Background: The activation of CD40 is essential for the development of humoral and cellular immune responses.
Results: CD40 dimerizes through the extracellular region via the CRD1, and recombinant CRD1 potentiates CD40L activity.
Conclusion: CD40-CRD1 participates to dimerization and is required for efficient receptor expression.
Significance: CD40 self-assembly modulates signaling, possibly by maintaining the receptor in a quiescent state.
Keywords: Membrane Proteins, Protein Cross-linking, Protein Folding, Protein Self-assembly, Receptor Structure-Function, CD40, Oligomerization, PLAD, Signaling, TNFRSF
Abstract
The activation of CD40 on B cells, macrophages, and dendritic cells by its ligand CD154 (CD40L) is essential for the development of humoral and cellular immune responses. CD40L and other TNF superfamily ligands are noncovalent homotrimers, but the form under which CD40 exists in the absence of ligand remains to be elucidated. Here, we show that both cell surface-expressed and soluble CD40 self-assemble, most probably as noncovalent dimers. The cysteine-rich domain 1 (CRD1) of CD40 participated to dimerization and was also required for efficient receptor expression. Modelization of a CD40 dimer allowed the identification of lysine 29 in CRD1, whose mutation decreased CD40 self-interaction without affecting expression or response to ligand. When expressed alone, recombinant CD40-CRD1 bound CD40 with a KD of 0.6 μm. This molecule triggered expression of maturation markers on human dendritic cells and potentiated CD40L activity. These results suggest that CD40 self-assembly modulates signaling, possibly by maintaining the receptor in a quiescent state.
Introduction
Ligand-induced oligomerization is an important process for activating cell surface receptors like cytokine receptors, G protein-coupled receptors, protein-tyrosine kinase receptors, Toll-like receptors, and tumor necrosis factor receptor superfamily members (TNFRSF)6 in a number of biological events (1, 2). TNF family ligands form noncovalent homotrimers that can bind three receptor molecules (3). Although this ligand-receptor complex is generally accepted to be the minimal requirement for signaling, some members of the family are only efficiently triggered by at least two adjacent trimeric ligands (4, 5). The form under which ligand-free TNFRSF members exist is still not fully understood. Indeed, unliganded TNFR1 crystallized as a parallel dimer (6, 7) whereas cell-based experiments rather proposed that receptors may preassemble as trimers in the absence of ligand (8). Interestingly, DcR3 (TNFRSF6B) assembled as parallel dimers when crystallized alone, but as trimers when co-crystallized with its ligand TL1A, each receptor in this trimeric form contacting a different ligand (9). Thus, receptors may exist both as dimers or trimers depending on the context. Gathering more structural information on unliganded TNF receptors will help to better understand the mechanism of ligand-induced receptor signaling.
The extracellular region of TNFRSF members is characterized by the presence of cysteine-rich domains (CRDs) which typically contain six cysteine residues engaged in the formation of three disulfide bonds. The number of CRDs in a given receptor usually varies from one to four (3). TNFR1 and CD40 possess four CRDs, of which only CRD2 and 3 are directly involved in ligand binding. Crystallography of unliganded TNFR1 (TNFRSF1A) (6) showed homodimerization in either parallel or anti-parallel manners (7). Anti-parallel dimers associate through an interface that overlaps with CRD1 and 2, whereas parallel dimers associate mainly via CRD1 and leave ligand binding sites accessible. Cell-based studies identified a preligand-binding assembly domain at the N terminus of TNFR1 and 2 that is required for preassembly of TNFR complexes in a fully ligand-responsive conformation (8). In any case, TNF receptors appear to exist as preformed complexes and not as monomeric receptors that would only oligomerize upon ligand binding. In line with these results, the CRD1 of Fas (TNFRSF6) was shown to be essential for the formation of homotypic, ligand-independent receptor complexes (10). The situation is more complex for the TNF-related apoptosis-inducing ligand (TRAIL), another TNF family member that binds five different receptors: two fully functional death receptors, TRAIL-R1 (TNFRSF10A) and TRAIL-R2 (TNFRSF10B), two membrane bound “decoy” receptors, TRAIL-R3 (TNFRSF10C) and TRAIL-R4 (TNFRSF10D), and a soluble decoy receptor, OPG (TNFRSF11B). Decoy receptors inhibit cell death by competing for TRAIL binding. However, inhibition of TRAIL-induced apoptosis by TRAIL-R4 critically depends on its association with TRAIL-R2 via the N-terminal domain containing the first partial CRD of both receptors. Thus, in contrast to homotypic TNFR1 or Fas complexes, TRAIL-R2 and TRAIL-R4 form mixed complexes as a means to regulate TRAIL-induced apoptosis (11).
The present study focuses on CD40 (TNFRSF5). Engagement of CD40 by CD40L leads to CD40 clustering, recruitment of TNFR-associated factors, and activation, among others, of the transcription factor NF-κB (12, 13). CD40 is expressed mainly on professional antigen-presenting cells, i.e. dendritic cells, macrophages, and B cells (14), and its activation is essential for the development of humoral and cellular immune responses. Thus, selective blockade or activation of CD40 is of pharmacological interest. The crystal structure of CD40-CD40L was recently solved showing that CRD2 and CRD3 are both involved in CD40L binding (15); however, little is known about the function of CRD1 and CRD4.
Here, we show that CD40 exists as preformed, noncovalent dimers on the cell surface and that dimerization is dependent on the extracellular region. Mutation of K29 in CRD1 impaired CD40 self-assembly. Recombinant CD40-CRD1 bound the extracellular domain of CD40 with a KD of approximately 0.6 μm and displayed an agonist-like activity on CD40-expressing cells such as human dendritic cells. Co-incubation of CD40-CRD1 with CD40L potentiated the NF-κB response induced by CD40L alone, suggesting that CRD1 interactions participate in CD40 signaling. These observations provide a novel means to manipulate CD40 signaling.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
Human embryonic kidney HEK293 cells were grown in RPMI 1640 medium (Lonza, Basel) supplemented with 10% fetal calf serum and 5 μg/ml penicillin and streptomycin. Anti-CD40 (clone C20) monoclonal antibody from Santa Cruz Biotechnology and horseradish peroxidase-coupled anti-myc antibody from Life Technologies were used for Western blot analysis. Two different CD40L (CD154) were used as indicated: muCD8-CD40L (Ancell) and Mega-CD40L (Adipogen). Antibodies for flow cytometry analysis were purchased from BD Biosciences: Pacific blue CD14 and PerCp5.5 HLA-DR were used for monocyte purification control; PE-Cy7 CD86, PerCp5.5 HLA-DR, and APC CD83 were used to follow monocyte-derived dendritic cell maturation.
Molecular Cross-linking at the Cell Surface
Approximately 2 × 107 HEK293 cells stably expressing CD40 or BJAB cells were washed three times with ice-cold PBS (pH 8) within the culture flask (HEK cells) or in 15-ml tubes (BJAB cells). BS3 cross-linker (Thermo Scientific) was added at a final concentration of 3 mm for 1 h at 4 °C. The reaction was quenched by the addition of Tris at a final concentration of 20 mm for 15 min. Cells were then harvested and analyzed by Western blotting.
Recombinant Proteins
myc-H-tagged CD40 was produced in HEK293 cells using the mammalian expression vector pSectag2-Hy (Life Technologies) and purified from cell culture supernatant by nickel affinity chromatography. The encoded mature protein sequence was DAAQPAMNS, followed by amino acids 20–193 of human CD40 and the sequence QAAARGGPEQKLISEEDLNSAVDHHHHHH. The CRD1 of CD40 was expressed in Escherichia coli BL21 Rosseta-gami 2 using the expression vector pET22b+ (Merck). The encoded protein sequence was MDIGINSDPNS, followed by amino acids 20–62 of human CD40 and the sequence AAALEHHHHHH. Mutations K29A, K29E, C26Q, or the triple mutation C37G/C38G/C41G was introduced by conventional molecular biology techniques. Bacteria were induced for production of the proteins at an A600 nm of 0.6 with 0.5 mm isopropyl β-d-thiogalactoside for 16 h at 20 °C. Soluble periplasmic proteins were extracted and purified by nickel affinity chromatography as described in Ref. 16.
Mass Spectrometry
Prior to mass analysis, protein samples were dialyzed against 200 mm ammonium acetate on Amicon Ultra-4 centrifugal filter unit-10000NMWL (Millipore). Mass spectrometry was performed on an ESI-TOF mass spectrometer Q-TOF micro. Purity and homogeneity were verified by mass analysis in denaturing conditions: the protein was diluted to a concentration of 0.3 μm in water-acetonitrile (1:10, v/v) acidified with 1% formic acid.
In nondenaturing conditions, the mass measurement of the protein was performed in 50 mm ammonium acetate with 10% isopropyl alcohol, to preserve the native conformation. The protein was injected at 2.0 μm. Clusters of Cs(n+1)In were used for the calibration of the extended masse range in the high m/z region. Mass data were acquired in the positive ion mode on the mass range 1000–10,000 m/z. CD40 deglycosylation was performed using the deglycosylation mix from New England Biolabs following the manufacturer's instructions.
Molecular Modeling
All procedures were performed with Discovery Studio 2.5 software from Accelrys (San Diego, CA). The crystal structure of the ligand-free TNFR1 (Protein Data Bank (PDB) ID code 1NCF) was used as template to build a model of dimeric CD40. The receptor monomer corresponds to the crystal structure of the complex CD40-CD40L (PDB ID code 3QD6). A fixed atom constraint was applied to both C-terminal residues, and the dimeric CD40 model was subjected to an initial minimization step (maximum 500), root-mean-square (RMS) gradient 0.1 kcal/mol by conjugated gradient; followed by a second minimization step (maximum 500), RMS gradient 0.0001 kcal/mol by conjugated gradient. The structure was then submitted to a dynamic simulation including heating (2000 steps), equilibration (1000 steps), and production (1000 steps) at 300 K with a time step of 0.001 ps under a distance-dependent, dielectric constant, implicit solvent model. A negative intermolecular energy of −361.5 kcal/mol was obtained for the complex under these conditions. In silico mutation K29A produced an increase on the interaction energy to −218.0 kcal/mol, indicating a less stable complex. All images were generated using the PyMOL Molecular Graphics System, version 1.0 (Schrödinger, LLC).
FRET Experiments
CD40-ECFP, CD40-EYFP, and the negative control ERBB2-EYFP C-terminal fusion proteins all contained the full-length receptor sequences. Mutations K29A and K29E were introduced by site-directed mutagenesis. Experiments were performed using a LSRII (BD Biosciences) flow cytometer instrument. EYFP signal was recorded using the 488-nm laser with a 530/30 filter; the ECFP signal was recorded using the 405-nm laser with a 450/50 filter, and FRET signal was recorded using the 405-nm laser with a 585/42 filter. For each condition we evaluated a minimum of 10,000 ECFP-EYFP double-positive cells. HEK293 cells were transiently transfected with ECFP and EYFP fusion receptors and analyzed at 16 h after transfection. Positive FRET cells were gated using an ECFP-EYFP fusion protein as positive control and a co-transfection of EFCP and EYFP as negative control according to Ref. 17.
Surface Plasmon Resonance
The Biacore 3000 system, sensor chip CM5, surfactant P20, amine coupling kit containing N-hydroxysuccinimide and EDAC and ethanolamine were from Biacore (Uppsala, Sweden). Biosensor assays were performed with HBS-EP buffer as running buffer (10 mm HEPES, 150 mm sodium chloride, 3 mm EDTA 0.005% surfactant P20 (pH 7.4)). The low carboxylated dextran matrix (B1) was activated with 35 μl of a mixture 0.2 m EDAC and 0.05 m N-hydroxysuccinimide at 5 μl/min. The anti-myc antibody (Life Technologies) was immobilized with the standard Biacore protocol at ∼2000 resonance units (RU) per channel. Soluble CD40-myc tag fusion protein was injected for 30 s, then increasing concentrations of recombinant CRD1 were injected for 2 min, followed by a dissociation phase of 3 min at a flow rate of 30 μl/min. All dilutions were performed in running buffer. The sensor chip surface was regenerated after each experiment by injecting 20 μl of 3 m MgCl2. The kinetic parameters were calculated using BIAeval 4.1 software. Analysis was performed using the simple Langmuir binding model. The specific binding profiles were obtained after subtracting the response signal from the control anti-myc antibody alone channel and from blank-buffer injection. The fitting to each model was judged by the chi square (χ2) value and randomness of residue distribution compared with the theoretical model. Active concentration was determined under mass transfer condition where the slope of the curves is directly correlated with the active concentration of the protein (18).
Luciferase Reporter Assay
HEK293 cells stably expressing either wt CD40 or CD40-K29A were cultured in 24 well plates at 3 × 105 cell/ml. After 24 h cells were transfected with control luciferase vectors or promoter specific luciferase reporter vectors (Panomics, Fremont, CA), using FuGENE6 transfection reagent (Roche Applied Science). Reporter luciferase assays were performed for STAT3, AP1, CRE, NF-κB, and SRE that all are CD40L-activated transcription factors. After 24h, cells were treated with CD40L or a recombinant CD40-CRD1 protein at the indicated concentrations in the presence of 20 μg/ml polymyxin B (Fluka, St. Louis, MO). After 16 h cells were lysed and luciferase activity was determined using a Wallac Victor2 1420 multilabel counter (PerkinElmer Life Sciences) according to instructions of the Promega luciferase assay system. Dual luciferase assay was performed using NF-κB reporter plasmid together with Renilla luciferase plasmid for normalization. Detection of firefly luciferase and Renilla luciferase expression was done using the dual luciferase assay detection kit (Promega).
Monocyte-derived Dendritic Cell Differentiation and Maturation
Elutriated monocytes were obtained from the Etablissement Français du Sang (EFS; Strasbourg, France). Monocytes were cultured in RPMI 1640 medium with Ultraglutamine (Lonza) supplemented with 10 μg/ml gentamycin, 2 mm HEPES, and 10% heat-inactivated fetal calf serum. At day 0, cells were seeded at a density of 4 × 106 cells/well in a 6-wells plate, and differentiation into CD209-positive dendritic cells was induced by addition of 5 ng/ml GM-CSFHuXp and 5 ng/ml IL-4HuXp (Humanzyme, Chicago, IL). At day 5, when >80% of the cells expressed CD209 (data not shown), cells were incubated with the indicated stimuli in the presence or absence of 100 μg/ml polymyxin B (Fluka). After 24 h of culture, cells were harvested and washed with 2 ml of cold PBS, and maturation maker expression was analyzed by flow cytometry.
Flow Cytometry Analysis
Cells were stained in PBS containing 2% fetal calf serum at 4 °C for 20 min with the various antibodies used at concentration recommended by the manufacturer. After two washes in PBS, cells were analyzed by flow cytometry with a Gallios flow cytometer (Beckman Coulter). At least 10,000 events were acquired for each experiment, and the data were processed with the Kaluza® Flow Analysis Software (Beckman Coulter).
Statistical Analysis
FRET experiments were analyzed using one-way ANOVA with Tukey post test, and luciferase NF-κB and monocyte-derived dendritic cell maturation assays were analyzed using two-way ANOVA with Bonferroni post test, using GraphPad Prism version 5.00 for Windows (GraphPad Software).
RESULTS
CD40 Dimerizes through the Extracellular Region
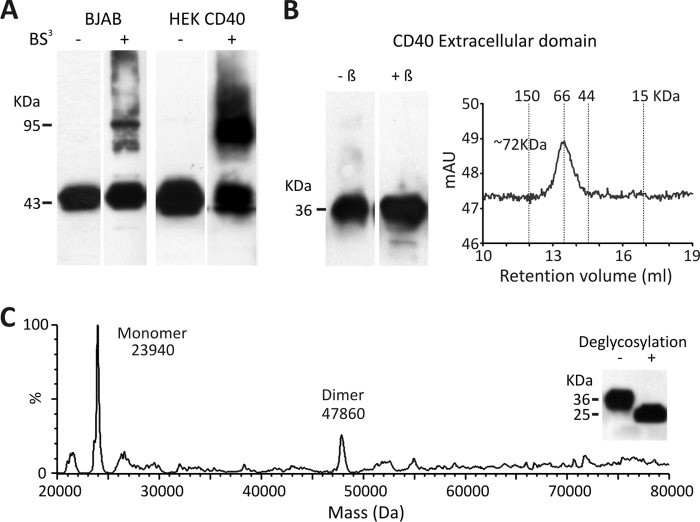
To define the oligomerization state of CD40 in the absence of ligand, HEK cells stably transfected with full-length WT CD40 were chemically cross-linked with the nonpermeable reagent BS3. Western blot analysis revealed that a portion of CD40, migrating at ∼45 kDa, shifted to a dimer of ∼90 kDa in cross-linked cell lysates (Fig. 1A). Some CD40 dimer was also observed in the B cell lymphoma BJAB cell line that expresses endogenous CD40 (Fig. 1A). To define the contribution of the extracellular region to dimerization, soluble CD40 fused to the nonoligomerizing myc and H tags was produced in mammalian cells. This soluble CD40 bound CD154 in a surface plasmon resonance assay (data not shown). Under denaturing SDS-PAGE, soluble CD40 migrated as a band of ∼36 kDa under both reducing and nonreducing conditions, but the same protein eluted with an apparent molecular mass of ∼72 kDa from a size exclusion chromatography column, suggesting that it may form roughly globular dimers (Fig. 1B). Although glycosylation interfered with ionization of soluble CD40 in native mass spectrometry (supplemental Fig. 1), the deglycosylated protein behaved well in this assay which allowed detection of two peaks at masses of 23.94 and 47.86 kDa, corresponding relatively closely to expected theoretical values for the nonglycosylated monomeric (23.4 kDa) and dimeric (46.7 kDa) CD40 (Fig. 1C). Taken together, these results indicate that at least a fraction of CD40 can self-assemble in the absence of ligand, most probably as a noncovalent homodimer.
FIGURE 1.
CD40 dimerizes through the extracellular region. A, BJAB cells and stably transfected CD40 HEK cells were treated or not with the nonpermeable chemical cross-linker BS3. CD40 was then detected by Western blotting with an anti-CD40 antibody. B, the recombinant extracellular region of CD40 was analyzed by SDS-PAGE in the presence or absence of β-mercaptoethanol (left) and by size exclusion chromatography (right). C, native mass spectrometry analysis of the recombinant extracellular region of CD40 is shown. The protein was deglycosylated before native mass analysis. The inset shows a Western blot analysis of soluble CD40 before and after deglycosylation.
CD40-CRD1 Mediates Self-interaction of CD40
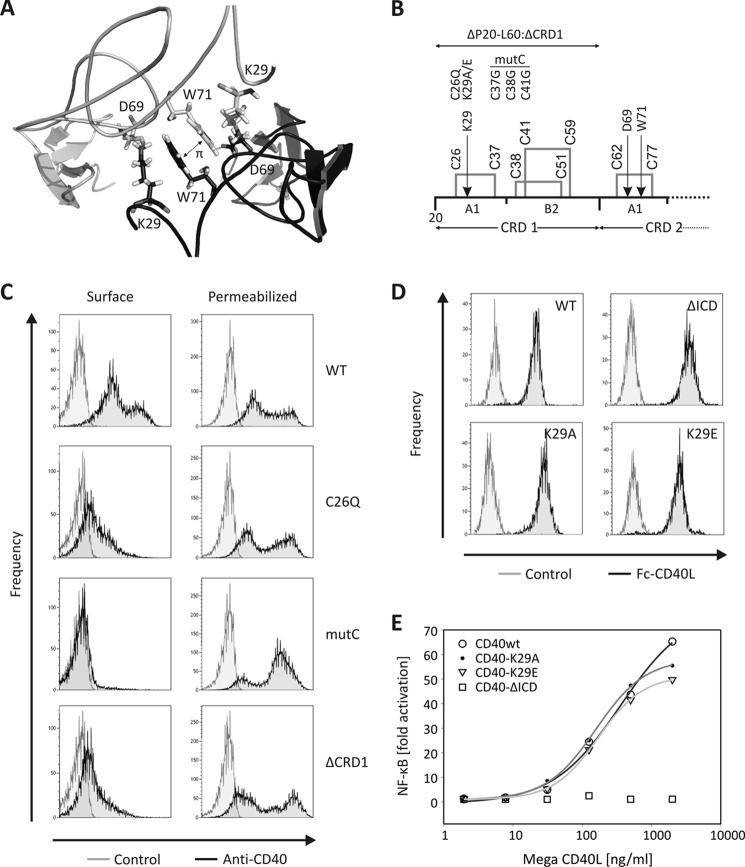
To define the site required for CD40 self-assembly, we first constructed a model of a CD40 dimer based on the structures of the parallel TNFR1 dimer (PDB ID code1NCF) and ligand-bound CD40 (PDB ID code 3QD6). The rational was that (i) the structure of monomeric (liganded) and dimeric (unliganded) TNFR1 differ strongly in CRD1, suggesting that this domain is important for self-assembly (supplemental Fig. 2A); (ii) lymphotoxin α-TNFR1 is the closest available crystal structure to CD40-CD40L (15); and given that (iii) CRD1 and 2 of TNFR1 and CD40 superimpose with a Cα root mean square deviation of 1.05 Å, chances of getting a relevant model were good (supplemental Fig. 2B).
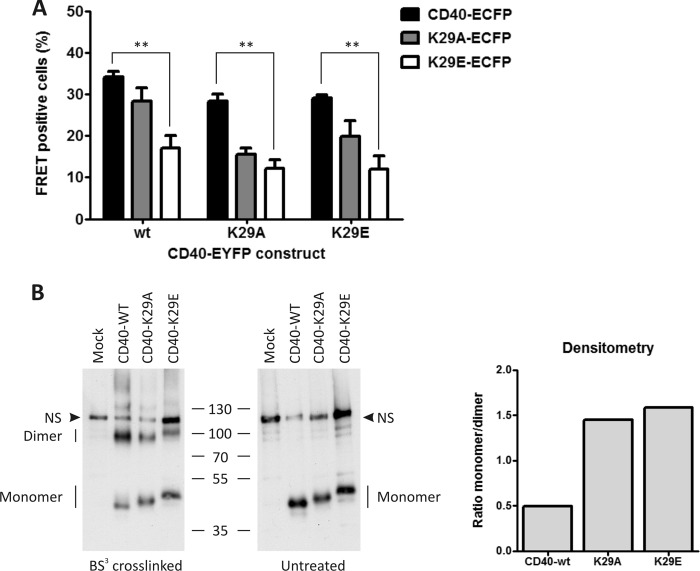
The model suggested the involvement of the CRD1 domain in CD40 self-assembly, with an important contribution of K29 (Fig. 2A). We therefore generated full-length CD40 constructs with mutation K29A, K29E, or with mutations predicted to disrupt folding of the first A1 sub-domain (C26Q) or of the entire CRD1 (triple cysteine mutant C37G/C38G/C41G: mutC), or lacking CRD1 but retaining the signal peptide (CD40ΔCRD1) (Fig. 2B). With the exception of the K29A and K29E mutants, all other untagged or EYFP-tagged CD40 CRD1 mutants were retained in the endoplasmic reticulum, failed to reach the cell surface efficiently, and did not respond to exogenously added CD40L (Fig. 2C and supplemental Figs. 3, 4, and 5A), indicating that CD40 is sensitive to mutations affecting normal folding of its CRD1. In line with these results, steered molecular dynamic simulation showed strong local perturbations in CD40-CRD1 in the case of C26Q and mutC mutations and strong local perturbations in CD40-CRD2 in the case of the ΔCRD1 construct (supplemental Fig. 5B). As mentioned above, the CD40 dimer model suggested a contribution of K29 in receptor dimerization, and it is noteworthy that this residue is conserved in human and mouse CD40. In silico mutation of this residue to an alanine increased the interaction energies from −361 kcal/mol to −219 kcal/mol, highlighting its importance in receptor dimerization. We also introduced the mutation K29E to obtain a stronger effect by charge repulsion. Neither the K29A nor K29E mutation altered the surface expression of CD40 (Fig. 2D). Furthermore, CD40L-induced NF-κB activity was similar in the WT CD40 cell line compared with the mutant cell lines K29A and K29E (Fig. 2E). Under the same conditions, no activity was observed in the CD40 cell line lacking the intracellular domain (CD40-ΔICD). However, the ability of the CD40 K29A or K29E mutant to produce FRET signals with either themselves or with WT CD40 was reduced by approximately 50%, supporting the notion that K29 is involved in CRD1-mediated dimerization of CD40 (Fig. 3A). Finally, chemical cross-linking of untagged mutant or WT CD40 showed a marked reduction of dimeric species at the cell surface when using both mutant receptors compared with WT CD40 (Fig. 3B).
FIGURE 2.
CD40-CRD1 is required for normal surface expression. A, detail of the CD40 dimer model in the region of CRD1-CRD1 interaction, showing side chains of K29, D49, and W71. B, schematic representation of the CRD1 domain of CD40 with disulfide bridges highlighted. Mutations tested are indicated. C, flow cytometry analysis of HEK293 cells transfected with untagged WT or mutant CD40 constructs. Staining was performed with an anti-CD40 antibody in permeabilized or nonpermeabilized (surface) conditions. D, flow cytometry analysis of different CD40-HEK293 stable cell lines used in this study. Staining was performed with Fc-CD40L followed by anti-human Fc-PE. E, dose response of CD40L in a NF-κB luciferase activity assay in different CD40-expressing HEK cell lines (CD40-WT, CD40-K29A, CD40-K29E, and CD40-ΔICD).
FIGURE 3.
Mutations of lysine 29 disrupt CD40 self-assembly. A, cytometric FRET analysis was performed of CD40 interaction with WT CD40, CD40-K29A, or CD40-K29E, using both ECFP and EYFP fusion combinations. Mean values with S.E. (error bars) of three independent experiments are shown (**, p < 0.01). B, CD40 stable cell lines (WT, K29A, and K29E) were treated or not with the nonpermeable chemical cross-linker BS3. CD40 was then detected by Western blotting with an anti-CD40 antibody. Densitometry analysis is shown on the right as a ratio between monomeric and dimeric receptors. NS, nonspecific signal.
Recombinant CD40-CRD1 Binds to CD40
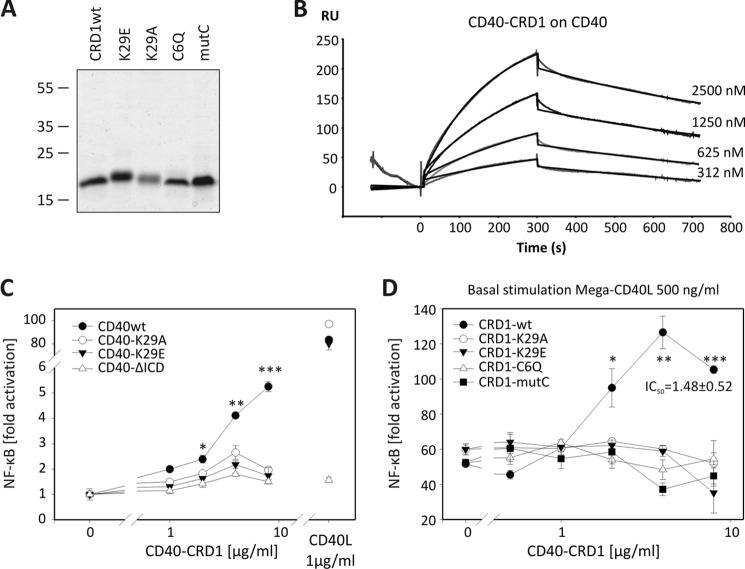
Although the isolated CD40-CRD1 was poorly secreted in HEK cells, it was successfully produced in a prokaryotic expression system with human-compatible tRNAs and enhanced ability to form disulfide bridges. CD40-CRD1 was also expressed with the K29A, K29E, and CRD1-disrupting mutations C26Q or mutC (Fig. 4A). All CRD1 molecules migrated on SDS-PAGE with an apparent molecular mass of ∼16 kDa despite their theoretical molecular mass of ∼8 kDa. Wild-type CD40-CRD1 bound to the extracellular domain of CD40 with a KD of 5.7 × 10−7 m (kon 2.04 × 103 m−1 s−1 and koff 1.16 × 10−3 s−1), as determined by surface plasmon resonance analysis, whereas no binding was detected for both cysteine mutants (Fig. 4B and supplemental Fig. 6). These data show the importance of an adequate CRD1 folding for binding to CD40. Of note, using surface plasmon resonance, the active concentration of CD40-CRD1 was estimated to be 4.7% of the total protein. In all experiments, the mentioned concentration of CD40-CRD1 is that of the active protein. The concentration of CRD1 mutants was estimated also as 4.7% of the total protein although it was not possible to estimate its active fraction because there is no detectable binding to CD40.
FIGURE 4.
Recombinant CD40-CRD1 binds to CD40 and potentiates CD40L activity. A, purified CD40-CRD1, CRD1-K29E, CRD1-K29A, CRD1-C6Q, and CRD1-mutC (20 μg of protein/lane) were resolved by SDS-PAGE and stained with Coomassie Blue. B, Biacore sensorgram of CD40-CRD1 binding to the entire extracellular domain of CD40. C, luciferase reporter assay for NF-κB activity in different CD40 HEK cell lines (CD40wt, CD40-K29A, CD40-K29E, and CD40-ΔICD) treated with recombinant soluble CD40-CRD1. Untreated cells are shown on the left, and Mega-CD40L-stimulated cells are shown on the right. D, luciferase reporter NF-κB assay in HEK CD40 cell line treated with 500 ng/ml CD40L and increasing concentrations of recombinant soluble CD40-CRD1, CRD1-K29A, CRD1-K29E, CRD1-C6Q, and CRD1-mutC. Mean values with S.E. (error bars) of three independent experiments are shown (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Recombinant CD40-CRD1 Induces a Mild Ligand-like Activity and Potentiates CD40L Signals
As expected, an overnight stimulation of CD40-expressing cells with CD40L activated the NF-κB transcription factor and induced expression of a NF-κB luciferase reporter gene (Fig. 4C). Surprisingly, soluble CD40-CRD1 showed a weak agonistic activity on WT CD40-expressing HEK cells, but not on cells expressing CD40-K29A, CD40-K29E, or CD40 lacking the intracellular domain (CD40-ΔICD) (Fig. 4C). Furthermore, WT CRD1, but not the mutants, potentiated CD40L-induced NF-κB activation by up to 2-fold, with an IC50 of approximately 1.5 ± 0.5 μg/ml (Fig. 4D). In line with the observation that WT CD40-CRD1 could induce NF-κB activity in cells expressing WT CD40, but not in cells expressing mutant CD40, recombinant mutant CD40-CRD1 proteins were unable to increase CD40L activity in WT CD40 HEK cells. Because the CRD1 of CD40 does not interact with CD40L in the crystallographic structure of the CD40-CD40L complex (15), we believe that the observed signal potentiation results from independent effects of CD40-CRD1 and CD40L on membrane CD40.
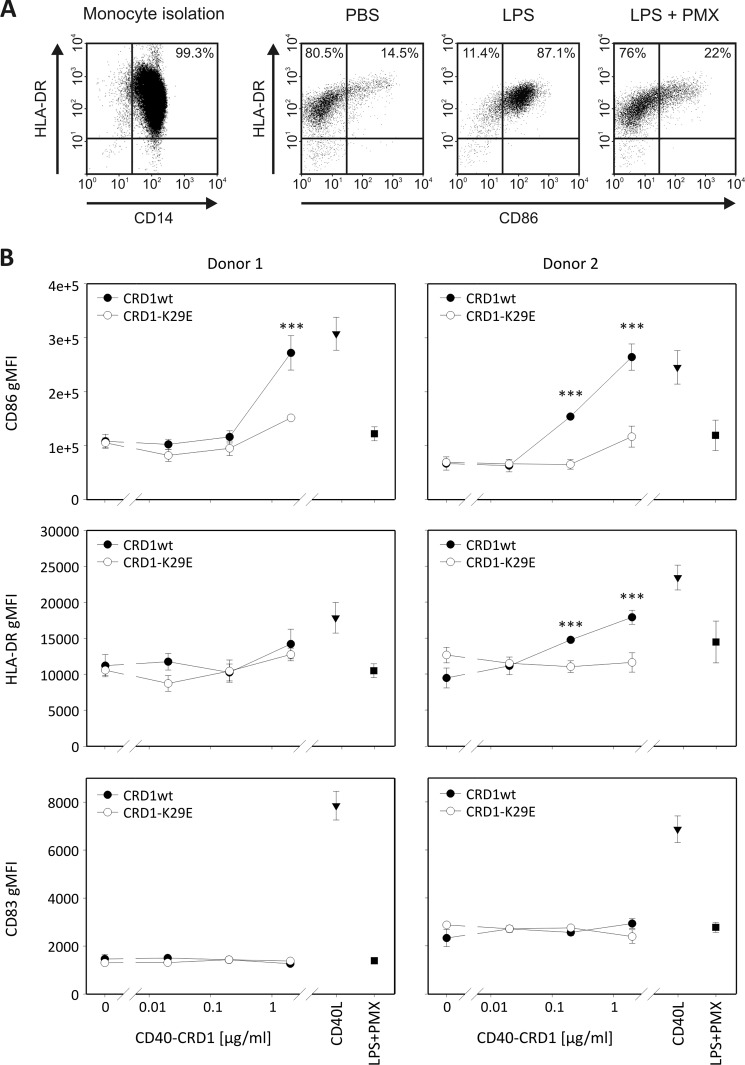
When human monocyte-derived dendritic cells from different donors were treated with increasing concentrations of WT CD40-CRD1, the surface maturation markers CD86 and HLA-DR were up-regulated (Fig. 5). This effect occurred in the presence of the LPS inhibitor polymyxin B (19) at a dose sufficient to fully inhibit up to 10 μg/ml LPS (Fig. 5A), making it unlikely that it was endotoxin-mediated. Moreover, CD40-CRD1 K29E was markedly less potent than its WT counterpart at stimulating dendritic cells. Unlike CD40L, WT CD40-CRD1 taken alone failed to up-regulate the activation marker CD83, suggesting that it is less potent than CD40L (Fig. 5B). When dendritic cells were treated simultaneously with WT CD40-CRD1 and CD40L, a modest potentiation of CD40L-induced activation was observed at low doses of recombinant CD40-CRD1 (supplemental Fig. 7, A and B). Taken together, our results suggest that the CRD1 of CD40 can increase CD40-dependent signals in target cells, alone or in cooperation with CD40L.
FIGURE 5.
Recombinant CD40-CRD1 induces maturation of human monocyte-derived dendritic cells. A, CD14-positive cells (scattergram on the left) were cultured in the presence of IL-4 and GM-CSF for 5 days. Cells were then treated for 24 h with PBS (vehicle), LPS (10 μg/ml), or LPS plus polymyxin B (100 μg/ml) (LPS+PMX) and analyzed for expression of activation markers CD86 and HLA-DR on CD209-positive cells. B, expression of maturation markers CD86, HLA-DR, and CD83 (geo-mean, gMFI) was assessed after treatment with the indicated concentrations of recombinant WT CD40-CRD1 or CD40-CRD1-K29E in the presence of polymyxin B (100 μg/ml). Control treatments CD40L (Mega-CD40L 200 ng/ml) and LPS plus polymyxin B (100 μg/ml) are also shown for each donor. Mean value with S.E. (error bars) of triplicates are shown (***, p < 0.001). Data are representative of two independent experiments performed on a total of three different donors.
DISCUSSION
Although it is well established that CD154 (CD40L) assembles as a trimer, not much research has been conducted on CD40 oligomerization before ligand engagement and on the potential functional effect of this oligomerization. Reports from Mourad and colleagues suggested that CD40 at the cell surface exist as disulfide-linked dimers and that disulfide-linked homodimerization was increased by CD40L (20–22). However, all cysteine residues in the extracellular region of CD40 and other crystallized TNFR family members are engaged in intrachain disulfide bridges, making it unlikely that CD40 would dimerize through interchain extracellular covalent bonds.
In the present study, we show that CD40 can self-assemble at the cell surface in a noncovalent manner, probably as dimers. Soluble CD40 also dimerizes, suggesting that the extracellular domain plays an important role in dimer formation. In addition, in silico modelization and site-directed mutagenesis confirmed the role of CD40-CRD1 in receptor-receptor interactions. Until now, only TNFR1 and DcR3 have been crystallized as ligand-free receptors. TNFR1 crystallized as both parallel and anti-parallel dimers (6). The parallel dimer was stabilized by interactions involving CRD1 and exposed fully accessible TNF binding sites in CRD2 and CRD3. In contrast, the anti-parallel dimer was stabilized by interactions involving CRD1 and CRD2 that occluded the TNF binding sites. Parallel TNFR dimers are favored at neutral pH, whereas anti-parallel TNFR dimers are favored at acidic pH (7). This observation led to the hypothesis that parallel dimers may occur at the cell surface and could serve to create an activation network in the presence of ligand, in contrast to anti-parallel dimers that may form upon acidification of endocytic vesicles and release the ligand from the complex (7). Unliganded DcR3 was also crystallized as parallel dimers, with the main contact area in CRD1 and 4 and an accessible ligand binding site to its ligand TL1A (9). Sedimentation equilibrium analyses of DcR3 showed weak self-association, with an estimated equilibrium dissociation constant greater than 2.4 × 10−3 m, a low value if we consider that the physiological concentration of this soluble receptor is in the range of 10−12 pm (23). Interestingly, the CRD1-CRD1 interaction surface modeled for CD40 in our study superimposed relatively well with the interaction surface found between the CRD1 of the TNF receptor HVEM and the immunomodulatory protein BTLA in the HVEM-BTLA complex (24). Therefore, a CRD1 seems to use the same interface to interact with another CRD1 (as in the TNFR1 dimer) or with a regulatory protein. The CRD1 could therefore be considered as an interaction domain that regulates receptor activity by interacting either with itself or with other proteins such as BTLA.
Using surface plasmon resonance, an equilibrium dissociation constant of 5.7 × 10−7 m was determined for the interaction between CD40 and recombinant CD40-CRD1. Although this equilibrium constant is rather low, it may still be relevant because local protein concentrations on the cell membrane can be high, especially in some microdomains, and because other interaction sites outside CRD1 may also participate to dimerization. These results raise the question of whether the active form of the receptor is the dimeric or the monomeric form and whether receptor dimers persist after ligand engagement. Further analyses are warranted to address these questions.
The CRD1 of TNFR1 fused to glutathione S-transferase as been described previously as an inhibitor of TNFR1-mediated signaling (25). This molecule could block the effects of TNF in vitro and was shown to inhibit arthritis in mice (25). Unfortunately, this study was not designed to address the molecular basis of these effects, nor did it test the potential impact of glutathione S-transferase-induced dimerization on CRD1 activity. In the case of Fas, the study of dominant interfering mutations associated with autoimmune lymphoproliferative syndrome revealed that Fas CRD1 was fully preserved in all dominant interfering mutations, including mutations that truncate Fas after 57 or 62 amino acids of the mature Fas protein and that would be unable to bind FasL (10). In addition, deletion of CRD1 in dominant negative inhibitory receptors restored susceptibility to FasL-induced apoptosis, indicating that mutant proteins must physically interact with wild-type proteins to create a nonfunctional complex. Although the phenotype of patients provides strong genetic evidence for a dominant negative function of the N-terminal portion of Fas, results obtained with N-terminal Fas deletion mutants must be taken with caution in view of our results demonstrating that CRD1 disruption prevents CD40 surface expression. At least for CD40, N-terminal truncation or severe perturbations of CRD1 structures are not appropriate to study CRD1 function. In any case, the studies mentioned above indicate that the N-terminal portions of TNFR1 and Fas taken alone antagonize receptor signaling.
Because of the described inhibitory activity of TNFR1 and Fas CRD1, the ability of recombinant CD40-CRD1 to induce a weak CD40 activation rather than an inhibition of NF-κB signaling was unexpected. A gross artifact can probably be excluded because NF-κB activation was CD40-dependent and because CRD1 disruption mutants were ineffective. The agonist activity of CD40-CRD1 could be explained in several ways: (i) if CD40 is inhibited by dimerization, then recombinant CRD1 may activate CD40 by disrupting the dimers; (ii) recombinant CRD1 could oligomerize CD40 on the cell surface, inducing an agonist-like signaling; and (iii) recombinant CRD1 may induce conformational changes in the receptor similar to those induced by the ligand upon signal activation. Importantly, the agonist activity of CD40-CRD1 was not restricted to cells overexpressing CD40, but was also observed in primary, monocyte-derived dendritic cells. However, the observation that CD40-CRD1 was unable to induce the full set of activation markers in dendritic cells probably indicates a weak agonist activity compared with CD40L.
Altogether, our result have revealed the ability of CD40 to self-assemble through its cysteine-rich domain 1 and the ability of recombinant CD40-CRD1 to potentiate CD40L in HEK CD40 cells and to induce a ligand-like activity in human dendritic cells. Our data indicate that CD40 homo-dimerization controls signaling, possibly by maintaining the receptor in a quiescent state, although further studies are needed to elucidate mechanisms mediating receptor activation.
Acknowledgments
We thank Dr. Sylviane Muller (UPR 9021 Strasbourg, France) for constant support and encouragement; Laure Willen (University of Lausanne) for expert technical assistance in plasmid cloning; Marie-Christine Rio and Fabien Alpy for kindly providing the ERBB2-EYFP construct; and Pedro Romero and Petra Baumgaertner (University of Lausanne) for helpful discussions, advice, and assistance in monocyte-derived dendritic cell assay.
This work was supported by the Centre National de la Recherche Scientifique (to S. F. and G. G.), the Swiss National Science Foundation (to P. S.), and the Agence Nationale Recherche Grant ANR-08-PCVI-0034-01) (to G. G.).

This article contains supplemental Figs. 1–7.
- TNFRSF
- TNF receptor superfamily
- BS3
- (bis)sulfosuccinimidyl suberate
- CRD
- cysteine-rich domain
- EDAC
- N-ethyl-N′-(3-dimethylaminopropyl)-carbodiimide
- TRAIL
- TNF-related apoptosis-inducing ligand.
REFERENCES
- 1. Tetsch L., Jung K. (2009) How are signals transduced across the cytoplasmic membrane? Transport proteins as transmitter of information. Amino Acids 37, 467–477 [DOI] [PubMed] [Google Scholar]
- 2. Heldin C. H. (1995) Dimerization of cell surface receptors in signal transduction. Cell 80, 213–223 [DOI] [PubMed] [Google Scholar]
- 3. Bodmer J. L., Schneider P., Tschopp J. (2002) The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27, 19–26 [DOI] [PubMed] [Google Scholar]
- 4. Holler N., Tardivel A., Kovacsovics-Bankowski M., Hertig S., Gaide O., Martinon F., Tinel A., Deperthes D., Calderara S., Schulthess T., Engel J., Schneider P., Tschopp J. (2003) Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 23, 1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haswell L. E., Glennie M. J., Al-Shamkhani A. (2001) Analysis of the oligomeric requirement for signaling by CD40 using soluble multimeric forms of its ligand, CD154. Eur. J. Immunol. 31, 3094–3100 [DOI] [PubMed] [Google Scholar]
- 6. Naismith J. H., Devine T. Q., Brandhuber B. J., Sprang S. R. (1995) Crystallographic evidence for dimerization of unliganded tumor necrosis factor receptor. J. Biol. Chem. 270, 13303–13307 [DOI] [PubMed] [Google Scholar]
- 7. Naismith J. H., Devine T. Q., Kohno T., Sprang S. R. (1996) Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure 4, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 8. Chan F. K., Chun H. J., Zheng L., Siegel R. M., Bui K. L., Lenardo M. J. (2000) A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288, 2351–2354 [DOI] [PubMed] [Google Scholar]
- 9. Zhan C., Patskovsky Y., Yan Q., Li Z., Ramagopal U., Cheng H., Brenowitz M., Hui X., Nathenson S. G., Almo S. C. (2011) Decoy strategies: the structure of TL1A:DcR3 complex. Structure 19, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siegel R. M., Frederiksen J. K., Zacharias D. A., Chan F. K., Johnson M., Lynch D., Tsien R. Y., Lenardo M. J. (2000) Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288, 2354–2357 [DOI] [PubMed] [Google Scholar]
- 11. Clancy L., Mruk K., Archer K., Woelfel M., Mongkolsapaya J., Screaton G., Lenardo M. J., Chan F. K. (2005) Preligand assembly domain-mediated ligand-independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 102, 18099–18104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pullen S. S., Miller H. G., Everdeen D. S., Dang T. T., Crute J. J., Kehry M. R. (1998) CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry 37, 11836–11845 [DOI] [PubMed] [Google Scholar]
- 13. Pullen S. S., Labadia M. E., Ingraham R. H., McWhirter S. M., Everdeen D. S., Alber T., Crute J. J., Kehry M. R. (1999) High-affinity interactions of tumor necrosis factor receptor-associated factors (TRAFs) and CD40 require TRAF trimerization and CD40 multimerization. Biochemistry 38, 10168–10177 [DOI] [PubMed] [Google Scholar]
- 14. van Kooten C., Banchereau J. (2000) CD40-CD40 ligand. J. Leukoc. Biol. 67, 2–17 [DOI] [PubMed] [Google Scholar]
- 15. An H. J., Kim Y. J., Song D. H., Park B. S., Kim H. M., Lee J. D., Paik S. G., Lee J. O., Lee H. (2011) Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 286, 11226–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smulski C., Labovsky V., Levy G., Hontebeyrie M., Hoebeke J., Levin M. J. (2006) Structural basis of the cross-reaction between an antibody to the Trypanosoma cruzi ribosomal P2β protein and the human β1 adrenergic receptor. FASEB J. 20, 1396–1406 [DOI] [PubMed] [Google Scholar]
- 17. Banning C., Votteler J., Hoffmann D., Koppensteiner H., Warmer M., Reimer R., Kirchhoff F., Schubert U., Hauber J., Schindler M. (2010) A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5, e9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richalet-Sécordel P. M., Rauffer-Bruyère N., Christensen L. L., Ofenloch-Haehnle B., Seidel C., Van Regenmortel M. H. (1997) Concentration measurement of unpurified proteins using biosensor technology under conditions of partial mass transport limitation. Anal. Biochem. 249, 165–173 [DOI] [PubMed] [Google Scholar]
- 19. Cardoso L. S., Araujo M. I., Góes A. M., Pacífico L. G., Oliveira R. R., Oliveira S. C. (2007) Polymyxin B as inhibitor of LPS contamination of Schistosoma mansoni recombinant proteins in human cytokine analysis. Microb. Cell Fact. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reyes-Moreno C., Girouard J., Lapointe R., Darveau A., Mourad W. (2004) CD40/CD40 homodimers are required for CD40-induced phosphatidylinositol 3-kinase-dependent expression of B7.2 by human B lymphocytes. J. Biol. Chem. 279, 7799–7806 [DOI] [PubMed] [Google Scholar]
- 21. Girouard J., Reyes-Moreno C., Darveau A., Akoum A., Mourad W. (2005) Requirement of the extracellular cysteine at position six for CD40/CD40 dimer formation and CD40-induced IL-8 expression. Mol. Immunol. 42, 773–780 [DOI] [PubMed] [Google Scholar]
- 22. Reyes-Moreno C., Sharif-Askari E., Girouard J., Léveillé C., Jundi M., Akoum A., Lapointe R., Darveau A., Mourad W. (2007) Requirement of oxidation-dependent CD40 homodimers for CD154/CD40 bidirectional signaling. J. Biol. Chem. 282, 19473–19480 [DOI] [PubMed] [Google Scholar]
- 23. Wu Y., Han B., Sheng H., Lin M., Moore P. A., Zhang J., Wu J. (2003) Clinical significance of detecting elevated serum DcR3/TR6/M68 in malignant tumor patients. Int. J. Cancer 105, 724–732 [DOI] [PubMed] [Google Scholar]
- 24. Compaan D. M., Gonzalez L. C., Tom I., Loyet K. M., Eaton D., Hymowitz S. G. (2005) Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J. Biol. Chem. 280, 39553–39561 [DOI] [PubMed] [Google Scholar]
- 25. Deng G. M., Zheng L., Chan F. K., Lenardo M. (2005) Amelioration of inflammatory arthritis by targeting the pre-ligand assembly domain of tumor necrosis factor receptors. Nat. Med. 11, 1066–1072 [DOI] [PubMed] [Google Scholar]