Background: TGF-β promotes tumor metastasis by inducing SNAI2.
Results: TGF-β induces SNAI2 and promotes EMT by repressing miR-203.
Conclusion: The SNAI2 and miR-203 feedback loop plays integral roles in EMT and tumor metastasis.
Significance: Our findings provide new insights into molecular mechanisms of EMT and tumor metastasis and identify a therapeutic target for tumor invasion and metastasis.
Keywords: Cancer Biology, Epithelial-Mesenchymal Transition, Gene Expression, Transcription, Transforming Growth Factor β (TGFβ), Tumor Metastases
Abstract
TGF-β promotes tumor invasion and metastasis by inducing an epithelial-mesenchymal transition (EMT). Understanding the molecular and epigenetic mechanisms by which TGF-β induces EMT may facilitate the development of new therapeutic strategies for metastasis. Here, we report that TGF-β induced SNAI2 to promote EMT by repressing miR-203. Although miR-203 targeted SNAI2, SNAI2 induced by TGF-β could directly bind to the miR-203 promoter to inhibit its transcription. SNAI2 and miR-203 formed a double negative feedback loop to inhibit each other's expression, thereby controlling EMT. Moreover, we found that miR-203 was significantly down-regulated in highly metastatic breast cancer cells. The restoration of miR-203 in highly metastatic breast cancer cells inhibited tumor cell invasion in vitro and lung metastatic colonization in vivo by repressing SNAI2. Taken together, our results suggest that the SNAI2 and miR-203 regulatory loop plays important roles in EMT and tumor metastasis.
Introduction
Tumor metastasis is the main cause of mortality in patients with solid tumors (1). Epithelial-mesenchymal transition (EMT)2 has been considered as an early step in tumor metastasis (2). EMT is a process in which cells lose their epithelial features and acquire mesenchymal properties, including cytoskeleton remodeling, epigenetic changes, and increased invasiveness (3). A hallmark of EMT is the loss of E-cadherin (CDH1), a defining marker of epithelial cells. E-cadherin is a transmembrane glycoprotein that forms the core of adherens junctions between the adjacent epithelial cells and plays a critical role in cell-to-cell adhesion (4). Both in vitro and in vivo studies have shown that the loss of E-cadherin is linked to cancer progression and metastasis (5–7). A family of E-box-binding transcription factors, including SNAI1, SNAI2, ZEB1, ZEB2, and TWIST1, have been found to promote EMT and tumor metastasis by repressing CDH1 (8). Although SNAI2 is considered a homologue of SNAI1, several studies suggest that SNAI2 may play unique roles in tumor invasive growth and metastasis. For example, SNAI2 inhibited apoptosis by repressing p53-mediated transcription (9). SNAI2, but not SNAI1, was found to inversely correlate with the expression of E-cadherin in breast cancer cells (10). Clinical studies found that SNAI2 was highly expressed in metastatic breast cancer and correlated with poor prognosis of breast cancer (11). Moreover, overexpression of SNAI2 has been found to induce ZEB1 and/or ZEB2, suggesting that SNAI2 may cross-talk with other regulators of EMT (12). TGF-β is considered as the major upstream inducer of EMT, and increasing evidence implicates TGF-β signaling as a key effector of EMT in cancer progression and metastasis (13). However, the precise molecular mechanism by which TGF-β signaling induces EMT and how TGF-β signaling is regulated are still obscure. Therefore, understanding the TGF-β signaling network may help to develop novel therapeutic strategies for metastasis.
MicroRNAs (miRNAs) are small non-coding RNAs that regulate a variety of biological processes by modulating gene expression at the post-transcriptional level (14). Growing evidence shows that miRNAs play an important role in the control of cancer cell invasion and metastasis (15–17). For example, miR-335 was found to inhibit metastasis by targeting SOX4 and tenascin C (16). miR-10b induced by TWIST stimulated tumor cell migration and invasion (17). The miR-200 family, including miR-200b, miR-200a, miR-409, miR-200c, and miR-141, formed a feedback loop with ZEB1 and ZEB2 to regulate EMT and tumor metastasis (18, 19). miR-203 inhibited breast cancer invasion by targeting SNAI2 (20). Interestingly, using a combination of bioinformatics and functional analyses, we found that TGF-β induced SNAI2 to suppress miR-203 in EMT. SNAI2 and miR-203 formed a double negative feedback loop to inhibit each other's expression, thereby controlling EMT and tumor invasive growth and metastasis. Repression of miR-203 was required for TGF-β-induced EMT. Moreover, the induction of SNAI2 by TGF-β controlled the miR-200 and ZEB1/2 regulatory loop that is an important regulator of EMT, suggesting that SNAI2 plays an integral role in tumor invasion and metastasis. Our clinical analysis and experimental models demonstrate that the SNAI2 and miR-203 loop is an attractive therapeutic target for tumor metastasis.
EXPERIMENTAL PROCEDURES
Cell Culture and Viral Transduction
Hs-578T, MCF7, MDA-MB-468, MDA-MB-231, T47D, HEK293T, and MDCK cells were maintained in DMEM containing 10% FBS and antibiotics. BT-549 and BT-474 were maintained in RPMI 1640 medium containing 10% FBS and antibiotics. SUM159 was purchased from Asterand Co. and cultured according to the manufacturer's instructions. For TGF-β treatment, MDCK was treated with 5 ng/ml recombinant human TGF-β1 (PeproTech) for the different days. During TGF-β treatment, cells were split every 2 or 3 days depending on the cell confluence.
For viral transduction, retroviruses were generated by co-transfection of pQNCX2 EV or pQNCX2-SNAI2 with packaging plasmids into HEK293T cells as described previously (21, 22). MDA-MB-468 cells were transduced with these virus particles and subsequently selected with 60 μg/ml neomycin for at least 10 days. Lentiviruses overexpressing miR-203 were packaged and generated in 293T cells according to the manufacturer's instructions (System Biosciences). MDA-MB-231 and MDCK cells were infected and subsequently subjected to FACS for green fluorescent protein (GFP) to obtain the stable cell lines. The target sequence for SNAI2 shRNA was 5′-GCATTTGCAGACAGGTCAAAT-3′, and the shRNA was subcloned into pLKO lentiviral vectors. The lentiviruses expressing SNAI2 shRNA were packaged and generated in 293T cells as described previously (20, 21). A sponge insert containing 8× tandem “bulged” (at positions 9–12) miR-203 binding sites in PUC57 was purchased from Genescript. The pBabe-d2eGFP miRNA sponge backbone was made by digesting pBabe-d2eGFP-miR-9 (Addgene) with XhoI and ApaI (removing the miR-9 sponge). The miR-203 sponge inserts were subcloned into the pBabe-d2eGFP vector.
Plasmids and Luciferase Reporter Assays
SNAI2 ORF was amplified from the MDA-MB-231 complementary DNA (cDNA) library and then cloned into pQNCX2 with or without an HA tag in the C terminus. The full-length 3′-UTR of SNAI2 was amplified from the MDA-MB-231 cDNA library and then cloned into the pmiR-REPORT luciferase construct (Ambion). The promoter region of miR-203, miR-200b, and miR-200c was amplified from MCF7 genome DNA and subcloned into pGL3-Basic vector (Promega). Mutations in the above constructs were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The pmiR-203 construct and the corresponding empty vector were purchased from System Biosciences. All of these constructs were verified by DNA sequencing. For transient transfections, cells of 50% confluence in 24-well plates were transfected using Lipofectamine 2000 reagents according to the manufacturer's protocol (Invitrogen). For the 3′-UTR reporter assay, 10 ng of reporter plasmids together with 150 ng of control plasmids or pmiR-203 plasmids and 2 ng of pmiR-β-gal plasmids (Applied Biosystems) were co-transfected into 293T cells. For the miRNA promoter-reporter assays, 50 ng of reporter plasmids together with 5–50 ng of pQNCX2-SNAI2 plasmids and 2 ng of β-pmiR-β-gal plasmids were co-transfected into 293T cells. The total amount of DNA in each individual well was kept constant by adding pQNCX2 empty vector. Luciferase and galactosidase activities were measured 24–48 h after transfection using the Dual-Luciferase reporter assay system (Promega) and GalactoStar kits (Tropix), respectively. All experiments were performed in triplicate with data pooled from at least two independent experiments.
Transfection of siRNA and miRNA and Real Time RT-PCR
For transient knockdown of SNAI2, 30 nm siRNA or control siRNA (Dharmacon) was transfected using RNAiMAX (Invitrogen) following the manufacturer's instructions. For transient overexpression of miR-203, 30 nm pre-miR-203 (Ambion) was transfected using RNAiMAX. In each case, total RNA and protein were collected for assay 3 days post-transfection. For functional assays, cells were plated for Matrigel invasion assays 2 days after transfection.
Total RNA from cultured cells was isolated using the mirVana miRNA isolation kit according to the manufacturer's instructions (Ambion). Mature miRNA expression was quantified using TaqMan miRNA assays according to the manufacturer's instructions (Applied Biosystems). miRNA expression was normalized to U6 small nuclear expression. Real time PCR was performed as described previously (21).
Western Blotting Analysis and Immunofluorescence
Cells were lysed in radioimmune precipitation assay buffer as described previously (22). The samples were separated on a 4–7.5% SDS-polyacrylamide gel. After transfer to PVDF membrane using a semidry transfer apparatus (Bio-Rad), probing was carried out with primary antibodies and subsequent secondary antibodies. Primary antibodies were purchased from the following commercial sources: anti-CDH1 (1:1000; BD Biosciences), anti-SNAI2 (1:500; Cell Signaling Technology), anti-vimentin (1:1000; Cell Signaling Technology), anti-ZEB1 (1:500; Santa Cruz Biotechnology, C-20), and anti-α-tubulin (1:100,000; Sigma-Aldrich). Membranes were exposed using the ECL method (GE Healthcare) according to the manufacturer's instructions. For immunofluorescence, cells were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and then stained with anti-CDH1 (1:100; BD Biosciences), anti-vimentin (1:100; Cell Signaling Technology), and anti-SNAI2 (1:100; Cell Signaling Technology). The primary antibodies were then detected with Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Nuclei were visualized by co-staining with DAPI.
ChIP Assay and Human Affymetrix Microarray
ChIP assays were carried out using a ChIP assay kit based on the manufacturer's instructions (Upstate Biotechnology). Cells were incubated with a dimethyl 3,3′-dithiobispropionimidate HCl (Pierce) solution (5 mm) for 10 min at room temperature, and then formaldehyde was added. For each ChIP reaction mixture, 2 × 106 cells were used. The resulting precipitated DNA samples were quantified by real time PCR with specific primer sets (supplemental Methods). The antibodies for ChIP assays were purchased from the following sources: anti-H3AC, anti-H3TriMeK4, anti-H3DiMeK9 (Millipore, CS200587), and anti-HA (Abcam, ab9110). Data are expressed as the percentage of input DNA.
For microarray analysis, we extracted total RNA using an miRNeasy kit based on the manufacturer's protocol (Qiagen). 5-μg aliquots of total RNAs from each sample were transcribed to double-stranded cDNA using SuperScript II reverse transcriptase (Invitrogen) with an oligo(dT) primer and then used to generate single-stranded RNAs. The biotin-labeled RNAs were fragmented and hybridized with an Affymetrix Human Genome U133 Plus 2.0 Array. We scanned the arrays with the GeneArray scanner (Affymetrix) and used the robust multichip average method to normalize the raw data. Microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE44239.
Matrigel Invasion Assays and Mouse Xenografts
Matrigel invasion assays were performed using BD BioCoat Matrigel invasion chambers as described previously (21). Briefly, 1 × 105 cells suspended in 0.5 ml of serum-free medium were plated in the upper chamber. In the lower chamber, 0.75 ml of medium containing 0.1% FBS was added as a chemoattractant. After 24–48 h, Matrigel was removed, and the cells that had invaded the lower chamber were stained with a HEMA-3 kit (Fisher). Cell numbers from four random fields were counted and averaged. Each group was performed in triplicates. All animal experiments were performed in accordance with a protocol approved by the UCLA Animal Research Committee. For lung metastasis, cells (1 × 106) were injected into the tail vein of mice (five mice per group). After 2 months, mice were euthanized, the lungs were resected, and nodules were counted.
Bioinformatics and Statistical Analysis
To search genes co-expressed with CDH1, the normalized data of mRNA and miRNA were downloaded from the Genomics and Bioinformatics Group website and combined into a single database. The GeneNeighbours module in the GenePattern program (23) was used to perform the search for genes co-expressing with CDH1 in our combined database. For gene expression correlation analysis in clinical samples, two independent databases containing both mRNA and miRNA normalized expression data were used. The first one had 91 samples from different cancer types, and the second one had 139 samples from prostate cancer. The Spearman correlation coefficient was calculated between CDH1 and miR-203 in an individual data set using SPSS 17.0 software. To compare miR-203 expression in primary and metastatic breast cancers, the raw data of a published data set (E-TABM-971) (24) was downloaded from European Bioinformatics Institute ArrayExpress. The global median method in BRB-ArrayTools (Richard Simon and Amy Peng Lam, National Cancer Institute, National Institutes of Health, Bethesda, MD) was used to normalize the raw data. The above mentioned prostate data set was also used to compare the miR-203 expression between prostate primary and metastatic cancers. For relapse-free survival analysis, the prostate data set, which provide detailed patient follow-up information, was used again. Patients were dichotomized into two groups as above or below median for the miR-203 expression value. A Kaplan-Meier survival plot was performed for relapse-free survival, and the log rank test was applied to test the survival differences and calculate the p value using SPSS 17.0 software. TargetScan (25) and miRDB (26) were used to predict the target genes of miR-203.
RESULTS
miR-203 Is Repressed in TGF-β-induced EMT
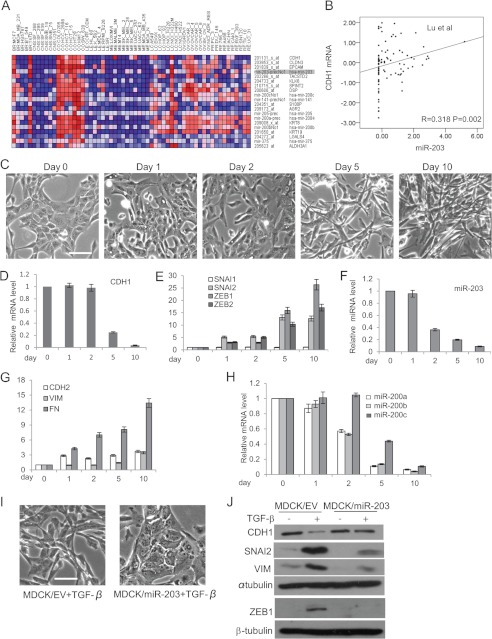
Because CDH1 is suppressed by TGF-β-SNAI2 signaling in EMT (8), it is possible that genes that were regulated by TGF-β might be co-expressed with CDH1. We interrogated the array data set composed of mRNA and miRNA of NCI60 cells using the GeneNeighbours module in the GenePattern program (23, 27, 28). The top 20 candidate genes, consisting of 13 coding mRNA genes and seven miRNA genes, were selected for further analysis (Fig. 1A). We found that the expression profile of miR-203 was closest to the expression of CDH1 among all miRNA genes in the NCI60 cells, including the miR-200 family. Correlation analysis showed a significant positive correlation between the expression of miR-203 and the expression of CDH1 at both mRNA and protein levels (supplemental Fig. S1, A and B). To confirm this finding, we examined the expression of miR-203 in eight breast cancer cell lines using quantitative real time PCR. Interestingly, the expression of miR-203 in four lowly invasive breast cancer cell lines (MCF7, MDA-MB-468, BT474, and T47D) with high CDH1 expression was several hundred times higher than in four highly invasive breast cancer cell lines (MDA-MB-231, BT549, Hs578T, and SUM159) with low CDH1 expression (supplemental Fig. S1C). Furthermore, to investigate whether this correlation also existed in human tumor tissues, we analyzed two published human tumor tissue expression data sets containing both mRNA and miRNA expression data (29, 30). The first data set consisted of tumors from six different tissues, including breast, whereas the second included only prostate tumors. In both data sets, we found that the expression of miR-203 was positively correlated with the expression of CDH1 (Fig. 1B and supplemental Fig. S1D). Altogether, these data indicated that miR-203 was co-expressed with CDH1 in both cancer cell lines and primary cancer tissues.
FIGURE 1.
miR-203 in MDCK cells is repressed in TGF-β-induced EMT. A, heat map showing the top 20 candidate genes co-expressed with CDH1 in the NCI60 database. B, the co-expression of CDH1 mRNA and miR-203 in human tumor tissues. The Spearman correlation coefficients and significance levels are indicated. C, phase-contrast microscopy of MDCK cells treated with TGF-β for the indicated days. Scale bar, 50 μm. D, real time RT-PCR showing the CDH1 expression level in MDCK cells treated with TGF-β for the indicated days. E, TGF-β induced the expression of SNAI2, ZEB1, and ZEB2 in MDCK cells. F, TGF-β repressed miR-203 in MDCK cells as determined by TaqMan miRNA assays. G, TGF-β induced the expression of CDH2, VIM, and FN1 in MDCK cells as determined by real time RT-PCR. H, TGF-β repressed miR-200a, miR-200b, and miR-200c in MDCK cells. I, the restoration of miR-203 prevented the morphology change induced by TGF-β. Scale bar, 50 μm. J, the restoration of miR-203 prevented the up-regulation of SNAI2 and the down-regulation of CDH1 induced by TGF-β. Error bars represent S.D.
To test whether the expression of miR-203 could be suppressed upon TGF-β treatment, we used a classical EMT model system: the MDCK cell, which can be induced to undergo EMT in response to TGF-β exposure. TGF-β treatment in MDCK cells led to a gradual morphological change (Fig. 1C) that was accompanied by the loss of CDH1 and induction of CDH2, vimentin (VIM), and FN1 (Fig. 1, D and G). TGF-β also potently induced the expression of SNAI2, ZEB1, and ZEB2 but not SNAI1 (Fig. 1E). These hallmark shifts at the morphological and molecular levels indicate that EMT was induced in MDCK cells by TGF-β. Importantly, we found that the level of miR-203 in MDCK cells was significantly reduced 2 days after TGF-β treatment, which is earlier than the reduction of CDH1 and later than the induction of SNAI2 (Fig. 1F). The miR-200 family was also down-regulated after TGF-β treatment (Fig. 1H). While our work was in progress, it was shown that loss of miR-203 was associated with cell invasion (31). To investigate whether the down-regulation of miR-203 was required for TGF-β-induced EMT, we stably expressed miR-203 in MDCK cells. Interestingly, we found that overexpression of miR-203 prevented the morphology change induced by TGF-β (Fig. 1I). Western blot analysis showed that ectopic expression of miR-203 potently prevented the induction of SNAI2 and the inhibition of CDH1 mediated by TGF-β. Interestingly, we found that miR-203 also inhibited the induction of ZEB1 induced by TGF-β (Fig. 1J). Taken together, these results suggest that the inhibition of miR-203 was required for TGF-β-induced EMT.
miR-203 Targets the SNAI2-E-cadherin Axis and Inhibits Cancer Metastasis
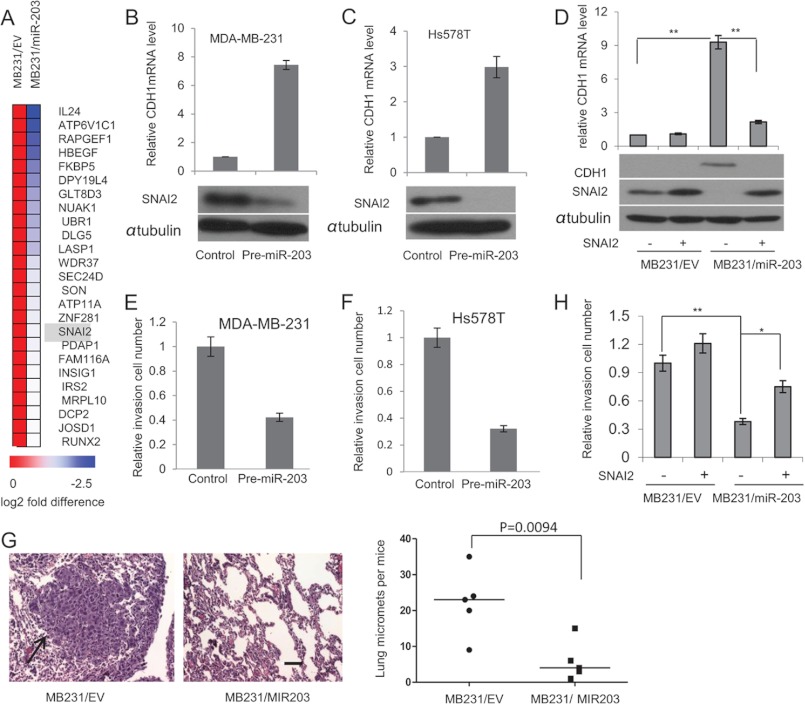
To investigate the mechanism through which miR-203 was co-expressed with CDH1 and regulated the TGF-β-induced EMT, we searched for targets of miR-203 that might mediate this effect using two different searching programs, TargetScan (25) and miRDB (26). Meanwhile, we made a stable cell line (MB231/miR-203) that overexpressed miR-203 and compared the gene expression profile between MB231/EV and MB231/miR-203 cells using a microarray. We found that 177 genes were down-regulated in MB231/miR-203 cells compared with MB231/EV cells (only a >0.5-fold difference was considered). 25 among these 177 genes were found to be the predicted targets by two different programs (Fig. 2A). The most interesting predicted target by the searching program and by the microarray analysis was SNAI2, which is one of the key CDH1 repressors. A 7-mer site recognized by miR-203 was found in the human SNAI2 3′-UTR that was evolutionarily conserved among diverse species (supplemental Fig. S2A). Consistent with the findings of Zhang et al. (20), our luciferase assay also confirmed that SNAI2 is a direct target of miR-203 (supplemental Fig. S2, B and C). We transiently expressed pre-miR-203 in both MDA-MB-231 and Hs578T cells in which SNAI2 is highly expressed and both CDH1 and miR-203 are repressed. Western blot analysis revealed that the SNAI2 proteins were significantly decreased after transient transfection of pre-miR-203 in both MDA-MB-231 and Hs-578T cells (Fig. 2, B and C). Consistently, real time RT-PCR showed that transient transfection of pre-miR-203 also restored CDH1 expression. To further examine whether the repression of SNAI2 by miR-203 mediated these effects, we made MDA-MB-231 cells stably expressing miR-203 alone (MB231/miR-203) or together with SNAI2. Although a significantly increased CDH1 expression accompanied by decreased SNAI2 was observed in MB231/miR-203, miR-203-induced CDH1 was abolished by the reintroduction of SNAI2 (Fig. 2D). Taken together, our results demonstrate that mi-R203 targets SNAI2 to regulate CDH1 expression.
FIGURE 2.
miR-203 targets SNAI2 and inhibits breast cancer metastasis. A, heat map showing 25 predicted targets that were suppressed by miR-203 through microarray analysis. B, transfection of pre-miR-203 in MDA-MB-231 cells inhibited SNAI2 and induced CDH1 as determined by Western blot and real time RT-PCR, respectively. C, transfection of pre-miR-203 in Hs578T cells inhibited SNAI2 and induced CDH1 as determined by Western blot and real time RT-PCR, respectively. D, miR-203 up-regulated CDH1 by targeting SNAI2. Both MB231/miR-203 cells and MB231/EV cells were transduced with retroviruses expressing SNAI2 without the 3′-UTR or control vector. The expression of SNAI2 was examined by Western blot, and the expression of CDH1 was examined by Western blot and real time RT-PCR. *, p < 0.05; **, p < 0.01. E and F, transfection of pre-miR-203 inhibited breast cancer invasion in both MDA-MD-231 and Hs578T cells as determined by Matrigel invasion assays. The data are mean ± S.D. of two independent experiments preformed in triplicates. G, the restoration of miR-203 in MDA-MB-231 cells inhibited lung metastasis in nude mice. Metastatic tumors were examined by H&E staining. Arrow, metastatic tumor tissues. The graph shows the quantification of the total number of nodules in individual lungs (n = 5). Student's t test was used for the significance calculation. Scale bar, 50 μm. H, the restoration of SNAI2 partially rescued miR-203-mediated inhibition. Error bars represent S.D. micromets, microscopic metastases.
The invasive growth of solid tumors is critical for distant metastasis. The hallmarks of EMT are the functional loss of CDH1 and increased invasion. SNAI2 can promote cell migration and invasion through suppression of CDH1 and an increase of matrix metalloproteinase expression (32). Interestingly, recent studies by Korpal et al. (33) showed that the miR-200 family promoted metastatic colonization by inducing E-cadherin and targeting the Sec23a-mediated tumor cell secretome, casting doubt on the potential therapeutic utility of miR-200s. They showed that overexpression of miR-200 promoted lung metastasis in a tail vein injection assay. To directly confirm the repressive role of miR-203 in metastasis, we examined whether the restoration of miR-203 could inhibit human breast cancer cell metastasis using the same model. Overexpression of miR-203 in both MDA-MB-231 and Hs578T cells significantly decreased their invasion as determined by Matrigel invasion assays (Fig. 2E). Also, the tail vein injection revealed that, unlike miR-200, miR-203 strongly inhibited MDA-MB-231 cell lung colonization in vivo (Fig. 2F). Interestingly, we noted that the restoration of SNAI2 only partially increased cell invasion in MB231/miR-203 cells by Matrigel invasion assays (Fig. 2G), suggesting that miR-203 may also inhibit invasion by targeting other molecules in addition to the SNAI2-E-cadherin axis. Two recent reports showed that miR-203 suppresses cell migration and invasion of breast cancer and squamous cell carcinoma through targeting LASP1 (34, 35). Interestingly, we found that LASP1 is on the potential target gene list based on our microarray (Fig. 2A).
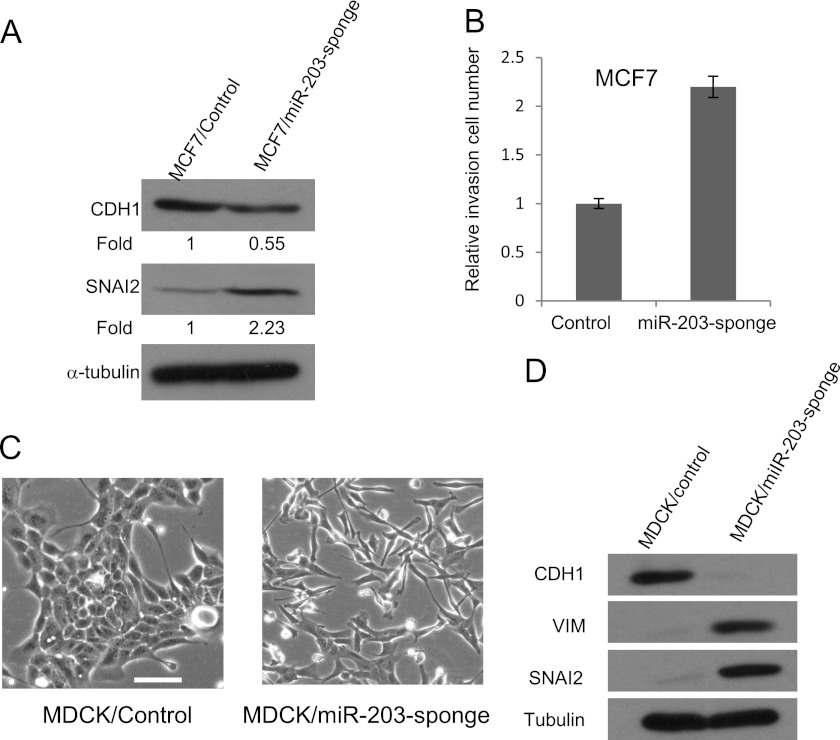
Because TGF-β inhibited miR-203 in EMT, we examined whether miR-203 was required for maintaining the epithelial property. We used miRNA sponges to stably knock down miR-203 in MCF7 cells, which have high levels of miR-203. Western blot showed that knockdown of miR-203 by miR-203 sponges in MCF7 significantly increased the expression of SNAI2 and decreased the expression of CDH1 (Fig. 3A). The knockdown of miR-203 also increased the invasion capability of MCF7 (Fig. 3B). The knockdown of miR-203 in MDCK cells drastically induced morphological changes. Western blot analysis revealed that the knockdown of miR-203 resulted in the induction of VIM and SNAI2 and the loss of CDH1 (Fig. 3, C and D), suggesting that miR-203 is required for maintaining the epithelial property of MDCK cells.
FIGURE 3.
Knockdown of miR-203 induces EMT in MCF7 cells. A, knockdown of miR-203 increased SNAI2 expression and decreased CDH1 expression in MCF7 cells. B, knockdown of miR-203 increased the invasion capability of MCF7 cells. C, knockdown of miR-203 in MDCK cells induced morphological changes. D, knockdown of miR-203 increased SNAI2 and VIM and decreased CDH1 in MDCK cells. Error bars represent S.D.
SNAI2 Directly Inhibits miR-203 to Promote EMT
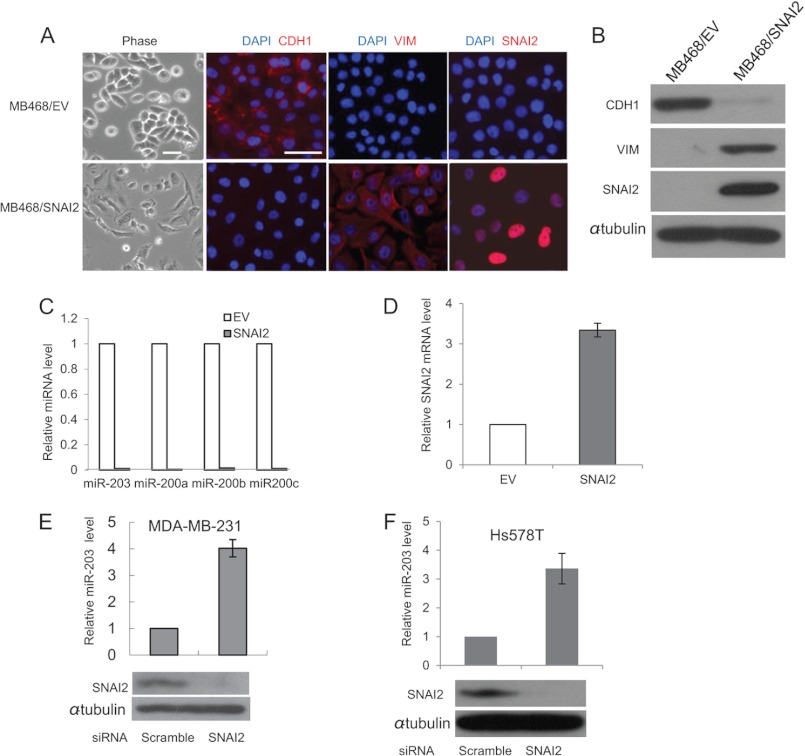
Increasing evidence has shown that a feedback circuit exists between miRNA and the transcription factor target to enhance the robustness of gene regulation in mammalian genomes (40). For unknown reasons, we were unable to efficiently knock down miR-203 in MDA-MB-468 cells. To investigate whether SNAI2 repressed miR-203 in a reciprocal regulatory circuit containing SNAI2 and miR-203, we overexpressed the full-length SNAI2 cDNA, lacking the 3′-UTR, in lowly metastatic MDA-MB-468 cells. Overexpression of SNAI2 in MDA-MB-468 cells (MB468/SNAI2) resulted in a morphological change from a rounded compact shape to a loose spindle shape indicative of EMT. This was accompanied by a loss of CDH1 and gain of VIM as revealed by immunofluorescence staining (Fig. 4A). The reduction of CDH1 and induction of VIM were further confirmed by Western blot analysis (Fig. 4B). A dramatic decrease of miR-203 was observed in SNAI2-expressing MDA-MB-468 cells, indicating that miR-203 might be the target of SNAI2 (Fig. 4C). Interestingly, overexpression of SNAI2 also induced the expression of endogenous SNAI2 as determined by real time RT-PCR with a specific set of primers targeting the 3′-UTR of SNAI2 mRNA (Fig. 4D). To determine whether endogenous SNAI2 could repress the expression of miR-203, we knocked down the expression of SNAI2 in both MDA-MB-231 and Hs578T cells using siRNA. Compared with scrambled siRNA, we found that the knockdown of SNAI2 potently stimulated miR-203 expression in both MDA-MB-231 and Hs578T cells (Fig. 4, E and F).
FIGURE 4.
SNAI2 induces EMT by repressing miR-203. A, overexpression of SNAI2 in MDA-MB-468 cells induced EMT. The expression of CDH1, VIM, and SNAI2 was examined by immunofluorescence staining. MB468/EV, MDA-MB-468 cells expressing empty vector; MB468/SNAI2, MDA-MB-468 cells expressing SNAI2. Scale bar, 50 μm. B, overexpression of SNAI2 induced VIM and inhibited CDH1 by Western blot. C, SNAI2 repressed miR-203 and miR-200s as determined by TaqMan miRNA assays. D, overexpression of SNAI2 induced endogenous SNAI2 as determined by real time RT-PCR. E, knockdown of SNAI2 induced miR-203 in MDA-MD-231 cells. F, the knockdown of SNAI2 induced miR-203 in Hs578T cells. Error bars represent S.D.
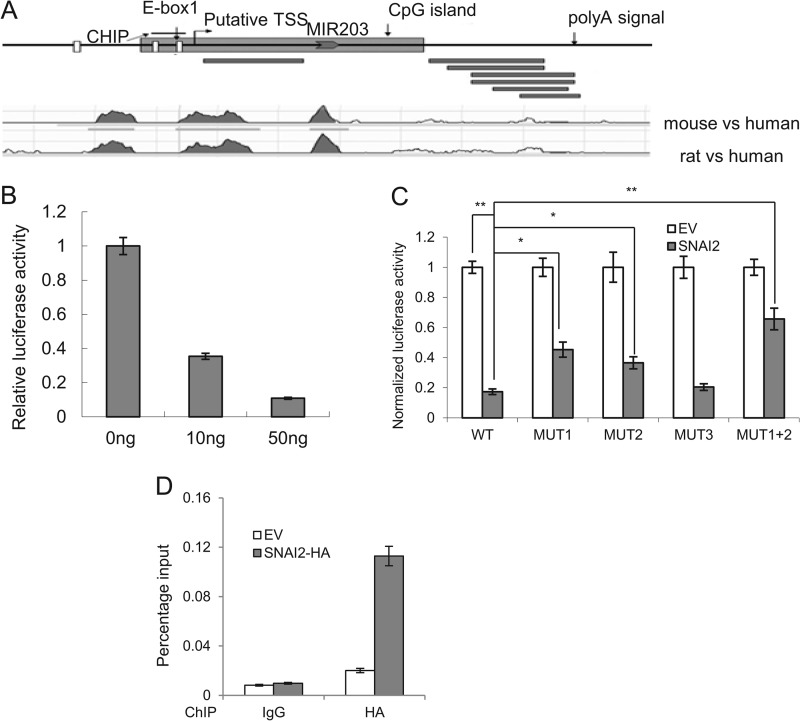
To investigate how TGF-β inhibited miR-203, we characterized the primary precursor transcript of miR-203. The miR-203 gene was localized on human chromosome 14 surrounded by several ESTs spanning around 2000 bp as annotated on the UCSC Genome Browser (36) (Fig. 5A). Six overlapping ESTs matched around 500 bp downstream of the miR-203 hairpin were found, and a canonical poly(A) signal, AATAAA, was found in EST BE551807, indicating that the 3′ boundary of this primary transcript could be identified. A transcription start site (TSS) predicted by the Eponine method (37) was found close to the 5′ terminus of the EST that matched the upstream region of the miR-203 hairpin. Comparative genomic analysis using the VISTA program (38) showed that the region around the predicted TSS is highly conserved among human, mouse, and rat (Fig. 5A). Furthermore, the predicted TSS is also supported by the presence of an overlapping CpG island (39) that is known to co-localize with the TSS. Collectively, through our analysis, the primary transcript of miR-203 is around 2 kb in length with no significant open reading frame (ORF) in it.
FIGURE 5.
SNAI2 directly inhibits miR-203 expression. A, schematic representations of miR-203 gene indicating the CpG island, three E-boxes, putative TSS, and the poly(A) signals. Black bars represent the reported ESTs. The ChIP assay region is shown. A comparative genomic plot shows the conserved regions in the miR-203 gene among human, mouse, and rat. B, SNAI2 inhibited the miR-203 promoter activity. C, SNAI2 inhibited miR-203 transcription through the E-box. The data are mean ± S.D. of three independent experiments. WT, wild type E-box; MUT1, mutant of E-box 1; MUT2, mutant E-box 2; MUT3, mutant E-box 3; MUT1+2, mutant of E-boxes 1 and 2. D, SNAI2 directly bound to the miR-203 promoter by ChIP assays. Error bars represent S.D.
To investigate whether SNAI2 directly repressed the transcription of miR-203, we cloned the putative promoter of miR-203, which spans from 700 bp upstream to 300 bp downstream of the predicted TSS, into a luciferase reporter construct. Co-transfection experiments showed that SNAI2 suppressed the luciferase activity of the reporter construct in a dose-dependent manner (Fig. 5B). The promoter region of miR-203 contains three putative binding sites for SNAI2 (E-boxes). Mutation of the first two E-boxes, but not the third E-box, significantly rescued the repression effect by SNAI2, indicating that these two E-boxes are responsible for SNAI2 binding (Fig. 5C). To further confirm this binding, we performed ChIP assays. Because of the lack of commercial ChIP-grade SNAI2 antibody, we made MDA-MB-468 cells stably expressing SNAI2-HA (MB468/SNAI2-HA). ChIP assays using HA antibodies confirmed that SNAI2 bound to the paired E-box region near the TSS of miR-203 (Fig. 5D). These results suggest that SNAI2 can directly suppress the expression of miR-203 by binding to the paired E-box in the promoter region of miR-203. Taken together, miR-203 and SNAI2 could repress each other and constitute a double negative feedback loop during EMT.
SNAI2 Directly Inhibits miR-200
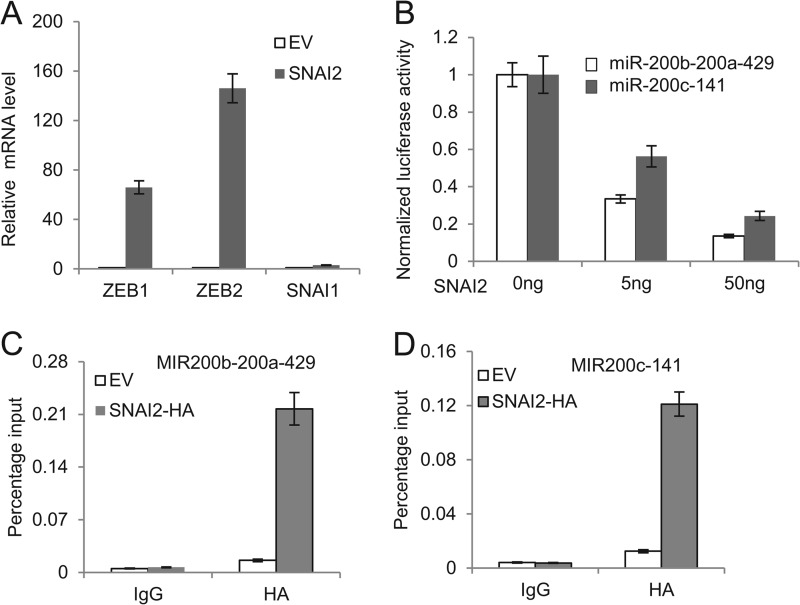
Recently, several reports demonstrated that the miR-200 family was an important epithelial marker and was lost during the EMT process. The miR-200 family targets ZEB1 and ZEB2, and inhibition of the miR-200 family has been found to induce EMT through up-regulation of ZEB1 and ZEB2 (18, 19, 41–43). Interestingly, a recent report showed that SNAI2 also formed the feedback loop with miR-200 to regulate EMT in prostate cancer (44). Therefore, we examined whether SNAI2 also inhibited miR-200 in breast cancer cells. Real time RT-PCR showed a substantial reduction in miR-200 family members after the induction of EMT in MDA-MB-468 cells by SNAI2 (Fig. 4C). Consistently, the expression of ZEB1 and ZEB2 was increased in SNAI2-expressing MDA-MB-468 cells (Fig. 6A). To examine whether SNAI2 could directly repress the miR-200 family, we performed both luciferase and ChIP assays. Luciferase assays showed that SNAI2 could repress the promoter activity of the miR-200 family in a dose-dependent manner (Fig. 6B). ChIP assays also showed that SNAI2 could bind to the promoter of the miR-200 family (Fig. 6, C and D). Our results suggest that these factors may act in concert to elicit the EMT process.
FIGURE 6.
SNAI2 directly inhibits the miR-200 family transcription. A, overexpression of SNAI2 induced the expression of ZEB1 and ZEB2. B, SNAI2 inhibited the promoter activity of miR-200b-200a-429 and miR-200c-141 in 293T cells. The data are mean ± S.D. of two independent experiments. C, SNAI2 bound to the promoter of miR-200b-200a-429. ChIP assays were performed using IgG or anti-HA antibodies. EV, MDA-MB-468 cells expressing empty vector; HA-SNAI2, MDA-MB-468 cells expressing HA-SNAI2. D, SNAI2 bound to the promoter of miR-200c-141 as determined by ChIP assays. Error bars represent S.D.
TGF-β Inhibits miR-203 through the Induction of SNAI2
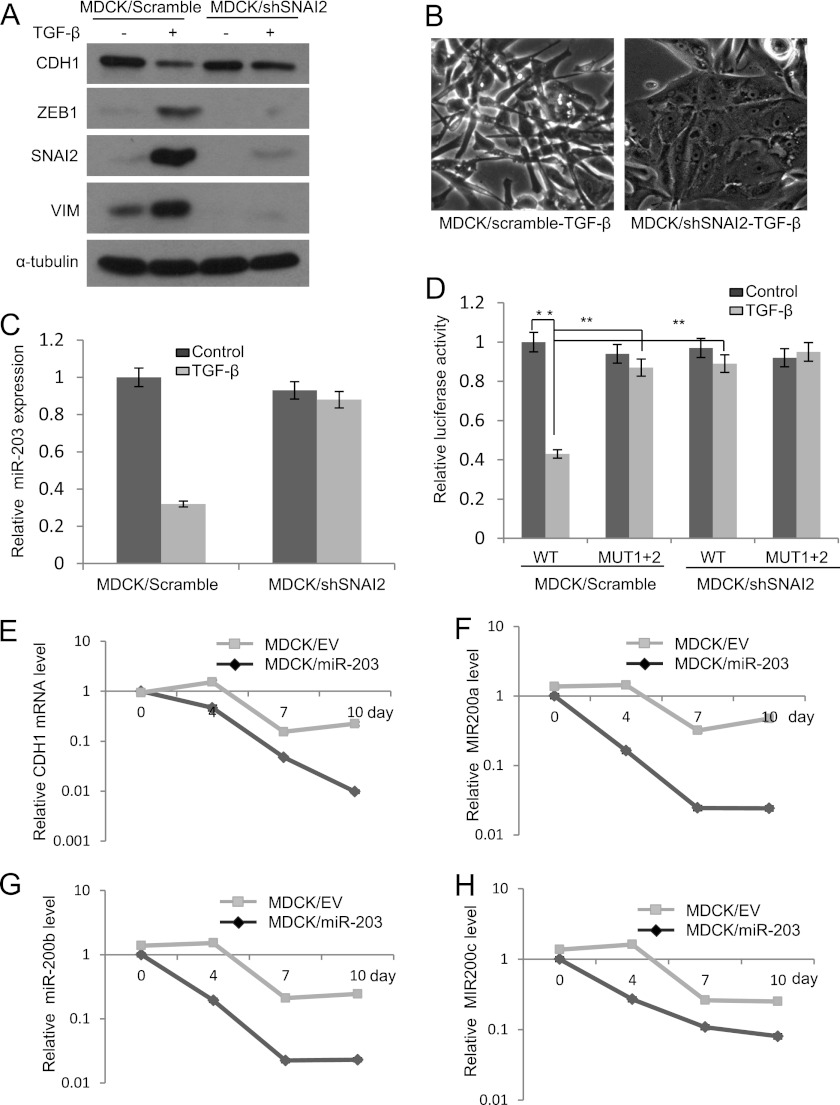
Based on the aforementioned observation that SNAI2 could repress the expression of miR-203, we asked whether the down-regulation of miR-203 by TGF-β was dependent on the induction of SNAI2. We stably knocked down SNAI2 in MDCK cells using shRNA. As shown in Fig. 7, A and B, knockdown of SNAI2 prevented the EMT induced by TGF-β, suggesting that induction of SNAI2 by TGF-β is essential for TGF-β-induced ΕΜΤ in MDCK cells. The knockdown of SNAI2 also inhibited TGF-β-mediated suppression of miR-203 (Fig. 6C). Luciferase assays also showed that the promoter activity of miR-203 was inhibited upon TGF-β treatment and that both mutation of the E-box and knockdown of SNAI2 could prevent this inhibition (Fig. 6D), suggesting that the integrity of the E-box in the promoter of miR-203 is important for the down-regulation of miR-203 by SNAI2 upon TGF-β treatment.
FIGURE 7.
SNAI2 is required for TGF-β-mediated inhibition of miR-203. A, knockdown of SNAI2 prevented the induction of VIM and ZEB1 and the repression of CDH1 by TGF-β in MDCK cells. B, knockdown of SNAI2 prevented the morphological change induced by TGF-β in MDCK cells. C, knockdown of SNAI2 prevented the inhibition of miR-203 by TGF-β in MDCK cells. D, knockdown of SNAI2 reversed TGF-β-mediated inhibition of the miR-203 promoter activity. E, the restoration of miR-203 attenuated TGF-β inhibition of CDH1 in MDCK cells. EV, MDCK cells expressing empty vector; miR-203, MDCK cells expressing miR-203. F–H, the restoration of miR-203 attenuated TGF-β inhibition of miR-200a, miR-200b, and miR-200c. The data are the mean of three independent experiments. Error bars represent S.D.
Because our result showed that the restoration of miR-203 in MDCK cells inhibited TGF-β-induced EMT, we examined whether miR-203 could prevent the down-regulation of the miR-200 family by TGF-β. Real time RT-PCR found that the ectopic expression of miR-203 suppressed the down-regulation of CDH1 and the miR-200 family induced by TGF-β (Fig. 7, E–H). Taken together, our results suggest that the miR-203 and SNAI2 double negative feedback loop is a master regulator of EMT and tumor metastasis.
miR-203 Is Lost in Human Metastatic Tumor Samples
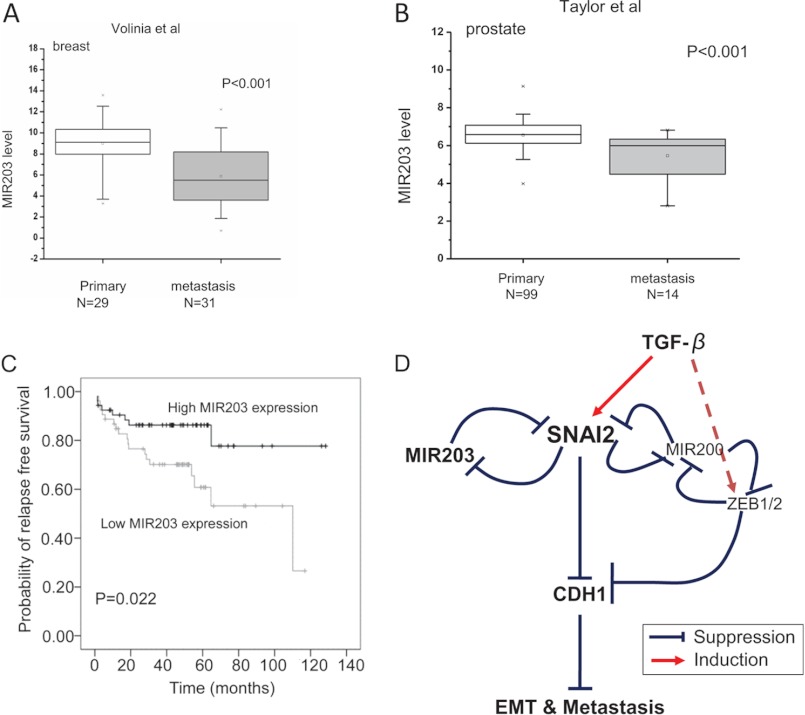
Our data established that the SNAI2-miR-203 double negative feedback loop regulated CDH1 expression and tumor invasion and metastasis in breast cancer cells. To determine whether loss of miR-203 was associated with human tumor metastasis, we compared the miR-203 expression between the primary and metastatic samples in a publicly available breast tumor data set (24). miR-203 expression was significantly decreased in human metastatic breast tumor samples compared with human primary breast tumor samples (Fig. 8A). Finally, we explored whether loss of miR-203 was involved in the metastasis of other tumors and associated with cancer progression and prognosis. We interrogated a prostate tumor data set (30) that provided detailed clinical information and found that down-regulation of miR-203 also occurred in the metastatic prostate tumors (Fig. 8B). Interestingly, survival analysis showed that loss of miR-203 was significantly correlated with reduced recurrence-free survival (Fig. 8C).
FIGURE 8.
Loss of miR-203 is associated with human tumor metastasis and tumor prognosis. A, loss of miR-203 was associated with human breast cancer metastasis (based on studies reported by Volinia et al. (24)). The p value was calculated by Mann-Whitney U test. x above error bars represents the maximal value; x below error bars represents the minimal value. B, miR-203 was down-regulated in human prostate cancer metastasis (based on studies reported by Taylor et al. (30)). The p value was calculated by Mann-Whitney U test. x above error bars represents the maximal value; x below error bars represents the minimal value. C, Kaplan-Meier curve showing the relapse-free survival of 107 prostate samples with high or low miR-203 expression. The p value was calculated by log rank test. D, a proposed model showing TGF-β signaling, the SNAI2-miR-203 double negative feedback loop, and SNAI2-miR-200-ZEB1/2-CDH1 feed-forward loop in metastasis.
DISCUSSION
In this study, we identified that SNAI2 and miR-203 form a new feedback regulatory loop that plays an important role in TGF-β-induced EMT and tumor metastasis. Although miR-203 targeted SNAI2 (20), SNAI2 repressed miR-203 by directly binding to its promoter. The loss of miR-203 and gain of SNAI2 were associated with human breast cancer invasion and metastasis. Moreover, we demonstrated that repression of miR-203 was indispensable for EMT induced by TGF-β. The SNAI2 and miR-203 loop also controlled the expression of miR-200 and ZEB1/2. Given the fact that miR-200 and ZEB1/2 have been found to play a critical role in EMT, our results suggest that the miR-203 and SNAI2 double negative feedback loop is a critical regulator of EMT and tumor metastasis.
A recent study showed that miR-203 could regulate EMT by forming a feedback loop with SNAI1 in the breast cancer cell line HTB129 (46). Overexpression of miR-203 down-regulated SNAI1 and promoted epithelial-like properties. In contrast, we found that miR-203 formed a feedback loop with SNAI2 in TGF-β-induced EMT. SNAI2 was inversely associated with the expression of miR-203. This difference may be cell context-dependent. Nevertheless, these results suggest that miR-203 plays an important role in maintaining the epithelial properties of the cells. Interestingly, miR-203 was found to be significantly down-regulated in the mesenchymal component of endometrial carcinosarcoma, which represents a bona fide example of EMT in vivo (47), and in claudin-low type breast cancer, which contains a majority of mesenchymal-like cells (48). Previously, the miR-200 family was found to inhibit EMT by targeting ZEB1 and ZEB2, suggesting that the miR-200 family has therapeutic value. On the contrary, Korpal et al. (33) recently found that the miR-200 family promoted metastatic colonization by inducing E-cadherin and targeting the Sec23a-mediated tumor cell secretome using the tail vein injection model. Their findings cast doubts on the potential therapeutic utility of miR-200s (33). Using the same model, we demonstrated that the restoration of miR-203 in human breast cancer cells could inhibit metastasis. Although both miR-200s and miR-203 inhibit EMT, our results suggest that miR-203 may be a more attractive target for tumor metastasis. Although EMT is required for the early steps of tumor metastasis, mesenchymal-to-epithelial transition plays a critical role in metastatic tumor growth (48). Therefore, miR-203 expression should be restored in metastatic tumors. However, analysis of publicly available breast tumor data sets revealed that miR-203 was significantly decreased in human metastatic breast tumors. Currently, we can provide an explanation for this inconsistency. It is possible that miR-203 may not be required for mesenchymal-to-epithelial transition during metastatic tumor growth. As shown in our microarray, miR-203 might target heparin-binding EGF, a potent mitogen for tumor growth (Fig. 2A). Wang et al. (34) showed that miR-203 inhibited baculoviral IAP repeat-containing protein 5 (BIRC5) to reduce cancer proliferation. Therefore, the down-regulation of miR-203 is important for metastatic tumor growth.
The inhibition of CDH1 by SNAI2 plays a critical role in EMT and tumor metastasis. However, repression of CDH1 could not fully explain tumor invasive growth and metastasis induced by SNAI2. Our studies demonstrated that the inhibition of miR-203 by SNAI2 is also critical for developmental EMT, providing novel insights into SNAI2-induced EMT. Given the pleiotropic roles of miRNAs, the inhibition of miR-203 by SNAI2 could affect a cohort of genes associated with tumor invasive growth and metastasis. Thus, although SNAI2 is a transcriptional repressor, it could directly up-regulate gene expression by inhibiting miR-203, thereby promoting tumor metastasis. Previous reports showed that overexpression of SNAI1/2 up-regulates ZEB1 by unknown mechanisms (12). A recent study showed that SNAI2, but not SNAI1, induces ZEB1 expression by directly binding to the promoter of ZEB1 (49). We found that SNAI2 also induced ZEB1 and ZEB2. However, given the fact that SNAI2 is a transcriptional repressor, it is unlikely that SNAI2 directly induces ZEB1 transcription. Interestingly, we demonstrated that the miR-200 family members, which are negative regulators of ZEB1 and ZEB2, were directly repressed by SNAI2, further amplifying EMT. Therefore, an alternative mechanism was that SNAI2 indirectly up-regulated ZEB1 and ZEB2 by repressing the miR-200 family. Moreover, we found that the restoration of miR-203 suppressed the down-regulation of miR-200 and up-regulation of ZEB1 mediated by TGF-β. Our results suggest that the SNAI2 and miR-203 regulatory loop may be in concert with miR-200 and ZEB1/2 to form a feed-forward loop to regulate EMT and gene expression (18, 19, 41, 42). In summary, the SNAI2-miR-203 double negative feedback loop identified here provided new insights into the molecular mechanisms of EMT and tumor metastasis. This feedback loop holds promise as a therapeutic target for tumor invasion and metastasis.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA132134 and R37 DE013848 and R01 DE15964 from the NIDCR (to C.-Y. W.).

This article contains supplemental Methods and Figs. S1 and S2.
- EMT
- epithelial-mesenchymal transition
- miRNA
- microRNA
- MDCK
- Madin-Darby canine kidney
- EV
- empty vector
- EST
- expressed sequence tag
- TSS
- transcription start site
- VIM
- vimentin.
REFERENCES
- 1. Chaffer C. L., Weinberg R. A. (2011) A perspective on cancer cell metastasis. Science 331, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 2. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 3. Nieto M. A. (2011) The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 27, 347–376 [DOI] [PubMed] [Google Scholar]
- 4. Gumbiner B. M. (2005) Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6, 622–634 [DOI] [PubMed] [Google Scholar]
- 5. Perl A. K., Wilgenbus P., Dahl U., Semb H., Christofori G. (1998) A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 392, 190–193 [DOI] [PubMed] [Google Scholar]
- 6. Onder T. T., Gupta P. B., Mani S. A., Yang J., Lander E. S., Weinberg R. A. (2008) Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 [DOI] [PubMed] [Google Scholar]
- 7. Derksen P. W., Liu X., Saridin F., van der Gulden H., Zevenhoven J., Evers B., van Beijnum J. R., Griffioen A. W., Vink J., Krimpenfort P., Peterse J. L., Cardiff R. D., Berns A., Jonkers J. (2006) Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 [DOI] [PubMed] [Google Scholar]
- 8. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 9. Pérez-Caro M., Bermejo-Rodríguez C., González-Herrero I., Sánchez-Beato M., Piris M. A., Sánchez-García I. (2008) Transcriptomal profiling of the cellular response to DNA damage mediated by Slug (Snai2). Br. J. Cancer 98, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hajra K. M., Chen D. Y., Fearon E. R. (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 11. Martin T. A., Goyal A., Watkins G., Jiang W. G. (2005) Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann. Surg. Oncol. 12, 488–496 [DOI] [PubMed] [Google Scholar]
- 12. Guaita S., Puig I., Franci C., Garrido M., Dominguez D., Batlle E., Sancho E., Dedhar S., De Herreros A. G., Baulida J. (2002) Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 277, 39209–39216 [DOI] [PubMed] [Google Scholar]
- 13. Bruna A., Darken R. S., Rojo F., Ocaña A., Peñuelas S., Arias A., Paris R., Tortosa A., Mora J., Baselga J., Seoane J. (2007) High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell 11, 147–160 [DOI] [PubMed] [Google Scholar]
- 14. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 15. Nicoloso M. S., Spizzo R., Shimizu M., Rossi S., Calin G. A. (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat. Rev. Cancer 9, 293–302 [DOI] [PubMed] [Google Scholar]
- 16. Tavazoie S. F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P. D., Gerald W. L., Massagué J. (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma L., Teruya-Feldstein J., Weinberg R. A. (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 [DOI] [PubMed] [Google Scholar]
- 18. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 19. Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Z., Zhang B., Li W., Fu L., Fu L., Zhu Z., Dong J. T. (2011) Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2, 782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramadoss S., Li J., Ding X., Al Hezaimi K., Wang C. Y. (2011) Transducin β-like protein 1 recruits nuclear factor κB to the target gene promoter for transcriptional activation. Mol. Cell. Biol. 31, 924–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park B. K., Zhang H., Zeng Q., Dai J., Keller E. T., Giordano T., Gu K., Shah V., Pei L., Zarbo R. J., McCauley L., Shi S., Chen S., Wang C. Y. (2007) NF-κB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat. Med. 13, 62–69 [DOI] [PubMed] [Google Scholar]
- 23. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 24. Volinia S., Galasso M., Costinean S., Tagliavini L., Gamberoni G., Drusco A., Marchesini J., Mascellani N., Sana M. E., Abu Jarour R., Desponts C., Teitell M., Baffa R., Aqeilan R., Iorio M. V., Taccioli C., Garzon R., Di Leva G., Fabbri M., Catozzi M., Previati M., Ambs S., Palumbo T., Garofalo M., Veronese A., Bottoni A., Gasparini P., Harris C. C., Visone R., Pekarsky Y., de la Chapelle A., Bloomston M., Dillhoff M., Rassenti L. Z., Kipps T. J., Huebner K., Pichiorri F., Lenze D., Cairo S., Buendia M. A., Pineau P., Dejean A., Zanesi N., Rossi S., Calin G. A., Liu C. G., Palatini J., Negrini M., Vecchione A., Rosenberg A., Croce C. M. (2010) Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 20, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X. (2008) miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 14, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shankavaram U. T., Reinhold W. C., Nishizuka S., Major S., Morita D., Chary K. K., Reimers M. A., Scherf U., Kahn A., Dolginow D., Cossman J., Kaldjian E. P., Scudiero D. A., Petricoin E., Liotta L., Lee J. K., Weinstein J. N. (2007) Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol. Cancer Ther. 6, 820–832 [DOI] [PubMed] [Google Scholar]
- 28. Blower P. E., Verducci J. S., Lin S., Zhou J., Chung J. H., Dai Z., Liu C. G., Reinhold W., Lorenzi P. L., Kaldjian E. P., Croce C. M., Weinstein J. N., Sadee W. (2007) MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol. Cancer Ther. 6, 1483–1491 [DOI] [PubMed] [Google Scholar]
- 29. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 30. Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., Gerald W. L. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saini S., Majid S., Yamamura S., Tabatabai L., Suh S. O., Shahryari V., Chen Y., Deng G., Tanaka Y., Dahiya R. (2011) Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin. Cancer Res. 17, 5287–5298 [DOI] [PubMed] [Google Scholar]
- 32. Shih J. Y., Tsai M. F., Chang T. H., Chang Y. L., Yuan A., Yu C. J., Lin S. B., Liou G. Y., Lee M. L., Chen J. J., Hong T. M., Yang S. C., Su J. L., Lee Y. C., Yang P. C. (2005) Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin. Cancer Res. 11, 8070–8078 [DOI] [PubMed] [Google Scholar]
- 33. Korpal M., Ell B. J., Buffa F. M., Ibrahim T., Blanco M. A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., Wei Y., Hu G., Garcia B. A., Ragoussis J., Amadori D., Harris A. L., Kang Y. (2011) Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang C., Zheng X., Shen C., Shi Y. (2012) MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 31, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takeshita N., Mori M., Kano M., Hoshino I., Akutsu Y., Hanari N., Yoneyama Y., Ikeda N., Isozaki Y., Maruyama T., Akanuma N., Miyazawa Y., Matsubara H. (2012) miR-203 inhibits the migration and invasion of esophageal squamous cell carcinoma by regulating LASP1. Int. J. Oncol 41, 1653–1661 [DOI] [PubMed] [Google Scholar]
- 36. Karolchik D., Baertsch R., Diekhans M., Furey T. S., Hinrichs A., Lu Y. T., Roskin K. M., Schwartz M., Sugnet C. W., Thomas D. J., Weber R. J., Haussler D., Kent W. J. (2003) The UCSC Genome Browser Database. Nucleic Acids Res. 31, 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Down T. A., Hubbard T. J. (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 12, 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rice P., Longden I., Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 40. Shalgi R., Brosh R., Oren M., Pilpel Y., Rotter V. (2009) Coupling transcriptional and post-transcriptional miRNA regulation in the control of cell fate. Aging 1, 762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 43. Korpal M., Lee E. S., Hu G., Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y. N., Yin J. J., Abou-Kheir W., Hynes P. G., Casey O. M., Fang L., Yi M., Stephens R. M., Seng V., Sheppard-Tillman H., Martin P., Kelly K. (2013) MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32, 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deleted in proof
- 46. Moes M., Le Béchec A., Crespo I., Laurini C., Halavatyi A., Vetter G., Del Sol A., Friederich E. (2012) A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS One 7, e35440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castilla M. Á., Moreno-Bueno G., Romero-Pérez L., Van De Vijver K., Biscuola M., López-García M. Á., Prat J., Matías-Guiu X., Cano A., Oliva E., Palacios J. (2011) Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J. Pathol. 223, 72–80 [DOI] [PubMed] [Google Scholar]
- 48. Herschkowitz J. I., Zhao W., Zhang M., Usary J., Murrow G., Edwards D., Knezevic J., Greene S. B., Darr D., Troester M. A., Hilsenbeck S. G., Medina D., Perou C. M., Rosen J. M. (2012) Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2778–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wels C., Joshi S., Koefinger P., Bergler H., Schaider H. (2011) Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 131, 1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]