Background: PglL-like enzymes catalyze protein O-linked glycosylation in many bacteria.
Results: The catalytic efficiency of this enzyme correlates with the length and conformation of the acyl chain of glycan donors. PglL also catalyzed glycan transfer from UDP-activated sugar.
Conclusion: Lipid carriers are not essential but influence glycosylation.
Significance: This work provides insights into the functionality of PglL.
Keywords: Glycolipids, Glycomics, Glycoprotein, Microbiology, Oligosaccharide, Oligosaccharyltransferase
Abstract
Oligosaccharyltransferases (OTases) are enzymes that catalyze the transfer of an oligosaccharide from a lipid carrier to an acceptor molecule, commonly a protein. OTases are classified as N-OTases and O-OTases, depending on the nature of the glycosylation reaction. The N-OTases catalyze the glycan transfer to amide groups in asparagines in a reaction named N-linked glycosylation. The O-OTases are responsible for protein O-linked glycosylation, which involves the attachment of glycans to hydroxyl groups of serine or threonine residues. These enzymes exhibit a relaxed specificity and are able to transfer a variety of glycan structures to different protein acceptors. This property confers OTases with great biotechnological potential as these enzymes can produce glycoconjugates relevant to the pharmaceutical industry. Furthermore, OTases are thought to be involved in pathogenesis mechanisms. Several aspects of the functionality of OTases are not fully understood. In this work, we developed a novel approach to perform kinetic studies on PglL, the O-OTase from Neisseria meningitidis. We investigated the importance of the acyl moiety of the lipid glycan donor substrate on the functionality of PglL by testing the efficiency of glycosylation reactions using synthetic substrates carrying the same glycan structure but different acyl moieties. We found that PglL can function with many lipids as glycan donors, although the length and the conformation of the lipid moiety significantly influenced the catalytic efficiency. Interestingly, PglL was also able to transfer a monosaccharide employing its nucleotide-activated form, acting as a Leloir glycosyltransferase. These results provide new insights on the function and the evolution of oligosaccharyltransferases.
Introduction
Bacterial cell surfaces are covered by carbohydrates in the form of different glycoconjugates. These molecules perform important roles during the interaction of the bacteria with other organisms and with the environment. In many bacterial species, N- or O-glycoproteins contribute to the repertoire of such surface glycoconjugates (1). In some bacteria, N- and O-linked protein glycosylation take place in the bacterial cytoplasm preceding the export of the glycoproteins to the surface. In this case, cytosolic Leloir (nucleotide-dependent) glycosyltransferases transfer the sugars from nucleotide-activated donors to the protein acceptors. For example, bacterial species such as Clostridium difficile, Haemophilus influenzae, and Helicobacter pylori glycosylate adhesins and flagellins using specialized N- and O-glycosyltransferases in the bacterial cytoplasm (2–4). Alternatively, glycosylation may occur via an en bloc mechanism, in which the sugars are assembled onto a lipid carrier before being transferred to acceptor proteins through the activity of specialized non-Leloir glycosyltransferases collectively known as “oligosaccharyltransferases” (OTases).4 The en bloc mechanism for N-glycosylation is a fundamental process in eukaryotes and archaea (5–7). However, in bacteria, this mechanism is only employed by species within the ϵ-proteobacteria, such as Campylobacter jejuni, and the δ-proteobacteria, particularly within the genus Desulfovibrio (8, 9). On the contrary, multiple proteins are O-glycosylated employing the en bloc mechanism by means of an O-OTase in various species such as Neisseria meningitidis, Neisseria gonorrheae, Acinetobacter baumannii, Bacteroides fragilis, Vibrio cholerae, Francisella tularensis, and Burkholderia thailandensis (10–15). The protein targets of O-linked glycosylation systems in bacteria are implicated in activities as varied as protein folding, adhesion, disulfide bond formation, and solute uptake, as well as both aerobic and anaerobic respiration (16). Lack of glycosylation may affect biofilm formation and virulence (12, 17, 18).
PglL from N. meningitidis is the best studied O-OTase to date. In vivo, PglL shows a remarkable lack of glycan specificity and is able to transfer virtually any glycan from the undecaprenylpyrophosphate carrier to proteins (19). This important feature makes this enzyme a promising tool for glycoengineering novel glycan-based vaccines, therapeutics, and diagnostics tools (20, 21). In this context, understanding of the molecular mechanisms of O-OTases is relevant to improve their biotechnological potential. Furthermore, the presence of O-OTases in many important bacterial pathogens and the observation that in some of them O-glycosylation appears to play a role during infection, suggests that they could be targets for inhibitors with antimicrobial properties (12).
In this work, we developed a quantitative analysis of glycosylation that allowed us to perform in vitro kinetic studies on PglL. We applied this method to assay the relevance of the aliphatic chains present in the glycan donor carriers in the glycosylation reaction. We show that PglL can catalyze glycosylation from all the assayed lipid carriers. However, the efficiency of the glycosylation reaction is influenced by the lipid carrier structure. Surprisingly, PglL can act as a Leloir glycosyltransferase, being also able to use a nucleotide-activated monosaccharide as substrate.
EXPERIMENTAL PROCEDURES
PglL and Pilin Purification
For PglL purification, Escherichia coli CLM24 cells transformed with pAMF10 plasmid (19), (which overexpresses C-His10-tagged PglL) were cultured at 37 °C with shaking until an A600 nm of 0.4–0.6 was reached, after which the cells were induced by isopropyl 1-thio-β-d-galactopyranoside (0.1 mm) at 30 °C during 6 h. Then, the cells were harvested, and the pellets were washed with 50 mm Tris-HCl buffer (pH 7.5), 0.25 m NaCl, 5% glycerol (v/v) (buffer A), and resuspended in the same buffer containing complete EDTA-free protease inhibitor mixture (Roche Applied Science). The cells were disrupted by French press, and 1 mg/liter of DNase I was added. After 15 min of incubation in ice, the cell extract was centrifuged (10,000 × g, 10 min) to remove large particles and cell debris. The membrane fraction was isolated by ultracentrifugation of the supernatant for 1 h at 200,000 × g and 4 °C. Membrane proteins were solubilized by mild tumbling of the obtained pellet with buffer A containing 1% (w/v) n-dodecyl-β-d-maltopyranoside (Affymetrix) and protease inhibitor mixture overnight at 4 °C. The solution was centrifuged (1 h at 200,000 × g and 4 °C), and the supernatant was loaded in a nickel-nitrilotriacetic acid column equilibrated with 50 mm Tris-HCl buffer (pH 7.5), 0.25 m NaCl, 5% glycerol (v/v), 0.5% (w/v) n-dodecyl-β-d-maltopyranoside, and 20 mm imidazole. After loading, the column was washed with 5 volumes of the same buffer and finally eluted with 50 mm Tris-HCl buffer (pH 7.5), 0.25 m NaCl, 5% glycerol (v/v), 0.5% n-dodecyl-β-d-maltopyranoside (w/v), and 200 mm imidazole. The purification of pilin was performed from E. coli CLM24 transformed with pAMF15 (19) (which overexpresses C-His10-tagged PilE) following the same procedure with the exception that 15 mm β-mercaptoethanol was added to all the buffers.
Synthesis of E. coli O Antigen O86 Linked to Different Lipid Carriers and UDP-diNAcBac
The synthesis of the different synthetic organic compounds was performed following published methods (22–24), uridine diphosphate N′-diacetylbacillosamine (UDP-diNAcBac) was kindly provided by Martin Young (Institute for Biological Sciences, Ottawa, Ontario, Canada).
Organic Extraction of Heptasaccharide of Campylobacter jejuni Linked to Undecaprenol Pyrophosphate (cLLO)
C. jejuni lipid-linked heptasacharide was isolated from E. coli cells containing the plasmid pACYCpglBmut (8), which carries C. jejuni protein glycosylation locus (pgl) containing mutations W458A and D459A in PglB, essentially as described previously (25).
Quantification of PglL and Pilin
PglL and pilin were quantified by SDS-PAGE and Coomassie staining. Different volumes of PglL and pilin purification aliquots were loaded in a 15% polyacrylamide gel simultaneously with 0.2, 0.4, 0.6, 0.8, 1.0, and 1.5 μg of BSA standard. After staining with Coomassie dye, the intensity of the bands of PglL and pilin were quantified by using the Odyssey software (Li-Cor Bioscences). The intensities of BSA bands were depicted versus μg of BSA, and the data were fitted to a linear curve. The μg of PglL and pilin were estimated by lineal interpolation.
Assay of Requirement of Metal Divalent Cations, Optimal Temperature, and pH
The requirement of metal divalent cations of PglL to catalyze the glycan transfer from cLLO to pilin protein acceptor was assayed in 50 μl of reaction medium containing 50 mm Tris-HCl (pH 7.5); 100 mm sucrose; 0.45 μm PglL, 200 μm pilin, 1.25 μl of cLLO organic extraction, and 1 mm of one of the following components: ZnCl2, CoCl2, MnCl2, CaCl2, MgCl2, NiCl2, or 20 mm EDTA (pH 7.8). Respective controls without cations, PglL and cLLO were simultaneously assayed. After overnight incubation at 30 °C, 20 μl of the reaction were loaded in a 15% SDS-PAGE to separate glycosylated and non-glycosylated pilin using the standard protocols. Then, the proteins were transferred to nitrocellulose for detection of glycosylation with Western blotting. Primary antibodies were monoclonal anti-pilin from N. meningitis MC58 (SM1, mouse antiserum) (19) and the HR6, rabbit antiserum, kindly provided by Markus Aebi (ETH, Zürich, Switzerland) (26). As secondary antibodies, either a goat anti-mouse or goat anti-rabbit IgG fluorescent dye-labeled were applied. The intensities of the glycosylation signals at different cation conditions were quantified by using the Odyssey software (Li-Cor Biosciences), which allows for direct infra red fluorescence detection and quantification at 685 and 785 nm simultaneously. The optimal temperature for the in vitro glycosylation was investigated by performing the activity of PglL in 50 μl of reaction medium containing 50 mm Tris-HCl (pH 7.0); 100 mm sucrose; 1 mm MnCl2; 0.5 μm PglL, 200 μm pilin, and 1.25 μl of cLLO organic extraction at 15, 25, 30, 37, and 42 °C. After overnight incubation, 20 μl of the different conditions were loaded in a 15% SDS-PAGE gel, and the optimal temperature was estimated according to the maximal glycosylation intensity by using quantitative Western blot as described above. A similar procedure was followed to find the optimal pH with the difference that the reactions were performed at 50 mm MES (pH 5.5, 6.0, and 6.5) or 50 mm Tris (pH 7.0, 7.5, 8.0, 8.5, and 8.8) at 30 °C, respectively.
Time Course Analysis and Optimal Enzyme Concentration
The optimal time and enzyme concentration to perform kinetic measurements under steady-state conditions were determined. Thus, to know the optimal time to perform the in vitro assays, 0.45 μm PglL, 200 μm pilin, and 1.25 μl of cLLO organic extraction in 50 μl of reaction medium (50 mm Tris-HCl (pH 7.0), 100 mm sucrose, and 1 mm MnCl2) were incubated at 30 °C during 0, 1, 5, 10, 15, 30, 45, 60, 120, and 240 min. The reactions were stopped by adding 1× SDS-PAGE loading buffer and further incubation at −20 °C. The glycosylation of pilin as time function was followed by Western blot analysis as described previously. A time course graph was plotted, and the optimal time was selected from the linear region of the graph to approximate a steady-state condition.
The activity of PglL was also analyzed at different enzyme concentration. Thus, in vitro glycosylation assays were carried out at 0, 0.015, 0.06, 0.12, 0.24, 0.27, 0.36, and 0.45 μm PgL in 50 μl of reaction medium (50 mm Tris-HCl (pH 7.0), 100 mm sucrose, and 1 mm MnCl2) during 15 min at 30 °C. The pilin glycosylation at each condition was calculated as described formerly, and a graph of relative pilin glycosylation versus μg of PglL was plotted. The optimal enzyme concentration was selected from the linear region of the graph.
Enzymatic Assays
The OTase activity of PglL was determined in 50 μl of reaction medium (50 mm Tris-HCl, pH 7.0; 100 mm sucrose, 1 mm MnCl2) containing 0.32 μm PglL, 300 μm pilin, and 0, 1.0, 2.5, 5.0, 10, 50, 100, 150, and 250 μm glycan donor substrates during 15 min at 30 °C. Fractions of 10 μl were loaded in 15% SDS-PAGE gels to separate glycosylated and non-glycosylated pilin and transferred to nitrocellulose for detection with Western blotting. SM1 were used as primary antibody and goat anti-mouse dye-labeled was the secondary antibody. The intensity of the glycosylated and non-glycosylated pilin were quantified by Odyssey infrared image system each substrate concentration. The percentages of glycosylated pilin were calculated at each substrate concentration considering the sum of both signal intensities as the 100% of intensity. Finally, the μmol of glycosylated pilin were calculated per time unit and mg of PglL. A similar procedure was applied to detect OTase activity of PglL with 0.15, 15, and 150 mm UDP-diNAcBac, UDP-GlcNAc, and UDP-GalNAc. In the assays with UDP-diNAcBac SM1 and UOS-2 (kindly provided by Michael Koomey, University of Oslo, Norway) were primary antibodies and goat anti-mouse and goat anti-rabbit IgG fluorescent dye-labeled were the secondary antibodies, respectively. MS analysis was performed to assay pilin glycosylation from UDP-GlcNAc, UDP-GalNac, and UDP-diNAcBac.
The OTase activity of PglB was determined in 50 μl of reaction medium (50 mm Tris-HCl, pH 8.0; 100 mm sucrose, 10 mm MnCl2) containing 0.32 μm PglB, 180 μm AcrA, and 1.25 μl of cLLO during 2 h at 30 °C. Fractions of 20 μl were loaded in 15% SDS-PAGE gels to separate glycosylated and non-glycosylated AcrA and transferred to nitrocellulose for detection with Western blotting. The same conditions were applied to assay the glycosylation of AcrA from 10 mm UDP-diNAcBac. Experimental data were fitted to theoretical curves using Sigmaplot considering a Michaelis-Menten model (Systat Software, Inc., Point Richmond, CA).
Digestion of Pilin
Isolated pilin bands were processed as described previously (27) with minor modification. Briefly, gel separated pilin bands were excised and destained in a 50:50 solution of 50 mm NH4HCO3:100% ethanol for 20 min at room temperature with shaking at 750 rpm. Destained bands were then washed with 100% ethanol, vacuum-dried for 20 min, and rehydrated in 10 mm DTT in 50 mm NH4HCO3. Reduction was carried out for 60 min at 56 °C with shaking. The reducing buffer was then removed and the gel bands washed twice in 100% ethanol for 10 min to ensure the removal of remaining DTT. Reduced ethanol washed samples were sequentially alkylated with 55 mm iodoacetamide in 50 mm NH4HCO3 in the dark for 45 min at room temperature. Alkylated samples were then washed with 2 rounds of 100% ethanol and vacuum dried. Alkylated samples were then rehydrated with 12 ng μl−1 trypsin (Promega, Madison, WI) in 40 mm NH4HCO3 at 4 °C for 1 h. Excess trypsin was removed, gel pieces were covered in 40 mm NH4HCO3 and incubated overnight at 37 °C. Peptides were concentrated and desalted using C18 stage tips (28, 29) and stored on tip at 4 °C. Peptides were eluted in buffer B (0.5% acetic acid, 80% MeCN) and dried before analysis by LC-MS.
Identification of Glycopeptides Using Reversed Phase LC-MS, CID MS/MS, and HCD MS/MS
Purified peptides were resuspend in buffer A (0.5% acetic acid) and separated using a in 25-cm house packaged, 50-μm inner diameter, 360-μm outer diameter, 2.4-μm ReproSil-Pur C18 AQ (Dr. Maisch, Ammerbuch-Entringen, Germany) reverse-phase analytical column coupled to a 2-cm-long, 100-μm-inner diameter fused silica trap column containing 5.0-μm Aqua C-18 beads (Phenomenex, Torrance, CA). Samples were loaded onto the trap using an Agilent 1290 Series HPLC (Agilent Technologies, Mississauga, ON) at 5 μl/min and eluted at 0.1 μl/min using passive splitting (30). The eluting peptides were infused into an LTQ-Orbitrap Velos (Thermo Scientific, San Jose, CA) using a 20-μm inner diameter fused silica gold-coated spray tip prepared in-house. A 90-min gradient was run from 0% buffer B to 32% B for 51 min, and then from 32% B to 40% B in the next 5 min, and then increased to 100% B over 2-min period, held at 100% B for 2.5 min, and then dropped to 0% B for another 20 min. The LTQ-Orbitrap Velos were operated using Xcalibur (version 2.2, Thermo Scientific) with a capillary temperature of 200 °C in a data-dependent mode automatically switching between MS, CID MS/MS, and HCD MS/MS as described previously (31).
Database Interrogation of Identified Glycopeptides
Raw files were processed as described previously (31, 32). Briefly, Proteome Discoverer (version 1.2, Thermo Scientific) was used to generate .mgf files, and the resulting file searched using MASCOT (version 2.2). Searches were performed against the database “proteobacteria” using the following parameters: peptide mass accuracy, 10 ppm; fragment mass accuracy, 0.02 Da; trypsin enzyme specificity; fixed modifications, carbamidomethyl; variable modifications, methionine oxidation and deamidated Asn and Gln, Q. The instrument setting of MALDI-QUAD-TOF was chosen due to previous studies showing quadrupole-like fragmentation within HCD spectra (33). Scan events that did not result in peptide identifications from MASCOT searches were exported to Microsoft Excel (Microsoft, Redmond, WA). To identify possible glycopeptides within exported non-match scans, the MS/MS module of GPMAW (version 8.2) was called an “.mgf graph.” Scan events containing the Bac diagnostic oxonium 229.12 m/z ion were manually inspected and identified as possible glycopeptides based on the presence of the deglycosylated peptide ion, corresponding to the parent mass −229.12 Da within a tolerance of 10 ppm. To facilitate glycopeptide assignments from HCD scan events, ions below the mass of the predicted deglycosylated peptides were extracted with Xcalibur (version 2.2) using the Spectrum list function. MASCOT searches were conducted again as described above, and the resulting glycopeptides manually annotated according to Roepstorff et al. (34).
RESULTS
PglL Transferase Activity Is Independent of Divalent Cations
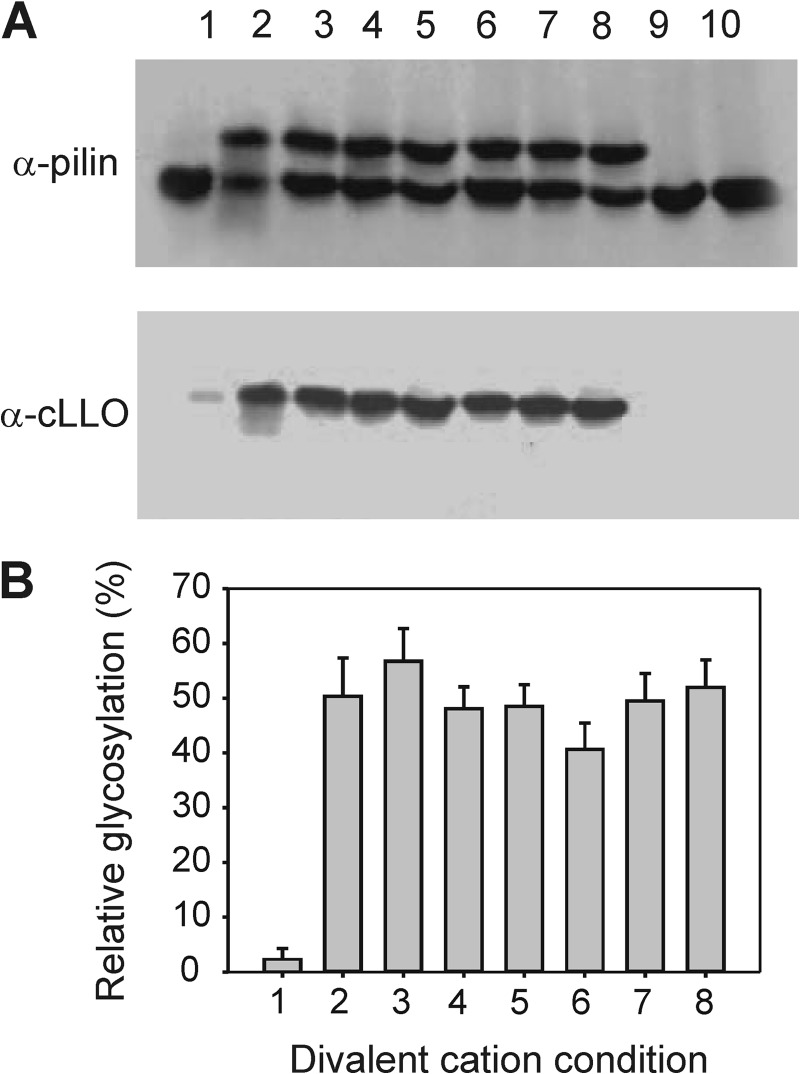
To optimize conditions required for performing kinetic studies, we employed PilE as protein acceptor, one of the natural glycosylation targets of PglL (35), and C. jejuni LLO served as glycan donor (36). This glycan donor, which has been shown to be a good substrate for PglL homologs (13), was chosen because it contains diBacNAc at the reducing end, which is the same sugar found at the reducing end of the endogenous Neisseria glycans (35, 36). We assayed the activity of PglL with different divalent metal cations to determine their relevance for in vitro activity of the enzyme (Fig. 1). PglL was active with all cations tested, with the exception of ZnCl2. Protein precipitation was observed upon this condition, which accounted for the low yield of glycosylated product. In all other conditions, the results showed that the presence of divalent cations does not result in a significant increase in the activity of PglL. Moreover, the presence of EDTA, a chelating agent of metal divalent cations, did not affect enzyme activity, suggesting that PglL activity is metal ion-independent.
FIGURE 1.
Influence of metal divalent cations on the activity of PglL in vitro. The capability of PglL to glycosylate pilin from cLLO was assayed at 30 °C in a medium containing 100 mm sucrose, 50 mm Tris-HCl (pH 7.5), and 1 mm ZnCl2 (lane 1), 1 mm CoCl2 (lane 2), 1 mm MnCl2 (lane 3), 1 mm CaCl2 (lane 4), 1 mm MgCl2 (lane 5), 1 mm NiCl2 (lane 6), 20 mm EDTA (lane 7), and without cations (lane 8). A, Western blot showing the results of the in vitro glycosylation assays. Lanes 9 and 10 correspond to the controls without PglL and cLLO, respectively. The upper bands correspond to the glycosylated pilin acceptor, as indicated by the glycan channel (lower panel). B, quantification of glycosylation as percentage of the total pilin signal at each condition. The experiment was done in duplicate.
PglL Activity Is Optimal at 30 °C and pH 7.0
To identify the ideal conditions for the in vitro glycosylation reaction, the activity of PglL was analyzed as a function of temperature (data not shown). PglL was active at 15, 25, 30, 37 °C, and slightly at 42 °C, but the maximal glycosylation activity was detected at 30 °C. Thus, the optimal temperature of 30 °C was applied to all of the subsequent reactions assayed in this work. The activity of PglL was also tested at different pH values, ranging from 5.5 to 8.8. The enzyme was active over the analyzed pH range; however, PglL displayed maximal activity at approximately pH 7.0, which may reflect the pH of the periplasm, the natural environment of the active site of this enzyme (data not shown). Therefore, for subsequent experiments, this pH was employed.
Time Course Analysis
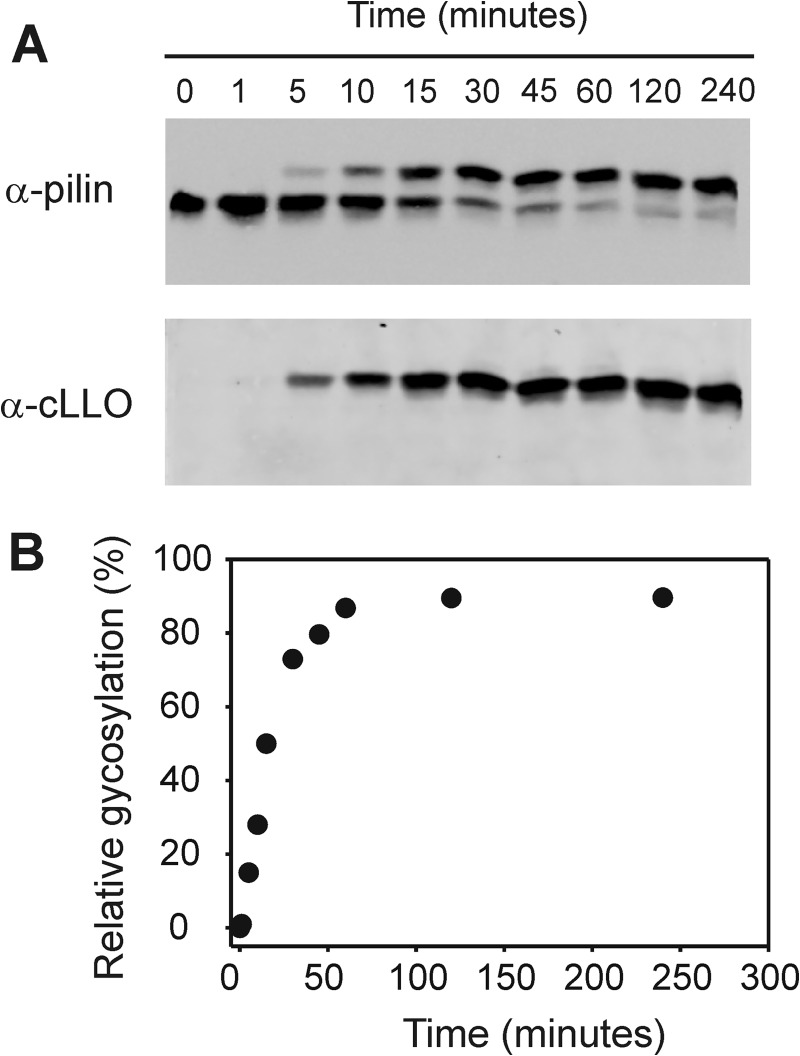
To perform kinetic analyses of PglL activity, we developed a quantitative assay for in vitro glycosylation by using fluorescence-based Western blot analysis (Fig. 2A). This technology, based on the quantification of an indefinitely stable fluorescent signal arising from fluorescent dye-labeled antibodies, has been validated previously in multiple studies (37–40). Additionally, we validated the method in our lab by demonstrating the proportionality of fluorescence signals to a wide range of concentrations of BSA (supplemental Fig. S1).
FIGURE 2.
Time course analysis of in vitro activity of PglL. A, Western blot of in vitro glycosylation assay after ending the reaction at the times indicated by adding 1× sample loading buffer containing 0.2% (w/v) SDS and immediate transfer to −20 °C. B, graph showing the relative glycosylation intensity as function of time.
We then performed a time course analysis of in vitro activity of PglL. The intensities of the signals corresponding to the glycosylated forms of pilin at different times were quantified and plotted as a function of time (Fig. 2B). To be certain that steady-state conditions are reached, the initial velocity measurements should be made when the velocity is linear regarding both time and enzyme concentration. From our experiments, we concluded that 15 min is the optimal time for the assays, and 0.32 μm PglL was the selected concentration for subsequent kinetic studies (supplemental Fig. S2).
Importance of the Lipid Carrier in PglL Activity
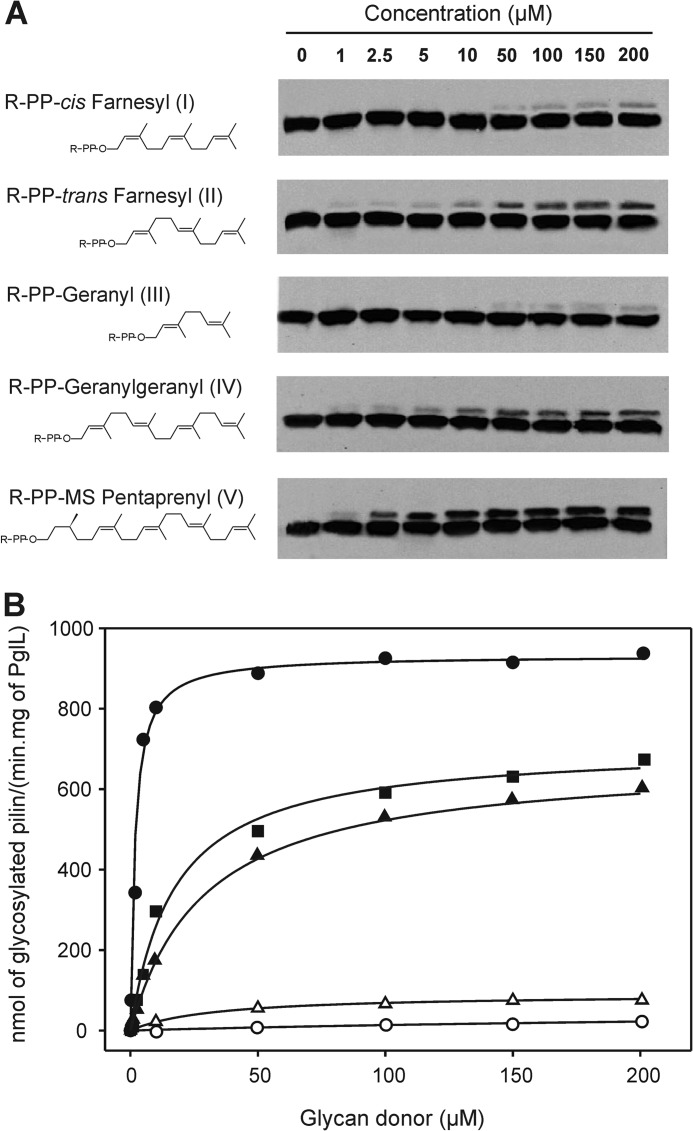
The lack of glycan specificity of PglL initially suggested that recognition of the lipid by the O-OTase could be crucial for catalysis, but we have previously shown that PglL can transfer a glycan from a synthetic farnesyl carrier in place of the undecaprenyl lipid used in vivo (19). However, our study was qualitative and did not address the quantitative effect of isomerization and length of the lipid carrier on the reaction efficiency. We therefore analyzed the influence of these two substrate features by employing synthetic glycolipidic substrates containing the same glycan portion (the subunit of E. coli O86 O antigen) but lipid carriers with different acyl moieties (Fig. 3A) in the reaction catalyzed by PglL. A similar strategy was recently employed to study WaaL glycan-donor specificity in vitro by Han et al. (22). The glycosylation reaction was followed via quantification of the relative intensity of the signals of the bands corresponding to glycosylated and unglycosylated pilin. PglL was able to glycosylate the pilin acceptor using all the lipid carriers tested (Fig. 3A). Taking into consideration the reaction time and the amounts of PglL and pilin used in the assays, it was possible to calculate the OTase activity at different concentrations of lipid-linked sugar substrates. Fig. 3B shows the progression of the glycosylation reactions over time, following a hyperbolic trend for all the substrates. Considering a Michaelian behavior for PglL upon the assayed conditions, the kinetic parameters associated to the reaction were calculated and are shown in Table 1. The influence of the cis-trans isomerization of the lipid carrier can be observed comparing the efficiency of PglL to glycosylate pilin from cis-farnesyl and trans-farnesyl as lipid carriers (Fig. 3A). We found that the affinity of PglL to both substrates is similar, as suggested by the estimated Km values (Table 1). However, the Vmax for the trans substrate was one order of magnitude higher than for the cis substrate. The parameter kcat/Km provides an estimation of the catalytic efficiency of the enzyme, so that higher kcat/Km values imply higher catalytic efficiency. Thus, at similar length of isoprenoid chain, the catalytic efficiency was higher with the substrate with trans isomerization, indicating that the geometry of the polyisoprenoid chain influences the course of the reaction (Table 1). The influence of the length of the lipid carrier was analyzed by comparing glycosylation from carriers consisting of geranyl, farnesyl, and geranylgeranyl, containing two, three, and four isoprenoid units, respectively. All three substrates were in their trans conformation. In this set of three substrates, geranylgeranyl (substrate IV) has the longest polyisoprenoid chain of the three lipids assayed, and PglL exhibited the lowest Km, i.e. the highest affinity, toward this substrate. On the contrary, with the glycolipid containing the geranyl chain (the shortest of the lipid carriers tested), the highest Km and the lowest Vmax values were observed. Following the trend correlating length of the aliphatic chain with increasing catalytic efficiency, farnesyl, the lipid of intermediate length displayed intermediate values of Km and Vmax.
FIGURE 3.
Activity of PglL with glycan donor substrates carrying the same glycan portion (E. coli O86 O antigen) but polyisoprenoid chains varying in length and geometry. A, structures of the substrates and Western blot analysis of the resultant in vitro glycosylation assays. The observed signals correspond to glycosylated and non-glycosylated pilin (upper and lower bands, respectively; antibody anti-pilin SM1). R-PP denotes the sugar pyrophosphate attached to each lipid. B, activity plots and kinetic adjustments for all the glycan donors. Black circles, substrate V; black squares, substrate IV; black triangles, substrate II; white triangles, substrate I; and white circles, substrate III.
TABLE 1.
Kinetic parameters of glycan transfer to pilin protein acceptor catalyzed by PglL
Each parameter value represents the average of three independent measurements.
| Substratea | Length of acyl chainb and isomerizationc | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|
| μm | nmol/min·mgr of PglL | 1/min | 1/min·mm | ||
| I | 3 (cis) | 32.7 ± 2.9 | 90.9 ± 8.7 | 0.006 | 0.1 |
| II | 3 (trans) | 31.7 ± 3.2 | 632.2 ± 59.8 | 0.04 | 1.3 |
| III | 2 (trans) | 371.7 ± 4.0 | 63.2 ± 5.4 | 0.004 | 0.01 |
| IV | 4 (trans) | 16.0 ± 2.1 | 719.3 ± 69.2 | 0.04 | 2.7 |
| V | 5 | 1.7 ± 0.9 | 932.7 ± 9.4 | 0.06 | 36.4 |
a The substrates analyzed correspond to different glycan donors constituted by the same glycan portion but different polyisoprenoid chains. The substrates were identified following the same nomenclature as in Fig. 3A.
b Lengths of the polyisoprenoid chains of the substrates according to the number of their constituent isoprenoid units.
c Isomerization of the first isoprenoid subunit bound to pyrophosphate.
The natural lipid carrier for bacterial OTases is undecaprenol, whereas for eukaryotes and archaea, it is dolichol. We additionally analyzed the capability of PglL of functioning with a dolichol-like substrate by testing pilin glycosylation from a monosaturated pentaprenyl lipid carrier (substrate V, Fig. 3A). Interestingly, for this substrate, which in addition has a longer poliprenoid chain than the other substrates analyzed, PglL displayed the highest catalytic activity (Table 1).
PglL Can Use UDP-diNacBAc as Substrate but Not UDP-GlcNAc or UDP-GalNAc
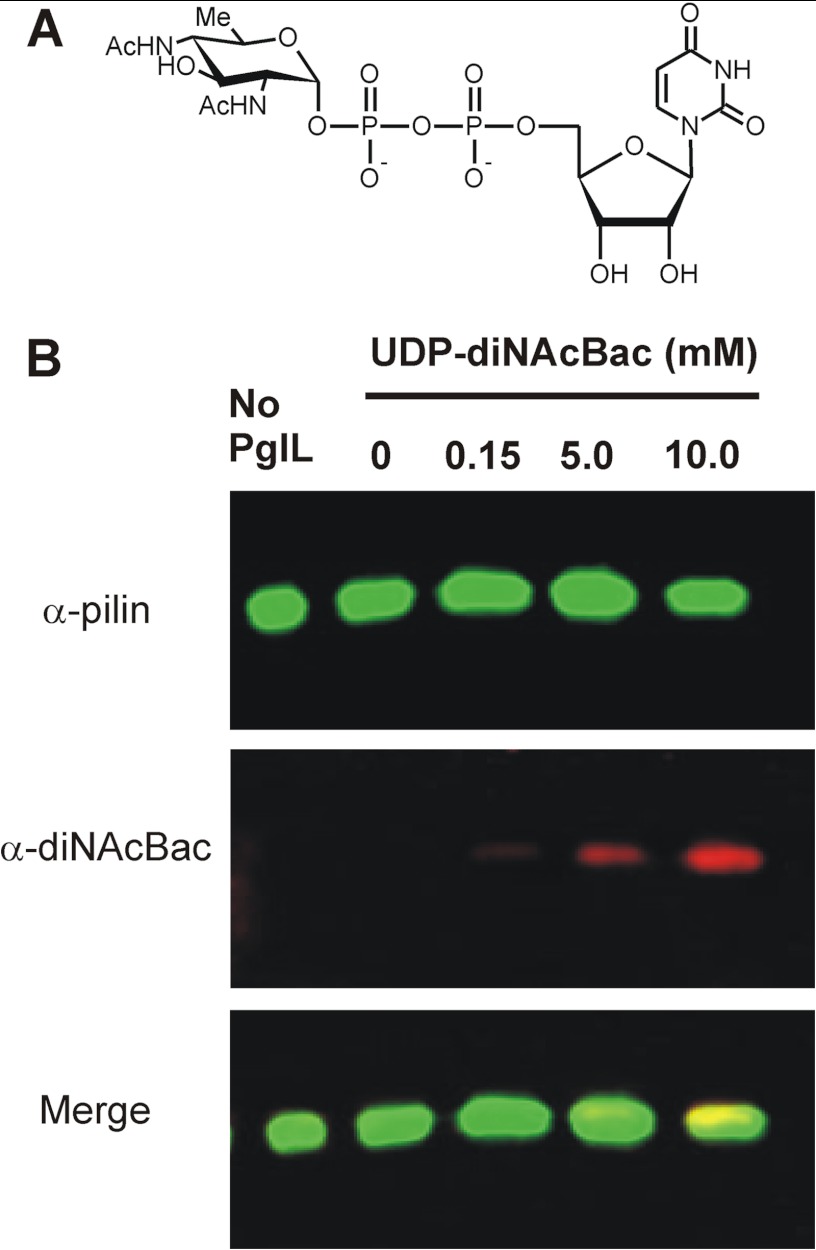
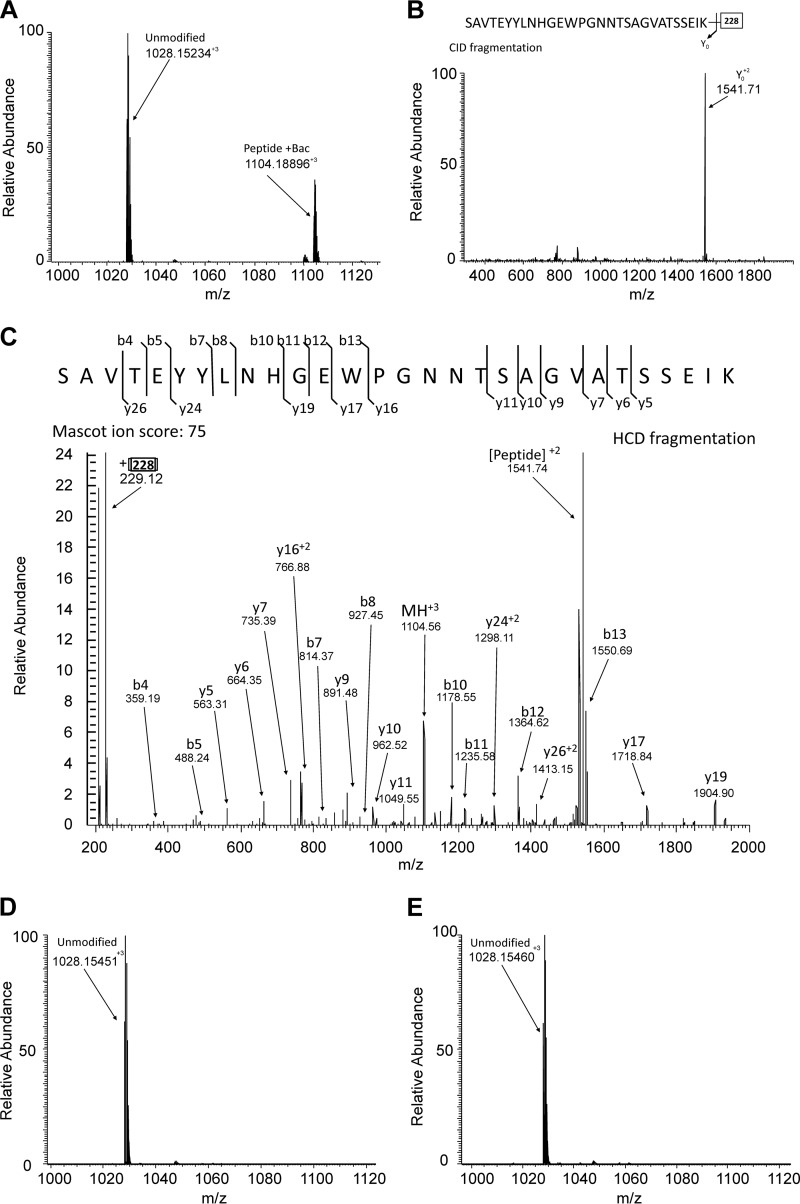
Although exhibiting different efficiencies, PglL transferred the oligosaccharide from all the lipid donors tested. We then wondered whether an aliphatic chain in the lipid donor is indeed required for the glycosylation reaction. We thus assayed the capability of PglL to glycosylate pilin from a nucleotide activated sugar, UDP-diNAcBac (Fig. 4A). UDP-diNAcBac was chosen because diNAcBac is the sugar at the reducing end of the glycan in the native host (35), and a powerful monoclonal antibody specific for this sugar was available (9, 41). For this assay, purified PglL was incubated with pilin in the presence of UDP-diNAcBac at different concentrations. Glycosylation was detected using concentrations of UDP-diNAcBac as low as 150 μm (Fig. 4B). Because glycosylation of pilin with a single sugar molecule does not change pilin electrophoretic mobility, the glycosylation levels could not be accurately quantified. However, using two different antibodies directed against the protein and the glycan moiety, respectively, glycosylated and unglycosylated pilin were detected in separate fluorescent channels and could be distinguished despite of the co-migration. To confirm this result, we analyzed the glycosylated pilin using MS. After incubation with UDP-diNAcBac and PglL, pilin was digested with trypsin, and the resulting peptides were analyzed by reversed phase LC-MS/MS. Consistent with previous reports of glycosylation occurring at Ser-63 (42) trypsinization of UDP-diNAcBac in vitro-glycosylated pilin revealed both modified and unmodified forms of the tryptic peptide spanning Ser-63 (Fig. 5A). The modification status and identity of these peptides were confirmed using CID and HCD MS/MS fragmentation (Fig. 5, B and C), confirming the addition of a single diNAcBac and the identity of the peptide substrate as SAVTEYYLNHGEWPGNNTSAGVATSSEIK.
FIGURE 4.
PglL can glycosylate pilin using UDP-diNAcBac as substrate. A, structure of UDP-diNAcBac. B, Western blot analysis of in vitro-glycosylated pilin by PglL and using UDP-diNAcBac as glycan donor. The signal corresponds to the antibody anti-diNAcBac located at the same position as the pilin acceptor.
FIGURE 5.
Detection of in vitro glycosylation of pilin within the known glycosylation region 52SAVTEYYLNHGEWPGNNTSAGVATSSEIK80 using mass spectrometry. A, MS analysis of in vitro-glycosylated PilE with UDP-diNAcBac revealed the presence of both diNAcBac modified (1104.18896+3) and unmodified (1028.15234+3) 52SAVTEYYLNHGEWPGNNTSAGVATSSEIK80. B, CID fragmentation supported the assignment of 1104.18896+3 as the diNAcBac-modified form of 1028.15234+3 due to the dominant loss of 228 Da upon fragmentation. C, HCD fragmentation of 1104.18896+3 enabled the confirmation of the peptide component of this diNAcBac modified peptide as the expected 52SAVTEYYLNHGEWPGNNTSAGVATSSEIK80, with a MASCOT ion score of 75. Unmodified 1028.15234+3 was also confirmed by HCD, with a MASCOT ion score 74 (not shown). In contrast, only the unmodified peptide could be detected after in vitro glycosylation assays with both UDP-GalNAc (D) and UDP-GlcNAc (E).
To test whether other UDP sugars can be used as substrates, we carried out a similar experiment using UDP-GlcNAc and UDP-GalNac as substrates. Lectin blots using wheat germ agglutinin (lectin from Triticum vulgaris) and soybean agglutinin (lectin from Glycine max), which are specific for GlcNAc and GalNAc, respectively, failed to show any glycosylated product (data not shown). Additionally, MS analysis only detected the unmodified form of SAVTEYYLNHGEWPGNNTSAGVATSSEIK for in vitro glycosylation reactions using UDP-GlcNAc and UDP-GalNac (Fig. 5, D and E). These results indicate that PglL is able to catalyze the glycan transfer from a nucleotide diphosphate carrier showing Leloir enzyme behavior (43) with diNAcBac but is unable to transfer significant amounts with UDP-GlcNAc or UDP-GalNAc. To test whether this feature is a general property of bacterial OTases, we analyzed whether C. jejuni PglB could also be active using UDP-diNAcBac as substrate. PglB was active in vitro using the cLLO substrate. However, the enzyme did not transfer diBacNAc to its acceptor protein AcrA at the UDP-diNAcBac concentrations sufficient for glycosylation with PglL (supplemental Fig. S3).
DISCUSSION
The bacterial OTases involved in en bloc glycosylation processes catalyze the glycan transfer from the undecaprenylpyrophosphate carrier to several protein acceptors, fulfilling a relevant function for bacterial adhesion and virulence (1). These enzymes also have a great potential to produce glycoconjugates interesting to the biotechnological industry (20, 21, 44–46). In this context, knowledge about the functionality of OTases is valuable for improving vaccines and therapeutics against bacterial infections.
Methods employing radioactive sugars have been applied to kinetically characterize N-OTases (47). However, little knowledge about the kinetic parameters of O-OTases is available to date. In this work, we developed a quantitative Western blot assay to study the functionality of the best characterized O-OTase, N. meningitidis PglL. This approach consists in separating the non-glycosylated and glycosylated forms of the protein acceptor by SDS-PAGE and subsequent quantification of both forms by measuring the integrated signals of secondary fluorescent-labeled antibodies in Western blots. These signals are indefinitely stable, proportional to the amount of inmunoreactant and can be quantified, constituting an excellent method to follow enzyme activities. Similar quantification procedures have been applied in different studies, and our technique could easily be adapted to investigate the functionality of other O-OTases or similar enzymes such as N-OTases and O antigen ligases or polymerases (37–40).
We have shown that PglL does not require metal divalent cations to function optimally (Fig. 1). Moreover, the enzyme did not change its activity in the presence of 20 mm EDTA, a chelator of metal divalent cations. This result is in agreement with data obtained by Ruan et al. (48) for the O antigen ligase WaaL from E. coli, which is homologous to PglL and catalyzes the glycan transfer from the undecaprenylpyrophosphate lipid carrier to lipid A. These authors showed that WaaL does not require a divalent metal cation for its activity (48).
We analyzed the activity of PglL at different concentrations of various synthetic glycolipidic substrates containing the same glycan portion but lipidic moities differing in their chain lengths and isoprenyl geometry (cis and trans) (Fig. 3A). PglL showed activity with all of the assayed substrates, but differences in the efficiencies were evident (Fig. 3B). This relaxed specificity toward the lipid moiety of the glycan donor displayed by PglL has also been reported for the enzyme WaaL (22). The conserved functional and mechanistic features between WaaL ligases and O-OTases strengthen the idea of a tightly evolutionary relationship between these two families of enzymes, which basically catalyze the same reaction but toward different acceptors.
A correlation between polyisoprenoid chain length and catalytic efficiency was observed, so that a higher polyisoprenoid chain length leads to a higher catalytic efficiency (Table 1). When substrates containing identical length but different isomerization of the first isoprene unit were analyzed, PglL displayed similar Km values but a higher catalytic efficiency with the trans substrate (Table 1), which is the isomerization of the undecaprenylpyrophosphate natural carrier in the inner membrane of Gram-negative bacteria. Thus, the geometry of the lipid might be relevant to fit the substrate optimally into the active site of PglL. In agreement, it has been proposed that the isoprenoid moiety of the glycan donor is embedded in a hydrophobic groove on the N-OTase PglB of C. lari, pointing into the lipid bilayer and allowing the orientation of the glycan portion toward the active site (49). In addition, it has been hypothesized that the roles of the lipid carrier are the translocation of the oligosaccharide from the cytoplasm into the periplasm and the positioning of the pyrophosphate group and glycan into the periplasmic lumen, adjacent to the PglL catalytic site (19). Therefore, following a similar argument, a longer aliphatic chain of the glycan donor implies higher hydrophobicity that might allow for a better interaction between the polyisoprenoid chain and the transmembrane portion of PglL and may therefore enhance the chances of fitting the substrate in a productive orientation into the catalytic site. It is tempting to speculate that the active site of PglL could be distributed between the lipid bilayer and the periplasmic space. This hypothesis is supported by biochemical evidences that suggest that the polyisoprenyl carrier plays a more specific chemical role in substrate enzyme interactions rather than a simple physical role as a hydrophobic membrane anchor (50).
Despite the different efficiencies, PglL could transfer a subunit of the E. coli O86 antigen from all the assayed lipid donors. Considering that the sugar structure was previously found to be irrelevant for activity, we concluded that the only common structure to all PglL substrates was the pyrophosphate linking the sugar to the lipid. This motif is also present in nucleotide-activated sugars. When nucleotide-activated sugars were assayed as substrates, PglL could glycosylate pilin using UDP-diNAcBac (Fig. 4) as substrate. Glycosylation was detectable at concentrations as low as 150 μm UDP-diNAcBac. This concentration is close to the Km of some Leloir glycosyltransferases and can be considered physiological (51, 52). However, UDP-diNAcBac is not present in the bacterial periplasm, and therefore, it cannot be encountered by PglL in vivo. Intriguingly, in contrast to what has been shown for lipid-linked glycans, PglL displayed sugar specificity when the nucleotide sugars were assayed as substrates. Under identical experimental conditions, PglL employed UDP-diNAcBac as substrate but not UDP-GlcNac or UDP-GalNac. This indicates that the binding of the lipid carrier to the enzyme may play a role in positioning the sugar residue in a catalytically productive conformation. This may not be achieved if instead of the lipid, a nucleotide is attached to these HexNAc sugars. diBacNAc is found at the reducing end of the native trisaccharide recognized by PglL and it can be transferred even from a nucleotide carrier, suggesting that this sugar fits better into the active site of the enzyme. Our results showed that in the absence of a lipid carrier, the chemical nature of the glycan influences the glycosylation reaction. In contrast to PglL, the N-OTase PglB cannot use UDP-diNAcBac as substrate, even though this sugar is also present at the reducing end of its native glycan. The active site of PglB could be located in a region not accessible for freely diffusible substrates, requiring a lipid carrier to position the substrate at the interface between the membrane and the periplasmic space, whereas the active site of PglL could be more open. Alternatively, the nucleotide may not allow the sugar to reach the active site of PglB due to sterical hindrance. Finally, it is tempting to speculate that the capacity of PglL of using a nucleotide-activated sugar could be a vestigial remnant related to the evolution of O-OTases from an ancestral Leloir glycosyltransferase. To date, no sequence similarity at the amino acid level supports this hypothesis. However, the sequence similarities between O-OTases, and more generally, among membrane glycosyltransferases are very low. Structural information on this kind of enzymes will be crucial to further our understanding of O-OTases. Such studies may lead to the identification of novel antimicrobials and possibilities to employ these enzymes for biotechnological purposes.
Acknowledgments
We thank Markus Aebi (ETH Swizterland) and Michael Koomey (University of Oslo) for kindly supplying us with antibodies and Martin Young (NRC, Ottawa) for providing the UDP-diNAcBac employed in this study. We thank Eduardo Ceccarelli and Brent Weber for critical reading of the manuscript and Amy McLeod for technical assistance.
This work was supported in part by grants from the Natural Sciences and Engineering Research Council of Canada and the Alberta Glycomics Center (to M. F. F.).

This article contains supplemental Figs. S1–S3.
- OTases
- oligosaccharyltransferases
- cLLO
- C. jejuni lipid-linked oligosaccharide
- UDP-diNAcBac
- uridine 5′-diphosphate N′-diacetylbacillosamine
- UDP-GlcNac
- uridine 5′-diphospho-N-acetylglucosamine
- CID MS/MS
- collision-induced dissociation-tandem mass spectrometry
- HCD MS/MS
- higher energy C-trap dissociation tandem mass spectrometry.
REFERENCES
- 1. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 2. Twine S. M., Reid C. W., Aubry A., McMullin D. R., Fulton K. M., Austin J., Logan S. M. (2009) Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191, 7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grass S., Lichti C. F., Townsend R. R., Gross J., St Geme J. W., 3rd. (2010) The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asakura H., Churin Y., Bauer B., Boettcher J. P., Bartfeld S., Hashii N., Kawasaki N., Mollenkopf H. J., Jungblut P. R., Brinkmann V., Meyer T. F. (2010) Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol. Microbiol. 78, 1130–1144 [DOI] [PubMed] [Google Scholar]
- 5. Weerapana E., Imperiali B. (2006) Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology 16, 91R-101R [DOI] [PubMed] [Google Scholar]
- 6. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 7. Calo D., Kaminski L., Eichler J. (2010) Protein glycosylation in Archaea: sweet and extreme. Glycobiology 20, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 8. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., Aebi M. (2002) N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 9. Ielmini M. V., Feldman M. F. (2011) Desulfovibrio desulfuricans PglB homolog possesses oligosaccharyltransferase activity with relaxed glycan specificity and distinct protein acceptor sequence requirements. Glycobiology 21, 734–742 [DOI] [PubMed] [Google Scholar]
- 10. Power P. M., Seib K. L., Jennings M. P. (2006) Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 347, 904–908 [DOI] [PubMed] [Google Scholar]
- 11. Aas F. E., Vik A., Vedde J., Koomey M., Egge-Jacobsen W. (2007) Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65, 607–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwashkiw J. A., Seper A., Weber B. S., Scott N. E., Vinogradov E., Stratilo C., Reiz B., Cordwell S. J., Whittal R., Schild S., Feldman M. F. (2012) Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8, e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gebhart C., Ielmini M. V., Reiz B., Price N. L., Aas F. E., Koomey M., Feldman M. F. (2012) Characterization of exogenous bacterial oligosaccharyltransferases in Escherichia coli reveals the potential for O-linked protein glycosylation in Vibrio cholerae and Burkholderia thailandensis. Glycobiology 22, 962–974 [DOI] [PubMed] [Google Scholar]
- 14. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomas R. M., Twine S. M., Fulton K. M., Tessier L., Kilmury S. L., Ding W., Harmer N., Michell S. L., Oyston P. C., Titball R. W., Prior J. L. (2011) Glycosylation of DsbA in Francisella tularensis subsp. tularensis. J. Bacteriol. 193, 5498–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joshua G. W., Guthrie-Irons C., Karlyshev A. V., Wren B. W. (2006) Biofilm formation in Campylobacter jejuni. Microbiology 152, 387–396 [DOI] [PubMed] [Google Scholar]
- 18. Jennings M. P., Jen F. E., Roddam L. F., Apicella M. A., Edwards J. L. (2011) Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol. 13, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faridmoayer A., Fentabil M. A., Haurat M. F., Yi W., Woodward R., Wang P. G., Feldman M. F. (2008) Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J. Biol. Chem. 283, 34596–34604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feldman M. (2009) Microbial Glycobiology (Moran A., ed) pp. 904–914, Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 21. Iwashkiw J. A., Fentabil M. A., Faridmoayer A., Mills D. C., Peppler M., Czibener C., Ciocchini A. E., Comerci D. J., Ugalde J. E., Feldman M. F. (2012) Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microbial Cell Factories 11, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han W., Wu B., Li L., Zhao G., Woodward R., Pettit N., Cai L., Thon V., Wang P. G. (2012) Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J. Biol. Chem. 287, 5357–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Woodward R., Yi W., Li L., Zhao G., Eguchi H., Sridhar P. R., Guo H., Song J. K., Motari E., Cai L., Kelleher P., Liu X., Han W., Zhang W., Ding Y., Li M., Wang P. G. (2010) In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M., Shen J., Liu X., Shao J., Yi W., Chow C. S., Wang P. G. (2008) Identification of a new α1,2-fucosyltransferase involved in O-antigen biosynthesis of Escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry 47, 11590–11597 [DOI] [PubMed] [Google Scholar]
- 25. Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., Aebi M. (2006) N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 [DOI] [PubMed] [Google Scholar]
- 26. Schwarz F., Lizak C., Fan Y. Y., Fleurkens S., Kowarik M., Aebi M. (2011) Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21, 45–54 [DOI] [PubMed] [Google Scholar]
- 27. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 28. Ishihama Y., Rappsilber J., Mann M. (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 29. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 30. Cristobal A., Hennrich M. L., Giansanti P., Goerdayal S. S., Heck A. J., Mohammed S. (2012) In-house construction of a UHPLC system enabling the identification of over 4000 protein groups in a single analysis. Analyst 137, 3541–3548 [DOI] [PubMed] [Google Scholar]
- 31. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell Proteomics 10, M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 34. Roepstorff P., Fohlman J. (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601. [DOI] [PubMed] [Google Scholar]
- 35. Stimson E., Virji M., Makepeace K., Dell A., Morris H. R., Payne G., Saunders J. R., Jennings M. P., Barker S., Panico M. (1995) Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17, 1201–1214 [DOI] [PubMed] [Google Scholar]
- 36. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y. V., Wade M., Wong E., Li Y. C., Rodewald L. W., Wahl G. M. (2007) Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc. Natl. Acad. Sci. U.S.A. 104, 12365–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wishart T. M., Paterson J. M., Short D. M., Meredith S., Robertson K. A., Sutherland C., Cousin M. A., Dutia M. B., Gillingwater T. H. (2007) Differential proteomics analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wlds) gene. Mol. Cell Proteomics 6, 1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Markovic D., Punn A., Lehnert H., Grammatopoulos D. K. (2008) Intracellular mechanisms regulating corticotropin-releasing hormone receptor-2β endocytosis and interaction with extracellularly regulated kinase 1/2 and p38 mitogen-activated protein kinase signaling cascades. Mol. Endocrinol. 22, 689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramsay A. J., Dong Y., Hunt M. L., Linn M., Samaratunga H., Clements J. A., Hooper J. D. (2008) Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J. Biol. Chem. 283, 12293–12304 [DOI] [PubMed] [Google Scholar]
- 41. Børud B., Aas F. E., Vik A., Winther-Larsen H. C., Egge-Jacobsen W., Koomey M. (2010) Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192, 2816–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Faridmoayer A., Fentabil M. A., Mills D. C., Klassen J. S., Feldman M. F. (2007) Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J. Bacteriol. 189, 8088–8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 44. Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., Morris H. R., Dell A., Valvano M. A., Aebi M. (2005) Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langdon R. H., Cuccui J., Wren B. W. (2009) N-linked glycosylation in bacteria: an unexpected application. Future Microbiol. 4, 401–412 [DOI] [PubMed] [Google Scholar]
- 46. Valderrama-Rincon J. D., Fisher A. C., Merritt J. H., Fan Y. Y., Reading C. A., Chhiba K., Heiss C., Azadi P., Aebi M., DeLisa M. P. (2012) Nat. Chem. Biol. 8, 434–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glover K. J., Weerapana E., Numao S., Imperiali B. (2005) Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem. Biol. 12, 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruan X., Loyola D. E., Marolda C. L., Perez-Donoso J. M., Valvano M. A. (2012) The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology 22, 288–299 [DOI] [PubMed] [Google Scholar]
- 49. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 50. Chen M. M., Weerapana E., Ciepichal E., Stupak J., Reid C. W., Swiezewska E., Imperiali B. (2007) Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 46, 14342–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laughlin M. R., Petit W. A., Jr., Dizon J. M., Shulman R. G., Barrett E. J. (1988) NMR measurements of in vivo myocardial glycogen metabolism. J. Biol. Chem. 263, 2285–2291 [PubMed] [Google Scholar]
- 52. Ciesla W. P., Jr., Bobak D. A. (1998) Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J. Biol. Chem. 273, 16021–16026 [DOI] [PubMed] [Google Scholar]