Background: Syk is a tyrosine kinase with both tumor promoting and tumor suppressing activities in cancer cells.
Results: Protein kinase A is phosphorylated on a C-terminal tyrosine by Syk.
Conclusion: The phosphorylation of PKA inhibits its activity and its ability to activate CREB.
Significance: The phosphorylation by Syk of PKA inhibits its participation in downstream signaling pathways.
Keywords: Bcl-2, CREB, Cyclic AMP (cAMP), Molecular Dynamics, Phosphorylation, Protein Kinase A (PKA), Tyrosine-protein Kinase (Tyrosine Kinase)
Abstract
The Syk protein-tyrosine kinase can have multiple effects on cancer cells, acting in some as a tumor suppressor by inhibiting motility and in others as a tumor promoter by enhancing survival. Phosphoproteomic analyses identified PKA as a Syk-specific substrate. Syk catalyzes the phosphorylation of the catalytic subunit of PKA (PKAc) both in vitro and in cells on Tyr-330. Tyr-330 lies within the adenosine-binding motif in the C-terminal tail of PKAc within a cluster of acidic amino acids (DDYEEEE), which is a characteristic of Syk substrates. The phosphorylation of PKAc on Tyr-330 by Syk strongly inhibits its catalytic activity. Molecular dynamics simulations suggest that this additional negative charge prevents the C-terminal tail from interacting with the substrate and the nucleotide-binding site to stabilize the closed conformation of PKAc, thus preventing catalysis from occurring. Phosphoproteomic analyses and Western blotting studies indicate that Tyr-330 can be phosphorylated in a Syk-dependent manner in MCF7 breast cancer cells and DT40 B cells. The phosphorylation of a downstream substrate of PKAc, cAMP-responsive element-binding protein (CREB), is inhibited in cells expressing Syk but can be rescued by a selective inhibitor of Syk. Modulation of CREB activity alters the expression of the CREB-regulated gene BCL2 and modulates cellular responses to genotoxic agents. Thus, PKA is a novel substrate of Syk, and its phosphorylation on Tyr-330 inhibits its participation in downstream signaling pathways.
Introduction
Syk is a nonreceptor protein-tyrosine kinase best known as a regulator of signaling from immune recognition receptors and thus is a well known modulator of immune cell development and signaling (1, 2). A role for Syk in tumorigenesis has been more enigmatic. In some cancer cells, such as breast and melanoma, Syk has been reported to be a tumor suppressor (3, 4). Syk is expressed in normal breast tissue and benign breast lesions but is frequently absent from invasive breast cancer tissues and cell lines (3, 5, 6). However, Syk also can protect breast cancer cells from apoptosis induced by TNF-α (7) and, in fact, acts in many cancer cells as a proto-oncogene with a major role in the promotion of cell survival (8–13). This has led to trials of Syk inhibitors as therapeutic agents for the treatment of hematopoietic malignancies (14).
The mechanisms by which Syk suppresses breast cancer metastasis or promotes cell survival are unclear but are undoubtedly a product of the substrates that it phosphorylates. Thus, the specific substrates that Syk phosphorylates are of considerable interest. To identify candidate substrates for Syk, we have carried out a series of mass spectrometry-based phosphoproteomic screens to identify proteins phosphorylated in cells in a manner dependent on the expression of Syk (15, 16). These screens included one where phosphopeptides derived from cellular phosphoproteins were dephosphorylated and then rephosphorylated in solution by recombinant Syk (15). These studies confirmed a strong preference of Syk for the phosphorylation of tyrosines located within highly acidic sites. Interestingly, included among the substrates identified in these screens was PKA, which was phosphorylated on a tyrosine (Tyr-330) in the C-terminal tail of the catalytic subunit that is located within a primary sequence that matches well the substrate specificity of Syk.
PKA is a serine/threonine kinase that is involved in diverse cellular functions including but not limited to the regulation of metabolism, gene transcription, cell cycle progression, and apoptosis. PKA normally exists in the form of a heterotetramer comprising two catalytic and two regulatory subunits that is activated by the binding of cAMP to the regulatory subunits, resulting in the release of active catalytic subunits from the complex (17).
The catalytic subunit (PKAc)3 also is regulated by cis-regulatory elements that are located in its C-terminal tail: the N-terminal lobe tether, the active site tether (AST), and the C-terminal lobe tether. The AST, which acts as a gate for nucleotide entry and egress, is conserved in sequence and function among members of the AGC family of serine/threonine kinases. However, although PKAc has a tyrosine within the AST at position 330, the other AGC kinases have a phenylalanine in the analogous location (18, 19). When ATP binds to PKAc, the AST becomes ordered such that Tyr-330 is positioned close to the adenine-binding pocket, where it is important for recognition of both the nucleotide and peptide substrates and for optimal catalytic efficiency (20–22) (Fig. 1).
FIGURE 1.
PKAc has a C-terminal tail that extends around the catalytic domain. PKAc, shown in cartoon representation, has an N-domain (top) and a larger C-domain (bottom). The C-terminal tail comprises the N-terminal lobe tether (purple), the AST (magenta), and the C-terminal lobe tether (gold). Tyr-330 and ATP are shown in vector representation. ATP is coordinated by two manganese ions shown as green spheres.
Among the many substrates of PKAc is the cAMP response element-binding protein (CREB), a ubiquitously expressed transcription factor. CREB, which mediates many of the effects of cAMP on gene transcription, binds to the conserved cAMP response element on DNA and is activated when phosphorylated on Ser-133 by PKAc (23). Phosphorylation of CREB recruits the transcriptional co-activator and histone acetyltransferase CREB-binding protein (CBP) to induce the transcription of multiple target genes (24, 25). Included among these target genes is BCL2, which encodes the anti-apoptotic protein Bcl-2.
In this study, we identified and characterized PKAc as a substrate of Syk. Through phosphorylating Tyr-330, Syk strongly inhibits the activity of PKAc, attenuates the phosphorylation of CREB in the nucleus of breast cancer cells, and down-regulates the expression of Bcl-2. This study reveals a new mechanism by which Syk can regulate signaling pathways in cells.
EXPERIMENTAL PROCEDURES
Antibodies and Cell Lines
Antibody against Syk was obtained from Santa Cruz Biotechnology. Antibodies for CREB, CREB phosphorylated on Ser-133, and poly(ADP-ribose) polymerase (PARP) were from Cell Signaling Technology. Anti-GAPDH was from Ambion. Antibodies against PKAc were from BD Biosciences or Santa Cruz Biotechnology. The Bcl-2 antibody was from BD Biosciences. Phosphotyrosine antibodies (4G10 and PT-66) were from Millipore. The PKAc substrate LRRASLG (Kemptide), GST-PKAc, and GST-Syk were purchased from Sigma.
Plasmids expressing Syk-EGFP and Syk-EGFP-NLS were constructed as described previously (26, 27). The His-PKAc expression plasmid was a generous gift from Dr. Susan Taylor (University of California, San Diego). His-PKAc(Y330F) and His-PKAc(Y330E) expression plasmids were generated by site-directed mutagenesis. A line of MCF7 cells lacking endogenous Syk was purchased from BD Biosciences as described previously (7). These cells are referred to as MCF7-B cells. MCF7 cells expressing endogenous Syk were purchased from ATCC (MCF7-A). All MCF7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/ml penicillin G, and 100 μg/ml streptomycin. To generate stable Syk-EGFP-NLS-expressing cells, Syk-deficient MCF7-B cells (7) were transfected with a plasmid expressing Syk-EGFP-NLS, selected by treatment with G418, and screened for Syk-EGFP-NLS expression by fluorescence microscopy and Western blotting. MCF7-B cells with tetracycline-regulated Syk-EGFP expression were constructed previously using the T-REXTM system (Invitrogen) (28). The cells were treated with 1 μg/ml tetracycline to induce Syk-EGFP expression where indicated. Wild-type DT40 B cells, Syk-deficient DT40 cells, Syk- and Lyn-deficient DT40 cells, and Syk- and Lyn-deficient DT40 cells transfected to stably express Syk-EGFP were as described previously (27).
Immunoprecipitation Assays
For immunoprecipitation assays, the cells were collected in a lysis buffer containing 50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1 mm PMSF, 10 μg/ml each of aprotinin and leupeptin, and 1 mm Na3VO4. The lysates were incubated with protein A-Sepharose beads preconjugated with primary antibody for 4 h at 4 °C. The protein/bead mixture was washed four times with lysis buffer, and then the bound proteins were separated by SDS-PAGE. PKAc or phosphotyrosine were detected by Western blotting using the appropriate horseradish peroxidase-conjugated secondary antibodies and chemiluminescence reagents (PerkinElmer Life Sciences).
Protein Kinase Assays
For the phosphorylation of PKAc in vitro, 1 μg of GST-Syk and 1 μg of GST-PKA were incubated together in a 50-μl reaction containing 25 mm HEPES, pH 7.2, 5 mm MnCl2, 0.5 mm Na3VO4, 0.02 mg/ml leupeptin, 0.02 mg/ml aprotinin, and 100 μm [γ-32P]ATP for the indicated times. Reactions were terminated by the addition of SDS sample buffer. Phosphoproteins were separated by SDS-PAGE and detected by autoradiography. To assess changes in PKAc activity, aliquots of the initial reaction were diluted into a reaction buffer containing 50 μm LRRASLG, 10 mm MgCl2, 50 mm Tris/HCl, pH 7.5, 5 mm p-nitrophenylphosphate, 50 μm [γ-32P]ATP, and 0.1 mg/ml BSA. Phosphopeptides were isolated on P81 phosphocellulose paper squares (Millipore) and counted using a scintillation counter. The reactions were run under conditions in which the rate of peptide phosphorylation was linear with respect to both time and PKAc concentration.
In some experiments, PKAc phosphorylated by Syk was isolated by adsorption to beads containing immobilized anti-phosphotyrosine 4G10 (Millipore) in a buffer containing 50 mm Tris/HCl, pH 7.5, 100 mm NaCl, 1% Triton X-100, 0.1 mg/ml BSA, 1 mm PMSF, 10 μg/ml each of aprotinin and leupeptin, 1 mm NaF, and 1 mm Na3VO4 for 4 h at 4 °C. The protein/bead mixture was washed four times with the same buffer. PKAc was eluted with the same buffer containing 50 mm phenylphosphate and analyzed by SDS-PAGE and Western blotting. The concentration of phosphorylated PKAc was determined by Western blotting and image analysis (ImageJ) in comparison with known concentrations of PKAc.
For PKA assays from cell lysates, the cells were lysed in buffer as described above. Aliquots were incubated in a 50-μl reaction buffer containing 50 μm LRRASLG, 10 μm cAMP, 10 mm MgCl2, 50 mm Tris/HCl, pH 7.5, 5 mm p-nitrophenylphosphate, and 50 μm [γ-32P]ATP at 30 °C for 3 min. The reactions were terminated by the addition of 70 μl of 3.2% TCA. Phosphopeptides were isolated using P81 paper. His-PKAc, His-PKA-Y330F, and His-PKA-Y330E were isolated from Bl21 Escherichia coli cell lysates by affinity purification using the MagneHis purification system (Promega). The elution products were assayed for kinase activity as described above for PKAc phosphorylated by Syk.
CREB Phosphorylation Assays
MCF7-B cells or MCF7-B cells expressing Syk-EGFP or Syk-EGFP-NLS were treated with 25 μm forskolin or vehicle for the indicated times and then lysed on ice in 10 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1% sodium deoxycholate, 1% Triton x-100, 0.1% SDS, 5 mm EDTA, 1 mm PMSF, 10 μg/ml each of aprotinin and leupeptin, and 1 mm Na3VO4. Proteins in supernatants collected following centrifugation at 18,000 × g for 10 min were separated by SDS-PAGE, transferred to PVDF membranes, and analyzed by Western blotting with the indicated antibodies. Where indicated, the cells were pretreated with vehicle, 20 μm H-89, or 25 μm piceatannol for 2 h before the addition of forskolin.
Bcl-2 Expression and Apoptosis Assays
For comparisons of Bcl-2 mRNA levels, the cells were lysed in Qiazol lysis reagent (Qiagen) for RNA extraction as described in the manufacturer's instructions. RNA (2 μg) was reverse transcribed using the iScript one-step RT-PCR kit (Bio-Rad). cDNA products were amplified for 30 cycles using Phusion high fidelity DNA polymerase (New England Biolabs) and the following primers: 5′-ACTTGTGGCCCAGATAGGCACCCA and 5′-CGACTTCGCCGAGATGTCCAGCCAG.
For the analysis of caspase-dependent PARP cleavage, cells, treated as indicated in each experiment, were lysed in 10 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 5 mm EDTA, 1 mm PMSF, 10 μg/ml each of aprotinin and leupeptin, and 1 mm Na3VO4 and sheared by passage through a 22-gauge needle. Proteins in supernatants collected following centrifugation at 18,000 × g for 10 min were separated by SDS-PAGE, transferred to PVDF membranes, and analyzed by Western blotting with a PARP antibody. The relative intensities of the bands of cleaved and uncleaved forms of PARP were quantified using ImageJ (National Institutes of Health).
Mass Spectrometric Analyses
MCF7-B cells lacking Syk or expressing Syk-EGFP-NLS were lysed in 1 ml of lysis buffer containing 50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1 mm sodium orthovanadate, 1× phosphatase inhibitor mixture (Sigma), and 10 mm sodium fluoride for 20 min on ice. The various DT40 B cell lines were pretreated with 100 μm pervanadate for 30 min at 37 °C prior to lysis. The cell debris was cleared by centrifugation at 16,100 × g for 10 min, and the supernatant containing soluble proteins was collected. The concentration of the cell lysate was determined using the BCA assay, and the samples were normalized to 5 mg of protein each. The proteins were denatured and reduced by incubating the lysates in 50 mm trimethylammonium bicarbonate containing 0.1% RapiGest and 5 mm dithiothreitol for 30 min at 50 °C. The samples were cooled to room temperature and incubated with 30 mm iodoacetamide for 1 h in the dark to alkylate the cysteines. The pH was adjusted to 8.0, and the samples were digested with 1:100 ratio of trypsin to proteins for 14 h at 37 °C. Following digestion, RapiGest was removed by decreasing the pH to below 3.0 with 1 n hydrochloric acid, incubating the samples at 37 °C for 40 min, centrifuging the sample for 10 min at 16,100 × g, and collecting the supernatant. The pH of the samples was adjusted to 7.4 with 1 m Tris/HCl, pH 8.0. 100 μl of the PT66 phosphotyrosine antibody beads slurry (Sigma) was added to the peptide samples and incubated overnight at 4 °C with agitation (29). The supernatant was carefully removed, and the beads were washed twice with 500 μl of the lysis buffer and twice with water. Tyrosine phosphopeptides were eluted by incubating the beads three times with 100 μl of 0.1% TFA with 10 min of vigorous agitation, twice with 100 μl of 0.1% TFA in 50% acetonitrile (10 min each) (30), and twice with 50 μl of 100 mm glycine, pH 2.5 (30 min each with vigorous agitation). All eluates for each sample were combined and dried completely in a SpeedVac. The resulting peptides were then further enriched by the PolyMAC method to isolate phosphopeptides, as described previously (16).
The eluted phosphopeptides were dried, redissolved in 8 μl of 0.5% formic acid and injected into an Eksigent two-dimensional Ultra nanoflow HPLC system. The reverse phase C18 was performed using an in-house C18 capillary column packed with 5-μm C18 Magic beads resin (Michrom; 75-μm inner diameter and 30-cm bed length). The mobile phase buffer consisted of 0.1% HCOOH in ultra pure water with the eluting buffer of 100% CH3CN run over a shallow linear gradient over 60 min with a flow rate of 0.3 μl/min. The electrospray ionization emitter tip was generated on the prepacked column with a laser puller (Model P-2000; Sutter Instrument Co.). The Eksigent Ultra HPLC system was coupled online with a high resolution hybrid linear ion trap Orbitrap mass spectrometer (LTQ-Orbitrap Velos; Thermo Fisher). The mass spectrometer was operated in the data-dependent mode in which a full scan MS (from m/z 300–2000 with the resolution of 30,000) was followed by 20 MS/MS scans of the most abundant ions. Ions with charge state of +1 were excluded. The mass exclusion time was 90 s. The LTQ-Orbitrap raw files were searched directly against the Homo sapiens database with no redundant entries (91,464 entries; human International Protein Index version 3.87) using a combination of SEQUEST algorithm and MASCOT on Proteome Discoverer (version 1.3; Thermo Fisher). For peptides derived from DT40 cells, files were searched against the Gallus gallus database. Peptide precursor mass tolerance was set at 10 ppm, and MS/MS tolerance was set at 0.8 Da. Search criteria included a static modification of cysteine residues of +57.0214 Da and variable modifications of +15.9949 Da to include potential oxidation of methionines and a modification of +79.996 Da on serine, threonine, or tyrosine for the identification of phosphorylation. Searches were performed with full tryptic digestion and allowed a maximum of two missed cleavages on the peptides analyzed from the sequence database. False discovery rates were set below 1% for each analysis. Proteome Discoverer generates a reverse “decoy” database from the same protein database, and any peptides passing the initial filtering parameters that were derived from this decoy database are defined as false positive identifications. The minimum cross-correlation factor filter was then readjusted for each individual charge state separately to optimally meet the predetermined target false discovery rate of 1% based on the number of random false-positive matches from the reversed “decoy” database. Thus, each data set had its own passing parameters. The most likely phosphorylation site localization from CID mass spectra was determined by PhosphoRS algorithm within the Proteome Discoverer 1.3 software.
For the analysis of phosphopeptides derived from DT40 cells, equivalent amounts of a standard peptide library containing 160 peptides4 were spiked into each sample before LC-MS/MS for label free quantification. Sample peptides were normalized to the adjusted intensities of the peptide standards.
Molecular Dynamics Simulation
Two systems, one with and one without Tyr-330 phosphorylation, were built based on Protein Data Bank structure 3FJQ (31), in which PKAc takes the closed and active conformation. Both systems included the protein, ATP, two Mn2+ ions, and water. For the phosphorylated system, the phosphate group of Tyr-330 was placed by calculating the potential energy of the protein complex solvated with the GBSW implicit solvent model (32) as a function of the torsion angle between the ζ-carbon and the bridging oxygen and the torsion angle between the bridging oxygen and the phosphorus atom. The combination that yielded the overall minimum of the system potential energy was chosen, with the result that the phosphate group is oriented toward the protein surface and induces no steric clashes with the rest part of the protein. Both structures were solvated in an 84 Å cubic box of TIP3P water molecules. Na+ and Cl− ions were added to neutralize the system and to generate a salt concentration of 150 mm. After 1000 steps of minimization with an adopted basis Newton-Raphson method, five molecular dynamics trajectories were generated for each system with different initial velocities. Each dynamics trajectory was propagated for 20 ns using the CHARMM 27 force field (33, 34) and the Langevin integrator in the NAMD program (35) with a friction coefficient of 2 ps−1 and a time step of 2 fs. Periodic boundary conditions were imposed, and the Ewald method was used to calculate the electrostatic interactions. The van der Waals interactions were cut off at 10 Å. All of the simulations were carried out at 300 K and 1 atm. The solvent-accessible surface area of the Tyr-330 side chain was measured with a probe radius of 1.6 Å.
RESULTS
Phosphorylation of PKAc by Syk in Vitro
Our previous phosphoproteomic screen using mass spectrometry demonstrated that PKAc could be phosphorylated on tyrosine in Syk-expressing cells and that a tryptic peptide derived from PKAc that contained this tyrosine was a substrate for Syk in vitro (15). This tyrosine, Tyr-330, is located in a region surrounded by acidic residues (DDYEEEE) that matches well the consensus sequence determined for optimal Syk substrates (15). To confirm the ability of Syk to directly phosphorylate PKAc, we conducted an in vitro kinase assay using [γ-32P]ATP with GST-PKA as a substrate and GST-Syk as a catalyst. In this experiment, proteins from the reaction mixture were separated by SDS-PAGE and transferred to a PVDF membrane, which was then treated with 1 m KOH at 65 °C for 2 h to eliminate the phosphates from phosphoserines or phosphothreonines that formed from a low level of PKAc autophosphorylation. GST-Syk catalyzed both an autophosphorylation reaction and the phosphorylation of GST-PKAc (Fig. 2A). The phosphorylation of PKAc was robust (it should be noted for comparative purposes that GST-Syk autophosphorylation leads to its modification on 10 different tyrosines (36)). To determine the stoichiometry of this reaction, we incubated GST-Syk and GST-PKAc in the kinase reaction buffer for varying periods of time up to 40 min. The reaction mixture was fractionated by SDS-PAGE, and bands corresponding to phosphorylated PKAc were excised and counted by liquid scintillation spectrometry. The percentage of PKAc that was tyrosine-phosphorylated was estimated to be 24% for a reaction of 40-min duration (Fig. 2B). Thus, PKAc was efficiently phosphorylated by Syk in vitro.
FIGURE 2.
Phosphorylation of PKAc by Syk in vitro. A, GST-PKA and/or GST-Syk were incubated in a kinase reaction buffer containing [γ-32P]ATP for the indicated times. The proteins were separated by SDS-PAGE and transferred to PVDF membranes, which were incubated in 1 m KOH prior to autoradiography. B, GST-PKAc was incubated with GST-Syk in a kinase reaction buffer containing [γ-32P]ATP for the indicated times. Phosphoproteins were separated by SDS-PAGE and counted in a scintillation counter to determine stoichiometry.
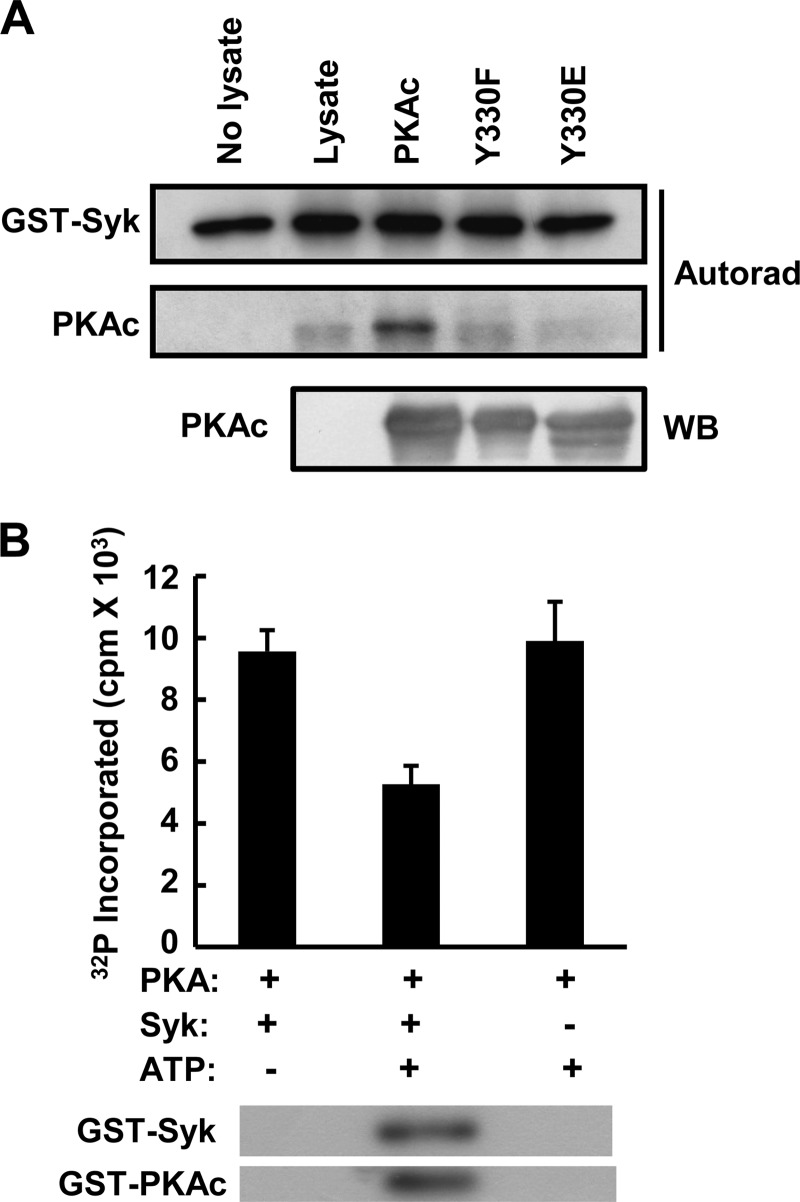
To confirm that PKAc was phosphorylated on Tyr-330 by Syk, we carried out site-directed mutagenesis on a murine His-PKAc expression plasmid to change Tyr-330 to either phenylalanine (His-PKAc(Y330F)) or glutamate (His-PKAc(Y330E)). These plasmids were transformed into Bl21 E. coli cells, and expression was induced by IPTG. Roughly equal expression of His-PKAc, His-PKAc(Y330F), and His-PKAc(Y330E) in the cell lysates was verified by Western blotting (Fig. 3A). These bacterial cell lysates were then incubated with GST-Syk in an in vitro kinase assay using buffer containing [γ-32P]ATP. Syk was able to catalyze the phosphorylation of His-PKAc even in the presence of a large excess of bacterial proteins. Neither His-PKAc(Y330F) nor His-PKAc(Y330E) was detectably phosphorylated by Syk (Fig. 3A). Thus, Tyr-330 was the only site on murine PKAc that was phosphorylated to a significant extent by Syk in vitro.
FIGURE 3.
Syk phosphorylates PKAc on Tyr-330 and inhibits its activity. A, lysates from BL21 cells (Lysate) or from BL21 cells induced to express His-PKAc (PKAc), His-PKAc(Y330F) (Y330F), or His-PKAc(Y330E) (Y330E) were incubated with GST-Syk in a kinase reaction buffer containing [γ-32P]ATP. The reaction products were separated by SDS-PAGE and detected by autoradiography (Autorad). PKAc or PKAc mutants present in the bacterial lysates were detected by Western blotting (WB) with anti-PKAc (WB). B, His-PKAc (PKAc) was preincubated with (+) or without (−) GST-Syk (Syk) and with (+) or without (−) [γ-32P]ATP for 2 h. Aliquots were removed and assayed for PKA activity using LRRASLG as a substrate and for verification of protein phosphorylation by SDS-PAGE and autoradiography (lower panels). The data represent the means and standard errors of experiments performed in triplicate.
Inhibition of PKA Activity by Phosphorylation
To study the effect of Tyr-330 phosphorylation on the activity of PKAc, we first incubated GST-PKAc in a kinase reaction buffer containing 500 μm ATP (and a trace of [γ-32P]ATP) in the presence or absence of GST-Syk for 2 h. The phosphorylation of GST-PKAc by GST-Syk in the presence of ATP was confirmed by autoradiography. Aliquots of each reaction were then taken and assayed using LRRASLG as a substrate to assess the activity of PKAc. The inclusion of GST-Syk in the initial kinase reaction resulted in a significant decrease in the activity of PKAc (Fig. 3B). No phosphorylation of LRRASLG was observed in reactions containing Syk but lacking PKAc (data not shown). Thus, the phosphorylation of PKAc on tyrosine resulted in a loss of activity.
Because GST-PKAc likely was not phosphorylated to 100% in the initial reaction with GST-Syk, the peptide (LRRASLG) phosphorylation assay reflected the activity of a mixture of unphosphorylated and phosphorylated PKAc. To measure more accurately the activity of only the tyrosine-phosphorylated fraction of PKAc, we isolated phospho-GST-PKAc from a reaction containing ATP, GST-Syk, and GST-PKAc using immobilized antibodies against phosphotyrosine. Bound, phosphorylated GST-PKAc was eluted with buffer containing phenylphosphate. As expected, only GST-PKAc that had been preincubated in a kinase reaction with GST-Syk could be specifically eluted from anti-phosphotyrosine beads with phenylphosphate (Fig. 4A). The concentration of phosphorylated GST-PKAc was quantified by the analysis of Western blots in comparison with a set of GST-PKAc standards of known concentration (not shown). Its activity was then compared with that of an equal amount of unphosphorylated enzyme again using LRRASLG as the substrate (Fig. 4B). No PKAc activity could be detected in the in vitro tyrosine-phosphorylated fraction. Thus, the activity of GST-PKAc was inhibited completely by its phosphorylation on Tyr-330.
FIGURE 4.
The phosphorylation of PKAc by Syk abolishes catalytic activity. A, GST-PKAc was incubated with (+Syk) or without (−Syk) GST-Syk in the presence of ATP for 2 h. The reaction products were adsorbed to beads containing immobilized antiphosphotyrosine and then eluted with phenylphosphate. Aliquots of the unbound (lane F), bound (lane B) and increasing amounts of the eluted (E1, 1 μl; E2, 5 μl; and E3, 20 μl) fractions were analyzed by Western blotting with anti-PKAc. B, equal amounts of tyrosine-phosphorylated PKAc (pPKAc) and unphosphorylated PKAc (PKAc) were assayed for catalytic activity using LRRASLG. C, recombinant His-PKAc (PKAc), His-PKAc(Y330F) (Y330F), or His-PKAc(Y330E) (Y330E) were isolated from bacterial lysates and assayed for catalytic activity using LRRASLG as a substrate. Isolation of each kinase was verified by Western blotting (inset). The data represent the means and standard error of three experiments.
To confirm the importance of Tyr-330 for the catalytic activity of PKA, we compared the activities of His-tagged versions of wild-type PKAc with those of His-PKAc(Y330F) and His-PKAc(Y330E). The Y330F mutation was shown previously to reduce, but not abolish, the activity of PKA (20). The Y330E mutation was generated to permanently position an acidic residue at position 330 to mimic a phosphotyrosine. Each was expressed in bacteria, isolated on nickel-chelated beads, and then eluted and incubated in a kinase reaction buffer containing [γ-32P]ATP and LRRASLG. The substitution of Tyr-330 with phenylalanine resulted in a decrease in the specific activity of PKAc as expected. However, the replacement of Tyr-330 with glutamate produced an enzyme with no detectable activity (Fig. 4C). Thus, the presence of an acidic residue, either phosphotyrosine or glutamate, at position 330 of PKAc completely abrogates kinase activity.
Modeling the Structural Effect of Tyr-330 Phosphorylation
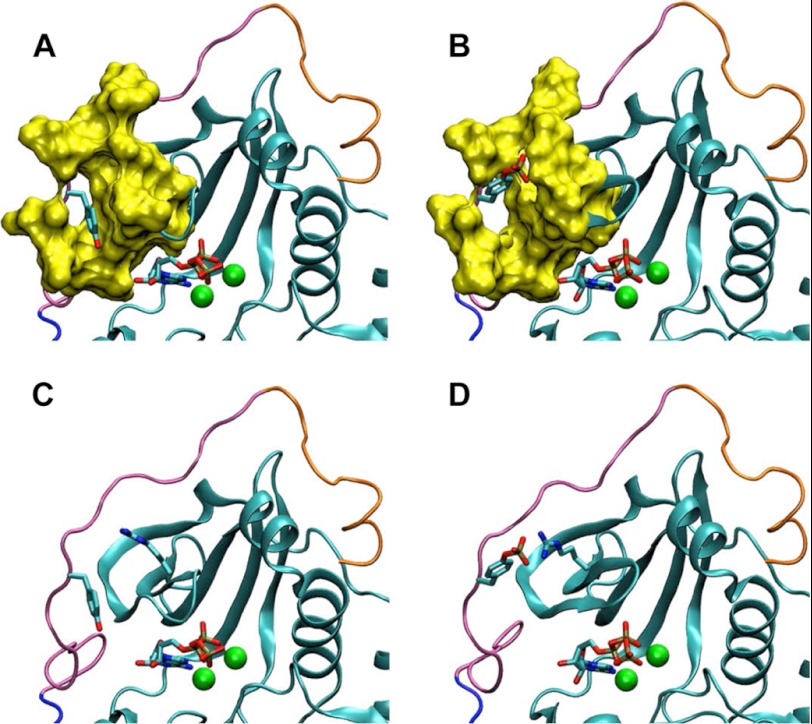
Given the inhibitory effect of Tyr-330 phosphorylation on the activity of PKA, we considered possible structural consequences of phosphorylation by modeling the PKAc-ATP complex with and without the Tyr-330 phosphorylation. We used molecular dynamics simulations to investigate the structural perturbation to PKAc induced by the phosphate group. Computer simulation of a solvated protein generates a more accurate representation of the protein conformational state than a single, static model and thus is a better approach for such an investigation. Five independent molecular dynamics trajectories each of 20 ns were calculated for both the phosphorylated and the unphosphorylated systems starting from the closed, active conformation of PKAc. The use of multiple simulations of shorter duration gives more extensive conformational sampling relative to a single, longer trajectory (37). The phosphate group does induce significant local changes in the structural environment of Tyr-330; however, large scale conformational change was observed for neither the backbone of the C-terminal tail nor the rest of PKAc. The alternative orientation of the Tyr-330 side chain in the two systems is noteworthy. In the unphosphorylated system, the tyrosine side chain is primarily oriented as observed in the initial Protein Data Bank structure, with the phenyl ring tightly packed against the cavity formed by the β-1,2 strands, the nucleotide, and the flanking residues of the AST (Fig. 5, A and C). The solvent-accessible surface area of the side chain averaged over all trajectories measures 78 ± 4 Å2. In contrast, the phosphorylated tyrosine side chain tends to orient perpendicular to the protein surface, resulting in an increased time-averaged solvent-accessible surface area of 115 ± 19 Å2 (Fig. 5, B and D), a substantial increase considering that the exposed surface area of an isolated Tyr side chain is 256 Å2. In two of the five phosphorylated simulations, this more highly solvated orientation is stabilized by a salt bridge interaction with the side chain of Arg-56 from the β-2 strand (Fig. 5D). These results suggest phosphorylation of Tyr-330 leads to an altered side chain conformation and promotes interaction of Tyr-330 with Arg-56.
FIGURE 5.
The phosphorylation of PKAc is predicted to alter the orientation of Tyr-330. Typical orientations of the Tyr-330 side chain in molecular dynamics simulations are shown for the unphosphorylated (A and C) and phosphorylated (B and D) forms of PKAc. The N-terminal lobe and C-terminal tail, colored as in Fig. 1, of PKAc are shown in cartoon representations. Tyr-330 and ATP are shown in vector representations. The manganese ions are colored in green. A and B, the molecular surface formed by residues surrounding Tyr-330 (residues 327–329, 331–334, 48–51, and 56–60) is shown in yellow. C and D, Arg-56, which forms a salt bridge with phosphorylated Tyr-330, is shown in vector representation.
Phosphorylation of PKAc in Cells
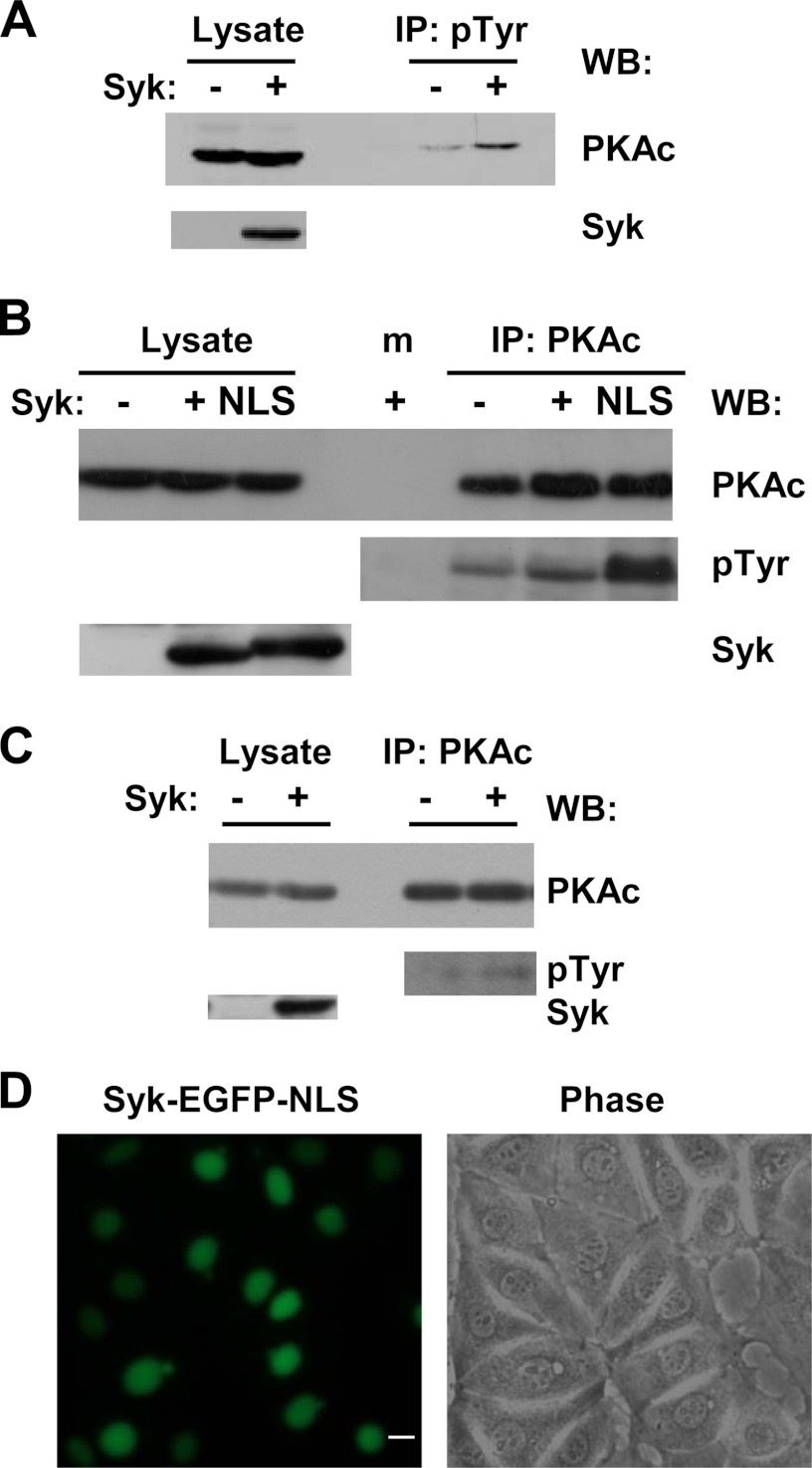
To explore a possible role for Syk in the phosphorylation of PKA in cells, we used MCF7 breast cancer cells as a model. We had previously identified a Syk-deficient line of MCF7 cells (MCF7-B) and had constructed a line of these cells that express a Syk-EGFP fusion protein upon treatment with tetracycline (7, 28). Tyrosine-phosphorylated proteins were immunoprecipitated with antibodies against phosphotyrosine from lysates of cells lacking Syk or induced to express Syk that had been treated with hydrogen peroxide to inhibit tyrosine phosphatases. Immune complexes were separated by SDS-PAGE and probed with antibodies against PKAc. The appearance of PKAc in the anti-phosphotyrosine immune complexes was enhanced in cells induced to express Syk-EGFP, consistent with its phosphorylation by Syk on tyrosine (Fig. 6A). To determine whether this phosphorylation was direct, we immunoprecipitated PKAc from MCF7-B cells expressing or lacking Syk and probed the resulting blot with antibodies against phosphotyrosine. Again, PKAc was found to be phosphorylated on tyrosine, and this phosphorylation was enhanced in cells expressing Syk-EGFP (Fig. 6B). Similarly, PKAc phosphorylation was enhanced in MCF7-A cells that contained normal levels of endogenous Syk as compared with cells lacking the kinase (Fig. 6C).
FIGURE 6.
Phosphorylation of PKA on tyrosine in MCF7 cells. A, phosphotyrosine-containing proteins were immunoprecipitated (IP) from lysates of Syk-deficient MCF7-B (−) or MCF7-B cells induced to express Syk-EGFP (+) with antibodies against phosphotyrosine (pTyr). Proteins in the lysates and immune complexes (IP) were probed with antibodies against PKAc or Syk. B, PKAc was immunoprecipitated from lysates of Syk-deficient MCF7-B (−) or MCF7-B cells expressing Syk-EGFP (+) or Syk-EGFP-NLS (NLS). Proteins in the lysates and immune complexes (IP) were probed with antibodies against PKAc, phosphotyrosine (pTyr), or Syk. A mock IP (m) was carried out in the absence of added antibody to detect possible nonspecific interactions. C, PKAc was immunoprecipitated from lysates of Syk-deficient MCF7-B (−) or MCF7-A cells. Proteins in the lysates and immune complexes (IP) were probed with antibodies against PKAc, phosphotyrosine (pTyr), or Syk. D, MCF7 cells stably expressing Syk-EGFP-NLS were viewed by fluorescence (left panel) or phase contrast microscopy (right panel). WB, Western blotting.
When released from its associated regulatory subunits by cAMP, the catalytic subunit (PKAc) is active and capable of catalyzing the phosphorylation of numerous protein substrates. These substrates include CREB, which PKAc phosphorylates after it translocates into the nucleus (23). A fraction of Syk also is present in the nuclear compartment of B cells and breast epithelial cells (26, 28). To look for phosphorylation of PKAc in the nucleus, we generated an MCF7-B cell line that stably expressed Syk-EGFP-NLS, a form of Syk-EGFP with a nuclear localization sequence derived from SV40 large T antigen appended to the C terminus (26). An examination of these cells by fluorescence microscopy confirmed a localization of Syk-EGFP-NLS primarily to the nucleus (Fig. 6D). Western blot analysis of anti-PKAc immune complexes isolated from cells expressing this nuclear localized Syk revealed a robust phosphorylation of PKAc on tyrosine (Fig. 6B).
To confirm the site of phosphorylation on PKAc as Tyr-330, we digested proteins isolated from MCF7-B cells and from cells expressing Syk-EGFP-NLS with trypsin, isolated the total population of phosphotyrosine-containing peptides, and searched for peptides derived from PKAc by mass spectrometry. Only one phosphotyrosine-containing peptide originating from PKAc was identified, and it corresponded to the tryptic peptide that contained Tyr-330 (GPGDTSNFDDYEEEEIR) (Fig. 7). No other phosphopeptides derived from PKAc were identified, and no PKAc-derived phosphopeptides could be found in samples isolated from MCF7-B cells lacking Syk.
FIGURE 7.
MS/MS spectrum coverage of PKAc peptide GPGDTSNFDDpYEEEEIR. The spectrum includes b-ions and y-ions. The spectrum ion coverage phosphoRS software (Thermo) assigned 100% probability to Tyr-11 as the site of phosphorylation.
To further examine the selectivity and specificity of PKA phosphorylation by Syk, we compared by mass spectrometric analysis the phosphorylation of PKAc Tyr-330 in DT40 B cells. In this study, we compared quantitatively wild-type DT40 cells that express endogenous Syk and the Src family kinase, Lyn, with engineered DT40 cells lacking Syk or both Syk and Lyn because of gene disruption. The Tyr(P)-330-containing phosphopeptide was identified in lysates from wild-type DT40 cells, but not from cells either lacking the expression of Syk or of both Syk and Lyn. Restoration of Syk to the double knock-out cells by transfection of an expression plasmid coding for Syk-EGFP (27) restored the phosphorylation of PKA on Tyr-330.
Effect of Syk on the Phosphorylation of CREB
Active PKAc in the nucleus catalyzes the phosphorylation of CREB on Ser-133 to promote the recruitment of CBP to stimulate gene transcription (23, 24). To determine whether the phosphorylation of CREB was affected by the presence of Syk, we treated MCF7-B cells lacking Syk or expressing either Syk-EGFP or Syk-EGFP-NLS with forskolin for different periods of time to activate adenylate cyclase to generate cAMP and activate PKA. The phosphorylation status of CREB was then analyzed by Western blotting (Fig. 8A). Both the basal and forskolin-stimulated phosphorylation of CREB was lower in cells expressing Syk-EGFP or Syk-EGFP-NLS than in Syk-deficient cells. The densities of the bands obtained from three separate experiments comparing Syk-deficient to Syk-EGFP-expressing cells were quantified, and the ratios of phospho-CREB to CREB were compared (Fig. 8B). The expression of Syk-EGFP led to a statistically significant decrease in both the basal and forskolin-induced phosphorylation of CREB.
FIGURE 8.
CREB phosphorylation is inhibited by Syk in MCF7 cells. A, Syk-deficient MCF7-B cells (Syk−) or MCF7-B cells expressing Syk-EGFP or Syk-EGFP-NLS were stimulated with forskolin for the indicated times. The lysates were fractionated by SDS-PAGE and probed with antibodies against phosho-CREB (pCREB), CREB, Syk, GAPDH, and PKAc. B, relative levels of pCREB in lysates of forskolin-treated MCF7-B cells lacking Syk (Syk−) or expressing Syk-EGFP were compared, and the ratios of the intensities of pCREB to CREB were plotted. The p values were calculated from Student's t tests from three separate experiments. IB, immunoblot.
To confirm that changes in the phosphorylation of CREB were Syk- and PKA-dependent, we treated the tetracycline-inducible MCF7-B cells with either H89 (PKA inhibitor) or piceatannol (Syk inhibitor) for 2 h prior to the addition of forskolin and analysis of CREB phosphorylation. A significant increase in CREB phosphorylation was seen in Syk-deficient MCF7-B cells following treatment with forskolin, a response that was blunted in cells induced to express Syk-EGFP (Fig. 9A). Pretreatment of either Syk-deficient or Syk-expressing cells with H89 inhibited the appearance of phospho-CREB, consistent with the inhibition of PKA. In contrast, pretreatment with piceatannol had no significant effect on forskolin-stimulated CREB phosphorylation in Syk-deficient cells but enhanced CREB phosphorylation in cells induced to express Syk-EGFP.
FIGURE 9.
CREB phosphorylation is enhanced by a Syk inhibitor. A, MCF7-B cells (Syk−) or MCF7-B cells expressing Syk-EGFP were pretreated with H-89 or piceatannol (PIC) and then stimulated with forskolin for the indicated times. The lysates were fractionated by SDS-PAGE and probed with antibodies against phospho-CREB (pCREB), CREB, and Syk. B, lysates from Syk-deficient MCF7-B cells (Syk−) or MCF7-B cells expressing Syk-EGFP were assayed for kinase activity against LRRASLG. The data represent the means and standard errors of three experiments.
To determine whether the loss of CREB phosphorylation in Syk-expressing cells correlated with a decrease in the activity of PKA, we examined the ability of lysates from cells lacking Syk or expressing Syk-EGFP to catalyze the phosphorylation of LRRASLG. As shown in Fig. 9B, PKA activity was reduced significantly in cells expressing Syk-EGFP as compared with Syk-deficient cells.
Effect of Syk on Transcription of the CREB-regulated Protein, Bcl-2
The promoter region of the BCL2 gene, which codes for the anti-apoptotic protein, Bcl-2, contains a cAMP response element and is regulated by CREB (38–40). To explore the effect of Syk on CREB-regulated transcription, we examined by Western blotting the level of Bcl-2 in cells expressing or lacking the kinase. Bcl-2 protein was readily detectable in the Syk-deficient MCF7-B cells but was reduced substantially in these cells when they were transfected to stably express Syk-EGFP (Fig. 10A). A similar difference was observed between the MCF7-B Syk-deficient cells and either MCF7-A cells that express endogenous Syk or MCF7-B cells transfected to stably express Syk-EGFP-NLS. Treatment of the MCF7-B with the PKA inhibitor H89 reduced the level of Bcl-2, whereas treatment of cells expressing Syk-EGFP-NLS with the Syk inhibitor, piceatannol, increased the level of Bcl-2 (Fig. 10A). To determine whether the regulation of this differential expression of Bcl-2 occurred at the level of transcription, we compared the amount of Bcl-2 mRNA in MCF7-B Syk-deficient cells with that in MCF7-B cells stably expressing either Syk-EGFP or Syk-EGFP-NLS. The level of Bcl-2 message was much higher in the Syk-deficient cells than in either Syk-expressing cell line, consistent with differences in the levels of Bcl-2 protein (Fig. 10B). Treatment of MCF7-B cells with forskolin to activate adenylate cyclase further increased the level of Bcl-2 mRNA, whereas treatment with H89 reduced transcription (Fig. 10C). Treatment of Syk-EFGP-expressing cells with forskolin enhanced the transcription of Bcl-2 mRNA. Together, these data support a role for Syk in the inhibition of the PKA and CREB-stimulated expression of the BCL2 gene.
FIGURE 10.
Syk regulates the expression of Bcl-2 via PKA and CREB. A, lysates from Syk-deficient MCF7-B cells (−), MCF7-B cells expressing Syk-EGFP (+) or Syk-EGFP-NLS (NLS), or MCF7-A cells (ATCC) were analyzed by Western blotting with antibodies against Bcl-2, Syk, or GAPDH as a loading control. Where indicated, MCF7-B cells or MCF7-B cells expressing Syk-EGFP-NLS were treated with H89 (20 μm) or piceatannol (PIC) (25 μm) for 24 h prior to lysis. B, RNA was extracted from MCF7-B cells (−) or MCF7-B cells expressing Syk-EGFP (+) or Syk-EGFP-NLS (NLS) and analyzed by RT-PCR using primers specific for Bcl-2 or GAPDH. C, MCF7-B cells (−) or MCF7-B cells expressing Syk-EGFP (+) were treated with or without forskolin (Fsk) (10 μm) in the presence of absence of H89 (20 μm) for 24 h prior to RNA extraction and analysis. D, MCF7-B cells (−) or MCF7-B cells expressing Syk-EGFP (+) were treated with or without doxorubicin (DOX) (5 μm) or etoposide (VP16) (2 μm) for 24 h prior to lysis. The lysates were probed with antibodies against PARP, Bcl-2, and GAPDH. E, MCF7-B cells (Syk-Def) or MCF7-B cells expressing Syk-EGFP-NLS (Syk-NLS) were treated with or without etoposide (VP16) (2 μm), forskolin (Fsk) (10 μm), H89 (20 μm), or piceatannol (PIC) (25 μm) for 24 h prior to lysis. The lysates were probed with antibodies to PARP. The ratio of the uncleaved PARP to the cleaved protein is indicated under each lane.
It is interesting that Syk functions as a pro-survival factor in many cancer cells, a fact that appears to conflict with its negative effects on Bcl-2 expression. To explore this, we compared the responses of MCF7 cells expressing or lacking Syk to treatment with either doxorubicin or etoposide by monitoring the cleavage of PARP, a well characterized substrate for effector caspases. A decreased level of PARP cleavage was observed in the cells expressing Syk-EGFP as compared with the Syk-deficient cells consistent with a pro-survival function for the kinase (Fig. 10D). This occurred despite the higher level of expression of Bcl-2 in the MCF7-B cells. Similarly, cells expressing Syk-EGFP-NLS were less sensitive to etoposide than were MCF7-B cells (Fig. 10E). Treatment with piceatannol sensitized the Syk-EGFP-NLS cells to etoposide-induced PARP cleavage but did not affect the response of Syk-deficient cells. The activation of PKA with forskolin to induce Bcl-2 expression protected both cell types from etoposide in a manner that could be blocked by treatment with H89 (Fig. 10E). Thus, elevated levels of PKA activity, which are associated with enhanced Bcl-2 expression, can protect cells from genotoxic stress. Although the expression of Syk reduces the expression of Bcl-2, it also protects cells from external stress stimuli.
DISCUSSION
Mass spectrometric analyses of protein substrates of Syk identified a consensus sequence for substrate recognition comprising a stretch of acidic amino acids located both proximal and distal to the phosphorylatable tyrosine (15). Among the substrates identified in this screen was PKAc, which was phosphorylated on Tyr-330, a residue located within a sequence (DDYEEEE) that matches well this consensus for an excellent Syk substrate. Consistent with this, PKAc is readily phosphorylated in vitro by Syk to high stoichiometry and can become phosphorylated in intact cells, particularly in cells containing nuclear-localized Syk. Of the 15 tyrosines in PKAc, only Tyr-330 is present within a highly acidic region. Correspondingly, substitution of Tyr-330 with either Phe or Glu effectively eliminates the ability of PKAc to serve as a substrate for Syk. Tyr-330 also is the only site of tyrosine phosphorylation on PKAc identified in our previous proteomic analyses (15), in MCF7 cells expressing Syk-EGFP-NLS or in DT40 cells expressing endogenous Syk. No phosphorylation was observed in DT40 cells that lacked Syk because of gene disruption. However, because some tyrosine phosphorylation of PKAc was observed by Western blotting in MCF7 cells lacking Syk, it is likely that other kinases also can catalyze this phosphorylation, which is consistent with a recent report that PKAc can be phosphorylated by both the EGF and PDGF receptors on Tyr-330 (41).
The core of PKAc is composed of an N-terminal lobe and a C-terminal lobe with residues required for ATP binding, substrate binding, and catalysis located at their interface (42). The C-terminal tail of PKAc contributes important cis-acting regulatory elements and, through direct interactions with the PKAc core, contributes to the formation of the ATP-binding site and influences substrate recognition (19–22). The stretch of acidic amino acids that contains Tyr-330 constitutes part of an adenosine-binding motif within the AST that acts as a gate to allow nucleotide movement into and out of the active site (22). The AST is flexible and solvent-exposed in the open conformation of PKAc but is ordered in the active, closed conformation where Tyr-330 makes contacts with the p-3 arginine of the peptide substrate (21). The importance of the hydroxyl group of Tyr-330 is revealed by the substantial loss of activity that results from the replacement of this tyrosine by phenylalanine, a change that increases the Km of PKAc for both ATP and peptide substrate and lowers its catalytic efficiency (20). Our studies indicate that an acidic residue at this position in the form of either a phosphotyrosine or glutamate completely blocks the catalytic activity of PKAc. The molecular dynamics simulation revealed that the addition of the phosphate group may substantially increase the solvation of the Tyr-330 side chain. The increased solvation perturbs the hydrophobic packing against the conserved ATP binding site and could jeopardize polar interactions with the substrate peptide, both of which are thought to be important in stabilizing the ATP and peptide bound in the active site of PKA. It should be noted that the current modeling study did not rule out the possibility that the Tyr-330 phosphorylation may induce large scale conformational changes of PKAc. Molecular dynamics simulations on orders of magnitude longer time scales are needed to explore such possibilities. The modeling results support the notion that the additional negative charge prevents the AST from interacting with the substrate and the nucleotide-binding site in a manner that stabilizes the closed conformation of PKAc, thus preventing catalysis from occurring. The presence of a tyrosine within the AST is unique to PKA among the AGC kinases and thus may reflect a mode of regulation of special importance to this particular family member.
Our results are in contrast to a previous report that the phosphorylation of PKAc on Tyr-330 by EGF receptor enhances its activity (41). The reasons for this discrepancy are unclear. However, the stoichiometry of tyrosine phosphorylation of PKAc by EGF receptor was not reported, and it is possible that the results reflected only a partial phosphorylation of the kinase.
Our mass spectrometric analyses and Western blotting studies indicate that PKAc also is phosphorylated in intact cells. Similar conclusions were reached previously for cells treated with EGF, PDGF, and fibroblast growth factor 2 (41). Because tyrosine-phosphorylated PKAc is inactive, the consequences of its covalent modification in cells would be an inhibition of its ability to phosphorylate intracellular substrates. Consistent with this, we observe in Syk-expressing cells a reduction in the phosphorylation of CREB on Ser-133, a site phosphorylated by PKAc. CREB, a bZIP transcription factor, is resident in the nucleus where it is phosphorylated by PKA catalytic subunits that traffic into the nucleus following activation of the holoenzyme in the cytosol (23). PKAc is phosphorylated on tyrosine in cells expressing Syk, which can traffic into and out of the nucleus (26) and most robustly in cells expressing a form of Syk with an attached NLS that localizes it primarily to the nucleus. This indicates that the free catalytic subunit in the nucleus constitutes a form of PKAc that is subject to regulation by Syk. The effects of elevated cAMP on cells are pleiotropic and context-dependent (43, 44). An association of active PKA with reduced cell growth can occur through the phosphorylation of CREB and enhancement of specific gene transcription and via the direct phosphorylation of various regulators of cell growth and survival such as Raf-1, Bim, and Par-4 (45–47). Thus, the inhibition of PKA by Syk would be expected to protect cells from apoptosis. Syk also has pro-survival activities that are mediated through the activation of Akt leading to the stabilization of the anti-apoptotic protein, Mcl-1 (9, 48). In contrast to a pro-apoptotic function for PKA, activated CREB is a pro-survival factor in many cancer cells and up-regulates Bcl-2 expression (38–40, 49–52). This activity is seen in the MCF7-B cells, which have elevated PKA activity and enhanced CREB phosphorylation. In these cells, the re-expression of Syk reduces the activity of PKA, reduces CREB phosphorylation, and down-regulates Bcl-2 expression. However, the Syk-expressing cells are still more resistant to genotoxic stress. Thus, it is clear that Syk can also function by mechanisms other than inhibition of PKA to promote cell survival.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI098132 (to R. L. G.) from the NIAID, R01CA115465 (to R. L. G. and W. A. T.) from the NCI, and R01GM039478 (to C. B. P.) and R01GM088317 (to W. A. T. and R. L. G.) from the NIGMS.
L. Xue and A. W. Tao, submitted for publication.
- PKAc
- catalytic subunit of cAMP-dependent protein kinase
- AST
- active site tether
- CREB
- cAMP response element-binding protein
- CBP
- CREB-binding protein
- EGFP
- enhanced GFP
- NLS
- nuclear localization sequence
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1. Geahlen R. (2009) Syk and pTyr'd. Signaling through the B cell antigen receptor. Biochim. Biophys. Acta 1793, 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mócsai A., Ruland J., Tybulewicz V. L. (2010) The Syk tyrosine kinase. A crucial player in diverse biological functions. Nat. Rev. Immunol. 10, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coopman P. J., Do M. T., Barth M., Bowden E. T., Hayes A. J., Basyuk E., Blancato J. K., Vezza P. R., McLeskey S. W., Mangeat P. H., Mueller S. C. (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406, 742–747 [DOI] [PubMed] [Google Scholar]
- 4. Bailet O., Fenouille N., Abbe P., Robert G., Rocchi S., Gonthier N., Denoyelle C., Ticchioni M., Ortonne J. P., Ballotti R., Deckert M., Tartare-Deckert S. (2009) Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res. 69, 2748–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toyama T., Iwase H., Yamashita H., Hara Y., Omoto Y., Sugiura H., Zhang Z., Fujii Y. (2003) Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 189, 97–102 [DOI] [PubMed] [Google Scholar]
- 6. Moroni M., Soldatenkov V., Zhang L., Zhang Y., Stoica G., Gehan E., Rashidi B., Singh B., Ozdemirli M., Mueller S. C. (2004) Progressive loss of Syk and abnormal proliferation in breast cancer cells. Cancer Res. 64, 7346–7354 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Q., Geahlen R. L. (2009) The protein-tyrosine kinase Syk interacts with TRAF-interacting protein TRIP in breast epithelial cells. Oncogene 28, 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young R. M., Hardy I. R., Clarke R. L., Lundy N., Pine P., Turner B. C., Potter T. A., Refaeli Y. (2009) Mouse models of non-Hodgkin lymphoma reveal Syk as an important therapeutic target. Blood 113, 2508–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J., Benavente C. A., McEvoy J., Flores-Otero J., Ding L., Chen X., Ulyanov A., Wu G., Wilson M., Wang J., Brennan R., Rusch M., Manning A. L., Ma J., Easton J., Shurtleff S., Mullighan C., Pounds S., Mukatira S., Gupta P., Neale G., Zhao D., Lu C., Fulton R. S., Fulton L. L., Hong X., Dooling D. J., Ochoa K., Naeve C., Dyson N. J., Mardis E. R., Bahrami A., Ellison D., Wilson R. K., Downing J. R., Dyer M. A. (2012) A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hahn C. K., Berchuck J. E., Ross K. N., Kakoza R. M., Clauser K., Schinzel A. C., Ross L., Galinsky I., Davis T. N., Silver S. J., Root D. E., Stone R. M., DeAngelo D. J., Carroll M., Hahn W. C., Carr S. A., Golub T. R., Kung A. L., Stegmaier K. (2009) Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell 16, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchner M., Fuchs S., Prinz G., Pfeifer D., Bartholomé K., Burger M., Chevalier N., Vallat L., Timmer J., Gribben J. G., Jumaa H., Veelken H., Dierks C., Zirlik K. (2009) Spleen tyrosine kinase is overexpressed and represents a potential therapeutic target in chronic lymphocytic leukemia. Cancer Res. 69, 5424–5432 [DOI] [PubMed] [Google Scholar]
- 12. Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. (2009) A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baudot A. D., Jeandel P. Y., Mouska X., Maurer U., Tartare-Deckert S., Raynaud S. D., Cassuto J. P., Ticchioni M., Deckert M. (2009) The tyrosine kinase Syk regulates the survival of chronic lymphocytic leukemia B cells through PKCδ and proteasome-dependent regulation of Mcl-1 expression. Oncogene 28, 3261–3273 [DOI] [PubMed] [Google Scholar]
- 14. Friedberg J. W., Sharman J., Sweetenham J., Johnston P. B., Vose J. M., Lacasce A., Schaefer-Cutillo J., De Vos S., Sinha R., Leonard J. P., Cripe L. D., Gregory S. A., Sterba M. P., Lowe A. M., Levy R., Shipp M. A. (2010) Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood 115, 2578–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue L., Wang W.-H., Iliuk A., Hu L., Galan J. A., Yu S., Hans M., Geahlen R. L., Tao W. A. (2012) Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iliuk A. B., Martin V. A., Alicie B. M., Geahlen R. L., Tao W. A. (2010) In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell Proteomics 9, 2162–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson D. A., Akamine P., Radzio-Andzelm E., Madhusudan M., Taylor S. S. (2001) Dynamics of cAMP-dependent protein kinase. Chem Rev. 101, 2243–2270 [DOI] [PubMed] [Google Scholar]
- 18. Romano R. A., Kannan N., Kornev A. P., Allison C. J., Taylor S. S. (2009) A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 18, 1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kannan N., Haste N., Taylor S. S., Neuwald A. F. (2007) The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. U.S.A. 104, 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chestukhin A., Litovchick L., Schourov D., Cox S., Taylor S. S., Shaltiel S. (1996) Functional malleability of the carboxyl-terminal tail in protein kinase A. J. Biol. Chem. 271, 10175–10182 [DOI] [PubMed] [Google Scholar]
- 21. Narayana N., Cox S., Shaltiel S., Taylor S. S., Xuong N. (1997) Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5–24) and adenosine. Biochemistry 36, 4438–4448 [DOI] [PubMed] [Google Scholar]
- 22. Narayana N., Cox S., Nguyen-huu X., Ten Eyck L. F., Taylor S. S. (1997) A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility. Structure 5, 921–935 [DOI] [PubMed] [Google Scholar]
- 23. Mayr B., Montminy M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 24. Goodman R. H., Smolik S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 25. Zhang X., Odom D. T., Koo S.-H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou F., Hu J., Harrison M. L., Geahlen R. L. (2006) Nucleocytoplasmic trafficking of the Syk protein tyrosine kinase. Mol. Cell Biol. 26, 3478–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma H., Yankee T. M., Hu J., Asai D. J., Harrison M. L., Geahlen R. L. (2001) Visualization of Syk-antigen receptor interactions using green fluorescent protein. Differential roles for Syk and Lyn in the regulation of receptor capping and internalization. J. Immunol. 166, 1507–1516 [DOI] [PubMed] [Google Scholar]
- 28. Zhang X., Shrikhande U., Alicie B. M., Zhou Q., Geahlen R. L. (2009) Role of the protein tyrosine kinase Syk in regulating cell-cell adhesion and motility in breast cancer cells. Mol. Cancer Res. 7, 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 30. Luo Y., Yang C., Jin C., Xie R., Wang F., McKeehan W. L. (2009) Novel phosphotyrosine targets of FGFR2IIIb signaling. Cell Signal. 21, 1370–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson E. E., Kornev A. P., Kannan N., Kim C., Ten Eyck L. F., Taylor S. S. (2009) Comparative surface geometry of the protein kinase family. Protein Sci. 18, 2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Im W., Lee M. S., Brooks C. L. (2003) Generalized born model with a simple smoothing function. J. Comput. Chem. 24, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 33. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., III, Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 34. Foloppe N., MacKerell A. D., Jr. (2000) All-atom empirical force field for nucleic acids. I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21, 86–104 [Google Scholar]
- 35. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furlong M. T., Mahrenholz A. M., Kim K.-H., Ashendel C. L., Harrison M. L., Geahlen R. L. (1997) Identification of the major sites of autophosphorylation of the murine protein-tyrosine kinase Syk. Biochim. Biophys. Acta 1355, 177–190 [DOI] [PubMed] [Google Scholar]
- 37. Caves L. S., Evanseck J. D., Karplus M. (1998) Locally accessible conformations of proteins. Multiple molecular dynamics simulations of crambin. Protein Sci. 7, 649–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson B. E., Mochon E., Boxer L. M. (1996) Induction of Bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol. Cell Biol. 16, 5546–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Riccio A., Ahn S., Davenport C. M., Blendy J. A., Ginty D. D. (1999) Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286, 2358–2361 [DOI] [PubMed] [Google Scholar]
- 40. Pugazhenthi S., Miller E., Sable C., Young P., Heidenreich K. A., Boxer L. M., Reusch J. E. (1999) Insulin-like growth factor-I induces bcl-2 promoter through the transcription factor cAMP-response element-binding protein. J. Biol. Chem. 274, 27529–27535 [DOI] [PubMed] [Google Scholar]
- 41. Caldwell G. B., Howe A. K., Nickl C. K., Dostmann W. R., Ballif B. A., Deming P. B. (2012) Direct modulation of the protein kinase A catalytic subunit α by growth factor receptor tyrosine kinases. J. Cell Biochem. 113, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 43. Insel P. A., Zhang L., Murray F., Yokouchi H., Zambon A. C. (2012) Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol. (Oxf.) 204, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Naviglio S., Caraglia M., Abbruzzese A., Chiosi E., Di Gesto D., Marra M., Romano M., Sorrentino A., Sorvillo L., Spina A., Illiano G. (2009) Protein kinase A as a biological target in cancer therapy. Expert Opin. Ther. Targets 13, 83–92 [DOI] [PubMed] [Google Scholar]
- 45. Dhillon A. S., Pollock C., Steen H., Shaw P. E., Mischak H., Kolch W. (2002) Cyclic AMP-dependent kinase regulates Raf-1 kinase mainly by phosphorylation of serine 259. Mol. Cell Biol. 22, 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moujalled D., Weston R., Anderton H., Ninnis R., Goel P., Coley A., Huang D. C., Wu L., Strasser A., Puthalakath H. (2011) Cyclic-AMP-dependent protein kinase A regulates apoptosis by stabilizing the BH3-only protein Bim. EMBO Rep. 12, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gurumurthy S., Goswami A., Vasudevan K. M., Rangnekar V. M. (2005) Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol. Cell Biol. 25, 1146–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gobessi S., Laurenti L., Longo P. G., Carsetti L., Berno V., Sica S., Leone G., Efremov D. G. (2009) Inhibition of constitutive and BCR-induced Syk activation downregulates Mcl-1 and induces apoptosis in chronic lymphocytic leukemia B cells. Leukemia 23, 686–697 [DOI] [PubMed] [Google Scholar]
- 49. Shankar D. B., Cheng J. C., Kinjo K., Federman N., Moore T. B., Gill A., Rao N. P., Landaw E. M., Sakamoto K. M. (2005) The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 7, 351–362 [DOI] [PubMed] [Google Scholar]
- 50. Aggarwal S., Kim S. W., Ryu S. H., Chung W. C., Koo J. S. (2008) Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res. 68, 981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng J. C., Kinjo K., Judelson D. R., Chang J., Wu W. S., Schmid I., Shankar D. B., Kasahara N., Stripecke R., Bhatia R., Landaw E. M., Sakamoto K. M. (2008) CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood 111, 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cougot D., Wu Y., Cairo S., Caramel J., Renard C.-A., Lévy L., Buendia M. A., Neuveut C. (2007) The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J. Biol. Chem. 282, 4277–4287 [DOI] [PubMed] [Google Scholar]