrane and function of outer membrane proteins (54). Through this interaction the synthesis of outer membrane proteins is inhibited and the expression of stress response proteins is induced through an unknown mechanism (54). Currently, no mechanistic analysis on DptB has been performed except for a role in the humoral response to bacterial infections and temperature stress (55). Other proline-rich antibacterial peptides in the drosocin family have been shown to bind to bacterial proteins such as DnaK inhibiting their function (56).
Fig 6.
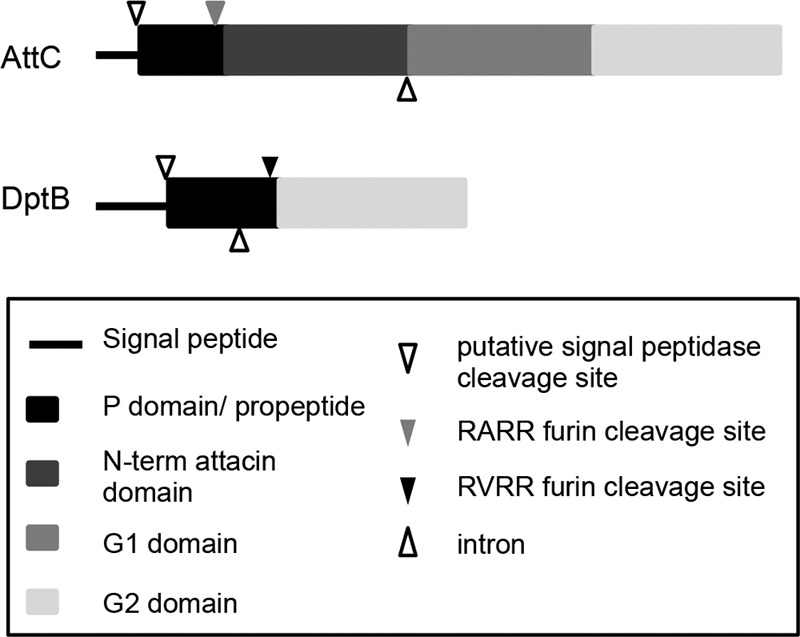
Protein domains of AttC and DptB. AttC and DptB are both glycine-rich, secreted peptides. Both contain a signal peptide sequence, which is cleaved at a putative signal peptidase sequence. The propeptide (or P domain) is proline-rich and followed by a furin-like cleavage site (RARR or RVRR). Both proteins have the glycine-rich G2 domain. In addition to these, AttC also has an N-terminal attacin domain and an additional glycine-rich domain, G1.
Due to the similar protein structure of these peptides, it seems reasonable to hypothesize that they may have similar functions and possible redundancy. Previously reported work indicates that similar peptides are able to interact with lipopolysaccharide of Gram-negative bacteria or intracellular bacterial proteins (54, 56). A possible protein-binding role of AttC and DptB could occur with a viral protein at any stage of the cell cycle: with the nonstructural replication complex, with the structural proteins during assembly, or with the spike proteins on the virus particle in the extracellular environment. Because SINrep replication increased in an attC heterozygous mutant in the absence of viral structural proteins, we can hypothesize that AttC interacts with a viral or cellular component of the replication complex, thereby inhibiting replication. The effect is amplified when virus injections are used in attC knockdown flies. More work is needed to determine whether this is a downstream effect of AttC's inhibition of replication or whether AttC interacts with other viral proteins and processes.
Hyalophora attacin has previously been reported to remain in the extracellular environment to exert its antibacterial effect on bacterial cells (54). However, the case may be different due to processing of attacin and/or the type of pathogen. Mutations in dptB did not affect the levels of SINrep replication but did decrease the level of virus production. It is possible that while the expression of dptB is induced by virus infection, DptB does not directly influence virus replication, but another aspect of the virus life cycle directly or indirectly. This fits with our data showing no change in nsP1-encoding RNA levels during SINrep replication but an increased viral load in dptB mutants. In addition, there is a developmental defect in dsRNA-dptB knockdown flies in the presence of SINrep. This may indicate an additional role for DptB in antiviral immunity during development. More work is needed to determine the function and localization of AttC and DptB during SINV infection and their possible interactions with viral proteins and processes.
In summary, we have identified a list of candidate genes from our microarray analysis that are SINrep sensitive and regulated by the innate immune pathways of Drosophila melanogaster. Among these genes, we demonstrated the antiviral role of attC and dptB in both the replicon fly system and whole virus injections. These results display the power of Drosophila genetics in aiding in identification and characterization of arthropod host factors that interact with the virus. Further characterization of upstream signaling cascades and the molecular mechanisms of effector molecules will provide additional insights into how innate immunity responds to viral infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Justin Kumar for critical reading of the manuscript and Jeanette McClintick of the Indiana University-Purdue University Indianapolis Center for Medical Genomics for assistance with analysis of the microarray data.
This study was supported by grant R01 AI090077 from the National Institutes of Health to R.W.H.
Footnotes
Published ahead of print 30 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03360-12.
REFERENCES
- 1. Enserink M. 2007. Infectious diseases. Chikungunya: no longer a third world disease. Science 318:1860–1861 [DOI] [PubMed] [Google Scholar]
- 2. Ligon BL. 2006. Reemergence of an unusual disease: the Chikungunya epidemic. Semin. Pediatr. Infect. Dis. 17:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laine M, Luukkainen R, Toivanen A. 2004. Sindbis viruses and other alphaviruses as cause of human arthritic disease. J. Intern. Med. 256:457–471 [DOI] [PubMed] [Google Scholar]
- 4. Taylor RM, Hurlbut HS, Work TH, Kingston JR, Frothingham TE. 1955. Sindbis virus: a newly recognized arthropod-transmitted virus. Am. J. Trop. Med. Hyg. 4:844–862 [DOI] [PubMed] [Google Scholar]
- 5. Bowers DF, Abell BA, Brown DT. 1995. Replication and tissue tropism of the alphavirus Sindbis in the mosquito Aedes albopictus. Virology 212:1–12 [DOI] [PubMed] [Google Scholar]
- 6. Bowers DF, Coleman CG, Brown DT. 2003. Sindbis virus-associated pathology in Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 40:698–705 [DOI] [PubMed] [Google Scholar]
- 7. Agaisse H, Perrimon N. 2004. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198:72–82 [DOI] [PubMed] [Google Scholar]
- 8. Ferrandon D, Imler JL, Hetru C, Hoffmann JA. 2007. The Drosophila systemic immune response: sensing and signaling during bacterial and fungal infections. Nat. Rev. Immunol. 7:862–874 [DOI] [PubMed] [Google Scholar]
- 9. Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697–743 [DOI] [PubMed] [Google Scholar]
- 10. Hoffmann JA. 2003. The immune response of Drosophila. Nature 426:33–38 [DOI] [PubMed] [Google Scholar]
- 11. Rodriguez-Andres J, Rani S, Varjak M, Chase-Topping ME, Beck MH, Ferguson MC, Schnettler E, Fragkoudis R, Barry G, Merits A, Fazakerley JK, Strand MR, Kohl A. 2012. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 8:e1002977 doi:10.1371/journal.ppat.1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. 2009. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7:590–597 [DOI] [PubMed] [Google Scholar]
- 14. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–1150 [DOI] [PubMed] [Google Scholar]
- 15. Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925 [DOI] [PubMed] [Google Scholar]
- 16. Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, Andino R. 2006. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Gene Dev. 20:2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zamore PD, Tuschl T, Sharp PA, Bartel DP. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25–33 [DOI] [PubMed] [Google Scholar]
- 19. Zambon RA, Vakharia VN, Wu LP. 2006. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 8:880–889 [DOI] [PubMed] [Google Scholar]
- 20. Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 17:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. 2009. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138:340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. 2008. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat. Immunol. 9:1425–1432 [DOI] [PubMed] [Google Scholar]
- 23. Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6:946–953 [DOI] [PubMed] [Google Scholar]
- 24. Souza-Neto JA, Sim S, Dimopoulos G. 2009. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U. S. A. 106:17841–17846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mudiganti U, Hernandez R, Brown DT. 2010. Insect response to alphavirus infection: establishment of alphavirus persistence in insect cells involves inhibition of viral polyprotein cleavage. Virus Res. 150:73–84 [DOI] [PubMed] [Google Scholar]
- 26. Zambon RA, Nandakumar M, Vakharia VN, Wu LP. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102:7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xi Z, Ramirez JL, Dimopoulos G. 2008. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4:e1000098 doi:10.1371/journal.ppat.1000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. 2009. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 5:e1000582 doi:10.1371/journal.ppat.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Costa A, Jan E, Sarnow P, Schneider D. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4:e7436 doi:10.1371/journal.pone.0007436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barillas-Mury C, Han YS, Seeley D, Kafatos FC. 1999. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J. 18:959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298:159–165 [DOI] [PubMed] [Google Scholar]
- 32. Dimopoulos G. 2003. Insect immunity and its implication in mosquito-malaria interactions. Cell. Microbiol. 5:3–14 [DOI] [PubMed] [Google Scholar]
- 33. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edgar R, Domrachev M, Lash AE. 2002. Gene Expr. Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bremer M, Doerge RW. 2010. Statistics at the bench. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. 2007. A κB sequence code for pathway-specific innate immune responses. EMBO J. 26:3826–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr 1996. Identification of a Stat gene that functions in Drosophila development. Cell 84:421–430 [DOI] [PubMed] [Google Scholar]
- 39. Carlsson A, Engstrom P, Palva ET, Bennich H. 1991. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect. Immun. 59:3040–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cudic M, Bulet P, Hoffmann R, Craik DJ, Otvos L., Jr 1999. Chemical synthesis, antibacterial activity and conformation of diptericin, an 82-mer peptide originally isolated from insects. Eur. J. Biochem. 266:549–558 [DOI] [PubMed] [Google Scholar]
- 41. Wicker C, Reichhart JM, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann JA. 1990. Insect immunity. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J. Biol. Chem. 265:22493–22498 [PubMed] [Google Scholar]
- 42. Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, Wang P, Krishnan MN, Higgs S, Fikrig E. 2011. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue, and yellow fever viruses. PLoS Pathog. 7:e1002189 doi:10.1371/journal.ppat.1002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, Gill SS. 2005. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 35:1293–1307 [DOI] [PubMed] [Google Scholar]
- 44. Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. 2008. Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc. Natl. Acad. Sci. U. S. A. 105:11881–11886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levashina EA, Ohresser S, Bulet P, Reichhart JM, Hetru C, Hoffmann JA. 1995. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur. J. Biochem. 233:694–700 [DOI] [PubMed] [Google Scholar]
- 46. Zaidman-Remy A, Poidevin M, Herve M, Welchman DP, Paredes JC, Fahlander C, Steiner H, Mengin-Lecreulx D, Lemaitre B. 2011. Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity, and function. PLoS One 6:e17231 doi:10.1371/journal.pone.0017231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. 2003. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5:441–450 [DOI] [PubMed] [Google Scholar]
- 48. Ekengren S, Hultmark D. 2001. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 284:998–1003 [DOI] [PubMed] [Google Scholar]
- 49. Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. 2000. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 97:11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo H, Rose PE, Roberts TM, Dearolf CR. 2002. The Hopscotch Jak kinase requires the Raf pathway to promote blood cell activation and differentiation in Drosophila. Mol. Genet. Genomics 267:57–63 [DOI] [PubMed] [Google Scholar]
- 51. Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ. 2012. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U. S. A. 109:18915–18920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hedengren M, Borge K, Hultmark D. 2000. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem. Biophys. Res. Commun. 279:574–581 [DOI] [PubMed] [Google Scholar]
- 53. Rabel D, Charlet M, Ehret-Sabatier L, Cavicchioli L, Cudic M, Otvos L, Jr., Bulet P. 2004. Primary structure and in vitro antibacterial properties of the Drosophila melanogaster attacin C Pro-domain. J. Biol. Chem. 279:14853–14859 [DOI] [PubMed] [Google Scholar]
- 54. Carlsson A, Nystrom T, de Cock H, Bennich H. 1998. Attacin–an insect immune protein–binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 144(Pt 8):2179–2188 [DOI] [PubMed] [Google Scholar]
- 55. Kim K, Lin YR, Park Y. 2010. Enhancement of stress resistances and downregulation of Imd pathway by lower developmental temperature in Drosophila melanogaster. Exp. Gerontol. 45:984–987 [DOI] [PubMed] [Google Scholar]
- 56. Kragol G, Hoffmann R, Chattergoon MA, Lovas S, Cudic M, Bulet P, Condie BA, Rosengren KJ, Montaner LJ, Otvos L., Jr 2002. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin. Eur. J. Biochem. 269:4226–4237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.