Background: MicroRNA 375 (miR-375) is expressed in the pituitary gland, but its functions and the related mechanisms have not been studied.
Results: miR-375 mediates the signaling pathway of CRF regulating POMC expression by targeting MAP3K8.
Conclusion: miR-375 negatively regulates POMC expression and related hormone secretion.
Significance: These new data suggest that miRNAs play important roles in regulating pituitary hormone secretion.
Keywords: Cell signaling, Endocrinology, Gene regulation, MicroRNA, Pituitary gland, MAP3K8, POMC, miR-375
Abstract
Pro-opiomelanocortin (POMC) is a common precursor of melanocortin-related peptides in the pituitary and primarily regulated by corticotropin- releasing factor (CRF). Our results show that miR-375 is highly expressed in the mouse pituitary gland and located specifically in the intermediate lobe of pituitary. The functional studies show that the forced inhibition of endogenous miR-375 in AtT-20 mouse pituitary tumor cells and in the intermediate lobe of the pituitary gland significantly increases POMC expression, whereas miR-375 overexpression down-regulates POMC expression and ACTH secretion stimulated by CRF. This function of miR-375 is accomplished by its binding to the 3′-UTR of mitogen-activated protein kinase kinase kinase-8. Our results here have demonstrated that miR-375 acts as a negative regulating molecule mediating the signaling pathway of CRF and affecting POMC expression by targeting mitogen-activated protein kinase kinase kinase-8, which subsequently down-regulates ERK1/2 phosphorylation and nerve growth factor-induced clone B (NGFI-B) transcription activity. Taken together, our results show that miR-375 is a novel negative regulator of POMC expression and related hormone secretion.
Introduction
The pro-opiomelanocortin (POMC)2 is a common precursor of the melanocortin-related peptides, including adrenocorticotrophin (ACTH), β-lipotropin, α-melanocyte-stimulating hormone, β-melanotropin, and β-endorphin. ACTH and β-lipotropin are primarily produced in the intermediate and anterior lobes of the pituitary gland (1) and α-melanocyte-stimulating hormone (α-MSH) and β-endorphin are mainly produced in the intermediate lobe (2, 3). These peptides affect metabolic (4), immune (5), and inflammatory responses (6). It has long been acknowledged that the corticotropin-releasing factor (CRF) from the hypothalamus acts as a key regulator of POMC gene expression in vivo and in vitro (7, 8). CRF enhances POMC transcription (8, 9) and ACTH secretion (10) by binding to the G protein-coupled CRF receptor, thus increasing intracellular cAMP (11) and calcium levels (12) and activating protein kinase A (PKA). PKA stimulates ERK1/2 activity and increases the transcriptional activity of the POMC promoter (13). In addition, some transcription factors, such as TBX19 (14), PITX1 (15), NGFI-B (16), and NURR1 (17) are involved in regulating POMC gene expression and transcription (18–20). The activities of these transcriptional factors are also affected by their cross-talks with CRF signaling molecules; however, their interactions and the related molecular mechanisms of CRF affecting POMC gene transcription remain unclear.
MicroRNAs (miRNAs) are small noncoding RNAs with regulatory functions of gene expressions. There are scant data about the function of microRNA in the pituitary gland, although it has been reported that certain miRNAs have been identified as predictive signatures of pituitary adenoma (21). One such miRNA, miR-26b, regulates pituitary development by specifically targeting the lymphoid enhancer factor 1, which modulates the pituitary transcription factor 1 (22). miR-325-3p is involved in regulating LH secretion (23), and miR-375 has been shown to regulate insulin secretion by targeting myotrophin in the MIN6 cell line and to inhibit insulin mRNA transcription in the INS-1 cell line by targeting PDK1 (24, 25). In addition, miR-375 functions to enhance estrogrn receptor α (ERα) signaling activity through the regulation of its target, RASD1 (26). Although miR-375 expression has been detected in the pituitary gland (27), the function of miR-375 in the pituitary gland is unknown.
Mitogen-activated protein kinase kinase kinase-8 (MAP3K8) is a serine-threonine kinase with crucial physiological roles in G protein-coupled receptor-mediated ERK, tumor necrosis factor, interleukin-1, CD40, and Toll-like receptor signaling transductions (28). Previous studies revealed that the overexpression of MAP3K8 promotes cell proliferation in a variety of cells and induces cell transformation (29–32) by activating ERK, JNK, p38 MAPK, nuclear factor of activated T cells (NFAT), and nuclear factor-κ B (NF-κB) (31). However, MAP3K8 is phosphorylated by casein kinase II, PKA, or Cas-phosphorylating kinase, it then acts as a functional kinase protein (32). In PC12 phenochromocytoma cells, MAP3K8 is a crucial intermediate for CRF-induced norepinephrine production (33).
In the present study, we initially examined miR-375 expression in the mouse pituitary gland and demonstrated that miR-375 is highly expressed and is mainly located in the intermediate lobe. Upon further research, we have shown that miR-375 functions as a mediator of the CRF signaling pathway by targeting MAP3K8 to inhibit POMC expression and transcription. These new findings suggest that miR-375 plays a key role in regulating POMC expression by targeting MAP3K8 and affects the synthesis and secretion of pituitary hormones by manipulating POMC gene transcription.
EXPERIMENTAL PROCEDURES
Animals and Tissue Collections
Adult male and female (6–8 months) Kunming white mice were purchased from the Animal Institute of the Chinese Medical Academy (Beijing, China) and were raised in a controlled temperature of 25 ± 1 °C and humidity (60–70%) on a 12-h light, 12-h dark cycle. The animal experiments were approved by the Chinese Association for Laboratory Animal Sciences. The mice were killed by cervical dislocation. The pituitary glands were removed and separated into anterior lobes, intermediate lobes, and posterior lobes. We also dissected whole pituitary glands, hypothalamus, brain, lung, liver, and heart and divided them into three sections per group for RT-PCR. For in situ hybridization and immunohistochemistry, the collected mice pituitary glands were fixed for 1 h in 4% paraformaldehyde at room temperature and then placed in 30% sucrose at 4 °C overnight. The tissues were embedded in Tissue Tek O.C.T. compound (TaKaRa Biotechnology, Dalian, China), and 10-μm sections were cut using a cryostat.
In Situ Hybridization (ISH)
miR-375 ISH was carried out by using digoxigenin-labeled locked nucleic acid (LNA) probes. Mus musculus miR-375-3p miRCURY LNA microRNA detection probes, and scrambled probes were purchased from Exiqon (Woburn, MA). We labeled the LNA probes with digoxigenin using a digoxigenin oligonucleotide tailing kit (Roche Diagnostics, Indianapolis, IN) following the manufacturer's instructions. miR-375 ISH assays were carried out as described previously (34). Briefly, the dried slides were fixed in 4% paraformaldehyde for 10 min at room temperature and then washed twice for 3 min in 1× PBS. The fixed sections were subjected to acetylation for 10 min, followed by PBS washes. The slides were prehybridized for 8 h at room temperature, and the hybridization was carried out at 54 °C overnight. After stringency washings using 5× SSC for 10 min and 0.2× SSC for 1 h at 60 °C, the slides were incubated in a blocking solution for 1 h at room temperature, which was followed by incubation with anterior lobe of the pituitary-conjugated antibody to digoxigenin at 4 °C overnight. After PBS and alkaline phosphates buffer washes, the slides were incubated in Nitrotetrazolium Blue chloride (NBT) and 5-Bromo-4-chloro-3-indolyl phosphate p-toluidine salt (BCIP) in the dark until the prospective intensity of staining was reached.
ISH and Immunohistochemistry (IHC) Dual Staining
The ISH-stained sections were treated with 10% normal donkey serum in PBS and incubated with anti-POMC antibody (1:50; Abcam, Cambridge, UK) at 4 °C overnight. The sections were incubated with donkey anti-goat IgG-TRITC (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) for 3 h. After washing with PBS for three times, the slides were observed under a fluorescence microscope (IX71, Olympus, Japan) and photographed.
Cell Culture and Treatments
The pituitary glands were removed and separated into anterior lobes and neurointermediate lobes from adult mice. The neurointermediate lobes were placed in sterile PBS. Preparation of the primary intermediate pituitary cells assay was carried out as described previously (35). AtT-20 cells originating from a mouse pituitary corticotrope tumor and 293T cell line were grown in DMEM (Invitrogen) containing 10% (v/v) FBS (Invitrogen) and 1% penicillin streptomycin. The cells were also incubated at 37 °C in a humidified atmosphere of 5% CO2. Primary intermediate lobe cells were transfected after being cultured for 48 h, and AtT-20 cells were passaged and cultured for 12 h.
MicroRNA and siRNA Transfections
M. musculus miR-375 mimic (forward, 5′-UUUGUUCGUUCGGCUCGCGUGA-3′ and reverse, 5′-ACGCGAGCCGAACGAACAAAUU-3′) and M. musculus miR-375 inhibitor (5′-UCACGCGAGCCGAACGAACAAA-3′) were commercially synthesized from Gene Pharma (Shanghai, China). Stable negative control (nc mimic) (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′) and nc inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′) were used as negative controls. PKA siRNA and signal silence control siRNA duplex were purchased from Cell Signaling Technology (Beverly, MA). The MAP3K8 siRNA kit was purchased from RibBio (Guangzhou,China). The X-tremeGENE siRNA transfection reagent (Roche Diagnostics, Penzberg, Germany) was used according to the manufacturer's instructions.
Analysis of Real-time PCR
According to the manufacturer's instructions (TaKaRa), the total RNA of the tissues and cells was isolated using the RNAiso Plus. The microRNA expression assay was based on the method described previously (36). U6 RNA was used for normalization of microRNA expression. Reverse transcriptase reactions contained the purified total RNA and 50 nm RT primer (the RT-miR-375 stem-loop primer, CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCGCACT; RT-miR-25 primer, CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCAATTGC; U6 RT primer, AACGCTTCACGAATTTGCGT). M-MLV reverse transcriptase (Promega, Madison, WI) was used. The 15-μl reactions were incubated in a DNA Thermal Cycler 4800 for 30 min at 16 °C, 30 min at 42 °C, and 5 min at 85 °C. Real-time PCR was performed using a standard Takara SYBR Premix Ex Taq protocol on an Applied Biosystems 7500 Real-time PCR system (Applied Biosystems, Foster City, CA). The primer sequences are listed in Table 1, and the conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative abundance of the genes was determined using the ABI PRISM 7500 equipped software (Applied Biosystems). All of the experiments were performed in at least triplicate.
TABLE 1.
Primers used in this study
| Primer names | Primer sequences (5′ to 3′) |
|---|---|
| miR-375 | |
| Sense | AGTGTCGTCAGAAAGAACGAACGGC |
| Antisense | CTCAACTGGTGTCGTGGAGTC |
| miR-U6 | |
| Sense | CTCGCTTCGGCAGCACA |
| Antisense | AACGCTTCACGAATTTGCGT |
| POMC | |
| Sense | GCAACGGAGATGAACAGCC |
| Antisense | CTTGTCCTTGGGCGGGTT |
| NGFI-B | |
| Sense | CTGGCATACCGATCTAAAC |
| Antisense | GGCGGG AACATCAACAC |
| GR | |
| Sense | TTTCCAGACTATTTTCGGTCAG |
| Antisense | GTATTTAGGAGGGTATTTT CAT |
| PITX-1 | |
| Sense | TCAACCCGTGAACTGAATGT |
| Antisense | TCCTCAGCCAGGCGTAAA |
| Tpit | |
| Sense | TTTATCTTGGCCACGCTTAGG |
| Antisense | CCCAGAACGGCTTGAGAGTAA |
| NeuroD1 | |
| Sense | TTTCGATAGCCATTCGCATCAT |
| Antisense | GGACAGTCACTGTACGCACAGT |
| SRC2 | |
| Sense | AAAGG GAGCAGATAGAAC |
| Antisense | GTGGGAGATTGGATGAA |
| MAP3K8 | |
| Sense | AGCCCTCACAAGATAGTAACCTCA |
| Antisense | GCTACCATACAATACACACCAGAA |
| KLF4 | |
| Sense | GCAATACACACGTAAAGATCACC |
| Antisense | TTAGAACCACGACTCACCAAGCA |
| Dusp6 | |
| Sense | GATTTGCTCCATTCATTGTTTT |
| Antisense | TCCCTATTCCTATGCTACCCTCT |
| Smad7 | |
| Sense | CTGAGCTCTGTAGACCAGCG |
| Antisense | ATACGAGCGAGCGTATGAGC |
| GAPDH | |
| Sense | GGTTGTCTCCTGCGACTTCA |
| Antisense | GGGTGGTCCAGGGTTTCTTA |
ELISA
ACTH concentrations in cultured AtT-20 cell supernatants were measured with mouse ACTH ELISA kit (Rapidbio, West Hills, CA) according to the manufacturer's instructions.
Luciferase Reporter Assay
The Dual-Luciferase reporter genes were constructed using the psiCHECKTM-2 vector (Promega, Madison, WI) and the 3′-UTR sequences of mouse Kruppel-like factor 4 (KLF4) or MAP3K8. Their 3′-UTR fragments were cloned using an overlap PCR. The sequences were introduced between the NotI and XhoI sites to Renilla luciferase 3′-UTR. The firefly luciferase vector was used for internal reference. Constructs with mutated 3′-UTR of KLF4 and MAP3K8 were used as negative controls. The 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum. A total of 4 × 104 cells per well were seeded in 24-well plates. After 24 h in culture, the cells were transfected using the Lipofectamine 2000 agent (Invitrogen) with a mixture containing 200 ng/ml of the Dual-Luciferase reporter plasmid and 40 nm miR-375 mimic or nc mimic. The cells transfected with the mutation in 3′-UTR of KLF4 or MAP3K8 vectors (p-Luc-3′-UTR MUT MAP3K8 or KLF4) served as controls for normalization. When the cells were transfected for 24 h, the luciferase activity was measured by a ModulusTM II microplate multimode reader (Promega) using a Dual Luc Reporter Assay Kit (Vigorous Biotechnology, Beijing, China). All transfections were repeated independently at least three times.
Western Blotting
The AtT-20 cells were lysed with cell lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 1 mm PMSF). The protein concentration of each sample was determined by the BCA assay reagent (Vigorous Biotechnology) according to the manufacturer's instructions. Equal amounts of proteins (100 μg) were electrophoresed on a 15% SDS-PAGE for POMC, ERK1/2, p-ERK1/2, MAP3K8 and GAPDH, and the bands were transferred to a PVDF membrane (Bio-Rad Laboratories). The membranes were blocked with 5% (w/v) BSA in 0.05 m TBS (pH 7.4) for 3 h and incubated at 4 °C overnight with a purified goat polyclonal IgG anti-MAP3K8 (1:500; Santa Cruz Biotechnology), goat polyclonal IgG anti-POMC (1:2500; Abcam), rabbit polyclonal IgG anti-phospho-ERK1/2 and total ERK1/2 antibody (1:1000; Cell Signaling Technology), and monoclonal mouse IgG anti-GAPDH (1:10000; R&D Systems, Minneapolis, MN) in TBST. The PVDF membrane was then washed three times for 30 min in TBST (0.1% Tween-20 in TBS) and incubated for 2 h with an HRP-conjugated donkey anti-goat IgG, goat anti-rabbit IgG (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA), or goat anti-mouse IgG (1:50,000; Jackson ImmunoResearch Laboratories) at room temperature. Finally, the membranes were washed for 1 h and treated with the SuperSignal West Pico kit (Thermo Scientific, Waltham, MA) substrate at room temperature from 10 s to 10 min.
Chromatin Immunoprecipitation and Quantitative PCR
EZ-ChIP chromatin immunoprecipitation kit was from Millipore, a rat anti-NGFI-B antibody was from Abcam. The ChIP experiment was performed as our report. Input and immunoprecipitated DNA were amplified by real-time PCR using primers as follows: POMC-P1, GGAGGTTGAAGACAGAAGTGTATTA (sense) and GACTTCCTGCTGTCCATCTTGTATC (antisense); POMC-P2, TCCATTGCCCACCACAGAGCGC (sense) and GCCTAGTTCTGAGATCTTGCAG (antisense). Real-time PCR products quantified by comparison with the PCR products of a dilution series of relevant input DNA.
Bioinformatic Analysis of MicroRNA Target Genes
Two algorithms were used to computationally predict targets of miR-375. They included Sanger's miRNA target database and the TargetScan database. The minimum free energy of hybridization between the target gene and miR-375 was predicted by RNAhybrid (37). Co-expressed genes with NGFI-B were analyzed by Coxpressiondb database.
Statistical Analysis
Results are presented as means ± S.E. single comparisons were performed by Student's t test, whereas multiple comparisons were performed by analysis of variance (ANOVA). A value of p < 0.05 was considered to be statistically significant.
RESULTS
miR-375 Is Expressed Specifically in the Intermediate Lobe of Mouse Pituitary Gland
We detected miR-375 levels in the whole mouse pituitary gland, the anterior lobe of the pituitary, the intermediate lobe, hypothalamus, brain, lung, liver, heart, and AtT-20 and LβT2 cell lines. The results showed that the miR-375 expression level was the highest in the pituitary gland among all the tissues and cell lines examined (Fig. 1A). We also found that miR-375 level was much higher in the intermediate lobe than in the anterior lobe of the pituitary and in the whole pituitary gland. AtT-20 and LβT2 are two types of pituitary adenoma cell lines, and there was a higher expression of miR-375 in AtT-20 cells than in LβT2 cells. To detect the miR-375 amount in the pituitary, we used absolute quantification real-time PCR. The results showed that there were ∼2.0 × 1013 copy numbers of miR-375 in 0.5 μg of total pituitary RNA. (Fig. 1C) We also detected miR-375 expression in the pituitary gland by the locked nucleic acid LNA-ISH method. It was observed that the miR-375 LNA-ISH signal was much stronger in the intermediate lobe of the pituitary gland (Fig. 1D). miR-375 LNA-ISH and POMC IHC dual staining in the intermediate lobe were performed. We observed that the POMC IHC staining was much stronger in areas with a weaker miR-375 LNA-ISH expressing signal and vice versa (Fig. 1E). These results confirm that miR-375 is highly expressed in mouse pituitary gland and is specifically located in the intermediate lobe. These results suggest that miR-375 negatively regulates POMC gene transcription and the related hormone secretion.
FIGURE 1.
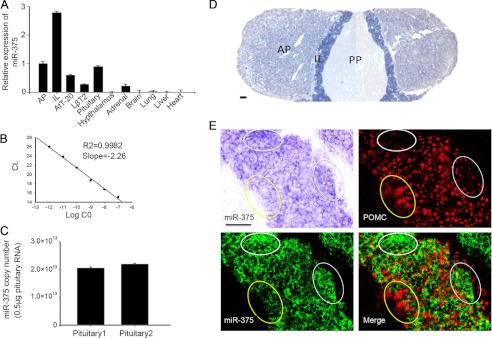
miR-375 expressed in the mouse pituitary gland. A, relative expression levels of miR-375 in different tissues of adult mice analyzed by real-time PCR (with U6 small nuclear RNA amplified as an internal normalized reference). Results are means ±S.E. of three independent experiments. B, absolute quantification standard curve of miR-375 was constructed by real-time PCR. C, absolute expression level of miR-375 detected in 0.5 μg of pituitary. Copy numbers per 0.5 μg of total RNA were calculated using standard curves. Results are means ± S.E. of three independent experiments. D, LNA-ISH detection of miR-375 in adult mouse pituitary. Scale bar, 200 μm, (PP, posterior lobe of pituitary; IL, intermediate lobe; AP, anterior lobe). E, miR-375 and POMC staining of the intermediate lobe of the pituitary gland. Scale bar, 50 μm.
miR-375 Negatively Regulates POMC Expression
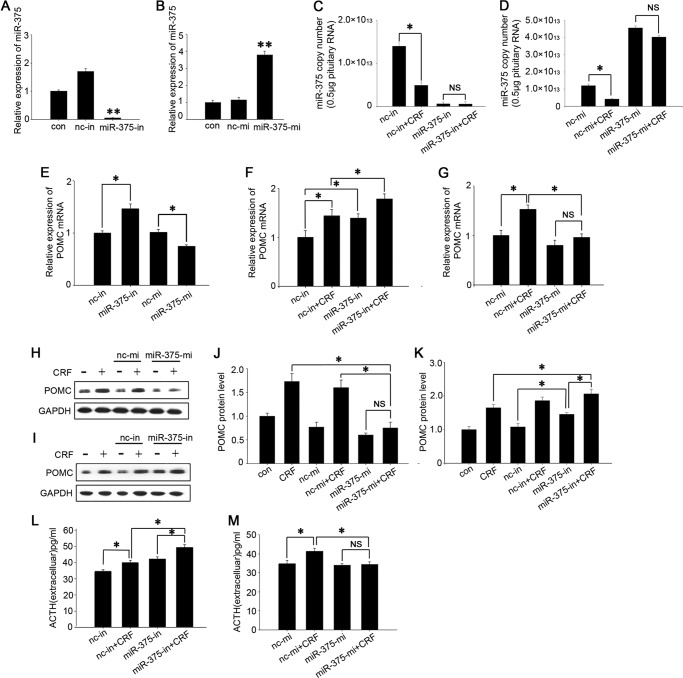
Because miR-375 expression was mainly localized in the intermediate lobe that primarily produces POMC, we deduced that miR-375 was involved in regulating POMC expression. To confirm this hypothesis, we transfected miR-375-inhibitors (miR-375-in) and miR-375-mimics (miR-375-mi) into the primary cultured cells and AtT-20 cells. The relative and absolute quantification of miR-375 results showed that miR-375 inhibitors dramatically down-regulated the endogenous miR-375 (Fig. 2, A and C) and the mimics sharply up-regulated miR-375 expression (Fig. 2, B and D), and these effects were not affect by CRF. Meanwhile, we assessed the effects of the miR-375 inhibitors and mimics on POMC expression in cultured primary cells from the intermediate lobe of the pituitary gland. The results showed that miR-375-in increased the POMC mRNA levels by ∼40% and miR-375-mi decreased POMC mRNA levels by ∼30% (Fig. 2E). These results demonstrate that miR-375 negatively regulates pituitary POMC expression.
FIGURE 2.
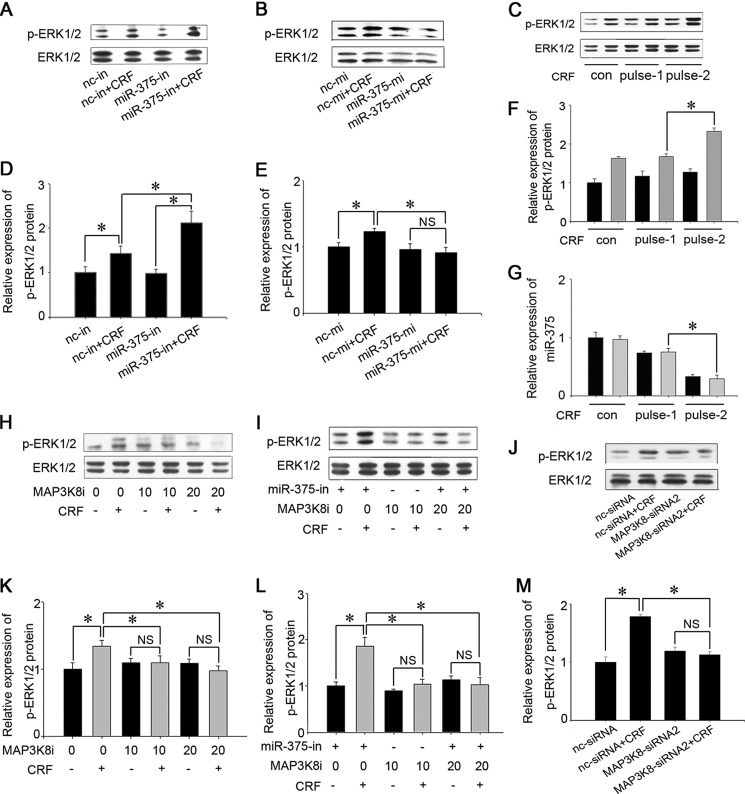
miR-375 negatively regulates POMC expression and ACTH secretion. A and B, AtT-20 cells were separately transfected with 100 nm miR-375-in and miR-375-mi for 24 h. The control (con) was a vehicle (PBS) and no transfection control. Quantification of intracellular miR-375 levels was done by real-time PCR. Results are means ± S.E. of four independent experiments. **, p < 0.01 versus cells transfected with nc inhibitor (nc-in) or nc mimic (nc-mi; t test). C and D, absolute quantification of miR-375 after the AtT-20 cells were transfected with miR-375-in and miR-375-mi for 24 h and then added 100 nm CRF for 12 h. The data are means ± S.E. for multiple separate transfections (n = 3). *, p < 0.05. NS, not significant (p > 0.05) (ANOVA). E, quantification of POMC mRNA levels in the primary cell culture of pituitary intermediate lobes transfected with miR-375-in and miR-375-mi for 24 h. Results are means ± S.E. of three independent experiments. *, p < 0.05 (t test). The data are means ± S.E. for multiple separate transfections (n = 3). F and G, quantification of POMC mRNA levels in AtT-20 cells. The cells were separately transfected with miR-375-in and miR-375-mi. Twenty-four hours later, the cells were treated with 100 nm CRF for 6 h. H and I, analysis of POMC protein. AtT-20 cells were transfected with miR-375-mi and miR-375-in, respectively, for 24 h, and we added 100 nm CRF and harvested the cells after 6 h. The control (con) was a vehicle control (PBS). Protein expression levels were analyzed by Western blotting. J and K, quantification of POMC protein level. L and M, Determination of ACTH concentrations in the cell culture medium. AtT-20 cells were transfected with nc inhibitor, miR-375-in, nc mimic, and miR-375-mi, respectively, for 24 h. We then added 100 nm CRF and collected the culture medium after 6 h. Data are presented as means ± S.E. (n = 3). *, p < 0.05; NS, not significant (p > 0.05) (ANOVA).
miR-375 Mediates CRF Signaling Pathway Influencing POMC Expression and ACTH Secretion
It is generally thought that CRF is the key factor influencing POMC expression and ACTH secretion. To confirm that miR-375 is involved in the CRF signaling pathway, cultured AtT-20 cells were transfected with miR-375-mi and miR-375-in for 24 h and then treated with 100 nm CRF for 6 h. POMC expression was detected both at the gene and protein levels. The results showed that miR-375-in increased both basal and CRF-enhanced POMC mRNA levels (Fig. 2F) and protein levels (Fig. 2, I and K). We also found that, as expected, CRF up-regulated POMC gene expression in cells transfected with negative control mimic, but this effect vanished in the cells that overexpressed miR-375 (Fig. 2G). The POMC protein level stimulated by CRF was also reduced by miR-375-mi (Fig. 2, H and J). These demonstrate that miR-375 negatively regulates the CRF stimulatory action on POMC expression both at the mRNA and protein levels. We were also interested in evaluating whether miR-375 is involved in regulating ACTH secretion stimulated by CRF. AtT-20 cells were separately transfected with miR-375-mi and miR-375-in and treated with 100 nm CRF for 6 h. The ACTH concentration in the culture medium was then assessed using an ACTH ELISA kit. The results showed that miR-375-in increased ACTH concentration in the culture medium (Fig. 2L), whereas miR-375-mi decreased the CRF-stimulated ACTH (Fig. 2M) parallel to the change of POMC expression. These results suggest that miR-375 participates in the CRF signaling pathway as a negative regulator.
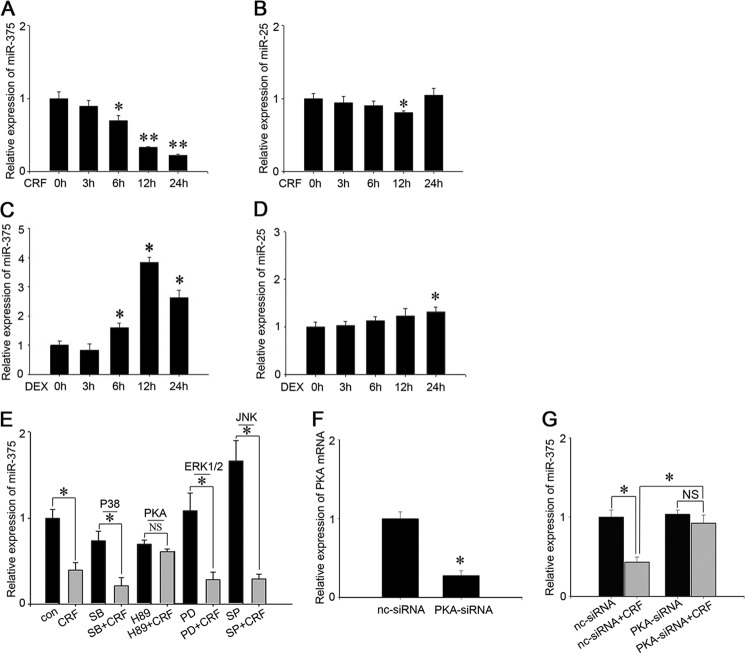
CRF Specifically Decreases miR-375 Expression through the PKA Pathway
To confirm that miR-375 is involved in the CRF signaling pathway that regulates POMC expression and ACTH secretion, we initially tested the effects of CRF and dexamethasone (DEX), a glucocorticoid analog, on miR-375 expressions. The results showed that miR-375 was down-regulated ∼30, 70, and 80% after 6-, 12-, and 24-h treatments with 100 nm CRF treatments, respectively (Fig. 3A). The opposite was true with DEX in that DEX induced a 3× increase of miR-375 expression after a 24-h treatment (Fig. 3C). From our previous work,3 we have shown that miR-25 is expressed in AtT-20 cells. As a control, we assessed the effects of CRF and DEX on miR-25 along with miR-375 expression level to determine whether CRF specifically influences miR-375 expression. The results showed that both CRF and DEX had no significant effects on miR-25 expression (Fig. 3, B and D). It is known that CRF influences POMC expression and ACTH secretion through different signal pathways, such as PKA, ERK1/2, p38, and JNK (38). To determine the signaling pathway in which miR-375 is involved, we separately added 20 μm of several signaling pathway inhibitors to AtT-20 cells. The inhibitors were SB 203580 (p38), H-89 (PKA), PD 98059 (ERK), and SP 600125 (JNK). The cells were then additionally treated with CRF for 12 h. The results show that miR-375 repression was only relieved by H-89 (PKA) (Fig. 3E). Furthermore, PKA siRNA and nc siRNA were transfected in AtT-20 cells, and the interference efficiency of PKA siRNA was ∼70% (Fig. 3F). At the same time, the repression of miR-375 was vanished after the cells were transfected with PKA siRNA (Fig. 3G). These results demonstrate that CRF specifically decreases miR-375 expression through the PKA pathway.
FIGURE 3.
miR-375 is specifically regulated by CRF and DEX in AtT-20 cells. A and B, AtT-20 cells were treated with 100 nm CRF for 0, 3, 6, 12, and 24 h, respectively. The expressions of miR-375 and miR-25 were analyzed by real-time PCR and normalized to the U6 transcript level. C and D, AtT-20 cells were treated with 100 nm DEX for 0, 3, 6, 12, and 24 h, respectively. The expressions of miR-375 and miR-25 were analyzed by real-time PCR and normalized to the U6 transcript level. Results are means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; NS, not significant (p > 0.05) versus control (t test). E, the relative level of miR-375 in AtT-20 cells treated with CRF and several different signal pathway inhibitors. The control (con) was a vehicle control (PBS). The inhibitors include, SB-203680 (SB), H89, PD98059 (PD), and SP-600125 (SP). The data are means ± S.E. for multiple separate transfections (n = 3); *, p < 0.05; NS, not significant (p > 0.05) (ANOVA). F, transfection of PKA-specific siRNA detect the PKA mRNA level by real-time PCR. Results are means ± S.E. of three independent experiments; **, p < 0.01 versus cell transfected with nc-siRNA (t test). G, AtT-20 cells were transfected with PKA siRNA or nc-siRNA for 24 h and then added 100 nm CRF and 12 h later detected miR-375 expression level. Data presented as means ± S.E. (n = 3). *, p < 0.05; NS, not significant (p > 0.05) (ANOVA).
MAP3K8 Is a Direct Target Gene of miR-375 in AtT-20 Cells
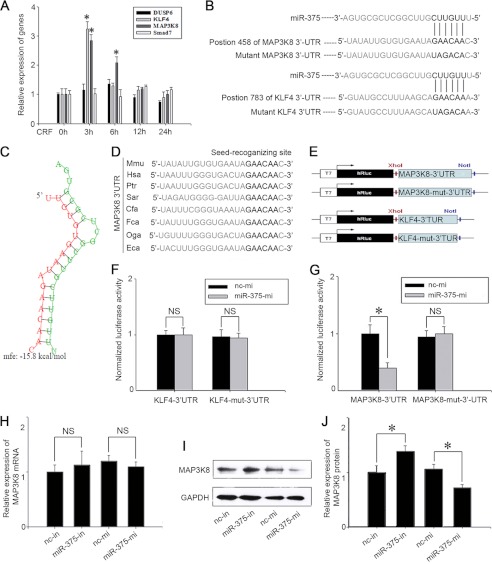
To find the target genes of miR-375, we initially predicated the top 300 genes co-expressed with NGFI-B according to Bioinformatics 4.1. NGFI- B is known to be a key transcription factor that regulates POMC gene transcription (16). To identify the target molecules of miR-375, we used the computational algorithm from the MicroRna website, which was designed to predict mRNA targets of microRNAs, and 11 of these 300 genes were predicted as target genes of miR-375. Finally, we narrowed the possibilities to four putative genes, which included Dusp6 (dual specificity phosphatase 6), KLF4, MAP3K8, and Smad7 (SMAD family member 7).
To identify whether CRF has stimulating effects on these four predicated miR-375 target genes, 100 nm CRF was added to cultured AtT-20 cells, and the mRNA levels of Dusp6, KLF4, MAP3K8 and Smad7 were detected after 3-, 6-, 12-, and 24-h treatments. The results show that CRF significantly increased KLF4 and MAP3K8 mRNA levels at 3 h, and the increase on MAP3K8 mRNA expression persisted until 6 h. However, CRF treatment did not have an obvious influence on Dusp6 and Smad7 expression at any time (Fig. 4A). We then identified the interactions between miR-375 and its putative target genes, MAP3K8 and KLF4 respectively. Since the predicted binding site of miR-375 is in the 3′-UTR of MAP3K8 and KLF4 mRNA (Fig. 4B). The minimum free energy of hybridization between the target gene and miR-375 was predicted by RNAhybrid (39), and this also supported the possibility that miR-375 could bind at the site (Fig. 4C). We used a psiCHECKTM-2 vector and cloned the putative 3′-UTR target site downstream of a luciferase reporter gene (Fig. 4E). We then co-transfected into 293T cells the psiCHECKTM-2 vector (wild-type or mutant) together with miR-375-mi or nc mimic. The results showed that the luciferase activity of the transfected cells with miR-375-mi and p-Luc-3′-UTR MAP3K8 decreased by ∼60% compared with the cells co-transfected with nc mimic and p-Luc-3′-UTR MAP3K8. The negative control construct of mutations in the 3′-UTR of MAP3K8 (p-Luc-3′-UTR MUT MAP3K8), showed no obvious change in luciferase activity (Fig. 4F). p-Luc-3′-UTR KLF4 construct luciferase activity did not change compared with mutations in the 3′-UTR of MAP3K8 (p-Luc-3′-UTR MUT MAP3K8). (Fig. 4G). Further functional analysis indicated that miR-375 overexpression in AtT-20 cells resulted in a reduction of MAP3K8 protein levels, whereas the inhibition of miR-375 significantly increased MAP3K8 protein levels (Fig. 4, I and J) without affecting MAP3K8 mRNA levels (Fig. 4H). We also observed that the nucleotides in seed-recognizing sites of the MAP3K8 3′-UTR were completely conserved in several species using the TargetScan database (Fig. 4D). These results demonstrate that MAP3K8 is the direct target gene of miR-375.
FIGURE 4.
miR-375 binds to MAP3K8 3′-UTR and down-regulates its protein level. A, the expression levels of Dusp6, KLF4, MAP3K8, and Smad7 in AtT-20 cells treated with 100 nm CRF. Data are presented as means ± S.E. (n = 3). *, p < 0.05 versus control (t test). B, the predicted miR-375 binding site is in the MAP3K8 3′-UTR and KLF4 3′-UTR. The data were taken from Sanger's miRNA target database. C, there is a schematic representation of the hybridization between miR-375 and MAP3K8 3′-UTR and theirs minimum free energy (mfe). Green and red letters indicate miR-375 and MAP3K8, respectively. D, the seed regions of miR-375 and the seed-recognizing sites in the MAP3K8 3′-UTR are indicated in boldface type, and all nucleotides in the seed-recognizing sites are completely conserved in several species. Mmu, M. musculus; Hsa, Homo sapiens; Ptr, Pan troglodytes; Sar, Sorex araneus; Cfa, Canis lupus familiaris; Fca, Felis catus; Oga, Otolemur garnettii; Eca, Equus caballus. E, schematic of inserted MAP3K8 3′-UTR and KLF-4 3′-UTR sequences. F and G, relative luminescence intensity detected by the ModulusTMII microplate multimode reader after miR-375-mi or nc mimic (nc-mi) and Dual-Luciferase vectors co-transfected into 293T cells. The data are means ± S.E. for multiple separate transfections (n = 4). *, p < 0.05. H, relative quantification of MAP3K8 mRNA levels. AtT-20 cells were transfected with nc inhibitor (nc-in), miR-375-in, nc mimic, miR-375-mi. The data are means ± S.E. for multiple separate transfections (n = 3). I, analysis of MAP3K8 protein. AtT-20 cells were transfected as shown above and protein extracts were analyzed by Western blotting. J, quantification of MAP3K8 protein levels. Results are means ± S.E. of three independent experiments; *, p < 0.05; NS, not significant (p > 0.05) versus cells transfected with nc inhibitor or nc mimic (t test). Data are presented as means ± S.E. (n = 3). *, p < 0.05.
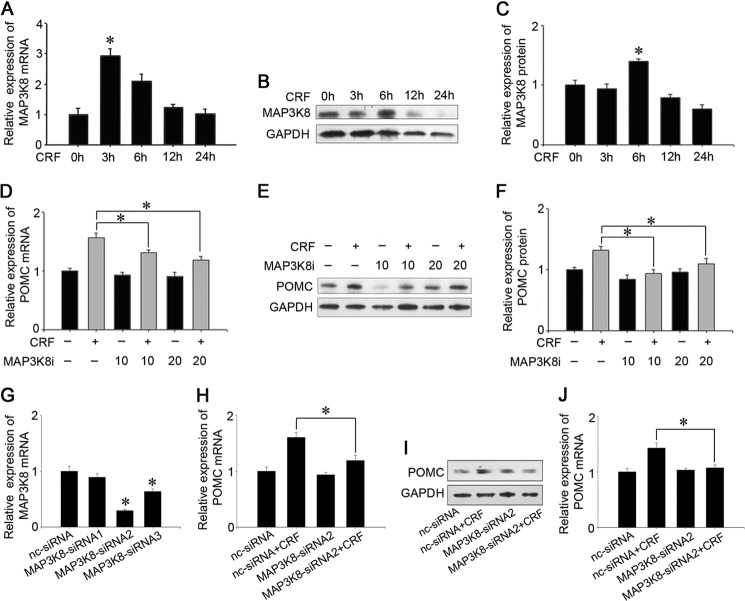
MAP3K8 Is Involved in the Signaling Pathway of CRF That Regulates POMC Expression
To identify whether MAP3K8 is involved in the signaling pathway of CRF that regulates POMC expression, cultured AtT-20 cells were treated with 100 nm CRF for 3, 6, 12, and 24 h, and MAP3K8 expression was assayed. The results show that CRF sharply increased MAP3K8 mRNA levels after a 3-h treatment, which persisted at 6 h, followed by no obvious influence at 12 and 24 h (Fig. 5A). However, Western blot analysis showed that CRF significantly up-regulated MAP3K8 protein level only at 6 h (Fig. 5, B and C). In addition, we treated AtT-20 cells with 20 μm 4-(3-chloro-4-fluorophenylamino)-6-(pyridin-3-yl-methylamino)-3-cyano[1,7]naphthyridine (MAP3K8i) (40), a MAP3K8 inhibitor from Merck Biosciences, and then added 100 nm CRF for 6 h. The results showed that MAP3K8i erased the up-regulating effect of CRF on POMC expression, and decreased POMC mRNA and protein levels by ∼26% (Fig. 5D) and 38% (Fig. 5, E and F), respectively. Then, we carried out three kinds of MAP3K8 siRNA and chose MAP3K8 siRNA2, which had higher inhibition on MAP3K8 mRNA (Fig. 5G). MAP3K8 siRNA2 also inhibited the effect of CRF on POMC mRNA and protein levels ∼40% (Fig. 5H) and 35% (Fig. 5, I and J). These preliminary results demonstrate that MAP3K8 plays an important role in mediating the regulating regulatory effect of CRF on POMC transcription and translation.
FIGURE 5.
MAP3K8 functions in regulating POMC stimulated by CRF. A, AtT-20 cells were treated with 100 nm CRF for 0, 3, 6, 12, and 24 h. Expression levels of MAP3K8 were analyzed by real time PCR and normalized to GAPDH transcript levels. Data are presented as means ± S.E. (n = 3). *, p < 0.05 versus control (t test). B, MAP3K8 protein expression levels were analyzed by Western blotting. C, quantification of MAP3K8 protein levels. Data are presented as means ± S.E. (n = 3). *, p < 0.05 versus control (t test). D, quantification of POMC mRNA levels after the addition of 20 μm MAP3K8i and 100 nm CRF into AtT-20 cells and treated for 6 h. E, POMC protein levels were analyzed by Western blotting. F, quantification of POMC protein levels. Data are presented as means ± S.E. (n = 3). *, p < 0.05; NS, not significant (p > 0.05) (ANOVA). G, quantification of intracellular MAP3K8 mRNA level after the AtT-20 cells were transfected with MAP3K8 siRNA1–3. Data are presented as means ± S.E. (n = 3). *, p < 0.05 versus nc siRNA (t test). H, quantification of POMC mRNA levels after transfecting nc siRNA or MAP3K8 siRNA2 and 24 h later, added 100 nm CRF into AtT-20 cells and were treated for 6 h. I, POMC protein expression levels were analyzed by Western blotting. The cells were treated as above. J, quantification of POMC protein levels. Data are presented as means ± S.E. (n = 3). *, p < 0.05 (p > 0.05) (ANOVA).
miR-375 and MAP3K8 Decrease ERK1/2 Phosphorylation Stimulated by CRF
Because the effect of the CRF/PKA pathway on POMC expression relies on up-regulating ERK1/2 phosphorylation and increasing the activities of transcription factors, including NGFI-B (13), we speculated that ERK1/2 was the downstream molecule of miR-375 and MAP3K8. To confirm this hypothesis, miR-375-mi and miR-375-in were, respectively, transfected into AtT-20 cells, and the transfected cells were treated with 100 nm CRF for 10 min. The influence of miR-375 on ERK1/2 phosphorylation was then detected. The results showed that miR-375-in up-regulated the level of ERK1/2 phosphorylation stimulated by CRF (Fig. 6, A and D), whereas miR-375 overexpression inhibited ERK1/2 phosphorylation induced by CRF (Fig. 6, B and E). Further, in order to demonstrate that the changes of miR-375 expression level in the CRF treated AtT-20 cell could alter the ERK response, we carried out CRF pulse experiments as reported by Becquet et al. (41). Briefly, AtT-20 cells were cultured for 45 min with CRF, and the cells were then cultured in the CRF-free medium for 5 h, followed by a 45-min treatment with CRF (pulse 1). The cells were cultured in the CRF-free medium for another 5 h (pulse 2), and one more CRF treatment was applied, and the levels of miR-375 expression and ERK1/2 phosphorylation were detected. The results showed that both CRF pulse treatments decreased miR-375 expressions and increased ERK1/2 phosphorylation levels, but the effects of the second CRF pulse treatment were much obvious than that of the first CRF pulse treatment. (Fig. 6, C, F, and G). Meanwhile, to assess the effect of MAP3K8 on ERK1/2, we added 10 μm or 20 μm MAP3K8i into the cells. The data shows that MAP3K8i blocked the function of CRF on ERK1/2 phosphorylation (Fig. 6, E and G). To prove that MAP3K8 was the only mediating factor between miR-375 and ERK1/2, miR-375-in was transfected, and 20 μm MAP3K8i was added to AtT-20 cells, respectively. One hour later, the cells were treated with 100 nm CRF for 10 min, and ERK1/2 phosphorylation levels were measured. The results showed that miR-375-in increased the level of ERK1/2 phosphorylation, but this effect was blocked by the addition of MAP3K8i (Fig. 6, F and H). We then used siRNA against MAP3K8 to confirm its implication in ERK1/2 activation in response to CRF (Fig. 6, I and J). MAP3K8 siRNA and MAP3K8i have the similar effect on ERK1/2 phosphorylation stimulated by 100 nm CRF. These results suggest that MAP3K8 acts as a mediating molecule between miR-375 and ERK1/2.
FIGURE 6.
miR-375 and MAP3K8 regulate ERK1/2 phosphorylation. A, analysis of ERK1/2 and P-ERK1/2 proteins. AtT-20 cells were transfected with miR-375 inhibitors and cultured with 100 nm CRF for 10 min, and protein expression levels were analyzed by Western blotting. B, analysis of ERK and P-ERK1/2 proteins. AtT-20 cells were transfected with miR-375-mi for 24 h and 100 nm CRF for 10 min. C, effect of CRF (100 nm) plus stimulation on ERK1/2 phosphorylation. G, effect of CRF (100 nm) pulse stimulation on miR-375 expression. H, analysis of ERK and p-ERK1/2 proteins. AtT-20 cells were treated with MAP3K8i. CRF was added 1 h later and cultured for 10 min. I, analysis of ERK and p-ERK1/2 proteins. AtT-20 cells were transfected with miR-375-in for 24 h, and then 20 μm MAP3K8i was added for 1 h. The cells were then treated with 100 nm CRF for 10 min. J, analysis of ERK and p-ERK1/2 proteins. AtT-20 cells were transfected with MAP3K8 siRNA2 for 24 h and then treated with 100 nm CRF for 10 min. D–F and K–M, quantification of p-ERK1/2 protein levels. Data are presented as means ± S.E. (n = 3). *, p < 0.05; NS, not significant (p > 0.05) (ANOVA). con, control; nc-mi, nc mimic; nc-in, nc inhibitor.
miR-375 and MAP3K8 Decrease Expression of NGFI-B Enhanced by CRF
In addition, it is well known that the signaling transduction of CRF regulates POMC transcription through ERK1/2 phosphorylation, which in turn enhances NGFI-B activity. We thus assessed the NGFI-B expression in AtT-20 cells after being transfected with miR-375-in or miR-375-mi, respectively, and then treating the cells with 100 nm CRF. The results showed that miR-375-in significantly increased NGFI-B and POMC mRNA expression (Fig. 7A), whereas miR-375-mi down-regulated NGFI-B mRNA levels by ∼70% (Fig. 7B). In addition, 10 and 20 μm of MAP3K8i down-regulated NGFI-B expression by ∼30 and 80%, respectively (Fig. 6C). Similarly, MAP3K8 siRNA2 reduced the NGFI-B mRNA level ∼75% (Fig. 7D). To confirm that NGFI-B stimulated by 100 nm CRF for 1 h regulated the POMC by its direct interactions with the POMC gene promoter, the mouse POMC sequence analysis was performed. The area from −703 to −332 bp containing the putative NGFI-B binding sites (NurRE/GTGATATTTACCTCCAAATGCCAG) were used to analyze NGFI-B binding site in precipitation of genomic DNA as previous reports (18, 20). The proximal region from −1449 to −1157 bp, which did not contain the NurRE was used as negative control (Fig. 7E). The ChIP experiment was then performed using NGFI-B antibody, and two pairs of primers were designed to amplify these regions. Of the two areas, only the region from −703 to −332 bp containing the NurRE were identified to be occupied by NGFI-B that stimulated by 100 nm CRF for 1 h. These data indicate that CRF increased the expression of NGFI-B, which directly binds to the NurRE of POMC gene promoter (Fig. 7F). These results are in agreement with the previous reports (18, 20) that NGFI-B stimulated by CRF directly binds to the NurRE of POMC gene promoter. All of these data demonstrate that miR-375 targets MAP3K8, which decreases ERK1/2 phosphorylation and NGFI-B activity, thus negatively regulating POMC expression.
FIGURE 7.
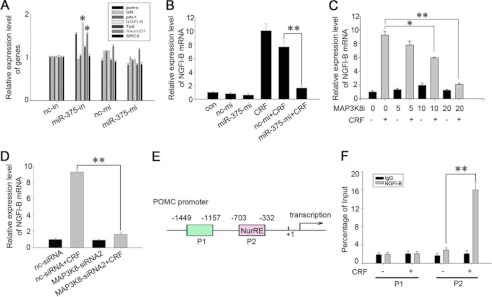
miR-375 and MAP3K8 regulate expression of NGFI-B. A, quantification of the mRNA level of POMC, Glucocorticoid receptor (GR), pitx1, NGFI-B, Tpit, NeuroD1, and SRC2 after transfection with either 100 nm miR-375-in or miR-375-mi. Data are presented as means ± S.E. (n = 3). *, p < 0.05 versus cells transfected with nc inhibitor (nc-in) or nc mimic (nc-mi) (t test). B, quantification of NGFI-B mRNA levels. AtT-20 cells were transfected with miR-375-mi and nc mimic. 24 h later, they were treated with 100 nm CRF for 1 h. C, analysis of NGFI-B mRNA levels. AtT-20 cells were treated with MAP3K8i (5, 10, and 20 μm) for 1 h. 100 nm CRF was added, and the cells were harvested after 1 h. D, quantification of NGFI-B mRNA levels. AtT-20 cells were transfected with MAP3K8 siRNA2 or nc siRNA. 24 h later, they were treated with 100 nm CRF for 1 h. E, a schematic representation of the POMC gene promoter. Putative NGFI-B-binding sequences are shown as rectangles. NGFI-B-binding sites and flanking sequences are depicted. F, ChIP analysis of NGFI-B to the POMC promoter after the AtT-20 cells were treated with 100 nm CRF for 1 h. Data are presented as means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 (ANOVA).
DISCUSSION
miR-375 expression has been detected in the mouse pituitary gland (42), but its location in the pituitary gland and its function have not been elucidated. Our results presented here show that miR-375 is mostly located in the intermediate lobe of the pituitary gland, inferring that miR-375 plays an important role in regulating the functions of POMC expression and the related pituitary hormone secretions. Using a gain and loss of function approach, we demonstrate that miR-375 dramatically inhibits POMC expression both at the gene and protein levels by targeting MAP3K8 and mediating the CRF signaling pathway. These new findings suggest that miR-375 serves as a key factor in regulating the synthesis and secretion of pituitary hormones such as ACTH. However, detailed in vivo experiments are required to confirm the physiological function of miR-375 in the pituitary gland.
miR-375 expression has been detected in pancreatic islets where it negatively regulates insulin synthesis and secretion by targeting the myotrophin (Mtpn) gene (24) and the 3-phosphoinositide-dependent protein kinase-1 (Pdpk1) gene in pancreatic β-cells (25). We have shown that miR-375 is highly expressed in the pituitary gland, especially in the intermediate lobe by real-time PCR and ISH. Further functional studies showed that miR-375 significantly inhibits POMC expression both at the mRNA and protein levels in cultured primary intermediate lobe cells and AtT-20 cells. miR-375 also decreases the ACTH secretion in AtT-20 cells. In addition, POMC IHC and miR-375 ISH dual staining results showed that a much weaker POMC IHC staining was observed in the areas that had a stronger miR-375 ISH signal, and areas with stronger miR-375 ISH signals showed weaker POMC IHC staining. These results demonstrate that miR-375 negatively regulates the synthesis and secretion of the pituitary hormones dependent on POMC in the pituitary. This is similar with what was found in the pancreatic islets (24), suggesting that miR-375 acts as a negative regulating molecule in hormone synthesis and secretion.
CRF from the hypothalamus has long been considered a key factor regulating POMC gene expression and transcription. The CRF signal is transduced mainly through the PKA pathway after binding the G protein-coupled receptor CRF receptor (8). PKA activates the MAPK pathway and enhances ERK/1/2 phosphorylation and the activity of NGFI-B, a major transcription stimulating factor of POMC (13). The present study has demonstrated that miR-375 mediates the PKA/ERK1/2 signaling pathway of CRF, influencing POMC expression. CRF significantly inhibits miR-375 expression through the PKA pathway, simultaneously enhances POMC expression and ACTH secretion. Dexamethasone (DEX), a glucocorticoid analog, increased miR-375 expression and inhibited POMC expression. In addition, H-89, a PKA inhibitor and PKA siRNA blocks the influences of CRF on miR-375 expression and ERK1/2 phosphorylation. Furthermore, miR-375 overexpression significantly inhibits the positive effects of CRF on POMC expression and ERK1/2 phosphorylation, whereas the inhibition of miR-375 up-regulated the phosphorylation level of ERK1/2. These data collectively suggest that miR-375 acts as a mediating molecule between PKA and ERK1/2. However, it is still needed to be elucidated about the mechanism of CRF influencing miR-375 expression.
To identify the target gene of miR-375, we initially used an algorithm from the microrna website that searches for matching base pairs between the miRNA and its target in combination with a thermodynamically based evaluation of miRNA: mRNA duplex interactions (43). From our search results, four putative miR-375 target genes, Dusp6, KLF4, MAP3K8, and Smad7, were predicted. Further in vitro experiments have shown that MAP3K8 is the target gene of miR-375. Furthermore, our in vitro experiments have shown that CRF down-regulates miR-375 and up-regulates MAP3K8 expression in AtT-20 cells. In addition, the luciferase reporter gene results showed that miR-375 reduces the luciferase activity by binding MAP3K8 3′-UTR sequence. Thirdly, miR-375-in up-regulated MAP3K8 expression, whereas miR-375-mi down-regulated MAP3K8 expression. These data demonstrate that miR-375 affects MAP3K8 expression by directly targeting the 3′-UTR of the MAP3K8 gene.
It has been reported that MAP3K8 is involved in ERK1/2 phosphorylation (31) after it is phosphorylated by PKA (32). The results of the present study have shown that the inhibition of miR-375 increases ERK1/2 phosphorylation, and MAP3K8i and MAP3K8 siRNA annul this effect of miR-375 on ERK1/2 phosphorylation. These results show that MAP3K8 is a mediating molecule between miR-375 and ERK1/2 in the signaling pathway of CRF to increase POMC expression. These new findings are critical for our understanding of the regulating mechanisms of POMC expression and its related hormone secretion effects.
In addition, it has been reported that a number of transcription factors are involved in regulating POMC gene expression and transcription (18–20). In the present study, we have assessed the relationships among miR-375, ERK1/2, and several transcription factors such as Tpit, pitx, NGFI-B, NeuroD1, GR, and SRC2. The analysis of their functions and expression levels demonstrate that only NGFI-B among the above transcription factors is affected by miR-375 overexpression, MAP3K8i and MAP3K8 siRNA. In addition, the enhancement of ERK1/2 phosphorylation increases NGFI-B and POMC expression, which is stimulated by CRF (13). These indicate that NGFI-B acts as a downstream molecule of ERK1/2 in the signaling pathway of CRF inducing POMC expression and the related pituitary hormone secretions of POMC. However, their interactions and related molecular mechanisms need to be elucidated in future studies.
The focus of the present work was on the miR-375 expression and its roles in mediating the signal pathway of CRF regulating POMC expression in adult mouse. However, it has been reported that the dicer deletion using Pitx2-cre only has obvious effect on pituitary somatotropes (22), although Pitx2 is highly expressed in the Rathkes pouch at embryonic day 10.5 (44) and transiently expresses in the precursors of POMC-producing cells of the anterior gland and intermediate lobes (45). However, the dicer deletion using POMC-cre results in the disappearance of the intermediate lobe and reduction of the pituitary corticotropes (46); this suggests that microRNAs play important roles in regulating the proliferation and differentiation of POMC-positive cells. However, the expressing pattern and function of miR-375 in the developing pituitary gland should be elucidated in future studies.
In conclusion, we have shown that miR-375 is highly expressed in mouse and is densely located in the intermediate lobe of the pituitary. It acts as a negative mediator in regulating the effect of CRF on POMC transcription. An additional contribution of this study is that we have shown that miR-375 is a PKA downstream molecule, which targets MAP3K8, and subsequently down-regulates ERK1/2 phosphorylation and the expression of NGFI-B. This novel signaling pathway mediates the effect of CRF, thus regulating POMC expression and the related hormone secretion in mouse pituitary gland (Fig. 8). In light of the results of our study, the tissue-specific miRNAs such as miR-375 play critical roles in regulating pituitary hormone synthesis and secretion and can be used as novel targets for physiological or pharmacological intervention in diseases resulting from pituitary hormone secretion abnormalities.
FIGURE 8.
The function of miR-375 in the CRF-enhanced POMC signaling pathway. CRF increases the intracellular cAMP through binding CRF receptor 1, which leads to the activation of PKA, which in turn enhances ERK1/2 phosphorylation levels. The phosphorylation of ERK1/2 activates NGFI-B and further promotes the transcription of POMC. Our results have shown that MAP3K8 mediates the CRF signal pathway through regulating ERK1/2 phosphorylation and POMC expression. However, miR-375 functions as a negative regulator in this pathway by down-regulating MAP3K8 protein levels in AtT-20 cells. CRF also inhibits the expression of miR-375 through PKA. This pathway indicates that miR-375 is a downstream molecule of PKA, which targets MAP3K8, and subsequently down-regulates ERK1/2 phosphorylation and the expression of NGFI-B. This is a novel signaling pathway that mediates the effect of CRF, which regulates the expression of POMC, thus regulating the POMC-related hormone secretion in the pituitary gland. Arrows indicate stimulation; T bars indicate inhibition.
Acknowledgments
The authors thank Professor A. F. Parlow (Harbor-UCLA Medical Center, Los Angeles, CA) for providing CRF powder.
This work was supported by the National Basic Research Program of China (2009CB941702 and 2012CB944703) and the Natural Science Foundation of China (31172287 and 31172289).
N. Zhang, J.-k. Lin, J. Chen, X.-f. Liu, J.-l. Liu, H.-S. Luo, Y.-q. Li, and S. Cui, unpublished data.
- POMC
- Pro-opiomelanocortin
- MAP3K8
- mitogen-activated protein kinase kinase kinase-8
- NGFI-B
- nerve growth factor-induced clone B
- ACTH
- adrenocorticotrophin
- PKA
- activating protein kinase A
- miRNAs
- microRNAs
- ISH
- in situ hybridization
- LNA
- locked nucleic acid
- IHC
- immunohistochemistry
- ANOVA
- analysis of variance
- MAP3K8i
- 4-(3-chloro-4-fluorophenylamino)-6-(pyridin-3-yl-methylamino)-3-cyano[1,7]naphthyridine.
REFERENCES
- 1. Raymond V., Lépine J., Giguère V., Lissitzky J. C., Côté J., Labrie F. (1981) Parallel stimulation of ACTH, β-LPH + β-endorphin and α-MSH release by α-adrenergic agents in rat anterior pituitary cells in culture. Mol. Cell. Endocrinol. 22, 295–303 [DOI] [PubMed] [Google Scholar]
- 2. Bertagna X. (1994) Proopiomelanocortin-derived peptides. Endocrinol. Metab. Clin. North Am. 23, 467–485 [PubMed] [Google Scholar]
- 3. Stevens A., White A. (2010) ACTH: cellular peptide hormone synthesis and secretory pathways. Results Probl. Cell Differ. 50, 63–84 [DOI] [PubMed] [Google Scholar]
- 4. Tsigos C., Chrousos G. P. (2002) Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J. Psychosom. Res. 53, 865–871 [DOI] [PubMed] [Google Scholar]
- 5. Heijnen C. J., Zijlstra J., Kavelaars A., Croiset G., Ballieux R. E. (1987) Modulation of the immune response by POMC-derived peptides. I. Influence on proliferation of human lymphocytes. Brain Behav. Immun. 1, 284–291 [DOI] [PubMed] [Google Scholar]
- 6. Getting S. J., Gibbs L., Clark A. J., Flower R. J., Perretti M. (1999) POMC gene-derived peptides activate melanocortin type 3 receptor on murine macrophages, suppress cytokine release, and inhibit neutrophil migration in acute experimental inflammation. J. Immunol. 162, 7446–7453 [PubMed] [Google Scholar]
- 7. Knight R. M., Farah J. M., Bishop J. F., O'Donohue T. L. (1987) CRF and cAMP regulation of POMC gene expression in corticotrophic tumor cells. Peptides 8, 927–934 [DOI] [PubMed] [Google Scholar]
- 8. Lundblad J. R., Roberts J. L. (1988) Regulation of proopiomelanocortin gene expression in pituitary. Endocr. Rev. 9, 135–158 [DOI] [PubMed] [Google Scholar]
- 9. Affolter H. U., Reisine T. (1985) Corticotropin releasing factor increases proopiomelanocortin messenger RNA in mouse anterior pituitary tumor cells. J. Biol. Chem. 260, 15477–15481 [PubMed] [Google Scholar]
- 10. Rivier C., Vale W. (1983) Modulation of stress-induced ACTH release by corticotropin-releasing factor, catecholamines and vasopressin. Nature 305, 325–327 [DOI] [PubMed] [Google Scholar]
- 11. Litvin Y., PasMantier R., Fleischer N., Erlichman J. (1984) Hormonal activation of the cAMP-dependent protein kinases in AtT20 cells. Preferential activation of protein kinase I by corticotropin releasing factor, isoproterenol, and forskolin. J. Biol. Chem. 259, 10296–10302 [PubMed] [Google Scholar]
- 12. Luini A., Lewis D., Guild S., Corda D., Axelrod J. (1985) Hormone secretagogues increase cytosolic calcium by increasing cAMP in corticotropin-secreting cells. Proc. Natl. Acad. Sci. U.S.A. 82, 8034–8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovalovsky D., Refojo D., Liberman A. C., Hochbaum D., Pereda M. P., Coso O. A., Stalla G. K., Holsboer F., Arzt E. (2002) Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: involvement of calcium, protein kinase A, and MAPK pathways. Mol. Endocrinol. 16, 1638–1651 [DOI] [PubMed] [Google Scholar]
- 14. Liu J., Lin C., Gleiberman A., Ohgi K. A., Herman T., Huang H. P., Tsai M. J., Rosenfeld M. G. (2001) Tbx19, a tissue-selective regulator of POMC gene expression. Proc. Natl. Acad. Sci. U.S.A. 98, 8674–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamolet B., Pulichino A. M., Lamonerie T., Gauthier Y., Brue T., Enjalbert A., Drouin J. (2001) A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell 104, 849–859 [DOI] [PubMed] [Google Scholar]
- 16. Drouin J., Maira M., Philips A. (1998) Novel mechanism of action for Nur77 and antagonism by glucocorticoids: a convergent mechanism for CRH activation and glucocorticoid repression of POMC gene transcription. J. Steroid Biochem. Mol. Biol. 65, 59–63 [DOI] [PubMed] [Google Scholar]
- 17. Murphy E. P., Conneely O. M. (1997) Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol. Endocrinol. 11, 39–47 [DOI] [PubMed] [Google Scholar]
- 18. Philips A., Lesage S., Gingras R., Maira M. H., Gauthier Y., Hugo P., Drouin J. (1997) Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol. Cell. Biol. 17, 5946–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Philips A., Maira M., Mullick A., Chamberland M., Lesage S., Hugo P., Drouin J. (1997) Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol. Cell. Biol. 17, 5952–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maira M M. C., Philips A, Drouin J. (1999) Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 19, 7549–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mao Z. G., He D. S., Zhou J., Yao B., Xiao W. W., Chen C. H., Zhu Y. H., Wang H. J. (2010) Differential expression of microRNAs in GH-secreting pituitary adenomas. Diagn. Pathol. 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z., Florez S., Gutierrez-Hartmann A., Martin J. F., Amendt B. A. (2010) MicroRNAs regulate pituitary development, and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression. J. Biol. Chem. 285, 34718–34728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nemoto T., Mano A., Shibasaki T. (2012) Increased expression of miR-325-3p by urocortin 2 and its involvement in stress-induced suppression of LH secretion in rat pituitary. Am. J. Physiol. Endocrinol. Metab. 302, E781–787 [DOI] [PubMed] [Google Scholar]
- 24. Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230 [DOI] [PubMed] [Google Scholar]
- 25. El Ouaamari A., Baroukh N., Martens G. A., Lebrun P., Pipeleers D., van Obberghen E. (2008) miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic β-cells. Diabetes 57, 2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Souza Rocha Simonini P., Breiling A., Gupta N., Malekpour M., Youns M., Omranipour R., Malekpour F., Volinia S., Croce C. M., Najmabadi H., Diederichs S., Sahin O., Mayer D., Lyko F., Hoheisel J. D., Riazalhosseini Y. (2010) Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor α in breast cancer cells. Cancer Res. 70, 9175–9184 [DOI] [PubMed] [Google Scholar]
- 27. Poy M. N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. (2009) miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatziapostolou M., Polytarchou C., Panutsopulos D., Covic L., Tsichlis P. N. (2008) Proteinase-activated receptor-1-triggered activation of tumor progression locus-2 promotes actin cytoskeleton reorganization and cell migration. Cancer Res. 68, 1851–1861 [DOI] [PubMed] [Google Scholar]
- 29. Makris A., Patriotis C., Bear S. E., Tsichlis P. N. (1993) Genomic organization and expression of Tpl-2 in normal cells and Moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J. Virol. 67, 4283–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patriotis C., Makris A., Bear S. E., Tsichlis P. N. (1993) Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. U.S.A. 90, 2251–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patriotis C., Makris A., Chernoff J., Tsichlis P. N. (1994) Tpl-2 acts in concert with Ras and Raf-1 to activate mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 91, 9755–9759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ceci J. D., Patriotis C. P., Tsatsanis C., Makris A. M., Kovatch R., Swing D. A., Jenkins N. A., Tsichlis P. N., Copeland N. G. (1997) Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 11, 688–700 [DOI] [PubMed] [Google Scholar]
- 33. Dermitzaki E., Tsatsanis C., Gravanis A., Margioris A. N. (2012) The calcineurin-nuclear factor of activated T cells signaling pathway mediates the effect of corticotropin releasing factor and urocortins on catecholamine synthesis. J. Cell. Physiol. 227, 1861–1872 [DOI] [PubMed] [Google Scholar]
- 34. Obernosterer G., Martinez J., Alenius M. (2007) Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nature Protoc. 2, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 35. Sei C., Toneff T., Aaron W., Hook V. Y. (2002) Regulation of cellular α-MSH and β-endorphin during stimulated secretion from intermediate pituitary cells: involvement of aspartyl and cysteine proteases in the control of cellular levels of α-MSH and β-endorphin. Peptides 23, 1409–1418 [DOI] [PubMed] [Google Scholar]
- 36. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krüger J., Rehmsmeier M. (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arzt E., Holsboer F. (2006) CRF signaling: molecular specificity for drug targeting in the CNS. Trends Pharmacol. Sci. 27, 531–538 [DOI] [PubMed] [Google Scholar]
- 39. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gavrin L. K., Green N., Hu Y., Janz K., Kaila N., Li H. Q., Tam S. Y., Thomason J. R., Gopalsamy A., Ciszewski G., Cuozzo J. W., Hall J. P., Hsu S., Telliez J. B., Lin L. L. (2005) Inhibition of Tpl2 kinase and TNF-α production with 1,7-naphthyridine-3-carbonitriles: synthesis and structure-activity relationships. Bioorg. Med. Chem. Lett. 15, 5288–5292 [DOI] [PubMed] [Google Scholar]
- 41. Becquet D., Guillaumond F., Bosler O., François-Bellan A. M. (2001) Long-term variations of AP-1 composition after CRH stimulation: consequence on POMC gene regulation. Mol. Cell. Endocrinol. 175, 93–100 [DOI] [PubMed] [Google Scholar]
- 42. Bak M., Silahtaroglu A., Møller M., Christensen M., Rath M. F., Skryabin B., Tommerup N., Kauppinen S. (2008) MicroRNA expression in the adult mouse central nervous system. RNA 14, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rajewsky N., Socci N. D. (2004) Computational identification of microRNA targets. Dev. Biol. 267, 529–535 [DOI] [PubMed] [Google Scholar]
- 44. Hjalt T. A., Semina E. V., Amendt B. A., Murray J. C. (2000) The Pitx2 protein in mouse development. Dev. Dyn 218, 195–200 [DOI] [PubMed] [Google Scholar]
- 45. Reyes R., Martínez S., González M., Tramu G., Bello A. R. (2008) Origin of adenohypophysial lobes and cells from Rathke's pouch in Swiss albino mice. Proliferation and expression of Pitx 2 and Calbindin D28K in corticotropic and somatotropic cell differentiation. Anat Histol. Embryol. 37, 263–271 [DOI] [PubMed] [Google Scholar]
- 46. Schneeberger M., Altirriba J., García A., Esteban Y., Castaño C., García-Lavandeira M., Alvarez C. V., Gomis R., Claret M. (2012) Deletion of miRNA processing enzyme Dicer in POMC-expressing cells leads to pituitary dysfunction, neurdegeneration and development of obesity. Mol. Metab., in press [DOI] [PMC free article] [PubMed] [Google Scholar]