Background: IMPACT inhibits GCN2, a kinase that directs stress remedial responses by attenuating translation and controls feeding behavior and memory.
Results: Neuronal IMPACT is developmentally up-regulated, promoting protein synthesis and neuritogenesis, opposing GCN2.
Conclusion: GCN2 and IMPACT modulate an early step in neuronal differentiation.
Significance: A neuron-specific developmental program is controlled by two evolutionarily conserved translational regulators.
Keywords: Neurite Outgrowth, Neurodifferentiation, Protein Synthesis, Ribosomes, Translation Control, GCN2, IMPACT, eIF2
Abstract
The product of the mouse Imprinted and Ancient gene, IMPACT, is preferentially expressed in neurons. We have previously shown that IMPACT overexpression inhibits the activation of the protein kinase GCN2, which signals amino acid starvation. GCN2 phosphorylates the α-subunit of eukaryotic translation initiation factor 2 (eIF2α), resulting in inhibition of general protein synthesis but increased translation of specific messages, such as ATF4. GCN2 is also involved in the regulation of neuronal functions, controlling synaptic plasticity, memory, and feeding behavior. We show here that IMPACT abundance increases during differentiation of neurons and neuron-like N2a cells, whereas GCN2 displays lowered activation levels. Upon differentiation, IMPACT associates with translating ribosomes, enhances translation initiation, and down-regulates the expression of ATF4. We further show that endogenous IMPACT promotes neurite outgrowth whereas GCN2 is a strong inhibitor of spontaneous neuritogenesis. Together, these results uncover the participation of the GCN2-IMPACT module of translational regulation in a highly controlled step in the development of the nervous system.
Introduction
Translational control allows for a quick adaptation of cells to changes in environmental conditions. One such regulatory pathway involves the phosphorylation of the α-subunit of the eukaryotic translation initiation factor 2 (eIF2α) mediated by a family of protein kinases that are activated by a variety of stress conditions (1). This phosphorylation event at residue Ser51 of eIF2α in mammals or a corresponding residue in other eukaryotes results in the attenuation, or a complete inhibition, of translation, depending on the strength or duration of the stress. The GTP-bound form of eIF2 participates in translation initiation by delivering the initiator methionyl-tRNA to the 40 S ribosomal subunit, which then binds to 5′ cap of mRNAs with the aid of other factors. This complex scans the mRNA, and upon encountering the initiator codon, eIF2 is released as eIF2-GDP. The 60 S ribosomal subunit joins the complex and elongation of the polypeptide chains ensues. The guanine exchange factor eIF2B mediates the exchange of GDP to GTP on eIF2 necessary for a subsequent round of initiation. Phosphorylated eIF2α (P-eIF2α)7 reduces the availability of eIF2-GTP by acting as a competitive inhibitor of eIF2B. Increased levels of P-eIF2α result in inhibition of general translation but, paradoxically, enhance the translation of specific messages that contain upstream open reading frames in their mRNA leader sequence, such as GCN4 in yeast and ATF4(CREB2) in mammals. Both transcription factors direct a cellular response of recovery to the condition that initiated this signal (2–4). This response is critical for cell survival, and its malfunction can lead to diseases (1).
There are four eIF2α kinases in mammals. GCN2, present in all eukaryotes, is activated by amino acid starvation and other stress conditions, such as proteasome inhibition and UV irradiation (5–7). PERK senses the accumulation of unfolded proteins in the endoplasmic reticulum. PKR responds to dsRNA and other stresses. HRI controls translation in reticulocytes in response to heme (for review, see Ref. 8).
The mechanism of GCN2 activation has been dissected by extensive studies in Saccharomyces cerevisiae, where GCN2 is the sole eIF2α kinase (4). Uncharged tRNAs that accumulate under amino acid starvation conditions bind to a region in GCN2 with similarity to histidyl tRNA synthetases, causing a conformational modification that results in the autophosphorylation of a threonine residue in the kinase catalytic domain and its ability to bind to and phosphorylate its substrate, eIF2α. The C terminus of GCN2 contains a dimerization and ribosome binding domain, with both activities being required for the activation of the kinase. The N-terminal region contains an RWD domain (present in RING finger proteins, WD-repeat containing proteins, and DEAD-like helicases) that binds to its effector protein GCN1, an interaction that is essential for GCN2 activation (9–11). It is thought that GCN1 promotes the transfer of uncharged tRNA from the ribosome to the histidyl tRNA synthetase domain of GCN2, when both proteins are tethered to the ribosome.
GCN2 controls behavioral and physiologically relevant events in mammals. GCN2 modulates the immune system controlling tumors, autoimmunity, and transplant tolerance (12, 13). In the brain GCN2 directs feeding behavior toward amino acid sources (14, 15). GCN2 is activated in the anterior piriform cortex shortly after mice are fed a diet poor in essential amino acids. Animals lacking GCN2 activity, contrary to wild type mice, do not show aversion to these diets. In these examples, GCN2 activation seems to be achieved by the low availability of amino acids. GCN2 also participates in memory consolidation. Mice devoid of GCN2 show decreased threshold for late long term potentiation and long term memory, phenotypes also displayed by mice heterozygous for a S51A mutation in eIF2α (16, 17). It is unclear how GCN2 activity is modulated in this case.
We have previously described that mammalian GCN2 is regulated by the protein IMPACT, the product of an imprinted gene in rodents (18, 19). IMPACT is a cytoplasmic protein preferentially expressed in neurons and is highly abundant in the hypothalamus and in scattered neurons in other areas, such as the hippocampal interneurons (20). IMPACT overexpression in mouse embryonic fibroblasts (MEFs) inhibits GCN2 activation induced by leucine starvation (19). IMPACT and its yeast ortholog Yih1 contain an RWD domain in their N-terminal half that interacts with GCN1 and competes with GCN2 for GCN1 binding. When overexpressed in yeast, both IMPACT and Yih1 inhibit Gcn2 activation (19, 21). IMPACT/Yih1 are found in most eukaryotes. Although overexpression studies in yeast and in mammalian cells have clearly indicated that IMPACT and Yih1 act as inhibitors of GCN2, no physiological function for endogenous neuronal IMPACT has been identified.
Here, we show that IMPACT is a developmentally regulated protein that promotes neuritogenesis, whereas GCN2 strongly inhibits spontaneous neurite outgrowth. Increased abundance of IMPACT in differentiated neuronal cells prevents GCN2 signaling, enhancing translation initiation. Our results provide novel neuronal functions for two highly conserved proteins and indicate that translational control mediated by GCN2 and its modulation by IMPACT are key determinants of a neuronal developmental process.
EXPERIMENTAL PROCEDURES
Animals
Mice (C57BL/6J background) were housed under standard laboratory conditions. All experimental protocols were approved by the Animal Care and Use Ethics Committee of UNIFESP. Gcn2−/− (129Sv background) animals were a kind gift from Dr. Douglas Cavener (22). The Gcn2-Neo allele was transferred to the C57BL/6J background by crosses for 13 generations.
siRNAs
The following siRNAs were used (sense strand): 5′-GCAAGAACGCGCAGACUUATT-3′ (siIMPACT), 5′-GCAGCAAACGCCAGGAUUATT-3′ (siControl, scrambled for siIMPACT), 5′-GCAUGGACGAGCUGUACAATT-3′ (siEGFP), and 5′-GGAAAUGGCUAAGCAGGAATT-3′ (siGCN2).
Cell Culture, N2a Cell Differentiation, Transfection, and Stress Conditions
N2a cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS and 1 mm sodium pyruvate. Differentiation was induced by incubation in DMEM containing 30% Opti-MEM (Invitrogen). Transfection with siRNA was performed with Lipofectamine 2000 (Invitrogen) for 6 h in Opti-MEM. Cells were starved for l-leucine by incubation for the indicated times in DMEM lacking l-leucine (Emcare, Brazil) containing 10% dialyzed FCS (Invitrogen) and sodium pyruvate. Immortalized MEF cells (19) were grown in DMEM with 10% FCS.
Primary Hippocampal Cultures and Neurite Outgrowth Assays
Primary cultures of hippocampal neurons were derived from embryonic day 16–18 brain of Gcn2+/+ or Gcn2−/− mice (C57BL/6J background) (22) and performed as described (23). Neuritogenesis assays were performed as described previously (24).
Cell Extracts and Immunoblotting
Cell extracts and brain homogenates were prepared in ice-cold lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EDTA, 16 μg/ml benzamidine HCl, 10 μg/ml phenanthroline, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 1 mm PMSF, 10 mm sodium fluoride, 1 mm sodium orthovanadate, 17.5 mm sodium β-glycerophosphate, 6 mm sodium pyrophosphate) for 10 min on ice and centrifuged at maximum speed in a refrigerated microcentrifuge for 15 min. Total proteins in supernatants were quantified by Bradford (Sigma) and subjected to SDS-PAGE followed by blotting to Hybond-C Extra membranes (GE Healthcare). Following incubation with primary antibodies, these were recognized by HRP-conjugated goat anti-rabbit or anti-mouse IgG (1:2000, Santa Cruz Biotechnology) or by HRP-conjugated protein A (1:2000, GE Healthcare; for anti-GCN2), and detected by ECL (GE Healthcare).
Antibodies
Primary antibodies were as follows. Affinity-purified rabbit anti-IMPACT, anti-GCN1, and guinea pig anti-GCN2 were previously described (19, 20). Anti-Thr(P)899-GCN2 was from Abcam. Anti-Ser(P)51-eIF2α and anti-eIF2α were from BIOSOURCE. Anti-ATF4, anti-PSD-95, anti-Tau, and anti-CHOP were from Santa Cruz Biotechnology. Anti-ribosomal protein S6 and anti-PERK were from Cell Signaling. Anti-MAP2 and anti-actin were from Sigma. Anti-fragile X mental retardation protein and anti-NeuN were from Millipore. Anti-GAPDH was from Ambion.
Polysome Profiles
N2a cells grown on two 150-mm dishes to 70% confluence (undifferentiated) or induced to differentiate for 3 days were treated with 100 μg/ml cycloheximide for 5 min at 37 °C, washed with PBS containing 100 μg/ml cycloheximide, sedimented by centrifugation, incubated for 10 min in lysis buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1% Triton X-100, 1 mm DTT, 100 μg/ml cycloheximide, 40 units/ml RNasin (Promega), and EDTA free-protease inhibitor mixture (Thermo)) on ice, and centrifuged for 10 min at 10,000 × g on a refrigerated microcentrifuge. The supernatant was applied to 7–47% sucrose gradients, prepared in 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT. Gradients were centrifuged in a SW41 Ti rotor (Beckman) at 38,000 rpm for 2:20 h, collected from the top, and the absorbance was measured in a continuous flow. For the polysome profiles of cells transfected with siRNAs, one 150-mm plate was used for each condition, and extracts were subjected to centrifugation on 7–47% sucrose gradients at 39,000 rpm for 2.5 h.
RESULTS
IMPACT Inhibits GCN2 in Neuron-like N2a Cells
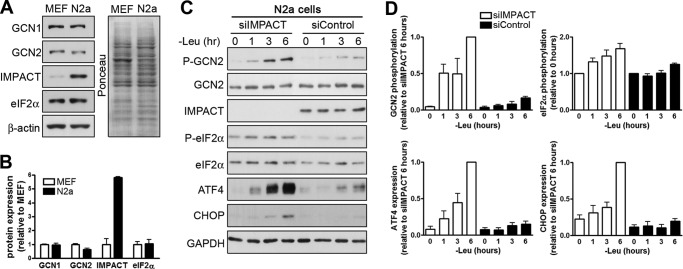
We found that the mouse neuroblastoma-derived N2a cell line expresses IMPACT in levels much above those of MEF cells (Fig. 1, A and B). We then used the N2a cells to study the role of endogenous IMPACT on GCN2 signaling by knocking down IMPACT expression with small interfering RNA (siRNA). Initially, we studied the response to leucine starvation, a specific signal for GCN2 activation. Cells transfected with siRNA against IMPACT mRNA (siIMPACT) showed a significant reduction in the amount of IMPACT compared with cells transfected with a control, scrambled siRNA (siControl) (Fig. 1C). The transfected cells were then subjected to leucine starvation for the indicated times and the cell extracts used to analyze the stress response. Compared with the control cells containing IMPACT, cells depleted of IMPACT were highly responsive to leucine withdrawal, showing a pronounced increase in GCN2 activation that can be detected by its autophosphorylation at residue Thr898 (equivalent to residue Thr882 in yeast GCN2 (25)) (Fig. 1, C and D). This was accompanied by an increase in phosphorylation of its substrate, eIF2α (Fig. 1, C and D). The expression of ATF4, a protein whose translation is increased when eIF2α is phosphorylated, was largely augmented. In addition, there was also a significant change in the expression of CHOP, a gene that is transcriptionally up-regulated by ATF4 (Fig. 1, C and D). Thus, endogenous IMPACT attenuates GCN2 activation upon amino acid starvation in cells with neuronal characteristics. These results are in agreement with, and complement, our previous data on the overexpression of IMPACT in MEF cells.
FIGURE 1.
Endogenous IMPACT in neuron-like N2a cells inhibits GCN2 signaling. A, extracts of undifferentiated N2a cells and of MEFs were subjected to immunoblotting to detect the indicated proteins. The Ponceau staining of the membrane from a 10% SDS-PAGE is presented. B, the graph represents the quantification of the immunoblots shown in A. The intensity of each band was normalized by the intensity of the Ponceau staining of the respective lane. GCN1 and GCN2 were normalized by the Ponceau staining of the membrane from a 6% SDS-PAGE (data not shown). Data are presented as mean ± S.E. from four independent experiments. C, undifferentiated N2a cells were transfected with siIMPACT or with a scrambled siRNA (siControl) and incubated in medium lacking leucine to activate GCN2 for the indicated times. Extracts were used in immunoblotting to detect the indicated proteins and their respective phosphorylated forms. The figures are representative of at least three independent experiments. D, immunoblots were quantified, with data presented as mean ± S.E. (error bars) from at least three independent experiments. Phosphorylated GCN2 was normalized against total GCN2, and the P-GCN2/GCN2 ratio at 6 h of leucine starvation in siIMPACT-transfected cells was set to 1. Phosphorylated eIF2α was normalized against total eIF2α, and the P-eIF2α/eIF2α ratio in unstarved condition was set to 1. ATF4 and CHOP were normalized against GAPDH, and their expression at 6 h of leucine starvation in siIMPACT transfected cells was set to 1.
Increased Abundance of IMPACT in Differentiating Neuronal Cells Inhibits GCN2
In the adult mouse, IMPACT is abundant in neurons of the central nervous system (CNS) (20). In embryos, IMPACT is also predominantly expressed in the CNS (Fig. 2A). Importantly, we found that its abundance strongly increases during brain development (Fig. 2, B and C). GCN1, GCN2, and eIF2α, on the other hand, remained unchanged relative to each other when extracts were normalized against the Ponceau staining of the membranes (Fig. 2, B and C).
FIGURE 2.
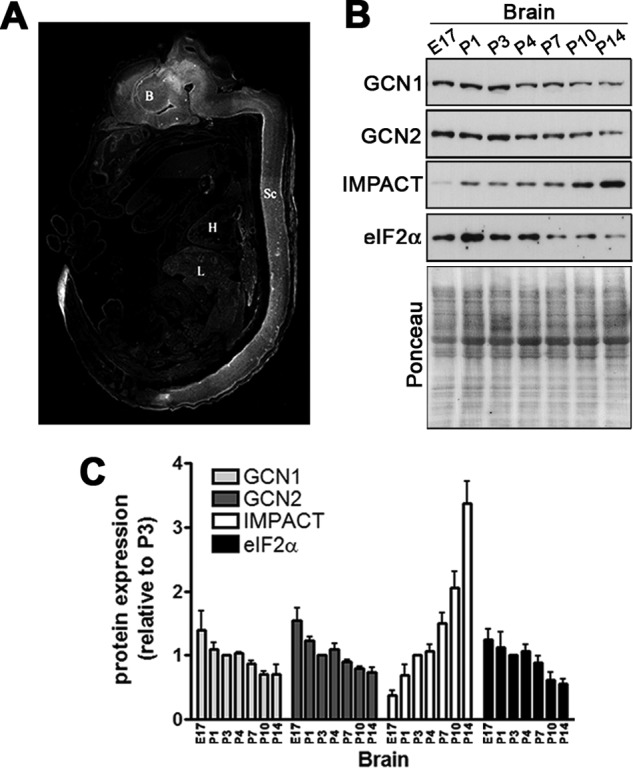
The abundance of IMPACT increases during brain development. A, IMPACT distribution in day 16 mouse embryo visualized by immunofluorescence using an anti-IMPACT antibody. Indicated in the image are brain (B), heart (H), liver (L), and spinal cord (Sc). B, immunoblots of brain extracts of mice of the indicated ages. The Ponceau-stained membranes of the 10% SDS-PAGE are shown at the bottom. E17, embryonic day 17. C, quantification of the immunoblots shown in B, normalized by the intensity of the Ponceau staining of the respective lanes. GCN1 and GCN2 were normalized by the Ponceau staining of the membrane from a 6% SDS-PAGE (data not shown). The expression of each protein in P3 was set to 1. Data are presented as mean ± S.E. (error bars) from at least four independent experiments.
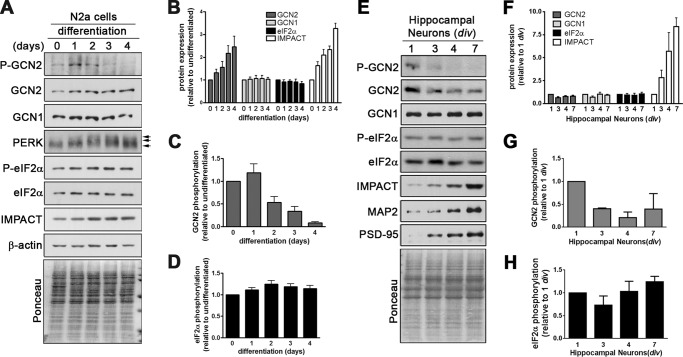
We found that in N2a cells the induction of differentiation by serum reduction was also accompanied by an increase in IMPACT abundance (Fig. 3, A and B). We then assessed the levels of active GCN2 during differentiation in N2a cells by evaluating its phosphorylation at Thr898. A small increase in activation of GCN2 was observed at day one of differentiation (Fig. 3, A and C), which is in agreement with previous reports that serum withdrawal activates GCN2 in other cell types (26). Importantly, however, we found that phosphorylated GCN2 decreases strongly afterward in the process of differentiation (Fig. 3, A and C). The expected effect on eIF2α phosphorylation was not detected, probably due to the activity of other eIF2α kinase (Fig. 3, A and D). To check this possibility we studied the activation of another eIF2α kinase, PERK. Upon its activation PERK becomes phosphorylated, which results in the slower migration of the protein on SDS-PAGE (27). We found that during N2a cell differentiation, the levels of slow migrating PERK increased (Fig. 3A). This could explain, at least partially, why we were not able to detect an overall reduction in the levels of phosphorylated eIF2α. Similarly, in primary cultures of hippocampal neurons there is also a strong increase in IMPACT levels upon differentiation which is accompanied by a concomitant reduction in GCN2 phosphorylation (Fig. 3, E–G). As we have seen in differentiating N2a cells, eIF2α phosphorylation levels remained apparently unchanged (Fig. 3, E and H). The elevation in IMPACT expression occurred in parallel with the increase of the neuronal differentiation markers MAP2 and PSD-95 (Fig. 3E).
FIGURE 3.
IMPACT expression increases during the differentiation of N2a cells and primary hippocampal neurons together with the lowering of active GCN2. A, N2a cells were induced to differentiate for the indicated number of days and the extracts used for immunoblots with antibodies against the indicated proteins or their phosphorylated forms. GCN1 and GCN2 were resolved on 6% gels; PERK on 7% gels; and eIF2α, IMPACT, and actin on 10% gels; the Ponceau-stained membrane of the latter is shown at the bottom. B, quantification of the expression of GCN1, GCN2, IMPACT, and eIF2α, normalized by the intensity of Ponceau staining of the respective lanes. The expression of each protein in undifferentiated conditions was set to 1. Data are presented as mean ± S.E. (error bars) from at least four independent experiments. C, quantification of GCN2 phosphorylated at Thr898 relative to total GCN2 levels. The ratio of P-GCN2/GCN2 in undifferentiated conditions was set to 1. Data are presented as mean ± S.E. from six independent experiments. D, quantification of phosphorylated eIF2α relative to total eIF2α levels. The ratio of P-eIF2α/eIF2α in undifferentiated conditions was set to 1. Data are presented as mean ± S.E. from six independent experiments. E, extracts of primary hippocampal neurons of the indicated number of days of culture in vitro (div) were used for immunoblotting with antibodies against the indicated proteins. The Ponceau-stained membrane of the 10% gel is shown at the bottom. F, quantification of the expression of GCN1, GCN2, IMPACT, and eIF2α, normalized by the intensity of Ponceau staining of the respective lanes. The expression of each protein at 1 div was set to 1. Data are presented as mean ± S.E. from at least four independent experiments. G, quantification of GCN2 phosphorylated at Thr898 relative to total GCN2 levels. The ratio of P-GCN2/GCN2 was set to 1. Data are presented as mean ± S.E. from three independent experiments. H, quantification of phosphorylated eIF2α relative to total eIF2α levels. The ratio of P-eIF2α/eIF2α at 1 div was set to 1. Data are presented as mean ± S.E. from at least three independent experiments.
IMPACT Depletion in Differentiated N2a Cells Results in the Activation of GCN2, Increased Phosphorylation of eIF2α, and Inhibition of Translation Initiation
The absence of the expected reduction in eIF2α phosphorylation levels following the inactivation of GCN2 along the process of differentiation could be a consequence of the activity of other eIF2α kinase. This prompted us to determine whether in differentiated cells the increased IMPACT expression and the resultant GCN2 inactivation may affect eIF2α phosphorylation. N2a cells differentiated for 1 day were transfected with siIMPACT or siControl, and extracts were prepared after 2 additional days in differentiation medium. This experimental design allowed us to compare the effect of the absence of IMPACT in the same context (i.e. differentiated cells) where, in theory, the rest of the system should remain unchanged. The knockdown of IMPACT resulted in increased phosphorylation of GCN2 and of its substrate eIF2α, compared with cells transfected with control siRNA (Fig. 4, A–C). A significant increase in ATF4 expression was also clearly detected (Fig. 4, A and D). These data should be compared with the results of the siRNA experiments in cells that were not subjected to differentiation, maintained in the presence of leucine shown in Fig. 1, time 0. In the latter case, no significant change in GCN2 phosphorylation or in ATF4 expression could be detected between cells depleted of IMPACT and control cells. Thus, the increased abundance of IMPACT in differentiating cells is capable of significantly down-regulating GCN2 activity.
FIGURE 4.
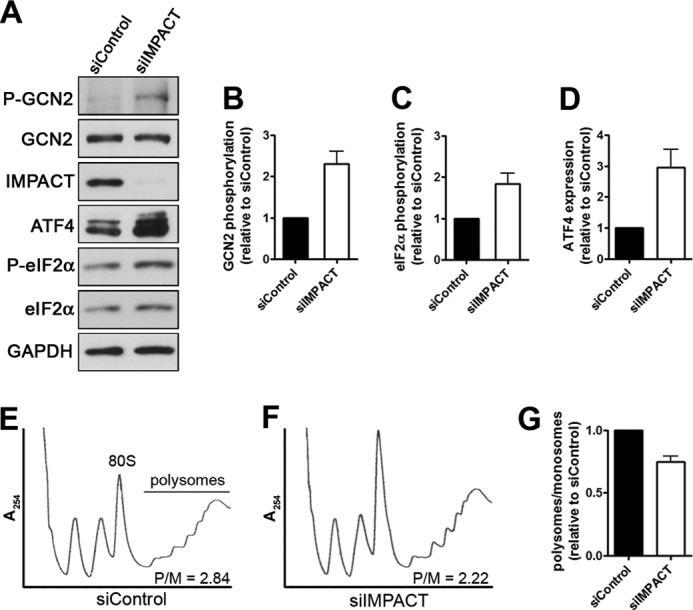
Depletion of IMPACT in differentiated N2a cells promotes GCN2 activation and decreases translation initiation. A, N2a cells transfected with siIMPACT or a scrambled siRNA (siControl) at 1 day of differentiation and left for 2 additional days in differentiating conditions. Extracts were used in immunoblotting to detect the indicated proteins and their respective phosphorylated forms. B–D, quantification of phosphorylated GCN2 relative to total GCN2, of phosphorylated eIF2α relative to total eIF2α, and of ATF4 relative to GAPDH. The ratios for siControl-transfected cells were set to 1. Data are presented as mean ± S.E. (error bars) from at least four independent experiments. E, representative polysome profiles of extracts of N2a cells transfected with siIMPACT or siControl, as described above, subjected to separation on 7–47% sucrose gradients. As a measure of the levels of translation initiation, the areas under the polysomes and 80 S peaks were determined, and the P/M was calculated for each condition. The P/M for this experiment is shown. F, quantification of the levels of translation initiation estimated by the P/Ms. The P/M for siControl-transfected cells was set to 1. Data represent the mean ± S.E. from three independent experiments.
The effect of IMPACT depletion on translation initiation was then analyzed by polysome profiles obtained by sucrose gradient fractionation of extracts from differentiated N2a cells transfected with siRNA in the same manner as described above. As expected from the previous results, IMPACT depletion resulted in a decrease of approximately 25% in the polysome/monosome ratio (P/M) compared with cells expressing IMPACT (Fig. 4, E and F). This reduction in P/M is a hallmark of impairment in the initiation step of protein synthesis. Thus, the data shown here strongly indicate that the increased expression of IMPACT in differentiating neuron-like cells promotes translation initiation by inhibiting GCN2.
IMPACT Associates with Polysomes upon N2a Cell Differentiation
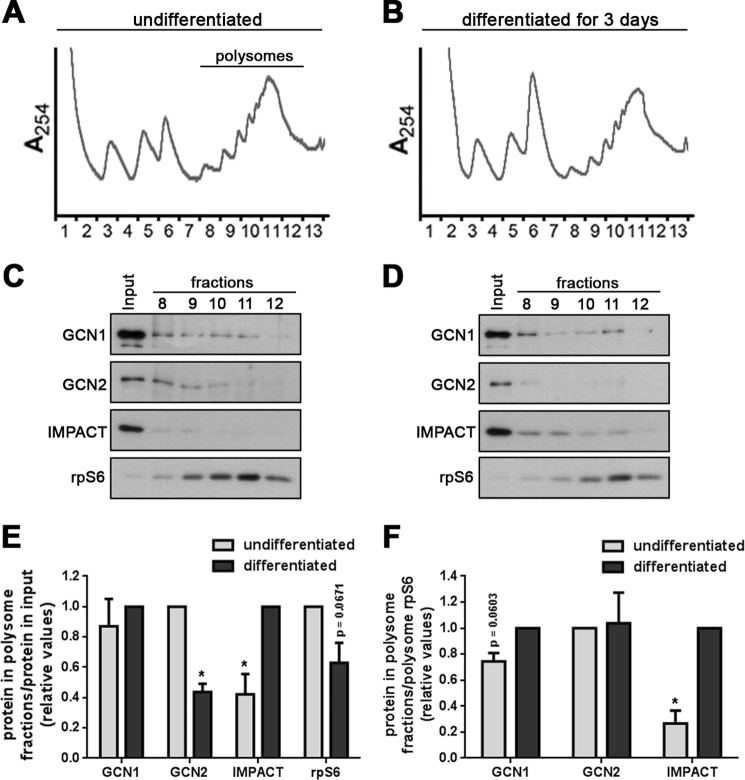
We next studied the association of IMPACT with the translation machinery by analyzing its distribution pattern on polysome profiles obtained from N2a cell extracts subjected to sucrose gradient centrifugation (Fig. 5 and supplemental Fig. S1). In addition, we also probed for the distribution of GCN2, GCN1, and the ribosomal protein S6 (rpS6). Impressively, in differentiated N2a cells the amount of IMPACT associated with polysomes was significantly higher than in undifferentiated cells (Fig. 5, C–F). On the other hand, GCN2 present in polysomal fractions was significantly reduced after differentiation (Fig. 5, C–E). We noticed that during differentiation there was a slight decrease in the number of polysomes as observed both by the polysome profiles (Fig. 5, A and B) and by the distribution of rpS6 (Fig. 5E). By using polysomal rpS6 for normalization, the observed increase in the polysome-associated IMPACT in differentiated cells compared with nondifferentiated cells was confirmed (Fig. 5F). However, using this normalization we found that the amount of GCN2 associated with polysomes remains practically unchanged between differentiated and undifferentiated cells (Fig. 5F). GCN1 present in polysome fractions remains unchanged during differentiation (Fig. 5, C–E). When taking into consideration the reduction in polysomes during differentiation, GCN1 displays a slight increase in its association with them (Fig. 5F). As controls, we probed for the fragile X mental retardation protein, described to associate with polyribosomes (28–30) and the extract of differentiated cells was treated with RNase A before separation on sucrose gradients (Supplemental Fig. S1). These data indicate that upon differentiation IMPACT increasingly associates with actively translating ribosomes, where it could interact with ribosome bound GCN1. Thus, during differentiation IMPACT not only increases in its expression levels, but also in its association with the translational machinery.
FIGURE 5.
Association of IMPACT with translating ribosomes. A and B, polysome profiles from extracts of N2a cells nondifferentiated (A) or differentiated for 3 days (B), fractionated on 7–47% sucrose gradients. C and D, proteins from the polysomal fractions of nondifferentiated (C) or differentiated for 3 days N2a cells (D), were precipitated with 15% TCA, applied to 4–15% gradient SDS-PAGE, and used for immunoblots to detect the indicated proteins. The first lane of the immunoblots corresponds to 1% of the total extract applied to the gradient (Input). E, quantification of the amount of IMPACT, GCN2, GCN1, and rpS6 present in the polysomal fractions. The sum of the band intensities of the fractions obtained by densitometry was divided by the intensity of the respective input. For GCN2 and rpS6, the value of the ratio for undifferentiated cells was set to 1. For IMPACT and GCN1, the value of the ratio for differentiated cells was set to 1. Data are presented as mean ± S.E. (error bars) from at least three independent experiments. *, p < 0.05 (Student's t test); p value for rpS6 is presented in the graph. F, quantification of the amount of IMPACT, GCN2, and GCN1 present in polysome fractions. The sum of the band intensities of the fractions obtained by densitometry was divided by the sum of the band intensities of rpS6 in polysome fractions, used as a measurement of the amount of polysomes. For GCN2, the value of the ratio for undifferentiated cells was set to 1. For IMPACT and GCN1, the value of the ratio for differentiated cells was set to 1. Data are presented as mean ± S.E. from at least three independent experiments. *, p < 0.05 (Student's t test); p value for GCN1 is presented in the graph.
IMPACT and GCN2 Regulate Neurite Outgrowth in Opposite Manners
The expression profile of IMPACT during neuronal differentiation and the decrease in active GCN2 during this process suggested an involvement of the two proteins in differentiation. We then addressed whether IMPACT and GCN2 were relevant for neurite outgrowth, an early step in neuronal differentiation. N2a cells were co-transfected with siIMPACT/siControl, siGCN2/siControl, siIMPACT/siGCN2, or siControl alone and were either maintained in growth medium or changed to differentiation medium. Depletion of both proteins simultaneously was as efficient as the single depletions (Fig. 6A). Incubation of cells transfected with siControl in differentiation medium led to a large increase in the number of cells with at least one neurite longer than the cell body diameter, as expected (Fig. 6, B and C). The depletion of IMPACT resulted in a strong inhibition of induced neuritogenesis (Fig. 6, B and C). As expected from the observed decrease in P-GCN2 during differentiation, the knockdown of GCN2 did not affect induced neurite outgrowth significantly. Surprisingly, however, the depletion of GCN2 caused a drastic increase in spontaneous neurite outgrowth in growth medium (Fig. 6, B and C). These results indicate that IMPACT facilitates N2a-induced neuritogenesis and that GCN2 is a strong determinant for the maintenance of the undifferentiated state of N2a cells. Furthermore, these data agree with the observed decrease in phosphorylated GCN2 promoted by IMPACT in differentiating N2a cells. Depletion of both GCN2 and IMPACT resulted in an intermediary phenotype in both growth and differentiation medium. This result may suggest that IMPACT could have roles important for N2a cells differentiation that are independent from GCN2 inhibition.
FIGURE 6.
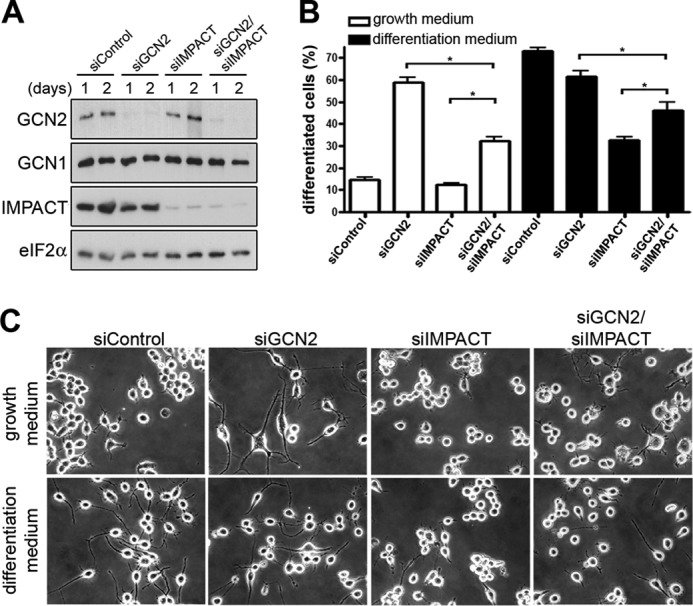
IMPACT and GCN2 regulate neurite outgrowth. A, immunoblots of extracts of N2a cells transfected with siControl (400 pmol of siEGFP), siGCN2 (200 pmol of siGCN2 and 200 pmol of siEGFP), siIMPACT (200 pmol of siIMPACT and 200 pmol of siEGFP), or siIMPACT/siGCN2 (200 pmol of siIMPACT and 200 pmol of siGCN2) were maintained in growth medium for the indicated number of days. B and C, 1 day after transfection, cells were maintained in growth medium or in differentiation medium for an additional day, and neurite outgrowth was analyzed. B, differentiation was quantified as the number of cells with at least one neurite longer than the cell body diameter. Data represent the mean of three independent experiments ± S.E. (error bars); *, p < 0.05 (ANOVA followed by the Newman-Keuls post hoc test). Significant differences (p < 0.05) not shown in the graph: siControl versus siGCN2 and siGCN2 versus siIMPACT in growth medium; siControl versus siIMPACT in differentiation medium. C, representative images of N2a cells transfected with the indicated siRNAs are shown.
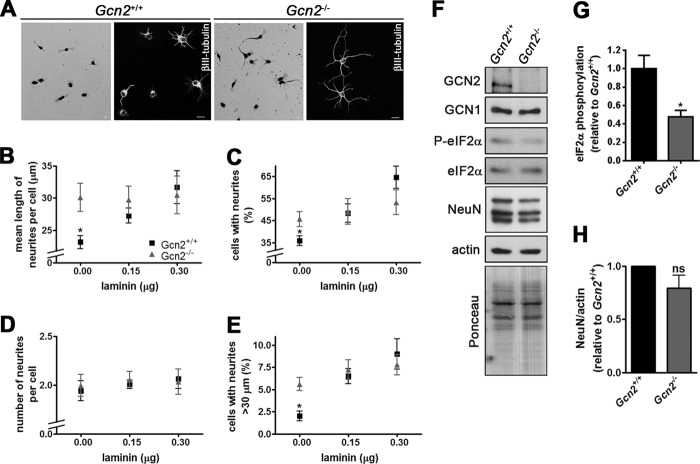
The striking inhibitory effect of GCN2 on N2a neurite outgrowth prompted us to analyze neuritogenesis in primary cultures of hippocampal neurons from Gcn2−/− mice. Primary neurons were plated on poly-l-lysine in the absence or presence of laminin, an inducer of differentiation, and after 1 day in culture, neuritogenesis was assessed by several parameters. In the absence of laminin, Gcn2−/− neurons showed an increased neurite outgrowth phenotype relative to Gcn2+/+ neurons, similar to the effect of the depletion of GCN2 in N2a cells (Fig. 7). Parameters such as mean length of neurites per cell, cells with neurites, and cells with neurites longer than 30 μm were significantly higher in Gcn2−/− neurons (0 μg of laminin in Fig. 7, B, C, and E). The number of neurites per cell was the only morphometric parameter that was not altered significantly in Gcn2−/− neurons (Fig. 7D). Importantly, the distinct phenotype of Gcn2−/− neurons correlated well with reduced levels of eIF2α phosphorylation (Fig. 7, F and G). Gcn2+/+ neurons responded efficiently to laminin stimulation, whereas neuritogenesis in Gcn2−/− neurons was not stimulated further by laminin probably because these neurons have already reached the neurite outgrowth levels achieved in wild type cells by this treatment (Fig. 7B). The effect of the lack of GCN2 seems to be restricted to neurite outgrowth because no significant difference was detected in the expression of NeuN, a transcription factor that is up-regulated during neuronal differentiation (Fig. 7, F and H).
FIGURE 7.
Gcn2−/− neurons show increased neuritogenesis. A, representative images of primary hippocampal neurons isolated from Gcn2+/+ or Gcn2−/− mice grown for 1 day in vitro (div) and stained with hematoxylin (left panels) or labeled with anti-βIII-tubulin (right panels). Scale bars, 10 μm. B–E, morphometric analysis of 1 div cultures of Gcn2+/+ or Gcn2−/− hippocampal neurons plated on the indicated amounts of laminin. Approximately 300 cells/sample were analyzed. Morphometric analyses were performed with ImageJ (National Institutes of Health, Bethesda, MD) and NeuronJ plugin. Data are presented as mean ± S.E. (error bars) from at least three independent experiments. *, p < 0.05 Gcn2+/+ versus Gcn2−/− for the indicated laminin amount (Student's t test). F, immunoblots of extracts of primary Gcn2+/+ or Gcn2−/− hippocampal neurons of 1 div for the indicated proteins. The Ponceau-stained membrane of the 10% SDS-PAGE is shown at the bottom. G, quantification of phosphorylated eIF2α relative to total eIF2α levels. The ratio of P-eIF2α/eIF2α in wild-type neurons was set to 1. Data are presented as mean ± S.E. from at least four independent experiments. *, p < 0.05 (Student's t test). H, quantification of NeuN levels normalized by actin. NeuN expression in wild type neurons was set to 1. ns, nonsignificant (Student's t test).
These results, together with the siRNA data in N2a cells, unambiguously showed that GCN2 mediates a strong negative control over neurite outgrowth in the absence of stimulation, whereas IMPACT down-regulates GCN2 promoting neuronal differentiation upon stimulation. Thus, IMPACT and GCN2 have opposite functions in neuritogenesis.
DISCUSSION
In this work we characterized endogenous neuronal IMPACT as an inhibitor of GCN2, and importantly, we provided evidence that these proteins participate in opposite manners in an early step in neuronal differentiation. We also determined that the expression of IMPACT increases in parallel with the expression of differentiation markers, such as MAP2 and PSD-95. The increased abundance of IMPACT may promote translation by maintaining low levels of active GCN2 in a timely manner to support neurite outgrowth. Interestingly, Impact is an imprinted gene in rodents, and imprinted genes are generally involved with developmental processes (18).
We have demonstrated that GCN2 is a strong negative regulator of neuritogenesis, in both a neuronal-like cell line and in primary neurons. Also, the response to a neuritogenic stimulus such as laminin was altered in Gcn2−/− neurons, with the neurons appearing as nonresponsive to low amounts of laminin. Likely then, the fine tuning of this phenomenon is altered in the absence of GCN2 and possibly in neurons that express high levels of IMPACT (20). Neurite outgrowth, a fundamental event in brain development, is accomplished by the integrated response to a multitude of signals that lead to an appropriate pattern of connections. Increasing evidence demonstrates that several pathways that control translation are the basis of the tight regulation of neurite outgrowth and regeneration. Especially, signals from the extracellular milieu have a special role in promoting or inhibiting neurite growth. The response to these cues has been shown to depend on translational control in several cases. For example, neurite growth in response to brain-derived neurotrophic factor, and insulin is dependent on mTOR-regulated translation (31). Although a large variety of molecules has been associated with neurite growth and retraction, very few are known inhibitors of neuritogenesis. A few examples are cytoskeleton-associated molecules such as Rho GTPases RhoA and Rac3 (32) and P-Rex1 (33). Interestingly, one noncytoskeleton-related protein described to inhibit neurite development is the transcription factor ATF5. ATF5 promotes neuroprogenitor cell expansion while inhibiting neurite outgrowth and differentiation induced by nerve growth factor (34, 35). This transcription factor is one of the few proteins whose mRNA translation is regulated by upstream ORFs and stimulated by eIF2α phosphorylation in a manner similar to ATF4 (36). ATF5 is also up-regulated by transcriptional activation mediated by ATF4. Both ATF4 and ATF5 interfere with cAMP-induced responses, one of the most important signaling pathways associated with neuronal differentiation. We have provided here ample evidence that GCN2 regulates ATF4 levels in N2a cells and that active GCN2 decreases upon differentiation. The lack of GCN2 or its inhibition by IMPACT may then promote neuritogenesis by lowering the expression of ATF4 and ATF5.
Along the differentiation process of both N2a cells and primary hippocampal neurons we were not able to observe the expected decrease in eIF2α phosphorylation that should follow GCN2 inactivation. It is possible that the other three eIF2α kinases (i.e. PERK, PKR, and HRI) could also be modulated during differentiation. Indeed, we found that during differentiation of N2a cells PERK was activated, which could mask the effect of decreased GCN2 activation on the overall levels of eIF2α phosphorylation. We also observed that the amount of polysomes was slightly reduced after differentiation (Fig. 5) in contrast to what would be expected from GCN2 inactivation. This case is more complicated to interpret because the translational level after differentiation is the result of the modulation of several regulatory mechanisms besides eIF2α phosphorylation (37, 38). However, the study of IMPACT depletion in differentiated cells indicated that IMPACT regulation of GCN2 results in reduced levels of eIF2α phosphorylation and improved translation initiation. Furthermore, reduced levels of eIF2α phosphorylation in Gcn2−/− neurons (Fig. 7, F and G) are consistent with the idea that GCN2 is functioning as a repressor of neuritogenesis by phosphorylating eIF2α. These observations taken together may suggest that a subpopulation of the eIF2α molecules may be the target of the GCN2-mediated effect on neuritogenesis and its regulation by IMPACT. Supporting this hypothesis, it has been shown that a subpopulation of the general translation regulator eIF4E-binding protein (4E-BP) molecules can be phosphorylated in neuronal growth cones after netrin-1 stimulation, without affecting 4E-BP molecules in other parts of the neuron (39).
Our findings that GCN2 functions as a strong negative regulator of neuritogenesis resembles the negative control exerted by GCN2 and eIF2α phosphorylation over the passage from short term to long term forms of synaptic plasticity and memory (16). Late long term potentiation and long term memory are prevented by pharmacologically induced increase in eIF2α phosphorylation (17, 40). The induction of late long term potentiation or long term memory results in decreased GCN2 activation (16, 17), and this indicates that there is an inhibitory barrier exerted by a basally activated GCN2. In this work, we also found an analogous inhibitory barrier dictated by a previously activated GCN2 that is inhibiting neuritogenesis and that is eventually overcome by IMPACT.
We demonstrated here that in differentiating cells IMPACT increasingly associates with polysomes. The shift in distribution of IMPACT was apparent despite there being more overall IMPACT in differentiating cells. It seems likely then that differentiation of N2a cells signals for the association of IMPACT with translating ribosomes. The mechanism that drives IMPACT molecules to associate with polysomes upon differentiation remains to be determined. The polysomal association of Yih1 in yeast has been recently demonstrated (41). Our data then further support the functional similarities of the two orthologs.
Based on studies in yeast, it has been proposed that GCN2 is activated with the aid of GCN1 when both proteins are bound to translating ribosomes (10, 42–44). Interestingly, we found that down-regulation of GCN2 activity in differentiating N2a cells was accompanied by a reduced fraction of GCN2 present in polysomal fractions (Fig. 5E). However, when taking into consideration the concomitant reduction in the amount of polysomes during differentiation, it seems that GCN2 remains associated with polysomes (Fig. 5F). This observation suggests that IMPACT may disrupt GCN1-GCN2 interaction on polysomes without GCN2 release. In fact, it has been demonstrated in yeast that Yih1 overexpression does not displace GCN2 from ribosomes (41). GCN1 distribution remained unchanged after differentiation; and when considering the reduction in polysomes, a slight increase in GCN1 association with translating ribosomes was noticed. The fact that GCN2 is not following the mentioned change in GCN1 distribution suggests that IMPACT might function by preventing GCN2 binding to GCN1 on polysomes.
Altogether, this work provided evidence that the pathway involving GCN2 and its inhibitor IMPACT contributes to the complex mechanism of proper differentiation of neuronal cells. Finding neuronal signals that regulate the GCN2-IMPACT module of translational control will be important for dissecting this pathway in more detail in the future.
Acknowledgments
We thank Dr. Evelyn Sattlegger for helpful discussions and for sharing unpublished data regarding Yih1, Dr. Douglas Cavener (Pennsylvania State University) for the gift of the Gcn2−/− animals, and Dr. Vilma R. Martins (International Research Center, Hospital A. C. Camargo) for the use of the laboratory facilities.
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (to B. A. C. and G. N. M. H.).

This article contains supplemental Fig. S1.
- P-eIF2α
- phosphorylated eIF2α
- MEF
- mouse embryonic fibroblast
- P/M
- polysome/monosome ratio
- rpS6
- ribosomal protein S6.
REFERENCES
- 1. Sonenberg N., Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 5. Deng J., Harding H. P., Raught B., Gingras A.-C., Berlanga J. J., Scheuner D., Kaufman R. J., Ron D., Sonenberg N. (2002) Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 12, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 6. Jiang H.-Y., Wek R. C. (2005) Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 280, 14189–14202 [DOI] [PubMed] [Google Scholar]
- 7. Jiang H.-Y., Wek R. C. (2005) GCN2 phosphorylation of eIF2α activates NF-κB in response to UV irradiation. Biochem. J. 385, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donnelly N., Gorman A. M., Gupta S., Samali A. (2013) The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marton M. J., Crouch D., Hinnebusch A. G. (1993) GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 13, 3541–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sattlegger E., Hinnebusch A. G. (2005) Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 280, 16514–16521 [DOI] [PubMed] [Google Scholar]
- 11. Kubota H., Ota K., Sakaki Y., Ito T. (2001) Budding yeast GCN1 binds the GI domain to activate the eIF2α kinase GCN2. J. Biol. Chem. 276, 17591–17596 [DOI] [PubMed] [Google Scholar]
- 12. Brenk M., Scheler M., Koch S., Neumann J., Takikawa O., Häcker G., Bieber T., von Bubnoff D. (2009) Tryptophan deprivation induces inhibitory receptors ILT3 and ILT4 on dendritic cells favoring the induction of human CD4+CD25+ Foxp3+ T regulatory cells. J. Immunol. 183, 145–154 [DOI] [PubMed] [Google Scholar]
- 13. Muller A. J., Sharma M. D., Chandler P. R., Duhadaway J. B., Everhart M. E., Johnson B. A., 3rd, Kahler D. J., Pihkala J., Soler A. P., Munn D. H., Prendergast G. C., Mellor A. L. (2008) Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3-dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 105, 17073–17078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hao S., Sharp J. W., Ross-Inta C. M., McDaniel B. J., Anthony T. G., Wek R. C., Cavener D. R., McGrath B. C., Rudell J. B., Koehnle T. J., Gietzen D. W. (2005) Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307, 1776–1778 [DOI] [PubMed] [Google Scholar]
- 15. Maurin A.-C., Jousse C., Averous J., Parry L., Bruhat A., Cherasse Y., Zeng H., Zhang Y., Harding H. P., Ron D., Fafournoux P. (2005) The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 1, 273–277 [DOI] [PubMed] [Google Scholar]
- 16. Costa-Mattioli M., Gobert D., Harding H., Herdy B., Azzi M., Bruno M., Bidinosti M., Ben Mamou C., Marcinkiewicz E., Yoshida M., Imataka H., Cuello A. C., Seidah N., Sossin W., Lacaille J.-C., Ron D., Nader K., Sonenberg N. (2005) Translational control of hippocampal synaptic plasticity and memory by the eIF2α kinase GCN2. Nature 436, 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., Krnjević K., Lacaille J.-C., Nader K., Sonenberg N. (2007) eIF2α phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagiwara Y., Hirai M., Nishiyama K., Kanazawa I., Ueda T., Sakaki Y., Ito T. (1997) Screening for imprinted genes by allelic message display: identification of a paternally expressed gene impact on mouse chromosome 18. Proc. Natl. Acad. Sci. U.S.A. 94, 9249–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pereira C. M., Sattlegger E., Jiang H.-Y., Longo B. M., Jaqueta C. B., Hinnebusch A. G., Wek R. C., Mello L. E., Castilho B. A. (2005) IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J. Biol. Chem. 280, 28316–28323 [DOI] [PubMed] [Google Scholar]
- 20. Bittencourt S., Pereira C. M., Avedissian M., Delamano A., Mello L. E., Castilho B. A. (2008) Distribution of the protein IMPACT, an inhibitor of GCN2, in the mouse, rat, and marmoset brain. J. Comp. Neurol. 507, 1811–1830 [DOI] [PubMed] [Google Scholar]
- 21. Sattlegger E., Swanson M. J., Ashcraft E. A., Jennings J. L., Fekete R. A., Link A. J., Hinnebusch A. G. (2004) YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 279, 29952–29962 [DOI] [PubMed] [Google Scholar]
- 22. Zhang P., McGrath B. C., Reinert J., Olsen D. S., Lei L., Gill S., Wek S. A., Vattem K. M., Wek R. C., Kimball S. R., Jefferson L. S., Cavener D. R. (2002) The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roffé M., Beraldo F. H., Bester R., Nunziante M., Bach C., Mancini G., Gilch S., Vorberg I., Castilho B. A., Martins V. R., Hajj G. N. (2010) Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc. Natl. Acad. Sci. U.S.A. 107, 13147–13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopes M. H., Hajj G. N., Muras A. G., Mancini G. L., Castro R. M., Ribeiro K. C., Brentani R. R., Linden R., Martins V. R. (2005) Interaction of cellular prion and stress-inducible protein 1 promotes neuritogenesis and neuroprotection by distinct signaling pathways. J. Neurosci. 25, 11330–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Romano P. R., Garcia-Barrio M. T., Zhang X., Wang Q., Taylor D. R., Zhang F., Herring C., Mathews M. B., Qin J., Hinnebusch A. G. (1998) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol. 18, 2282–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berlanga J. J., Santoyo J., De Haro C. (1999) Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem. 265, 754–762 [DOI] [PubMed] [Google Scholar]
- 27. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 28. Stefani G., Fraser C. E., Darnell J. C., Darnell R. B. (2004) Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khandjian E. W., Huot M.-E., Tremblay S., Davidovic L., Mazroui R., Bardoni B. (2004) Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. U.S.A. 101, 13357–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zalfa F., Achsel T., Bagni C. (2006) mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr. Opin. Neurobiol. 16, 265–269 [DOI] [PubMed] [Google Scholar]
- 31. Takei N., Inamura N., Kawamura M., Namba H., Hara K., Yonezawa K., Nawa H. (2004) Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J. Neurosci. 24, 9760–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hajdo-Milasinović A., Ellenbroek S. I., van Es S., van der Vaart B., Collard J. G. (2007) Rac1 and Rac3 have opposing functions in cell adhesion and differentiation of neuronal cells. J. Cell Sci. 120, 555–566 [DOI] [PubMed] [Google Scholar]
- 33. Waters J. E., Astle M. V., Ooms L. M., Balamatsias D., Gurung R., Mitchell C. A. (2008) P-Rex1: a multidomain protein that regulates neurite differentiation. J. Cell Sci. 121, 2892–2903 [DOI] [PubMed] [Google Scholar]
- 34. Angelastro J. M., Ignatova T. N., Kukekov V. G., Steindler D. A., Stengren G. B., Mendelsohn C., Greene L. A. (2003) Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J. Neurosci. 23, 4590–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greene L. A., Lee H. Y., Angelastro J. M. (2009) The transcription factor ATF5: role in neurodevelopment and neural tumors. J. Neurochem. 108, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., Wek R. C. (2008) Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 37. Park K. K., Liu K., Hu Y., Smith P. D., Wang C., Cai B., Xu B., Connolly L., Kramvis I., Sahin M., He Z. (2008) Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bidinosti M., Ran I., Sanchez-Carbente M. R., Martineau Y., Gingras A.-C., Gkogkas C., Raught B., Bramham C. R., Sossin W. S., Costa-Mattioli M., DesGroseillers L., Lacaille J.-C., Sonenberg N. (2010) Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol. Cell 37, 797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leung K.-M., van Horck F. P. G., Lin A. C., Allison R., Standart N., Holt C. E. (2006) Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 9, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bartsch D., Ghirardi M., Skehel P. A., Karl K. A., Herder S. P., Chen M., Bailey C. H., Kandel E. R. (1995) Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83, 979–992 [DOI] [PubMed] [Google Scholar]
- 41. Waller T., Lee S. J., Sattlegger E. (2012) Evidence that Yih1 resides in a complex with ribosomes. FEBS J. 279, 1761–1776 [DOI] [PubMed] [Google Scholar]
- 42. Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G. (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell. Biol. 17, 4474–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramirez M., Wek R. C., Hinnebusch A. G. (1991) Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu S., Wek R. C. (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J. Biol. Chem. 273, 1808–1814 [DOI] [PubMed] [Google Scholar]