Abstract
HIV-1 latency poses a major barrier to viral eradication. Canonically, latency is thought to arise from progressive epigenetic silencing of active infections. However, little is known about when and how long terminal repeat (LTR)-silent infections arise since the majority of the current latency models cannot differentiate between initial (LTR-silent) and secondary (progressive silencing) latency. In this study, we constructed and characterized a novel, double-labeled HIV-1 vector (Red-Green-HIV-1 [RGH]) that allows for detection of infected cells independently of LTR activity. Infection of Jurkat T cells and other cell lines with RGH suggests that the majority of integrated proviruses were LTR-silent early postinfection. Furthermore, the LTR-silent infections were transcriptionally competent, as the proviruses could be reactivated by a variety of T cell signaling agonists. Moreover, we used the double-labeled vector system to compare LTRs from seven different subtypes with respect to LTR silencing and reactivation. These experiments indicated that subtype D and F LTRs were more sensitive to silencing, whereas the subtype AE LTR was largely insensitive. Lastly, infection of activated human primary CD4+ T cells yielded LTR-silent as well as productive infections. Taken together, our data, generated using the newly developed RGH vector as a sensitive tool to analyze HIV-1 latency on a single-cell level, show that the majority of HIV-1 infections are latent early postinfection.
INTRODUCTION
Upon infection of target cells, HIV-1 integrates into the host genome and the subsequent transcription of the provirus by the cellular machinery leads to an accumulation of genomic and spliced viral RNAs. However, the HIV-1 long terminal repeat (LTR) can be transcriptionally inactive and, as a result, no viral proteins are produced. Transcriptionally inactive, latent HIV-1 proviruses can form as a result of multiple and progressive epigenetic mechanisms that silence otherwise productive infections (reviewed in references 17 and 54). These mechanisms include, but are not limited to, the following: (i) host cell activation state and transcription factor pools, (ii) HIV-1 LTR chromatin structure and modifications (e.g., nucleosome positioning, histone tail modification, DNA methylation), (iii) proviral genomic location (e.g., host chromatin environment, provirus orientation relative to host genes), (iv) threshold levels of the viral transactivator Tat, and (v) upstream control of HIV-1 activation by a currently unknown protein kinase (3). Cumulatively, these molecular mechanisms have led to the prevailing view that HIV-1 latency is largely the product of progressive epigenetic silencing of active infections.
Postintegration HIV-1 latency is one of the main obstacles to viral eradication, since latently infected cells are not detected by the immune system and are not affected by highly active antiretroviral therapy (HAART) (4–6). The latent reservoir is formed within days of initial infection (7–10) and is primarily composed of long-lived resting memory CD4+ T cells, although other cell types may exist (1). Latency makes lifelong HAART a requirement for HIV-1-infected individuals (11), as cessation of therapy results in viral rebound within weeks (12).
Eradication of HIV-1 requires novel pharmacotherapies directed at modulating the latent reservoir. Such strategies necessitate compounds that are able to potently and specifically induce latent HIV-1 expression from the viral LTR promoter in a broad range of epigenetic environments and cellular states. To date, efforts to purge the latent reservoir have focused on the usage of histone deacetylase inhibitors (HDACi) to induce LTR transcription. While studies have been able to induce HIV-1 expression ex vivo, early efforts with these drugs showed little impact on the latent reservoir in vivo (13–15). A recent study demonstrated, however, the induction of cell-associated HIV-1 RNA in patients treated with the HDACi suberoylanilide hydroxamic acid (SAHA) (16). A more comprehensive understanding of the processes determining the establishment of HIV-1 latency is essential to discovering efficacious new therapies.
Several model systems have been developed to study latency in cell culture, since studies in vivo are complicated because latently infected cells from patients are extremely rare (less than ∼1 × 106 per patient) and phenotypically indistinguishable from uninfected cells (6). Typically, HIV-1 latency models utilize a single marker of active viral infection, such as an LTR-driven fluorescent protein, such as green fluorescent protein (GFP), followed by selection and long-term cell culture to establish latency (reviewed in references 17, 18, and 19). However, this design inherently limits the ability to study early latency establishment because of the absence of a positive marker for latent infection. Additionally, the cell selection steps employed may polarize the model toward specific latent populations, i.e., either initial or secondary latency. Some evidence suggests that “silent infections” may play a role in HIV-1 latency (20); however, the frequency and regulation of these infections remain largely unknown.
In order to better study HIV-1 early latency establishment, we constructed a “double-labeled,” single-cycle HIV-1 vector suitable for studying HIV-1 transcription at the single-cell level (Red-Green HIV-1 [RGH]). We show that in infected Jurkat cells, a substantial proportion of infections (approximately 65%) result in an LTR-silent population very early after infection. The LTR-silent population is transcriptionally competent and can be reactivated by several HIV-1-activating compounds. In addition, we show that the frequency of early silent infections is variable with respect to specific HIV-1 subtype LTRs. Finally, we show that both LTR-silent and productive infections are also represented in activated primary human CD4+ T cells. In summary, the series of RGH vectors described here provides a new model system to effectively study different aspects of HIV-1 latency at the single-cell level without the need for selection steps or extensive culturing.
MATERIALS AND METHODS
Construction of a panel of RGH vectors.
To construct the RGH molecular clone, an ApaI-BssHII fragment containing gag-enhanced green fluorescent protein (eGFP) was cloned from the Gag-iGFP NL4-3 clone (21) into pLAI (22) to create pLAI-Gag-iGFP. Of note, the Envelope open reading frame was disrupted by the introduction of a frameshift at position 7136 by digestion with KpnI, blunting, and religation. The cytomegalovirus-mCherry (CMV-mCherry) (23) cassette was PCR amplified from pcDNA3.1+-mCherry and cloned into pLAI-Gag-iGFP using the BlpI-XhoI sites to create the final double-labeled construct. The ΔU3 RGH clone was created by cloning a ΔU3 linker from pTY-EFeGFP (24–27) into the KpnI-SacI sites of the 3′ LTR in the double-labeled clone. The ΔCMV RGH clone was created by removing a BsmBI-PmeI fragment containing the CMV promoter from the double-labeled construct.
The integrase mutant D116A was created by two-step PCR-mediated site-directed mutagenesis and cloning of the final amplicon into the ApaI-SalI sites of the RGH construct. The group M subtype promoters (n = 7; B, A, C, D, AE, F, and G) were cloned into the RGH construct by transferring a HindIII-XhoI fragment from subtype constructs (28) to the 3′ LTR of the RGH clone.
Cell culture, virion production, and transduction.
Jurkat E6-1 (29), SupT1 (30), U937 (ATCC), HeLa (ATCC), and HEK293T (ATCC) cells were cultured under standard conditions as previously described (31).
Vesicular stomatitis virus G (VSV-G) pseudotyped viral stocks were created by transfecting HEK293T cells with viral molecular clones and pHEF-VSVg (24) in a 10:1 ratio as previously described (31). Purified, concentrated viral stocks were prepared by centrifugation of clarified supernatants from transfected HEK293T cells through a 6% iodixanol–OptiPrep (Sigma) cushion. Concentrated virions were resuspended in complete media.
Jurkat E6-1 cells were infected by spinoculation as previously described (32), except that 5 × 105 cells were subjected to spinoculation for 1.5 h in 12-well plates in 1 ml complete media supplemented with 4 μg/ml Polybrene (500 × g, room temperature). A 25-μl volume of viral supernatant was added to the cells to yield an average infection rate of 10%, thereby ensuring single-copy integrations. Unless otherwise indicated, infected cells were utilized for experiments at 4 days postinfection.
Primary T cell infections.
Primary CD4+ T cells from three healthy donors (HC-8537, HC-8536, and HC-4142) were obtained from Sanguine Biosciences and thawed according to the company's recommendations. Cells were cultured for 48 h in a humidified atmosphere of 37°C and 5% CO2 in RPMI 1640–10% (vol/vol) fetal bovine serum–100 U/ml penicillin–100 mg/ml streptomycin–20 U/ml interleukin-2 (IL-2) (33). Cells (2.5 × 106) from each donor were incubated either with IL-2 alone (nonactivated) or with IL-2 and phytohemagglutinin (PHA-P [2 μg/ml]) (activated) for 48 h.
Activated and nonactivated primary CD4+ T cells were subjected to spinoculation with purified RGH viral stocks as described for Jurkat cells, except that 2.5 × 105 cells were subjected to spinoculation in 24-well plates in 0.5 ml of complete media supplemented with 2 μg/ml Polybrene. A 10- or 100-μl volume of 25×-concentrated viral supernatant was added to the activated or nonactivated cells, respectively. For the remainder of the experiment, cells were maintained in complete RPMI media supplemented with IL-2 (20 U/ml) and media was replenished every 2 days. Cells were analyzed by flow cytometry 6 days postinfection.
Flow cytometry and drug treatments.
Prior to flow cytometry analysis, cells were fixed in 1% (vol/vol) formaldehyde for 10 min at room temperature. A total of 10,000 to 20,000 live cell events (as determined on the basis of forward scatter channel-side scatter channel [FSC/SSC] profiles) were analyzed on a Becton Dickinson LSRII flow cytometer, and the data were analyzed using FlowJo. Statistical tests (Student's t test) were performed using R 2.15.1. Live cells were sorted by a Becton Dickinson Influx fluorescence-activated cell sorter (FACS).
Cells were treated with various drugs for the times and durations indicated in the individual experiments. Drugs were added at the listed concentrations to complete media. Unless otherwise stated, drugs were used at the following concentrations: raltegravir, 10 μM; tumor necrosis alpha (TNF-α), 10 ng/ml; SAHA, 1 μM (34); trichostatin A (TSA), 50 ng/ml; phorbol 12-myristate 13-acetate (PMA), 4 ng/ml; ionomycin (Iono), 1 μM; 5-aza-dC, 5 μM.
RESULTS
Characterization of the double-labeled HIV-1 molecular clone RGH.
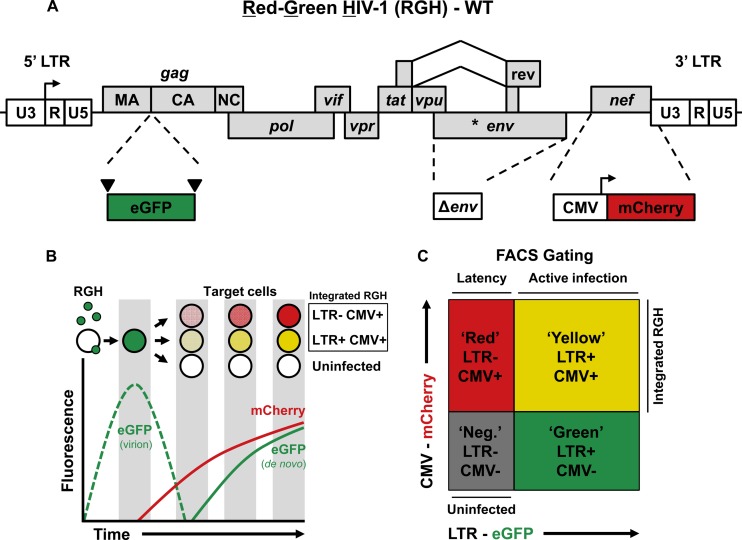
We constructed an HIV-1 LAI molecular clone containing two fluorescent markers to identify both silent and productive HIV-1 infections (Fig. 1A).
Fig 1.
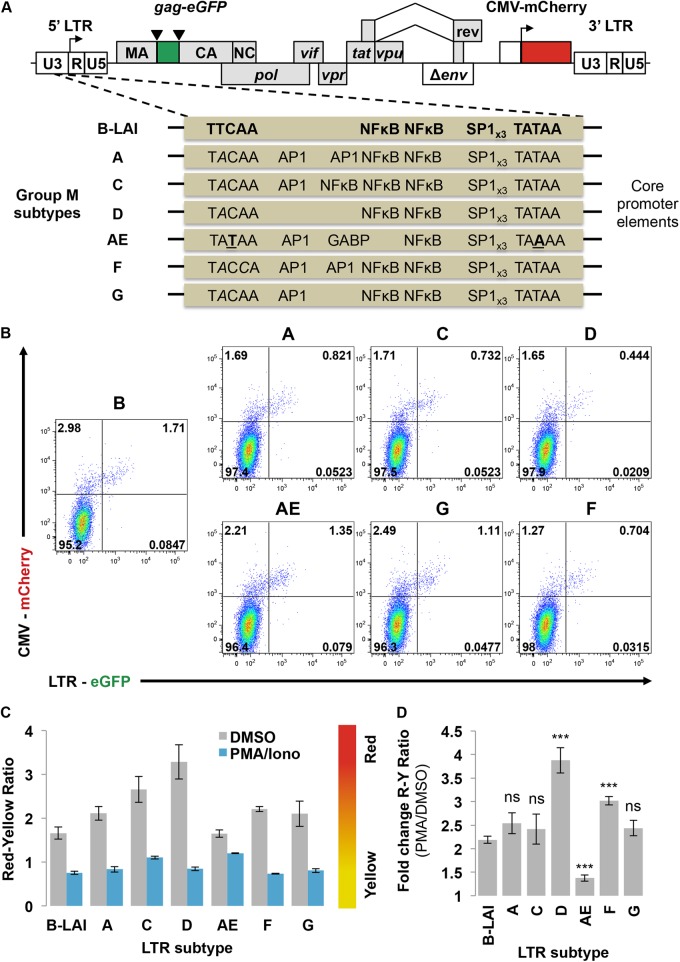
Schematic representation of the Red-Green-HIV-1 (RGH) vector. (A) HIV-1 B-LAI Δenv is labeled with eGFP as an in-frame gag fusion flanked by HIV-1 protease cleavage sites (inverted triangles), and by a CMVIE-driven mCherry cassette located in place of nef. WT, wild type. (B) Schematic depiction of RGH infection of target cells and the resultant fluorescent protein profiles over time. (C) Schematic depiction of HIV-1 RGH-infected cell populations detected by FACS analysis.
The eGFP protein is flanked by HIV-1 protease cleavage sites and placed in gag between matrix and capsid (21, 35). Each Gag protein contains eGFP, resulting in green virions, which, upon fusion with the target cell, release eGFP into the target cell (Fig. 1B). Upon integration, Gag-eGFP expression is under the control of the HIV-1 promoter and may serve as a quantitative marker for HIV-1 LTR gene activity. Moreover, Gag-eGFP is a late gene product and, hence, is likely indicative of HIV-1 virus production. Of note, low-level Gag expression in primary cells can occur in the absence of virion production (2), suggesting that certain Gag expression levels are required for efficient production of new virions.
The second reporter, inserted in the viral nef position, consists of a cassette containing the CMV immediate-early promoter expressing mCherry fluorescent protein (23). mCherry is constitutively expressed and allows detection of integrated virus regardless of the transcriptional state of the HIV-1 LTR.
The combination of the two fluorescent proteins under the control of the HIV-1 LTR and an internal CMV promoter (Red-Green-HIV-1 [RGH]) allows quantitative detection of active virus production (eGFP+ and mCherry+; “yellow”), but more importantly, also detects nonproductive HIV-1 (eGFP− and mCherry+; “red”) (Fig. 1C). In addition, the HIV-1 envelope is inactivated, which prevents spreading viral replication and superinfection, as well as envelope-induced/associated cell toxicity. The deletion in envelope also allows us to circumvent the use of antiretroviral drugs in cultures, which would be required to prevent multiple rounds of infections.
Red-Green-HIV-1 identifies LTR-silent infections, which are established early after infection and require integration.
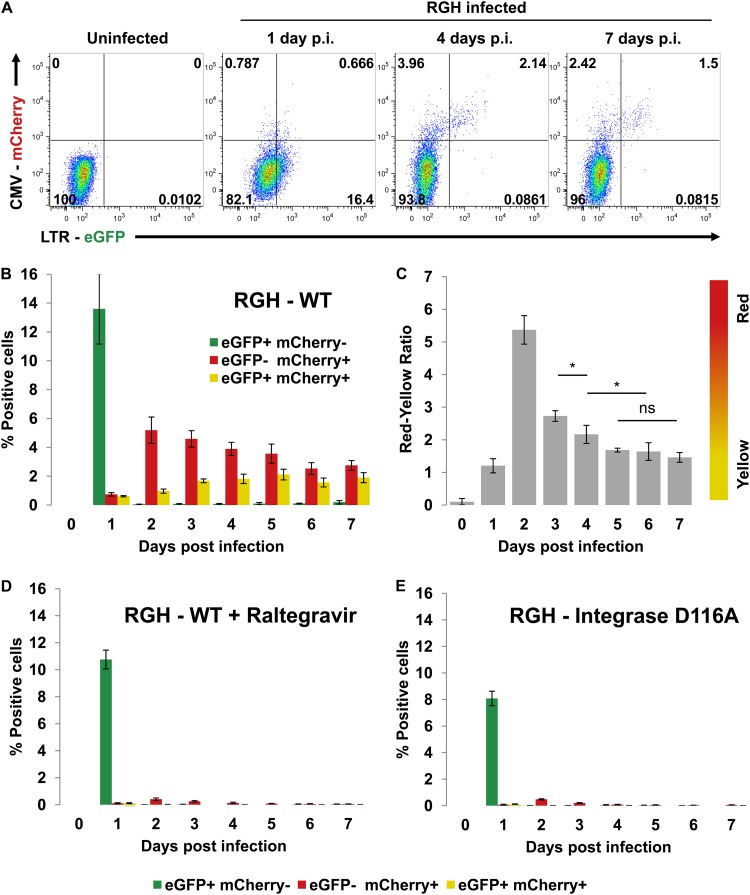
To test the double-labeled system (Fig. 1), we produced VSV-G pseudotyped RGH viral stocks in HEK293T cells and infected Jurkat T cells at a low multiplicity of infection (MOI) (∼0.2). At 1 day postinfection, distinct green, yellow, and red populations were observed, but only the yellow and red populations, representing integrated proviruses, persisted through later time points (Fig. 2A and B). The number of LTR-silent infected cells (red) consistently outnumbered the doubly positive HIV-1-producing cells (yellow), indicating that the majority of HIV-1 infections are silent early postinfection. Interestingly, day 1 postinfection was characterized by a high number of GFP+ mCherry− cells, which were no longer observed at later time points (Fig. 2A and B). This transient eGFP+ cell population is the result of the Gag-eGFP fusion proteins incorporated in the virus used to infect the cells and hence rapidly disappears after infection (Fig. 1B and references 21 and 35). Calculation of the ratio of red to yellow cells for all the time points of the infection (Fig. 2C) showed a transient increase of the number of red cells (eGFP-mCherry+) over the number of yellow cells (eGFP+/mCherry+) 2 days postinfection. This spike could reflect delayed late Gag-eGFP expression compared to more rapid mCherry accumulation, and/or the observed drop in mCherry+ cell numbers could result from CMV promoter silencing. Importantly, however, the red cell/yellow cell ratio stabilized approximately 4 to 5 days after infection (Fig. 2C).
Fig 2.
LTR silencing in RGH-infected Jurkat cells occurs early and requires integration. (A) Flow cytometry time course of RGH-infected Jurkat cells. Plots shown are representative of the results of triplicate infections. p.i., postinfection. (B) Plot of positive cells from each colored fraction (eGFP+ mCherry−, eGFP− mCherry+, eGFP+ mCherry+) over the course of infection. Error bars represent standard deviations of the results of triplicate experiments. (C) Data from the experiment represented by panel B are enumerated as ratios of red cells to yellow cells (proportion of eGFP− mCherry+ to eGFP+ mCherry+). Error bars represent standard deviations of the results of triplicate experiments. ns, nonsignificant; *, P < 0.05 (Student's t test). (D) Jurkat cells were infected with RGH in the presence of the integrase inhibitor raltegravir (10 μM). Cells were analyzed by flow cytometry over a period of 7 days. Error bars represent standard deviations of the results of triplicate experiments. (E) Jurkat cells were infected as described for panel D, except that an RGH variant bearing a catalytically defective integrase variant (D116A) was used. Additionally, raltegravir was excluded.
Nonintegrated 2-LTR circles have been described to express several HIV-1 nonstructural proteins, such as Tat, Rev, and Nef (36, 37). To exclude the possibility that the higher mCherry signal results from expression from 2-LTR circles, we infected Jurkat cells with RGH virus in the presence of the integrase inhibitor raltegravir (Fig. 2D) or with an RGH virus bearing a D116A inactivating mutation in integrase (Fig. 2E and reference 52). Both raltegravir and the integrase mutant prevented the formation of red and yellow cells (Fig. 2D and E), indicating that the red and yellow signals observed in RGH-infected cells represent integrated proviruses and not 2-LTR circles. The eGFP expression at day 1 postinfection, in contrast, was not affected by raltegravir or the integrase mutation, underscoring that the initially observed eGFP peak is caused by the virally packaged eGFP and not from newly expressed eGFP in the target cells. Thus, the RGH vector can efficiently discriminate between active and silent HIV-1 expression. Moreover, infection of Jurkat cells with RGH suggests that the majority of infections are latent early postinfection.
Identification of LTR-silent infections is dependent on the presence of both LTR and CMV promoters.
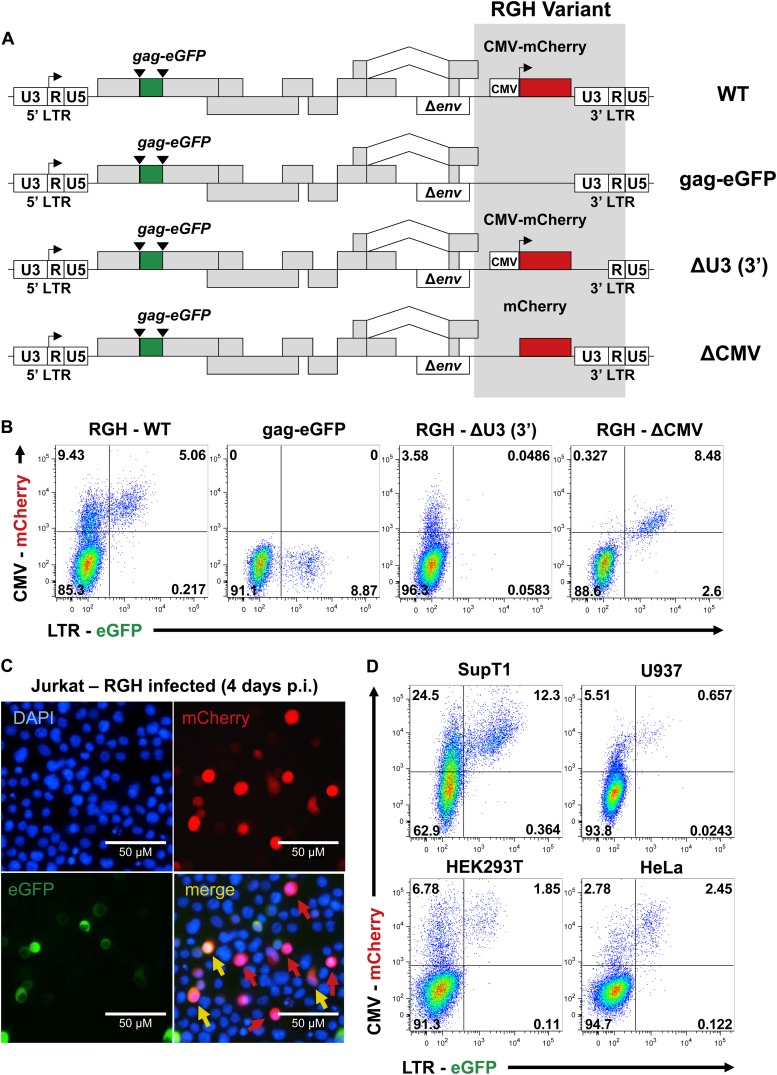
Infection of Jurkat cells with RGH indicates that the majority of integrated proviruses are LTR silent 4 to 5 days postinfection (Fig. 2). To characterize the effect of both the LTR and CMV promoters on this infection profile, we constructed a number of control RGH variant viruses that lack CMV-mCherry, the CMV promoter, or the HIV-1 promoter (Fig. 3A). As expected, CMV-mCherry-deleted virus produced only green cells (Fig. 3B). RGH lacking the U3 promoter region of the 3′ LTR results in an integrated virus devoid of the 5′ HIV-1 promoter and results in only red cells (Fig. 3B). Finally, the deletion of the CMV promoter in RGH renders mCherry dependent on the HIV-1 promoter and results in the simultaneous expression of eGFP and mCherry, thereby producing exclusively eGFP+ mCherry+ (yellow) cells (Fig. 3B).
Fig 3.
Identification of LTR-silent RGH infections is dependent on both the LTR and CMV promoters, and is not specific for Jurkat T cells. (A) Schematic representation of RGH variant viruses. Gag-eGFP lacks the CMV-mCherry construct, while ΔU3 and ΔCMV both contain both fluorescent coding sequences (eGFP and mCherry) but lack U3 in the 3′ LTR and the CMV promoter, respectively. Variants are otherwise isogenic. (B) Jurkat cells were infected with WT and variant RGH and analyzed by flow cytometry 4 days postinfection. Plots shown are representative of the results of triplicate experiments. (C) Fluorescence microscopy (×100) of RGH-infected Jurkat cells (4 days postinfection). eGFP− mCherry+ and eGFP+ mCherry+ cells are indicated in the merge panel by red and yellow arrows, respectively. Images shown are representative of the results of triplicate experiments. (D) SupT1, U937, HEK293T, and HeLa cells were infected with RGH and analyzed by flow cytometry 4 days postinfection. Plots are representative of the results of duplicate experiments.
Inspection of Jurkat cells infected with wild-type RGH by fluorescence microscopy showed two distinct cell populations: cells expressing both fluorescent proteins (eGFP+/mCherry+; yellow) and cells expressing only mCherry (eGFP−/mCherry+; red) (Fig. 3C). Taken together, these data confirm that having a virus with independent expression of both fluorescent reporters is necessary to detect early LTR-silent infections.
We next wanted to determine if the LTR-silent infections were specific for Jurkat T cells or a bona fide property of HIV-1 infection. To do this, we infected different human cell lines such as the T-cell line SupT1, the monocyte line U937, and the nonimmune cell lines HEK293T and HeLa with the RGH viral stocks. As shown in Fig. 3D, cell lines differed in their susceptibilities to infection (between 5% and 37% infection rate) (Fig. 3D) but most infected cell lines yielded at least twice as many eGFP−/mCherry+ (red) cells as eGFP+/mCherry+ (yellow) cells (for SupT1 cells, red, 24.5%, yellow, 12.3% [ratio, 1.99]; for U937 cells, red, 5.5%, yellow, 0.7% [ratio, 7.85]; for HEK293T cells, red, 6.8%, yellow, 1.9% [ratio, 3.58]; and for HeLa cells, red, 2.8%, yellow, 2.5% [ratio, 1.12]). These results indicate that LTR-silent infections are common and evident in many different cell types (Fig. 3D). Collectively, these data suggest that the independent expression of the two fluorescent reporters is necessary to detect early LTR-silent infections and that these infections are a bona fide feature of HIV-1 infection.
Silent LTRs in infected cells are transcriptionally competent.
The HIV-1 LTR contains binding sites for a multitude of transcription factors (such as NF-κB, nuclear factor of activated T-cells [NFAT], and AP-1) that act downstream of the T-cell signaling pathway and hence can be activated by a variety of signaling agonists (reviewed in references 17 and 53). For example, tumor necrosis factor alpha (TNF-α) and phorbol 12-myristate 13-acetate (PMA)-ionomycin (Iono) are both potent activators of HIV-1 (TNF-α in transformed T cells only) by stimulating the protein kinase C (PKC) and mitogen-activated protein (MAP) kinase/calcineurin pathways, respectively (39, 40). Additionally, general epigenetic modifying drugs such as the HDAC inhibitors SAHA and trichostatin A (TSA) cause hyperacetylation of LTR nucleosomes and activation of LTR-driven transcription (41, 42). The DNA methyltransferase inhibitor 5-aza-2′deoxycytidine (5-aza-dC) has been shown to activate HIV-1 transcription, although its effects depend on the model system used (43).
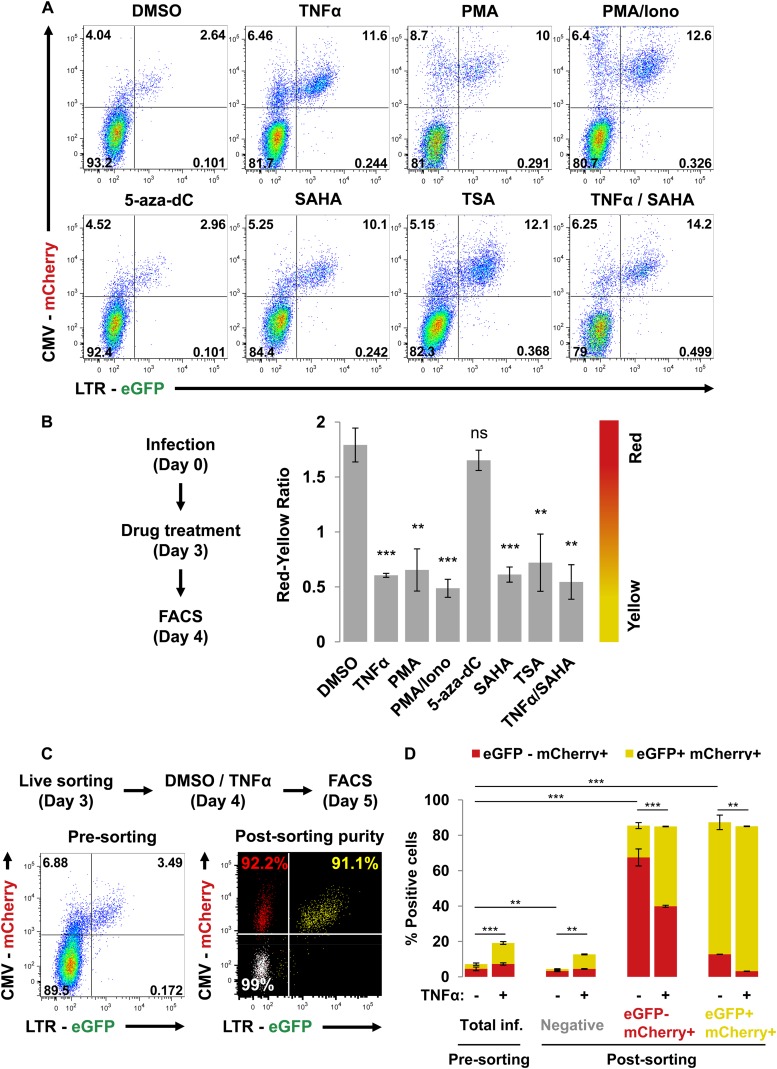
We next determined whether the majority of RGH-infected Jurkat cells, which are LTR silent 4 days after infection (Fig. 2), could be reactivated. We treated RGH-infected Jurkat cells 4 days postinfection with TNF-α, PMA, Iono, PMA/Iono, 5-aza-dC, SAHA, TSA, TNF-α/SAHA, or dimethyl sulfoxide (DMSO) (negative control) for 24 h followed by FACS analysis (Fig. 4A and B). TNF-α, PMA, PMA/Iono, SAHA, TSA, and TNF-α/SAHA activated silent LTRs compared to the DMSO control (approximately 3-fold decrease in the red/yellow ratio) (Fig. 4A and B). Interestingly, 5-aza-dC failed to activate silent LTRs (Fig. 4A and B). It has been proposed that DNA methylation is a late mark in HIV-1 proviral silencing and, thus, that 5-aza-dC would not be expected to play a role at an early time point after integration (38). Additionally, the combination of TNF-α and SAHA did not have an additive effect on LTR activation, suggesting that the maximal effect had been reached with either drug alone (Fig. 4A and B).
Fig 4.
Silent LTRs in RGH-infected cells are transcriptionally competent. (A) RGH-infected Jurkat cells (4 days postinfection) were treated with DMSO, TNF-α, PMA, PMA/Iono, 5-aza-dC, SAHA, TSA, or TNF-α/SAHA for 24 h prior to analysis by flow cytometry. Plots shown are representative of the results of triplicate experiments. (B) Data from panel A are enumerated as the red/yellow ratio of the infected population. Error bars represent standard deviations of the results of triplicate experiments. ns, nonsignificant; **, P < 0.01; ***, P < 0.001 (Student's t test). (C) RGH-infected Jurkat cells were sorted to greater than 90% purity 3 days postinfection. The left panel depicts the Jurkat cells infected with RGH at day 3 postinfection, and the right panel shows the purity of the individual cell populations after sorting (composite of three individual FACS plots). Cells were left to recover for 24 h prior to treatment with either DMSO or TNF-α for a further 24 h, followed by analysis by flow cytometry. (D) Data from the experimental outline in panel C. Cells were analyzed by flow cytometry. Error bars represent standard deviations of the results of duplicate experiments. inf., infected. **, P < 0.01; ***, P < 0.001 (Student's t test).
Interestingly, we consistently noticed a decrease in the double-negative cell population upon reactivation of RGH (eGFP− mCherry−) (lower left quadrant, Fig. 4A and B), which suggests that the double-negative cells may harbor infected cells with inactivated LTR and CMV promoters. To better understand the reactivation features of the different RGH-infected cell populations, we sorted RGH-infected Jurkat cells into their constituent populations (3 days postinfection). Each of the three sorted cell populations, as well as unsorted infected Jurkat cells, was activated with TNF-α (Fig. 4C and D). The presorted infected Jurkat cells showed the typical infection profile for both DMSO and TNF-α treatment, with red cells outnumbering the yellow cells for DMSO and a reversal of this distribution for TNF-α (Fig. 4D). TNF-α treatment of the sorted double-negative cells resulted in both red and yellow cells (4.5% red, 8.2% yellow; red/yellow ratio, 0.55) (Fig. 4D), indicating that a portion of the “uninfected” cell population, indeed, harbored fully competent proviruses with both promoters silenced. The sorted mCherry singly positive population remained predominantly mCherry positive and showed an increase in the numbers of yellow cells upon TNF-α treatment compared to the DMSO treatment (Fig. 4D). The sorted doubly positive yellow cell population comprised primarily LTR-active proviruses, and TNF-α activation resulted in only a minor increase in the numbers of yellow cells, thus confirming that the HIV-1 LTR is already fully activated in these cells (Fig. 4D).
Taken together, these data indicate that LTR-silent RGH proviruses in Jurkat cells are transcriptionally competent and can be reactivated by a variety of T-cell signaling agonists and HDAC inhibitors.
Group M subtype LTRs differ in their sensitivities to latency.
Given that the RGH vector can detect LTR-silent infections in Jurkat cells (Fig. 2, 3, and 4), we reasoned that the ratio of LTR-silent red cells to LTR-active yellow cells would likely be affected by intrinsic determinants within the HIV-1 LTR, such as transcription factor binding sites. Therefore, we cloned natural subtype LTR variants (28) into the RGH vector and assessed the effect of LTR variations on silent infections and reactivation potential. Jurkat cells were infected with each of the RGH subtype LTR viruses (subtypes B, A, C, D, AE, F, and G) (Fig. 5A) at low MOI and treated with PMA/Iono or DMSO 4 days postinfection to activate LTR-silent RGH (Fig. 5B and C). After PMA/Iono activation, the red/yellow ratio dropped substantially (approximately 2- to 4-fold) for most subtypes compared to the control DMSO treatment results. However, subtype AE exhibited the lowest red/yellow ratio difference between DMSO and PMA/Iono treatment, suggesting that the subtype AE promoter is constitutively expressed and thus cannot be further activated by PMA/Iono (Fig. 5C and D). Since the subtype AE promoter contains only one canonical NF-κB binding site (28, 44), limited activation by the PMA/Iono treatment could be the underlying reason (Fig. 5C and D).
Fig 5.
RGH recapitulates differences in latency observed between HIV-1 group M subtypes. (A) Schematic representation of RGH variants containing promoter sequences from the major group M viral subtypes. Viruses are isogenic except for a 209-bp BseAI-AflII fragment containing the core promoter region of the LTR that stretches from position −147 to position +68. (B) Jurkat cells were infected with the subtype RGH variants and analyzed by flow cytometry 4 days postinfection. Plots shown are representative of the results of triplicate experiments. (C) Jurkat cells were infected with the subtype RGH variants. At 4 days postinfection, cells were treated with either DMSO or PMA/ionomycin for 24 h and then analyzed by flow cytometry. Error bars represent standard deviations of the results of triplicate experiments. (D) Data from panel C are enumerated as fold change between the PMA and DMSO treatments. Error bars represent standard deviations of the results of triplicate experiments. R, red; Y, yellow; ns, nonsignificant; ***, P < 0.001 (Student's t test).
Interestingly, the fold change in latency ratios is significantly higher for subtype D and F LTRs, suggesting that these promoters are more prone to transcriptional silencing in Jurkat T cells (Fig. 5D). Of note, for subtype AE the fold difference in latency ratios between DMSO and PMO/Iono treatment is substantially lower than that seen with B-LAI (Fig. 5D).
Our results suggest that different HIV-1 subtypes display different levels of LTR silencing. Subtype AE was less likely to become silenced than the other subtypes, which is in full agreement with previous reports (28, 44), while subtypes D and F were more prone to silencing. Using the RGH Jurkat cell model, we observed differences in the occurrence of LTR-silent infections between subtypes, which recapitulated well the results observed in earlier studies using different models to study HIV-1 latency.
LTR-silent RGH infections occur in primary CD4+ T cells.
The majority of RGH infections in Jurkat cells result in direct LTR silencing (Fig. 2, 3, 4, and 5). Although Jurkat cells are widely used to study HIV-1 latency, they may not accurately represent the memory CD4+ T cells that make up the latent HIV-1 reservoir in vivo. To study RGH infection in a more relevant context, we infected human primary CD4+ T cells obtained from three healthy donors. We subjected activated (IL-2-plus-PHA-treated) and nonactivated (IL-2 alone) cells from the different donors to spinoculation with RGH and cultured the infected cells for 6 days prior to flow cytometry analysis (Fig. 6A and B).
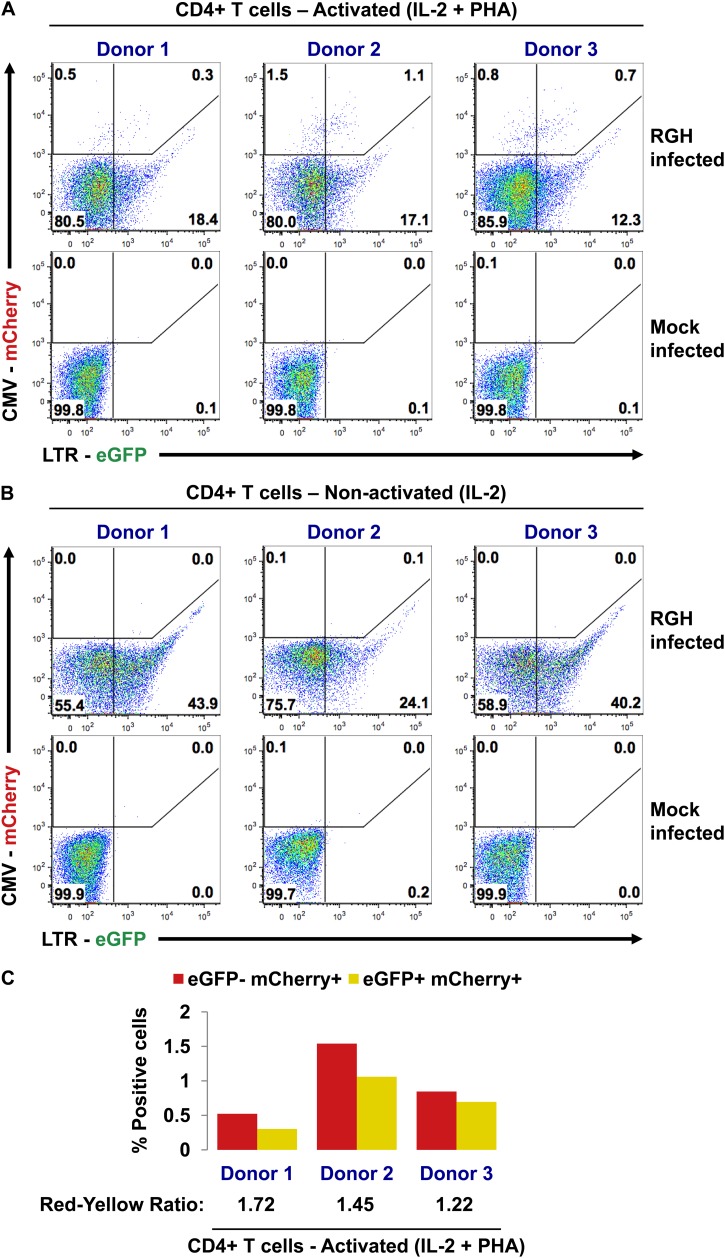
Fig 6.
LTR-silent RGH infections occur in primary CD4+ T cells. (A) Activated (IL-2 + PHA) primary CD4+ T cells from three healthy donors were infected with purified RGH. Infected cells were cultured in IL-2-containing media for 6 days prior to FACS analysis. (B) Nonactivated (IL-2) primary CD4+ T cells from three healthy donors were infected with 10-fold more virus than in the experiment represented by panel A. Infected cells were cultured in IL-2-containing media for 6 days prior to FACS analysis. (C) Data from panel A are displayed graphically, with the ratio of red cells to yellow cells indicated at the bottom.
The activated cells (IL-2 plus PHA) showed both red and yellow cells indicative of successful infections. Red cells outnumbered yellow cells in all donors (Fig. 6A and C), demonstrating the presence of LTR-silent infections in primary cells, similar to the results seen with Jurkat cells (Fig. 2, 3, 4, and 5). However, many cells were eGFP+ only (Fig. 6A; compare RGH infected to mock infected) and microscopy inspection of the cells shortly after infection revealed the presence of large intracellular eGFP aggregates that persisted for more than 6 days (data not shown). This eGFP signal represents the initial, virally derived eGFP signal that, in contrast to Jurkat cells (Fig. 2B), persists for a long time in primary CD4+ T cells.
Despite using 10-fold-more virus to infect nonactivated CD4+ T cells (IL-2 alone), we could not detect red or yellow infected cells (Fig. 6B), indicating that the RGH virus is unable to productively infect nonactivated cells. Of note, nonactivated CD4+ T cells are difficult to infect, a problem that may be overcome in some models by using a very high MOI (reviewed in references 18 and 19). Despite the absence of productive infections, we detected eGFP+-only cells at levels similar to those observed in activated cells from the same donor (Fig. 6B).
Collectively, these data suggest that both LTR-silent and productive infections occur in primary CD4+ T cells.
DISCUSSION
A better understanding of the dynamics of HIV-1 latency and the cellular mechanisms controlling these processes is instrumental to developing HIV-1 eradication strategies. In this study, we constructed and characterized a novel double-labeled HIV-1 vector (Red-Green-HIV-1 [RGH]) (Fig. 1), which reduces the selection bias inherent to traditional single-label models of latency. Additionally, this model allows examination of several phases of HIV-1 latency and reactivation, including establishment, maintenance, and expression. By incorporating the CMV-mCherry reporter, the RGH model can positively identify latently infected cells rather than utilizing excluding selection steps, which is a common denominator for many other latency systems. Moreover, utilization of CMV-mCherry allows the distinction of latently infected cells from uninfected ones, thereby allowing transcriptome and proteome comparisons, without the need for viral reactivation by T cell activation. Reactivation likely destroys the unique transcriptional properties of latently infected cells, thus preventing meaningful comparisons of the viral and cellular states of both latently and actively infected cells. Thus, with the RGH vector, all populations of infected cells, including those with initially suppressed LTRs, can be studied and quantified at both the single-cell and population levels. Additionally, the RGH vector may also be used to study canonical mechanisms of HIV-1 latency by continual passaging. These mechanisms, which contribute to the progressive epigenetic silencing of active infections, include both cis-acting factors (e.g., HIV-1 integration site and chromatin environment) and trans-acting factors (e.g., cellular activation state and transcription factor pools [reviewed in references 1 and 2]).
The major advantage of the RGH vector system is the LTR-independent marker of infection (mCherry), which requires a separate, HIV-1-independent promoter to drive its expression. We choose the CMV promoter for this purpose since it is widely used by the research community. Of note, the CMV promoter may also have its limitations, especially in primary cells (45–47). Activation of RGH-infected Jurkat cells (Fig. 4A and 4B, whole-cell population, and Fig. 4D, double-negative sorted cells) revealed that the eGFP−/mCherry− cell population also contains LTR- and CMV-silent proviruses, which can be activated by TNF-α treatment (Fig. 4). Inactive/silenced CMV promoters would, therefore, lead to underestimation of the true infection rate in our system. Moreover, the CMV promoter has been shown to be responsive to T cell activation signals and, consequently, to be expressed at much lower levels in nonactivated cells (45–47). Thus, an alternative constitutive nonviral promoter such as EF1α, PGK, or UbC may be useful alternatives to the CMV promoter used in the current RGH vector. Despite these CMV promoter-associated potential limitations, we observed that the majority of RGH CMV-mCherry+ infections were LTR silent (Fig. 2, 3, and 4) in different cell lines (in T cell lines, Jurkat, Sup-T1, myeloid, epithelial, etc.). In this light, utilizing a stronger mCherry promoter would lower the frequency of double-silent promoter proviruses, thereby ensuring that the detectable cells (eGFP− and mCherry+) better represent the silenced HIV-1 provirus.
Importantly, the RGH infection experiments suggest that transcriptionally competent, LTR-silent proviruses are common in T cell lines, as well as in other transformed cell types (Fig. 2). Moreover, we also observed LTR-silent infections in primary CD4+ T cells, suggesting that RGH vector may also be suitable for studying direct-silent infections in primary cells (Fig. 6). However, some shortcomings of the RGH vector became more apparent in primary CD4+ T cell infections. First, a substantial and persistent eGFP+-only signal, reflecting eGFP-labeled incoming particles, was present in both resting and activated CD4+ T cells (Fig. 6A and B). This was also observed for RGH-infected Jurkat cells, but this virally packaged eGFP signal disappeared rapidly in the Jurkat cells (Fig. 2). Apparently, the virally packaged eGFP is much more stable in primary cells, possibly because the CD4+ T cells divide less, and thus, eGFP would not be lost by dilution. Importantly, the presence of the eGFP+-only signal may complicate the distinction of the red and yellow infected cell populations, as virally derived eGFP may cause red cells to shift into the yellow gate, thereby leading to underestimation of the LTR-silent population. In addition, some of the cells in the eGFP+ population could be infected with only the transcriptionally LTR-active (eGFP+ only) cells. Indeed, the CMV promoter has been observed to be expressed with lower efficiency in some primary lymphocyte systems (46, 48–50). Moreover, the low infectivity in CD4+ T cells is likely caused by the insertion of eGFP into Gag, which was reported to have cell type-specific effects on replication (21). Future versions of the vector could address these CD4+ T cell-specific limitations by using other constitutive promoters (EF1α, PGK, or UbC) driving mCherry and/or by replacing certain HIV-1 genes, such as the Vif or Vpr gene, with the eGFP gene, so that the fluorescent reporter would no longer be packaged into virions, and may be less detrimental to viral infectivity.
Despite these shortcomings, the RGH vector data suggest that direct-silent infections have been underappreciated so far and, thus, it is likely that these cells contribute to HIV-1 latency. To our knowledge, only one other study has identified silent infection as a contributor to HIV-1 latency. In that study, which used a singly labeled vector to infect a cell line, silent infections were correlated with NF-κB levels in cells at the time of infection, with transcriptional interference from host genes being the actual silencing mechanism (20). Further studies are needed to definitively address whether control of latency by NF-κB is indeed a causative factor and whether this mechanism applies to all sites of viral integration.
Differential mechanisms that lead to silent infection or silencing from canonically formed latency may require unique pharmacotherapeutics to reactivate latent proviruses. One of the most commonly proposed eradication strategies, called “shock and kill,” would utilize small molecules to induce HIV-1 expression from latently infected cells, thus allowing induced cells to be cleared by the immune system or killed by viral cytopathic effects (reviewed in references 11 and 51). However, any efficacious drug suitable to purging HIV-1 latency would have to target both forms of latent infection, as excluding any part of the latent reservoir would render such therapy less effective. Given its ability to minimize selection bias and include all populations of infected cells, the RGH vector model will be a very useful tool to screen for latency-modulating drugs that fulfill these criteria.
In conclusion, we generated and characterized a comprehensive panel of single-cycle vectors to study the very early events involved in establishment of HIV-1 latency. Our results suggest that the proviral LTR is silenced immediately upon integration in a high proportion of infections. HIV-1 latency, thus, may not be simply the result of progressive epigenetic silencing of active infections but rather represents a multifaceted process that can form very early postintegration and in the absence of previous LTR activity. This underappreciated and previously hard-to-measure population of initially latent cells may have profound consequences for the establishment and treatment of latency. A better understanding of the mechanisms controlling the fate of integrated proviruses will inform improved treatment strategies.
ACKNOWLEDGMENTS
We thank Andy Johnson and Justin Wong of the UBC Flow Cytometry Facility for performing FACS analysis as well as for assistance with flow cytometry. We also gratefully acknowledge Ben Berkhout for providing the molecular clone pLAI and HIV-1 subtype constructs, and Benjamin Chen for providing the molecular clone Gag-iGFP. We thank Vicente Planelles for critical review of the manuscript and helpful suggestions. We also thank Jacob Hodgson, Adam Chruscicki, Kevin Eade, Benjamin Martin, and Nicolas Coutin for helpful discussion throughout this project.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Jurkat clone E6-1 from Arthur Weiss, Sup-T1 from James Hoxie, raltegravir (catalog no. 11680) from Merck & Company, Inc., SAHA (Vorinostat) and pHEF-VSVG from Lung-Ji Chan, pTY-EFeGFP from Lung-Ji Chang, and human rIL-2 from Maurice Gately, Hoffmann-La Roche Inc.
This work was supported by Canadian Institute of Health Research (CIHR) grants to I.S. (MOP-77807, HOP-120237) and NIH/NIAID grants to V.S. (AI064001, AI089246, AI90935). M.S.D. is supported by a CIHR fellowship (CGD-96495).
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 95:8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pace MJ, Graf EH, Agosto LM, Mexas AM, Male F, Brady T, Bushman FD, O'Doherty U. 2012. Directly infected resting CD4+T cells can produce HIV Gag without spreading infection in a model of HIV latency. PLoS Pathog. 8:e1002818 doi:10.1371/journal.ppat.1002818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolschendorf F, Bosque A, Shishido T, Duverger A, Jones J, Planelles V, Kutsch O. 2012. Kinase control prevents HIV-1 reactivation in spite of high levels of induced NF-κB activity. J. Virol. 86:4548–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517 [DOI] [PubMed] [Google Scholar]
- 5. Richman DDD, Margolis DMM, Delaney M, Greene WCC, Hazuda D, Pomerantz RJJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304 doi:10.1126/science.1165706 [DOI] [PubMed] [Google Scholar]
- 6. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727–728 [DOI] [PubMed] [Google Scholar]
- 7. Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Wiegand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. 2012. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. U. S. A. 109:9523–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chun TW, Justement JS, Moir S, Hallahan CW, Maenza J, Mullins JI, Collier AC, Corey L, Fauci AS. 2007. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195:1762–1764 [DOI] [PubMed] [Google Scholar]
- 9. Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295 [DOI] [PubMed] [Google Scholar]
- 11. Choudhary S, Margolis DM. 2011. Curing HIV: pharmacologic approaches to target HIV-1 latency. Annu. Rev. Pharmacol. Toxicol. 51:397–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho DD, Zhang L. 2000. HIV-1 rebound after anti-retroviral therapy. Nat. Med. 6:736–737 [DOI] [PubMed] [Google Scholar]
- 13. Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, Bandarenko N, Schmitz JL, Bosch RJ, Landay AL, Coffin JM, Margolis DM. 2008. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS 22:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sagot-Lerolle N, Lamine A, Chaix ML, Boufassa F, Aboulker JP, Costagliola D, Goujard C, Pallier C, Delfraissy JF, Lambotte O. 2008. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS 22:1125–1129 [DOI] [PubMed] [Google Scholar]
- 15. Siliciano JD, Lai J, Callender M, Pitt E, Zhang H, Margolick JB, Gallant JE, Cofrancesco J, Moore RD, Gange SJ, Siliciano RF. 2007. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J. Infect. Dis. 195:833–836 [DOI] [PubMed] [Google Scholar]
- 16. Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colin L, Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111 doi:10.1186/1742-4690-6-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hakre S, Chavez L, Shirakawa K, Verdin E. 2012. HIV latency: experimental systems and molecular models. FEMS Microbiol. Rev. 36:706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pace MJ, Agosto L, Graf EH, O'Doherty U. 2011. HIV reservoirs and latency models. Virology 411:344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hübner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81:12596–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672 [DOI] [PubMed] [Google Scholar]
- 23. Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotech. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 24. Chang LJ, Urlacher V, Iwakuma T, Cui Y, Zucali J. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715–728 [DOI] [PubMed] [Google Scholar]
- 25. Cui Y, Iwakuma T, Chang LJ. 1999. Contributions of viral splice sites and cis-regulatory elements to lentivirus vector function. J. Virol. 73:6171–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwakuma T, Cui Y, Chang LJ. 1999. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology 261:120–132 [DOI] [PubMed] [Google Scholar]
- 27. Zolotukhin S, Potter M, Hauswirth W, Guy J, Muzyczka N. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeeninga RRE, Hoogenkamp M, Armand-Ugon M, De Baar M, Verhoef K, Berkhout B. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weiss A, Wiskocil RL, Stobo JD. 1984. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J. Immunol. 133:123–128 [PubMed] [Google Scholar]
- 30. Smith SD, Shatsky M, Cohen PS, Warnke R, Link MP, Glader BE. 1984. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res. 44:5657–5660 [PubMed] [Google Scholar]
- 31. Dahabieh MS, Ooms M, Malcolm T, Simon V, Sadowski I. 2011. Identification and functional analysis of a second RBF-2 binding site within the HIV-1 promoter. Virology 418:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Doherty U, Swiggard W, Malim M. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lahm HW, Stein S. 1985. Characterization of recombinant human interleukin-2 with micromethods. J. Chromatogr. 326:357–361 [DOI] [PubMed] [Google Scholar]
- 34. Marks PA. 2007. Discovery and development of SAHA as an anticancer agent. Oncogene 26:1351–1356 [DOI] [PubMed] [Google Scholar]
- 35. Hübner W, Mcnerney GP, Chen P, Dale BM, Gordon RE, Chuang FYS, Li X, Asmuth DM, Huser T, Chen BK. 2009. Quantitative 3D video microscopy of HIV transfer Across T cell virological synapses. Science 323:1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gelderblom HC, Vatakis DN, Burke SA, Lawrie SD, Bristol GC, Levy DN. 2008. Viral complementation allows HIV-1 replication without integration. Retrovirology 5:60 doi:10.1186/1742-4690-5-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gillim-Ross L, Cara A, Klotman ME. 2005. Nef expressed from human immunodeficiency virus type 1 extrachromosomal DNA downregulates CD4 on primary CD4+ T lymphocytes: implications for integrase inhibitors. J. Gen. Virol. 86:765–771 [DOI] [PubMed] [Google Scholar]
- 38. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi:10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. U. S. A. 86:2365–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Folks TM, Justement J, Kinter A, Schnittman S, Orenstein J, Poli G, Fauci AS. 1988. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol. 140:1117–1122 [PubMed] [Google Scholar]
- 41. Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses 25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. 2009. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fernandez G, Zeichner SL. 2010. Cell line-dependent variability in HIV activation employing DNMT inhibitors. Virol. J. 7:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van der Sluis RMRM, Pollakis G, Van Gerven ML, Berkhout B, Jeeninga RE. 2011. Latency profiles of full length HIV-1 molecular clone variants with a subtype specific promoter. Retrovirology 8:73 doi:10.1186/1742-4690-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hunninghake GW, Monick MM, Liu B, Stinski MF. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J. Virol. 63:3026–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reeves M, Sinclair J. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:297–313 [DOI] [PubMed] [Google Scholar]
- 47. Sambucetti LC, Cherrington JM, Wilkinson GW, Mocarski ES. 1989. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan YJ, Chiou CJ, Huang Q, Hayward GS. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lympho. J. Virol. 70:8590–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sinclair J, Sissons P. 1996. Latent and persistent infections of monocytes and macrophages. Intervirology 39:293–301 [DOI] [PubMed] [Google Scholar]
- 50. Sinzger C, Jahn G. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302–319 [DOI] [PubMed] [Google Scholar]
- 51. Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berthoux L, Sebastian S, Muesing MA, Luban J. 2007. The role of lysine 186 in HIV-1 integrase multimerization. Virology 364:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2:a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb. Perspect. Med. 1:a007096 doi:10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]