Background: θ-Defensins are antimicrobial peptides comprising a cyclic backbone and three disulfide bonds.
Results: θ-Defensins retain antibacterial and membrane-binding activity, but lose structure and stability when the disulfide bonds are removed.
Conclusion: Disulfide bonds are important for the stability and structure of θ-defensins, but not antibacterial activity.
Significance: Understanding the role of disulfide bonds and cyclization in θ-defensins facilitates applications as peptide drug scaffolds.
Keywords: Antimicrobial Peptides, Defensins, Disulfide, Nuclear Magnetic Resonance, Peptides, Cyclic Cystine Ladder, Cyclic Peptides, θ-Defensins
Abstract
θ-Defensins are ribosomally synthesized cyclic peptides found in the leukocytes of some primate species and have promising applications as antimicrobial agents and scaffolds for peptide drugs. The cyclic cystine ladder motif, comprising a cyclic peptide backbone and three parallel disulfide bonds, is characteristic of θ-defensins. In this study, we explore the role of the cyclic peptide backbone and cystine ladder in the structure, stability, and activity of θ-defensins. θ-Defensin analogues with different numbers and combinations of disulfide bonds were synthesized and characterized in terms of their NMR solution structures, serum and thermal stabilities, and their antibacterial and membrane-binding activities. Whereas the structures and stabilities of the peptides were primarily dependent on the number and position of the disulfide bonds, their antibacterial and membrane-binding properties were dependent on the cyclic backbone. The results provide insights into the mechanism of action of θ-defensins and illustrate the potential of θ-defensin analogues as scaffolds for peptide drug design.
Introduction
θ-Defensins were first isolated from the leukocytes of rhesus macaques (1) and are currently the only backbone-cyclic peptides known in animals (2). These 18-residue peptides display antifungal, antibacterial, and antiviral activities and are thought to have an important role in the innate immunity of the primates in which they are found (3, 4). Whereas ribosomally synthesized cyclic peptides from plants, fungi, and bacteria are products of single genes (5, 6), θ-defensins are biosynthesized by the head-to-tail ligation of two separate translation products (1). Although the biosynthetic mechanism of θ-defensins has not yet been elucidated, several mutants have been chemically synthesized and are being developed as therapeutics (7).
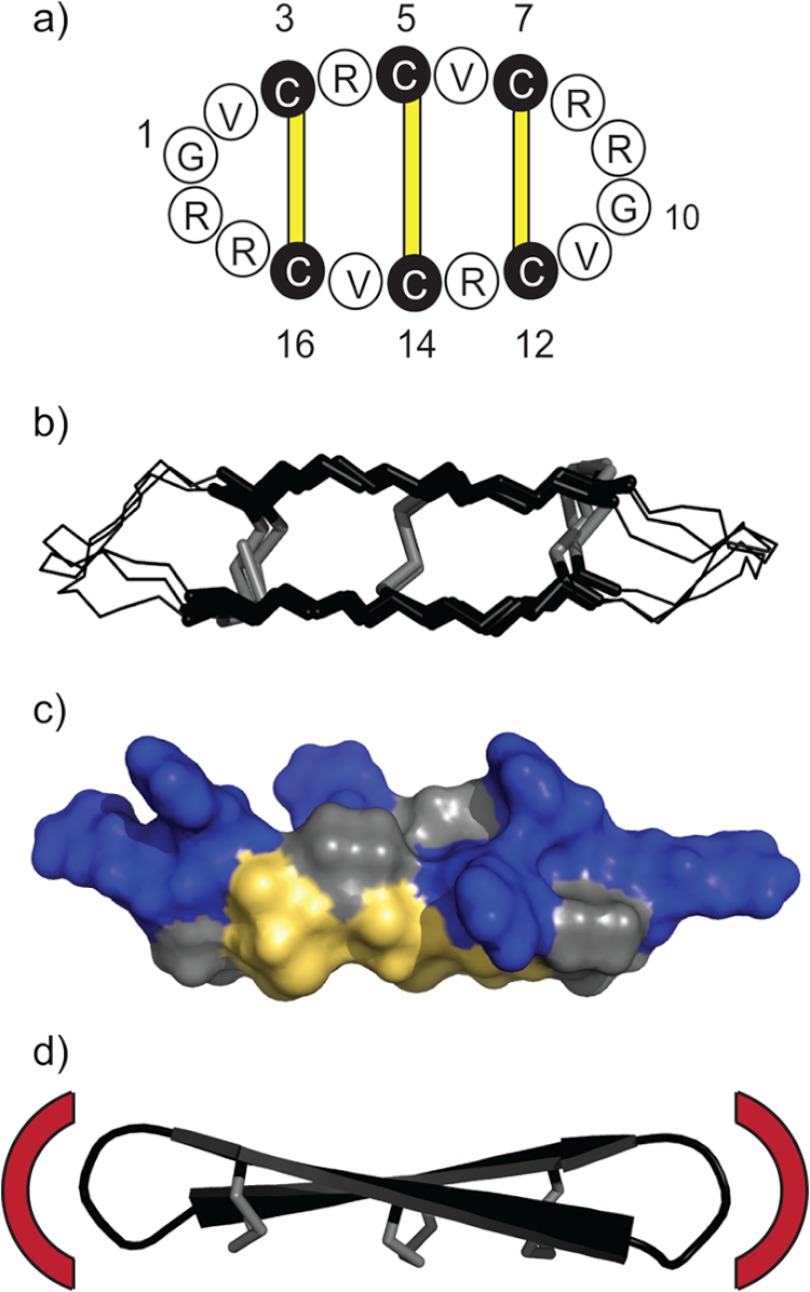
The characteristic structural feature of θ-defensins is the cyclic cystine ladder, comprising a head-to-tail cyclic peptide backbone and three parallel disulfide bonds (Fig. 1a) (8). NMR solution structures have shown that θ-defensins have a highly constrained symmetrical conformation made up of two antiparallel β-strands joined by two turns (Fig. 1b) (9, 10). The three disulfide bonds are all on one face of the molecule with the cationic arginine side chains exposed on the other face (Fig. 1c). NMR relaxation measurements and derived order parameters suggested that the cyclic cystine ladder imparts rigidity to the θ-defensin backbone (8). However, the functional role of the cyclic cystine ladder in θ-defensins has not yet been established.
FIGURE 1.
The cyclic cystine ladder of θ-defensins. a, the cyclic backbone of θ-defensins comprises 18 residues and is cross-braced by three disulfide bonds (yellow bars). Amino acids are represented by one-letter codes, and the six cysteine residues are shaded black. Residues 1 and 10 are marked, as are the positions of the disulfide bonds (3–16, 5–14, and 7–12). b, overlay of the NMR solution structures of RTD-1, BTD-2, and human θ-defensin-2 (HTD-2) shows the highly constrained and symmetrical conformation of the cyclic cystine ladder. The cyclic cystine ladder is shown in stick representation with the disulfide bonds in gray. c, molecular surface of BTD-2 shows the three disulfide bonds on one face and the arginine side chains on the opposite face. Arginine residues are colored blue, cystine residues are colored yellow, and glycine and valine residues are colored gray. d, drawing represents the solution structure of BTD-2, with disulfide bonds shown in stick representation, illustrating the potential of θ-defensins as a scaffold for peptide drug design. Red arcs illustrate how the residues in the two turn regions could be replaced with a bioactive epitope to impart a desired bioactivity to the cyclic cystine ladder framework.
Disulfide bonds have both structural and functional roles in peptides, and cysteine residues are highly conserved relative to other amino acids (11). In the cyclotides, a class of backbone-cyclic peptides from plants, the cyclic cystine knot has a crucial role in the thermal, enzymatic, and chemical stability of the peptides (12, 13) and is also important for their activity (14, 15). The single disulfide bond in sunflower trypsin inhibitor-1 (SFTI-1)6 helps to maintain the hydrogen-bonding network of the cyclic peptide and is essential for the structural integrity of acyclic analogues (16). Disulfide bonds can also have a direct functional role in proteins, being involved in catalysis or redox reactions in the active site (11). In contrast, allosteric disulfide bonds are not found in the active site of a protein, but mediate the function of a protein by causing a structural change when they are oxidized or reduced (11).
Three families of defensins, α-, β-, and θ-defensins, are found in mammals. Although all three families are antimicrobial and have three disulfide bonds, their disulfide connectivities and three-dimensional structures vary (4). In α-defensins the disulfide bonds are connected Cys1-Cys6, Cys2-Cys4, and Cys3-Cys5. A study in which the disulfide bonds of the α-defensin, mouse cryptdin-4, were mutated, showed that the antibacterial activity is not affected by removal of the disulfide bonds (17). However, the structures of disulfide mutants were highly compromised and susceptible to cleavage by the processing enzyme matrix metalloproteinase-7, suggesting that the disulfide bonds have a functional role in protecting α-defensins during biosynthesis and activation (17). β-Defensins naturally have a Cys1-Cys5, Cys2-Cys4, and Cys3-Cys6 disulfide connectivity, but synthetic human β-defensin 3 with a variety of disulfide connectivities was generated by folding in vitro (18). Strikingly, the disulfide bond isomers, and disulfide mutants, of human β-defensin 3 all had similar antibacterial activity, but the chemotactic activity was shown to be dependent on the presence of disulfide bonds (18). Furthermore, human β-defensin 1 is poorly antimicrobial in its oxidized state, but a remarkable increase in antibacterial potency was observed when the disulfide bonds were reduced, suggesting that redox regulation might have a role in innate immunity (19).
Backbone cyclization and inclusion of disulfide bonds are both established methods for increasing the stability of proteins and peptides (20), making the cyclic cystine ladder a potentially useful scaffold for drug design and protein engineering applications (Fig. 1d). The cyclic cystine knot has already been used as a stable core scaffold onto which bioactive peptide sequences can be inserted (21). Whereas the cyclic cystine knot provides a scaffold for bioactive epitopes in helical or loop conformations, the cyclic cystine ladder could be a complementary scaffold for peptides requiring a more extended β-sheet structure. Determining whether the number and positions of the disulfide bonds in the cyclic cystine ladder are likely to affect the stability and activity of a potential drug molecule will increase the potential of θ-defensins as a scaffold for peptide drugs.
The aim of this study was to determine the role of the cyclic cystine ladder in the structure, stability, and antibacterial activity of θ-defensins to gain insight into the mechanism of action of θ-defensins and to provide information about structural constraints that have to be considered when designing new functionalities into the scaffold. Baboon θ-defensin-2 (BTD-2) (22) was selected for this study because of its symmetry and potent antibacterial activity, and a series of analogues was designed in which different numbers and combinations of disulfide bonds were replaced with α-aminobutyric acid. The structures, stabilities, and antibacterial activities of the θ-defensin analogues were then compared to determine the role of the disulfide bonds and cyclic backbone.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and NMR Spectroscopy
Cyclic θ-defensin analogues BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, [Aba5,7,12,14]BTD-2, [Aba3,5,7,12,14,16]BTD-2, and a linear analogue, l-BTD-2, were synthesized on a 0.25-mmol scale by manual solid phase peptide synthesis using tert-butyloxycarbonyl (Boc)-chemistry as described previously (8). The acyclic θ-defensin analogue, BTD-2[3,4], in which the cyclic peptide backbone was broken between residues Cys3 and Arg4, was synthesized using Fmoc chemistry on an automated synthesizer (Symphony; Protein Technologies, Inc.). The peptide chain was assembled on 2-chlorotrityl chloride resin (Novabiochem), piperidine was used for the deprotection step, and coupling of the Fmoc-protected amino acids was carried out as described for Boc synthesis.
The θ-defensin analogues were synthesized from C to N terminus, with the starting point for synthesis chosen to give an N-terminal cysteine residue (Cys14 for BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, and l-BTD-2, and Cys16 for [Aba5,14]BTD-2 and [Aba5,7,12,14]BTD-2), to allow for cyclization by native chemical ligation (23). [Aba3,5,7,12,14,16]BTD-2 was synthesized with Gly1 as the N terminus. After removal of the final N-terminal Boc group with trifluoroacetic acid (TFA), the resin was washed with dimethylformamide and dichloromethane and dried under nitrogen. The peptide was cleaved from the resin with an HF/p-cresol/p-thiocresol mixture (20:1:1, v:v:v) at 0 °C for 2 h. Residual HF was removed by evaporation, and the peptide was precipitated with cold diethyl ether, then dissolved in a 50% acetonitrile/water mixture, and lyophilized. The acyclic peptide BTD-2[3,4], synthesized with Fmoc chemistry, was cleaved from the resin using a TFA/triisopropylsilane/water mixture (90:5:5) for 2 h, followed by washing with cold diethyl ether. Kalata B1, used as a control in the hemolytic, antibacterial, and membrane-binding studies, was extracted and purified from Oldenlandia affinis according to the method described by Colgrave and Craik (12).
Purification of the crude peptide was carried out by RP-HPLC using a 1%/min gradient of 90% acetonitrile in 0.05% aqueous TFA on a preparative C18 column (Phenomenex). Electrospray mass spectrometry was used to confirm the mass and purity of the peptides. Cyclization by native chemical ligation (23) and oxidation were carried out concurrently in NH4HCO3 buffer (0.1 m, pH 8.5) overnight for peptides BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, and [Aba5,7,12,14]BTD-2. The thioester linkers were removed from [Aba3,5,7,12,14,16]BTD-2 and l-BTD-2 by stirring overnight in NH4HCO3 buffer (0.1 m, pH 8.5). Cyclization of [Aba3,5,7,12,14,16]BTD-2 was achieved by stirring with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and diisopropylethylamine in dimethylformamide overnight. All of the θ-defensin analogues were then further purified by RP-HPLC, using a 0.5%/min gradient of 90% acetonitrile in 0.05% aqueous TFA with absorbance detection at 215 nm, and lyophilized. The concentrations of the peptide solutions were determined by comparison of the intensities of the RP-HPLC trace with that of a standard that had been quantified by amino acid analysis (24).
NMR spectroscopy and structure calculations were carried out as described previously (8). In brief, 1H and 13C chemical shifts were assigned from two-dimensional NMR spectra (1H-1H TOCSY and NOESY; 1H-13C HSQC spectra) of peptide samples in either a 90% H2O/10% D2O mixture or 99.9% D2O at ∼ 0.6 mm and pH ∼ 4. 3JHN-Hα and 3JHα-Hβ coupling constants were measured from one-dimensional and two-dimensional DQF-COSY and ECOSY spectra and, together with intraresidual NOE intensities, were used for stereospecific assignments and to confirm backbone angle predictions. Predictions of the φ and ψ backbone angles were also generated using TALOS+ (25). Hydrogen-bonding constraints were derived from amide proton temperature sensitivities and deuterium exchange experiments. Interproton distance constraints were derived from the intensity of NOE cross-peaks and were used for preliminary structure calculations with CYANA 3.0 (26), using torsion angle dynamics. Protocols from the RECOORD database (27) were then used to calculate an ensemble of 50 structures and to refine the structures in a water shell within the program CNS (28). A set of 20 structures with the lowest energy, no violations greater than 0.2 Å or 2°, and the best MolProbity scores, was selected for the final ensemble.
Thermal Stability
Thermal stability experiments were carried out on Bruker-Avance-500 or Bruker-Avance-600 NMR spectrometers using peptide samples in a 90% H2O/10% D2O mixture at ∼0.6 mm and pH ∼4. 1H one-dimensional and TOCSY experiments were recorded at temperatures of 298, 308, 318, 328, 338, 348, and 353 K after a temperature equilibration time of 30 min. The temperature was maintained at 353 K for 2 h and then lowered to 298 K, and a final set of spectra was recorded. Calibration of the temperature-control unit was carried out according to standard Bruker protocols.
Serum Stability
Incubation of the θ-defensin analogues in serum was used to determine their resistance to biodegradation. Human serum, male type AB (Sigma) was centrifuged at 13,000 rpm for 15 min, and the lipid layer was removed. Peptide stocks (∼1 mm) were prepared in sterile phosphate-buffered saline (PBS) and diluted 10-fold in each of serum and PBS. Three aliquots of the peptide/serum and peptide/PBS mixtures were made for each of the time points and incubated at 37 °C. Time points for the peptide/serum mixture were 0, 1, 3, 6, 9, 12, and 24 h; and time points for the peptide/PBS mixture were 0, 12, and 24 h. At each time point serum proteins were denatured by adding equal volumes of 6 m urea and 6% TFA to the aliquot. The peptide/PBS mixtures were treated in the same way. After centrifugation at 13,000 rpm for 15 min, the supernatant was removed and stored at 4 °C until analysis. Analytical RP-HPLC was used to determine the amount of peptide remaining at each time point, relative to the amount present at the start of the experiment. The height of the UV absorbance peak at 214 nm on the HPLC trace was recorded, and the average ± S.D. were calculated over the three replicates. The peptide/PBS mixture was used as a negative control, and a linear analogue was included as a positive control in each set of experiments.
Antibacterial Activity
The antimicrobial activity of the θ-defensin analogues was examined by bacterial growth inhibition and bactericidal activity. The susceptibility of the Gram-negative Escherichia coli ATCC 25922 and of the Gram-positive Staphylococcus aureus ATCC 25923 was studied using a microtiter broth dilution method (29), using both Luria broth (LB, medium that contains ∼180 mm NaCl) and Muller-Hinton broth (medium that does not contain NaCl). Briefly, overnight cultures were subcultured in medium at 37 °C with vigorous shaking until mid-log phase (∼2 h). Bacterial suspensions (5 × 105 colony-forming units (cfu)/ml were incubated with 2-fold dilutions of the peptides ranging from 0.03 to 64 μm, when tested with LB, or 0.015 to 32 μm, when tested with Muller-Hinton broth, in 96-well nonbinding surface plates (Corning), and incubated at 37 °C for 24 h. Standard antibiotic solutions (collestin and tetracycline for E. coli, and kanamycin and tetracycline for S. aureus) were also tested in concentrations ranging from 0.03 to 64 μg/ml. Growth inhibition was measured by absorbance at 600 nm, and the minimum inhibitory concentration (MIC) was the lowest concentration showing no visible growth.
The minimal bactericidal concentration (MBC) was determined by the addition of resazurin dye (30 μl, 0.01% w/v) to each well after measurement of the MIC. The plates were incubated at 37 °C for a further 18–24 h. Wells in which the dye turned pink indicated live bacteria, whereas wells in which the dye remained blue indicated dead bacteria. The reported MBC was the lowest concentration with blue coloration.
Hemolytic Activity
The hemolytic activity of the θ-defensin analogues was determined using human red blood cells (RBCs) as previously reported (30). Peptides were dissolved in PBS and tested in triplicate, with final concentrations ranging from 0.5 to 64 μm. Melittin, a peptide with known hemolytic properties, was also included as a positive control, with final concentrations ranging from 0.25 to 8 μm. Triton X-100 (0.1% v/v) and PBS were used as positive and negative controls to establish 100 and 0% hemolysis, respectively. The percentage hemolysis was quantified as before (30).
Membrane Binding Studies
Peptide solutions for membrane-binding studies were prepared in HEPES (10 mm, pH 7.4) containing 150 mm NaCl, and eight concentrations were tested (64, 32, 16, 8, 4, 2, 1, and 0 μm). Synthetic lipids palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoylphosphatidylglycerol (POPG), and palmitoyloleolylphosphatidylethanolamine (POPE), and extracted lipids from E. coli (E. coli polar lipids extract, containing phosphatidylethanolamine/phosphatidylglycerol/cardiolipin (67:23.2:9.8 weight ratio)) were purchased from Avanti Polar Lipids. A suspension of small unilamellar vesicles (0.5 mm lipid) was prepared in HEPES buffer (10 mm, pH 7.4) containing 150 mm NaCl, using a previously described method (31). Surface plasmon resonance measurements were carried out on a Biacore 3000 instrument using an L1 sensor chip according to the method described previously (31). Four model membrane systems (E. coli lipids, POPC, POPC/POPE (1:1 molar ratio), and POPC/POPG (1:1 molar ratio) were tested. The affinity of each peptide for the membrane was compared based on the peptide to lipid ratio (P/L) obtained at a reporting point (t = 176 s) at the end of the association phase, as previously detailed (31, 32) and calculated for each peptide and lipid system tested.
RESULTS
A set of θ-defensin analogues in which pairs of cysteine residues were replaced with α-aminobutyric acid was designed to investigate the role of the cyclic cystine ladder in defining structure and stability. Fig. 2a shows the disulfide arrangements of the native BTD-2 θ-defensin, the disulfide mutants, and the acyclic analogue that were synthesized by solid phase peptide synthesis. In the disulfide mutants, α-aminobutyric acid was chosen to replace cysteine because it has steric properties similar to cysteine (33, 34), but does not form disulfide bonds. Cyclization of BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, and [Aba5,7,12,14]BTD-2 proceeded smoothly by native chemical ligation with concurrent oxidation in NH4HCO3 buffer, and the desired disulfide isomers were formed as the major product. The three-disulfide mutant [Aba3,5,7,12,14,16]BTD-2 was cyclized using a peptide coupling reaction with HBTU and diisopropylethylamine in dimethylformamide (1), but the yield of cyclic peptide was lower than that of the peptides cyclized by native chemical ligation. The acyclic analogue BTD-2[3,4] was synthesized by Fmoc solid phase peptide synthesis and oxidized in NH4HCO3 buffer to form the three disulfide bonds.
FIGURE 2.
Topologies, secondary Hα shifts and NMR solution structures of θ-defensin analogues. a, the topologies of BTD-2 and its disulfide bond mutants are represented by circles, with the disulfide bonds shown as gray bars. Residues identical to those in native BTD-2 are shown as white circles, cysteine residues are shown as black circles, and α-aminobutyric acid residues are shown as gray circles. b, secondary Hα chemical shifts of the θ-defensin analogues are calculated as the difference between the measured chemical shift of each Hα proton and its respective random coil shift. Whereas θ-defensin analogues with large secondary Hα chemical shifts are highly structured, analogues with small secondary Hα chemical shifts are disordered. Regions with positive secondary Hα shifts indicate β-sheet secondary structures. c, NMR solution structures of the θ-defensin analogues are shown in line representation, with disulfide bonds in gray. The structures are shown as an ensemble of the 20 lowest energy structures after refinement.
To investigate the structural consequences of removing disulfide bonds or the circular backbone, each analogue was studied by solution NMR spectroscopy. The data were generally of good quality with sharp resonance lines, but significant differences in the degree of spectral dispersion were observed for the different peptides, consistent with differences in structure. The secondary Hα chemical shifts of the θ-defensin analogues, i.e. the difference between the Hα chemical shift of each residue and its respective random coil shift (35), are presented in Fig. 2b. The Hα random coil shift of α-aminobutyric acid (4.20 ppm) was not available in the literature and was determined in our laboratory according to the method of Wishart et al. (35). The magnitudes of the secondary shifts give an indication of the degree of structure and their sign is used to predict the secondary structural elements; positive secondary shifts indicate residues in β-sheet conformations whereas negative secondary shifts indicate helices or turns. As shown in Fig. 2b, the θ-defensin analogues comprise β-sheet regions joined by turns. The magnitudes of the secondary Hα shifts decrease as successive disulfide bonds are removed, suggesting that the cyclic cystine ladder contributes to the θ-defensin ordered structure.
NMR solution structures of the θ-defensin analogues (Fig. 2c) were calculated from NMR data using CYANA and CNS and showed that the well defined β-strand and β1′-type turn conformations are maintained even when one or two disulfide bonds are removed. The three-dimensional structures are consistent with the secondary Hα chemical shifts (Fig. 2b). Hydrogen bonds and NOE interactions characteristic of θ-defensins (8) were observed for [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, and [Aba5,7,12,14]BTD-2. Although several long range NOE interactions were observed for [Aba3,5,7,12,14,16]BTD-2, the lack of angle, disulfide bond, and hydrogen bond restraints resulted in a poorly defined structure. The two turns of the acyclic analogue BTD-2[3,4] are well defined in themselves, but the break in the backbone allows the two hairpin structures to rotate relative to each other. The secondary Hα shifts of the linear analogue l-BTD-2 are all less than 0.12 ppm in magnitude, consistent with an unstructured peptide, thus no structure calculations were performed for this peptide. The structural statistics are summarized in Fig. 3, and the coordinates and chemical shifts have been deposited in the Protein Data Bank and the Biological Magnetic Resonance Bank with accession numbers BTD-2 (2LYE, 18722), [Aba5,14]BTD-2 (2M1P, 18875), [Aba3,16]BTD-2 (2M2G, 18913), [Aba3,7,12,16]BTD-2 (2M2H, 18914), [Aba5,7,12,14]BTD-2 (2M2S, 18931), [Aba3,5,7,12,14,16]BTD-2 (2M2X, 18937), and BTD-2[3,4] (2M2Y, 18938).
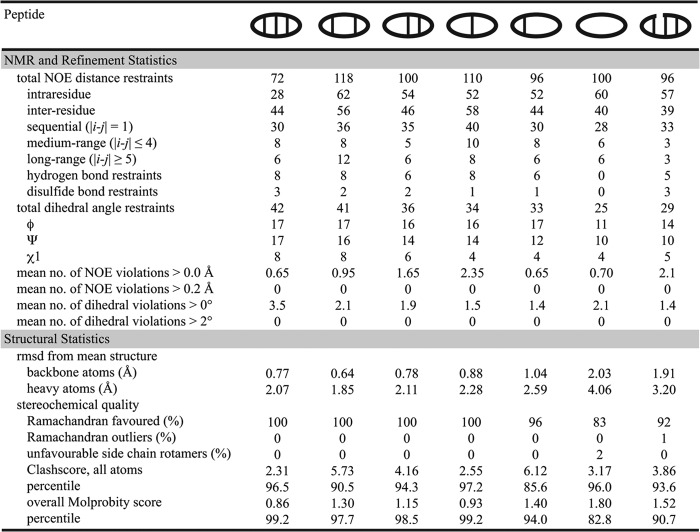
FIGURE 3.
Structural statistics for the NMR solution structures of BTD-2 and its analogues. Structures were refined in CNS and the 20 structures with lowest energy and best scores (according to MolProbity) were selected for analysis. Structural statistics are given as the mean for the 20 best structures. The clashscore is defined as the number of steric overlaps > 0.4 Å/1000 atoms.
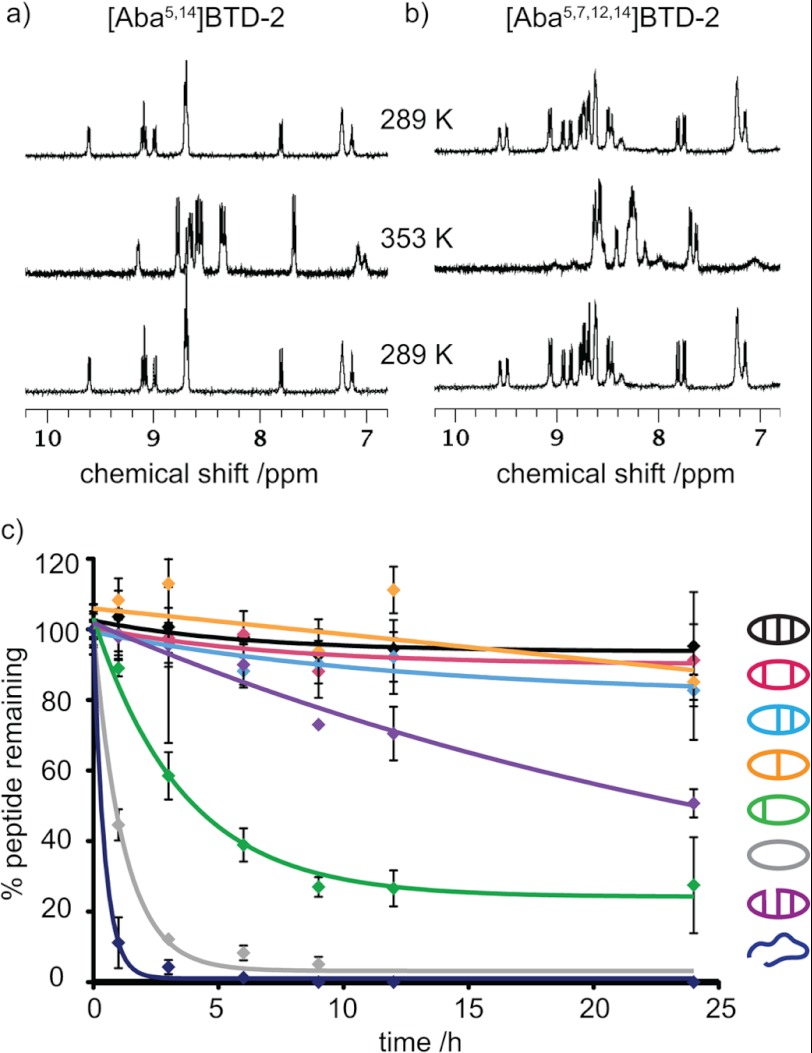
The thermal stabilities of the θ-defensin analogues were determined by comparing the 1H NMR spectra acquired at temperatures between 298 and 353 K. Although changes in the chemical shifts of the amide peaks were observed, the amide peaks of BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba3,7,12,16]BTD-2, and BTD-2[3,4] remained sharp and well dispersed (Fig. 4a). In contrast, the amide peaks of [Aba5,7,12,14]BTD-2 and [Aba3,5,7,12,14,16]BTD-2 became broad and merged together as the temperature increased (Fig. 4b), consistent with thermal unfolding. The deviation from linearity of the amide temperature dependence above 328 K also reflects the lower thermal stability of [Aba5,7,12,14]BTD-2 and [Aba3,5,7,12,14,16]BTD-2. Furthermore, the decrease in structural stability of the analogues [Aba5,7,12,14]BTD-2 and [Aba3,5,7,12,14,16]BTD-2 is reflected in a general increase in amide exchange rate for these two peptides. For example, whereas the slowly exchanging amides in native BTD-2 can be observed after >12 h, all of the signals for [Aba3,5,7,12,14,16]BTD-2 exchange within 4 h. When the temperature was cycled back to 298 K, all of the peptides gave 1H spectra identical to the original spectrum, showing that the temperature-induced changes in [Aba5,7,12,14]BTD-2 and [Aba3,5,7,12,14,16]BTD-2 are reversible.
FIGURE 4.
Thermal and serum stabilities of θ-defensin analogues. a, 1H NMR spectra (amide region) of [Aba5,14]BTD-2 at 298 and 353 K. The amide proton peaks remain sharp and well dispersed as the temperature is increased from 298 to 353 K and then cycled back to 298 K, showing that the peptide is thermally stable. b, 1H NMR spectra (amide region) of [Aba5,7,12,14]BTD-2 at 298 and 353 K. The amide proton peaks become broad and overlap as the temperature is increased from 298 to 353 K, showing that the peptide becomes unstructured at high temperatures. c, percentage of θ-defensin analogue remaining after incubation in human serum at 37 °C over 24 h.
The θ-defensin analogues were incubated in human serum for 24 h to determine their resistance to proteolytic degradation. Fig. 4c shows the percentage of each peptide remaining after incubation at 37 °C in serum for 0–24 h. More than 90% of BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, and [Aba3,7,12,16]BTD-2 remained after 24 h, showing that these peptides are highly resistant to degradation by enzymes in serum. Peptides [Aba5,7,12,14]BTD-2 and BTD-2[3,4] degraded slowly, but [Aba3,5,7,12,14,16]BTD-2 and the linear peptide were completely degraded in 6 h. All peptides were stable in PBS for 24 h. These results illustrate the importance of the cyclic backbone and the number and position of the disulfide bonds for proteolytic stability.
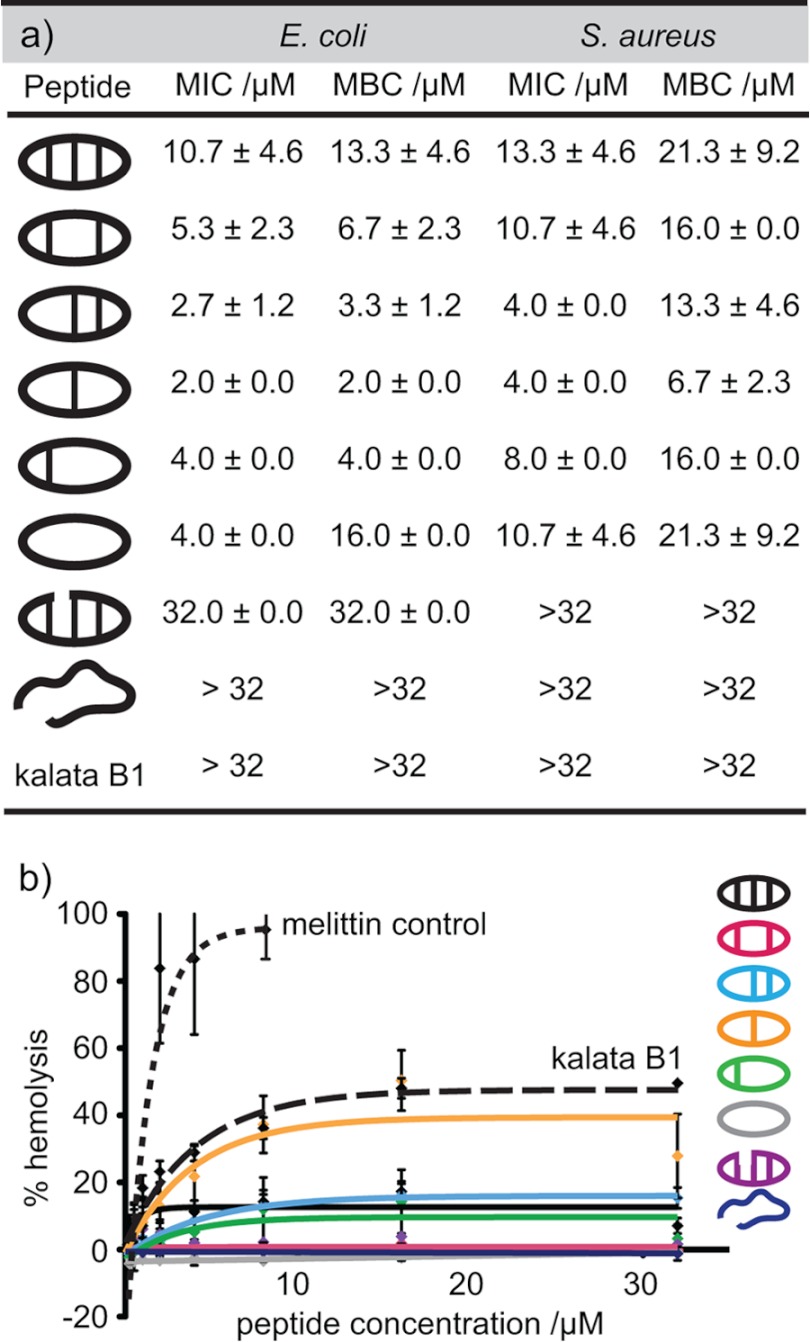
The role of the cyclic cystine ladder in the biological activity of θ-defensins was investigated by measuring the MIC and MBC of the θ-defensin analogues against E. coli and S. aureus. Fig. 5a shows the MIC and MBC for each of the peptides and illustrates that the disulfide bonds are not essential for antibacterial activity. In fact, [Aba3,7,12,16]BTD-2, with one disulfide bond, shows the highest activity of the peptides tested. Although the MBCs are higher than the corresponding MICs, the results show that, in addition to being bacteriostatic, the peptides are also bactericidal. The peptides have higher activity against E. coli (Gram-negative) than S. aureus (Gram-positive), possibly because of a higher ability to target and disrupt Gram-negative than Gram-positive plasma membranes, as reported for other antimicrobial peptides (36). The cyclic peptide backbone also contributes to the antibacterial activity, as shown by the lack of activity of the acyclic analogue BTD-2[3,4]. The hemolytic activities of the θ-defensin analogues were measured (Fig. 5b) and compared with melittin, a well known hemolytic peptide, as a positive control. All of the θ-defensin analogues were found to have low hemolytic activity (HC50 >64 μm). By comparison, the cystine knot peptide kalata B1 did not show any antibacterial activity up to 32 μm, although the peptide induced ∼40% hemolysis at this concentration.
FIGURE 5.
Antibacterial and hemolytic activities of θ-defensin analogues. a, MIC and MBC of the θ-defensin analogues and kalata B1 against E. coli and S. aureus in Mueller-Hinton broth. Concentrations are shown as the average ± S.D. of three replicates. b, percentage hemolytic activity (relative to Triton X-100) against red blood cells for the θ-defensin analogues, kalata B1, and melittin. Error bars, S.D.
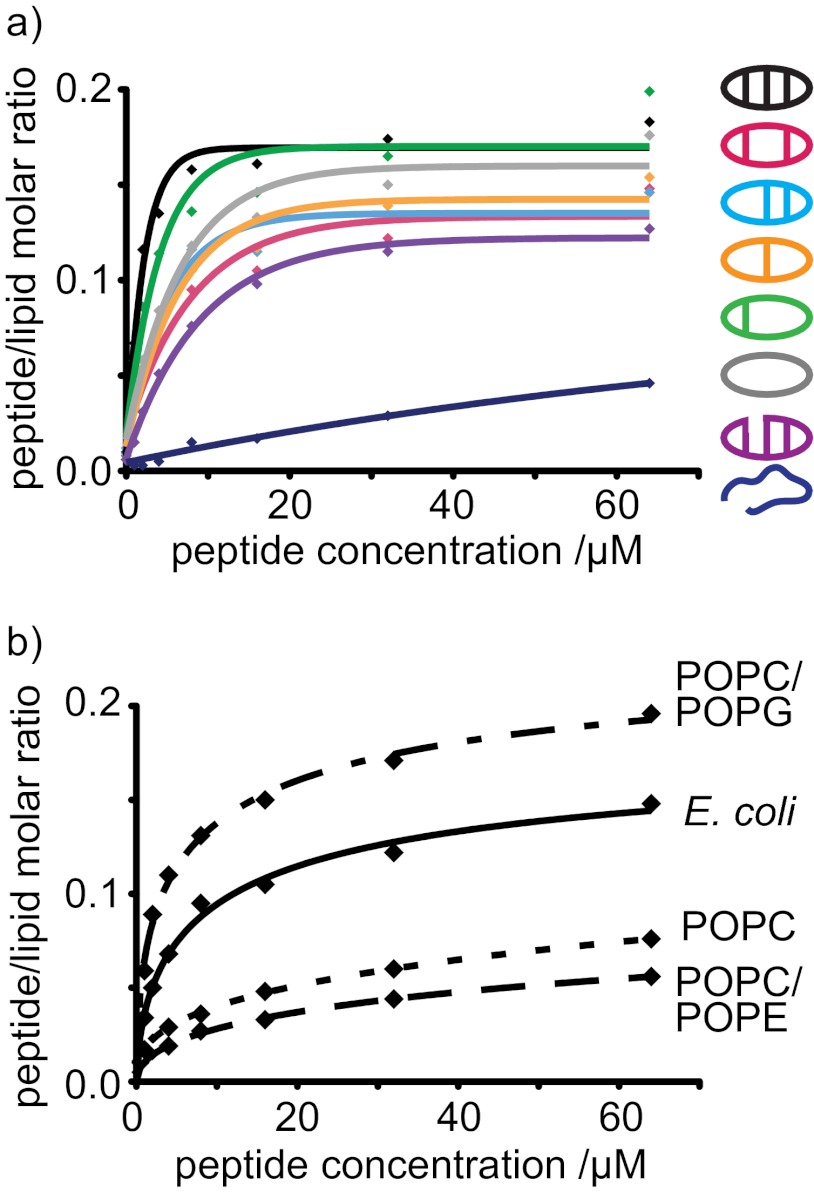
Surface plasmon resonance was used to compare binding of the θ-defensin analogues with model membranes. Diverse lipid compositions were examined to evaluate whether binding to the membrane is involved in the antibacterial activity of the θ-defensin analogues. Binding of the θ-defensin analogues to each of the membrane systems was compared by P/L, as shown in Fig. 6. All the θ-defensin analogues, except for the linear analog, bound to a model membrane prepared from an E. coli polar lipid extract (Fig. 6a), suggesting that membrane binding is involved in antimicrobial activity. E. coli membranes contain high levels of phospholipids with phosphatidylethanolamine and phosphatidylglycerol headgroups. To distinguish possible contributions of the zwitterionic phosphatidylethanolamine phospholipids from those of the negatively charged phosphatidylglycerol phospholipids, the affinity of the θ-defensin analogues was compared for model membranes composed with POPC/POPE, POPC/POPG, and POPC.
FIGURE 6.
Binding of θ-defensin analogues to model membrane systems. a, P/L for the binding of θ-defensin analogues to a model membrane comprising an extract of phospholipids from E. coli. b, P/L ratio (mol/mol) for the binding of [Aba5,14]BTD-2 to four membrane systems. The 50:50 POPC/POPG mixture and extract of E. coli lipids are anionic and model bacterial membranes. POPC and the 50:50 POPC/POPE mixture are neutral and model mammalian membranes.
The θ-defensin analogues showed affinity for the membrane systems in the order POPC/POPG > E. coil lipids ≫ POPC > POPC/POPE, as illustrated for [Aba5,14]BTD-2 in Fig. 6b. The peptides have higher affinity for anionic membranes than for membranes that are globally neutral, suggesting that membrane binding is improved by electrostatic interactions. Although there are small differences in affinity, all of the cyclic analogues bind more strongly to membranes than the acyclic analogue BTD-2[3,4] for all of the lipid systems tested. The linear peptide showed the lowest binding for all four membrane systems, consistent with the lack of antimicrobial activity for this peptide, again confirming that membrane binding is not determined by the peptide sequence alone, nor by the overall charge of the peptide.
DISCUSSION
In this study we have determined the structural and functional consequences of disrupting the disulfide array and/or circular backbone of the θ-defensin BTD-2. Solid phase peptide synthesis and native chemical ligation were used to synthesize BTD-2 and a series of disulfide bond mutants. Investigation of the role of the cyclic cystine ladder was facilitated by the 2-fold symmetry of BTD-2, which allowed all disulfide bond combinations to be represented by six BTD-2 analogues. Formation of the desired disulfide pairing as the major product during cyclization and oxidation illustrated the thermodynamic stability of the β-sheet structure and highlighted the potential of the cyclic cystine ladder as a drug scaffold.
The ease with which the disulfide bonds form in the θ-defensins might be due to the CXCXC motif, which places the cysteine side chains in proximity on the same face of the molecule. Furthermore, cysteine spacing has been found to be important for cysteine pairing, and inclusion of a CXC motif has recently been proposed as a method for orthogonal disulfide pairing in peptides with multiple disulfide bonds (37). The high yields of cyclic product obtained by native chemical ligation, compared with traditional peptide coupling methods, suggests that inclusion of disulfide bonds in the design of a cyclic peptide framework could increase the synthetic accessibility as well as the stability.
The Role of the Disulfide Bonds and the Circular Backbone in Maintaining Structure
NMR analysis revealed that the characteristic β-sheet and turn structure of the θ-defensins (8) was retained even when one or two disulfide bonds were removed. In contrast, the α-defensin mouse cryptdin-4 lost β-sheet structure when either Cys4-Cys29 or Cys11-Cys28 was mutated (17), highlighting the role of the hydrogen bonds in θ-defensins in holding the β-sheets together in the absence of one or two of the disulfide bonds. In the variants with only one disulfide bond, four hydrogen bond pairs across the sheet were observed for [Aba3,7,12,16]BTD-2, whereas only three were observed for [Aba5,7,12,14]BTD-2, suggesting that a sole disulfide is better at maintaining the hydrogen-bonding network when in the central position.
The structure of [Aba3,5,7,12,14,16]BTD-2 is poorly defined and is likely to be flexible due to the lack of disulfide and hydrogen bond constraints. In a similar study, the single disulfide bond in SFTI-1 was found to have a role in maintaining the hydrogen bonds, especially in the acyclic SFTI-1 analogues (16). Whereas the 14-residue disulfide bond mutant, [Aba3,11]SFTI-1, retained the β-sheet structure of the native peptide (16), the 18-residue disulfide bond mutant in the current study, [Aba3,5,7,12,14,16]BTD-2, did not have defined β-sheet regions. A report on the β-sheet periodicity in small cyclic peptides (38) concluded that cyclic peptides with 2(2n + 1) residues would form β-hairpin structures (38); however, the maximum length tested was a 14-residue peptide. The lack of β-sheet structure in [Aba3,5,7,12,14,16]BTD-2, the next peptide in the predicted series, suggests that there might be a limit to the length of the β-sheet if there are no cross-bracing disulfide bonds.
Although the mechanism by which two precursor peptides are ligated to form the cyclic backbone of θ-defensins is not yet known, the acyclic analogue BTD-2[3,4] is a possible intermediate in the biosynthesis (39). The structure of BTD-2[3,4] shows that the two β-hairpins are well structured in themselves and that the cross-strand disulfide bonds hold the termini in proximity for a possible peptide ligation reaction. Thus the current study supports the suggestion that the biosynthesis might involve a structured acyclic 18-mer peptide. It does not, however, exclude the possibility of a concerted double ligation reaction of two 9-mers.
The Role of the Disulfide Bonds and the Circular Backbone in Maintaining Thermal Stability
The thermal stability of cyclic cystine-rich peptides has facilitated their discovery and characterization and makes them attractive as scaffolds for peptide drugs (40). Traditional use of the cyclotide kalata B1 as a component of a medicinal tea illustrates that the cyclic cystine knot imparts stability to this peptide, even after boiling (12). The NMR spectra in Fig. 4a show that θ-defensins also have high thermal stability, with no change in structure up to 80 °C. θ-Defensin analogues BTD-2, [Aba5,14]BTD-2, [Aba3,16]BTD-2, [Aba37,12,16]BTD-2, and BTD-2[3,4] are stable up to 80 °C, and it is expected that they would withstand boiling without denaturation. In contrast, [Aba5,7,12,14]BTD-2 and [Aba3,5,7,12,14,16]BTD-2 become unstructured above ∼65 °C. It appears that the single central disulfide bond in [Aba3,7,12,16]BTD-2 is sufficient to hold the β-strands together when the hydrogen bonds are weakened as the temperature increases. However, all of the peptides were able to re-fold on cooling, showing that the unfolding is a reversible process. The results shown in Fig. 4 also highlight the importance of the inclusion and correct positioning of disulfide bonds in the design of stable frameworks.
The Role of the Disulfide Bonds and the Circular Backbone in Serum Stability
A major limitation in the development of peptides as therapeutic agents is their susceptibility to degradation by proteases (41). Peptide cyclization can reduce susceptibility to exoproteases because it removes the free N and C termini and induces rigidity to the peptide backbone. The resulting serum stability is one of the main advantages of cyclic peptides as therapeutics and drug scaffolds (40, 42). The faster degradation of BTD-2[3,4] in serum, compared with BTD-2, illustrates the role of the cyclic backbone in serum stability. However, cyclization alone might not result in sufficient stability, as shown by the degradation of [Aba3,5,7,12,14,16]BTD-2 within 6 h. The θ-defensin analogues with two disulfide bonds, or one central disulfide bond, all have stability comparable with that of BTD-2, suggesting that inclusion of only one or two disulfide bonds in the design of a peptide could allow the accommodation of longer bioactive sequences in the θ-defensin framework. However, the positioning of a single disulfide bond appears to have a role in serum stability, as illustrated by the faster degradation of [Aba5,7,12,14]BTD-2 compared with [Aba3,7,12,16]BTD-2.
The Role of the Disulfide Bonds and the Circular Backbone in Antibacterial and Hemolytic Activity
The trends in antibacterial activities of the θ-defensin analogues, shown in Fig. 5a, suggest that the cyclic backbone, but not the disulfide bonds, are required for inhibition of bacterial growth. A study on the function of the three disulfide bonds in the α-defensin, mouse cryptdin-4, also showed that antibacterial activity was not affected when the disulfide bonds were mutated (17), and a similar result was obtained for human β-defensin-3 (18). In contrast, human β-defensin-1 is only weakly antimicrobial, but displayed an increase in potency on reduction of the three disulfide bonds (19). The MIC and MBC values in Fig. 5a are higher than those previously reported for BTD-2 (22); however, in the current study, the inhibition of bacterial growth was measured in nutrient media, rather than in buffer. Although θ-defensins have been reported to be antibacterial at physiological salt concentrations (1), in our hands no antibacterial activity was observed in LB at concentrations up to 64 μm.
The low hemolytic activity of the θ-defensin analogues indicates that they are selective for bacterial cells and highlights their potential as drug scaffolds. The number of disulfide bonds and the presence of the cyclic backbone have little effect on the hemolytic activity. The trends in hemolytic activity are interesting in that all peptides, except for [Aba3,7,12,16]BTD-2, are nonhemolytic. The low hemolytic activity for [Aba3,7,12,16]BTD-2 possibly reflects greater flexibility near the turn region, with a central anchoring disulfide bond allowing a symmetrical flexing motion. However, the peptide [Aba3,7,12,16]BTD-2 also showed the highest antibacterial activity, so lower concentrations would be required if it were to be used as a therapeutic. In the design of peptide drugs, the hemolytic activity could be reduced by residue substitutions, as has been illustrated for kalata B1 (30).
The Role of the Disulfide Bonds and the Cyclic Backbone in Binding to Model Membranes
Preferential binding of the θ-defensin analogues to anionic membranes, compared with zwitterionic membranes, correlates with high activity against bacterial cells and low hemolytic properties and suggests that electrostatic attractions between the cationic peptide and the anionic bacterial membrane are important for the antibacterial activity. Whereas kalata B1 has been shown to have an affinity for phospholipids containing phosphatidylethanolamine headgroups in membranes (31), the results in Fig. 6b indicate that θ-defensins have a preference for anionic membranes rather than specific headgroups. Small angle x-ray scattering and differential scanning calorimetry experiments have also shown that θ-defensins have affinity for anionic bacterial membranes rather than eukaryotic membranes and do not target individual lipid species (43, 44).
A mechanism of action dependent on electrostatic interactions is consistent with the high proportion of arginine residues in θ-defensins (45). Although all of the peptides in this study have the same net charge, the spatial distribution of the charges appears to affect the ability to bind to and disrupt membranes, as shown by the lower antibacterial activity of the acyclic and linear BTD-2 analogues compared with the cyclic peptides. The solution structure of BTD-2 (8) shows that the arginine side chains are evenly distributed over one side of the molecule, forming a positively charged face and suggesting that the peptides might bind parallel to the membrane. The “carpet” model of membrane disruption describes peptides that insert into the membrane in a parallel orientation and cause membrane thinning and leakage (46). Oriented circular dichroism, x-ray diffraction, and solid state NMR studies of rhesus θ-defensin-1 (RTD-1) in lipid bilayers have also been used to support a parallel membrane binding mechanism followed by pore formation (47, 48).
The cyclic θ-defensin analogues all bind to anionic membranes with similar affinities and have similar antibacterial activities; however, small differences might be due to structural flexibility of the peptides. Antimicrobial peptides that maintain some conformational flexibility have been reported to have higher antimicrobial activities than rigid peptides containing several proline residues (49). The single disulfide bond in [Aba3,7,12,16]BTD-2 might allow the peptide to insert into the bacterial membrane more easily than the more rigid BTD-2, resulting in higher antibacterial activity of [Aba3,7,12,16]BTD-2 compared with BTD-2.
Although the antibacterial activity of θ-defensins appears to be primarily through membrane binding and disruption, they might also have other mechanisms of action inside the cell, such as DNA binding or enzyme inhibition (46). θ-Defensins also have antiviral and anti-inflammatory activities that are mediated by specific peptide-receptor interactions. Human θ-defensin-1 (retrocyclin-1) inhibits HIV-1 entry by binding to CD4 and gp120 receptors (50, 51), and studies have indicated that the disulfide bonds and cyclic backbone have a role in receptor binding (52). θ-Defensins are also able to inhibit adhesion and entry of herpes simplex virus by binding a glycoprotein (53), and influenza A virus by inducing viral aggregation (54). Potent anti-inflammatory and antisepsis activities of RTDs have recently been reported, and studies suggest that RTD-1 inhibits the release of proinflammatory cytokines and tumor necrosis factors (42). The above examples illustrate the variety of functions that θ-defensins are likely to have in the immune system and their potential for development as therapeutics for a wide range of conditions.
In conclusion, we have shown that the cyclic cystine ladder has an important role in defining the structure and stability of θ-defensins but is not required for their antibacterial activity or membrane binding. However, the role of the cyclic cystine ladder in maintaining the structure and stability of θ-defensins is likely to underpin their antimicrobial activity by allowing them to remain stable and active in a wide range of environments. The present study also highlights the stability and plasticity of θ-defensins, properties that make them promising scaffolds for peptide drug design. Furthermore, knowledge of the role of the number and positions of disulfide bonds in cyclic peptides will more broadly facilitate the use of disulfide-rich scaffolds in the design of stable peptide drugs.
- SFTI-1
- sunflower trypsin inhibitor-1
- Boc
- tert-butyloxycarbonyl
- BTD
- baboon θ-defensin
- Fmoc
- fluorenylmethyloxycarbonyl
- HBTU
- 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- MBC
- minimal bactericidal concentration
- MIC
- minimal inhibitory concentration
- P/L
- peptide to lipid ratio
- POPC
- palmitoyloleoylphosphatidylcholine
- POPE
- palmitoyloleolylphosphatidylethanolamine
- POPG
- palmitoyloleoylphosphatidylglycerol
- RTD
- rhesus θ-defensin.
REFERENCES
- 1. Tang Y.-Q., Yuan J., Osapay G., Osapay K., Tran D., Miller C. J., Ouellette A. J., Selsted M. E. (1999) A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286, 498–502 [DOI] [PubMed] [Google Scholar]
- 2. Craik D. J. (2006) Seamless proteins tie up their loose ends. Science 311, 1563–1564 [DOI] [PubMed] [Google Scholar]
- 3. Lehrer R. I. (2004) Primate defensins. Nat. Rev. Microbiol. 2, 727–738 [DOI] [PubMed] [Google Scholar]
- 4. Selsted M. E., Ouellette A. J. (2005) Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 5. Göransson U., Burman R., Gunasekera S., Strömstedt A. A., Rosengren K. J. (2012) Circular proteins from plants and fungi. J. Biol. Chem. 287, 27001–27006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montalbán-López M., Sánchez-Hidalgo M., Cebrián R., Maqueda M. (2012) Discovering the bacterial circular proteins: bacteriocins, cyanobactins, and pilins. J. Biol. Chem. 287, 27007–27013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehrer R. I., Cole A. M., Selsted M. E. (2012) θ-Defensins: cyclic peptides with endless potential. J. Biol. Chem. 287, 27014–27019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conibear A. C., Rosengren K. J., Harvey P. J., Craik D. J. (2012) Structural characterization of the cyclic cystine ladder motif of θ-defensins. Biochemistry 51, 9718–9726 [DOI] [PubMed] [Google Scholar]
- 9. Trabi M., Schirra H. J., Craik D. J. (2001) Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from rhesus macaque leukocytes. Biochemistry 40, 4211–4221 [DOI] [PubMed] [Google Scholar]
- 10. Daly N. L., Chen Y. K., Rosengren K. J., Marx U. C., Phillips M. L., Waring A. J., Wang W., Lehrer R. I., Craik D. J. (2007) Retrocyclin-2: structural analysis of a potent anti-HIV θ-defensin. Biochemistry 46, 9920–9928 [DOI] [PubMed] [Google Scholar]
- 11. Azimi I., Wong J. W., Hogg P. J. (2011) Control of mature protein function by allosteric disulfide bonds. Antioxid. Redox Signal. 14, 113–126 [DOI] [PubMed] [Google Scholar]
- 12. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 13. Wang C. K., Hu S. H., Martin J. L., Sjögren T., Hajdu J., Bohlin L., Claeson P., Göransson U., Rosengren K. J., Tang J., Tan N. H., Craik D. J. (2009) Combined x-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as drug scaffold. J. Biol. Chem. 284, 10672–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daly N. L., Craik D. J. (2000) Acyclic permutants of naturally occurring cyclic proteins: characterization of cystine knot and β-sheet formation in the macrocyclic polypeptide kalata B1. J. Biol. Chem. 275, 19068–19075 [DOI] [PubMed] [Google Scholar]
- 15. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., Craik D. J. (2011) Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korsinczky M. L., Clark R. J., Craik D. J. (2005) Disulfide bond mutagenesis and the structure and function of the head-to-tail macrocyclic trypsin inhibitor SFTI-1. Biochemistry 44, 1145–1153 [DOI] [PubMed] [Google Scholar]
- 17. Maemoto A., Qu X., Rosengren K. J., Tanabe H., Henschen-Edman A., Craik D. J., Ouellette A. J. (2004) Functional analysis of the α-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279, 44188–44196 [DOI] [PubMed] [Google Scholar]
- 18. Wu Z., Hoover D. M., Yang D., Boulègue C., Santamaria F., Oppenheim J. J., Lubkowski J., Lu W. (2003) Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human β-defensin 3. Proc. Natl. Acad. Sci. U.S.A. 100, 8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schroeder B. O., Wu Z., Nuding S., Groscurth S., Marcinowski M., Beisner J., Buchner J., Schaller M., Stange E. F., Wehkamp J. (2011) Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469, 419–423 [DOI] [PubMed] [Google Scholar]
- 20. Chakraborty K., Thakurela S., Prajapati R. S., Indu S., Ali P. S., Ramakrishnan C., Varadarajan R. (2005) Protein stabilization by introduction of cross-strand disulfides. Biochemistry 44, 14638–14646 [DOI] [PubMed] [Google Scholar]
- 21. Henriques S. T., Craik D. J. (2010) Cyclotides as templates in drug design. Drug Discov. Today 15, 57–64 [DOI] [PubMed] [Google Scholar]
- 22. Garcia A. E., Osapay G., Tran P. A., Yuan J., Selsted M. E. (2008) Isolation, synthesis, and antimicrobial activities of naturally occurring θ-defensin isoforms from baboon leukocytes. Infect. Immun. 76, 5883–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Synthesis of proteins by native chemical ligation. Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 24. Conibear A. C., Daly N. L., Craik D. J. (2012) Quantification of small cyclic disulfide-rich peptides. Biopolymers 98, 518–524 [DOI] [PubMed] [Google Scholar]
- 25. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 27. Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) Recoord: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 29. Wiegand I., Hilpert K., Hancock R. E. (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 [DOI] [PubMed] [Google Scholar]
- 30. Huang Y. H., Colgrave M. L., Clark R. J., Kotze A. C., Craik D. J. (2010) Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J. Biol. Chem. 285, 10797–10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henriques S. T., Huang Y.-H., Castanho M. A. R. B., Bagatolli L. A., Sonza S., Tachedjian G., Daly N. L., Craik D. J. (2012) Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 287, 33629–33643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henriques S. T., Castanho M. A., Pattenden L. K., Aguilar M. I. (2010) Fast membrane association is a crucial factor in the peptide Pep-1 translocation mechanism: a kinetic study followed by surface plasmon resonance. Biopolymers 94, 314–322 [DOI] [PubMed] [Google Scholar]
- 33. Hoover D. M., Wu Z., Tucker K., Lu W., Lubkowski J. (2003) Antimicrobial characterization of human β-defensin 3 derivatives. Antimicrob. Agents Chemother. 47, 2804–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. (1989) Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245, 616–621 [DOI] [PubMed] [Google Scholar]
- 35. Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) 1H,13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest neighbor effects. J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 36. Torcato I. M., Huang Y. H., Franquelim H. G., Gaspar D., Craik D. J., Castanho M. A., Troeira Henriques S. (2013) Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against gram-negative and gram-positive bacteria. Biochim. Biophys. Acta 1828, 944–955 [DOI] [PubMed] [Google Scholar]
- 37. Wu C., Leroux J.-C., Gauthier M. A. (2012) Twin disulfides for orthogonal disulfide pairing and the directed folding of multicyclic peptides. Nat. Chem. 4, 1044–1049 [DOI] [PubMed] [Google Scholar]
- 38. Gibbs A. C., Kondejewski L. H., Gronwald W., Nip A. M., Hodges R. S., Sykes B. D., Wishart D. S. (1998) Unusual β-sheet periodicity in small cyclic peptides. Nat. Struct. Biol. 5, 284–288 [DOI] [PubMed] [Google Scholar]
- 39. Selsted M. E. (2004) θ-Defensins: cyclic antimicrobial peptides produced by binary ligation of truncated α-defensins. Curr. Protein Pept. Sci. 5, 365–371 [DOI] [PubMed] [Google Scholar]
- 40. Craik D. J., Cemazar M., Daly N. L. (2006) The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 9, 251–260 [PubMed] [Google Scholar]
- 41. Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. (2010) Synthetic therapeutic peptides: science and market. Drug Discov. Today 15, 40–56 [DOI] [PubMed] [Google Scholar]
- 42. Schaal J. B., Tran D., Tran P., Ösapay G., Trinh K., Roberts K. D., Brasky K. M., Tongaonkar P., Ouellette A. J., Selsted M. E. (2012) Rhesus macaque θ-defensins suppress inflammatory cytokines and enhance survival in mouse models of bacteremic sepsis. PLoS ONE 7, e51337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmidt N. W., Mishra A., Lai G. H., Davis M., Sanders L. K., Tran D., Garcia A., Tai K. P., McCray P. B., Ouellette A. J., Selsted M. E., Wong G. C. (2011) Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133, 6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abuja P. M., Zenz A., Trabi M., Craik D. J., Lohner K. (2004) The cyclic antimicrobial peptide RTD-1 induces stabilized lipid-peptide domains more efficiently than its open-chain analogue. FEBS Lett. 566, 301–306 [DOI] [PubMed] [Google Scholar]
- 45. Chan D. I., Prenner E. J., Vogel H. J. (2006) Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta 1758, 1184–1202 [DOI] [PubMed] [Google Scholar]
- 46. Brogden K. A. (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 47. Weiss T. M., Yang L., Ding L., Waring A. J., Lehrer R. I., Huang H. W. (2002) Two states of cyclic antimicrobial peptide RTD-1 in lipid bilayers. Biochemistry 41, 10070–10076 [DOI] [PubMed] [Google Scholar]
- 48. Tang M., Waring A. J., Lehrer R. I., Hong M. (2006) Orientation of a β-hairpin antimicrobial peptide in lipid bilayers from two-dimensional dipolar chemical-shift correlation NMR. Biophys. J. 90, 3616–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vermeer L. S., Lan Y., Abbate V., Ruh E., Bui T. T., Wilkinson L. J., Kanno T., Jumagulova E., Kozlowska J., Patel J., McIntyre C. A., Yam W. C., Siu G., Atkinson R. A., Lam J. K., Bansal S. S., Drake A. F., Mitchell G. H., Mason A. J. (2012) Conformational flexibility determines selectivity and antibacterial, antiplasmodial, and anticancer potency of cationic α-helical peptides. J. Biol. Chem. 287, 34120–34133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Münk C., Wei G., Yang O. O., Waring A. J., Wang W., Hong T., Lehrer R. I., Landau N. R., Cole A. M. (2003) The θ-defensin, retrocyclin, inhibits HIV-1 entry. AIDS Res. Hum. Retroviruses 19, 875–881 [DOI] [PubMed] [Google Scholar]
- 51. Gallo S. A., Wang W., Rawat S. S., Jung G., Waring A. J., Cole A. M., Lu H., Yan X., Daly N. L., Craik D. J., Jiang S., Lehrer R. I., Blumenthal R. (2006) θ-Defensins prevent HIV-1 env-mediated fusion by binding gp41 and blocking 6-helix bundle formation. J. Biol. Chem. 281, 18787–18792 [DOI] [PubMed] [Google Scholar]
- 52. Wang W., Cole A. M., Hong T., Waring A. J., Lehrer R. I. (2003) Retrocyclin, an antiretroviral θ-defensin, is a lectin. J. Immunol. 170, 4708–4716 [DOI] [PubMed] [Google Scholar]
- 53. Yasin B., Wang W., Pang M., Cheshenko N., Hong T., Waring A. J., Herold B. C., Wagar E. A., Lehrer R. I. (2004) θ-Defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 78, 5147–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Doss M., White M. R., Tecle T., Gantz D., Crouch E. C., Jung G., Ruchala P., Waring A. J., Lehrer R. I., Hartshorn K. L. (2009) Interactions of α-, β-, and θ-defensins with influenza A virus and surfactant protein D. J. Immunol. 182, 7878–7887 [DOI] [PubMed] [Google Scholar]