Abstract
Of the various kinds of cell division, the most common mode is binary fission, the division of a cell into two morphologically identical daughter cells. However, in the case of asymmetric cell division, Caulobacter crescentus produces two morphologically and functionally distinct cell types. Here, we have studied cell cycle progression of the human pathogen Helicobacter pylori using a functional green fluorescent protein (GFP) fusion of FtsZ protein and membrane staining. In small cells, representing newly divided cells, FtsZ localizes to a single cell pole. During the cell cycle, spiral intermediates are formed until an FtsZ ring is positioned with very little precision, such that central as well as acentral rings can be observed. Daughter cells showed considerably different sizes, suggesting that H. pylori divides asymmetrically. Fluorescence recovery after photobleaching (FRAP) analyses demonstrate that the H. pylori FtsZ ring is about as dynamic as that of Escherichia coli but that polar assemblies show less turnover. Strikingly, our results demonstrate that H. pylori cell division follows a different route from that in E. coli and Bacillus subtilis. It is also different from that in C. crescentus, where cytokinesis regulation proteins like MipZ play a role. Therefore, this report provides the first cell-biological analysis of FtsZ dynamics in the human pathogen H. pylori and even in epsilonproteobacteria to our knowledge. In addition, analysis of the filament architecture of H. pylori and E. coli FtsZ filaments in the heterologous system of Drosophila melanogaster S2 Schneider cells revealed that both have different filamentation properties in vivo, suggesting a unique intrinsic characteristic of each protein.
INTRODUCTION
Helicobacter pylori is a Gram-negative, highly motile, microaerophilic, spiral-shaped organism which belongs to the class of the epsilonproteobacteria. The natural habitat of this pathogen is the human gastric mucosa, and infection of humans results in persistent gastritis, which can develop into peptic ulcer disease and adenocarcinoma (1, 2). Today, at least half of the world's human population is infected (3). Although extensive research has been conducted on H. pylori, remarkably little is known about the molecular basis of cell division in this important human pathogen. The comparison of the complete genome sequences of two H. pylori strains revealed that 14 homologs of Escherichia coli cell division and chromosome segregation genes have been recognized (4), and it was suggested that the basic mechanisms of replication and cell division are similar to those of E. coli. These are genes such as ftsZ, ftsA, and the ring inhibitor genes minC, minD, and minE. However, some orthologues of cell division proteins like ZipA and all periplasmic connector proteins are missing in H. pylori. These differences may be attributed to the smaller genome size of H. pylori as well as its life style; i.e., H. pylori is adapted to its unique niche in gastric mucus with a fixed temperature and slow doubling time, whereas E. coli is a free-living organism with fast proliferation under different temperatures (5).
In most organisms, cell division occurs after placement of a septum through the midpoint of the dividing cell and equal distribution of the cellular components into the two daughter cells (6). Division site determination is accomplished by FtsZ ring formation at the future septum. The Z ring is usually positioned at midcell early during the division process (7, 8) and serves as a scaffold for the assembly of the other cell division proteins. FtsZ assembly is tightly regulated, and a diverse repertoire of accessory proteins contributes to the formation of a functional division machinery that is responsive to cell cycle status. In rod-shaped bacteria like E. coli or Bacillus subtilis, FtsZ localizes either diffusely in the cell, in a helical pattern underneath the cell membrane, or as a FtsZ ring at the beginning of the division process. The positioning of the Z ring is dependent on the so-called Min and nucleoid occlusion systems (9), which prevent the assembly of Z rings at the cell poles and over chromosomal DNA. In particular, in E. coli, Min proteins oscillate from pole to pole, resulting in the formation of a zone of FtsZ inhibition at the cell poles. Protection of the replicated nucleoid DNA near the midcell from bisection by the Z ring is ensured by Noc in B. subtilis and by SlmA in E. coli (10). Both the Z ring and the helical localization of FtsZ are highly dynamic, with a high turnover rate (11, 12). In contrast to E. coli and B. subtilis, FtsZ of Caulobacter crescentus is clustered at a single cell pole before it is induced to assemble at midcell, a process that is regulated by MipZ (13).
In this study, we investigated cell cycle progression of the human pathogen H. pylori by monitoring FtsZ. To this end, we used our previously developed system (14) that permits in vivo localization of individual proteins in H. pylori. Our results indicate that the H. pylori FtsZ ring is positioned with very little precision, resulting in daughter cells showing considerably different sizes. FtsZ-ring formation and disassembly are also different from the processes in E. coli and B. subtilis. Thus, this report provides the first cell-biological analysis of FtsZ dynamics in the human pathogen Helicobacter pylori and even in epsilonproteobacteria to our knowledge. Furthermore, our results demonstrated that H. pylori and E. coli FtsZ (HpFtsZ and EcFtsZ, respectively) filaments have different filamentation properties in vivo in the heterologous system of Drosophila melanogaster S2 Schneider cells, suggesting a unique intrinsic characteristic of each protein despite the common function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are listed in Table 1. H. pylori strains were routinely cultivated on Dent blood agar in a microaerobic atmosphere as described earlier (19). Growth experiments were performed in brucella broth with 5% fetal calf serum (BBF). Growth rate was assessed by optical density at 600 nm (OD600). E. coli strains were grown aerobically at 37°C in Luria-Bertani medium. When appropriate, growth media were supplemented with 50 μg/ml ampicillin (Ap) or 20 μg/ml chloramphenicol (Cm).
Table 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Bethesda Research Laboratories |
| H. pylori | ||
| 26695 | Wild type containing the entire cag pathogenicity island | 15 |
| 1061 | Wild type | 16 |
| 1061-FtsZ-GFP | 1061 ftsZ-gfp (at original locus) | This study |
| KE88-3887 | Piglet-passaged strain 26695 | 17 |
| Plasmids | ||
| pSG1164 | bla cat Pxyl-gfpmut1 | 18 |
| pSG1164-FtsZ | bla Pxyl-ftsZ-gfpmut1 cat | This study |
| pFD1 | pRmHa3 containing a multiple cloning site as well as the coding sequence of the fluorescent protein gene of pSG1164 | P. L. Graumanna |
| pFD323 | pFD1 EcFtsZ-YFP | This study |
| pFD320 | pFD1 HpFtsZ-YFP | This study |
| pFD321 | pFD1 EcFtsZ | This study |
| pFD319 | pFD1 HpFtsZ | This study |
| Primers | ||
| FtsZup | ATTGGGCCCCTACGATTATCACCAAACCC | This study |
| FtsZdw | CTTGAATTCCCCTCCACCATCTTGTTGGATTCTTATGGTTG | This study |
| EcftsZ up | TCAGGGCCCATGTTTGAACCAATGGAACTTAC | This study |
| EcftsZ down NS | TCAATCGATATCAGCTTGCTTACGCAGGAAT | This study |
| EcftsZ down S | TCAATCGATTTAATCAGCTTGCTTACGCAGG | This study |
| HpftsZ up | TCAGGGCCCATGGTTCATCAATCAGAGATGG | This study |
| Hp ftsZ down NS | TCAATCGATGTCTTGCTGGATTCTCATGG | This study |
| Hp ftsZ down S | TCAATCGATTCAGTCTTGCTGGATTCTCATG | This study |
DNA techniques and mutagenesis of H. pylori.
Restriction and modifying enzymes (New England BioLabs) were used according to the manufacturer's instructions. Cloning was performed in E. coli according to standard protocols. Plasmids were isolated with a QIAprep Spin Miniprep Kit from Qiagen (Qiagen 27104). For the generation of a C-terminal green fluorescent protein (GFP) fusion of FtsZ, the ftsZ gene was amplified by PCR using the primer pair FtsZup/FtsZdw (Table 1), and the resulting 511-bp fragment was cloned at ApaI and EcoRI restriction sites on the pSG1164 vector (18). Integration of the fluorescent tag vector pSG1164-FtsZGFP into the H. pylori chromosome was achieved via single crossover using electroporation according to standard procedures (20). H. pylori 1061-FtsZGFP clones carrying the FtsZ-GFP fusion were selected on Dent blood agar with 20 mg/liter chloramphenicol. The correct placement of the integration was verified by PCR.
To create C-terminal fusions of E. coli and H. pylori FtsZ with enhanced yellow fluorescent protein(eYFP) for transfection of the S2 cells, the coding sequence of FtsZ was amplified by PCR using the primer pairs EcftsZ_up/EcftsZ_down_NS and HpftsZ_up/HpftsZ_down_NS, respectively. Additionally, E. coli and H. pylori FtsZ proteins were amplified by PCR using the primer pairs EcftsZ_up/EcftsZ_down_S and HpftsZ_up/HpftsZ_down_S, respectively. Plasmid pFD1 was constructed by combining the multiple cloning site as well as the coding sequence of the fluorophore of the plasmid pSG1164 (18) with the plasmid pRmHa3 (21) using KpnI and SpeI. Subsequently amplicons were cloned into the vector pFD1 using ApaI and ClaI, giving rise to the plasmids pFD323 (EcFtsZ-YFP), pFD320 (HpFtsZ-YFP), pFD321 (EcFtsZ), and pFD319 (HpFtsZ), respectively.
Immunoblotting and determination of GFP stability.
Cells were grown until log phase and then treated with puromycin dihydrochloride (50 mg/ml) for 10 min to 1 h. After protein determination, equal amounts of the samples were lysed in SDS-PAGE loading buffer by boiling for 5 min. SDS-PAGE and transfer of protein to polyvinylidene fluoride (PVDF) membrane were performed with standard procedures. Samples were probed with anti-GFP primary antibody. The same amount of wild-type protein extract and a verified GFP fusion protein served as negative and positive controls, respectively.
Cell culture of Schneider cells and transient transfection.
D. melanogaster S2 Schneider cells were grown in Schneider's Drosophila medium (Lonza Group, Ltd.) supplemented with 5 to 10% fetal calf serum (FCS) at 25°C without addition of CO2. Cells were passaged every 2 to 3 days to maintain optimal growth. S2 cells were transfected using FuGENE 6 transfection reagent (Roche). The S2 cells were spread in a six-well plate at 1 × 106 cells per well in 3 ml of medium with 5% FCS. Supercoiled plasmids (0.3 μg of each plasmid) were complexed with lipid (10 μl of FuGENE reagent) in 200 μl of serum-free medium. The complex was incubated at room temperature for 15 min and filled up with serum-free medium to 1 ml, and this was added to cells from which the growth medium had been removed (cells were washed once with serum-free medium). After 18 h, the supernatant was removed and replaced by 3 ml of medium containing 5% FCS. After further incubation for 24 h, the production of the proteins was induced by adding CuSO4 to a final concentration of 1 mM.
Immunofluorescence.
Immunofluorescence of H. pylori cells was performed as described earlier (22) with the following modifications: anti-C. glutamicum FtsZ antibody (1:100 dilution in 1× phosphate-buffered saline [PBS] and 100 μg/ml bovine serum albumin [BSA]) was used as a primary antibody, which was detected by the secondary antibody, goat anti-guinea pig coupled to fluorescein isothiocyanate (FITC; Invitrogen) (1:100 dilution in 1× PBS and 100 μg/ml BSA). For immunofluorescence of the S2 Schneider cells, 100 μl of transfected S2 cells was transferred onto a poly-l-lysine-treated glass slide and left to settle for 15 min. For fixation the slide was placed for 5 min in cooled methanol (−20°C) and afterwards for 30 s in cooled acetone (−20°C). After the sample was rehydrated with 1× PBS for 5 min at room temperature (RT), 100 μl of Image-iT Fx signal enhancer (Invitrogen) was applied and incubated overnight at 4°C. For visualization of FtsZ, anti-C. glutamicum FtsZ antibodies (1:100 dilution in 1× PBS; origin, guinea pig) were added and incubated for 90 min at RT. Unbound antibodies were removed by three washes with 1× PBS (5 min at RT), and FtsZ signals were visualized by the secondary antibody, goat anti-guinea pig coupled to FITC (Invitrogen) (1:100 dilution in 1× PBS; incubation for 1 h at RT). After samples were washed for 5 min at RT, cells were mounted with fluorescent mounting medium (Dako).
Scanning electron microscopy.
Infected monolayers grown on coverslips were fixed with 3% glutaraldehyde in PBS at room temperature. After several washing steps with PBS, samples were dehydrated with a gradient series of ethanol (30, 50, 60, 70, 80, 90, and 100%) at room temperature for 12 h for each step. Samples were then subjected to critical-point drying with liquid CO2 (CPD030; Bal-Tek). Dried samples were covered with gold film by sputter coating for 80 s (SCD 050; Bak-Tek). Examinations were performed in a field emission scanning electron microscope (FEI Quanta 250 FEG) using an Everhart Thornley detector and an acceleration voltage of 5 kV.
Fluorescence microscopy.
Fluorescence microscopy was performed on a Zeiss Axioobserver Z1 microscope using a 100× objective with a numerical aperture of 1.45. Cells were mounted on agarose gel pads containing brucella liquid medium on object slides. Images were acquired with a digital Cascade electron microscopy charge-coupled-device (EM-CCD) camera (Photometrix); signal intensities and cell length were measured using Metamorph, version 6.3, software (Universal Imaging Corp.). Membranes were stained with FM4-64 (final concentration, 1 nM). The following filters were used: for GFP, 460- to 495-nm excitation, dc505 (where dc is dichroic), and 510- to 550-nm emission; for FM4-64, 480- to 550-nm excitation, dc570, and 590-nm emission. For monitoring cell division under the microscope, we used a heating stage at 32°C and a CO2 atmosphere of 5% producing a microaerophilic milieu (Tokai Hit).
FRAP.
Cells were imaged, and fluorescence after photobleaching (FRAP) experiments were performed on a Zeiss Axioobserver Z1 microscope equipped with a 50-mW solid-phase laser. The laser was focused to 1 μm through a lens system, and the cell in question was positioned into the stationary 405-nm laser beam. Fluorescence of the selected region of interest was bleached for 0.1 s. All images and FRAP measurements were taken at room temperature. A prebleach image and postbleach images were acquired with 488-nm laser excitation. A postbleach image series was taken automatically every 0.1 s using the Metamorph program and statistically analyzed using ImageJ (NIH) in combination with Excel (Microsoft). Calculations were done according to Schulmeister et al. (23). Briefly, fluorescence intensity of the region of interest (ROI) was measured automatically in image sequences, using a custom-written ImageJ plug-in. Gradual bleaching of the image was compensated during scanning by normalizing the fluorescence of the ROI to the integral fluorescence of the entire cell in the same image. To facilitate comparison of multiple experiments with different bleaching depths and different cluster intensities, the relative fluorescence intensity of the ROI in the image sequence was normalized again to the relative ROI intensity before bleaching. Data were subsequently processed by using Excel.
RESULTS
FtsZ localization in H. pylori.
To analyze cell cycle progression in H. pylori, we generated a C-terminal GFP fusion of FtsZ that is expressed as the sole source of the protein in strain 1061. Analysis of the growth characteristics revealed that the fusion was fully functional at 32°C and partially functional at 37°C because cells displayed a filamentous phenotype at higher temperatures (see Fig. S1A in the supplemental material). We further confirmed fusion protein abundance and fusion protein stability by Western blotting of cell extracts with antibodies to GFP (Fig. 1A). To this end, H. pylori cells were treated with puromycin to inhibit protein biosynthesis. This treatment revealed that the FtsZ-GFP fusion is very stable because the fusion was still visible even after 60 min of incubation (Fig. 1A). In addition this experiment confirmed that GFP was not cleaved off.
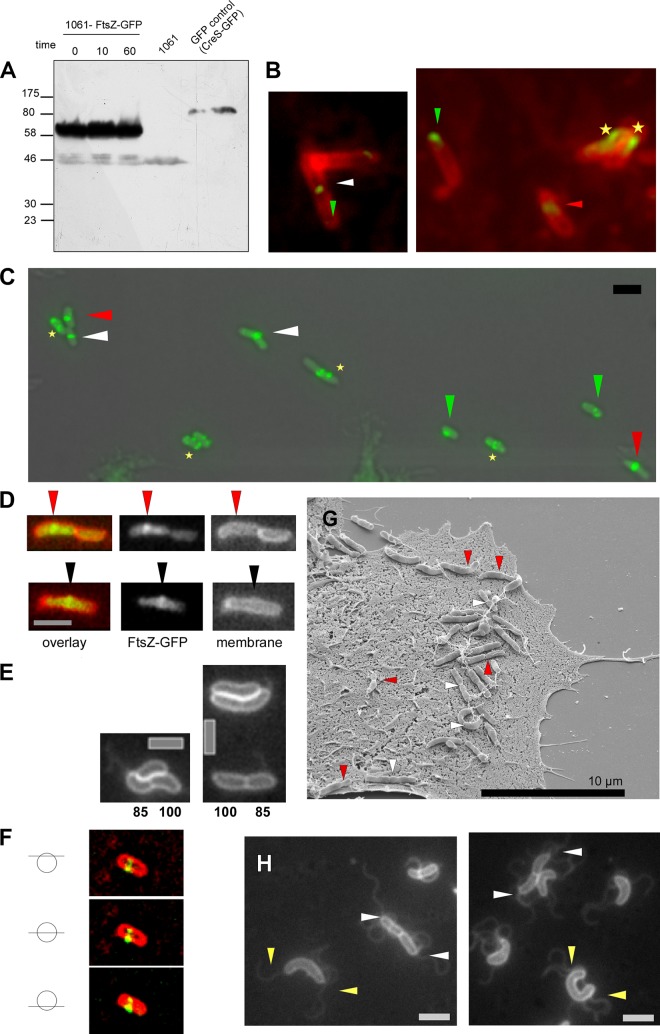
Fig 1.
(A) Western blot analysis of puromycin-treated H. pylori cells expressing FtsZ-GFP with antibodies to GFP. Incubation time of puromycin is indicated. GFP-control (GFP fusion protein of crescentin served as a positive control) and 1061 wild-type cells (negative control) are marked. (B) Immunofluorescence images with anti-C. glutamicum FtsZ antiserum. White triangles indicate localization of the FtsZ ring at midcell, red triangles indicate asymmetric localization of the FtsZ-ring, green triangles indicate polar foci of FtsZ in small cells, and asterisks indicate speckled/helical FtsZ pattern. (C) Fluorescence microscopy of H. pylori 1061 cells expressing the FtsZ protein as a FtsZ-GFP fusion. White triangles indicate localization of the FtsZ-ring at midcell, red triangles indicate asymmetric localization of the FtsZ-ring, green triangles indicate polar foci of FtsZ in small cells, and asterisks indicate speckled/helical FtsZ pattern. (D) Overlay of FtsZ-GFP (green) and membrane stain (red). Red triangles indicate asymmetric localization; black triangles indicate midcell localization. (E) Membrane stains of small (and thus young) cells supporting asymmetric division. Numbers indicate average sizes of smaller daughter cells relative to the larger cells. (F) Z stack of a straight H. pylori cell displaying different sections of the asymmetric localization of the FtsZ ring. (G) Scanning electron microscopy (SEM) of H. pylori cells during infection of AGS (human gastric adenocarcinoma epithelial cell line) cells. Asymmetric cell division is indicated by red triangles, and symmetric cell division is marked with white triangles. (H) Membrane staining of exponentially growing H. pylori wild-type cells. White triangles indicate localization of flagella in newly divided cells; yellow triangles indicate flagella in cells prior to septum formation. Scale bar, 2 μm.
Subsequently, we analyzed 1061-FtsZ-GFP cells grown in liquid culture until log phase using epifluorescence microscopy. In this phase H. pylori has a tight spiral shape, whereas coccoid cells start to appear at the earliest in late log phase (24). Single midcell bands of FtsZ were visible in large cells (>2.2 μm), verifying that the GFP tagging system can produce functional fusions in live cells (Fig. 1C, white triangles). Surprisingly, FtsZ bands were positioned off center in about 50% of the cells (85 cells analyzed) (Fig. 1C, red triangles), suggesting an asymmetric cell division. As spiral or mildly curved bacteria might introduce visual artifacts (e.g., as the entire bacterial body might not be in the same focal plane), we further confirmed our finding using 1061-FtsZ-GFP H. pylori cells which displayed a straighter phenotype. To this end, we performed Z stacks of the asymmetric localized Z ring demonstrating that the localization is indeed not a visual artifact (Fig. 1F). In this context it has to be noted that straight wild-type cells are always found to a small extent in H. pylori cell cultures.
To support the idea of asymmetric cell division, we measured the distance of the division septum to the two old cell poles in dividing cells (Fig. 1D and Table 2). The measurement of cells (n = 60) demonstrated that on average one daughter cell had 80.8% of the length of the other daughter (Table 2), in support of an asymmetric cell division. To clarify this finding, we split the cells into two groups according to their Z-ring positions: cells with symmetric cell division were defined to have a midcell position of the Z ring which varies less than 10% of the middle position, whereas the asymmetric localization was defined as more than 10% variation of the Z ring from the cell center. Thus, the average size of the small part as a percentage of the large part was 90.7% or 70.3% in the case of symmetric or asymmetric cell division, respectively. This shows that positioning of the Z ring during cell division is more relaxed in H. pylori than in E. coli or in B. subtilis, where the central positioning of the Z rings varies less than 5% (25). Strikingly, this is even true in the case of the symmetrically positioned H. pylori Z ring. Thus, cell division in H. pylori occurred in an asymmetric manner more similar to that in C. crescentus.
Table 2.
Distance of the division septum to the two old cell poles in dividing cells
| Z-ring position | Avg size of the small part of diving cell as a percentage of the large part | Total cell length (μm) | Distance to Z ring (μm) by cell section |
|
|---|---|---|---|---|
| Large part | Small part | |||
| Out of center | 70.3 | 3.8 ± 1.15 | 2.3 ± 0.6 | 1.6 ± 0.4 |
| Center (±10%) | 90.7 | 3.3 ± 1.0 | 1.8 ± 0.6 | 1.6 ± 0.5 |
| Total | 80.8 | 3.6 ± 1.17 | 2.0 ± 0.7 | 1.6 ± 0.5 |
Interestingly, small cells (up to 2.3 μm) predominantly contained clear FtsZ-GFP foci at a single cell pole (Fig. 1B, green triangles). These small cells constitute young cells just after complete cell division because these cells are predominantly found in the exponential growth phase. In contrast, coccoid cells displayed a complete delocalization of FtsZ (data not shown). In this context it is noteworthy that, because of polar FtsZ-GFP foci, viable cells were clearly distinguishable from cells which started dying and therefore became coccoid. Thus, this observation suggests that polar FtsZ-GFP foci relocate from the new pole to the midcell region during the course of the cell cycle.
In order to confirm these different localizations independently of the GFP fusion, we performed immunofluorescence analysis in strain 1061, using an anti-FtsZ antibody against Corynebacterium glutamicum FtsZ. This antibody was used because a BLAST search using the Comprehensive Microbial Resource (CMR) website (http//:cmr.jcvi.org) revealed that C. glutamicum FtsZ is more closely related to H. pylori FtsZ than E. coli FtsZ (58.9% similarity and 38% identity compared to 55.3% similarity and 33.1% identity, respectively). Indeed, immunofluorescence analysis verified symmetric and asymmetric positions of the Z ring as well as polar foci (Fig. 1C). As a further control, we performed immunofluorescence analysis with aztreonam-treated H. pylori wild-type cells. Unevenly spaced Z rings in the filamentous cells could be seen (see Fig. S1B in the supplemental material), supporting the asymmetric positioning of the Z ring.
Thus, our results show that there are two main patterns of localization of H. pylori FtsZ: at one single cell pole and in the Z ring, which can be positioned symmetrically as well as asymmetrically. This finding is strikingly dissimilar to the situation in E. coli and B. subtilis, and cell cycle progression is more reminiscent of that in C. crescentus. However, H. pylori does not contain homologs of cytokinesis regulation proteins that are abundant in C. crescentus, e.g., MipZ and TipN.
An asymmetrically positioned Z ring results in two daughter cells of different sizes.
Next, we wanted to determine if the asymmetric localization of the Z ring results in two daughter cells with different sizes or if our finding constitutes only a snapshot followed by increased growth of the smaller cell. Therefore, we measured the cell length of newly divided wild-type cells in H. pylori strains 1061, 26695, and KE88-3887 using a fluorescent membrane stain. Newly divided cells were defined as two cells which were separated by a membrane (which could be easily seen with the membrane stain) but which were still attached to each other. Our measurements demonstrate that the asymmetric position of the Z ring indeed resulted in two daughter cells of different sizes (Fig. 1E and Table 3). All three wild-type strains displayed the same relaxed type of cell division. In 50% of the dividing wild-type cells, the lengths of the cells were 95% ± 0.5% of each other, whereas the other 50% wild-type cells had one daughter which was only 80% (±1%) the length of the other (with about 80 cells analyzed for each strain).
Table 3.
Cell length of newly divided wild-type cells
| Strain | Cell division type | Avg size of the small cell as a percentage of the large cell | Total cell length (μm) | Distance to cell constriction (μm) in: |
|
|---|---|---|---|---|---|
| Large cell | Small cell | ||||
| 26695 | Symmetric | 95.7 | 4.5 ± 0.6 | 2.3 ± 0.3 | 2.2 ± 0.4 |
| Asymmetric | 81.0 | 4.6 ± 0.9 | 2.5 ± 0.6 | 2.0 ± 0.4 | |
| KE | Symmetric | 94.7 | 4.3 ± 0.5 | 2.2 ± 0.3 | 2.0 ± 0.3 |
| Asymmetric | 80.5 | 4.6 ± 0.7 | 2.5 ± 0.4 | 2.0 ± 0.3 | |
| 1061 | Symmetric | 94.6 | 4.6 ± 0.9 | 2.4 ± 0.5 | 2.3 ± 0.4 |
| Asymmetric | 78.5 | 4.0 ± 0.9 | 2.3 ± 0.5 | 2.0 ± 0.4 | |
In addition, we analyzed the division site placement by observing the cell constriction using scanning electron microscopy (SEM) of H. pylori cells. AGS cells (human gastric adenocarcinoma epithelial cell line) were infected with strain KE88-3887. SEM pictures clearly corroborated the findings that the imprecision of the Z-ring positioning causes asymmetric (Fig. 1G, red triangles) as well as symmetric (Fig. 1G, white triangles) cell constriction. Intriguingly, these results demonstrate that this mode of cell division also occurred during H. pylori infection. To rule out that growth conditions affect the precision of cell division, we grew E. coli K-12 wild-type cells under both aerobic and microaerophilic conditions and measured the cell lengths of newly divided cells according to the procedure in H. pylori. Cell lengths were identical, with an aberrance of less than 5% under both conditions (data not shown).
These experiments substantiate that the observed asymmetrically localized Z ring indeed generated daughter cells that differ in size. Furthermore, a possible wild-type strain-dependent effect could be ruled out.
H. pylori flagella do not act as polar markers during cell division.
We also analyzed the localization of the monopolar flagella of newly divided cells in order to determine whether there is a preferred positioning of flagella during cell division. H. pylori cells possess two to eight polar flagella, which are covered by a membranous sheath (26). Thus, these flagella are visible by the use of a fluorescent membrane stain. However, it must be pointed out that the flagella are not observed in all cells because they can become sheared during application of the cells onto the agarose-containing microscope slides or may be present in a different focal plane from that of the cell body. Analysis of H. pylori wild-type cells during the exponential growth phase revealed that the polar flagella were visible at both ends of newly divided cells irrespective of cell length (Fig. 1H, white triangles). In addition, we found some large cells which already possessed two polar bundles of flagella, indicating that the formation of flagella occurs prior to the placement of a septum (Fig. 1H, yellow triangles). It was not possible to define the precise time point of flagellum formation during cell cycle progression because of the fastidious growth requirements of this bacterium (see below).
FRAP of the Z ring and the polar FtsZ foci.
In E. coli and B. subtilis, the Z ring shows high turnover inside live cells, with fluorescence after photobleaching (FRAP) half-times of 6 to 9 s (27). In order to study the in vivo dynamics of the Z ring in H. pylori, we used FRAP to examine the dynamics of the H. pylori FtsZ polymers. To facilitate data analysis, the fluorescence intensity in the region of interest (ROI) was normalized to the fluorescence of the entire cell at each time point; these ratio values were subsequently renormalized to the prebleach ratios. One representative FRAP time series is shown in Fig. 2A. We bleached half of a Z ring and calculated a half-time of recovery of about 10 s (n = 5) (Fig. 2A), which is similar to half-times measured in E. coli and B. subtilis. Nevertheless, this result is interesting as H. pylori has a considerably slower cell cycle with a generation time of about at least 3 h (28) in comparison to a maximal doubling time of 20 min in E. coli. Therefore, our results indicate that the duration of the cell cycle is independent of the turnover rate of the Z ring.
Fig 2.
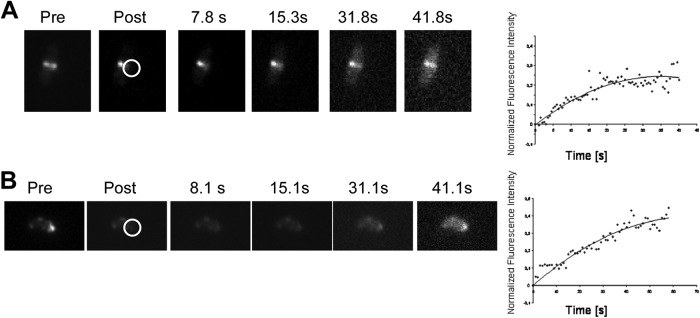
FRAP experiment on FtsZ-GFP-expressing H. pylori 1061 cells. (A) FRAP of the H. pylori Z ring. (B) FRAP of polar FtsZ foci. The right side shows one representative FRAP time series, and the left side demonstrates the mean value of the normalized fluorescence intensity plotted against time of at least five independent experiments. pre, before bleaching; post, after bleaching. Seconds after bleaching are indicated. Circles indicate the area of bleaching.
Next, we performed FRAP experiments of FtsZ at its polar location. We bleached a small area close to a cell pole and monitored the recovery of fluorescence. Figure 2B (left) shows an example of a FRAP experiment. Interestingly, we calculated a half-time of recovery of about 18 s (n = 5) (Fig. 2B, right). Therefore, we assume that these foci are at least ordered structures. Also, these structures were distinct from the filaments in the Z ring as the half-time was almost twice as long.
Monitoring of FtsZ during cell cycle progression in H. pylori.
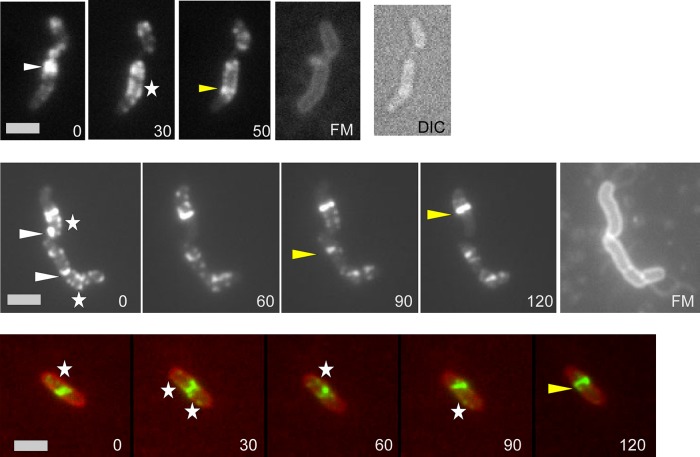
In order to visualize cell cycle progression in H. pylori, we monitored cell division under the microscope using a heating stage at 32°C and a CO2 atmosphere of 5% producing a microaerophilic milieu. Due to the fastidious environmental requirements of H. pylori, monitoring was not possible over a complete cell cycle in this human pathogen; however, we were able to perform time-lapse microscopy, in which we could follow FtsZ-GFP (Fig. 3) over different time periods. Thus, it was possible to observe FtsZ-GFP moving from the polar localization (Fig. 3, white triangles) to the localization where the Z ring was built (Fig. 3, yellow triangles). This confirms that the polar foci of FtsZ-GFP are functional and that the polar accumulation of FtsZ is indeed part of the cell cycle progression in H. pylori. Interestingly, this movement seemed to follow a spiral pattern (Fig. 3, white asterisks), which appears to be similar to results obtained in E. coli, B. subtilis, and Streptomyces coelicolor, in which FtsZ also localizes in a dynamic helical pattern associated with repositioning of the Z ring (29). These helical FtsZ patterns in H. pylori could be visualized as a clearly distinct pattern (Fig. 3) because there was very little background fluorescence within the cell. In the numerous time-lapse experiments, the FtsZ spirals condensed at central as well as at peripheral positions in a random manner.
Fig 3.
Time-lapse microscopy of FtsZ-GFP. White triangles indicate polar foci of FtsZ; yellow triangles indicate the Z ring. Spiral patterns are marked with white asterisks. Numbers indicate the time in minutes. FM, membrane stain FM4-64. The lower panel shows an overlay of the FtsZ-GFP fusion (green) and membrane stain FM4-64 (red). DIC, differential interference contrast. Scale bar, 2 μm.
FtsZ filaments of H. pylori and of E. coli differ in the heterologous system of S2 Schneider cells.
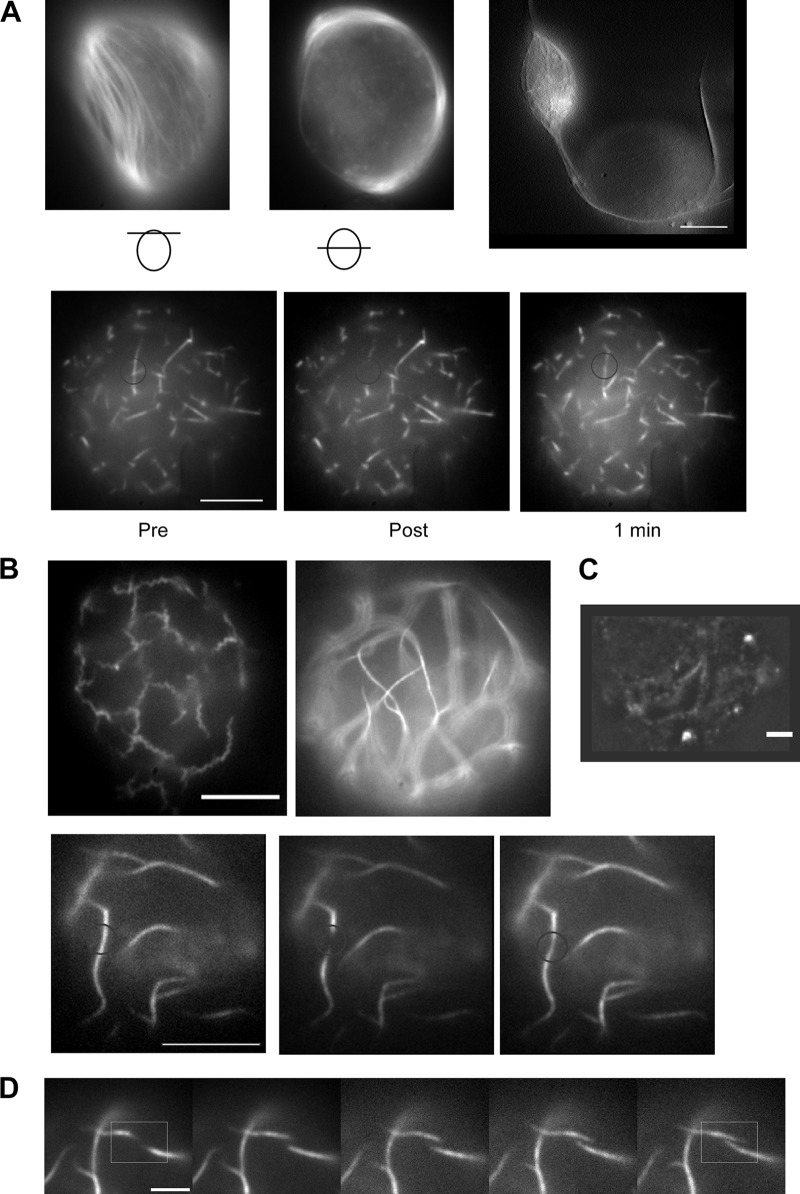
Previously, we have shown that the heterologous system of D. melanogaster S2 Schneider cells (derived from macrophages) is convenient to study filamentation properties of different cytoskeleton elements in vivo (14). In order to characterize filament architecture and filament dynamics, we transfected S2 cells with plasmids (Table 1) containing H. pylori or E. coli FtsZ-YFP in combination with the respective wild-type FtsZ. Coinduction of wild-type and tagged proteins was used to avoid tag artifacts. For E. coli, straight and rarely branched filamentous structures could be observed soon after induction of transcription (Fig. 4A, lower panel). Further induction resulted in long filaments that were ordered in parallel and exclusively found underneath the membrane, which was shown by imaging of different Z planes within cells (Fig. 4A, upper panel). Interestingly, some of these filaments reached a length of more than 20 μm, causing striking cell extrusions (Fig. 4A, upper panel). The localization underneath the membrane, which enables the highest possible straightness of these filaments as well as causing protrusions, supports the idea of a remarkable stiffness of E. coli FtsZ filaments in S2 Schneider cells. Qualitative FRAP experiments demonstrated that fluorescence recovery occurred within 1 min, indicating functional filaments with subunit turnover (Fig. 4A, lower panel).
Fig 4.
Expression of FtsZ-YFP in combination with wild-type FtsZ in S2 Schneider cells. (A and B) Upper panels demonstrate distinct morphologies, and lower panels show representative FRAP results for E. coli FtsZ (A) and H. pylori FtsZ (B) (C) Immunofluorescence images of H. pylori expressing S2 Schneider cells with anti-C. glutamicum FtsZ antiserum. (D) Time series follows filament polymerization of H. pylori FtsZ. Bars, 5 μm (A, B, and D) and 2 μm (C).
Contrarily, H. pylori FtsZ filaments were found to have two distinct patterns. Either FtsZ-YFP filaments were curled (Fig. 4B, upper panel, left image) or straight (Fig. 4B upper panel, right image). Interestingly, there were no cells with mixed types of filaments. Furthermore, even straight filaments differed from E. coli filaments as they were completely detached from the membrane and never caused cell extrusions. These filaments were also seen by immunofluorescence using the anti-FtsZ antibody against Corynebacterium glutamicum FtsZ, supporting the specificity of this serum (Fig. 4C). Qualitative FRAP experiments confirmed the subunit turnover of both kinds of filaments (Fig. 4B, lower panel; also data not shown). Surprisingly, it was possible in some cases to follow filament polymerization via time-lapse microscopy (Fig. 4D). Therefore, it was possible to calculate overall filamentation speed by measuring filament extension over time (n = 5). The average of polymerization/depolymerization was 0.35 ± 0.04 μm/min with a maximum of 0.4 μm/min.
Thus, our results demonstrated that H. pylori and E. coli FtsZ filaments have different filamentation properties in vivo in the heterologous system of D. melanogaster S2 Schneider cells, suggesting unique intrinsic characteristics of each protein despite their common functions.
DISCUSSION
This report provides the first cell-biological analysis of FtsZ dynamics in the human pathogen Helicobacter pylori and even in epsilonproteobacteria, a group of organisms that has hardly been studied at the cell-biological level so far. Indeed, recent research discerned that biological strategies employed by model organisms, which have contributed greatly to our knowledge of basic biology and pathogenesis, do not always represent those of other bacterial species as these model species represent only a small fraction of the known bacterial diversity (30).
By using an ftsZ-gfp fusion expressed from the original gene locus, we demonstrate that approximately 50% of the H. pylori cells show clearly asymmetrically localized FtsZ rings. However, even in cells with an apparent midcell Z ring, there was 10% variation of Z-ring positioning, which is much larger than the maximum 5% variation observed in E. coli or B. subtilis (25). Furthermore, measurement of the cell length of newly divided wild-type cells revealed that the asymmetrically positioned Z ring indeed resulted in daughter cells of different lengths. We confirmed our findings by observing the cell constriction of H. pylori cells by the use of scanning electron microscopy (SEM) during infection of a human gastric adenocarcinoma epithelial cell line. Moreover, these results demonstrate that asymmetric cell constriction also occurred during H. pylori infection in this model system. Thus, in half of the cases the H. pylori FtsZ ring localizes asymmetrically, giving rise to two daughter cells of different sizes, which leads to the suggestion of an asymmetric cell division. However, we cannot exclude the possibility that the asymmetry arises from differential growth of the daughter cell compartments. These resulting daughter cells are of different sizes but seem to have apparent identical functions. On the other hand, there could be a yet unidentified difference in the cell fate of these cells, which will be the subject of further studies.
In contrast to E. coli, which has flagella in a peritrichous arrangement, H. pylori has to define the localization of the formation of the new flagella. However, H. pylori does not contain a TipN homologue, which regulates cell polarity and therefore the placement of the flagellum in C. crescentus. In any case, it is clear that both large and small H. pylori daughter cells are indistinguishable in terms of obtaining the new flagellum machinery because flagella were always observed at both cell poles, even shortly before cell division. These findings suggest that asymmetry may be a stochastic process although it is possible that asymmetric factors exist which have not yet been identified.
In addition, small cells often displayed polar FtsZ-GFP foci. Time-lapse analyses following the movement of FtsZ from its polar position to midcell suggest that FtsZ relocates from the new pole to the midcell region, forming spiral-like intermediates, during the course of the cell cycle. This behavior is reminiscent of the dynamics of FtsZ in C. crescentus. However, H. pylori does not contain MipZ, which regulates FtsZ localization in C. crescentus (13), but instead has MinC, MinD, and MinE, which prevent the formation of a Z ring close to the cell poles in E. coli and B. subtilis (31). These observations show that cell division in H. pylori follows a different route from that in E. coli and B. subtilis and is also dissimilar to that of C. crescentus.
Previous research using FRAP (fluorescence recovery after photobleaching) analysis revealed that the Z ring is very dynamic and that subunits in the Z ring are exchanging with those in the cytoplasm on a time scale of 8 to 11 s (32). As the turnover rates were very similar in both E. coli and B. subtilis, it has been suggested that this rate is a common feature in bacteria. Consistent with this assumption, we found a half-time recovery of 10 s of the Z ring in H. pylori despite the considerably slower cell cycle (28) of this bacterium. In contrast to this, FRAP analysis of the Z ring in Mycobacterium smegmatis, which has a slower cell cycle as well, gave an average turnover half-time of 34 s, with a broad spread from 10 to 70 s (32). However, the authors of this study subsequently considered that the FRAP data for M. smegmatis were much more scattered than for E. coli, and they had to exclude 20% of the measurements (33). In addition, we also performed FRAP experiments on the polar FtsZ foci in H. pylori. Interestingly, we calculated a 2-fold slower turnover half-time for this ill-defined structure. In E. coli, the dynamics of FtsZ outside the Z ring are fast, indicating freely diffusing molecules (12, 34). These findings demonstrate that the situation of FtsZ outside the Z ring in H. pylori is different from that in E. coli and that these foci are at least partially ordered structures. However, whether these structures were distinct from the filaments in the Z ring or associated with a distinct set of proteins regulating their dynamics in a different manner will be the subject of further studies. Time-lapse experiments confirmed that the polar foci of FtsZ-GFP are functional and that the polar accumulation of FtsZ was indeed part of the cell cycle progression in H. pylori.
Furthermore, we characterized and compared filament architecture and filament formation of both H. pylori and E. coli FtsZ in the heterologous system of D. melanogaster S2 Schneider cells (14). Whereas E. coli FtsZ built up long filaments that were ordered in parallel and were exclusively found underneath the cell membrane, H. pylori FtsZ filaments were found in the cytoplasm detached from the membrane in two distinct patterns, which were either curled or straight. One possible explanation for these two distinct filament architectures might be a concentration-dependent switch in polymerization. Hence, single curled filaments were seen in the case of a small amount of fusion protein. By exceeding a threshold concentration, the same single filaments intercoiled and therefore gave rise to a bundle of filaments. This bundle would have appeared as a single smooth structure due to the resolution of fluorescence microscopy. The finding that, indeed, curled filaments displayed lower fluorescence intensity than the straight structures supports our model. Previous studies have shown that E. coli FtsZ requires a membrane tether to attach to the membrane (10). We therefore assume that localization underneath the membrane occurred because this position allows the highest possible straightness of FtsZ filaments which are longer than the cell diameter of the round S2 cell. Thus, this localization as well as the finding that some of these filaments caused striking cell extrusions suggests a remarkable stiffness of E. coli FtsZ filaments in S2 Schneider cells.
To summarize, we suggest a model of cell division in H. pylori in which FtsZ accumulates at one cell pole after a complete cell division and then starts moving from this pole to the next localization of cell division, building up the Z ring. Thereby, the FtsZ ring is positioned with little precision, such that central as well as acentral rings can be observed. Daughter cells showed considerably different sizes, suggesting that H. pylori divides asymmetrically. Overall, our results provide evidence that the cell cycle of H. pylori is clearly dissimilar to the E. coli cell cycle and more similar to that of C. crescentus in spite of a dissimilar division machinery, which has important implications for future research on the human pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marc Bramkamp for providing the anti-C. glutamicum FtsZ antibody, Maren Lingnau for technical assistance, Jihad El Andari for technical assistance concerning S2 Schneider cell culture, and Peter Graumann for helping with fluorescence microscopy and writing of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (WA2574/1-1, WA2574/1-2, and FOR 929).
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01490-12.
REFERENCES
- 1. Blaser MJ. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161: 626–633 [DOI] [PubMed] [Google Scholar]
- 2. Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i: 1311–1315 [DOI] [PubMed] [Google Scholar]
- 3. Go MF. 2002. Review article: natural history and epidemiology of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 16(Suppl. 1):3–15 [DOI] [PubMed] [Google Scholar]
- 4. Takeuchi H, Nakazawa T. 2001. Chromosomal replication, plasmid replication, and cell division, p 259–268 In Mobley HLT, Mendz GL, Hazell SL. (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 5. Nakazawa T, Takeuchi H. 2008. Replication, partitioning, segregation and cell division in Helicobacer pylori, p 179–192 In Yamaoka Y. (ed), Helicobacter pylori: molecular genetics and cellular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 6. Rothfield L, Taghbalout A, Shih YL. 2005. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3: 959–968 [DOI] [PubMed] [Google Scholar]
- 7. Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- 8. Moller-Jensen J, Lowe J. 2005. Increasing complexity of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 17: 75–81 [DOI] [PubMed] [Google Scholar]
- 9. Vats P, Yu J, Rothfield L. 2009. The dynamic nature of the bacterial cytoskeleton. Cell. Mol. Life Sci. 66: 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Boer PA. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thanedar S, Margolin W. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14: 1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters PC, Migocki MD, Thoni C, Harry EJ. 2007. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 64: 487–499 [DOI] [PubMed] [Google Scholar]
- 13. Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- 14. Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. 2009. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 5: e1000669 doi:10.1371/journal.ppat.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 16. Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28: 383–393 [DOI] [PubMed] [Google Scholar]
- 17. Hoffman PS, Vats N, Hutchison D, Butler J, Chisholm K, Sisson G, Raudonikiene A, Marshall JS, Veldhuyzen van Zanten SJ. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain of Helicobacter pylori. Infect. Immun. 71: 2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewis PJ, Marston AL. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227: 101–110 [DOI] [PubMed] [Google Scholar]
- 19. van Vliet AH, Kuipers EJ, Waidner B, Davies BJ, de, Penn VNCW, Vandenbroucke-Grauls CM, Kist M, Bereswill S, Kusters JG. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69: 4891–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ge Z, Jiang Q, Kalisiak MS, Taylor DE. 1997. Cloning and functional characterization of Helicobacter pylori fumarate reductase operon comprising three structural genes coding for subunits C, A and B. Gene 204: 227–234 [DOI] [PubMed] [Google Scholar]
- 21. Bunch TA, Grinblat Y, Goldstein LS. 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16: 1043–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mukherjee A, Lutkenhaus J. 1998. Purification, assembly, and localization of FtsZ. Methods Enzymol. 298: 296–305 [DOI] [PubMed] [Google Scholar]
- 23. Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. 2008. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnamurthy P, Phadnis SH, DeLoney CR, Rosenthal RS, Dunn BE. 2001. Biosynthetic pathways related to cell structure and function, p 159–166 In Mobley HLT, Mendz GL, Hazell SL. (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 25. Harry EJ. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40: 795–803 [DOI] [PubMed] [Google Scholar]
- 26. Geis G, Suerbaum S, Forsthoff B, Leying H, Opferkuch W. 1993. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J. Med. Microbiol. 38: 371–377 [DOI] [PubMed] [Google Scholar]
- 27. Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186: 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynolds DJ, Penn CW. 1994. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology 140: 2649–2656 [DOI] [PubMed] [Google Scholar]
- 29. Cabeen MT, Jacobs-Wagner C. 2010. The bacterial cytoskeleton. Annu. Rev. Genet. 44: 365–392 [DOI] [PubMed] [Google Scholar]
- 30. Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR. 2011. Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol. Mol. Biol. Rev. 75: 84–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Errington J, Daniel RA, Scheffers DJ. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74: 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Y, Anderson DE, Rajagopalan M, Erickson HP. 2007. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 282: 27736–27743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.