Abstract
Enviroxime is an antipicornavirus compound that targets host phosphatidylinositol 4-kinase III beta (PI4KB) activity for its antipicornavirus activity. To date, several antipoliovirus (PV) compounds similar to enviroxime that are associated with a common resistance mutation in viral protein 3A (a G5318A [3A-Ala70Thr] mutation in PV) have been identified. Most of these compounds have a direct inhibitory effect on PI4KB activity, as well as enviroxime (designated major enviroxime-like compounds). However, one of the compounds, AN-12-H5, showed no inhibitory effect on PI4KB and was considered to belong to another group of enviroxime-like compounds (designated minor enviroxime-like compounds). In the present study, we performed a small interfering RNA (siRNA) sensitization assay targeting PI4KB-related genes and identified oxysterol-binding protein (OSBP) as a target of minor enviroxime-like compounds. Knockdown of OSBP and OSBP2 increased the anti-PV activities of AN-12-H5 and a newly identified minor enviroxime-like compound, T-00127-HEV2, and also to T-00127-HEV1 to a minor extent, in the cells. A ligand of OSBP, 25-hydroxycholesterol (25-HC), acted as a minor enviroxime-like compound. Minor enviroxime-like compounds induced relocalization of OSBP to the Golgi apparatus in cells. Treatment of the cells with major or minor enviroxime-like compounds suppressed the expression of genes (HMGCS1 and SQLE) in the SREBP/SCAP regulatory pathway and diminished endogenous phosphatidylinositol 4-phosphate (PI4P) at the Golgi apparatus. Our results suggested that minor enviroxime-like compounds are phenotypically identical to 25-HC and that major and minor enviroxime-like compounds suppress the production and/or accumulation of PI4P in PV-infected cells by targeting PI4KB and OSBP family I activities, respectively.
INTRODUCTION
Poliovirus (PV) is a small nonenveloped virus with a single-stranded positive genomic RNA of about 7,500 nucleotides (nt) belonging to Human enterovirus species C in the genus Enterovirus, family Picornaviridae. PV is the causative agent of poliomyelitis, which is caused by the destruction of motor neurons by direct infection of cells by PV (1, 2). With established live attenuated oral PV vaccine (OPV) and inactivated PV vaccine (IPV) for PV (3, 4), the global eradication program for poliomyelitis has been continued by the Global Polio Eradication Initiative (GPEI) of the World Health Organization (WHO) since 1988. Currently, indigenous wild PVs are restricted to three countries where they are endemic, with drastic reduction of the number of cases due to wild PV (650 cases in 2011 and 193 cases as of November 2012). In the eradication program for poliomyelitis, antivirals for PV are anticipated to have roles in the posteradication era of PV in the control of a circulating vaccine-derived PV (cVDPV), along with IPV; for treatment of patients chronically infected with PV; and for persons exposed to PV (5, 6). However, there is currently no antiviral available for PV infection.
Compounds with anti-PV activity can be classified into capsid-binding inhibitors, replication inhibitors, and encapsidation inhibitors in terms of the target stages in PV infection. Capsid-binding inhibitors target hydrophobic pockets on the virion and inhibit the uncoating process by stabilizing the virion or the attachment process by inducing the conformational change of the virion (7, 8). Replication inhibitors could be classified into direct-acting antivirals and host-targeting antivirals. Viral proteins 2A, 2C, 3C, and 3D have been identified as the targets of direct-acting antivirals, including elastase inhibitors (9), guanidine hydrochloride (GuHCl) and related compounds (10–13), rupintrivir (AG7088) (14, 15), and gliotoxin (16), respectively. As host proteins, eIF4A, GBF1, and phosphatidylinositol 4-kinase III beta (PI4KB) have been identified as the targets of the host-targeting antivirals hippuristanol, brefeldin A, PIK93, and enviroxime (17–23). Hippuristanol, a natural product of the coral Isis hippuis, suppresses initiation of translation by inhibiting RNA binding of eIF4A and delayed the expression of viral proteins in PV replication for 2 h (17). Brefeldin A blocks membrane traffic between the cis- and trans-Golgi compartments by targeting a cellular guanine nucleotide exchange factor, GBF1, and inhibits PV replication, but not encephalomyocarditis virus (EMCV) replication (18, 19, 24, 25). PIK93 is considered to suppress the interaction of the phosphatidylinositol 4-phosphate (PI4P) produced with viral 3D polymerase on the reorganized membrane vesicle by inhibiting PI4KB (20). Enviroxime, which inhibits positive-strand RNA synthesis by preventing normal formation of the replication complex (21, 26), was recently identified as a nonspecific PI4KB inhibitor, as well as PIK93 (20, 22, 23). Direct interaction of enviroxime with viral protein 3A or 3AB was not detected (27); however, the inhibitory effect was antagonized by a resistant mutation in the 3A-encoding region (G5318A [3A-Ala70Thr] mutation) (26).
In a large-scale screening for antienterovirus compounds, most of the noncytotoxic compounds with potent antiviral activity are capsid-binding inhibitors or enviroxime-like compounds, which was defined as compounds that associate with a common resistance mutation in the 3A-encoding region (G5318A [3A-Ala70Thr] mutation) with little structural similarity to enviroxime (22, 26). As a candidate compound of capsid-binding inhibitors, the effectiveness of V-073 (previously designated SCH 48973) on in vitro PV infection and in a mouse infection model has been intensively analyzed in terms of the resistant mutation, pathogenicity of resistant mutants, and effects on immunization with IPV (28–32). To date, several enviroxime-like compounds have been identified, including TTP-8307 (33), some cellular protein kinase inhibitors (GW5074 and Flt3 inhibitor II) (34, 35), and a bifunctional antienterovirus compound, AN-12-H5, which targets the replication process of PV and enterovirus 71 (EV71) and also an early stage of EV71 infection (36).
Enviroxime-like compounds can be classified into at least 2 different groups; the majority of identified enviroxime-like compounds are PI4KB inhibitors (e.g., enviroxime, PIK93, GW5074, and T-00127-HEV1 {3-(3,4-dimethoxyphenyl)-2,5-dimethyl-N-[2-(4-morpholinyl)ethyl] pyrazolo [1,5-a]pyrimidin-7-amine}, designated major enviroxime-like compounds here), and the minority are non-PI4KB inhibitors (e.g., AN-12-H5, designated minor enviroxime-like compounds). Recently, itraconazole was identified as another minor enviroxime-like compound (37). The characteristic properties of minor enviroxime-like compounds are (i) weak resistance of a PV mutant with a G5318A mutation compared to that observed for the major group (around 5-fold increase of PV replication versus >50-fold increase of PV replication) and (ii) anti-hepatitis C virus (HCV) activity (22, 36). This suggested a common replication pathway inhibited by minor enviroxime-like compounds in enterovirus and HCV, possibly related to PI4P production, which depends on different PI4 kinases: PI4KB in enterovirus and PI4KA in HCV replication (20, 22, 38, 39).
In the present study, we searched for the target of minor enviroxime-like compounds by a small interfering RNA (siRNA) sensitization assay (22) targeting PI4KB-related genes with AN-12-H5, T-00127-HEV1 (a specific PI4KB inhibitor), and a newly identified minor enviroxime-like compound, T-00127-HEV2 {(3β,17β)-16,16-dimethyl-3-[(tetrahydro-2H-pyran-2-yl)oxy]-androst-5-en-17-ol}, as probe compounds. We identified members of oxysterol-binding protein (OSBP) family I as targets of AN-12-H5 and T-00127-HEV2. We found that a high-affinity ligand of OSBP, 25-hydroxycholesterol (25-HC), acted as a minor enviroxime-like compound. Our results suggested that major and minor enviroxime-like compounds suppress the production and/or accumulation of PI4P in PV-infected cells by targeting PI4KB and OSBP activities, respectively.
MATERIALS AND METHODS
Cells, viruses, and chemical library.
RD cells (a human rhabdomyosarcoma cell line) and HEK293 cells (human embryonic kidney cells) were cultured as monolayers in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). RD cells were used for titration of viruses and pseudoviruses and for screening of anti-PV compounds. HEK293 cells were used for siRNA screening to identify cellular targets of antienterovirus compounds. The Huh7.5.1 cell line was a kind gift from Frank Chisari (Scripps Research Institute). Huh7.5.1 cells were used for analysis of the inhibitory effects of anti-PV compounds on HCV replication. PV pseudoviruses (TE-PV-Fluc mc) (40), which encapsidated luciferase-encoding PV replicons with capsid proteins derived from PV1(Mahoney), were used for screening of anti-PV compounds. PV1(Mahoney) was used to analyze the inhibitory effects of the identified compounds on PV infection. PV pseudovirus mutants that have known drug resistance mutations, including G5318A (enviroxime and GW5074 resistance; 3A-Ala70Thr) (26, 35), U4614A (GuHCl resistance; 2C-Phe164Tyr) (41), and G4361A and C5190U (brefeldin A resistance; 2C-Val80Ile and 3A-Ala27Val) (42), were used for characterization of identified antienterovirus compounds. The expression vector for a fusion protein, CERT-enhanced green fluorescent protein (EGFP), was a kind gift from Kentaro Hanada (Department of Biochemistry and Cell Biology, National Institute of Infectious Diseases, Japan) (43).
A diverse subset of 59,200 compounds from a chemical library of the University of Tokyo was used for screening. The purity of compounds was determined by liquid chromatography-mass spectrometry (LC-MS) based on the signal of evaporative light-scattering detection (ELSD). The purity of T-00127-HEV2 was >99%. T-00127-HEV1 was supplied by Pharmeks Ltd. (Moscow, Russia) (purity, >99%). 25-HC was purchased from Sigma-Aldrich Co. LLC (purity, ≥98%). Enviroxime (purity, >99%) was a kind gift from Masanobu Agoh (Nagasaki Prefectural Institute for Environmental Research and Public Health, Japan).
siRNA libraries targeting human genes related to PI4KB, OSBP, and PI4P binding proteins were purchased from Thermo Fisher Scientific, Inc., as a form of siGenome Smart pools, which contain 4 sets of different siRNAs for each mRNA and silence target mRNA expression by at least 75%. As control siRNAs, siGenome nontargeting siRNAs numbers 1 and 2 were used in each experiment.
Screening of anti-PV compounds.
Screening of anti-PV compounds was performed as previously described (22). Briefly, 5 μl of compound solution (60 μM; final concentration, 10 μΜ) and 5 μl of PV pseudovirus solution (800 infectious units [IU]) were added to RD cells (5.0 × 103 cells per well in 20 μl medium) in 384-well plates (catalog no. 781080; Greiner Bio-One), and the luciferase activity of the infected cells was measured at 7 h postinfection (p.i.) with a Steady-Glo Luciferase Assay System (Promega). PV pseudovirus infection was calculated as a percentage of the luciferase activity of the infected cells, where the luciferase activity in the infected cells in the absence of compounds was taken as 100%. The cutoff value of the inhibitory effects of candidate compounds was set to <10% of PV pseudovirus infection in the treated cells. The cytotoxicity of compounds was evaluated from the viability of compound-treated cells by using a Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega). The cutoff value of screening for candidate compounds was set to >90% of the viability of treated cells compared to that of mock-treated cells. For candidate compounds, the anti-PV activity and cytotoxicity were further evaluated by determination of the 50% effective concentration (EC50) based on PV pseudovirus infection and the 50% cytotoxic concentration (CC50), respectively.
The inhibitory effects of enviroxime-like compounds on HCV replication were analyzed by using an HCV replicon (replicon clone pSGR-JFH-LucNeo-4) in Huh7.5.1 cells as previously described (22, 44).
siRNA transfection.
An RNA duplex of each siRNA (final concentration, 20 nM) was transfected into HEK293 cells (5.0 × 103 cells in 100 μl medium per well) in 96-well plates by using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. The transfection efficiency of siRNA in the cells was evaluated by the efficiency of incorporation of fluorescence-labeled siRNA (siGlo control siRNAs) in the transfected cells at 24 h posttransfection (p.t.) and by the efficiency of cell death in the cells transfected with siGenome Tox Transfection Control at 72 h p.t. (cells transfected with this control reagent die by apoptosis). siRNA-transfected cells were used for experiments at 72 h p.t.
TISS assay.
A target identification by siRNA sensitization (TISS) assay was performed as previously described (22). Briefly, siRNA-transfected cells were inoculated with 800 IU PV pseudovirus at 72 h p.t. in the presence of suboptimal concentrations of anti-PV compounds selected around their EC50s, which resulted in 39 to 84% PV pseudovirus infection. The cells were incubated at 37°C for 7 h, and then the luciferase activity in the cells was measured with a Steady-Glo Luciferase Assay System (Promega). To evaluate the specific inhibitory effect of siRNA treatment on PV infection, the net PV pseudovirus infection, which is a ratio of the PV pseudovirus infection in siRNA-transfected cells (%) to cell viability (%), was determined for each siRNA treatment. The net PV pseudovirus infection in mock-transfected cells was 1. To evaluate the effect of siRNA treatment on the sensitivity to each compound, normalized PV pseudovirus infection, which is a ratio of PV pseudovirus infection in siRNA-transfected cells in the presence of compounds (%) to PV pseudovirus infection in siRNA-transfected cells in the absence of compounds (%), was determined. The normalized PV pseudovirus infection in mock-transfected cells was 1. The sensitization effect of each siRNA was analyzed by a paired t test with normalized PV pseudovirus infection in mock-transfected cells and siRNA-transfected cells.
OSBP relocalization assay.
HEK293 cells stably expressing an OSBP-EGFP fusion protein were prepared as follows. Expression vectors for a human OSBP-EGFP fusion were constructed with pLEGFP-N1 (BD Biosciences Clontech). GP2-293 cells were cotransfected with OSBP-EGFP expression vector and pVSV-G (Clontech), and the cell culture supernatant of the transfected cells was collected at 72 h p.t. HEK293 cells were inoculated with the collected supernatant. HEK293 cells stably expressing OSBP-EGFP fusion protein were colony purified and used for the assay. The cells were incubated in the absence or presence of the compounds at 37°C for 1 h. Relocalization of OSBP-EGFP fusion protein from the cytoplasm to the Golgi apparatus was observed with a fluorescence microscope (BZ-8000; Keyence).
Quantitative real-time reverse transcription (RT)-PCR.
RD cells in 24-well plates were treated with compounds at 37°C for 6 h, and then the total RNA was extracted from the treated cells by using a High Pure RNA Isolation Kit (Roche). The isolated total RNAs were reverse transcribed using a Reverse Transcription System (Promega) with random hexamers. The relative expression levels of ACTB, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HMG-coenzyme A (CoA) synthase (HMGCS1), and squalene epoxidase (SQLE) mRNAs were determined by real-time PCR with primers and probes of the Solaris Human qPCR Gene Expression Assay (Thermo Fisher Scientific Inc.) and a Solaris qPCR Gene Expression Low ROX Master Mix kit (Thermo Fisher Scientific Inc.) using an Applied Biosystems 7500 Fast Real-Time PCR System. GAPDH mRNA was used as the endogenous control, and the expression levels of ACTB, HMGCS1, and SQLE mRNAs were normalized by the expression levels in the mock-treated cells.
Immunofluorescence microscopy.
Cells were fixed with 3% paraformaldehyde for 10 min at room temperature and then permeabilized with 20 μM digitonin in HBS (21 mM HEPES buffer [pH 7.4], 1.8 mM disodium hydrogen phosphate, 137 mM NaCl, 4.8 mM KCl) for 5 min as previously described (45). The cells were stained by indirect immunofluorescence with primary antibodies against PI4KB (rabbit antibody; Millipore) and PI4P (mouse IgM antibody; Echelon Biosciences), secondary antibodies (anti-rabbit IgG and anti-mouse IgM goat antibodies conjugated with Alexa Fluor 488 and 594 dyes, respectively; Molecular Probes), and Hoechst 33342 (Molecular Probes) for counterstaining of nuclei. Samples were observed with a confocal scanning laser microscope (FV1000; Olympus).
Statistical analysis.
The results of experiments are shown as averages with standard deviations. A one-tailed t test was performed with data obtained from 3 or 4 independent experiments, as indicated. P values of less than 0.05 were considered significant differences and are indicated by asterisks.
RESULTS
Identification of a novel minor enviroxime-like compound, T-00127-HEV2.
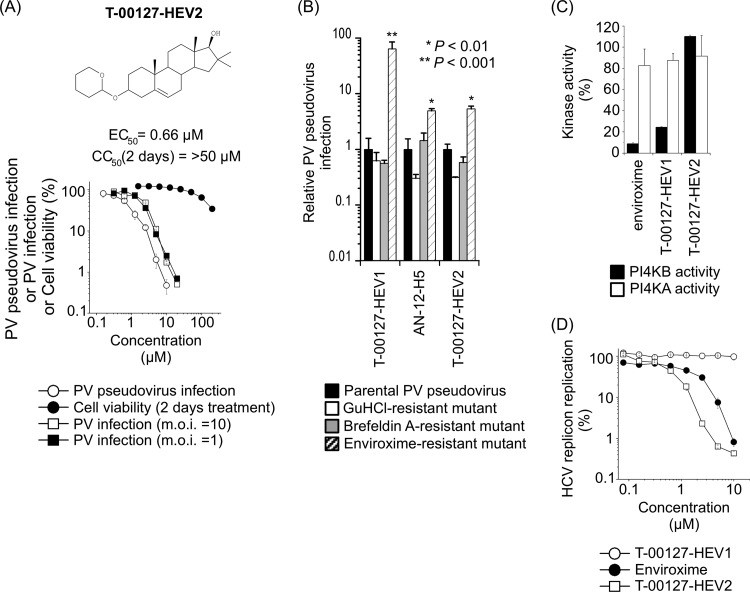
Previously, to identify potent anti-PV compounds that target conserved factors required for enterovirus replication, we performed a screening of 72,000 compounds and identified T-00127-HEV1, which is a major enviroxime-like compound and a specific PI4KB inhibitor (22). In the present study, we performed additional screening with 59,200 compounds and identified 3 compounds that meet the following criteria: (i) no apparent cytotoxicity in cells after 2 days of treatment with 10 μM compound (>90% viability with no morphological changes in the cells), (ii) targeting of the replication step (>90% inhibitory effect after the uncoating step), and (iii) inhibition of PV infection (inhibition of cytopathic effect [CPE]). Consistent with our previous results, all three identified compounds are enviroxime-like compounds, and 2 of them are PI4KB inhibitors (data not shown). One of the compounds, which was designated T-00127-HEV2, showed a potent inhibitory effect on PV pseudovirus infection with low cytotoxicity (EC50, 0.66 μM; CC50, >50 μM) and also on PV1(Mahoney) infection (Fig. 1). Partial resistance of a PV mutant with enviroxime-resistant mutation, no inhibitory effect on PI4KB activity, and anti-HCV activity were also observed for T-00127-HEV2. In contrast, T-00127-HEV1 did not show any anti-HCV activity, as observed in our previous report (22). Moderate anti-HCV activity was observed for enviroxime at high concentrations, consistent with a recent report (23), possibly due to its nonspecific anti-PI4KA activity. These results suggested that T-00127-HEV2 is a novel minor enviroxime-like compound, along with AN-12-H5 (22, 36).
Fig 1.
(A) Characterization of T-00127-HEV2. (Top) Structure of T-00127-HEV2. (Bottom) Inhibitory effect of T-00127-HEV2 on PV pseudovirus and viability of RD cells. PV pseudovirus infection (luciferase assay), viability of cells, or PV1(Mahoney) infection (number of copies of the viral genome at 7 h p.i. in RD cells) in the absence of compounds was taken as 100%. T-00127-HEV2 showed precipitation above 50 μM. (B) Specificity of resistance mutations to T-00127-HEV2. RD cells were infected with PV pseudovirus mutants that have resistance mutations to GuHCl (U4614A), brefeldin A (G4361A plus C5190U), and enviroxime (G5318A) in the presence of antienterovirus compounds: T-00127-HEV1 (3.1 μM), AN-12-H5 (25 μM), and T-00127-HEV2 (6.3 μM). Relative PV pseudovirus infection is shown, where parental PV pseudovirus infection in the presence of each compound was taken as 1. n = 3. (C) Inhibitory effects of enviroxime-like compounds on in vitro activity of PI4KB and PI4KA. In vitro kinase activities were analyzed in the presence of each compound (1 μM) and ATP (10 μM). (D) HCV replicon replication in the presence of enviroxime-like compounds. HCV replicon replication in the absence of compounds was taken as 100%. The error bars indicate standard deviations.
OSBP is a target of minor enviroxime-like compounds for its anti-PV activity.
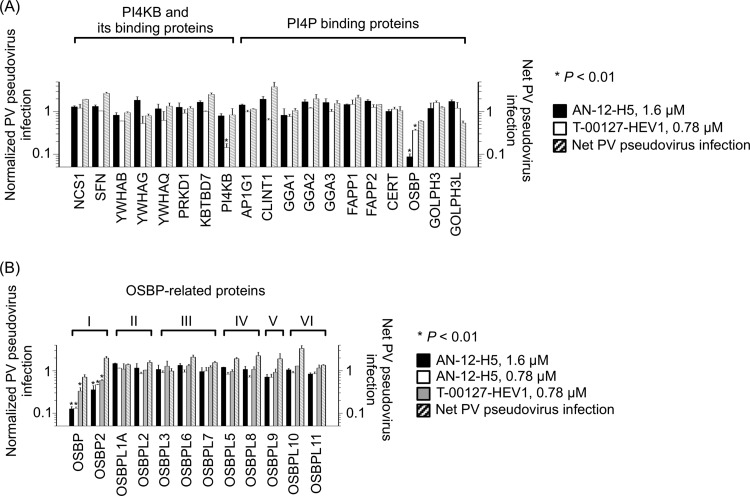
To identify the target of minor enviroxime-like compounds, we performed an siRNA sensitization (TISS) assay with a minor enviroxime-like compound, AN-12-H5, and also with a major enviroxime-like compound, T-00127-HEV1, as a control. First, we performed a TISS assay with an siRNA library that targets membrane-trafficking genes (140 genes) but could not identify genes that showed enhanced sensitivity to AN-12-H5 (see Table 1 in the supplemental material). Next, we analyzed PI4KB-related genes (genes for PI4KB binding proteins and PI4P binding proteins) (Fig. 2A). Knockdown of PI4KB caused enhanced sensitivity to T-00127-HEV1, but not to AN-12-H5, as previously observed (22). In this subset of genes, we finally found that knockdown of OSBP drastically enhanced the sensitivity of the cells to AN-12-H5, and also to T-00127-HEV1, although to a minor extent. Next, we analyzed the effect of knockdown of all the members of the human OSBP family on sensitivity to AN-12-H5 (Fig. 2B). Interestingly, among the 6 families of OSBP-related genes (46), only the knockdown of members of OSBP family I (OSBP and OSBP2) enhanced sensitivity to AN-12-H5, and also to T-00127-HEV1 to a minor extent. This suggested that OSBP family I is a target of enviroxime-like compounds in a PI4KB-related pathway in PV replication.
Fig 2.
siRNA sensitization assay for enviroxime-like compounds targeting PI4KB binding proteins and PI4P binding proteins. (A and B) Normalized PV pseudovirus infection in the presence of AN-12-H5 (1.6 and 0.78 μM) or T-00127-HEV1 (0.78 μM) and net PV pseudovirus infection in siRNA-transfected HEK293 cells targeting PI4KB binding proteins and PI4P binding proteins (A) or OSBP-related proteins (B). Normalized PV pseudovirus infection is the ratio of PV pseudovirus infection in compound-treated and siRNA-transfected cells (percent) to PV pseudovirus infection in siRNA-transfected cells in the absence of compounds (percent). Normalized PV pseudovirus infection in mock-transfected cells was taken as 1. n = 3. The error bars indicate standard deviations.
An OSBP ligand, 25-HC, acts as a minor enviroxime-like compound.
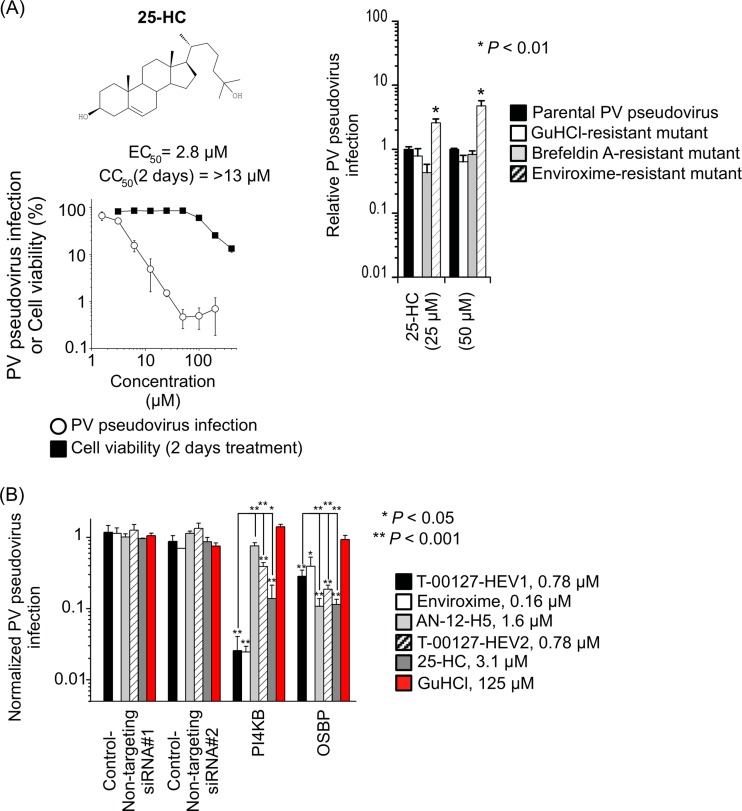
We examined the effect of a high-affinity ligand of OSBP, 25-HC (Kd [dissociation constant] = 8 nM) (47), on PV pseudovirus infection. We found that 25-HC suppressed PV pseudovirus infection (EC50, 2.8 μM; CC50, >13 μM), and an enviroxime-resistant mutant showed a weak resistant phenotype typical of minor enviroxime-like compounds, as observed for T-00127-HEV2 (a 2.6- to 4.8-fold increase of PV replication with the enviroxime-resistant mutation) (Fig. 3A). Next, we examined the effect of knockdown of PI4KB and OSBP on sensitivity to T-00127-HEV2 and 25-HC (Fig. 3B). The sensitivity of the cells transfected with siRNA targeting PI4KB to T-00127-HEV2 and 25-HC was slightly enhanced, but the effects were weaker than that to T-00127-HEV1. In contrast, enhanced sensitivities of the cells transfected with siRNA targeting OSBP to T-00127-HEV2 and 25-HC were significantly stronger than that to T-00127-HEV1 and similar to that to AN-12-H5. These results suggested that an OSBP ligand, 25-HC, also acts as a minor enviroxime-like compound.
Fig 3.
(A) Characterization of 25-HC. (Top) Structure of 25-HC. (Bottom) Inhibitory effect of 25-HC on PV pseudovirus and viability of RD cells. PV pseudovirus infection or viability of the cells in the absence of compounds was taken as 100%. 25-HC showed precipitation above 13 μM. (Right) Specificity of resistance mutations to 25-HC. RD cells were infected with PV pseudovirus mutants that have resistance mutations to GuHCl (U4614A), brefeldin A (G4361A plus C5190U), and enviroxime (G5318A) in the presence of 25-HC (25 and 50 μM). Relative PV pseudovirus infection is shown, where parental PV pseudovirus infection in the presence of 25-HC was taken as 1. n = 3. (B) siRNA sensitization assay for enviroxime-like compounds. HEK293 cells were transfected with siRNA targeting PI4KB or OSBP or with nontargeting siRNAs (nontargeting siRNAs 1 and 2), and the sensitization effect was analyzed in the presence of enviroxime-like compounds (T-00127-HEV1 [0.78 μM], enviroxime [0.16 μM], AN-12-H5 [1.6 μM], T-00127-HEV2 [0.78 μM], and 25-HC [3.1 μM]) and with GuHCl (125 μM) as a control compound. Normalized PV pseudovirus infection is shown, with P values. n = 4. The error bars indicate standard deviations.
Minor enviroxime-like compounds are phenotypically identical to 25-HC.
We compared the properties of AN-12-H5 and T-00127-HEV2 with those of 25-HC. Treatment with 25-HC causes relocalization of OSBP from the cytoplasm to the Golgi apparatus in cells (48). Interestingly, AN-12-H5 and T-00127-HEV2, but not T-00127-HEV1 and GuHCl, caused profound relocalization of OSBP much more clearly even than 25-HC (Fig. 4A). Relocalization of OSBP could be observed as early as 15 min after addition of the compounds. To evaluate the specificity of the effects of minor enviroxime-like compounds on OSBP, we also tested the effects of compounds on the relocalization of another lipid transfer protein, CERT, which has functional protein domain topology similar to that of OSBP (i.e., a PI4P binding pleckstrin homology [PH] domain, a FFAT motif, and a lipid binding domain) (49). In contrast to OSBP, we could not observe apparent relocalization of CERT to the Golgi apparatus in the compound-treated cells after 1 h of treatment. Next, we analyzed an effect of enviroxime-like compounds on the expression of genes in the SREBP/SCAP regulatory pathway, which modulates expression of genes related to cholesterol synthesis and uptake in response to cellular cholesterol levels and certain sterols (50). Expression of the genes in the SREBP/SCAP regulatory pathway, such as the HMGCS1 and SQLE genes, was immediately suppressed by 25-HC treatment (51). We found that treatment with T-00127-HEV1, AN-12-H5, and T-00127-HEV2, but not GuHCl, suppressed the expression levels of HMGCS1 and SQLE mRNAs, as well as 25-HC (Fig. 4B). Next, we analyzed the effective periods of the compounds for their anti-PV activity by a time-of-addition experiment in PV pseudovirus infection (Fig. 4C). Significant inhibitory effects of minor enviroxime-like compounds could be observed when added −3 to 5 h p.i. (for 25-HC) or −3 to 6 h p.i.(for T-00127-HEV2 and AN-12-H5). Addition of minor enviroxime-like compounds before PV infection (3 to 1 h preinfection) did not enhance its inhibitory effects compared to addition at 0 h p.i. This suggested that minor enviroxime-like compounds were indistinguishable from 25-HC in terms of their in vivo phenotypes and also that PI4KB is involved in the expression of genes in the SREBP/SCAP regulatory pathway.
Fig 4.
Effects of minor enviroxime-like compounds on OSBP relocalization and gene expression in the SREBP/SCAP regulatory pathway. (A) Effects of enviroxime-like compounds on relocalization of OSBP. HEK293 cells expressing OSBP-EGFP or CERT-EGFP were treated with 10 μM T-00127-HEV1, AN-12-H5, T-00127-HEV2, or 25-HC or with 2 mM GuHCl for 1 h. Scale bars, 20 μm. (B) Effects of enviroxime-like compounds on expression of HMGCS1 and SQLE mRNAs. RD cells were treated with 10 μM T-00127-HEV1, AN-12-H5, T-00127-HEV2, or 25-HC or with 2 mM GuHCl for 6 h. Total RNA was collected from the treated cells and subjected to quantitative real-time RT-PCR. Relative expression levels of ACTB, HMGCS1, and SQLE mRNAs normalized by GAPDH mRNA are shown. n = 3. (C) Evaluation of the effective period for addition of enviroxime-like compounds in PV infection. Enviroxime-like compounds (20 μM) and GuHCl (2 mM) were added to RD cells at the indicated times (−3 h to 6 h p.i.). PV pseudovirus was added at 0 h p.i., and the luciferase activity was measured at 7 h p.i. PV pseudovirus infection in the absence of compounds was taken as 100%. The times of drug addition, where significant suppression of PV pseudovirus infection was observed (P < 0.01), are indicated by lines. n = 3. The error bars indicate standard deviations.
Major and minor enviroxime-like compounds reduce accumulation of PI4P at the Golgi apparatus.
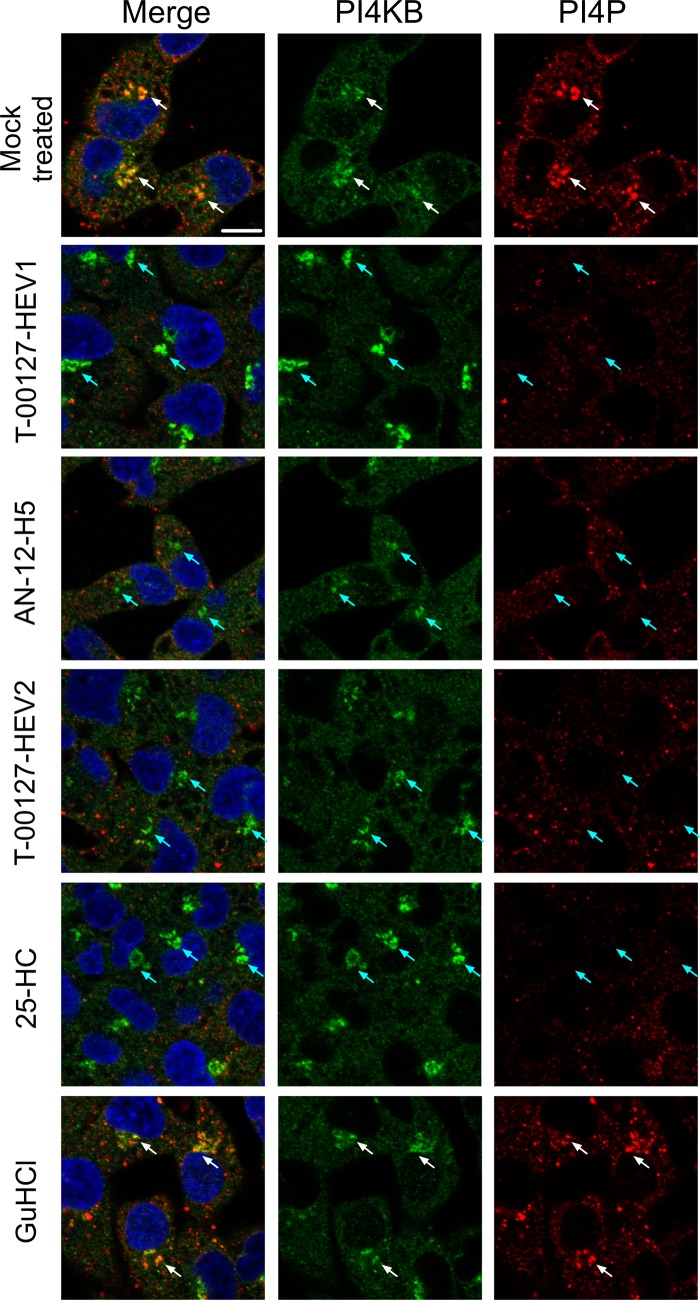
We analyzed the effects of enviroxime-like compounds on PI4P production at the Golgi apparatus (Fig. 5). Treatment with T-00127-HEV1 enhanced PI4KB localization at the Golgi apparatus and diminished PI4P, consistent with a recent report on major enviroxime-like compounds (52). Interestingly, enhancement of PI4KB localization at the Golgi apparatus was not observed in cells treated with minor enviroxime-like compounds, but significant reduction of PI4P at the Golgi apparatus was observed. This suggested that major and minor enviroxime-like compounds inhibit PI4P production and/or accumulation at the Golgi apparatus by targeting PI4KB and OSBP, respectively.
Fig 5.
Effects of enviroxime-like compounds on PI4P accumulation at the Golgi apparatus. Indirect immunofluorescence of PI4KB and PI4P in RD cells treated with 10 μM T-00127-HEV1, AN-12-H5, T-00127-HEV2, 25-HC, or 2 mM GuHCl overnight is shown. Blue, nuclear (staining with Hoechst 33342); green, PI4KB; red, PI4P. Scale bars, 10 μm. The white arrows indicate some of the colocalized sites, and the cyan arrows indicate some of the noncolocalized sites.
DISCUSSION
To identify further antiviral candidate compounds for PV, we performed a high-throughput screening with a large-scale chemical library (59,200 compounds) and identified a novel minor enviroxime-like compound, T-00127-HEV2 (Fig. 1). Consistent with our previous observations (22), all 3 candidate compounds identified, which showed potent anti-PV activity with low cytotoxicity, were enviroxime-like compounds, 2 major and 1 minor. T-00127-HEV2 was the 3rd minor enviroxime-like compound, following AN-12-H5 and itraconazole (36, 37). Conserved properties of minor enviroxime-like compounds (i.e., weak resistance to a PV mutant with a G5318A mutation and anti-HCV activity) suggested a common specific pathway or targets in their antiviral activities, possibly related to PI4KB activity or PI4P produced.
By using minor and major enviroxime-like compounds, we performed an siRNA sensitization assay targeting membrane-trafficking genes and PI4KB-related genes to identify the targets of minor enviroxime-like compounds (Fig. 2). Knockdown of members of the OSBP family did not significantly suppress PV replication in HEK293 cells (net PV pseudovirus infection of 0.7 to 3.3); however, knockdown of OSBP and OSBP2, family I of OSBP-related genes (46), increased sensitivity to minor enviroxime-like compounds. OSBP was originally identified as a high-affinity receptor for oxysterol, including 25-HC (53), and members of the OSBP family are involved in sterol signaling and/or sterol transport between cellular organelles (reviewed in reference 54). Consistent with the anti-HCV activity of 25-HC, inhibitory effects of OSBP knockdown on HCV replication and virus particle release from the infected cells have been reported (55, 56). It is interesting that only members of OSBP family I, among the 6 families of OSBP-related genes (46), act as targets of minor enviroxime-like compounds, despite potential functional redundancy of the members of this gene family (57).
Next, we analyzed the anti-PV activity of a high-affinity ligand of OSBP, 25-HC. 25-HC is known as an inhibitor of HCV replication, and its inhibitory effect on the transcription of genes in the SREBP/SCAP pathway has been suggested as the mechanism of its inhibitory effect on HCV replication (58, 59). The mechanism of the inhibitory effect of 25-HC is different from that of a cholesterol-depleting agent, methyl-β-cyclodextrin (MBCD), which directly disrupted the membranous web of the HCV replication complex (60). We found that 25-HC suppressed PV pseudovirus replication, and in fact, 25-HC acts as a minor enviroxime-like compound (Fig. 3). This suggested that minor enviroxime-like compounds might be defined as 25-HC-like compounds.
Relocalization of OSBP from the cytoplasm to the Golgi apparatus and transcriptional inhibition of the SREBP/SCAP pathway have been known as characteristic properties of 25-HC. We found that treatment with minor enviroxime-like compounds causes relocalization of OSBP, but not of another lipid transfer protein, CERT (49), in the treated cells, as well as 25-HC (Fig. 4A), suggesting specificity of the effects of minor enviroxime-like compounds on OSBP. Minor enviroxime-like compounds also suppressed transcription of the HMGCS1 and SQLE genes, which are genes in the SREBP/SCAP regulatory pathway (Fig. 4B). We also found that treatment with a major enviroxime-like compound, a PI4KB-specific inhibitor, T-00127-HEV1 (22), also suppressed transcription of HMGCS1 and SQLE genes, suggesting a functional link between PI4KB activity and the SREBP/SCAP transcriptional-regulatory pathway. An inhibitory effect of a minor enviroxime-like compound could be observed when added even 5 h p.i., and pretreatment of the cells before PV infection did not enhance its inhibitory effect (Fig. 4C). PV1(Mahoney) infection was not severely impaired by actinomycin D treatment, consistent with a previous report (61), and at most 50% reduction of the infection could be observed in the presence of 20 to 0.15 μM actinomycin D (M. Arita, unpublished data). This suggested that the inhibitory effect of minor enviroxime-like compounds on PV replication does not depend on transcriptional activation or suppression during infection, but rather, occurs in a direct manner.
Interestingly, knockdown of OSBP also increased sensitivity to a major enviroxime-like compound, although to a minor extent compared with that observed for minor enviroxime-like compounds (Fig. 3B). Some of the minor enviroxime-like compounds (T-00127-HEV2 and 25-HC, but not AN-12-H5) also showed increased inhibitory effects on PV infection in PI4KB knockdown cells. This suggested that there is a functional link between PI4KB and members of OSBP family I, consistent with the resistance phenotype of PV with a G5318A mutation. The drastic resistant phenotype conferred by a G5318A mutation on PI4KB inhibitors might suggest direct involvement of viral proteins or a protein complex with host proteins in the production of PI4P via PI4KB. In HCV infection, NS5A has been identified as a PI4KA activator (62, 63). In Aichi virus infection, a host protein, ACBD3, might play the role of a PI4KB activator, along with viral proteins (45). In enterovirus infection, corresponding viral proteins, possibly other than 3A protein (52), remain to be identified. We found that treatment of both major and minor enviroxime-like compounds reduced accumulation of PI4P at the Golgi apparatus (Fig. 5). PI4KB is known as the main PI4P producer at the Golgi apparatus (52, 64). To our knowledge, direct interaction of OSBP with PI4KB has not been reported. Actually, the relocalizations of PI4KB and OSBP observed in compound-treated cells are different; T-00127-HEV1 caused accumulation of PI4KB, but not of OSBP, at the Golgi apparatus (Fig. 4 and 5). Activation of PI4K2A by OSBP has been observed (65), and an altered sterol environment produced by OSBP has been proposed as a mechanism for this activation (54). A yeast homologue of OSBP, Osh4p, directly activates PI4P phosphatase Sac1p (66). Recently, direct binding of PI4P, which competes with cholesterol binding to Osh4p and OSBP, has been shown (67, 68), and Osh4p was even able to exchange sterol and PI4P between membranes by itself (67). Considering that OSBP is a host factor for HCV replication and HCV replication utilizes PI4KA instead of PI4KB (38, 55), these observations suggest OSBP family I activates some PI4 kinases with broad specificity and/or helps in the accumulation of PI4P at the sites of viral RNA replication. The exact mechanism of OSBP activity on PI4P accumulation at the Golgi apparatus has yet to be determined; our results suggested that major and minor enviroxime-like compounds cooperatively inhibit PI4P production and/or accumulation at the Golgi apparatus by targeting PI4KB and members of OSBP family I, respectively.
In summary, we identified the members of OSBP family I as targets of minor enviroxime-like compounds and found that minor enviroxime-like compounds are phenotypically indistinguishable from 25-HC. Our results suggested that major and minor enviroxime-like compounds inhibit PI4P production and/or accumulation at the Golgi apparatus in PV infection.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Junko Wada for her excellent technical assistance. We are grateful to Masanobu Agoh for kindly providing enviroxime and to Kentaro Hanada and Masayoshi Fukasawa for kindly providing an expression vector for CERT-EGFP and also helpful discussions on lipid transfer proteins.
This study was supported in part by Grants-in-Aid for the Promotion of Polio Eradication and Research on Emerging and Reemerging Infectious Diseases from the Ministry of Health, Labor and Welfare, Japan, and by a grant from the World Health Organization for a collaborative research project of the Global Polio Eradication Initiative.
Footnotes
Published ahead of print 30 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03546-12.
REFERENCES
- 1. Bodian D. 1949. Histopathologic basis of clinical findings in poliomyelitis. Am. J. Med. 6:563–578 [DOI] [PubMed] [Google Scholar]
- 2. Couderc T, Christodoulou C, Kopecka H, Marsden S, Taffs LF, Crainic R, Horaud F. 1989. Molecular pathogenesis of neural lesions induced by poliovirus type 1. J. Gen. Virol. 70:2907–2918 [DOI] [PubMed] [Google Scholar]
- 3. Salk JE, Bazeley PL, Bennett BL, Krech U, Lewis LJ, Ward EN, Youngner JS. 1954. Studies in human subjects on active immunization against poliomyelitis. II. A practical means for inducing and maintaining antibody formation. Am. J. Public Health Nations Health 44:994–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabin AB. 1965. Oral poliovirus vaccine. History of its development and prospects for eradication of poliomyelitis. JAMA 194:872–876 [DOI] [PubMed] [Google Scholar]
- 5. Collett MS, Neyts J, Modlin JF. 2008. A case for developing antiviral drugs against polio. Antiviral Res. 79:179–187 [DOI] [PubMed] [Google Scholar]
- 6. Committee on Development of a Polio Antiviral and Its Potential Role in Global Poliomyelitis Eradication NRC 2006. Exploring the role of antiviral drugs in the eradication of polio: workshop report. National Academies Press, Washington, DC [Google Scholar]
- 7. Fox MP, Otto MJ, McKinlay MA. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pevear DC, Fancher MJ, Felock PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, Dutko FJ. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Molla A, Hellen CU, Wimmer E. 1993. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase-specific inhibitors. J. Virol. 67:4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caliguiri LA, Tamm I. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408–417 [DOI] [PubMed] [Google Scholar]
- 11. Eggers HJ, Tamm I. 1961. Spectrum and characteristics of the virus inhibitory action of 2-(alpha-hydroxybenzyl)-benzimidazole. J. Exp. Med. 113:657–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimizu H, Agoh M, Agoh Y, Yoshida H, Yoshii K, Yoneyama T, Hagiwara A, Miyamura T. 2000. Mutations in the 2C region of poliovirus responsible for altered sensitivity to benzimidazole derivatives. J. Virol. 74:4146–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Palma AM, Heggermont W, Lanke K, Coutard B, Bergmann M, Monforte AM, Canard B, De Clercq E, Chimirri A, Purstinger G, Rohayem J, van Kuppeveld F, Neyts J. 2008. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 82:4720–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, Fuhrman SA, Meador JW, Zalman LS, Matthews DA, Worland ST. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Palma AM, Purstinger G, Wimmer E, Patick AK, Andries K, Rombaut B, De Clercq E, Neyts J. 2008. Potential use of antiviral agents in polio eradication. Emerg. Infect. Dis. 14:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez PL, Carrasco L. 1992. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J. Virol. 66:1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 [DOI] [PubMed] [Google Scholar]
- 18. Irurzun A, Perez L, Carrasco L. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166–175 [DOI] [PubMed] [Google Scholar]
- 19. Maynell LA, Kirkegaard K, Klymkowsky MW. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wikel JH, Paget CJ, DeLong DC, Nelson JD, Wu CY, Paschal JW, Dinner A, Templeton RJ, Chaney MO, Jones ND, Chamberlin JW. 1980. Synthesis of syn and anti isomers of 6-[[(hydroxyimino)phenyl]methyl]-1-[(1-methylethyl)sulfonyl]-1H-benzimidaz ol-2-amine. Inhibitors of rhinovirus multiplication. J. Med. Chem. 23:368–372 [DOI] [PubMed] [Google Scholar]
- 22. Arita M, Kojima H, Nagano T, Okabe T, Wakita T, Shimizu H. 2011. Phosphatidylinositol 4-kinase III beta is a target of enviroxime-like compounds for antipoliovirus activity. J. Virol. 85:2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delang L, Paeshuyse J, Neyts J. 2012. The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem. Pharmacol. 84:1400–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216 doi:10.1371/journal.ppat.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cuconati A, Molla A, Wimmer E. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinz BA, Vance LM. 1995. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J. Virol. 69:4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown-Augsburger P, Vance LM, Malcolm SK, Hsiung H, Smith DP, Heinz BA. 1999. Evidence that enviroxime targets multiple components of the rhinovirus 14 replication complex. Arch. Virol. 144:1569–1585 [DOI] [PubMed] [Google Scholar]
- 28. Buontempo PJ, Cox S, Wright-Minogue J, DeMartino JL, Skelton AM, Ferrari E, Albin R, Rozhon EJ, Girijavallabhan V, Modlin JF, O'Connell JF. 1997. SCH 48973: a potent, broad-spectrum, antienterovirus compound. Antimicrob. Agents Chemother. 41:1220–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oberste MS, Moore D, Anderson B, Pallansch MA, Pevear DC, Collett MS. 2009. In vitro antiviral activity of V-073 against polioviruses. Antimicrob. Agents Chemother. 53:4501–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kouiavskaia DV, Dragunsky EM, Liu HM, Oberste MS, Collett MS, Chumakov KM. 2011. Immunological and pathogenic properties of poliovirus variants selected for resistance to antiviral drug V-073. Antivir. Ther. 16:999–1004 [DOI] [PubMed] [Google Scholar]
- 31. Kouiavskaia D, Collett MS, Dragunsky EM, Sarafanov A, Chumakov KM. 2011. Immunogenicity of inactivated polio vaccine with concurrent antiviral V-073 administration in mice. Clin. Vaccine Immunol. 18:1387–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu HM, Roberts JA, Moore D, Anderson B, Pallansch MA, Pevear DC, Collett MS, Oberste MS. 2012. Characterization of poliovirus variants selected for resistance to the antiviral compound v-073. Antimicrob. Agents Chemother. 56:5568–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Palma AM, Thibaut HJ, van der Linden L, Lanke K, Heggermont W, Ireland S, Andrews R, Arimilli M, Altel T, De Clercq E, van Kuppeveld F, Neyts J. 2009. Mutations in the non-structural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307. Antimicrob. Agents Chemother. 53:1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arita M, Wakita T, Shimizu H. 2008. Characterization of pharmacologically active compounds that inhibit poliovirus and enterovirus 71 infectivity. J. Gen. Virol. 89:2518–2530 [DOI] [PubMed] [Google Scholar]
- 35. Arita M, Wakita T, Shimizu H. 2009. Cellular kinase inhibitors that suppress enterovirus replication have a conserved target in viral protein 3A similar to that of enviroxime. J. Gen. Virol. 90:1869–1879 [DOI] [PubMed] [Google Scholar]
- 36. Arita M, Takebe Y, Wakita T, Shimizu H. 2010. A bifunctional anti-enterovirus compound that inhibits replication and early stage of enterovirus 71 infection. J. Gen. Virol. 91:2734–2744 [DOI] [PubMed] [Google Scholar]
- 37. Ulferts R, van der Linden L, Lanke KH, Thibaut HJ, Neyts J, van Kuppeveld FJ. 2012. New lead drug candidates for the treatment of enterovirus infections. abstr P-110. EUROPIC 2012, XVII Meeting of the European Study Group on the Molecular Biology of Picornaviruses, Saint Raphael, France, 3 to 7 June 2012 [Google Scholar]
- 38. Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vaillancourt FH, Brault M, Pilote L, Uyttersprot N, Gaillard ET, Stoltz JH, Knight BL, Pantages L, McFarland M, Breitfelder S, Chiu TT, Mahrouche L, Faucher AM, Cartier M, Cordingley MG, Bethell RC, Jiang H, White PW, Kukolj G. 2012. Evaluation of phosphatidylinositol-4-kinase IIIalpha as a hepatitis C virus drug target. J. Virol. 86:11595–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arita M, Nagata N, Sata T, Miyamura T, Shimizu H. 2006. Quantitative analysis of poliomyelitis-like paralysis in mice induced by a poliovirus replicon. J. Gen. Virol. 87:3317–3327 [DOI] [PubMed] [Google Scholar]
- 41. Baltera RF, Jr, Tershak DR. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J. Virol. 63:4441–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crotty S, Saleh MC, Gitlin L, Beske O, Andino R. 2004. The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein. J. Virol. 78:3378–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kawano M, Kumagai K, Nishijima M, Hanada K. 2006. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 281:30279–30288 [DOI] [PubMed] [Google Scholar]
- 44. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31:754–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lehto M, Olkkonen VM. 2003. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta 1631:1–11 [DOI] [PubMed] [Google Scholar]
- 47. Dawson PA, Van der Westhuyzen DR, Goldstein JL, Brown MS. 1989. Purification of oxysterol binding protein from hamster liver cytosol. J. Biol. Chem. 264:9046–9052 [PubMed] [Google Scholar]
- 48. Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. 1992. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116:307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426:803–809 [DOI] [PubMed] [Google Scholar]
- 50. Horton JD, Goldstein JL, Brown MS. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishimura T, Inoue T, Shibata N, Sekine A, Takabe W, Noguchi N, Arai H. 2005. Inhibition of cholesterol biosynthesis by 25-hydroxycholesterol is independent of OSBP. Genes Cells 10:793–801 [DOI] [PubMed] [Google Scholar]
- 52. van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Purstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. 2012. Coxsackievirus mutants that can bypass host factor PI4KIIIbeta and the need for high levels of PI4P lipids for replication. Cell Res. 22:1576–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kandutsch AA, Shown EP. 1981. Assay of oxysterol-binding protein in a mouse fibroblast, cell-free system. Dissociation constant and other properties of the system. J. Biol. Chem. 256:13068–13073 [PubMed] [Google Scholar]
- 54. Ridgway ND. 2010. Oxysterol-binding proteins. Subcell. Biochem. 51:159–182 [DOI] [PubMed] [Google Scholar]
- 55. Amako Y, Sarkeshik A, Hotta H, Yates J, III, Siddiqui A. 2009. Role of oxysterol binding protein in hepatitis C virus infection. J. Virol. 83:9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Amako Y, Syed GH, Siddiqui A. 2011. Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J. Biol. Chem. 286:11265–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beh CT, Cool L, Phillips J, Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157:1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 99:15669–15674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pezacki JP, Sagan SM, Tonary AM, Rouleau Y, Belanger S, Supekova L, Su AI. 2009. Transcriptional profiling of the effects of 25-hydroxycholesterol on human hepatocyte metabolism and the antiviral state it conveys against the hepatitis C virus. BMC Chem. Biol. 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sagan SM, Rouleau Y, Leggiadro C, Supekova L, Schultz PG, Su AI, Pezacki JP. 2006. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem. Cell Biol. 84:67–79 [DOI] [PubMed] [Google Scholar]
- 61. Schaffer FL, Gordon M. 1966. Differential inhibitory effects of actinomycin D among strains of poliovirus. J. Bacteriol. 91:2309–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J. Virol. 85:8870–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1:280–287 [DOI] [PubMed] [Google Scholar]
- 65. Banerji S, Ngo M, Lane CF, Robinson CA, Minogue S, Ridgway ND. 2010. Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the Golgi apparatus requires phosphatidylinositol 4-kinase IIalpha. Mol. Biol. Cell 21:4141–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144:389–401 [DOI] [PubMed] [Google Scholar]
- 67. de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195:965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goto A, Liu X, Robinson CA, Ridgway ND. 2012. Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Mol. Biol. Cell 23:3624–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.