Background: The mechanism of E6AP has been previously defined by the structure of a substrate-bound intermediate.
Results: Rate studies of polyubiquitin chain formation preclude the canonical model based on the crystal structure.
Conclusion: Kinetics define a mechanism requiring two functionally distinct E2∼ubiquitin thioester binding sites.
Significance: A general mechanism for Hect ligase polyubiquitin chain formation is defined for E6AP based on empirical rate measurements.
Keywords: Enzyme Kinetics, Enzyme Mechanisms, Ubiquitin, Ubiquitin Conjugating Enzyme (Ubc), Ubiquitin Ligase, E6AP, UBE3A, Polyubiquitin Chain
Abstract
By exploiting 125I-polyubiquitin chain formation as a functional readout of enzyme activity, we have quantitatively examined the mechanism of human E6AP/UBE3A for the first time. Initial rate studies identify UbcH7 as the cognate E2 carrier protein for E6AP, although related Ubc5 isoforms and the ISG15-specific UbcH8 paralog also support E6AP with reduced efficacy due to impaired binding and catalytic competence. Initial rates of polyubiquitin chain formation displayed hyperbolic kinetics with respect to UbcH7 concentration (Km = 57.6 ± 5.7 nm and kcat = 0.032 ± 0.001 s−1) and substrate inhibition above 2 μm. Competitive inhibition by an isosteric UbcH7C86S-ubiquitin oxyester substrate analog (Ki = 64 ± 18 nm) demonstrates that Km reflects intrinsic substrate affinity. In contrast, noncompetitive inhibition by a UbcH7C86A product analog (Ki = 7 ± 0.7 μm) and substrate inhibition at high concentrations require two functionally distinct E2∼ubiquitin substrate binding sites. The kinetics of polyubiquitin chain formation reflect binding at a cryptic Site 1 not previously recognized that catalyzes E6AP∼ubiquitin thioester formation. Subsequent binding of E2∼ubiquitin at the canonical Site 2 present in the extant crystal structure is responsible for polyubiquitin chain elongation. Other rate studies show that the conserved −4 Phe849 residue is required for polyubiquitin chain formation rather than target protein conjugation as originally suggested. The present studies unambiguously preclude earlier models for the mechanism of Hect domain-catalyzed conjugation through the canonical binding site suggested by the crystal structure and define a novel two-step mechanism for formation of the polyubiquitin degradation signal.
Introduction
The Hect2 (homologous to E6AP carboxyl terminus) domain ligases constitute a family of ∼100-kDa enzymes catalyzing the attachment of ubiquitin to specific target proteins as a signal for degradation by the 26 S proteasome or for directing target protein cellular localization and function, most recently reviewed by Rotin and Kumar (1). Ubiquitin conjugation proceeds through a canonical three-step pathway comprising ubiquitin activation by the Uba1 ubiquitin activating enzyme, transfer of the high energy Uba1∼ubiquitin thioester to a specific E2/Ubc3 carrier protein to form an E2∼ubiquitin thioester, and subsequent conjugation of the E2-bound ubiquitin to the target protein by ubiquitin ligase (E3)-catalyzed aminolytic cleavage of the ubiquitin carboxyl-terminal thioester bond (1, 2). The Hect domain ligases are distinguished from the larger superfamily of Ring-type ligases in forming an obligatory ubiquitin thioester to a conserved active site cysteine present in the catalytic domain as the immediate donor of activated ubiquitin for isopeptide bond formation (1). In humans, the Hect ligase family comprises 29 genes whose products occupy key regulatory roles within cells and share similar domain architectures comprised of a highly conserved 350-residue carboxyl-terminal Hect domain responsible for cognate E2∼ubiquitin thioester binding and subsequent target protein conjugation; in addition, one or more amino-terminal domains recruit target protein substrates for conjugation by the Hect catalytic domain (1, 3).
In addition to the intrinsic targeting mechanism encoded into the amino-terminal sequences of the Hect ligases, accumulating evidence indicates that adapter proteins are frequently employed to recruit substrates to the ligation machinery (4, 5). Viruses exploit the latter feature of target protein selection as integral steps in their replication cycle to enhance viral particle yield, reviewed by Isaacson and Ploegh (6). In the human papillomavirus virus type 8/16 replication cycle, viral E6 protein recruits host p53 to the Hect ligase E6AP/UBE3A for conjugation and targeted degradation by the 26 S proteasome as one step in cervical cell transformation (4, 7). Viral E6 protein recruits additional substrates to the E6AP ligase, including NFX1 in the activation of telomerase (8, 9) and the ADA3 transcriptional co-activator of GCN5 histone acetyltransferase (10). Hepatitis C virus NS5B protein serves a similar role in targeting retinoblastoma tumor suppressor protein for degradation, increasing risk for cirrhosis and hepatocellular carcinoma (5).
E6AP is the founding member of the Hect domain ligases (11) and has been identified as the Angelman syndrome gene, loss-of-function deletions/mutations of which result in an inherited neuropathy with a frequency of 1 in 10,000–20,000 births that is responsible for severe mental retardation, absence of speech, ataxia, and an unusually happy demeanor (12–15). UBE3A resides within the chromosome 15q11-13 region and is subject to genomic imprinting that results in a maternal-specific expression pattern in brain (16, 17). UBE3A is also a strong candidate for the autism phenotype based on imprint status and its causative role in Angelman syndrome in those instances of gain-of-function maternal-specific interstitial duplication in the 15q11-13 region (18, 19). The neuronal targets of E6AP have been identified as Arc, which regulates endocytic trafficking of the neuronal AMPA receptor, and Ephexin5, which is a RhoA guanine nucleotide exchange factor regulating excitatory synapse development, both of which are required for neuronal plasticity and long term memory (20–22). Recruitment of E6AP to aggresomes, degradation of polyglutamine expansion proteins, and induction by various stress mechanisms suggest the ligase serves additional functions as part of the cellular protein quality control response (23, 24). The E6AP ligase also serves regulatory functions as a dual function co-activator of steroid hormone receptors through targeted degradation of transcriptional complexes, reviewed in Ref. 25.
The crystal structure for the Hect domain of E6AP reveals two lobes arranged in an L shape with an amino-terminal lobe that can be subdivided into large and small subdomains, the latter of which binds E2 in a defined pocket, connected by a hinge peptide to a carboxyl-terminal lobe harboring the active site Cys820 to which the E2-bound ubiquitin thioester is transferred (26). The conventional mechanism of Hect ligase conjugation posits binding of E2∼ubiquitin thioester to the small amino-terminal subdomain, based on the crystal structure, then transfer of its activated polypeptide to the active site Hect domain cysteine to form an E3∼ubiquitin thioester, which serves as the immediate donor for target protein conjugation and subsequent sequential polyubiquitin chain elongation (26, 27). However, the structure of UbcH7 bound to the small amino-terminal subdomain of E6AP poses problems in accounting for the mechanism of Cys820 thioester formation from the bound E2-ubiquitin thioester, as the latter is separated from Cys820 by 41 Å, whereas thioester transfer requires the donor and acceptor sulfurs within the two sites to be within atomic distance for nucleophilic attack (26). Structures for the WWP1/AIP5 (28), Smurf2 (29), Nedd4–1 (30), Nedd4–2 (31), and HuWE1 (32) Hect domains reveal differences in orientation for the amino- and carboxyl-terminal lobes that suggest potential conformational changes during the catalytic cycle may close some of the distance required for nucleophilic attack (31) as mutation of residues in the hinge region of WWP1 that restricts mobility of the carboxyl-terminal domain also ablates ligase activity (28). A recent structure for a stable UbcH5B-ubiquitin oxyester bound to Nedd4–2/Nedd4L suggests a hypothetical structure for such a Michaelis complex; however, the sulfur atoms remain separated by 8 Å (31). Finally, early single-turnover studies of Hect domain function suggest that ligases may employ different mechanisms for polyubiquitin chain synthesis in which initial transfer of activated ubiquitin from the E2∼ubiquitin thioester to the active site Hect domain cysteine is followed by conjugation to the target protein and sequential elongation of the polyubiquitin chain in the case of KIAA10 or assembly of the polyubiquitin chain as an E6AP Cys820-linked thioester before its en bloc transfer to the target protein lysine (33).
In the present study we have exploited the intrinsic ability of E6AP to form free polyubiquitin chains in the absence of substrate (34) as a reporter function to study the core catalytic behavior of the enzyme (35). Kinetic analysis of free chain formation in biochemically defined functional assays has unambiguously defined the E2 specificity of the ligase. Unexpectedly, these results reveal for the first time that the E6AP Hect domain harbors a second cryptic E2∼ubiquitin thioester binding site that is directly required for formation of the Hect domain Cys820∼ubiquitin thioester intermediate, potentially resolving the long-standing question of active site geometry and E2-E3 transthiolation mechanism (26). Other observations suggest that the ubiquitin moiety of the E2∼ubiquitin thioester contributes significantly to overall binding affinity of the charged intermediate compared with uncharged E2 at the non-canonical binding site. These studies provide the first quantitative functional insights into the mechanistic details of the E6AP catalytic cycle and of interactions between the ligase and its E2-ubiquitin thioester substrate. Structural conservation among the Hect ligases in their catalytic domains suggest these mechanistic characteristics are shared by other members of the ligase superfamily.
MATERIALS AND METHODS
Bovine ubiquitin and creatine phosphokinase were purchased from Sigma. Ubiquitin was further purified to apparent homogeneity by FPLC chromatography and quantitated spectrophotometrically (36). Ubiquitin was radioiodinated by the chloramine-T procedure to yield specific radioactivities of ∼15,000 cpm/pmol using carrier free Na125I purchased from either GE Healthcare or PerkinElmer Life Sciences (37). Wild type Lys48-linked tetra-ubiquitin was purchased from Boston Biochemicals, a portion of which was reductively methylated as previously described by Hershko and Heller (38). Human erythrocyte Uba1 was purified to apparent homogeneity from outdated human blood (37). Active Uba1 was quantitated by the stoichiometric formation of 125I-ubiquitin thioester (39, 40). The amino-terminal acetylated Ac-AKGFGML peptide corresponding to the carboxyl-terminal heptapeptide of E6AP was synthesized in the Protein and Nucleic Acid Core Facility of Medical College of Wisconsin (Milwaukee, WI) and purified by HPLC. Peptide purity was confirmed by mass spectrometry, and peptide concentration was determined spectrophotometrically at 257.5 nm.
Generation and Purification of Recombinant E2 Paralogs
Human recombinant E2 proteins Ubc2b (UBE2B), Ubc5A (UBE2D1), Ubc5B (UBE2D2), Ubc5C (UBE2D3), UbcH6 (UBE2E1), UbE2E2 (UBE2E2), UbcM2 (UBE2E3), UbcH7 (EBE2L3), and UbcH8 (UBE2L6) were those described previously (41). The UbcH7C86A, UbcH7C86S, UbcH7F63A, UbcH7K100A, and UbcH8C86A point mutants were generated from pGEX4T1-HsUbcH7 or pGEX4T3-HsUbcH8, respectively, using the QuikChange mutagenesis kit (Stratagene). An analogous strategy was used to produce Ubc5AC85A from pGEX4T1-HsUbc5A. Coding regions for all clones were sequenced to preclude cloning artifacts and, where relevant, to ensure the correct point mutation. Recombinant wild type and mutant proteins were expressed in Escherichia coli BL21 (DE3) cells harboring the desired pGEX-E2 plasmid, purified to apparent homogeneity, and processed with thrombin to remove the GST tag as described (41). Active E2 concentrations were quantitated by the Uba1-dependent stoichiometric formation of 125I-ubiquitin thioester or, in the case of UbcH7C86S, the corresponding 125I-ubiquitin oxyester and compared with total E2 protein determined spectrophotometrically using their calculated 280-nm extinction coefficients (42). Active E2 protein relative to total protein varied in a consistent paralog-specific manner from ∼90% (Ubc2b) to ∼20% (wild type and C86S mutant of UbcH7). Because UbcH7C86A, UbcH8C86A, and Ubc5AC85A proteins are inactive and cannot be quantitated by 125I-ubiquitin thioester formation, the concentration of “active” protein was calculated from the corresponding active fraction of total protein found for wild type E2 under the assumption that the mutation does not alter wild type protein stability. The E2 proteins were stored at −80 °C in small aliquots and were stable for greater than 6 months, although they were subject to differential rates of activity loss with successive freeze-thaw cycles (35, 42).
Generation and Purification of Recombinant E6AP
Human E6AP isoform 3 (UBE3A; IMAGE clone NM00046.2) was subcloned into the BamH1/Not1 sites of pGEX4T1 to yield pGEX4T1-E6AP. The E6APF849A and E6APF849Y point mutants were generated from pGEX4T1-E6AP using the QuikChange protocol of Stratagene to yield pGEX4T1-E6APF849A and pGEX4T1-E6APF849Y, respectively.4 The E6APΔ847 truncation lacking the carboxyl-terminal pentapeptide of wild type enzyme was generated by inserting a STOP codon after codon 847 of pGEX4T1-E6AP to yield pGEX4T1-E6APΔ847. The coding regions for all E6AP clones were sequenced to preclude cloning artifacts and to verify the desired mutation. E. coli BL21 (DE3) cells harboring pGEX-E6AP plasmids were grown at 37 °C then induced at A600 of 0.6 by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm. After 3 h at 37 °C, cells were harvested by centrifugation at 6000 × g for 15 min then resuspended in 50 mm Tris-HCl (pH 7.5) containing 1 mm DTT (42). Cells were lysed by Emulsiflex (Avestin) then centrifuged at 30,000 × g for 30 min (42). The GST-E6AP fusion proteins were purified from the resulting supernatant by glutathione-Sepharose affinity chromatography (42). The GST moiety was not processed from recombinant E6AP fusion protein as the GST moiety enhanced the stability of the enzyme, and processing typically resulted in a 15–30% decrease in activity. Otherwise, processed and unprocessed E6AP appeared identical in their activities. The purification protocol consistently yielded 3–4 mg of GST-E6AP protein per liter of medium. Resolution by SDS-PAGE and detection by Coomassie staining versus Western blotting with anti-GST antibody revealed a series of GST-associated bands ranging from a relative molecular mass of ∼130 kDa for full-length protein to 25 kDa representing the free GST moiety; however, only the band of highest relative molecular weight, corresponding to full-length GST-E6AP, formed a 125I-ubiquitin thioester (not shown). The activities of GST-E6AP and its mutants were quantitated by stoichiometric 125I-ubiquitin thioester formation (42) and compared with protein of the full-length band, estimated densitometrically using BSA as a protein standard. Typically the E6AP preparations exhibited ∼1% active enzyme based on total full-length protein.
E6AP-catalyzed 125I-ubiquitin Conjugation Assay
The E3 ligase activity of recombinant E6AP5 was quantitated in kinetic assays under initial velocity conditions (35). Rates of E6AP-catalyzed 125I-polyubiquitin chain formation were measured at 37 °C in incubations of 25 μl final volume containing 50 mm Tris-HCl (pH 7.5), 1 mm ATP, 10 mm MgCl2, 1 mm DTT, 10 mm creatine phosphate, 1 IU of creatine phosphokinase, 5 μm 125I-ubiquitin (∼1.5 × 104 cpm/pmol), 50 nm human Uba1, and the indicated concentrations of E2 and E6AP (42, 43). Reactions were initiated by the addition of 125I-ubiquitin. After 10 min the reactions were quenched by the addition of 25 μl of 2× SDS sample buffer containing 0.3% (v/v) β-mercaptoethanol, then the samples were heated to 100 °C for 5 min. The polyubiquitin conjugates were resolved from free 125I-ubiquitin by 12% SDS-PAGE under reducing conditions at 4 °C and visualized by autoradiography of the dried gels (42, 43). Polyubiquitin chain formation was measured by excising lanes and quantitating associated 125I-ubiquitin by γ-counting (42, 43). Active Uba1, E2, and E6AP were independently determined in parallel by their stoichiometric formation of 125I-ubiquitin thioester (42). Hyperbolic kinetics were verified by the linearity of corresponding double reciprocal plots; however, kinetic values were calculated by nonlinear regression analysis using GraFit 5.0 (Erithacus Software Ltd.) (35). Replicate determinations of the kinetic values agreed within 10–20%.
RESULTS
UbcH7 Is the Cognate E2 for E6AP
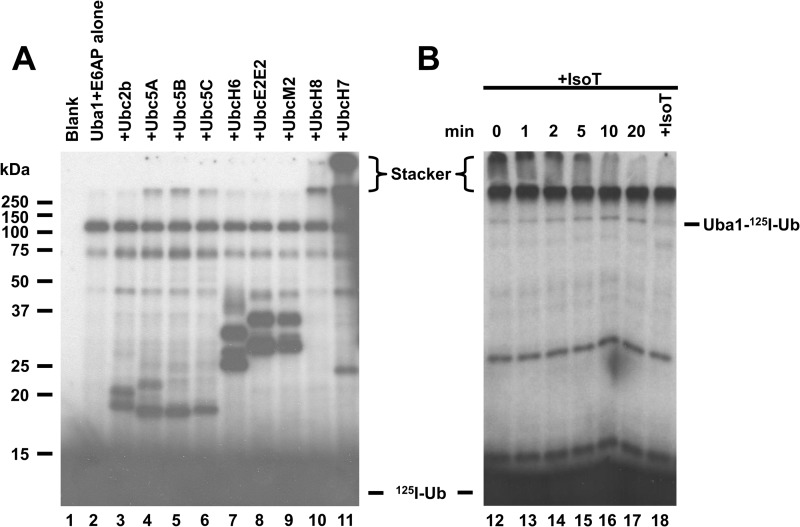
The E6AP ubiquitin ligase was initially suggested to function with members of the Ubc5 family of E2 ubiquitin conjugating enzymes (44); however, subsequent work proposed that E6AP activity is supported by several distinct E2 families, including UbcH6, UbcH7, and UbcH8 (45, 46). All four E2 families belong to the Ubc4/5 superclade of E2 paralogs (47), indicating that the apparent broad specificity is a function of conserved surface features among these distinct E2 species. Because E3 ligases typically function with only a single E2 family, we chose to resolve the question of E2 specificity through the functional screen shown in the autoradiogram of Fig. 1. The E3 functional assay relies on the ability of ligases to form 125I-ubiquitin chains in the absence of protein substrate (34, 35), shown by the accumulation of label at the top of the stacker and running gels, Fig. 1A. Each incubation was conducted in the absence or presence of 100 nm concentrations of the active E2 (panel A). The screen differs from similar studies reported previously by others in that the active E2 concentrations were determined in parallel by a functional stoichiometric assay based on the end point formation of a E2-125I-ubiquitin thioester rather than calculated from total protein (40), the latter of which leads to overestimations of E2 concentrations due to the lability of many of the Ubc families to spontaneous denaturation and active site cysteine oxidation, as discussed previously (35, 41). No bands were found in the presence of 125I-ubiquitin alone (Fig. 1, lane 1). In the absence of E2, a prominent band of mono-ubiquitinated Uba1 ubiquitin activating enzyme was observed at 120 kDa that results from partitioning of a fraction of the Uba1-125I-ubiquitin thioester to reaction with active site nucleophile(s) on the enzyme in addition to lower molecular weight adducts to Uba1 fragments (Fig. 1, lane 2). Several of the E2 paralogs showed mono- and di-autoubiquitination products below 50 kDa in the presence of E6AP (Fig. 1A) that also form in the absence of E6AP (not shown). Significant E6AP-catalyzed polyubiquitin chain formation was only observed in the presence of UbcH7 (Fig. 1, lane 11), whereas lower levels of chain formation were supported by equivalent concentrations of UbcH8 (lane 10) and each of the three Ubc5 isoforms (lanes 4–6). At longer exposure times, a low level of chain formation can be seen in the presence of UbcH6 (lane 7). Because the experimental conditions for the incubations and assay time were empirically set to be E6AP-limiting and within the initial velocity region for UbcH7-dependent polyubiquitin chain formation, the relative autoradiographic intensity with each E2 paralog is proportional to the initial velocity of the reporter reaction. Therefore, these results confirm that UbcH7 is the cognate E2 for E6AP, although other members of the Ubc4/5 superclade support the E6AP reaction at much lower rates.
FIGURE 1.
UbcH7 is the cognate E2 for E6AP. A, 125I-ubiquitin conjugation assays containing 16 nm GST-E6AP were conducted for 10 min in the absence (lane 2) or presence of 100 nm concentrations of the indicated E2 (lanes 3–11) then quenched with SDS sample buffer and resolved by 12% (w/v) SDS-PAGE under reducing conditions as described under “Materials and Methods.” The resulting gel was dried, and 125I-ubiquitin conjugates were visualized by autoradiography. B, incubation identical to that of lane 11 was depleted of ATP by the addition of 6 IU apyrase (0 min) then recombinant isopeptidase T (+IsoT) was added to a final concentration of 17 nm. Aliquots were taken at the indicated times and quenched with SDS sample buffer. Additional isopeptidase T was added at 20 min to a final concentration of 34 nm, then the reaction was allowed to proceed for an additional 20 min before being quenched with SDS sample buffer. Samples were resolved and visualized as in panel A. Mobility of molecular weight markers are shown to the left. Positions of the monoubiquitinated activating enzyme (Uba1-125I-Ub) and free ubiquitin (125I-Ub) are as indicated. The position of the 5% (w/v) stacker is shown by brackets.
That the autoradiographic density at the top of the stacker gel represents free polyubiquitin chains is demonstrated by the time-dependent loss of this signal on addition of Isopeptidase T, a ubiquitin-specific protease that sequentially disassembles free but not conjugated polyubiquitin chains from the proximal (free carboxyl-terminal) end to release free ubiquitin (48, 49) Fig. 1 ( panel B). Because the autoradiographic intensity at the top of the running gel is not similarly affected by isopeptidase T, these chains must be directly conjugated to protein (35). When a parallel conjugation reaction with UbcH7 was subsequently incubated with anti-E6AP antibody, nearly all of 125I radioactivity present at the top of the running gel, but not at the top of the spacer gel, was immunoprecipitated on the addition of Protein A-Sepharose (not shown), indicating partitioning of the growing polyubiquitin chain to autoubiquitination of the ligase.
UbcH7 Exhibits Hyperbolic Kinetics for E6AP-catalyzed Polyubiquitin Chain Formation
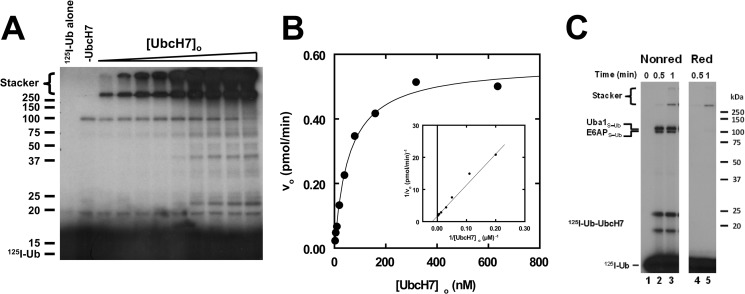
We have previously shown that the initial rate for E3-catalyzed 125I-ubiquitin conjugation can be used as a facile reporter activity for kinetic characterization of ligases (42, 43, 50). In similar kinetic studies, we examined the dependence of the initial rate for E6AP-catalyzed 125I-ubiquitin chain formation on [UbcH7]o (Fig. 2). The resulting autoradiogram shows increasing amounts of 125I-ubiquitin conjugate formation as [UbcH7]o is increased from 2 to 650 nm (Fig. 2, panel A). To quantify the absolute amount of total 125I-ubiquitin conjugated, lanes were excised above 25 kDa, and associated radioactivity was determined by γ-counting. Absolute 125I-ubiquitin conjugated was then calculated using the specific radioactivity of the labeled polypeptide (42, 43, 51). Separate control assays were conducted to ensure that the incubation time was under initial velocity conditions and that the rates were [E6AP]o-limiting, indicated by the independence of the initial rate on [Uba1]o, not shown (52).
FIGURE 2.
UbcH7 exhibits hyperbolic kinetics for E6AP-catalyzed polyubiquitin chain formation. A, shown is a representative autoradiogram of E6AP-catalyzed polyubiquitin chain formation in which 125I-ubiquitin conjugation assays containing 30 nm GST-E6AP were conducted under initial velocity conditions in the absence (−UbcH7) or presence of 2–650 nm UbcH7 as described under “Materials and Methods.” After autoradiography, 125I-ubiquitin conjugates larger than 25 kDa were excised and quantified by γ-counting, then the absolute rate was calculated using the specific radioactivity of the labeled ubiquitin (42, 43). B, shown is the concentration dependence of initial velocity versus [UbcH7]o from the assay shown in panel A. The solid line represents the nonlinear hyperbolic regression fit of the data for kinetic constants listed in Table 1. The inset shows the double reciprocal plot of the data. C, shown is the time course for 125I-ubiquitin thioester formation resolved by non-reducing (Nonred) and reducing (Red) conditions as described under “Materials and Methods” for assays containing 38 nm Uba1, 128 nm UbcH7, and 12 nm E6AP. Mobility of molecular weight markers (kDa) are shown to the left (panel A) or right (panel C). For panel C, positions of the various thioester are shown to the left. The UbcH7-125I-ubiquitin thioester migrates as two bands due to an artifact of incomplete denaturation under non-reducing conditions.
The initial rate of E6AP-catalyzed 125I-ubiquitin conjugation with respect to [UbcH7]o exhibits hyperbolic kinetics (Fig. 2B), as demonstrated by the linearity of the corresponding double reciprocal plot (inset); however, at a concentration above 2 μm, the initial rates of conjugation exhibit substrate inhibition that tends to zero rate at higher concentration (not shown). Nonlinear regression analysis of the data within the hyperbolic region resulted in a Km for UbcH7 of 58 ± 6 nm and a kcat of 0.031 ± 0.009 s−1, the latter defined as Vmax/[E6AP]o, where active E6AP concentration was determined independently by its stoichiometric formation of 125I-ubiquitin thioester (34). The Km for UbcH7 compares favorably with the Km for Ubc2b of 54 ± 18 nm in supporting human E3α/Ubr1 in the N-end Rule pathway (42). Because the corresponding UbcH7∼ubiquitin thioester represents the actual substrate for E6AP-catalyzed polyubiquitin chain formation, the Km reflects the affinity for binding of the cognate thioester intermediate to the ligase; therefore, the observed parity in affinities across distinct targeting pathways suggests a common functional requirement for high substrate affinity.
The kcat determined here is unlikely to reflect the rate of Hect domain Cys820 thioester formation during E6AP-catalyzed UbcH7-ubiquitin transthiolation. When the components of the incubations are assayed under non-reducing conditions, E6AP∼ubiquitin thioester formation reaches the end point within 30 s (Fig. 2C). Because we can easily distinguish differences in 125I-associated radioactivity in the E6AP∼ubiquitin thioester band of <0.2%, representing ∼900 cpm, this sets a lower limit for the first order rate constant for Cys820∼ubiquitin thioester formation of 3.5 s−1, 100-fold greater than the kcat for polyubiquitin chain formation calculated from Vmax (Fig. 2B) but in good agreement with a value of 1 s−1 estimated independently by Purbeck et al. (53).
Parallel kinetic studies were conducted with UbcH8 and Ubc5A to quantify their observed differences in catalytic efficiency suggested by the semiquantitative data of Fig. 1A. Both UbcH8 and Ubc5A also exhibited hyperbolic kinetics in E6AP-catalyzed polyubiquitin chain formation (not shown), from which values of Km and kcat could be determined by nonlinear regression analysis of the initial rate data, the results of which are summarized in Table 1. Human UbcH8∼125I-ubiquitin thioester exhibited lower affinity for binding to E6AP, indicated by the 6-fold higher Km compared with UbcH7 (ΔΔGbinding of 1.1 kcal/mol; Table 1). However, the similar kcat values for UbcH7∼ and UbcH8∼125I-ubiquitin thioesters suggest a common transition state geometry for polyubiquitin chain formation between the E2 paralogs, once bound to the ligase. In contrast, Ubc5A exhibited a similar Km to that of UbcH8 but a 10-fold lower kcat. The latter indicates comparable binding interactions for Ubc5A∼125I-ubiquitin thioester with the ligase but a loss of catalytic competence, presumably resulting from altered orientation of the bound intermediate in the transition state of the reaction. Overall, there is a 10-fold difference in catalytic specificity, defined by kcat/Km, for the cognate UbcH7 compared with UbcH8, and a 60-fold difference compared with Ubc5A, accounting for the semiquantitative results observed in Fig. 1A. In contrast, both Ubc5A and UbcH8 formed a Cys820∼ubiquitin thioester with kinetics similar to that shown by UbcH7 in Fig. 2C under the assay conditions, and rates of 125I-polyubiquitin chain formation were unaffected by increased [Uba1]o (not shown). Therefore, these quantitative data unambiguously demonstrate that UbcH7 is the cognate E2 for E6AP-catalyzed conjugation, supported by recent evidence that UbcH8 is the specific E2 for the conjugation of the interferon-induced ISG15 ubiquitin-like protein (54, 55).
TABLE 1.
Summary of kinetic constants
| Km | kcat | kcat/Km | Kda | |
|---|---|---|---|---|
| nm | s−1 | m−1s−1 | μm | |
| UbcH7 | 58 ± 6 | 0.031 ± 0.009 | 5.4 × 105 | 5.0 ± 0.5 |
| UbcH8 | 360 ± 100 | 0.021 ± 0.001 | 5.8 × 104 | 7.5 ± 0.7 |
| Ubc5A | 216 ± 83 | 0.0020 ± 0.0001 | 9.3 × 103 | 160 ± 30 |
| UbcH7F63A | NAb | NA | NA | 810 ± 550 |
| UbcH7K100A | 32 ± 18 | 0.0034 ± 0.0004 | 5.8 × 104 | 45 ± 15 |
a Fluorescence polarization binding data of Eletr and Kuhlman (56).
b Not active.
E6AP Harbors Two UbcH7∼ubiquitin Thioester Binding Sites
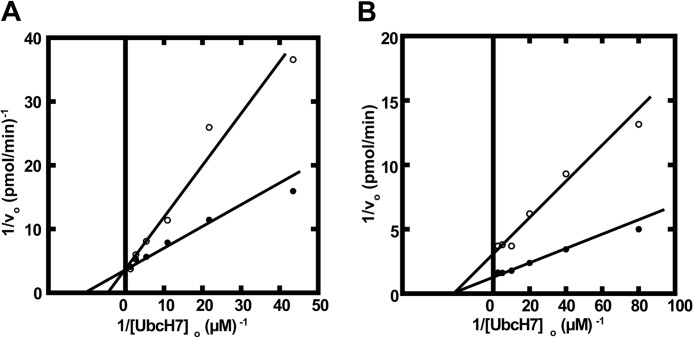
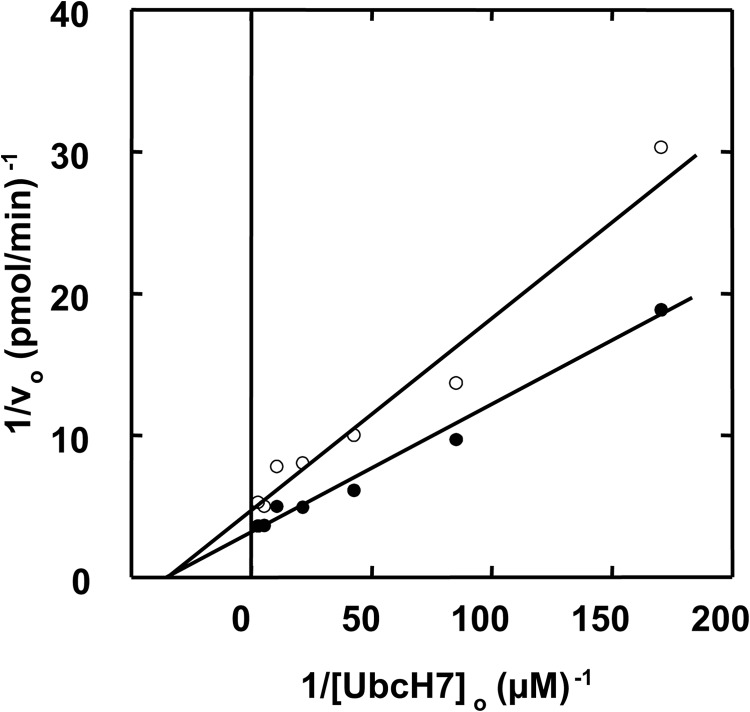
Previously Eletr and Kuhlman (56) employed fluorescence polarization to quantify the binding of various uncharged E2 paralogs to E6AP; however, the values of Kd from these equilibrium binding experiments are orders of magnitude larger than the Km values determined from the kinetic studies reported here; Table 1. Because the actual substrates for E3 ligases are the cognate E2∼ubiquitin thioesters rather than the uncharged carrier protein, the fluorescence polarization results of Eletr and Kuhlman must reflect enzyme-product binding. However, more recent studies by Purbeck et al. (53) using disulfide-linked UbcH7-UbG76C adducts to approximate the corresponding E2-ubiquitin thioester also demonstrate micromolar Kd values, suggesting that the ubiquitin moiety does not significantly contribute to substrate binding at the canonical small amino-terminal domain site. Earlier kinetic measurements with human E3α and Ubc2b demonstrated that the ligase followed rapid equilibrium kinetics in which the resulting Km values approximated the intrinsic Kd for binding of theUbc2b∼125I-ubiquitin thioester to the enzyme (42). To test this for E6AP, we generated a UbcH7C86S point mutant and used it to prepare UbcH7C86S-125I-ubiquitin oxyester as a stable, structurally orthologous analog of the corresponding UbcH7∼125I-ubiquitin thioester (42), as outlined under “Materials and Methods.” Such active site oxyesters can be formed at a reduced rate on the Cys → Ser active site E2 mutant by Uba1 but are incapable of supporting subsequent conjugation reactions catalyzed by ligases (42, 57, 58). When the kinetics of E6AP-catalyzed polyubiquitin chain formation are determined similar to that of Fig. 2 in the absence or presence of 400 nm FPLC-purified UbcH7C86S-125I-ubiquitin oxyester (42), the stable substrate analog exhibited competitive inhibition with respect to UbcH7, from which a Ki of 64 ± 18 nm could be calculated (Fig. 3A). Because the Ki for competitive inhibition represents an intrinsic Kd for binding of the inhibitor to the enzyme, the good agreement between the Ki and the Km for binding of the corresponding UbcH7∼ubiquitin thioester indicates that the enzyme follows rapid equilibrium kinetics for which the latter value is equivalent to the corresponding binding dissociation constant (Kd) (42). That the UbcH7-125I-ubiquitin oxyester is a competitive inhibitor of the corresponding UbcH7∼125I-ubiquitin thioester requires the two species bind at the same or overlapping sites; therefore, values of Km determined kinetically most reasonably represent binding of the cognate E2-ubiquitin thioester to the active site of E6AP.
FIGURE 3.
E6AP harbors two sites for UbcH7∼ubiquitin thioester binding. A, initial rates of 125I-ubiquitin conjugation were determined as in Fig. 2 under E6AP limiting conditions in assays containing 51 nm GST-E6AP and the indicated concentrations of wild type UbcH7 in the absence (closed circles) or presence (open circles) of 400 nm UbcH7C86S-125I-ubiquitin oxyester. B, the experiment of panel A was repeated with 22 nm GST-E6AP in the absence (close circles) or presence (open circles) of 8 μm UbcH7C86A.
We also generated a UbcH7C86A point mutant as a non-reactive product analog of the corresponding uncharged E2 (42). When the kinetics of E6AP-catalyzed polyubiquitin chain formation were determined similar to that of Fig. 3A but in the absence or presence of 8 μm UbcH7C86A (42), the non-reactive product analog exhibited noncompetitive inhibition with respect to wild type UbcH7, from which a Ki of 7 ± 0.7 μm could be calculated (Fig. 3B). As under these conditions the Ki is equivalent to the Kd for binding of UbcH7C86A to E6AP, the value of 7 ± 0.7 μm is in excellent agreement with the Kd of 5.0 ± 0.5 μm determined earlier by fluorescence polarization (56); Table 1. When the experiment of Fig. 3B was repeated with UbcH8C86A, noncompetitive inhibition with respect to wild type UbcH7 was again observed and yielded a Ki value of 2 ± 0.2 μm, in reasonable agreement with the Kd of 7.5 ± 0.7 μm determined by fluorescence polarization (56). Because the Ubc5A isoform also exhibited measurable activity in E6AP-catalyzed polyubiquitin chain formation (Fig. 1A and Table 1), we tested Ubc5AC85A and found it also to be a noncompetitive inhibitor with respect to wild type UbcH7, yielding a Ki of 3.6 ± 0.3 μm. Unlike the results with UbcH7C86A and UbcH8C86A, the Ki for Ubc5AC85A was significantly lower than the Kd of 160 ± 30 μm determined by fluorescence polarization (56); Table 1. Similar values of Ki were determined in independent kinetic assays using a different preparation of Ubc5AC85A. We have previously observed that the wild type Ubc5 isozymes are particularly labile to spontaneous inactivation on prolong storage or repeated freeze-thaw cycles compared with UbcH7 or UbcH8, as measured by end point 125I-ubiquitin thioester assays. Therefore, the significant difference between the Kd reported here as Ki based on the kinetic assay and the Kd determined previously by fluorescence polarization (56) probably reflects a much higher fraction of denatured protein in the earlier studies, leading to an overestimation of Kd.
The observation that UbcH7C86S-125I-ubiquitin oxyester and UbcH7C86A exhibit different inhibition patterns with respect to wild type UbcH7∼125I-ubiquitin thioester requires that the substrate and product analogs bind to different sites. Such a conclusion is inconsistent with the conventional model for Hect ligases in which E2∼ubiquitin thioester binds to the canonical site within the small amino-terminal subdomain before transferring the ubiquitin thioester to Cys820 (26). The good agreement between the Ki for UbcH7C86A determined kinetically and the Kd for wild type UbcH7 determined by fluorescence polarization (56) suggests that the two species likely bind to the same site, which mutation analysis assigns as the “canonical” binding site of the Hect domain (56), originally identified for UbcH7 interacting with the E6AP Hect domain (26). Binding through this site is strongly dependent on complementary interactions between the Hect domain and Loop 1/2 residues of the E2, chief among them Phe63 and Lys100 of UbcH7 (26, 56), respectively. The latter residues were individually mutated to alanine within human UbcH7. Both point mutants supported Uba1-catalyzed 125I-ubiquitin transthiolation (not shown); in addition, the resulting UbcH7∼125I-ubiquitin thioesters of both point mutants were capable of binding to and transferring their high energy intermediates to Cys820 of the Hect domain with kinetics qualitatively indistinguishable from wild type UbcH7 (not shown).
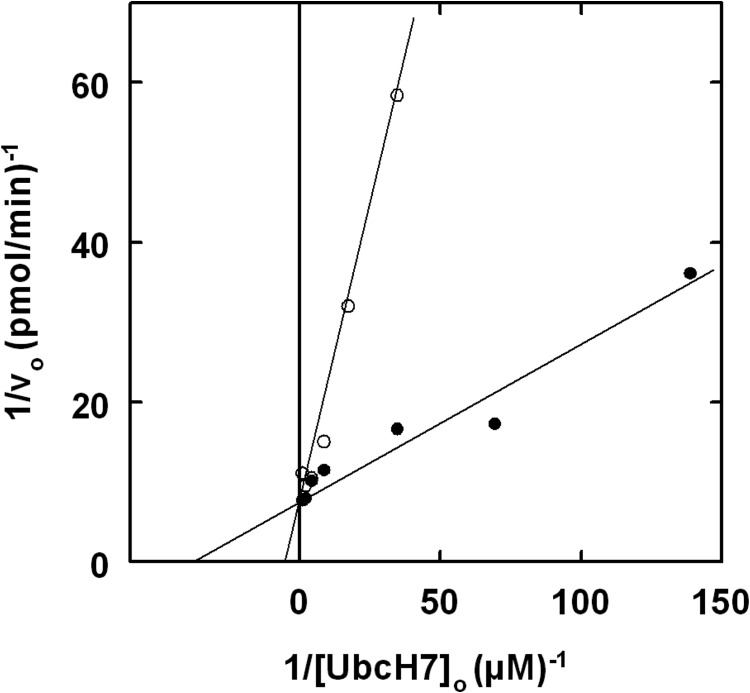
The UbcH7K100A mutant supported E6AP-catalyzed 125I-polyubiquitin chain formation with a Km (32 ± 18 nm) comparable with that of the wild type E2 but a 10-fold lower kcat (0.0034 ± 0.0004 s−1); Table 1. In contrast, equilibrium binding measured by fluorescence polarization previously showed that the UbcH7K100A mutation has a significant effect on affinity at the canonical E6AP binding site, increasing the Kd to 45 ± 15 μm (Table 1) (56). The difference in Kd corresponds to a ΔΔGbinding of 1.3 kcal/mol and predicts a Km of 530 nm for the kinetic assay if the latter value reflected binding at the canonical site. The marked difference in binding affinity for UbcH7K100A determined by fluorescence polarization versus enzymatic activity requires that the two methods monitor binding at different sites, consistent with the existence of two UbcH7∼ubiquitin thioester binding sites on E6AP. In contrast, the UbcH7F63A mutant failed to support E6AP-catalyzed 125I-ubiquitin chain formation even at 32 μm (not shown). The loss of activity accompanying mutation of Phe63 did not result from an effect on binding of the UbcH7F63∼ubiquitin thioester to E6AP but rather due to loss of catalytic competence, as shown in Fig. 4, for which UbcH7F63A at a concentration of 950 nm (present as its corresponding 125I-ubiquitin thioester) acts as a competitive inhibitor of wild type UbcH7 in supporting polyubiquitin chain formation, from which a Ki of 400 ± 216 nm could be calculated (Fig. 4) corresponding to a ΔΔGbinding of 1.2 kcal/mol. In contrast, the Kd for UbcH7F63A binding to E6AP determined by fluorescence polarization is 810 ± 550 μm (56), representing a ΔΔGbinding of 3.0 kcal/mol and predicting a Km of 10 μm if the kinetic assay monitored binding at the canonical site. The difference in binding effects for the UbcH7F63A mutant is consistent with the two methods again measuring binding affinities at distinct sites. Therefore, mutation of Phe63 on UbcH7 results in loss of catalytic competence of the corresponding 125I-ubiquitin thioester to support polyubiquitin chain formation rather than quantitative abrogation of E6AP binding.
FIGURE 4.
UbcH7F63A is a competitive inhibitor of E6AP polyubiquitin chain formation. Initial rates of 125I-ubiquitin conjugation were measured as in Fig. 2 under E6AP limiting conditions with 7 nm GST-E6AP in the absence (close circles) or presence (open circles) of 950 nm UbcH7F63A, present as its corresponding 125I-ubiquitin thioester.
The E6AP Carboxyl Terminus Is Required for Polyubiquitin Chain Formation
Hect domains contain a disordered 5–6-residue carboxyl-terminal extension peptide not resolved in the crystal structure (26), truncation of which demonstrates that it is essential for target protein polyubiquitination but not Hect domain thioester formation (28). Within the carboxyl-terminal sequence is an absolutely conserved phenylalanine residue at the −4 or −5 position whose mutation replicates the loss-of-function phenotype of peptide truncation (28). By introducing a premature STOP codon, we generated E6APΔ847 lacking the carboxyl-terminal pentapeptide extension. Truncation had no qualitative effect on the rate of E6APΔ847∼125I-ubiquitin thioester formation in an experiment similar to that of Fig. 2C; however, E6APΔ847 was unable to form 125I-polyubiquitin chains (not shown). Loss of polyubiquitin chain formation could be duplicated by mutating the conserved Phe849 within wild type E6AP to alanine (not shown). These results replicate analogous observations for target protein conjugation reported previously but now indicate that Phe849 is required for the step of polyubiquitin chain formation rather than direct target protein conjugation as originally proposed (59). Therefore, polyubiquitin chain formation recapitulates functional characteristics of target protein conjugation, validating this approach as a reporter function for ligase activity. In contrast, mutation of Phe849 to tyrosine results in active E6APF849Y that exhibits hyperbolic kinetics with respect to [UbcH7]o and yields a Km of 52 ± 8 nm that was nearly identical to wild type ligase, indicating that Phe849 does not participate in UbcH7∼125I-ubiquitin thioester binding. However, E6APF849Y displayed a kcat of 1.4 ± 0.1 × 10−3 s−1 (kcat/Km = 2.8 × 104 m−1 s−1), 22-fold lower than for wild type enzyme.
Collectively, the marked effect on kcat but not Km suggests that Phe849 functions in the catalytic step of polyubiquitin chain formation rather than target protein ligation; however, we cannot rule out that Phe849 is required to stabilize E6AP protein. The precise role of the carboxyl-terminal peptide and Phe849 is unclear in the absence of additional structural information. In either model, the carboxyl-terminal extension peptide must be physically anchored to the ligase to contribute to Hect domain activity as the corresponding acetylated synthetic peptide (Ac-AKGFGML) does not functionally complement polyubiquitin chain formation catalyzed by E6APΔ847 when added to the incubation (not shown). Instead, the data of Fig. 5 reveal that at 2 mm peptide, Ac-AKGFGML is a noncompetitive inhibitor of UbcH7∼ubiquitin thioester during wild type E6AP-catalyzed polyubiquitin chain formation (Ki = 2.6 ± 0.3 mm). At near-saturating concentrations, Ac-AKGFGML quantitatively blocks polyubiquitin chain formation catalyzed by E6AP (not shown), consistent with noncompetitive inhibition. Therefore, Ac-AKGFGML must bind to a site distinct from that of the UbcH7∼125I-ubiquitin substrate and ablate the apparent kcat for polyubiquitin chain formation.
FIGURE 5.
The E6AP carboxyl terminus contributes to polyubiquitin chain elongation. Initial rates of free 125I-polyubiquitin chain formation were measured as in Fig. 2 under E6AP limiting conditions with 8 nm GST-E6AP and the indicated concentrations of wild type UbcH7 in the absence (closed circles) or presence (open circles) of 2 mm Ac-AKGFGML peptide.
E6AP Can Bind and Elongate Free Lys48-linked Polyubiquitin Chains
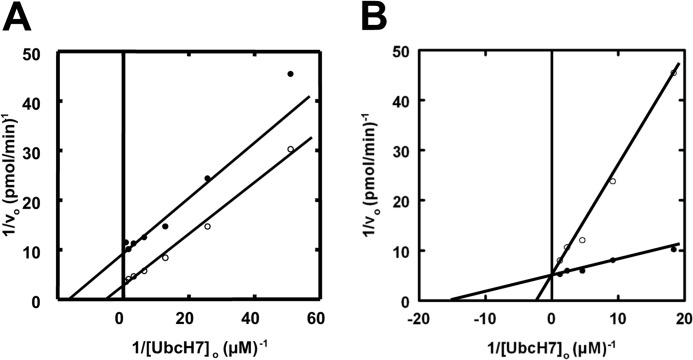
As will be more fully developed under “Discussion,” the current data are consistent with a model for E6AP comprising two sites, one of which is required for Cys820 thioester formation and one of which is required for polyubiquitin chain elongation. To test this, we asked whether E6AP was competent to elongate an unanchored chain. When free Lys48-linked Ub4 chains were added to an assay similar to that of Fig. 2, complex kinetics resulted, as shown in Fig. 6A. The presence of the free Ub4 chains resulted in parallel lines with respect to [UbcH7∼125I-ubiquitin]o, consistent with a pattern of apparent activation arising from an increase in kcat. Such an effect could arise by the free Ub4 chains serving either as sites for 125I-ubiquitin chain elongation or by their ability to allosterically activate E6AP. To distinguish between these alternate models, we reductively methylated free Ub4 chains to block conjugation at the lysine residues (38). When the experiment of Fig. 6A was repeated with reductively methylated Ub4, competitive inhibition with respect to UbcH7∼125I-ubiquitin was observed (Fig. 6B), yielding a Ki of 1.1 ± 0.3 μm. The latter observation suggests that E6AP can catalyze the elongation of unanchored chains as their presence increases the kcat for net polyubiquitin chain formation (Fig. 6A) and that the chains bind to the ligase before the elongation step (Fig. 6B). Consistent with this model, the apparent Vmax observed in the presence of Ub4 in Fig. 6A should be defined as Vmax(1 + [Ub4]o/Kd), where Vmax is the maximum velocity in the absence of Ub4, and Kd is the dissociation constant for binding of Ub4 to E6AP, equivalent to the Ki determined in Fig. 6B. The data of Fig. 6A predict a Kd value of 2.3 ± 0.2 μm that is in reasonably good agreement with the Ki of 1.1 ± 0.3 μm calculated from Fig. 6B.
FIGURE 6.
E6AP binds and elongates Lys48-linked tetra-ubiquitin chains. Double-reciprocal plots of initial rates of 125I-ubiquitin conjugation in the presence of the indicated concentrations of UbcH7 were measured as in Fig. 2 under E6AP limiting conditions with 33 nm GST-E6AP in the absence (closed circles) or presence (open circles) of either 6 μm Lys48-linked tetra-ubiquitin (panel A) or 7 μm reductively methylated Lys48-linked tetra-ubiquitin (panel B).
DISCUSSION
The original structure for UbcH7 bound to the Hect domain of E6AP reported by Huang et al. (26) has been extremely influential in shaping our understanding of this important family of ubiquitin ligases and the protein interactions that contribute to E2 recognition and the mechanism of ubiquitin conjugation to target proteins. However, the structure has proven problematic as it fails to reconcile the restricted spatial requirements of E2-E3 transthiolation with the observed 41 Å distance between the active site cysteines of the Hect domain and the bound UbcH7 (26). Recognition of significant mobility in the carboxyl-terminal subdomain among Hect ligase paralogs (28–32), empirical data demonstrating a marked loss of activity when such conformational flexibility is restricted by mutation (28), and a recent structure for UbcH5B-ubiquitin oxyester bound to the small amino-terminal subdomain of Nedd4L/Nedd4–2 (31) are thought in part to resolve these questions of geometry and the mechanistic requirements for E2-E3 transthiolation. Unrelated studies demonstrating that some Hect ligases conjugate their target proteins by first assembling an intact polyubiquitin chain on the Hect domain active site cysteine before en bloc transfer to the target protein pose additional questions regarding the mechanism of chain assembly for this class of enzymes (34, 60).
In the present work we have exploited enzyme kinetics to probe molecular details of E6AP function using the innate ability of the ligase to form polyubiquitin chains in the absence of substrate as a reporter function (Fig. 1). This approach provides a functional assay that allows us to address questions not experimentally accessible by other methods, as discussed previously (35). The results of these studies demonstrate for the first time that E6AP possesses a cryptic E2∼ubiquitin thioester binding site that is responsible for initial Cys820 thioester formation but that is not present in the canonical structure reported by Huang et al. (26). Other kinetic studies quantitatively identify the cognate E2 for the ligase as UbcH7, although a limited number of other E2 paralogs function with reduced efficacy that is defined by quantitative differences in Km, kcat, or both. Finally, structural conservation among the Hect domains for members of this superfamily suggests these conclusions are relevant for other paralogs.
The biochemically defined E2 screen of Fig. 1 unambiguously demonstrates that UbcH7 is the cognate E2 for E6AP, although other members of the Ubc4/5 superclade support the ligase with reduced efficacies, notably the Ubc5 isoforms and the ISG15-specific UbcH8. The ability of isopeptidase T to disassemble the resulting high molecular weight conjugates that accumulate in the stacker gel demonstrates that some of these adducts previously defined as “autoconjugates” are, in fact, free polyubiquitin chains as has been noted previously (35). Initial rate studies show that E6AP displays hyperbolic kinetics with respect to the concentration of the cognate UbcH7∼ubiquitin thioester (Fig. 2), the actual co-substrate for the ligase, and binds with substantially greater affinity than previously reported for the uncharged carrier protein as measured by equilibrium binding methods (56); Table 1. The Km of 58 ± 6 nm for binding of the UbcH7∼ubiquitin thioester to the E6AP Hect domain approximates the intrinsic Kd, as the corresponding isosteric substrate analog, UbcH7C86S-ubiquitin oxyester, shows competitive inhibition and yields a Ki of 64 ± 18 nm that agrees well with the empirical Km (Fig. 3A). Initial rate kinetics account for differences in efficacy of the other E2 paralogs in supporting E6AP-catalyzed chain formation and yield provocative conclusions regarding the basis of E2 specificity; Table 1.
The UbcH8 family of carrier proteins is closest in sequence to UbcH7 among the E2 families but is relatively specific for charging by UbE1L, the activating enzyme for the interferon-induced ISG15 ubiquitin-like protein, although it can be forced to function with Uba1 under nonphysiological conditions (54, 55). Surprisingly, UbcH8 is nearly as efficient as UbcH7 in supporting E6AP-catalyzed chain formation at saturation, as judged by their similar kcat values; Table 1. The discrimination against UbcH8 shown in Fig. 1 under identical conditions results from a significant difference in binding affinity between the ubiquitin thioesters of UbcH7 versus UbcH8; Table 1. Conversely, the ubiquitin-specific Ubc5A fails to function effectively with E6AP due to a 15-fold lower kcat even though the corresponding ubiquitin thioester binds with slightly better affinity than that of UbcH8; Table 1. These results demonstrate that E2 specificity is a combination of binding affinity (Km) and catalytic competence (kcat) of the resulting E6AP·E2∼ubiquitin thioester Michaelis complex, the latter presumably reflecting the degree to which the optimal geometry is achieved for the different E2 species.
The canonical structure of UbcH7 bound to the Hect domain and the subsequent transfer of the associated ubiquitin thioester to the Cys820 of the ligase predicted by the model (26) requires that the uncharged E2 should serve as a competitive inhibitor with respect to the corresponding E2∼ubiquitin thioester, as is observed for Ubc2b-dependent conjugation of Type 1 N-end rule substrates by human E3α/Ubr1 (42). Surprisingly, the dominant negative UbcH7C86A product analog exhibits noncompetitive inhibition with respect to UbcH7∼ubiquitin thioester and significantly reduced affinity (Ki = 7 ± 0.7 μm) relative to the Km of 58 ± 6 nm for the corresponding UbcH7∼ubiquitin thioester; Fig. 3B and Table 1. This observation requires a priori that UbcH7∼ubiquitin thioester and uncharged UbcH7, modeled as UbcH7C86A, bind to different sites on the Hect domain, a conclusion also consistent with substrate inhibition by UbcH7∼ubiquitin thioester above 2 μm (Fig. 2). Observation of substrate inhibition additionally requires ordered binding of the E2-ubiquitin thioesters to the two sites.
Kinetic data with the Lys100 and Phe63 mutants provide additional support for the presence of two functionally distinct E2∼ubiquitin thioester binding sites on E6AP. Lys100 of UbcH7 interacts directly with side chains donated by the canonical small amino-terminal subdomain (26, 56). Previous work shows that the K100A mutation has a significant effect on the Kd for binding of uncharged UbcH7 to E6AP as measured by fluorescence polarization (56), summarized in Table 1; however, our kinetic data demonstrate that the mutation has no effect on the affinity for binding of the corresponding UbcH7∼ubiquitin thioester to E6AP measured directly as Km, although the mutation lowers the kcat by 10-fold (Table 1). This result requires that the UbcH7∼ubiquitin thioester binding step monitored kinetically cannot involve the canonical site. In contrast, the F63A mutation abrogates polyubiquitin chain formation but is catalytically competent to form the E6AP Cys820 thioester with kinetics qualitatively similar to wild type E2. The effect of mutating Phe63 demonstrates that thioester formation at Cys820 of the Hect domain is functionally distinct from subsequent polyubiquitin chain formation. The inability of UbcH7F63A to support polyubiquitin chain formation is not due to impaired affinity for binding to E6AP, as the corresponding ubiquitin thioester acts as a competitive inhibitor of wild type UbcH7∼ubiquitin thioester and exhibits an affinity, measured as Ki, which is considerably greater than one would predict from the corresponding ΔΔG values derived from equilibrium fluorescence polarization studies; Table 1.
These observations are consistent with a model in which E6AP harbors two spatially and functionally separate E2∼ubiquitin binding sites as part of the mechanism of polyubiquitin chain formation. We propose that Site 1 (our designation) represents a cryptic binding site distinct from the canonical structure for E6AP·UbcH7 (26), as binding of the corresponding UbcH7∼ubiquitin thioester is unaffected by K100A and F63A point mutations that markedly ablate interaction at the canonical site (56) in addition to the observation that the latter mutation supports kinetically competent E6AP-ubiquitin thioester formation but not polyubiquitin chain formation. The functional consequence of the latter point mutant emphasizes that seemingly minor alterations in E2 can have profound consequences for overall function without significantly affecting binding affinity. In contrast, Site 2 (our designation) represents the canonical UbcH7 binding site identified in the crystal structure of Huang et al. (26) as there is excellent correspondence between the Kd for binding of uncharged UbcH7 and UbcH8 to this site, determined by fluorescence polarization (56), and the Ki for noncompetitive inhibition of the corresponding C86A point mutants with respect to wild type UbcH7∼ubiquitin thioester in the initial rate studies (Fig. 3B and text).
Kinetic identification of Site 1 potentially resolves the distance problem posed by the original structure for uncharged UbcH7 bound to E6AP (26). The present data do not address the possible location of Site 1; however, because association of E6AP with E2∼ubiquitin thioester is not constrained by the canonical UbcH7·Hect structural model, it is reasonable to propose that the binding site might be sterically close to the active site Cys820 of the Hect domain to allow efficient transthiolation. We note that the structure of E6AP and of other Hect ligases reveals a “ledge” adjacent to Cys820 that provides an attractive candidate for Site 1. Moreover, if this region represents Site 1, then one can easily envision how binding of E2∼ubiquitin thioester to the lower affinity Site 2 within the small amino-terminal subdomain might sterically occlude binding at Site 1, accounting for substrate inhibition at higher concentrations of UbcH7.
Why is the putative Site 1 not represented in the crystal structure for UbcH7 bound to the Hect domain? This potential caveat can be reconciled by proposing that the affinity for binding of E2∼ubiquitin thioester to Site 1 is determined by combinatorial interactions between the Hect domain and both the E2 and ubiquitin moieties rather than E2 alone, as appears to be the case for Site 2. Because there are significant binding contributions arising from entropic effects when linking otherwise independent binding sites (61),6 the difference in affinity between E2∼ubiquitin thioester and uncharged E2 can be substantial, accounting for Site 1 binding not being represented in the crystal structure compared with the lower affinity canonical Site 2. Moreover, the cryptic Site 1 is lost due to steric hindrance on formation of the Cys820∼ubiquitin thioester after transthiolation, accounting for sequential binding at the two sites. Our observation that UbcH7C86A is a noncompetitive inhibitor of the corresponding thioester and binds at the canonical Site 2, based on good agreement between the Ki determined here and the Kd measured previously by fluorescence polarization (56), is consistent with such a model as it requires the active site point mutant to have greater affinity for Site 2 than for Site 1. Finally, because the UbcH7F63A point mutation abrogates polyubiquitin chain formation but only minimally affects binding of the corresponding thioester to Site 1 (Fig. 4), Site 2 is likely responsible for polyubiquitin chain elongation from the Cys820-linked ubiquitin moiety formed at Site 1.
With these insights, a two-step minimal model emerges for the mechanism of E6AP and, presumably, other Hect ligases: an initial step of Hect domain∼ubiquitin thioester formation arising from binding at Site 1 followed by Site 2-dependent chain elongation. Rapid formation of the Cys820-ubiquitin thioester at Site 1 sets a lower limit for the first order rate constant for transthiolation of 3.5 s−1 (Fig. 2C) that is ∼100-fold greater than the kcat for polyubiquitin chain elongation determined under steady state conditions (Fig. 2B). This requires that Site 2-dependent chain elongation be rate-limiting under the kinetic assay conditions employed; however, other data have consistently indicated that the Km values determined here reflect binding at Site 1. The apparent differences in binding affinities and rates for the two sites suggest a burst-like mechanism in which a rapid initial formation of the Cys820∼ubiquitin thioester is followed by a slower steady state elongation of the polyubiquitin chain from Site 2, with both steps exhibiting hyperbolic binding of the E2-ubiquitin thioester substrate. Under these conditions, the rate for the chain elongation step is determined by the steady state concentration of the Cys820∼ubiquitin thioester, which in turn is a function of saturable binding at Site 1. Therefore, the Km measured in the present studies will reflect binding of the E2∼ubiquitin thioester at Site 1 whereas the kcat will reflect the subsequent step of chain elongation at Site 2. The data of Fig. 6 demonstrate that E6AP is capable of chain elongation and that the ligase harbors a low affinity polyubiquitin chain binding site with a Kd of 1 μm. Such a site is consistent with reports of a free ubiquitin binding site within the Hect domains of Nedd4 (30) and Rsp5 (62) corresponding to Kd values of 11 and 90 μm, respectively. The marked differences in affinities for polyubiquitin chain binding (this study) versus free ubiquitin (30, 61) suggest the Hect domain ubiquitin binding site may have a preference for the former. Our data do not address whether chain elongation occurs on a free chain, as modeled in the experiment of Fig. 6, or from a Cys820-anchored chain as proposed by Wang et al. (33). We favor the latter model because there is little mechanistic rationale for the presence of two functionally distinct E2∼ubiquitin binding sites in the former model for elongation of free chains, which requires initiation of chain formation from a bound monomeric ubiquitin. In addition, elongation of anchored Cys820∼polyubiquitin chains accounts for formation of free and conjugated chains by partitioning of this high energy intermediate.
Although Purbeck et al. (53) were the first to examine the rates of E6AP-ubiquitin thioester formation, the present studies represent the first comprehensive kinetic examination of the E6AP mechanism in polyubiquitin chain formation. The kinetics of polyubiquitin chain formation functionally mirrors properties of target protein conjugation, as demonstrated by the dependence of the former on an intact Hect domain C terminus and the absolute requirement for Phe849 within this sequence (Table 1 and Fig. 5). More important, the present observations are inconsistent with the conventional model of Hect domain mechanism (26) and structural models predicated on cysteine active site transthiolation from E2∼ubiquitin bound at the canonical site within the small amino-terminal subdomain (31). The present kinetic data provide an internally consistent empirically based model for the mechanism of polyubiquitin chain formation catalyzed by E6AP and, presumably, other Hect domain ligases.
This work was supported, in whole or in part, by National Institutes of Health Grant GM034009 (to A. L. H.).
We use E1 and E2 generically to refer to the paralogous activating enzymes and carrier proteins/conjugating enzymes, respectively, for Class 1 ubiquitin-like proteins.
Isoform 3 differs from isoform 1, from which the crystal structure of E6AP was determined, by an additional 20 resides at the amino terminus. To coincide with residues in the crystal structure, we refer to residue numbers in isoform 3 by their paralogous positions in isoform 1.
Unless otherwise stated, E6AP refers to the GST-E6AP fusion protein.
Similar entropic effects account for the enhanced binding of polyubiquitin chains to tandem ubiquitin binding domains compared with that of single ubiquitin moieties.
- Hect
- homologous to E6AP carboxyl terminus
- Uba1
- ubiquitin activating enzyme (gene name, UBE1)
- E1
- generic term for activating enzymes of Class 1 ubiquitin-like proteins
- E2/Ubc
- generic name for ubiquitin carrier protein/ubiquitin conjugating enzyme
- Ubc2b
- b isoform of the human/rabbit ortholog of Saccharomyces cerevisiae Rad6/Ubc2 (also termed E214Kb
- gene name
- UBE2B)
- Ubc5B
- B isoform of the human ortholog of the S. cerevisiae Ubc5 (also termed UbcH5B
- gene name
- UBE2D2).
REFERENCES
- 1. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2001) Mechanism underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 3. Ng C. C., Arakawa H., Fukuda S., Kondoh H., Nakamura Y. (2003) p53RFP, a p53-inducible RING-finger protein, regulates the stability of p21WAF1. Oncogene 22, 4449–4458 [DOI] [PubMed] [Google Scholar]
- 4. Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 [DOI] [PubMed] [Google Scholar]
- 5. Munakata T., Liang Y., Kim S., McGivern D. R., Huibregtse J., Nomoto A., Lemon S. M. (2007) Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 3, 1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Isaacson M. K., Ploegh H. L. (2009) Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scheffner M., Werness B. A., Huibregtse J. M., Levine A. J., Howley P. M. (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 8. Gao Q., Singh L., Kumar A., Srinivasan S., Wazer D. E., Band V. (2001) Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 75, 4459–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bedard K. M., Underbrink M. P., Howie H. L., Galloway D. A. (2008) The E6 oncoproteins from human betapapillomaviruses differentially activate telomerase through an E6AP-dependent mechanism and prolong the lifespan of primary keratinocytes. J. Virol. 82, 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Y., Ye F., Lu W., Hong D., Wan X., Xie X. (2009) HPV16 E6-induced and E6AP-dependent inhibition of the transcriptional coactivator hADA3 in human cervical carcinoma cells. Cancer Invest. 27, 298–306 [DOI] [PubMed] [Google Scholar]
- 11. Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lossie A. C., Whitney M. M., Amidon D., Dong H. J., Chen P., Theriaque D., Hutson A., Nicholls R. D., Zori R. T., Williams C. A., Driscoll D. J. (2001) Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J. Med. Genet. 38, 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams C. A., Beaudet A. L., Clayton-Smith J., Knoll J. H., Kyllerman M., Laan L. A., Magenis R. E., Moncla A., Schinzel A. A., Summers J. A., Wagstaff J. (2006) Angelman syndrome 2005. Updated consensus for diagnostic criteria. Am. J. Med. Genet. A 140, 413–418 [DOI] [PubMed] [Google Scholar]
- 14. Matsuura T., Sutcliffe J. S., Fang P., Galjaard R. J., Jiang Y. H., Benton C. S., Rommens J. M., Beaudet A. L. (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 15, 74–77 [DOI] [PubMed] [Google Scholar]
- 15. Kishino T., Lalande M., Wagstaff J. (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15, 70–73 [DOI] [PubMed] [Google Scholar]
- 16. Albrecht U., Sutcliffe J. S., Cattanach B. M., Beechey C. V., Armstrong D., Eichele G., Beaudet A. L. (1997) Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature Genetics 17, 75–78 [DOI] [PubMed] [Google Scholar]
- 17. Yamasaki K., Joh K., Ohta T., Masuzaki H., Ishimaru T., Mukai T., Niikawa N., Ogawa M., Wagstaff J., Kishino T. (2003) Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 12, 837–847 [DOI] [PubMed] [Google Scholar]
- 18. Cook E. H., Jr., Lindgren V., Leventhal B. L., Courchesne R., Lincoln A., Shulman C., Lord C., Courchesne E. (1997) Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am. J. Hum. Genet. 60, 928–934 [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Y. H., Sahoo T., Michaelis R. C., Bercovich D., Bressler J., Kashork C. D., Liu Q., Shaffer L. G., Schroer R. J., Stockton D. W., Spielman R. S., Stevenson R. E., Beaudet A. L. (2004) A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am. J. Med. Genet. A 131, 1–10 [DOI] [PubMed] [Google Scholar]
- 20. Chowdhury S., Shepherd J. D., Okuno H., Lyford G., Petralia R. S., Plath N., Kuhl D., Huganir R. L., Worley P. F. (2006) Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greer P. L., Hanayama R., Bloodgood B. L., Mardinly A. R., Lipton D. M., Flavell S. W., Kim T. K., Griffith E. C., Waldon Z., Maehr R., Ploegh H. L., Chowdhury S., Worley P. F., Steen J., Greenberg M. E. (2010) The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140, 704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Margolis S. S., Salogiannis J., Lipton D. M., Mandel-Brehm C., Wills Z. P., Mardinly A. R., Hu L., Greer P. L., Bikoff J. B., Ho H. Y., Soskis M. J., Sahin M., Greenberg M. E. (2010) EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell 143, 442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mishra A., Dikshit P., Purkayastha S., Sharma J., Nukina N., Jana N. R. (2008) E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J. Biol. Chem. 283, 7648–7656 [DOI] [PubMed] [Google Scholar]
- 24. Mishra A., Godavarthi S. K., Maheshwari M., Goswami A., Jana N. R. (2009) The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J. Biol. Chem. 284, 10537–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramamoorthy S., Nawaz Z. (2008) E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl. Recept. Signal. 6, e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Structure of an E6AP-UbcH7 complex. Insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 27. Kim H. C., Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. (2003) Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249–259 [DOI] [PubMed] [Google Scholar]
- 29. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 30. Maspero E., Mari S., Valentini E., Musacchio A., Fish A., Pasqualato S., Polo S. (2011) Structure of the HECT-ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandya R. K., Partridge J. R., Love K. R., Schwartz T. U., Ploegh H. L. (2010) A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J. Biol. Chem. 285, 5664–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang M., Cheng D., Peng J., Pickart C. M. (2006) Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 25, 1710–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang M., Pickart C. M. (2005) Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 24, 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ronchi V. P., Haas A. L. (2012) Measuring rates of ubiquitin chain formation as a functional readout of ligase activity. Methods Mol. Biol. 832, 197–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baboshina O. V., Haas A. L. (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2epf and Rad6 are recognized by the 26S proteasome subunit 5. J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 37. Haas A. L. (2005) Purification of E1 and E1-like enzymes. Methods Mol. Biol. 301, 23–35 [DOI] [PubMed] [Google Scholar]
- 38. Hershko A., Heller H. (1985) Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 39. Haas A. L., Rose I. A. (1982) The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 40. Haas A. L., Bright P. M. (1988) The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263, 13258–13267 [PubMed] [Google Scholar]
- 41. Tokgöz Z., Siepmann T. J., Streich F., Jr., Kumar B., Klein J. M., Haas A. L. (2012) E1-E2 interactions in the ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 287, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L. (2003) Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 43. Baboshina O. V., Crinelli R., Siepmann T. J., Haas A. L. (2001) N-end rule specificity within the ubiquitin/proteasome pathway is not an affinity effect. J. Biol. Chem. 276, 39428–39437 [DOI] [PubMed] [Google Scholar]
- 44. Scheffner M., Huibregtse J. M., Howley P. M. (1994) Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl. Acad. Sci. U.S.A. 91, 8797–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nuber U., Schwarz S., Kaiser P., Schneider R., Scheffner M. (1996) Cloning of the human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP AND RSP5. J. Biol. Chem. 271, 2795–2800 [DOI] [PubMed] [Google Scholar]
- 46. Kumar S., Kao W. H., Howley P. M. (1997) Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J. Biol. Chem. 272, 13548–13554 [DOI] [PubMed] [Google Scholar]
- 47. Haas A. L., Siepmann T. J. (1997) Pathways of ubiquitin conjugation. FASEB J. 11, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 48. Wilkinson K. D., Tashayev V. L., O'Connor L. B., Larsen C. N., Kasperek E., Pickart C. M. (1995) Metabolism of the polyubiquitin degradation signal. Structure, mechanism, and role of isopeptidase T. Biochemistry 34, 14535–14546 [DOI] [PubMed] [Google Scholar]
- 49. Reyes-Turcu F. E., Shanks J. R., Komander D., Wilkinson K. D. (2008) Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 283, 19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tokgöz Z., Bohnsack R. N., Haas A. L. (2006) Pleiotropic effects of ATP·Mg2+ binding in the catalytic cycle of ubiquitin activating enzyme. J. Biol. Chem. 281, 14729–14737 [DOI] [PubMed] [Google Scholar]
- 51. Haas A. L., Rose I. A. (1981) Hemin inhibits ATP-dependent ubiquitin-dependent proteolysis. Role of hemin in regulating ubiquitin conjugate degradation. Proc. Natl. Acad. Sci. U.S.A. 78, 6845–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Purbeck C., Eletr Z. M., Kuhlman B. (2010) Kinetics of the transfer of ubiquitin from UbcH7 to E6AP. Biochemistry 49, 1361–1363 [DOI] [PubMed] [Google Scholar]
- 54. Haas A. L. (2006) in Protein Degradation (Mayer R. J., Ciechanover A., Rechsteiner M., eds) pp. 103–131, Wiley-VCH Verlag, Weinheim, Germany [Google Scholar]
- 55. Durfee L. A., Kelley M. L., Huibregtse J. M. (2008) The basis for selective E1-E2 interactions in the ISG15 conjugation system. J. Biol. Chem. 283, 23895–23902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eletr Z. M., Kuhlman B. (2007) Sequence determinants of E2-E6AP binding affinity and specificity. J. Mol. Biol. 369, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sullivan M. L., Vierstra R. D. (1993) Formation of a stable adduct between ubiquitin and the Arabidopsis ubiquitin-conjugating enzyme, AtUBC1. J. Biol. Chem. 268, 8777–8780 [PubMed] [Google Scholar]
- 58. Sung P., Prakash S., Prakash L. (1990) Mutation of cysteine 88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. U.S.A. 87, 2695–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Salvat C., Wang G., Dastur A., Lyon N., Huibregtse J. M. (2004) The −4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J. Biol. Chem. 279, 18935–18943 [DOI] [PubMed] [Google Scholar]
- 60. You J., Pickart C. M. (2001) A HECT domain E3 enzyme assembles novel polyubiquitin chains. J. Biol. Chem. 276, 19871–19878 [DOI] [PubMed] [Google Scholar]
- 61. Jencks W. P. (1975) Binding energy, specificity, and enzymic catalysis. The Circe effect. Adv. Enzymol. Relat. Areas Mol. Biol. 43, 219–410 [DOI] [PubMed] [Google Scholar]
- 62. Kim H. C., Steffen A. M., Oldham M. L., Chen J., Huibregtse J. M. (2011) Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 12, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]