Background: TGFβ/BMP signaling plays an important role in palate development.
Results: Inactivation of Tak1 in the neural crest lineage leads to cleft palate associated with malformed tongue and micrognathia, resembling human Pierre Robin sequence clefting.
Conclusion: Cleft palate formation in Tak1 mutants is a secondary consequence of abnormal tongue development.
Significance: TAK1 could represent a candidate gene for human Pierre Robin sequence clefting.
Keywords: Craniofacial Development, Embryo, p38 MAPK, Transforming Growth Factor β (TGFβ), Transgenic Mice, Fgf10, TGFβ/BMP Signaling, Tak1, Cleft Palate, Tongue
Abstract
Cleft palate represents one of the most common congenital birth defects in humans. TGFβ signaling, which is mediated by Smad-dependent and Smad-independent pathways, plays a crucial role in regulating craniofacial development and patterning, particularly in palate development. However, it remains largely unknown whether the Smad-independent pathway contributes to TGFβ signaling function during palatogenesis. In this study, we investigated the function of TGFβ activated kinase 1 (Tak1), a key regulator of Smad-independent TGFβ signaling in palate development. We show that Tak1 protein is expressed in both the epithelium and mesenchyme of the developing palatal shelves. Whereas deletion of Tak1 in the palatal epithelium or mesenchyme did not give rise to a cleft palate defect, inactivation of Tak1 in the neural crest lineage using the Wnt1-Cre transgenic allele resulted in failed palate elevation and subsequently the cleft palate formation. The failure in palate elevation in Wnt1-Cre;Tak1F/F mice results from a malformed tongue and micrognathia, resembling human Pierre Robin sequence cleft of the secondary palate. We found that the abnormal tongue development is associated with Fgf10 overexpression in the neural crest-derived tongue tissue. The failed palate elevation and cleft palate were recapitulated in an Fgf10-overexpressing mouse model. The repressive effect of the Tak1-mediated noncanonical TGFβ signaling on Fgf10 expression was further confirmed by inhibition of p38, a downstream kinase of Tak1, in the primary cell culture of developing tongue. Tak1 thus functions to regulate tongue development by controlling Fgf10 expression and could represent a candidate gene for mutation in human PRS clefting.
Introduction
Cleft palate, a common birth defect in humans with a prevalence ranging between 1/700 and 1/1,000, arises from genetic or environmental perturbations in palate development. The definite mammalian palate forms through the union of a primary palate and two secondary palatal shelves. Formation of the secondary palate involves palatal shelf growth, elevation of the shelves, fusion between paired shelves, and the disappearance of the midline epithelial seam (1). Whereas disruptions to any of these steps could result in a cleft palate defect, it has been estimated that among those afflicted by isolated cleft palate, approximately 90% of the cases could be attributed to defective palate elevation (2). Palatal shelf elevation, a step that brings the palatal shelves from a vertical to a horizontal position above the tongue, is triggered by intrinsic forces within the palatal shelf and by the coordinated growth and movement of other craniofacial and oral structures (1). Intrinsic or extrinsic defects that cause failed or delayed palatal shelf elevation and ultimately cleft palate formation have been well illustrated in many mutant mouse models (3–16). Among these models, physical obstructions such as abnormal palatal shelf-mandible/tongue fusion, a small mandible, or steric hindrance by the tongue appear to be major factors. In humans, cleft palate attributed to physical obstruction to palatal shelf elevation by the tongue and micrognathia is clinically known as the Pierre Robin sequence (PRS)5 (17). Mutations in several genes, including SATB2 and SOX9, have been implicated in PRS-like clefting (18–20).
The tongue is a muscular organ derived from two mesenchymal cell lineages in addition to the covering epithelium: the mesoderm-derived myogenic progenitors and the cranial neural crest (CNC)-derived connective tissues (21–23). Genetic labeling experiments have shown that the mesoderm-derived myogenic progenitors primarily occupy the core of the tongue primordium, whereas the CNC-derived cells largely contribute to the region that surrounds the myogenic core as well as the tongue septum (21). Interactions between these two groups of cells are essential for tongue development, and TGFβ-mediated FGF signaling plays a critical role in such tissue-tissue interactions (21, 24).
Transforming growth factor β (TGFβ) signaling plays diverse roles in a variety of cellular processes during development (25). Signaling of the TGFβ superfamily, including bone morphogenetic proteins (BMPs), is transduced into cells by binding of ligands to the type I and type II serine/threonine kinase receptor complex. Canonical TGFβ/BMP signaling occurs via activation of receptor-regulated Smads (R-Smads) 2/3 and 1/5/8, respectively. The activated R-Smads then bind to common Smad (Smad4) and enter the nucleus to interact with other transcription factors and regulate downstream gene expression. In addition to the canonical Smad pathway, TGFβ/BMP receptors can also activate members of the mitogen-activated protein kinase (MAPK) pathway, known as the noncanonical pathway, including the TGFβ-activated kinase 1 (Tak1) (26, 27). Tak1, a member of the MAPKKK family, was originally as a key regulator of MAPK kinase activation in TGFβ signaling pathways (27). Upon activation, Tak1 phosphorylates MKK3/6 directly, leading to the activation of a number of downstream kinases including p38 and Jun kinases (28–30).
Many studies have demonstrated the critical roles of TGFβ/BMP signaling in every step of palatogenesis (31–35). However, most of these studies were focused on the ligands and receptors as well as Smad-dependent signaling pathways. Recent studies have implicated a role for p38 MAPK, a downstream kinase of Tak1, in TGFβ-induced palatal fusion and cell proliferation in the palatal mesenchyme (36, 37). In addition, Tak1-haploinsufficiency in CNC cells rescued the cleft palate defect in Tgfbr2 mutant mice (38). However, the role of Tak1 in palate development remains elusive. In this study, we examined Tak1 expression and investigated its function during palatogenesis by tissue-specific inactivation.
EXPERIMENTAL PROCEDURES
Animals
The generation and genotyping of Tak1F/F, Wnt1-Cre, K14-Cre, Osr2-Cre, and R26R reporter mice have been described previously (39–43). To generate Wnt1-Cre;Tak1F/F mice, Wnt1-Cre;Tak1F/+ mice were crossed with Tak1F/F mice. K14-Cre;Tak1F/F and Osr-2Cre;Tak1F/F mice were obtained by crossing K14-Cre;Tak1F/+ or Osr2-Cre;Tak1F/+ mice with Tak1F/F mice, respectively. Fgf10 conditional transgenic mice (pMes-Fgf10) were generated by pronuclear injection of a construct containing the full-length mouse Fgf10 cDNA inserted at the 3′ end of the LoxP-flanked STOP cassette and the 5′ end of the IRES-Egfp sequence under the control of the chick β-actin promoter. Embryos were harvested from timed-pregnant mice. Animals and procedures used in this study were approved by the IACUC of Tulane University.
Histology, Immunohistochemical Staining, and in Situ Hybridization
Staged embryos were harvested in ice-cold phosphate-buffered saline (PBS), and embryonic heads were removed and fixed in 4% paraformaldehyde/PBS overnight at 4 °C. For histological and in situ hybridization analyses, samples were dehydrated through a graded ethanol series and embedded in paraffin. Serial sections were made at 10 μm and subjected to standard hematoxylin/eosin staining or to section in situ hybridization using nonradioactive riboprobes, as described previously (44). Three independent experiments of in situ hybridization were performed for the expression of each gene. For immunohistochemical staining, samples were washed with 30% sucrose/PBS, then embedded in O.C.T. compound (Tissue-Tek) and cryo-sectioned. Sections were washed in PBS and blocked with 10% goat serum in PBS for 1 h prior to being incubated at 4 °C overnight with the primary antibodies, including anti-Tak1 (Santa Cruz Biotechnology), anti-FGF10 (Santa Cruz Biotechnology), anti-MyoD (Santa Cruz Biotechnology), and anti-phosphorylated-p38 (R&D Systems). For negative controls, the primary antibody was omitted. For 3,30-diaminobenzidine reaction, biotinylated anti-rabbit IgG (H+L) (Vector Laboratories) was used as the secondary antibody. Sections were then treated with streptavidin-HRP according to the manufacturer's instructions. Finally, the specimens were visualized using 3,30-diaminobenzidine reagent kit (Invitrogen). Slides were counterstained in 0.5% methyl green in 0.1 m sodium acetate for 1 min, washed in water, dehydrated in 100% n-butyl alcohol, rinsed in xylene, and mounted with Permount (Fisher Scientific). For fluorescent visualization, FITC-conjugated (Alexa Fluor 488; Invitrogen) and Texas Red-conjugated (Alexa Fluor 595; Invitrogen) antibodies were used.
Cell Proliferation Assay
BrdU labeling was conducted to determine the cell proliferation rate, as described previously (38, 45). BrdU was injected into timed pregnant mice peritoneally for 1 h prior to embryo harvest at a dose of 1.5 ml of labeling reagent/100 g of body weight using the BrdU Labeling and Detection Kit II from Roche Applied Science. Samples were fixed in Carnoy's fixative, ethanol-dehydrated, paraffin-embedded, and sectioned at 5 μm. The sections were subjected to immunodetection according to the manufacturer's protocol. Cell proliferation rates were measured by counting BrdU-positive cells and total cells in defined arbitrary areas. The outcome was presented as percentage of labeled cells out of the total number of cells in the defined arbitrary areas, and Student's t test was used to determine whether there was a significant difference between the mutants and wild type controls. Three independent BrdU-labeling experiments with minimal three samples of each genotype were performed.
In Vitro Roller Cultures
Roller cultures were performed essentially as described previously (7). Briefly, embryonic day (E) 13.5 mice were collected and decapitated individually in sterile ice-cold PBS. The tail of each embryo was subjected to DNA extraction for genotyping. Each embryonic head, either with removal of the tongue or removal of the mandible, was placed in a 20-ml glass bottle containing 2 ml of DMEM supplemented with 20% fetal calf serum. Bottles were fixed in a vertical position on a rotary apparatus rotating at a speed of 4 rpm in an incubator at 37 °C and 5% CO2. After 24 h in culture, samples were washed in PBS, fixed in 4% paraformaldehyde, and processed for histological examination, as described above.
Tongue Height Measurements
E13.5 and E14.5 embryonic heads (n = 3 for each genotype) were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned in the coronal plane at 10 μm. Littermates of wild type controls and transgenic or knock-out mice were processed in parallel and used for comparison. Sections were subjected to hematoxylin/eosin staining and photographed using an Olympus digital capture system with a digital caliper placed on the image. Sections of both wild type and transgenic/mutant embryos at comparable levels along the anterior to the posterior axis were selected, and the height of the tongue was measured from the bottom of the tongue to the most superior aspect of the mid-tongue epithelium along the midline. Three continuous sections at each level were measured. Statistical analysis of the measurements was performed using Excel and a paired Student's t test. Statistical significance was determined if p < 0.05.
Immunoblotting Analysis
Immunoblots were performed as described previously (46, 47). The antibodies used for immunoblotting were rabbit polyclonal antibody against FGF10 (Millipore) and mouse monoclonal antibody against GAPDH (Chemicon).
Tongue Mesenchymal Cell Culture
Primary tongue mesenchymal cells were obtained from E13.5 wild type embryos. Briefly, the tongues were isolated and trypsinized for 30 min at 37 °C in a CO2 incubator. After pipetting thoroughly, cells were cultured in DMEM containing 10% fetal bovine serum supplemented with penicillin, streptomycin, l-glutamate, sodium pyruvate, and nonessential amino acids. Tongue mesenchymal cells were treated with or without p38 MAPK inhibitor SB203580 (10 μm) for 24 h prior to being subjected for RNA or protein extraction.
Tongue Organ Culture
The tongues were microdissected from E13.5 embryos and cultured in serum-free chemically defined medium as described previously (36, 38). Agarose beads soaked with either BSA (1 mg/ml) or FGF10 (10 μg/ml) (R&D Systems) or FGF10 neutralizing antibody (FGF10 Nab; 10 μg/ml) (Millipore) were implanted onto explanted tongue samples. After 20 h in culture, explants were pulsed with BrdU labeling reagent (0.1 mg/ml) for 4 h prior to harvest, fixed in 4% paraformaldehyde/0.1 m phosphate buffer (pH 7.4), and processed as described previously (38, 47). Total and BrdU-positive cells around the beads were counted.
Quantitative RT-PCR
Total RNA was isolated from primary mouse tongue mesenchymal cells with the QIAshredder and RNeasy Micro extraction kit (Qiagen), as described previously (36, 47). The following PCR primers were used: Fgf10, 5′-AAGAACGGCAAGGTCAGCGGGA-3′ and 5′-CATTTGCCTGCCATTGTGCTGC-3′; Gapdh, 5′-AACTTTGGCATTGTGGAAGG-3′ and 5′-ACACATTGGGGGTAGGAACA-3′.
MicroCT Analysis
MicroCT analysis was performed using SCANCO μCT50 at the University of Southern California Molecular Imaging Center. The microCT images were acquired with the x-ray source at 70 kVp and 250 μA. The data were collected at a high resolution of 20 μm. The reconstruction was done with AVIZO 6 (Visualization Sciences Group), as described previously (38).
RESULTS
Expression of Tak1 in the Developing Palate and Tongue
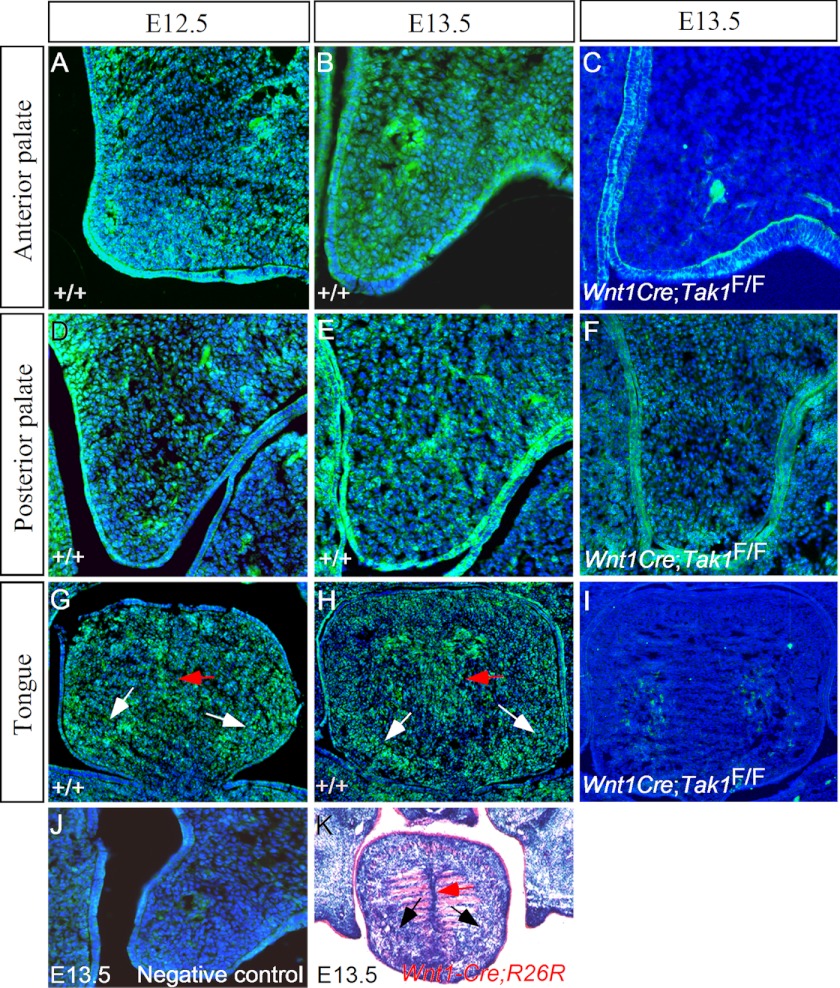
To study the potential function of Tak1 in palate development, we first examined Tak1 expression in the developing secondary palatal shelves using immunohistochemistry. Our results show that Tak1 expression was detected in the epithelium and mesenchyme of the developing palatal shelf at E12.5 and E13.5 with a relatively higher level of mesenchymal expression in the anterior portion (Fig. 1, A, B, D, and E). At the same stages, Tak1 expression was also detected in the surrounding mesenchymal cells circumscribing the myogenic core of the developing tongue (Fig. 1, G and H). In addition, Tak1 expression was observed in the tongue septum region. These Tak1-expressing domains overlap with areas populated by CNC-derived cells in the tongue primordium (Fig. 1K and Ref. 21).
FIGURE 1.
Expression of Tak1 in the developing palate and tongue. A, B, D, and E, Tak1 protein is present in the palatal epithelium and mesenchyme in the anterior and posterior domain of E12.5 and E13.5 wild type palatal shelves. G and H, Tak1 expression is also seen in the peripheral mesenchymal cells (white arrows) and lingual septum area (red arrows) of the wild type developing tongue at E12.5 and E13.5. C, F, and I, Tak1 protein is dramatically reduced or almost undetectable in the palatal mesenchyme and the tongue of E13.5 Wnt1-Cre;Tak1F/F embryo. Note that Tak1 protein remains in the palatal epithelium of the mutant. J, negative control for immunohistochemistry. K, LacZ staining shows population of CNC-derived cells in the peripheral mesenchymal region (black arrows) and the lingual septum domain (red arrow) in the tongue of E13.5 Wnt1-Cre;R26R embryo.
Tak1 Deficiency in the CNC Lineage Leads to Complete Clefting of the Secondary Palate
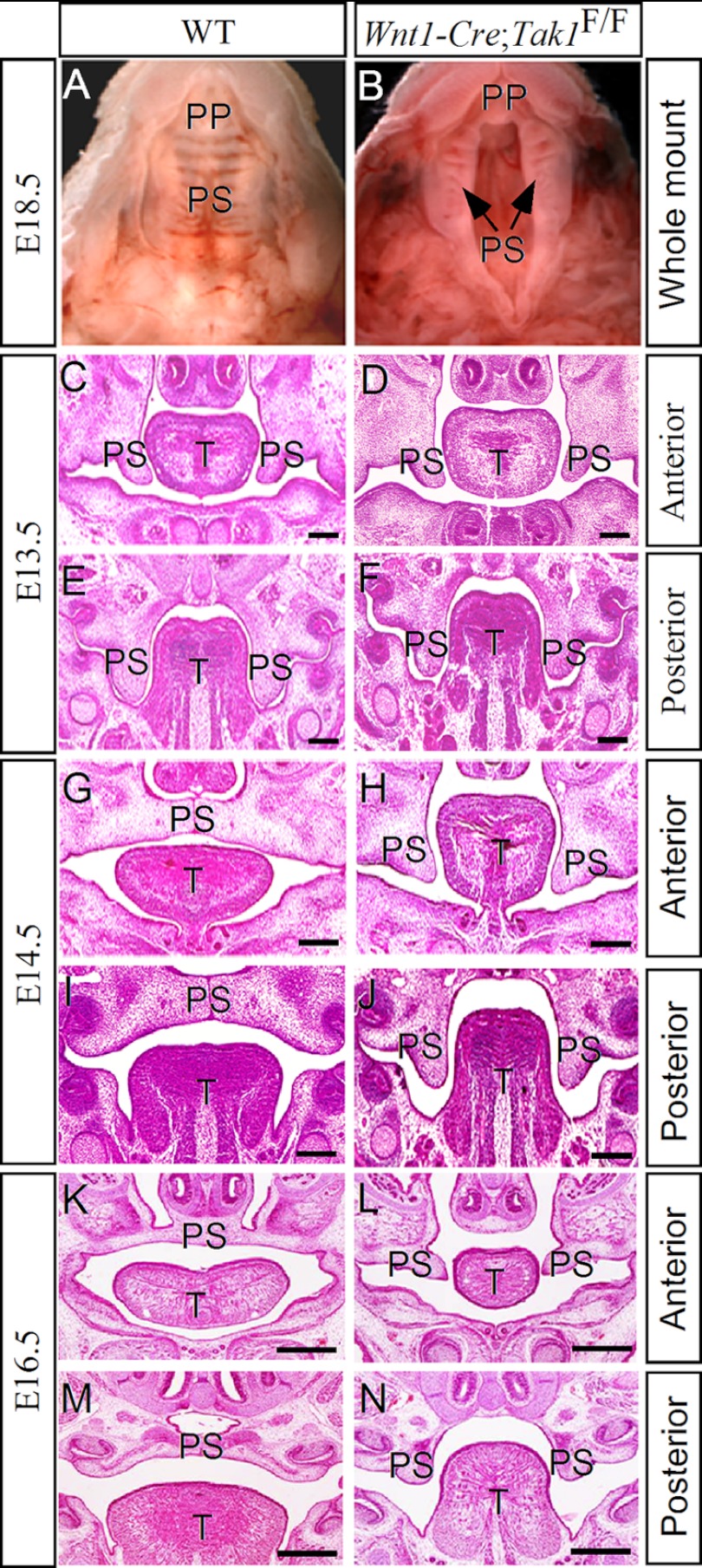
Because Tak1 is expressed in the CNC-derived palatal mesenchyme and the Tak1 downstream kinase p38 MAPK is implicated in proliferation of palatal mesenchyme (36), we investigated the role of Tak1 in palatal development by inactivating Tak1 in the CNC-derived cells using the Wnt1-Cre transgenic allele and floxed Tak1 mice. The efficiency of Tak1 gene deletion by the Wnt1-Cre allele in the palatal and tongue mesenchyme was shown by immunohistochemical staining on Tak1 protein in the mutants (Fig. 1, C, F, and I). Wnt1-Cre;Tak1F/F mice died perinatally, exhibiting a cleft palate defect with 100% penetrance, micrognathia, and hypoplastic calvarial bones (Fig. 2B and supplemental Fig. S1). Histological examinations revealed that morphological structures of the palatal shelves along the anterior-posterior axis in the mutants were comparable with the wild type controls up to E13.5 (Fig. 2, C and D). However, a pronounced aberration in palate development was observed in the mutants at E14.5, by which time the wild type palatal shelves have elevated to above the tongue and begun to fuse at the midline (Fig. 2, G and I). In Wnt1-Cre;Tak1F/F embryos, both the anterior and posterior portions of the palatal shelves failed to elevate and remained in the vertical position on both sides of a heightened tongue (Fig. 2, H and J). At E16.5, the palatal shelves have fused completely in the wild type controls, but the mutant palate shelves remained at the vertical position (Fig. 2, K–N). These observations suggest a role for Tak1 in the palatal mesenchyme during palatogenesis.
FIGURE 2.
Deletion of Tak1 in the CNC lineage leads to failed elevation of secondary palate and a complete clefting. A and B, whole mount views of E18.5 wild type (A) and Wnt1-Cre;Tak1F/F (B) mice show a completely clefting of the secondary palate in mutant. C–F, histological sections show comparable morphology of control (C and E) and mutant (D, F) palatal shelves at both the anterior and posterior domains at E13.5. G–N, histological sections show failed elevation of palatal shelves at E14.5 and E16.5 in Wnt1-Cre;Tak1F/F mice (H, J, L, and N) compared with stage-matched controls (G, I, K, and M). T, tongue; PP, primary palate; PS, palatal shelf. Scale bars, 100 μm (C–J), 500 μm (K–N).
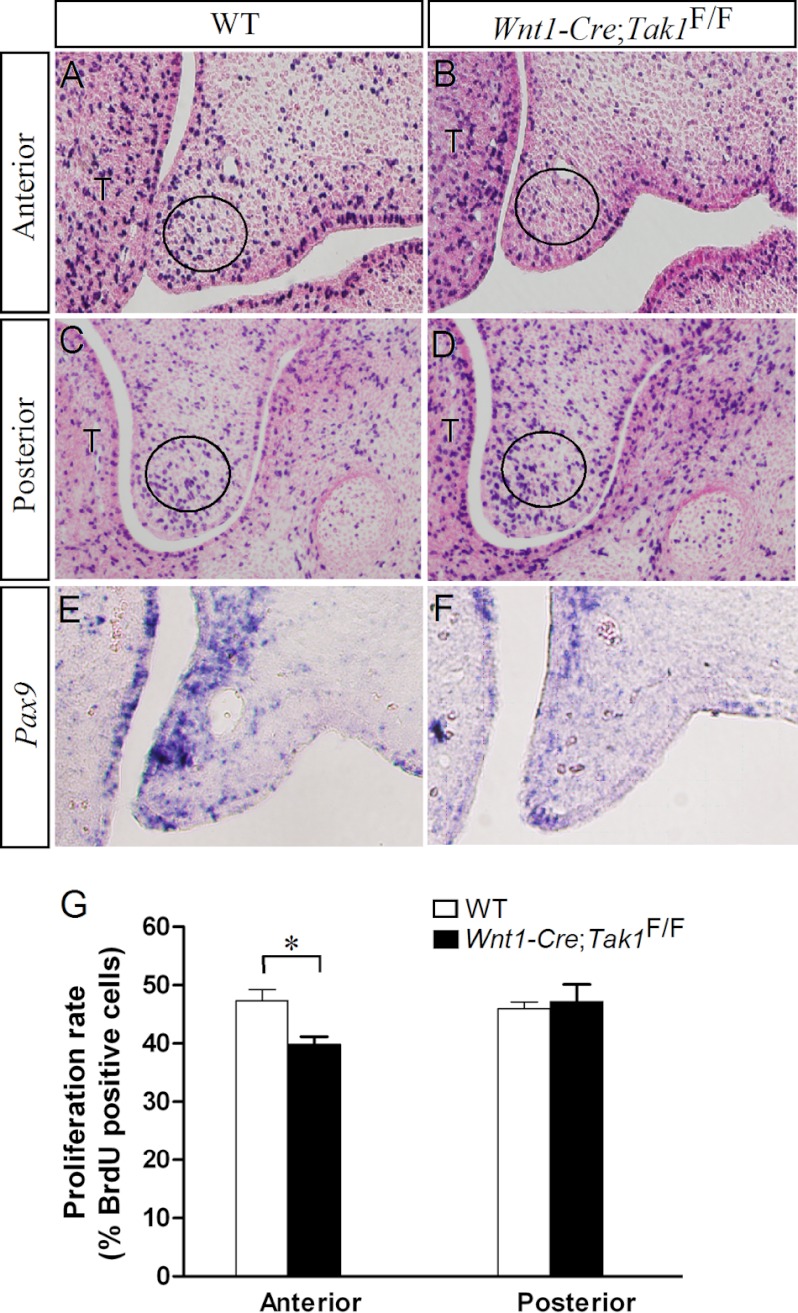
Because defective cell proliferation was found to be associated with an altered activation of p38 MAPK in the palatal mesenchyme (36), we performed a BrdU labeling experiment to determine whether the absence of Tak1 could cause defective cell proliferation in the palatal mesenchyme of Wnt1-Cre;Tak1F/F embryos at E13.5 before the obvious palatal defect became discernible. We found that the mutants exhibited a reduction of approximately 9% on the cell proliferation index in the anterior palatal mesenchyme compared with its wild type counterpart (Fig. 3, A, B, and G). However, the cell proliferation rate in the posterior palatal mesenchyme of the mutants was comparable with the wild type controls (Fig. 3, C, D, and G). This restricted change in cell proliferation rate in the anterior palatal mesenchyme is consistent with a higher level of Tak1 expression in the anterior palatal shelves (Fig. 1).
FIGURE 3.
Cell proliferation in the developing palate. A–D, BrdU labeling shows cell proliferation rate in E13.5 wild type control (A and C) and Wnt1-Cre;Tak1F/F (B and D) palatal shelves. The circles demarcate the arbitrary regions for counting total cells and BrdU-positive cells. E and F, in situ hybridization shows down-regulation of Pax9 in the palatal mesenchyme of E13.5 Wnt1-Cre;Tak1F/F embryo (E) compared with the control (F). G, comparison of the ratio of BrdU-labeled cells in the defined regions of the palatal shelves of E13.5 wild type and Wnt1-Cre;Tak1F/F embryos is shown. *, p < 0.05. T, tongue. Error bars, S.E.
We next compared the expression of several genes in the wild type and Wnt1-Cre;Tak1F/F palatal shelves that are known to be either downstream targets of TGFβ/BMP signaling or to be involved in cell proliferation regulation in the palatal mesenchyme during palatogenesis. These genes included Msx1, Bmp2, Bmp4, Osr2, and Pax9 (11, 45, 48). Because Tak1 is highly expressed in the anterior palatal mesenchyme and a defective cell proliferation was found there, we focused on the expression of these selected genes in the anterior palate. Our results showed that Bmp2, Bmp4, Msx1, and Osr2 were expressed normally in the mutant palate (supplemental Fig. S2). However, we indeed observed a down-regulation of Pax9 expression in the mutant (Fig. 3, E and F), suggesting that Tak1 acts as a mediator of Pax9 expression in the developing palate and that the down-regulation of Pax9 expression may contribute to the reduced cell proliferation rate in the Wnt1-Cre;Tak1F/F palatal shelves.
Tak1 Is Not an Intrinsic Factor Essential for Palatogenesis
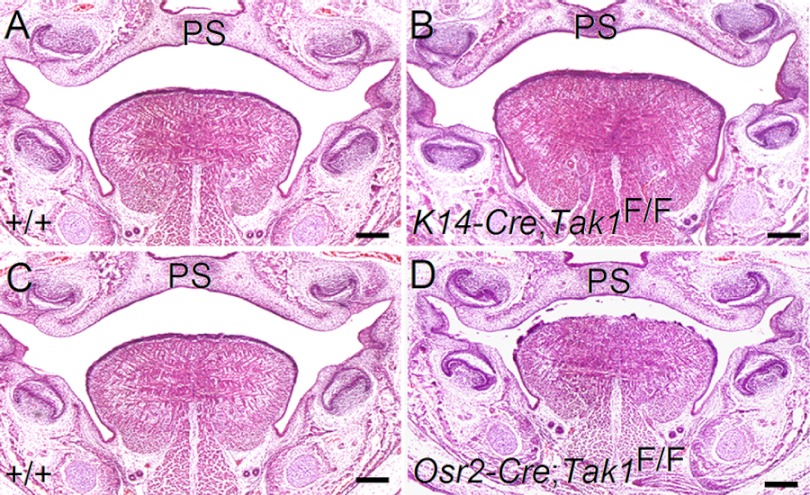
Because Tak1 protein is also present in the palatal epithelium as well, we sought to address the question of whether Tak1 is also required in the epithelium for palate development. We used a K14-Cre transgenic mouse line to inactivate Tak1 specifically in the embryonic epithelium, including the palatal epithelium. Histological examination showed normal palate formation in such mice (K14-Cre;Tak1F/F) (Fig. 4B), suggesting that Tak1 is dispensable in the epithelium for normal palatogenesis.
FIGURE 4.
Normal palate development in mice lacking Tak1 in either palatal epithelium or mesenchyme. Coronal sections through mid-level palate of E16.5 wild type control (A and C) and K14-Cre;Tak1F/F (B) and Osr2-Cre;Tak1F/F (D) mice show normal palate formation. Scale bars, 200 μm. PS, palatal shelf.
Because CNC cells contribute to the formation of a variety of craniofacial organs and Wnt-Cre;Tak1F/F mice exhibited a complete clefting of the secondary palate associated with other craniofacial anomalies, we asked whether the cleft palate defect in Wnt1-Cre;Tak1F/F mice is the direct consequence of loss of Tak1 in the palatal mesenchyme or whether it is instead secondary to other craniofacial organ defects. To address this question, we utilized the Osr2-Cre mouse line, which expresses Cre recombinase specifically in the palatal mesenchyme starting at E11.5, the time when mouse palatogenesis begins (41). Surprisingly, Osr2-Cre;Tak1F/F mice developed a normal palate (Fig. 4C) and could even survive to adulthood (n > 10, data not shown). These observations indicate that Tak1 is not an intrinsic factor in the palatal mesenchyme that is required for palate development. The failed palate elevation in Wnt1-Cre;Tak1F/F mice is a secondary consequence to extrinsic defects.
Malformed Tongue Obstructs Palate Elevation in Wnt1-Cre;Tak1F/F Mice
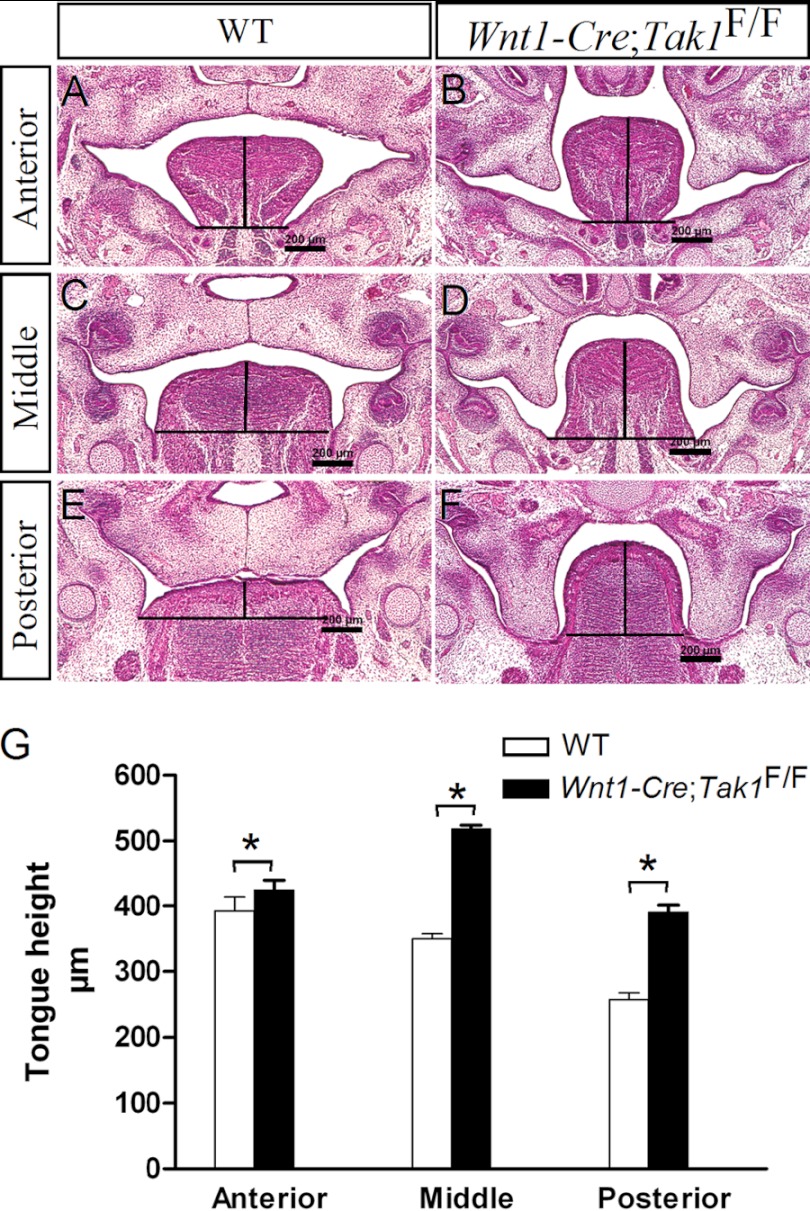
Steric hindrance of a malformed tongue represents a major extrinsic obstruction for palate elevation. We therefore set out to examine the histological phenotype of the tongue in Wnt1-Cre;Tak1F/F embryos. At E13.5, both wild type and mutant mice exhibited similar tongue morphology along the anterior-posterior axis (supplemental Fig. S3). However, at E14.5, although the width of the mutant tongue did not increase, the height of the tongue was increased significantly (p < 0.01) along the anterior-posterior axis compared with wild type littermates (Fig. 5).
FIGURE 5.
Comparison of tongue height in E14.5 control and Wnt1-Cre;Tak1F/F embryos. Measurement of tongue height along the anterior-posterior axis of E14.5 wild type control (A, C, and E) and Wnt1-Cre;Tak1F/F (B, D, and F) embryos shows a significantly heightened tongue (G) in Wnt1-Cre;Tak1F/F embryo. *, p < 0.01. Scale bars, 200 μm.
To determine whether the malformed tongue is responsible for the failed palate elevation in Wnt1-Cre;Tak1F/F mice, we performed in vitro organ culture on a rotary apparatus. E13.5 embryos were harvested from mating between Wnt1-Cre;Tak1F/+ and Tak1F/F mice. Embryonic heads were collected individually, and either the tongue or the mandible was removed. Each head without its mandible or tongue was placed in a bottle and subjected to roller culture for 24 h, then processed for histological examination after genotyping. As shown in Fig. 6, similar to the wild type controls (7 of 7; Fig. 6A), the palatal shelves of the mutant head without the mandible (8 of 8; Fig. 6B) were able to elevate after 24 h in culture. Furthermore, in both control (6 of 6) and mutant (6 of 6) samples with removal of the tongue, the palatal shelves elevated and made contact after 24 h in culture (Fig. 6, C and D). Identical results were also observed by other group.6 These observations indicate that the failure of palate elevation is indeed secondary to physical obstruction by the malformed tongue as well as the reduced size of the mandible in Wnt1-Cre;Tak1F/F mice, resembling human PRS cleft of the secondary palate (17).
FIGURE 6.
Tak1 is not an intrinsic regulator of palatal shelf elevation. A and B, histological sections show palatal shelf elevation in E13.5 wild type (A) and Wnt1-Cre;Tak1F/F (B) embryonic head with removal of the mandible after 24 h in roller culture. C and D, histological sections show elevation and contact of palatal shelves in wild type (C) and Wnt1-Cre;Tak1F/F (D) embryonic head with removal of the tongue after 24 h in roller culture. M, Meckel's cartilage; PS, palatal shelf.
Enhanced Cell Proliferation Is Associated with Overexpression of Fgf10 in the Developing Tongue of Wnt1-Cre;Tak1F/F Mice
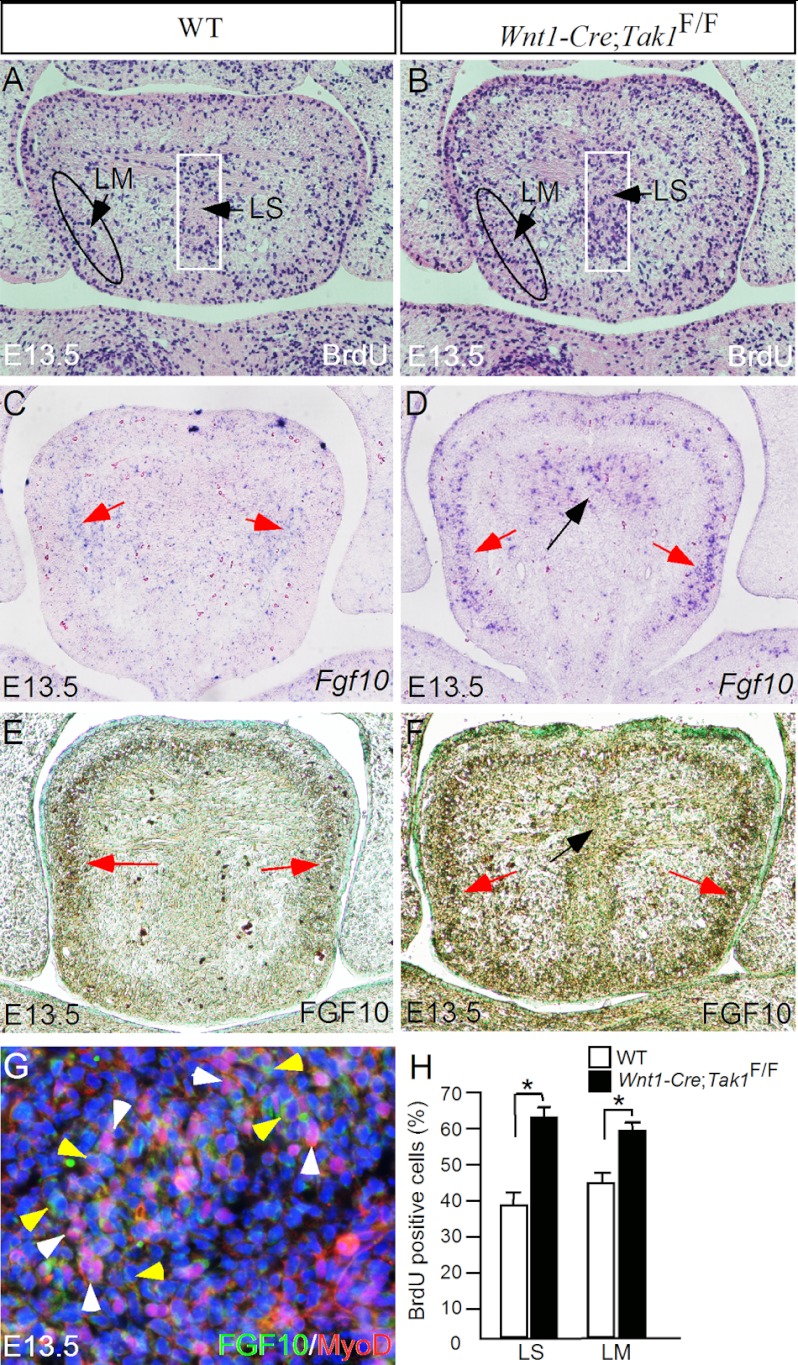
To determine whether the heightened tongue in Wnt1-Cre;Tak1F/F embryos resulted from an increased cell proliferation rate, we performed BrdU labeling experiments on the tongue primordia in both wild type controls and mutants at E13.5, before the time at which the tongue phenotype becomes obvious. We counted BrdU-positive cells and total cells in two arbitrarily defined regions populated by CNC-derived cells (Fig. 1K): the lingual septum area and the lower peripheral region where the longitudinal myogenic cells also reside (Fig. 7, A and B). The percentage of BrdU-positive cells within these two defined areas of the control and mutant tongues was compared. As summarized in Fig. 7H, the cell proliferation rate is indeed increased significantly (p < 0.01) in mutants in both regions, which appears to contribute to the malformed tongue in mutants.
FIGURE 7.
Enhanced cell proliferation in the tongue of Wnt1-Cre;Tak1F/F mice is associated with overexpression of Fgf10. A and B, BrdU labeling in the tongue of E13.5 control (A) and Wnt1-Cre;Tak1F/F embryo (B). BrdU-positive cells within the two arbitrary regions designated as LM (lingual septum) and LS (longitudinal muscle) were counted, and the percentage of the positive cells among the total cells within the arbitrary region was used for comparison between the control and mutant. C–F, in situ hybridization (C and D) and immunohistochemical staining (E and F) showing enhanced FGF10 mRNA and protein expression in the mutant tongue (D and F) at E13.5 compared with the controls (C and E). Red arrows point to in the surrounding mesenchymal tissue, and black arrows point to lingual septum domain where FGF10 expression is enhanced. G, double immunohistochemical staining showing expression of FGF10 (green color) and MyoD (red color) in distinct cell populations; white arrowheads, MyoD-positive cells, and yellow arrowheads, FGF10-expressing cells. H, comparison of BrdU-labeled cells in designated areas of the tongue in controls and mutants. *, p < 0.01. Error bars, S.E.
It has been demonstrated previously that FGF10 from the CNC-derived mesenchymal cells acts to induce proliferation of mesoderm-derived myogenic progenitors in the developing tongue (21, 24). Our double immunohistochemical staining confirmed distinct populations of FGF10-expressing cells and MyoD-positive cells in the developing tongue (Fig. 7G). We then determined whether inactivation of Tak1 could lead to overexpression of FGF10 in the CNC-derived mesenchymal cells in the developing tongue by in situ hybridization and immunohistochemical staining. We found that in the wild type controls, FGF10 mRNA and proteins were detected primarily in the peripheral mesenchyme at E13.5 and E14.5 (Fig. 7, C and E, and data not shown). In the Wnt1-Cre;Tak1F/F mutant tongue, we detected dramatically elevated levels of FGF10 mRNA and proteins at the same stages (Fig. 7, D and F, and data not shown).
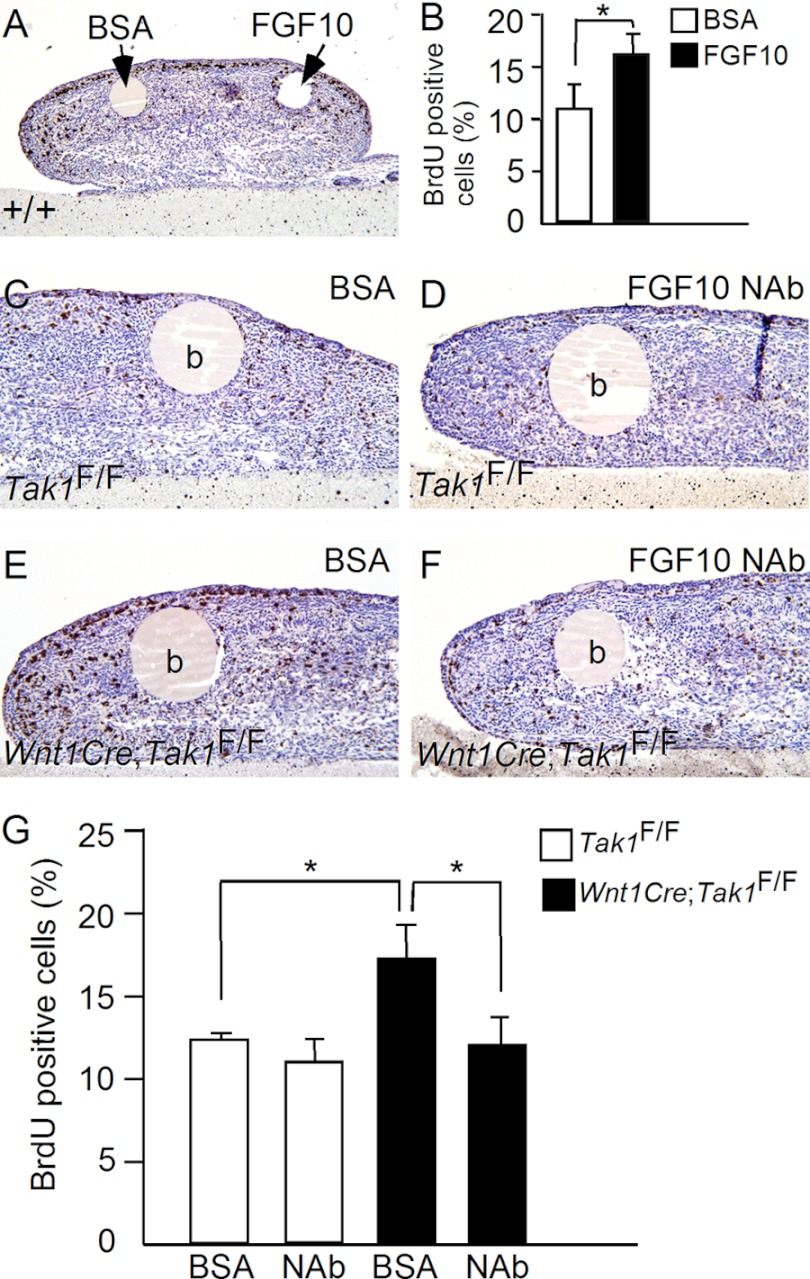
To confirm that the enhanced Fgf10 expression in the CNC-derived mesenchymal cells is responsible for the elevated cell proliferation rate in Wnt1-Cre;Tak1F/F tongue, we performed in vitro organ culture of developing tongue with protein-soaked agarose beads. We first tested whether exogenous applied FGF10 protein was able to stimulate cell proliferation in the tongue by implanting BSA and FGF10-soaked beads to the opposite side of E13.5 wild type tongue explants (n = 5) in Trowell organ culture (Fig. 8A). After 24 h in culture, explants were impulse with BrdU labeling reagent, and cell proliferation rates were examined. Indeed, exogenously applied FGF10 protein induced a significant amount of BrdU positive cells around the bead compared with BSA control (Fig. 8, A and B). We subsequently examined whether FGF10-neutralizing antibody would rescue aberrant cell proliferation in Wnt1Cre;Tak1F/F tongue. As shown in Fig. 8, whereas application of FGF10 antibody to E13.5 control tongue explants (n = 5) slightly reduced cell proliferation rate compared with BSA beads, FGF10 antibody was able to reduce cell proliferation rate significantly in E13.5 Wnt1Cre;Tak1F/F tongue explants (n = 5) compared with BSA controls (n = 5). Cell proliferation rate in the mutant tongue was resumed to the control level (Fig. 8G).
FIGURE 8.
FGF10 stimulates tongue cell proliferation in organ culture. A and B, BrdU labeling shows FGF10 significantly stimulates cell proliferation in E13.5 wild type tongue compared with BSA control. C–G, BrdU labeling shows inhibition of cell proliferation by FGF10-neutralizing antibody in E13.5 Wnt1Cre;Tak1F/F tongue explants. *, p < 0.05. b, bead; NAb, neutralizing antibody. Error bars, S.E.
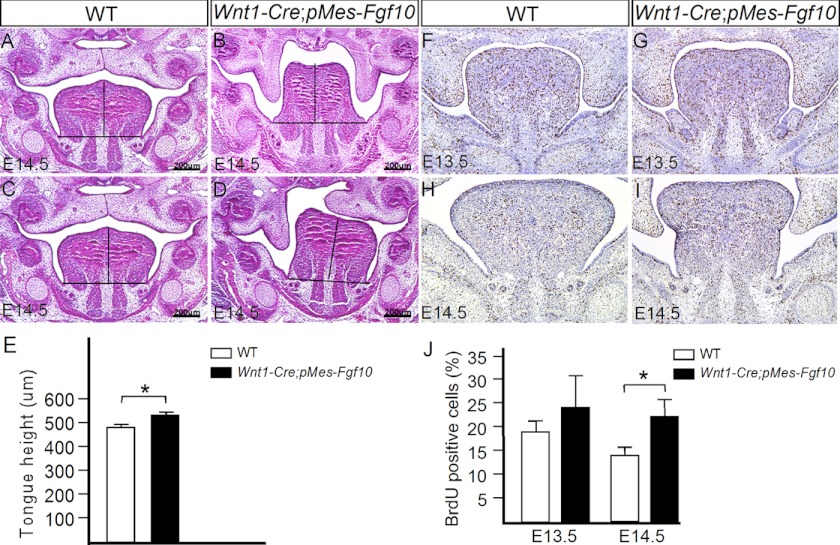
To test whether overexpression of Fgf10 in the CNC lineage could recapitulate the tongue and cleft palate phenotypes observed in Wnt1Cre;Tak1F/F mice, we created a conditional transgenic Fgf10 mouse line. Upon compounding with the Wnt1-Cre transgenic allele, the transgenic Fgf10 allele is activated in CNC-derived cells. Whereas this transgenic allele was not fully penetrant, we did observe complete cleft palate formation in 26% of Wnt1-Cre;pMes-Fgf10 mice (5 of 19). We then further analyzed the double transgenic embryos that exhibited abnormal palate development at E14.5 and compared them with their wild type littermates. We found that in these transgenic mice, the palatal shelves failed to elevate on one or both sides, associated with a significantly heightened (p < 0.01) tongue (Fig. 9, B–E). BrdU labeling assay further revealed an elevated cell proliferation rate in the Wnt1Cre;pMes-Fgf10 tongue (Fig. 9, F–J). Thus, overexpression of Fgf10 in CNC-derived cells recapitulates the cleft palate and tongue phenotypes in Wnt1-Cre;Tak1F/F mice.
FIGURE 9.
Failed palate elevation associated with heightened tongue in mice overexpressing Fgf10 in the tongue. A–E, comparison of tongue height through mid-level of tongues of E14.5 control (A and C) and Wnt1-Cre;pMes-Fgf10 (B and D) mice reveals a heightened tongue in the transgenic animal (E). F–J, BrdU labeling shows enhanced cell proliferation rate in E13.5 and E14.5 Wnt1-Cre;pMes-Fgf10 tongue compared with controls. Total and BrdU-positive cells in the entire tongue of nine sections from three samples for each stage were counted. *, p < 0.05. Scale bars, 200-μm. Error bars, S.E.
Inhibition of p38 MAPK Activity Up-regulates Fgf10 Expression in the Developing Tongue
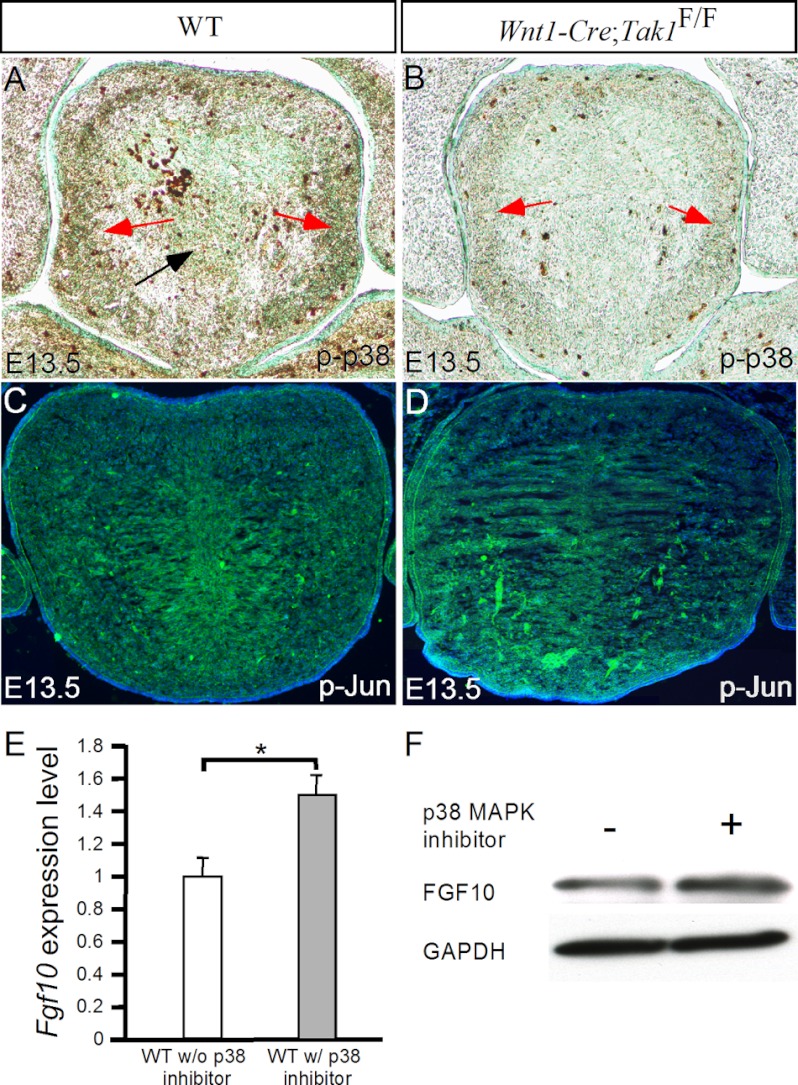
Tak1 acts through MKK3/6 to activate a number of downstream kinases including p38 and Jun kinases (28–30). It was demonstrated previously that the elevated activity of p38 MAPK, the downstream kinase of Tak1, leads to repression of Fgf9 expression in the developing palatal mesenchyme (36). We assumed that the same mechanism might be used in the developing tongue to regulate Fgf10 expression. To test this hypothesis, we first confirmed that the loss of Tak1 leads to a significantly reduced level of p38 phosphorylation (p-p38) but does not alter the level of phosphorylated Jun (p-Jun) in the developing tongue (Fig. 10, A–D). We then prepared primary mesenchymal cells derived from the tongue primordia of E13.5 wild type embryos and cultured them in the presence or absence of the p38 MAPK inhibitor SB203580. After 24 h in culture, the cells were subjected to analysis of Fgf10 expression by real-time RT-PCR and Western blotting. As shown in Fig. 10E, in the presence of p38 inhibitor, the tongue primary mesenchymal cells expressed a significantly higher level of Fgf10 mRNA than in the absence of p38 inhibitor (p < 0.01), as measured by quantitative RT-PCR. This enhanced FGF10 expression was further confirmed by Western blotting assay at protein level (Fig. 10F).
FIGURE 10.
Inhibition of p38 MAPK promotes Fgf10 expression in primary tongue mesenchymal cells. A and B, immunostaining on the tongue of E13.5 control (A) and Wnt1-Cre;Tak1F/F mutant (B) reveals a significantly reduced level of phosphorylated p38 MAPK (p-p38) in the mutant. C and D, immunostaining shows comparable levels of p-Jun in the tongue of E13.5 wild type and Wnt1-Cre;Tak1F/F embryos. E, quantitative RT-PCR shows an approximately 50% enhanced Fgf10 expression in primary tongue mesenchymal cells in the presence of p38 MAPK inhibitor compared with the control. F, Western blotting shows increased FGF10 expression in primary tongue mesenchymal cells in the presence of p38 MAPK inhibitor. *, p < 0.01. Error bars, S.E.
DISCUSSION
The importance of TGFβ/BMP signaling in palate development has been studied extensively. For example, inactivation of several TGFβ/BMP receptors, including BmprIa, Tgfbr1, and Tgfbr2, in the developing palate leads to a cleft palate formation (35). Similarly in humans, mutations in either TGFBR1 or TGFBR2 have been shown to underlie Loeys-Diestz syndrome, which presents craniofacial anomalies including cleft palate (49–51). Although TGFβ/BMP signals mainly through the Smad-dependent pathway, TGFβ also signals via a Smad-independent pathway that complements Smad action (25). Activation of the Smad-independent pathway could reinforce, attenuate, or modulate downstream cellular responses induced by Smad-dependent signaling (30). Thus, the balance between Smad-dependent and Smad-independent pathways appears to be critical for cellular-specific response to TGFβ. However, the extent to which the noncanonical pathway contributes to TGFβ signaling in palate development remains poorly understood. In this study, we investigated whether Tak1-mediated noncanonical TGFβ signaling plays a role in palatogenesis.
We show that Tak1 is expressed in both the epithelium and mesenchyme of the developing palatal shelves. Inactivation of Tak1 in the neural crest lineage leads to failed palatal shelf elevation and subsequently to a cleft palate formation. However, the fact that inactivation of Tak1 in either the palatal epithelium or palatal mesenchyme does not affect palate formation argues for a dispensable role for the Tak1-mediated noncanonical TGFβ signaling in palate development. We provide evidence demonstrating that the failed elevation of palatal shelves in Wnt1-Cre;Tak1F/F mice is not caused by an intrinsic defect but is rather a secondary consequence to the steric hindrance by the malformed tongue.
It is generally accepted that palatal shelf elevation is a rapid process controlled by both intrinsic erectile force and extrinsic influences from other craniofacial structures (1). Failed or delayed palatal shelf elevation caused by either intrinsic or extrinsic defects results in cleft palate, as is well illustrated in a number of mutant mouse models (31, 33). The failure of palate elevation due to physical obstruction by the tongue was reported in several mutant strains (4, 5, 9, 13). Interestingly, mutation in Prdm16, a transcriptional cofactor that regulate TGFβ signaling, causes failed palate elevation associated with an elevated tongue and smaller mandibular bone, mimicking human PRS clefting (5). Prdm16 is expressed in the developing craniofacial organs, including the palatal shelves, tongue, and mandibular arch. Loss of Prdm16 results in reduced TGFβ signaling in these craniofacial organs (5). Together with the similar clefting phenotype found in Wnt1-Cre;Tak1F/F mice, these observations suggest that mutations in TGFβ signaling pathways may be implicated in human PRS clefting and make TAK1 a candidate gene for such human congenital disease.
Whereas most of the muscles in the tongue originate from the occipital somites, CNC-derived cells form connective tissues. CNC cells may act to initiate and direct tongue development and function to regulate proliferation, survival, and differentiation of myogenic progenitors (24). It was shown previously that deletion of Tgfbr2 in CNC cells results in microglossia due to a reduced proliferation rate of myogenic cells associated with down-regulation of Fgf10 in CNC-derived cells, indicating a positive regulation of Fgf10 by TGFβ signaling (21). However, in Wnt1-Cre;Tak1F/F mutants, Fgf10 expression is up-regulated in CNC-derived tongue tissues. The repressive effect of the Tak1-mediated pathway on Fgf10 expression was further confirmed by elevated Fgf10 expression in primary tongue mesenchymal cells in the presence of p38 MAPK inhibitor. Because the inhibition of p38 did not enhance Fgf10 expression level as dramatically as was seen in Wnt1-Cre;Tak1F/F mutant, it is likely that other signaling cascade downstream from Tak1 also contributes to the inhibition of Fgf10 expression in the developing tongue. These observations suggest a dual role for TGFβ signaling in the regulation of Fgf10 expression in the developing tongue, with the Tak1-mediated noncanonical pathway as a negative regulator and other pathways, possibly the Smad-dependent pathway, as a positive regulator. This hypothesis is consistent with the notion that the Smad-independent pathways sometimes attenuate downstream cellular responses to TGFβ signaling (30).
Malformation of the tongue in Wnt1-Cre;Tak1F/F mice appears to be associated with dysregulated cell proliferation. The increased cell proliferation rate in the mutant tongue is very likely responsible for the formation of the heightened tongue. The smaller mandible could also contribute to this tongue phenotype in the mutant. Nevertheless, the abnormal tongue development in Wnt1-Cre;Tak1F/F mice apparently results from Fgf10 overexpression, as the phenotype of failed palate elevation associated with a malformed tongue is also found in the transgenic mouse model that bears Fgf10 overexpression in CNC cells. It should be pointed out that whereas Osr2-Cre activity is detected in peripheral mesenchyme of developing tongue (41), we did not observe a heightened tongue in Osr2-Cre;Tak1F/F mice (Fig. 4 and data not shown). The difference in tongue phenotype in Wnt1-Cre;Tak1F/F and Osr2-Cre;Tak1F/F mice could be explained by the much broader expression domain of Wnt1-Cre in the developing tongue.
In conclusion, our results demonstrate a nonintrinsic role for Tak1-mediated TGFβ signaling during palatogenesis. Inactivation of Tak1 in the CNC lineage leads to cleft palate because of failed palate elevation. This failed elevation is due to physical obstruction by a malformed tongue and possibly micrognathia as well, making it similar to human PRS clefting. TAK1 is therefore suggested as a candidate gene for mutation in human PRS clefting. Tak1 regulates tongue development by negatively controlling Fgf10 expression in CNC-derived cells.
Acknowledgments
We thank Dr. Zhijian Chen of University of Texas Southwestern Medical Center for providing access to Tak1F/F mice for this study. We also thank Dr. Vesa Kaartinen of University of Michigan for communicating results prior to publication.
This work was supported, in whole or in part, by National Institutes of Health Grants DE14044 (to Y.P. C.) and DE12711, DE14078, and DE17007 (to Y. C.).

This article contains supplemental Figs. S1–S3.
V. Kaartinen, unpublished observations.
- PRS
- Pierre Robin sequence
- BMP
- bone morphogenetic protein
- CNC
- cranial neural crest
- E
- embryonic day
- Tak1
- TGFβ-activated kinase 1.
REFERENCES
- 1. Ferguson M. W. (1988) Palate development. Development 103(Suppl), 41–60 [DOI] [PubMed] [Google Scholar]
- 2. Ferguson M. W. (1977) The mechanism of palatal shelf elevation and the pathogenesis of cleft palate. Virchows Arch. A Pathol. Anat. Histol. 375, 97–113 [DOI] [PubMed] [Google Scholar]
- 3. Alappat S. R., Zhang Z., Suzuki K., Zhang X., Liu H., Jiang R., Yamada G., Chen Y. (2005) The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev. Biol. 277, 102–113 [DOI] [PubMed] [Google Scholar]
- 4. Barrow J. R., Capecchi M. R. (1999) Compensatory defects associated with mutations in Hoxa1 restore normal palatogenesis to Hoxa2 mutants. Development 126, 5011–5026 [DOI] [PubMed] [Google Scholar]
- 5. Bjork B. C., Turbe-Doan A., Prysak M., Herron B. J., Beier D. R. (2010) Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 19, 774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casey L. M., Lan Y., Cho E. S., Maltby K. M., Gridley T., Jiang R. (2006) Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 235, 1830–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He F., Popkie A. P., Xiong W., Li L., Wang Y., Phiel C. J., Chen Y. (2010) Gsk3β is required in the epithelium for palatal elevation in mice. Dev. Dyn. 239, 3235–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He F., Xiong W., Wang Y., Matsui M., Yu X., Chai Y., Klingensmith J., Chen Y. (2010) Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev. Biol. 347, 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang X., Goudy S. L., Ketova T., Litingtung Y., Chiang C. (2008) Gli3-deficient mice exhibit cleft palate associated with abnormal tongue development. Dev. Dyn. 237, 3079–3087 [DOI] [PubMed] [Google Scholar]
- 10. Jiang R., Lan Y., Chapman H. D., Shawber C., Norton C. R., Serreze D.V., Weinmaster G., Gridley T. (1998) Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12, 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lan Y., Ovitt C. E., Cho E. S., Maltby K. M., Wang Q., Jiang R. (2004) Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development 131, 3207–3216 [DOI] [PubMed] [Google Scholar]
- 12. Matsumura K., Taketomi T., Yoshizaki K., Arai S., Sanui T., Yoshiga D., Yoshimura A., Nakamura S. (2011) Sprouty2 controls proliferation of palate mesenchymal cells via fibroblast growth factor signaling. Biochem. Biophys. Res. Commun. 404, 1076–1082 [DOI] [PubMed] [Google Scholar]
- 13. Murray S. A., Oram K. F., Gridley T. (2007) Multiple functions of Snail family genes during palate development in mice. Development 134, 1789–1797 [DOI] [PubMed] [Google Scholar]
- 14. Rice R., Spencer-Dene B., Connor E. C., Gritli-Linde A., McMahon A. P., Dickson C., Thesleff I., Rice D. P. (2004) Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 113, 1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Snyder-Warwick A. K., Perlyn C. A., Pan J., Yu K., Zhang L., Ornitz D. (2010) Analysis of a gain-of-function FGFR2 Crouzon mutation provides evidence of loss of function activity in the etiology cleft palate. Proc. Natl. Acad. Sci. U.S.A. 107, 2515–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiong W., He F., Morikawa Y., Yu X., Zhang Z., Lan Y., Jiang R., Cserjesi P., Chen Y. (2009) Hand2 is required in the epithelium for palatogenesis in mice. Dev. Biol. 330, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robin P. (1994) A fall of the base of the tongue considered as a new cause of nasopharyngeal respiratory impairment: Pierre Robin sequence, a translation. 1923. Plast. Reconstr. Surg. 93, 1301–1303 [PubMed] [Google Scholar]
- 18. Benko S., Fantes J. A., Amiel J., Kleinjan D. J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C. T., McBride D., Golzio C., Fisher M., Perry P., Abadie V., Ayuso C., Holder-Espinasse M., Kilpatrick N., Lees M. M., Picard A., Temple I. K., Thomas P., Vazquez M. P., Vekemans M., Roest Crollius H., Hastie N. D., Munnich A., Etchevers H. C., Pelet A., Farlie P. G., Fitzpatrick D. R., Lyonnet S. (2009) Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 41, 359–364 [DOI] [PubMed] [Google Scholar]
- 19. FitzPatrick D. R., Carr I. M., McLaren L., Leek J. P., Wightman P., Williamson K., Gautier P., McGill N., Hayward C., Firth H., Markham A. F., Fantes J. A., Bonthron D. T. (2003) Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet. 12, 2491–2501 [DOI] [PubMed] [Google Scholar]
- 20. Jakobsen L. P., Ullmann R., Christensen S. B., Jensen K. E., Mølsted K., Henriksen K. F., Hansen C., Knudsen M. A., Larsen L. A., Tommerup N., Tümer Z. (2007) Pierre Robin sequence may be caused by dysregulation of SOX9 and KCNJ1. J. Med. Genet. 44, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hosokawa R., Oka K., Yamaza T., Iwata J., Urata M., Xu X., Bringas P., Jr., Nonaka K., Chai Y. (2010) TGF-β mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341, 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noden D. M. (1983) The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev. Biol. 96, 144–165 [DOI] [PubMed] [Google Scholar]
- 23. Noden D. M., Francis-West P. (2006) The differentiation and morphogeneis of craniofacial muscle. Dev. Dyn. 235, 1194–1218 [DOI] [PubMed] [Google Scholar]
- 24. Parada C., Han D., Chai Y. (2012) Molecular and cellular regulatory mechanisms of tongue myogenesis. J. Dent. Res. 91, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massagué J. (2012) TGFβ signaling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mulder K. M. (2000) Role of Ras and MAPKs in TGFβ signaling. Cytokine Growth Factor Rev. 11, 23–35 [DOI] [PubMed] [Google Scholar]
- 27. Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. (1995) Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270, 2008–2011 [DOI] [PubMed] [Google Scholar]
- 28. Moriguchi T., Kuroyanagi N., Yamaguchi K., Gotoh Y., Irie K., Kano T., Shirakabe K., Muro Y., Shibuya H., Matsumoto K., Nishida E., Hagiwara M. (1996) A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J. Biol. Chem. 271, 13675–13679 [DOI] [PubMed] [Google Scholar]
- 29. Shirakabe K., Yamaguchi K., Shibuya H., Irie K., Matsuda S., Moriguchi T., Gotoh Y., Matsumoto K., Nishida E. (1997) TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J. Biol. Chem. 272, 8141–8144 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y. E. (2009) Non-Smad pathways in TGF-β signaling. Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bush J. O., Jiang R. (2012) Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chai Y., Maxson R. E., Jr. (2006) Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353–2375 [DOI] [PubMed] [Google Scholar]
- 33. Gritli-Linde A. (2007) Molecular control of secondary palate development. Dev. Biol. 301, 309–326 [DOI] [PubMed] [Google Scholar]
- 34. Hilliard S. A., Yu L., Gu S., Zhang Z., Chen Y. P. (2005) Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 207, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwata J., Parada C., Chai Y. (2011) The mechanisms of TGF-β signaling during palate development. Oral Dis. 17, 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwata J., Tung L., Urata M., Hacia J.G., Pellkan R., Suzuki A., Ramenzoni L., Chaudhry O., Parada C., Sanchez-Lara P. A., Chai Y. (2012) Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor β (TGFβ) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. J. Biol. Chem. 287, 2353–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu X., Han J., Ito Y., Bringas P., Jr., Deng C., Chai Y. (2008) Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev. Cell 15, 322–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwata J., Hacia J. G., Suzuki A., Sanchez-Lara P. A., Urata M., Chai Y. (2012) Modulation of noncanonical TGF-βsignaling prevents cleft palate in Tgfbr2 mutant mice. J. Clin. Invest. 122, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andl T., Ahn K., Kairo A., Chu E. Y., Wine-Lee L., Reddy S. T., Croft N. J., Cebra-Thomas J. A., Metzger D., Chambon P., Lyons K. M., Mishina Y., Seykora J. T., Creshaw E. B., 3rd, Millar S. E. (2004) Epithelial BmprIa regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 131, 2257–2268 [DOI] [PubMed] [Google Scholar]
- 40. Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 41. Lan Y., Wang Q., Ovitt C. E., Jiang R. (2007) A unique mouse strain expressing Cre recombinase for tissue-specific analysis of gene function in palate and kidney development. Genesis 45, 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu H. H., Xie M., Schneider M. D., Chen Z. J. (2006) Essential role of TAK1 in thymocyte development and activation. Proc. Natl. Acad. Sci. U.S.A. 103, 11677–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soriano P. (1999) Generalized LacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 44. St Amand T. R., Zhang Y., Semina E. V., Zhao X., Hu Y., Nguyen L., Murray J. C., Chen Y. (2000) Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 217, 323–332 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z., Song Y., Zhao X., Zhang X., Fermin C., Chen Y. (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129, 4135–4146 [DOI] [PubMed] [Google Scholar]
- 46. Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. (2006) Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 281, 4035–4041 [DOI] [PubMed] [Google Scholar]
- 47. Iwata J., Hosokawa R., Sanchez-Lara P. A., Urata M., Slavkin H., Chai Y. (2010) Transforming growth factor-β regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem. 285, 4975–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu W., Sun X., Braut A., Mishina Y., Behringer R. R., Mina M., Martin J. F. (2005) Distinct functions for Bmp signaling in lip and palate fusion in mice. Development 132, 1453–1461 [DOI] [PubMed] [Google Scholar]
- 49. Mizuguchi T., Collod-Beroud G., Akiyama T., Abifadel M., Harada N., Morisaki T., Allard D., Varret M., Claustres M., Morisaki H., Ihara M., Kinoshita A., Yoshiura K., Junien C., Kajii T., Jondeau G., Ohta T., Kishino T., Furukawa Y., Nakamura Y., Niikawa N., Boileau C., Matsumoto N. (2004) Heterozygous TGFBR2 mutations in Marfan syndrome. Nat. Genet. 36, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loeys B. L., Chen J., Neptune E. R., Judge D. P., Podowski M., Holm T., Meyers J., Leitch C. C., Katsanis N., Sharifi N., Xu F. L., Myers L. A., Spevak P. J., Cameron D. E., De Backer J., Hellemans J., Chen Y., Davis E. C., Webb C. L., Kress W., Coucke P., Rifkin D. B., De Paepe A. M., Dietz H. C., (2005) A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 36, 275–281 [DOI] [PubMed] [Google Scholar]
- 51. Loeys B. L., Schwarze U., Holm T., Callewaert B. L., Thomas G. H., Pannu H., De Backer J. F., Oswald G. L., Symoens S., Manouvrier S., Roberts A. E., Faravelli F., Greco M. A., Pyeritz R. E., Milewicz D. M., Coucke P. J., Cameron D. E., Braverman A. C., Byers P. H., De Paepe A. M., Dietz H. C. (2006) Aneurysm syndrome caused by mutations in the TGF-β receptor. N. Engl. J. Med. 355, 788–798 [DOI] [PubMed] [Google Scholar]