Background: Classical zinc finger proteins are extremely abundant and interact with DNA using a well defined recognition code.
Results: We solved the structure of ZNF217 bound to its cognate DNA.
Conclusion: ZNF217 presents a unique DNA interaction pattern including a new type of protein-DNA contact.
Significance: This study deepens our understanding of DNA recognition by classical zinc fingers.
Keywords: Crystal Structure, DNA-binding Protein, Oncogene, Protein Design, Zinc Finger, Protein-DNA Complex Structure, ZNF217
Abstract
Classical zinc fingers (ZFs) are one of the most abundant and best characterized DNA-binding domains. Typically, tandem arrays of three or more ZFs bind DNA target sequences with high affinity and specificity, and the mode of DNA recognition is sufficiently well understood that tailor-made ZF-based DNA-binding proteins can be engineered. We have shown previously that a two-zinc finger unit found in the transcriptional coregulator ZNF217 recognizes DNA but with an affinity and specificity that is lower than other ZF arrays. To investigate the basis for these differences, we determined the structure of a ZNF217-DNA complex. We show that although the overall position of the ZFs on the DNA closely resembles that observed for other ZFs, the side-chain interaction pattern differs substantially from the canonical model. The structure also reveals the presence of two methyl-π interactions, each featuring a tyrosine contacting a thymine methyl group. To our knowledge, interactions of this type have not previously been described in classical ZF-DNA complexes. Finally, we investigated the sequence specificity of this two-ZF unit and discuss how ZNF217 might discriminate its target DNA sites in the cell.
Introduction
Zinc finger (ZF)2 proteins are one of the largest superfamilies of proteins in mammals and are also widespread in other phyla. ZFs are small domains (usually less than ∼100 amino acids) characterized by the presence of one or more zinc ions that stabilize(s) the fold. Many different structural classes of ZF have been discovered, and different classes can have similar or unrelated functions (1). Their classification is based on their three-dimensional structure and the identity and spacing of the residues (typically cysteine and histidine, and occasionally other residues) that coordinate the zinc ion(s) (2–5). Several structurally distinct classes of ZF are able to act as nucleic acid-binding modules, including GATA-type ZFs (6), steroid hormone receptor ZFs (7), Gal4-type ZFs (8, 9), which bind double-stranded DNA, and tristetraprolin (TTP) family (10), RanBP2-type (11), and nucleocapsid (12) ZFs, which bind RNA.
By far the most abundant ZF domain, however, is the classical or C2H2 ZF, which is defined by a CX2–4CX12HX2–6H motif. Classical ZFs were the first discovered class of ZF (13) and are found in more than 700 human proteins (14), many of which harbor multiple ZF domains (up to 30 or more in some instances). In the majority of cases in which there are experimental data on classical ZFs, these domains are found in transcriptional regulatory proteins and act as sequence-specific DNA-binding modules, appearing in arrays of three of more domains in which each member contacts three base pairs of double-stranded DNA (15–17). Typically, these arrays bind promoter elements with dissociation constants in the nanomolar range, allowing associated transcriptional activation or repression domains to be targeted to the gene.
Several three-dimensional structures of ZF-DNA complexes have been determined (15–22), and these structures reveal a number of commonalities that have elicited talk of a recognition “code” for ZF-DNA interactions (23). Although a robust code has yet to be identified, some principles have emerged; the domains (which comprise a small β-hairpin and a single α-helix) insert the N-terminal end of their α-helix into the major groove of DNA, and typically 3–4 amino acid side chains in the helix make direct contacts with either the bases or the backbone of the DNA. Residues in the +2-, +3-, and +6-positions of the helix, together with the residue immediately preceding the helix (the −1-position), are most frequently found to make sequence-specific contacts (Fig. 1A and supplemental Fig. 1), whereas residues in the β-hairpin and in the conserved interdomain linker often make nonspecific electrostatic interactions with the phosphodiester backbone. In fact, the relatively conserved nature of these interactions, together with the robustness of the fold and the modularity of the interactions, have led to the creation of designer DNA-binding ZF proteins capable of delivering an associated activation, repression, or nuclease domain to a chosen genomic site (24–27).
FIGURE 1.
Sequence-specific base contacts in classical ZNF-DNA complexes. A, the canonical model derived from consideration of the known ZF-DNA structures. B, ZNF217_F67 (Protein Data Bank (PDB) ID: 4IS1, this work). C, TRAMTRACK (PDB ID: 2DRP) (18). D, Zif268 (PDB ID: 1AAY) (15). In the canonical model, the sequence-specific contacts are made from side chains located at key positions −1, +2, +3 and +6 along the α-helix. Base-specific protein-DNA interactions are represented by black arrows.
Despite these successes and the apparently well understood DNA binding properties of classical ZF domains, a number of questions remain. For example, several ZFs have been shown to act as protein recognition modules (28) or to bind to RNA (29, 30) rather than (or as well as) DNA, and currently we have no clear means by which to distinguish these different functional classes of ZF. Although DNA-binding ZFs tend to occur in clusters of three or more modules, there are hundreds of classical ZFs in the human genome that are found either as single isolated domains or as pairs, and the likelihood that these domains bind DNA is not known. Similarly, a significant number of proteins contain multiple ZF clusters distributed across a large amount of sequence, and the reasons underlying the presence of so many ZFs in a single protein are currently obscure.
ZNF217 is an ∼1000-residue protein that has been identified as a candidate oncoprotein. High levels of ZNF217 are associated with a poor prognosis in breast cancer (31), and overexpression of the ZNF217 gene in human mammary epithelial cells immortalizes those cells (32). ZNF217 contains seven predicted classical ZFs, found in two clusters in the N-terminal half of the protein. Although nothing is known about the function of the first cluster of five ZFs, a recent study demonstrated that the cluster comprising ZFs 6 and 7 (ZNF217_F67) can bind to double-stranded DNA (33). A site selection experiment identified the consensus sequence CAGAAY as the preferred in vitro recognition site for ZNF217_F67.
Subsequently, we identified an extended version of this consensus sequence to which stronger binding was observed in gel shifts: namely (T/A)(G/A)CAGAA(T/G/C) (34). Transient transfection experiments in which this sequence was incorporated into a promoter region showed that ZNF217 could act as a transcriptional repressor and that the activity was dependent on the structural integrity of the F67 unit. Surprisingly, we also observed that in vitro, the affinity of ZNF217_F67 for DNA was reduced by only 2-fold when the consensus sequence was extensively mutated, suggesting that this double ZF domain might have relatively low DNA binding specificity when compared with other transcriptional regulators that directly contact DNA.
To understand the basis for these observations, we have determined the three-dimensional structure of a ZNF217_F67-DNA complex by x-ray crystallography. Our data show that ZNF217 displays both similarities and significant differences from other ZF domains in the mechanism by which it recognizes DNA, making several base-specific contacts with DNA that have not previously been observed in classical ZF-DNA complexes. ZNF217 has an affinity of 25 nm for its target DNA site, and in solution, we also observed the formation of a lower affinity nonspecific complex between ZNF217_F67 and DNA at lower ionic strength. A combination of calorimetric and NMR data indicate that although this nonspecific complex involves the canonical DNA-binding surface of the protein, it is “looser” in nature, displaying less favorable bonding interactions. Interactions of this type might well play a role in the DNA recognition process in vivo for ZNF217 and other ZF proteins.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant ZNF217_F67
A construct encoding F67 of human ZNF217 (amino acids 467–523) was cloned into pMALC2 and pGEX2T vectors to allow the expression of MBP and GST fusion proteins, respectively. The MBP construct was expressed in Escherichia coli Rosetta2 cells overnight at 25 °C following the addition of 0.7 mm isopropyl-1-thio-β-d-galactopyranoside and 1 μm ZnSO4 to the log phase culture. Expression of the GST fusion construct was induced overnight at 22 °C by the addition of 0.4 mm isopropyl-1-thio-β-d-galactopyranoside to E. coli BL21 cells supplemented with 1 μm ZnSO4. Cells were lysed in a buffer containing 50 mm Tris-HCl (pH 8), 1 m NaCl, 1 mm DTT, and 1 mm PMSF. MBP and GST fusion proteins were recovered from the soluble fraction and purified by affinity chromatography. The fusion tags were cleaved using thrombin (3 h at room temperature) in 50 mm Tris (pH 8), 1 m NaCl, 10 mm CaCl2, and 1 mm DTT. F67 was then dialyzed into 50 mm Tris (pH 7), 1 mm DTT and further purified by cation-exchange chromatography (UnoS1, Bio-Rad). The construct identity and correct folding of F67 were confirmed by DNA sequencing and one-dimensional 1H NMR spectroscopy, respectively. 15N-labeled ZNF217_F67 was prepared following the procedure of Cai et al. (35) and purified as described above.
Design and Preparation of the Oligonucleotides Used in the Crystallization Trials
Three different double-stranded oligonucleotides, containing either one or two copies of the 8-bp consensus sequence TGCAGAAT, were used in efforts to crystallize a ZNF217_F67-DNA complex. All oligonucleotides were 20 residues in length with two complementary overhang nucleotides at the 5′ extremities of each strand. The first set of oligonucleotides contains two binding sites running in the same direction (forward, 5′-TTTGCAGAATCGTGCAGAAT-3′; reverse, 5′-ACGTCTTAGCACGTCTTAAA-3′). The second contains two binding sites running in opposite directions (forward, 5′-TTTGCAGAATCGATTCTGCA-3′; reverse, 5′-ACGTCTTAGCTAAGACGTAA-3′). The last contains a single binding site (forward, 5′-TTTCCATTGCAGAATTGTGG-3′; reverse, 3′-AGGTAACGTCTTAACACCAA-5′). ssDNA oligonucleotides were purchased from Sigma and heated at 95 °C for 15 min in a 50 mm Tris-HCl (pH 7.4) buffer containing 150 mm NaCl. Oligonucleotides were then annealed at room temperature overnight and purified by size exclusion chromatography (Sephadex-75, GE Healthcare).
Crystallization and Data Collection
Purified ZNF217_F67 and the different DNA duplexes were dialyzed in 20 mm Tris (pH 7), 50 mm NaCl. and 1 mm DTT before being mixed together (ZNF217_F67-DNA, 1:0.6 with the DNA duplexes that carried two binding sites and 1:1.2 with the oligonucleotide that contained a single site). The final protein concentration was 10 mg/ml. Initial crystallization trials were set up at 298 K as vapor diffusion hanging drops using a Mosquito robot (Molecular Dimensions) by mixing 400 nl of sample solution and 400 nl of reservoir solution and placing the resultant drop over 80 μl of reservoir solution in flat-bottom 96-well PS microplates (Greiner Bio-One). JSGC+ and PACT (Qiagen) screens were trialed. Large crystals in two different space groups were obtained with the oligonucleotide containing two binding sites running in opposite directions. Crystals in space group P6522 grew in the presence of 200 mm sodium acetate (pH 7), and 20% (w/v) polyethylene glycol (PEG) 3350 precipitant solution. Crystals in space group C2 grew in 100 mm MES (pH 6), 10 mm zinc chloride, and 20% (v/v) PEG 6000. Diffraction data were recorded on a mar345 image plate detector (Marresearch) using x-rays produced by a Rigaku RU200H rotating-anode generator (CuKα) focused with Osmic mirrors (MSC Rigaku). The diffraction data were integrated and scaled with HKL-2000 (36).
Solution and Refinement of the Crystal Structures
Phases for the P6522 crystal form were determined using the SIRAS technique with a lead derivative. Crystals were soaked for 2 h in crystallization buffer containing 10 mm trimethyl lead, and a 3.0-Å data set was collected. SIRAS phasing was realized using AutoSol (37), which identified two lead atoms and resulted in a mean figure of merit after density modification of 0.69. The resulting electron density was of sufficient quality to allow building of the oligonucleotide and peptide backbone. Successive rounds of model building were carried out using Coot (38), and refinement utilized REFMAC5 (39). Combined TLS (translation/libration/screw) and individual atomic displacement parameter refinement were also carried out in the final stages. The C2 crystal form phase was solved by molecular replacement with the P6522 model using PHASER (40). Model building and refinement were performed similarly to that described for the P6522 form. Four additional zinc ions were identified in the asymmetric unit. Due to their absence in the P6522 crystal form and their location on the surface of the protein-nucleic acid complex, we attribute these atoms to the presence of 10 mm ZnCl2 in the crystallization solution.
Fluorescence Anisotropy Titrations
Cleaved or GST-tagged ZNF217_F67 and 5′-fluorescein-labeled dsDNA oligonucleotides (WT sequence forward, 5′-Fl-TCCATTGCAGAATTGTGG-3′; mutated sequence, forward, 5′-Fl-TCCATCTGGAGTATGTGG-3′; poly(A), forward, 5′-Fl-(A)18-3′; the bold sequences correspond to the 8 bp consensus sequence and its mutated version recognized by ZNF217) were dialyzed into a 10 mm phosphate buffer, pH 7, containing 50 mm NaCl and 1 mm DTT. Fluorescence anisotropy titrations were performed at 25 °C on a Cary Eclipse fluorescence spectrophotometer with a slit width of 10 nm, and data were averaged over 15 s. The excitation and detection wavelengths were 495 and 520 nm, respectively. In each titration, the fluorescence anisotropy of a solution of 50 nm fluorescein-tagged dsDNA was measured as a function of the added protein concentration. Binding data were fitted to a simple 1:1 binding model by nonlinear least squares regression. Each titration was performed three times, and the final affinity was taken as the mean of these measurements.
Isothermal Titration Calorimetry (ITC)
ZNF217_F67 and the two DNA duplexes (see above) were dialyzed overnight against the same reservoir of buffer containing 10 mm Tris buffer, pH 7.0, 50 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine. Titrations were also carried out at 150 mm NaCl. ZNF217_F67 (200 μm) was titrated into DNA (20 μm). Titrations were carried out on a MicroCal i200 ITC microcalorimeter (GE Healthcare) at 25 °C. For each titration, an initial injection of 0.2 μl (data from which were discarded) and 20 injections of 2 μl of titrant were made at 120-s intervals. Data were corrected for heats of dilution from control experiments of the protein into buffer and analyzed using Origin7.0 (MicroCal Software, Northampton, MA).
The two-binding-event titration curve observed for the ZNF217 binding to the specific DNA sequence could not be fitted with confidence using a two-site model because the error associated with this fit was above 100%. We therefore made the assumption that the second, low affinity binding event was identical to the single binding event observed during the titration of the mutated sequence. Using this assumption, we subtracted from the first titration the data points observed for the latter titration and could then fit the remaining data to a single binding event with an associated error under 20%. The derived dissociation constant for the tight interaction was indistinguishable from that obtained with the two-site model, except that the uncertainty in the fit was substantially lower for the single-site fit.
NMR Spectroscopy
For 15N HSQC chemical shift perturbation experiments, purified 15N-labeled ZNF217_F67 and the different dsDNA oligonucleotides were extensively dialyzed into a buffer comprising 10 mm Na2HPO4 (pH 7.0), 50 mm NaCl, and 1 mm DTT and were concentrated to ∼300 μm. All NMR samples contained 5–10% D2O and 10 μm 2,2-dimethyl-2-silapentane-5-sulfonic acid as a chemical shift reference. All experiments were run at 298 K on either a 600-MHz or an 800-MHz Bruker AvanceIII spectrometer equipped with a cryoprobe. 15N HSQC spectra were recorded for the ZNF217_F67 alone and following the addition of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, 1, 1.2, and 1.5 molar eq of either wild type (WT) or nonspecific DNA. The interaction between ZNF217_F67 and nonspecific DNA was in fast exchange, allowing straightforward resonance assignment from the titration data. NOESY spectra were also recorded to confirm these assignments. NMR data were processed using Topspin (Bruker, Karlsruhe, Germany) and analyzed with SPARKY (76).
RESULTS
Overall Arrangement of the ZNF217-DNA Complex
To understand the DNA binding activity of the F67 double-ZF module of ZNF217, we set up crystallization trials with a recombinant protein containing residues 467–523 of human ZNF217 and several different dsDNA oligonucleotides. Each DNA fragment contained either one or two copies of the 8-bp sequence TGCAGAAT, derived from published systematic evolution of ligands by exponential enrichment (SELEX) data (33); we call this sequence WT DNA. Crystals were obtained with all the tested DNA fragments, but the biggest crystals with optimum diffraction were obtained for the oligonucleotide containing two binding sites running in opposite directions. The rationale for this approach to oligonucleotide design was that the consequent creation of a two-fold symmetry axis within the oligonucleotide and also within the protein-DNA complex might improve the likelihood of crystallization. Data were collected for two crystals that belonged to different space groups: P6522 and C2; these crystals diffracted to 2.6 and 2.1 Å, respectively. Phases for the P6522 form were obtained by SIRAS phasing using a heavy atom derivative created by soaking a crystal in trimethyl lead acetate, and a model was built. Phases for the second crystal form were obtained by molecular replacement using the model built from the P6522 crystal form. The Rcryst of the final model (of the C2 form) is 22.4%, and the Rfree is 25.6%. All residues were visible in the final electron density except for the N-terminal residues Gly-467–Ser-470 and C-terminal Lys-523, presumably due to disorder. All backbone φ and ψ angle values are located within allowed regions of the Ramachandran plot, and the statistics of data collection and refinement are given in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics for the ZNF217-DNA complex
Pb indicates lead derivative.
| P6522 (PDB ID:4F2J) | P6522–2 Pb | C2 (PDB ID: 4IS1) | |
|---|---|---|---|
| Data collection and processing | |||
| X-ray source | Rotating anode | Rotating anode | Rotating anode |
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418 |
| Resolution range (Å) | 48.31–2.64 | 50.00–3.04 | 15.0–2.10 |
| Highest resolution shells | 2.68–2.64 | 3.09–3.04 | 2.14–2.10 |
| Space group | P6522 | P6522 | C2 |
| Unit cell parameters | |||
| a (Å) | 56.93 | 56.54 | 124.67 |
| b (Å) | 56.93 | 56.54 | 40.98 |
| c (Å) | 242.54 | 243.02 | 52.04 |
| α (°) | 90 | 90 | 90 |
| β (°) | 90 | 90 | 97.21 |
| γ (°) | 120 | 120 | 90 |
| Matthews coefficient (Å3 Da−1) | 4.73 | 4.73 | 2.71 |
| Solvent content (%) | 74 | 74 | 54 |
| No. of complexes per asymmetric unit | 0.5 | 0.5 | 1 |
| Total reflections | 81,911 (2100)a | 45,782 (2246) | 123,593 (6067) |
| Unique reflections | 7517 (344) | 8287 (416) | 15,360 (768) |
| Completeness (%) | 98.5 (95.0) | 98.4 (99.8) | 99.9 (100.0) |
| Redundancy | 10.9 (6.1) | 5.5 (5.4) | 8.0 (7.9) |
| Rmerge (%)b | 9.1 (18.8) | 8.8 (76.4) | 8.6 (89.9) |
| Average I/σ(1) | 28.6 (3.6) | 16.6 (2.2) | 13.8 (2.2) |
| Phasing | |||
| f′ | −4.82 | ||
| f″ | 8.51 | ||
| Phasing power (centric/acentric) | 0.68/0.78 | ||
| Figure of merit after AutoSol | 0.69 | ||
| Structural refinement | |||
| Reflections (total) | 7467 | 15,288 | |
| Reflections (test set) | 344 | 769 | |
| Rcryst (%)c | 25.2 | 23.9 | |
| Rfree (%)d | 26.4 | 26.2 | |
| Coordinate error (Å) | 0.33 | 0.24 | |
| r.m.s.d. from ideal bonds/angles (Å/degree) | 0.016/2.3 | 0.010/1.4 | |
| No. of atoms | 857 | 1791 | |
| Protein | 451 | 920 | |
| DNA | 406 | 814 | |
| Ligand/ion | 0 | 57 | |
| Mean B-factor (Å2) | 96.5 | 54.6 | |
| Protein | 102.4 | 56.8 | |
| DNA | 90.0 | 52.0 | |
| Ligand/ion | NAe | 56.8 | |
| Ramachandran statistics (%) | |||
| Favored | 96 | 99 | |
| Allowed | 100 | 100 | |
| Disallowed | 0 | 0 | |
a Highest resolution shell data.
b Rmerge = 100 × Σh Σi|Ii(h) − bI(h)N|/Σh I(h), where Ii(h) is the ith measurement of the h reflection and bI(h)N is the average value of the reflection intensity.
c Rcryst = 100 × Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the structure factor amplitudes from the data and the model, respectively.
d Rfree is Rcryst on 5% of the test set structure factors.
e NA, not applicable.
In both structures, two protein molecules were found bound to a single double-stranded DNA oligonucleotide. However, in the P6522 crystal form, the inherent symmetry of this complex required only one of the protein molecules and one DNA strand to be built as the remaining DNA strand and protein molecule were generated by a crystallographic two-fold symmetry axis. It is also notable that the two copies of the protein-DNA complex in the C2 form are highly similar but not identical (r.m.s.d. = 0.37 Å over all heavy atoms), indicating that small distortions occurred due to crystal packing. However, no differences were observed in the protein-DNA contacts.
Fig. 2A shows the structure of the C2 crystal form with two protein molecules bound per DNA fragment, as anticipated. The protein-DNA interactions are indistinguishable in the two crystal forms, and the only significant difference is that the DNA in the P6522 form is bent by ∼24° midway between the two protein-DNA interaction sites (supplemental Fig. 2). Because all protein-DNA contacts were identical in the two structures, we focus here on the C2 form.
FIGURE 2.
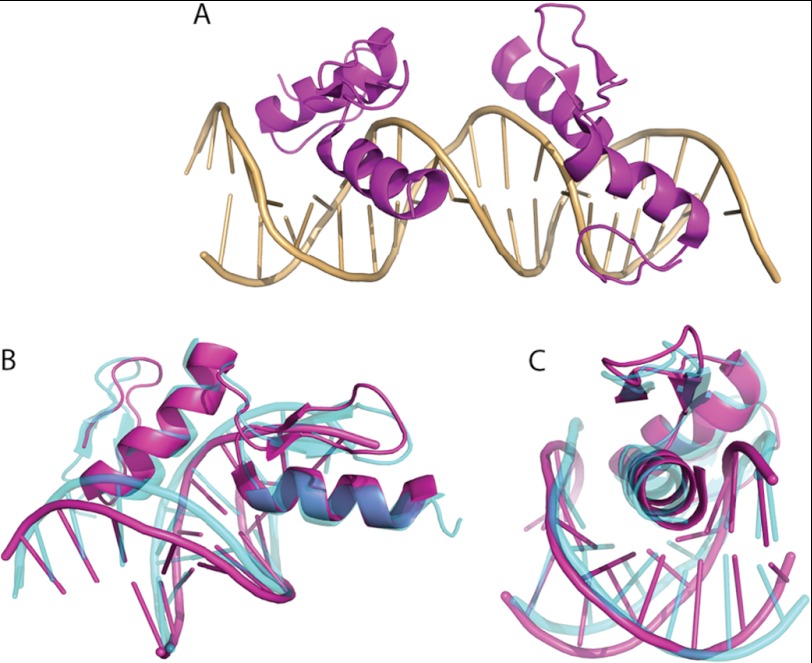
Structure of ZNF217_F67 in complex with a 20-bp oligonucleotide obtained from the C2 crystal form. A, ribbon diagram of the two DNA-bound protein molecules. The PDB code for the structure is 4IS1. B and C, structural alignment of ZNF217_F67 (magenta) and zinc fingers 2 and 3 from the Zif268-DNA complex (cyan) complexed to DNA (PDB 1AAY (75)). The alignment was made over all heavy atoms. Two views are shown.
Both F6 and F7 adopt the canonical ββα ZF fold, with a small β-hairpin packed against a α-helix and a single zinc ion coordinated by the side chains of two cysteine and two histidine residues. The two domains insert the N-terminal end of their α-helix into the DNA major groove, and the overall orientation of each domain relative to the DNA is essentially indistinguishable from other ZF-DNA complexes. An overlay of the complex with the Zif268-DNA complex (using ZFs 2 and 3 of Zif268), with which ZNF217_F67 shares 34% sequence identity, has an r.m.s.d. over all heavy atoms of 1.3 Å (Fig. 2, B and C).
ZNF217 Makes Noncanonical Interactions with DNA, Including a Methyl-π Interaction
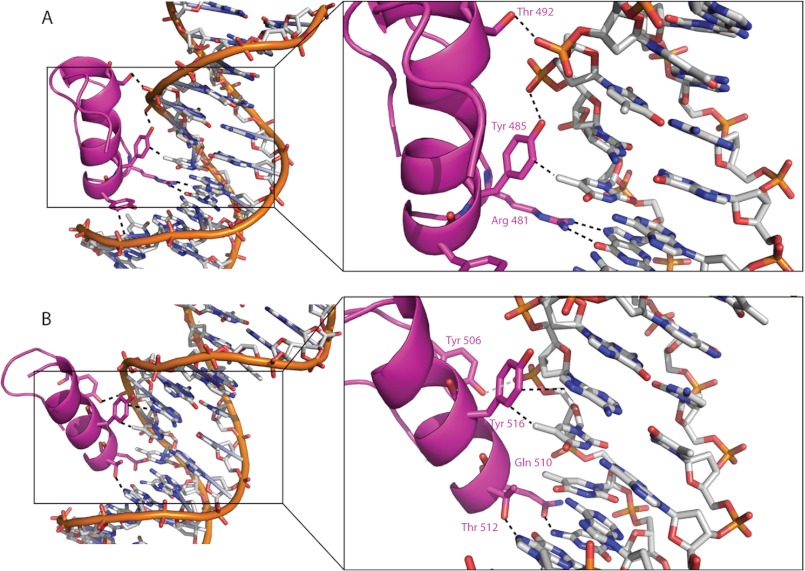
Two sequence-specific contacts with DNA are made by ZNF217_F6 (Fig. 3A). The guanidinium group of Arg-481, which is in the −2-position of the α-helix, makes a double hydrogen bond with O6 and N7 of G18. More surprisingly, the face of the hydrophobic ring of Tyr-485 is positioned 3.7 Å away from the methyl group of T17, making a methyl-π interaction. Interactions of this type occur in a number of protein-nucleic acid complexes, but to our knowledge, this DNA recognition strategy has not been described for classical ZF domains. In addition to these base-specific interactions, the side chains of Tyr-484, Tyr-485, and Thr-492 all make electrostatic interactions with the sugar-phosphate backbone (Fig. 3A).
FIGURE 3.
Protein-DNA interactions mediated by ZNF217_F67. A and B, phosphate backbone contacts and sequence-specific interactions observed between the protein and the DNA for finger 6 (A) and for finger 7 (B) are represented by black dashed lines.
The interaction of F7 with DNA is stabilized by four sequence-specific interactions, as shown in Fig. 3B. The side-chain carbonyl group of Gln-510 forms a hydrogen bond with N7 of C4, whereas the hydroxyl group of Thr-512 hydrogen-bonds to O7 of G7. The two other sequence-specific contacts involve the aromatic ring of Tyr-516, which forms a second methyl-π interaction with the methyl group of T14 and also a van der Waals contact with the C8H of A13. An additional interaction between the hydroxyl group of Tyr-506 and the sugar-phosphate DNA backbone is also observed.
ZNF217 Recognizes Its Cognate DNA Sequence Using a Unique Interaction Pattern When Compared with Other Classical ZNFs
The overall pattern of sequence-specific interactions observed between ZNF217_F67 and DNA is summarized in Fig. 1B and compared with the canonical interaction pattern observed for ZF proteins such as Zif268 (Fig. 1, A and D). Supplemental Fig. 1 shows the patterns observed for the solved structures of a number of classical ZNF-DNA complexes. Although ZNF217 and Zif268 ZFs are positioned in the DNA major groove in the same overall manner (including sharing the same conformation for the inter-ZF linker), the pattern of interacting side chains and the base positions involved in DNA binding are significantly different. The simple pattern of base-specific contacts observed for Zif268 involves residues at positions −1, +2, +3, and +6 of the recognition helix. The residue at position +6 contacts the 5′ base (of the three typically recognized by a single ZF) on the “noncoding” strand, the residue at position +3 contacts the central base, and the residue at position −1 contacts the 3′ base. The residue at position +2 often contacts the 5′-flanking base on the coding strand, giving rise to some overlap in the recognition of each 3-bp motif. This simple pattern breaks down in the ZNF217 complex; only two canonical DNA contacts are observed for ZNF217 (in terms of which helix position interacts with which base position in the target DNA). Indeed, when compared with all the known structures of natural classical ZF-DNA complexes, ZNF217 presents the lowest similarity with the canonical model in the way that it contacts DNA. Taken together with the unusual Tyr-base interactions, these data extend the scope of interactions that can be made by classical ZFs.
ZNF217_F67 Is a Sequence-specific DNA-binding Module
In previous work, we assessed the DNA binding affinity and specificity of ZNF217_F67 using fluorescence anisotropy measurements (34). Our measurements indicated that a GST fusion of ZNF217_F67 recognizes the WT DNA sequence with a relatively low affinity (Kd = 130 nm). Even more surprisingly, the affinity was reduced by only 2.5-fold when the core recognition sequence was extensively mutated (from TGCAGAAT to CTGGAGTA) (34). A similar affinity was observed for binding to a poly(AT) sequence (supplemental Fig. 3). These unusual observations prompted us to further examine the sequence specificity of ZNF217_F67 DNA binding.
Because all previous measurements had been made using GST-F67, which is known to dimerize at micromolar concentrations (41), we remeasured the affinities by fluorescence anisotropy using the isolated F67 module. Removal of the GST tag induced a 40-fold decrease in affinity and a complete loss of specificity as determined by fluorescence anisotropy, with all dissociation constants for both specific and unrelated sequences measured to be ∼5 μm (supplemental Fig. 3).
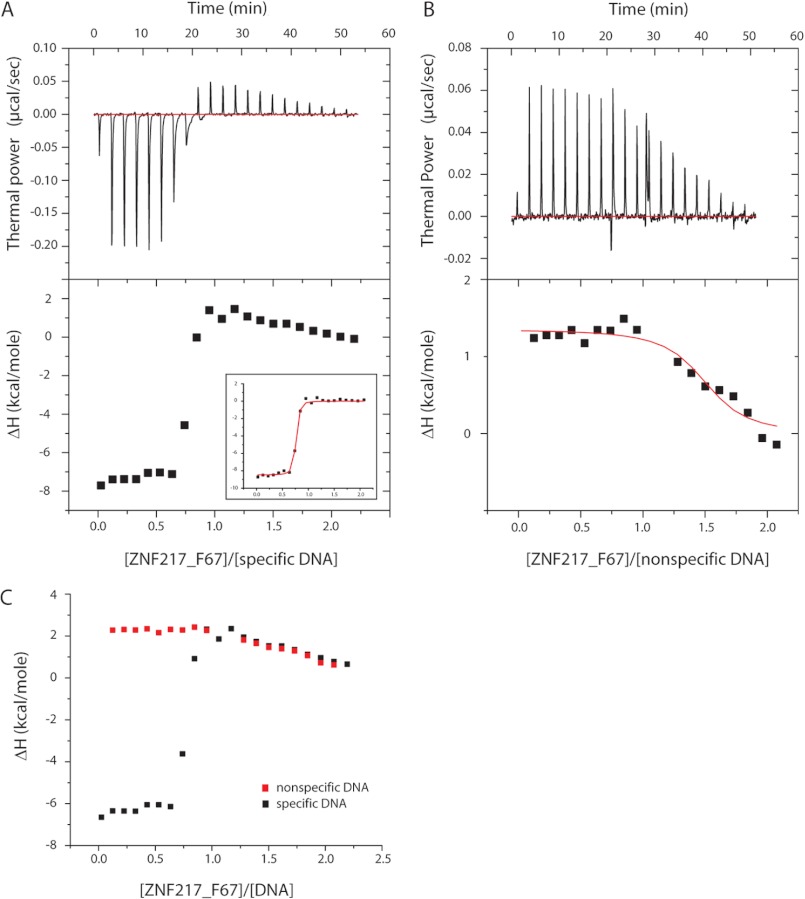
This low apparent specificity is inconsistent with the crystal structure of the ZNF217_F67 complex, which shows numerous specific interactions. We therefore used ITC as an independent means to remeasure the affinity and specificity of ZNF217_F67 for DNA. Fig. 4A shows the binding isotherm obtained for the interaction of ZNF217_F67 with WT DNA. The titration curve clearly indicates the presence of two binding events. The first is relatively tight and is exothermic, whereas the second binding event is weakly endothermic. In contrast, titration of the protein into the nonspecific DNA sequence (Fig. 4B) reveals a single endothermic binding event that perfectly overlays with the endothermic binding observed during the titration with WT DNA (Fig. 4C). A fit of the interaction with the nonspecific DNA to a simple binding model yields a dissociation constant of 1.2 ± 0.1 μm and a stoichiometry of 1.8:1 (ZNF217_F67:DNA).
FIGURE 4.
A and B, ITC data for the titration of ZNF217_F67 into specific (A) and nonspecific (B) DNA sequences. The upper panels represent the difference in heat release/uptake between the sample and the reference cell containing water. The lower panels correspond to the integrated enthalpy changes per mole of injected protein. The fit of the tight binding event observed for the specific DNA titration is detailed under “Experimental Procedures” and displayed in the inset (following subtraction of the nonspecific binding component). The experiment was conducted in 10 mm Tris buffer, pH 7.0, 50 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine at 25 °C. C, overlay of the integrated enthalpy changes calculated for the specific (black) and nonspecific (red) DNA titrations.
We analyzed the interaction with WT DNA by first subtracting the binding isotherm from the nonspecific DNA interaction and then fitting the resultant data to a single-site model. The data fitted well, giving a dissociation constant of 25 ± 5 nm and a stoichiometry of 0.67:1 (ZNF217_F67:DNA). The deviation of this latter value from 1 most likely reflects uncertainty in the concentration of protein and/or DNA (which we have observed for numerous other protein-nucleic acid complexes). Thus, under these conditions, our ITC data indicate that two binding events can take place: a high affinity interaction that corresponds to the interaction observed in the crystal structure, and a second, low affinity interaction that can also take place when no specific target site is present in the DNA sequence. The derived stoichiometry for the weak interaction suggests that more than one molecule of ZNF217_F67 can bind to DNA simultaneously in this mode. Titrations carried out at higher ionic strength ([NaCl] = 150 mm) showed only the tight binding event for the interaction with WT DNA (KD = 400 nm with protein:DNA stoichiometry = 1:1.1), whereas the nonspecific DNA exhibited an interaction with an unchanged dissociation constant of 2.5 μm.
Probing the Nonspecific Interaction of ZNF217_F67 with DNA
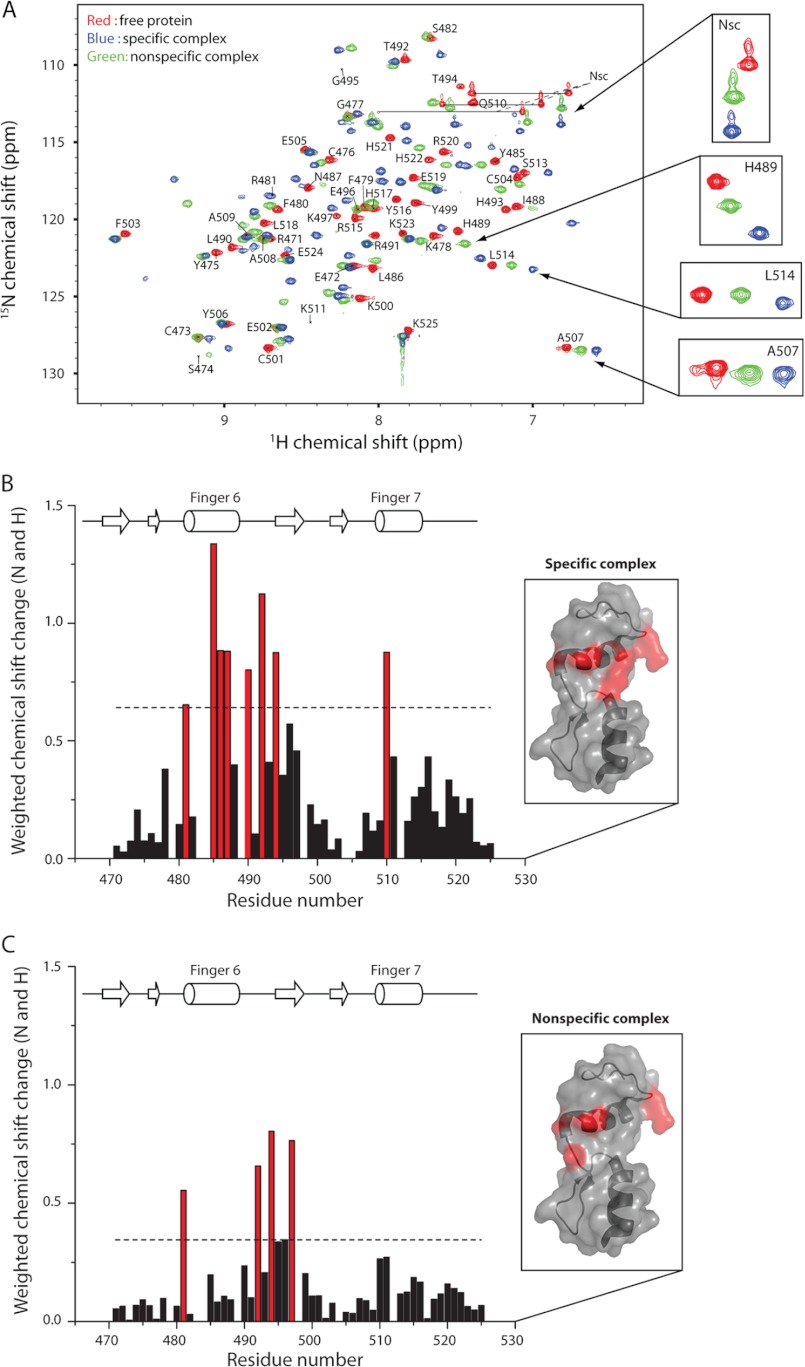
There is a growing appreciation that nonspecific protein-DNA interactions are relevant in vivo given the extremely high concentration of noncognate DNA in a eukaryotic nucleus. Attempts were therefore made to crystallize a complex of ZNF217 with several noncognate DNA sequences. Although crystals were formed, no good diffraction could be obtained. To gain more insight into the nature of the nonspecific interaction between ZNF217_F67 and DNA, we recorded 15N HSQC spectra of the protein in the absence and presence of either the WT or nonspecific DNA sequences (supplemental Fig. 4). As shown previously (34), substantial chemical shift changes are observed following the addition of WT DNA, and these changes are consistent overall with the structure of the complex. Fig. 5A shows spectra of ZNF217_F67 alone (red) and in complex with either nonspecific DNA (green) or WT DNA (blue). Overall, chemical shift changes are in the same direction for both complexes, indicating that the same surface of the protein contacts DNA in each case (Fig. 5, B and C). However, the changes associated with the nonspecific complex have uniformly smaller magnitudes, consistent with formation of a complex that is less enthalpically favorable. The kinetics of complex formation also differ in the two cases (supplemental Fig. 5); the WT DNA complex displays intermediate exchange on the chemical shift timescale, whereas the nonspecific DNA complex is in fast exchange, suggesting that the former complex is characterized by a slower dissociation rate constant.
FIGURE 5.
Chemical shift titration data for specific and nonspecific ZNF217_F67-DNA complexes. A, overlay of 15N HSQC spectra of the unbound protein (red) and bound to the specific (blue) or nonspecific DNA sequences (green). Assignments for the unbound protein are indicated (Nsc indicates assignments corresponding to asparagine side chain amide groups). B and C, weighted average chemical shift changes for backbone nuclei of ZNF217_F67 following the addition of specific (B) and nonspecific (C) DNA sequences. Chemical shift changes less than one standard deviation above the mean are shown in black, and those that are larger than one standard deviation are represented in red. The horizontal dashed line indicates one standard deviation above the mean of the chemical shift changes of all protein residues. Residues undergoing significant chemical shift changes in both complexes were also mapped onto the ZNF217 structure as displayed in the insets.
DISCUSSION
The ZNF217_F67-DNA Structure Extends the Classical ZF Paradigm
DNA-protein recognition is an essential aspect of gene regulation, and classical ZF domains are the most common of all DNA-binding modules in complex organisms. In this study, we have determined the structural basis for DNA recognition by a two-ZF module from the human oncoprotein ZNF217. Only one other structure currently exists of a two-ZF module: that of the Drosophila TRAMTRACK protein (18). The second TRAMTRACK ZF contacts DNA in a manner that closely resembles the accepted “standard” interaction mode that is also observed for the three ZFs of Zif268 (15) (Fig. 1), whereas in the first ZF, the −1 residue in the α-helix interacts with the DNA backbone rather than making a base-specific contact.
More than 20 structures have been determined of classical ZF-DNA complexes, and as shown in supplemental Fig. 1, these structures reveal a number of different interactions that can be made by side chains on the ZFs to specify the affinity and/or specificity of DNA binding. Even so, most of the base-specific interactions made by ZNF217_F67 either are uncommon or have not been observed previously. For example, one of the most frequently observed interactions in ZF-DNA complexes is the “double-headed” hydrogen bond from an arginine at position −1 to the O6 and N7 of a guanine at position 3 (supplemental Fig. 1). A unique variation of this interaction is observed in ZNF217_F6; an arginine at −2 forms an analogous pair of hydrogen bonds, but with a −2-position guanine. This is the only known example of base-specific recognition by an amino acid at the −2-position, and such an interaction opens new possibilities for the design of ZF proteins with tailored specificities. A great deal of both academic and commercial interest has centered on the creation of classical ZF variants that can recognize any chosen 3-bp sequence of dsDNA, and a library of variants has been built up over the last 15 years that can recognize ∼40 of the 64 possible triplets. These domains have been created primarily through the use of combinatorial methods such as phage display. However, the residues that are subjected to randomization are typically (although not exclusively (42)) restricted to −1-, +1-, +2-, +3-, +5-, and +6-positions of the α-helix. Our data demonstrate that the −2-position can make sequence-specific interactions and therefore might allow the range of addressable DNA triplets to be expanded.
The Methyl-π Interaction Specifies Thymine in DNA Targets
Our structure of the ZNF217_F67-DNA complex also revealed the existence of two methyl-π interactions (between Tyr-485 and T17 and between Tyr-516 and T14). Mutation of either of these tyrosines to alanine reduces the affinity of ZNF217 for DNA (34). The weakly polar methyl-π interaction has been the subject of debate for some time, and experimental proof of its existence has been demonstrated only very recently (43). It is now established that these noncovalent interactions are an important stabilizing force for many biomolecular structures and are common both in protein hydrophobic cores and in nucleic acids (in particular, between thymine methyl groups and neighboring adenine rings (44)). To our knowledge, however, no interactions between aromatic protein side chains and nucleic acid methyl groups have been described in the context of classical ZF-DNA complexes. We do note, however, that the very recently reported structure of ZFP57 bound to methylated DNA (45) includes an interaction between a methyl group of a methylated cytosine and the π-rich guanidino group of an arginine. The distance and angles between the methyl and guanidino group axes match those that characterize a methyl-π interaction (43), although the energetic contribution of this interaction is not known.
A published examination of amino acid diversity in classical ZFs (46) indicates that tyrosines are found at this position in ∼4% of all classical ZF domains (1156 instances in ∼28,000 ZFs from 12 species), making it the eighth most abundant residue in that position (tyrosine is the 15th most abundant amino acid overall in proteins listed in the UniProt database). As high throughput binding data become available for more of these proteins, through methods such as ChIP sequencing (47), it will be interesting to see whether tyrosine-thymine interactions are, as we would predict, conserved. It is also notable that tyrosines have not been selected from the randomly mutated Zif268 libraries that have been screened using phage display during efforts to create designer ZF domains (42, 48, 49), perhaps suggesting that only a limited number of sequence contexts are compatible with the methyl-π interaction observed in the ZNF217_F67 structure. In many cases, however, aromatic residues were explicitly excluded from design efforts by the use of VNS codons in the combinatorial library (50). This, however, is an avenue worth exploring given that specific recognition of thymine residues has been relatively problematic in the design of tailored ZF proteins (42, 48, 49).
It is also notable that the recognition of methyl groups in DNA by classical ZFs has been achieved through completely different mechanisms, as recently described for Kaiso (51) and ZFP57 (45). The structure of ZFP57 bound to DNA containing two methylated cytosines (45) reveals that one of the methyl groups is enclosed by a layer of ordered water molecules, whereas the second forms methyl-π interactions with the guanidino group of an arginine, as noted above, and the side-chain carboxylate group of a glutamate (which also contacts the N4 atom of this same methyl group via one of its carboxylate oxygens). Alanine and valine residues were also described to interact with methyl groups (linked to either thymine or cytosine) (52) by forming a tight hydrophobic interaction between the amino acid side chains and the methyl group. The structure of the classical 3-ZF protein Kaiso in complex with tetramethylated DNA shows that two methyl groups are recognized by glutamate side chain-mediated hydrogen bonds (CH–OH), whereas another is accommodated in a hydrophobic pocket created by a cysteine and a threonine residue. One of the methyl groups forms a methyl-π interaction with an arginine side chain (53). Overall, these data suggest that methyl recognition in protein-DNA interaction can be achieved by several different molecular mechanisms.
An alignment of ZNF217 with the most closely related human proteins (Fig. 6), ZNF219 and ZNF536, as well as with homologues from other species, indicates that Tyr-516 is strictly conserved, whereas Tyr-485 is uniformly replaced by histidine. Although it is difficult to draw inferences from the conservation of Tyr-516, given the extremely high similarity across the whole sequence, the substitution of Tyr-485 with histidine might have one of two consequences. First, the selectivity for thymine might be retained if the aromatic ring of histidine makes a comparable methyl-π interaction. Methyl interactions involving histidines have been seen, for example, in plastocyanin structures between histidine residues involved in copper coordination and the methyl groups of leucines or methionines (54–56). Second, the substitution might result in a change in DNA binding specificity. Histidines are seen at the +3-position in both Zif268 and a designed ZF protein (supplemental Fig. 1), where they instead make hydrogen bonds with N7 of either guanine or adenine, respectively (15, 57).
FIGURE 6.
Amino acid sequence alignment of human ZNF217_F67. The sequence alignment shows the conservation of the two tyrosine residues (gray boxes) involved in the methyl-π interactions with DNA. The interspecies sequence alignment of ZNF217 shows an extremely good conservation of the entire F67 domain across different species.
ZNF217 Is a Sequence-specific DNA-binding Protein
In this and our previous study (34), we have determined the sequence specificity of ZNF217 using fluorescence anisotropy and ITC. ITC data showed that ZNF217_F67 is able to discriminate its Cyclic Amplification and Selection of Targets (CAST)-derived consensus sequence with an affinity that is 50-fold higher than that measured for a mutated binding site. Surprisingly, this tight and specific binding event was not detected at all in our fluorescence anisotropy titrations, whereas the weak, nonspecific binding was clearly measureable. This discrepancy most likely highlights a pitfall of the fluorescence anisotropy approach that prevents the measurement of interactions that do not result in a substantial change in rotational motion of the fluorophore. In this case, it is the 18-bp oligonucleotide, which has a very extended shape (an axial ratio of ∼3) (58), that is fluorescently labeled, and the binding of the 6-kDa F67 polypeptide presumably does not alter the tumbling properties of this molecule sufficiently to give rise to a measureable change in fluorescence anisotropy. We have observed similar behavior before during our analysis of the DNA binding properties of the GATA-type ZF from MED-1 (6). In contrast, the second, weaker binding event, which introduces a second molecule of F67 to the DNA, must perturb the tumbling of the DNA to a greater extent. In this regard, it is notable that the derived stoichiometry of the nonspecific interaction is ∼1.8:1, indicating that more than one protein molecule might be contacting the DNA simultaneously (which could amplify the observed change in anisotropy). As always, it is prudent to make measurements using complementary techniques where possible.
The Nonspecific DNA Binding Properties of ZNF217
Our data show a nonspecific DNA binding activity with micromolar affinity for ZNF217 (although it is possible that other ZFs in ZNF217 might make additional contributions to this activity). Recently, nonspecific protein-DNA binding has received considerable attention and is likely to be an important aspect of protein-DNA recognition (59–61). It was pointed out many years ago that sequence-specific DNA recognition is extremely challenging in the cell (62, 63) given that specific binding sites are diluted by a huge molar excess of other DNA sequences that exhibit a strong overall geometric resemblance with the specific DNA site. Although the organization of eukaryotic DNA into nucleosomes and the packaging of a subset of this DNA into heterochromatin reduces the number of potential nonspecific binding sites, a large number of such sites will still exist. Jen-Jacobson (64) estimated that for a hypothetical protein that recognizes a unique DNA-binding site in the human genome and presents a 1000-fold difference in affinity between its specific and a nonspecific binding site, only 1 in 3 million protein molecules will be bound to the specific binding site. Different theories have been proposed to explain how proteins discriminate their cognate DNA sequence in the context of a cell, including (i) sliding (65–68); (ii) dissociation-reassociation; and (iii) intersegmental transfer (69). Computational predictions (70), x-ray crystallography (71), and a variety of experimental methods have been used recently to study nonspecific protein-DNA complexes (59, 72, 73), but the analysis of nonspecific protein-DNA complexes remains a major challenge.
We show here that the DNA binding mode of ZNF217_F67 for the specific and nonspecific DNA sequences is similar and that the observed exchange rate between the free and bound protein is faster for the nonspecific complex, indicative of a higher dissociation rate constant. These observations are consistent with a model in which ZNF217 in the cell probes open genomic DNA with its DNA-binding surface and, by virtue of its slower off-rate when bound to its cognate target sites, is more likely to induce recruitment of additional proteins at such sites. Similar observations have been made for other DNA-binding proteins, including the HoxD9 homeodomain (73, 74) and the ZF protein Zif268 (59). Our study joins a small but growing body of work that begins to provide insight into the mechanism by which proteins can efficiently find their target DNA-binding site immersed in a sea of nonspecific DNA. However, much still remains to be learned about how transcription factors can reach their cognate binding site so quickly in the presence of substantial molecular distractions.
This work was supported by grants from the National Health and Medical Research Council and the Australian Research Council (to J. P. M.).

This article contains supplemental Figs. 1–5 and references.
The atomic coordinates and structure factors (codes 4F2J and 4IS1) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ZF
- zinc finger
- ZNF
- Zinc finger protein
- MBP
- maltose binding protein
- ITC
- isothermal titration calorimetry
- SIRAS
- single isomorphous replacement with anomalous scattering
- HSQC
- heteronuclear single quantum correlation
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Krishna S. S., Majumdar I., Grishin N. V. (2003) Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31, 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klug A., Schwabe J. W. (1995) Protein motifs 5. Zinc fingers. FASEB J. 9, 597–604 [PubMed] [Google Scholar]
- 4. Mackay J. P., Crossley M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4 [DOI] [PubMed] [Google Scholar]
- 5. Sánchez-García I., Rabbitts T. H. (1994) The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 10, 315–320 [DOI] [PubMed] [Google Scholar]
- 6. Lowry J. A., Gamsjaeger R., Thong S. Y., Hung W., Kwan A. H., Broitman-Maduro G., Matthews J. M., Maduro M., Mackay J. P. (2009) Structural analysis of MED-1 reveals unexpected diversity in the mechanism of DNA recognition by GATA-type zinc finger domains. J. Biol. Chem. 284, 5827–5835 [DOI] [PubMed] [Google Scholar]
- 7. Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 75, 567–578 [DOI] [PubMed] [Google Scholar]
- 8. Johnston M. (1987) Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature 328, 353–355 [DOI] [PubMed] [Google Scholar]
- 9. Hansen A., Van Hoy M., Kodadek T. (1991) Spectroscopic studies of the DNA binding site of the GAL4 “zinc finger” protein. Biochem. Biophys. Res. Commun. 175, 492–499 [DOI] [PubMed] [Google Scholar]
- 10. Blackshear P. J. (2002) Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952 [DOI] [PubMed] [Google Scholar]
- 11. Nguyen C. D., Mansfield R. E., Leung W., Vaz P. M., Loughlin F. E., Grant R. P., Mackay J. P. (2011) Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 407, 273–283 [DOI] [PubMed] [Google Scholar]
- 12. De Guzman R. N., Wu Z. R., Stalling C. C., Pappalardo L., Borer P. N., Summers M. F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science 279, 384–388 [DOI] [PubMed] [Google Scholar]
- 13. Miller J., McLachlan A. D., Klug A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tadepally H. D., Burger G., Aubry M. (2008) Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes, and effector domains. BMC Evol. Biol. 8, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pavletich N. P., Pabo C. O. (1991) Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Ä. Science 252, 809–817 [DOI] [PubMed] [Google Scholar]
- 16. Pavletich N. P., Pabo C. O. (1993) Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261, 1701–1707 [DOI] [PubMed] [Google Scholar]
- 17. Foster M. P., Wuttke D. S., Radhakrishnan I., Case D. A., Gottesfeld J. M., Wright P. E. (1997) Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA. Nat. Struct. Biol. 4, 605–608 [DOI] [PubMed] [Google Scholar]
- 18. Fairall L., Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature 366, 483–487 [DOI] [PubMed] [Google Scholar]
- 19. Lee S., Doddapaneni K., Hogue A., McGhee L., Meyers S., Wu Z. (2010) Solution structure of Gfi-1 zinc domain bound to consensus DNA. J. Mol. Biol. 397, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 20. Schuetz A., Nana D., Rose C., Zocher G., Milanovic M., Koenigsmann J., Blasig R., Heinemann U., Carstanjen D. (2011) The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 68, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Omichinski J. G., Pedone P. V., Felsenfeld G., Gronenborn A. M., Clore G. M. (1997) The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat. Struct. Biol. 4, 122–132 [DOI] [PubMed] [Google Scholar]
- 22. Segal D. J., Crotty J. W., Bhakta M. S., Barbas C. F., 3rd, Horton N. C. (2006) Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J. Mol. Biol. 363, 405–421 [DOI] [PubMed] [Google Scholar]
- 23. Choo Y., Klug A. (1997) Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 7, 117–125 [DOI] [PubMed] [Google Scholar]
- 24. Dreyer A. K., Cathomen T. (2012) Zinc-finger nucleases-based genome engineering to generate isogenic human cell lines. Methods Mol. Biol. 813, 145–156 [DOI] [PubMed] [Google Scholar]
- 25. Cai C. Q., Doyon Y., Ainley W. M., Miller J. C., Dekelver R. C., Moehle E. A., Rock J. M., Lee Y. L., Garrison R., Schulenberg L., Blue R., Worden A., Baker L., Faraji F., Zhang L., Holmes M. C., Rebar E. J., Collingwood T. N., Rubin-Wilson B., Gregory P. D., Urnov F. D., Petolino J. F. (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol. Biol. 69, 699–709 [DOI] [PubMed] [Google Scholar]
- 26. Durai S., Mani M., Kandavelou K., Wu J., Porteus M. H., Chandrasegaran S. (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 33, 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu J., Kandavelou K., Chandrasegaran S. (2007) Custom-designed zinc finger nucleases: what is next? Cell. Mol. Life Sci. 64, 2933–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P. (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70 [DOI] [PubMed] [Google Scholar]
- 29. Brown R. S. (2005) Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15, 94–98 [DOI] [PubMed] [Google Scholar]
- 30. Mackay J. P., Font J., Segal D. J. (2011) The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 18, 256–261 [DOI] [PubMed] [Google Scholar]
- 31. Vendrell J. A., Thollet A., Nguyen N. T., Ghayad S. E., Vinot S., Bièche I., Grisard E., Josserand V., Coll J. L., Roux P., Corbo L., Treilleux I., Rimokh R., Cohen P. A. (2012) ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 72, 3593–3606 [DOI] [PubMed] [Google Scholar]
- 32. Nonet G. H., Stampfer M. R., Chin K., Gray J. W., Collins C. C., Yaswen P. (2001) The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 61, 1250–1254 [PubMed] [Google Scholar]
- 33. Cowger J. J., Zhao Q., Isovic M., Torchia J. (2007) Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene 26, 3378–3386 [DOI] [PubMed] [Google Scholar]
- 34. Nunez N., Clifton M. M., Funnell A. P., Artuz C., Hallal S., Quinlan K. G., Font J., Vandevenne M., Setiyaputra S., Pearson R. C., Mackay J. P., Crossley M. (2011) The multi-zinc finger protein ZNF217 contacts DNA through a two-finger domain. J. Biol. Chem. 286, 38190–38201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai M., Huang Y., Sakaguchi K., Clore G. M., Gronenborn A. M., Craigie R. (1998) An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. J. Biomol. NMR 11, 97–102 [DOI] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D. Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Riley L. G., Ralston G. B., Weiss A. S. (1996) Multimer formation as a consequence of separate homodimerization domains: the human c-Jun leucine zipper is a transplantable dimerization module. Protein Eng. 9, 223–230 [DOI] [PubMed] [Google Scholar]
- 42. Segal D. J., Dreier B., Beerli R. R., Barbas C. F., 3rd. (1999) Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl. Acad. Sci. U.S.A. 96, 2758–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plevin M. J., Bryce D. L., Boisbouvier J. (2010) Direct detection of CH/π interactions in proteins. Nat. Chem. 2, 466–471 [DOI] [PubMed] [Google Scholar]
- 44. Umezawa Y., Nishio M. (2002) Thymine-methyl/π interaction implicated in the sequence-dependent deformability of DNA. Nucleic Acids Res. 30, 2183–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y., Toh H., Sasaki H., Zhang X., Cheng X. (2012) An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 26, 2374–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emerson R. O., Thomas J. H. (2009) Adaptive evolution in zinc finger transcription factors. PLoS Genet. 5, e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lefrançois P., Zheng W., Snyder M. (2010) ChIP-Seq using high-throughput DNA sequencing for genome-wide identification of transcription factor binding sites. Methods Enzymol. 470, 77–104 [DOI] [PubMed] [Google Scholar]
- 48. Dreier B., Fuller R. P., Segal D. J., Lund C. V., Blancafort P., Huber A., Koksch B., Barbas C. F., 3rd. (2005) Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 280, 35588–35597 [DOI] [PubMed] [Google Scholar]
- 49. Dreier B., Beerli R. R., Segal D. J., Flippin J. D., Barbas C. F., 3rd (2001) Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 276, 29466–29478 [DOI] [PubMed] [Google Scholar]
- 50. Choo Y., Klug A. (1994) Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc. Natl. Acad. Sci. U.S.A. 91, 11163–11167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buck-Koehntop B. A., Stanfield R. L., Ekiert D. C., Martinez-Yamout M. A., Dyson H. J., Wilson I. A., Wright P. E. (2012) Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl. Acad. Sci. U.S.A. 109, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choo Y. (1998) Recognition of DNA methylation by zinc fingers. Nat. Struct. Biol. 5, 264–265 [DOI] [PubMed] [Google Scholar]
- 53. Liu Y., Zhang X., Blumenthal R. M., Cheng X. (January 23, 2013) A common mode of recognition for methylated CpG. Trends Biochem. Sci. 10.1016/j.tibs.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bond C. S., Bendall D. S., Freeman H. C., Guss J. M., Howe C. J., Wagner M. J., Wilce M. C. (1999) The structure of plastocyanin from the cyanobacterium Phormidium laminosum. Acta Crystallogr. D. Biol. Crystallogr. 55, 414–421 [DOI] [PubMed] [Google Scholar]
- 55. Schmidt L., Christensen H. E., Harris P. (2006) Structure of plastocyanin from the cyanobacterium Anabaena variabilis. Acta Crystallogr. D. Biol. Crystallogr. 62, 1022–1029 [DOI] [PubMed] [Google Scholar]
- 56. Shibata N., Inoue T., Nagano C., Nishio N., Kohzuma T., Onodera K., Yoshizaki F., Sugimura Y., Kai Y. (1999) Novel insight into the copper-ligand geometry in the crystal structure of Ulva pertusa plastocyanin at 1.6-Ä resolution: structural basis for regulation of the copper site by residue 88. J. Biol. Chem. 274, 4225–4230 [DOI] [PubMed] [Google Scholar]
- 57. Kim C. A., Berg J. M. (1996) A 2.2 Ä resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 3, 940–945 [DOI] [PubMed] [Google Scholar]
- 58. Clegg R. M., Murchie A. I., Zechel A., Lilley D. M. (1993) Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 90, 2994–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zandarashvili L., Vuzman D., Esadze A., Takayama Y., Sahu D., Levy Y., Iwahara J. (2012) Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. U.S.A. 109, E1724–E1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peterson S. N., Dahlquist F. W., Reich N. O. (2007) The role of high affinity non-specific DNA binding by Lrp in transcriptional regulation and DNA organization. J. Mol. Biol. 369, 1307–1317 [DOI] [PubMed] [Google Scholar]
- 61. Roberts M. A., Wadhams G. H., Hadfield K. A., Tickner S., Armitage J. P. (2012) ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 6698–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Von Hippel P. H., McGhee J. D. (1972) DNA-protein interactions. Annu. Rev. Biochem. 41, 231–300 [DOI] [PubMed] [Google Scholar]
- 63. Lin S., Riggs A. D. (1975) The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell 4, 107–111 [DOI] [PubMed] [Google Scholar]
- 64. Jen-Jacobson L. (1997) Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers 44, 153–180 [DOI] [PubMed] [Google Scholar]
- 65. Halford S. E., Marko J. F. (2004) How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dahlquist F. W. (2006) Slip sliding away: new insights into DNA-protein recognition. Nat. Chem. Biol. 2, 353–354 [DOI] [PubMed] [Google Scholar]
- 67. Pingoud A., Wende W. (2007) A sliding restriction enzyme pauses. Structure 15, 391–393 [DOI] [PubMed] [Google Scholar]
- 68. Givaty O., Levy Y. (2009) Protein sliding along DNA: dynamics and structural characterization. J. Mol. Biol. 385, 1087–1097 [DOI] [PubMed] [Google Scholar]
- 69. de la Rosa M. A., Koslover E. F., Mulligan P. J., Spakowitz A. J. (2010) Dynamic strategies for target-site search by DNA-binding proteins. Biophys. J. 98, 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao M., Skolnick J. (2009) From nonspecific DNA-protein encounter complexes to the prediction of DNA-protein interactions. PLoS Comput. Biol. 5, e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kalodimos C. G., Biris N., Bonvin A. M., Levandoski M. M., Guennuegues M., Boelens R., Kaptein R. (2004) Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 305, 386–389 [DOI] [PubMed] [Google Scholar]
- 72. Iwahara J., Schwieters C. D., Clore G. M. (2004) Characterization of nonspecific protein-DNA interactions by 1H paramagnetic relaxation enhancement. J. Am. Chem. Soc. 126, 12800–12808 [DOI] [PubMed] [Google Scholar]
- 73. Iwahara J., Zweckstetter M., Clore G. M. (2006) NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 15062–15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iwahara J., Clore G. M. (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 75. Elrod-Erickson M., Rould M. A., Nekludova L., Pabo C. O. (1996) Zif268 protein-DNA complex refined at 1.6 Ä: a model system for understanding zinc finger-DNA interactions. Structure 4, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 76. Goddard T. D., Kneller D. G. (2007) SPARKY 3, University of California, San Francisco [Google Scholar]