Abstract
Many microbial species form multicellular structures comprising elaborate wrinkles and concentric rings, yet the rules governing their architecture are poorly understood. The opportunistic pathogen Pseudomonas aeruginosa produces phenazines, small molecules that act as alternate electron acceptors to oxygen and nitrate to oxidize the intracellular redox state and that influence biofilm morphogenesis. Here, we show that the depth occupied by cells within colony biofilms correlates well with electron acceptor availability. Perturbations in the environmental provision, endogenous production, and utilization of electron acceptors affect colony development in a manner consistent with redox control. Intracellular NADH levels peak before the induction of colony wrinkling. These results suggest that redox imbalance is a major factor driving the morphogenesis of P. aeruginosa biofilms and that wrinkling itself is an adaptation that maximizes oxygen accessibility and thereby supports metabolic homeostasis. This type of redox-driven morphological change is reminiscent of developmental processes that occur in metazoans.
INTRODUCTION
The ubiquity of multicellularity—a property observed in all three domains of life—underscores the advantages conferred on organisms that assume this lifestyle (1, 2). Multicellularity allows division of labor, as well as protection from environmental insults, but also presents a significant challenge by exacerbating limitations for growth substrates. Eukaryotic macroorganisms alleviate this problem in part through (i) internal circulation that allows delivery of substrates to specific locations and (ii) metabolic differentiation. Although the significance of the multicellular lifestyle for metabolism and pathogenicity of microorganisms is well recognized (3–5), the mechanisms enabling microbial communities to cope with substrate limitation are poorly defined.
Pseudomonas aeruginosa is a leading causative agent of nosocomial infections that forms biofilms (i.e., surface-attached communities) on indwelling medical devices or tissues within the host. It is also the primary cause of morbidity and mortality among people with cystic fibrosis, in whom it aggregates within accumulated mucus, causing chronic lung infections (6). The fact that P. aeruginosa forms biofilms in a variety of model laboratory systems has allowed researchers to identify mechanisms important for biofilm and aggregate formation. Our research has focused on the intricate structures formed by communities of P. aeruginosa as they grow on the surfaces of rich media solidified with agar (“colony biofilms”). Colony morphogenesis is highly dependent on the presence of endogenously produced redox-active small molecules called phenazines; colonies that produce phenazines are relatively smooth as they develop, while mutants that are unable to produce phenazines are more rugose and start wrinkling earlier in the incubation period (7) (Fig. 1A). Similar results have been obtained for flow cell biofilms (8). The phenazines produced by P. aeruginosa vary in structure and chemical properties (9, 10), but their redox potentials are such that they all can be reduced by the bacterial cell and react extracellularly with higher-potential oxidants, such as ferric iron and oxygen, acting as electron shuttles between the bacterium and an external substrate (11).
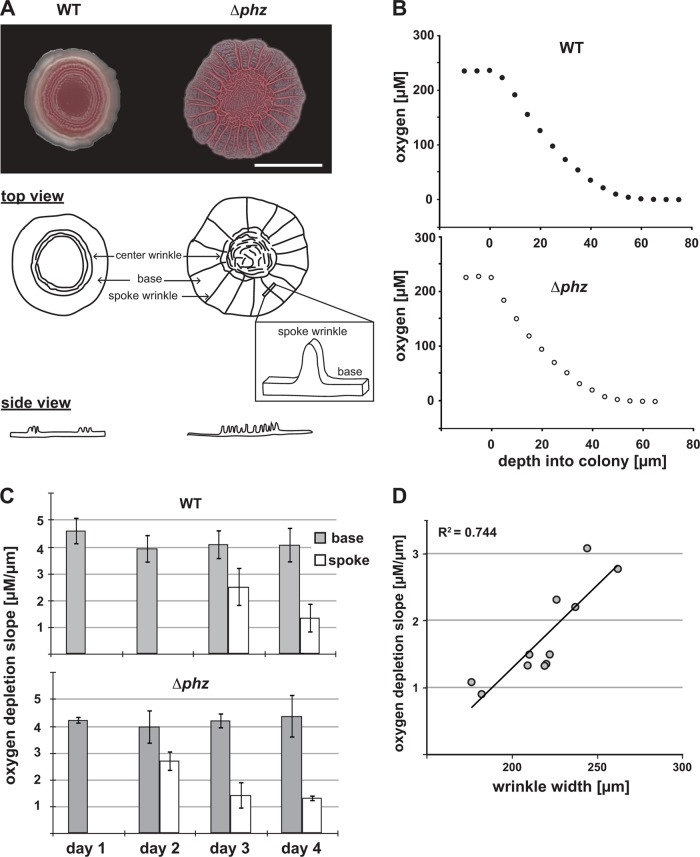
Fig 1.
P. aeruginosa colony morphotypes and oxygen profiles. (A) Graphical representation of terms used to describe features within P. aeruginosa colonies. WT, wild type; Δphz, phenazine-null mutant. The scale bar represents 1 cm, and the images were taken after 3 days of colony development. (B) Oxygen profiles in colony biofilms. Oxygen concentrations were measured in P. aeruginosa colonies on day 3 as a function of microelectrode depth in the colony. (C) Oxygen depletion (calculated as the initial slope, typically between 0- and 20-μm depth, of oxygen profiles over depth in the colony, as depicted in panel B) for the base and a wrinkle (spoke) during colony development. Wrinkles appear in Δphz colonies 1 day earlier than in WT colonies. The error bars represent the standard deviations of this measurement in the bases or wrinkles of 5 independent colonies. (D) Oxygen depletion slopes from Δphz colonies (representing data plotted in panel C) shown as a function of wrinkle thickness.
In the early 20th century, E. S. Guzman Barron, Ernst Friedheim, and others postulated that redox-cycling compounds such as phenazines are “accessory respiratory pigments” that can sustain bacterial “respiration” based on their ability to stimulate oxygen consumption in suspensions of many different types of cells (12–14). They speculated that these compounds can extend the depth of respiration for cells deprived of oxygen, such as those found in normal tissues and tumors (15, 16). This work was carried out before respiratory pathways were fully understood and well before the importance of microbial biofilms in nature and disease was widely recognized. In this regard, Barron, Friedheim, and their colleagues were both ahead of their time and handicapped by a lack of information. In the interval between these pioneering studies and the present work, attention shifted to exploring the roles of phenazines as virulence factors (17). Over a decade ago, we revived the “respiratory pigment” hypothesis in a biofilm context, speculating that the capacity for extracellular electron transfer might provide a physiological benefit for oxidant-limited cells (9, 18). While it has long been appreciated that biofilms are metabolically heterogeneous and that oxygen availability defines different metabolic zones (19, 20), to the best of our knowledge, no study has yet demonstrated that endogenous electron shuttles such as phenazines increase the habitability zone for biofilm cells. Evidence in support of this hypothesis, until now, has been indirect (21). Here, we test the hypothesis directly and go beyond it to demonstrate that the intracellular redox state, not phenazines per se, correlates with colony morphological development.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. For routine liquid cultures, P. aeruginosa PA14 was grown in 3 ml lysogeny broth (LB) (22) in 12- by 100-mm tubes at 37°C with shaking at 250 rpm. Growth conditions for colonies are described below.
Table 1.
Strains and plasmids used in this studya
| Strains/plasmids | Characteristics | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PA14 | Clinical isolate UCBPP-PA14 | 42 |
| PA14 Δphz | PA14 with deletions of operons phzA1-G1 and phzA2-G2 | 27 |
| PA14 YFP | PA14 with chromosomally integrated constitutive eYFP | 8 |
| PA14 Δphz YFP | PA14 Δphz with chromosomally integrated constitutive eYFP | 8 |
| BigBlue | DKN370; PA14 containing two copies of phzM | 26 |
| Δpel | PA14 with a deletion of pelB-pelG | G. Squyres |
| Δphz Δpel | PA14 Δphz with a deletion of pelB-pelG | G. Squyres |
| Denitrification | ||
| PA14 ΔnapA | PA14 with a deletion of napA | This study |
| PA14 ΔnapA Δphz | PA14 with deletions of napA, phzA1-phzG1, and phzA2-phzG2 | This study |
| PA14 ΔnarG | PA14 with a deletion of narG | This study |
| PA14 ΔnarG Δphz | PA14 with deletions of narG, phzA1-phzG1, and phzA2-phzG2 | This study |
| PA14 nirS::Tn | nirS::MAR2XT7; Genr | 43 |
| PA14norB::Tn | norB::MAR2XT7; Genr | 43 |
| PA14 nosF::Tn | nosF::MAR2XT7; Genr | 43 |
| E. coli | ||
| UQ950 | E. coli DH5α λ(pir) host for cloning; F− Δ(argF-lac)169 φ80dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96(Nalr) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | D. Lies |
| BW29427 | Donor strain for conjugation: thrB1004 pro thi rpsL hsdS | B. Wanner |
| lacZ ΔM15RP4–1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | ||
| Saccharomyces cerevisiae | ||
| InvSc1 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | 23; Invitrogen |
| Plasmids | ||
| pMQ30 | Yeast-based allelic-exchange vector; sacBa CEN/ARSH URA3+ Genr | 23 |
| pMQ72 | Yeast-based expression vector 2μm; URA3+ Genr | 23 |
| pLD314 | napA deletion fragments cloned into pMQ30 | This study |
| pLD263 | narG deletion fragments cloned into pMQ30 | This study |
| pAPW1 | pMQ72 with napEFDABC operon insert | This study |
Transposon mutants used in this study were confirmed by PCR or sequencing.
Construction of deletion and complementation plasmids.
Unmarked deletions were generated for the genes napA and narG in PA14 wild-type and Δphz backgrounds. Deletion plasmids were generated using yeast gap repair cloning. Flanking regions (∼1 kb in length) for napA and narG were generated using primers listed in Table 2. The flanking regions and the linearized allelic-replacement vector pMQ30 were assembled by gap repair cloning using the yeast strain InvSc1 (23). The resulting deletion plasmid was transformed into Escherichia coli BW29427 and mobilized into PA14 using biparental conjugation. PA14 single recombinants were selected on LB agar containing 100 μg/ml gentamicin. Potential napA or narG deletion mutants were generated by selecting for double recombinants by identifying strains that grew in the presence of 10% sucrose. Strains with properties of double recombination were further analyzed by PCR for the desired deletion.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| Transposon check | |
| nirS-1 | ATTTGGCAAGCCACTGGT |
| nirS-2 | GTCGTGCTGGGTGTTGTAGA |
| norB-1 | AATGGCTCCCTGAAATTCG |
| norB-2 | GGAAGCTCAGCAGGTAGGC |
| nosF-1 | CCATGAGCCTGGTCGAGAT |
| nosF-2 | ATGTGGTGGCGATAGAGGTC |
| Deletion plasmids | |
| pLD314 | |
| ΔnapA 5′ flank-1 | GGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTAAGCCAGGCTCTTCCTGTTC |
| ΔnapA 5′ flank-2 | CAGTCAGCAGAGGTTTCATGGATTCACGACGGGTGAGGTT |
| ΔnapA 3′ flank-1 | ACCTCACCCGTCGTGAATCCATGAAACCTCTGCTGACTG |
| ΔnapA 3′ flank-2 | CCAGGCAAATTCTGTTTTATCAGACCGCTTCTGCGTTCTGATACTCGAAGTTGTGGCAGTTG |
| pLD263 | |
| ΔnarG 5′ flank-1 | GGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTGATCTGGGTGCCGTTCATC |
| ΔnarG 5′ flank-2 | TCCAGTTCAGGGTGATCTCCGGCTCTCGTTGCTGGTCTC |
| ΔnarG 3′ flank-1 | GAGACCAGCAACGAGAGCCGGAGATCACCCTGAACTGGA |
| ΔnarG 3′ flank-2 | CCAGGCAAATTCTGTTTTATCAGACCGCTTCTGCGTTCTGATGGACCTGGTCGAAGTTCTTG |
| Complementation plasmid | |
| pAPW1 | |
| napA-1 | GAATTCTCCAAGCGCTTCAC |
| napA-2 | CAGCACGTCGTAGAGGGTCT |
For construction of the nap operon complementation plasmid pAPW1, primers were designed using the P. aeruginosa PA14 genome sequence to anneal 490 bp upstream of napE and to the last 19 bases of napC, yielding a PCR product including the napEFDABC operon and a putative promoter region. The amplified DNA was digested using restriction sites (HindIII and NheI) engineered within the primers. It was then ligated into plasmid pMQ72 digested with the same restriction enzymes and treated with calf intestinal phosphatase (Sigma). The resulting plasmid, pAPW1, contains the napEFDABC operon under the control of its native promoter.
Colony morphology assay.
Agar plates for colony morphology experiments were prepared as follows. A mixture of 1% agar (Teknova) and 1% tryptone (Teknova) was autoclaved and cooled to 60°C before 20 μg/ml Coomassie blue (EMD) and 40 μg/ml Congo red (EMD) were added. Sixty milliliters of medium was poured per 10-cm-square plate (Simport; D210-16) and allowed to dry with closed lids at room temperature for 24 h.
For colony spotting and developmental studies, precultures were inoculated from single streak plate colonies and grown in LB medium for 12 h. Ten microliters of the preculture was spotted on plates and incubated for up to 6 days at 23 to 25°C. Colony images were taken daily with a digital microscope (Keyence; VHX-1000).
For colony development at different oxygen concentrations (15%, 21%, and 40%), we incubated plates in C-Chambers (BioSpherix; C274). Oxygen concentrations were regulated by mixing pure nitrogen and oxygen (TechAir) using the gas controller ProOx P110 (BioSpherix). Each chamber contained an open 10-cm round plate filled with 25 ml of water to keep the chambers humid. Humidity was monitored using an iButton Humidity Data Sensor (Maxim) and maintained at >90%. For colony growth under anoxic conditions, plates were stored in an anaerobic glove box filled with 80% N2, 15% CO2, and 5% H2.
Preparation and imaging of colony thin sections.
Colonies were covered with 4% paraformaldehyde and allowed to fix for 10 min. The colonies were then lifted from the agar by gently shaking the plate and allowed to fix for an additional 10 min. Following this, the colonies were transferred into a wash basket, and the fixative was removed by washing the colonies three times in phosphate-buffered saline (PBS). Excess PBS was removed by washing in 25%, 50%, and 75% Tissue-Tek OCT (Sakura; no. 4583) in PBS before the colonies were transferred to disposable embedding molds (Electron Microscopy Sciences; no. 70182) and overlaid with Tissue-Tek OCT. After flash freezing in a dry-ice–ethanol slurry, the samples were stored at −80°C. The frozen samples were cut into 10-μm-thick sections using a Leica CM1850 microtome at −16°C. The sections were collected on Thermo Superfrost Plus slides (no. 6776214) and stored at −80°C until imaging.
Thin sections were photographed using a Zeiss Axio Imager 2. All images were obtained at ×10 magnification using a Zeiss EC-Plan Neofluar objective with differential interference contrast (DIC) and fluorescence optics. The images were taken at an exposure time of 328 ms, false colored, and processed in Photoshop CS4 (Adobe).
Oxygen measurements.
Oxygen profiles were taken using a miniaturized Clark-type oxygen sensor (Unisense; 10-μm tip diameter). The electrode was connected to a picoampere amplifier multimeter (Unisense) and polarized with −0.8 V. The sensor was calibrated using a two-point calibration system. The atmospheric oxygen reading was obtained by placing the electrode in a calibration chamber (Unisense) that contained well-aerated deionized water. Complete aeration was achieved by constantly bubbling the water with air. The zero reading was obtained by bubbling water in the calibration chamber with ultra-high-purity nitrogen gas (TechAir). All calibration readings and profile measurements were obtained using SensorTrace pro 2.0 software (Unisense).
NADH/NAD+ assay.
Extraction and quantification of NADH and NAD+ were carried out according to the methods described by San et al. (24) and Bernofsky and Swan (25). For cultures grown in LB, two 1-ml samples of culture were placed in two separate microcentrifuge tubes and centrifuged at 16,000 × g for 1 min. Colonies grown on 1% tryptone and 1% agar plates amended with 40 μg/ml Congo red and 20 μg/ml Coomassie blue dyes were scraped off the agar plate at the indicated time points using sterile razor blades and resuspended in 1 ml of 1% tryptone. The colonies were disrupted using a pellet disrupter. For each resuspended colony, two 450-μl samples were placed into two separate microcentrifuge tubes. NADH and NAD+ were then extracted from the liquid culture- or colony-derived samples, and relative or absolute quantification was carried out using an enzyme-cycling assay, as described by Price-Whelan et al. (26).
RESULTS AND DISCUSSION
In this study, we set out to characterize the relationship between redox metabolism/electron acceptor availability and P. aeruginosa biofilm development. This work supports the hypothesis that phenazine production and colony rugosity are adaptations that facilitate survival by mitigating electron acceptor limitation. In homogeneous liquid cultures of P. aeruginosa, phenazines affect gene expression and oxidize the intracellular redox state (26–28). Under conditions where no other oxidant is available, phenazine-dependent electron transfer between cells and an oxidizing electrode supports survival (21). Given that cells in biofilms experience steep gradients in oxygen availability, leading to hypoxia or anoxia at a distance from the surface (19, 20), we reasoned that the morphological switch observed in phenazine-deficient colonies is related to their limited ability to access oxidants.
To test this hypothesis, we first characterized the depth of oxygen penetration within our colony biofilm system. P. aeruginosa phenazine-null (Δphz) biofilms characteristically increase their surface area relative to wild-type communities. Δphz colonies also produce more exopolysaccharides than wild-type colonies (7). We therefore asked whether dissolved oxygen concentrations differed in the wild-type and Δphz biofilms when they were grown under atmospheric (21%) oxygen on a nutrient-rich complex medium (1% tryptone, 1% agar containing 20 μg/ml Coomassie blue and 40 μg/ml Congo red). Measurements taken with a Clark oxygen electrode (10-μm tip) revealed steep gradients, with oxygen becoming undetectable 60 μm into the colony when the base area was profiled (Fig. 1B). Qualitatively, this is consistent with oxygen microelectrode measurements of P. aeruginosa biofilms grown in flow cells showing oxygen depletion at depth due to the combined effects of consumption and diffusion limitation (20). The extent of oxygen depletion in the base remained constant over 4 days (Fig. 1C). When we measured oxygen depletion in emerging wrinkles, its slope was lower and decreased over time compared to the base. We found that wrinkle thickness correlated with oxygen abundance: thinner wrinkles were less oxygen depleted than thicker wrinkles (Fig. 1D).
While respiratory versatility is a hallmark of some bacterial species, P. aeruginosa is relatively limited in this regard. It can grow by aerobic respiration and denitrification and poorly by arginine fermentation (29); therefore, the major energy-generating metabolism contributing to growth of colonies on 1% tryptone is aerobic respiration. Moreover, it is generally believed that in the organic-rich environment of infections—such as the mucus that collects on the lungs of individuals with cystic fibrosis—growth and survival of Pseudomonas is not carbon/electron limited but constrained by oxygen availability (30). Microelectrode measurements showed oxygen depletion for both wild-type and Δphz colonies 60 μm from the surface (Fig. 1B), although wild-type colonies were 100 μm tall. We asked if cell distributions in the colonies were comparable, expecting the cells to be confined to the region within 60 μm from the colony surface, where oxygen was available. For these experiments, we generated versions of the wild type and the Δphz mutant that constitutively expressed a stable yellow fluorescent protein (YFP). Colonies were fixed under oxic conditions with paraformaldehyde and embedded in Tissue-Tek OCT, frozen, and sectioned at a thickness of 10 μm (Fig. 2A to F). Because YFP can fully mature and fluoresce posttranslationally in the presence of oxygen (31, 32), our thin-section preparation method ensured that YFP could be seen even in regions of the sections that were previously anoxic.
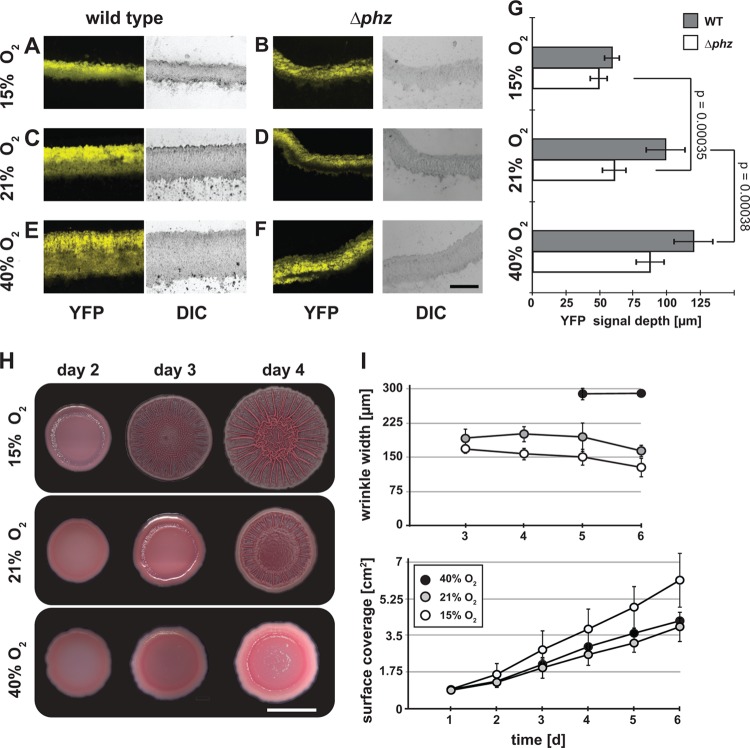
Fig 2.
Oxygen and phenazine availability modulates colony morphology. (A to F) Oxygen and phenazines affect cell layer thickness in P. aeruginosa colonies. Colonies expressing constitutive (PA1/04/03-driven [41]) YFP were grown at 15% (hypoxic), 21% (atmospheric), and 40% (hyperoxic) oxygen for 3 days and then embedded in Tissue-Tek OCT. The images were taken using fluorescence (YFP) and DIC microscopy. The scale bar represents 200 μm. (G) Depth at which the YFP signal was detected in the sample. The error bars represent standard deviations for the means of measurements for 15 bases or wrinkles from 3 colonies. P values were calculated using the one-tailed heteroscedastic Student's t test comparing YFP signal depth in WT or Δphz colonies at different oxygen concentrations. The least significant differences are shown. (H) Colony morphology. Representative images of Δphz colonies grown at various atmospheric oxygen concentrations over 3 days. The scale bar represents 1 cm. (I) Colony wrinkle width (top) and surface coverage (bottom) for Δphz colonies grown at various oxygen concentrations, with nitrogen comprising the atmospheric balance. The error bars represent the standard deviations of widths for 15 wrinkles from 3 colonies (top) or of measurements taken from 5 colonies (bottom).
In sections taken from Δphz colonies, cells were found within 60 μm from the surface, in the oxic zone (Fig. 2G). In sections taken from wild-type colonies, however, cells were found up to 100 μm from the surface (Fig. 2G). Some of these cells, therefore, inhabited the 40-μm-thick anoxic zone. Reasoning that phenazines enable this survival by acting as alternate electron acceptors in the absence of oxygen, we altered the ambient oxygen concentration and predicted that the extent of the oxic zone within Δphz colonies would determine the cell colonization depth (i.e., wider zones of habitation would correspond to higher atmospheric oxygen levels). We grew colonies under hyperoxic and hypoxic conditions (40% and 15% oxygen, respectively) and observed that the depth at which cells could be detected in both wild-type and Δphz colonies correlated with the concentration of oxygen provided (Fig. 2A to F). Quantification of the cell layer thickness showed that under all conditions the Δphz mutant was significantly thinner than the wild type, confirming that the presence of phenazines increases the habitable zone (Fig. 2G). We further probed this correlation using a mutant that overproduces the phenazine pyocyanin and found that the habitable zone increased beyond that of the wild type (see Fig. S1 in the supplemental material).
We next evaluated whether oxygen accessibility affects other aspects of colony morphology by quantifying the surface coverage and wrinkle thickness of colonies grown with various concentrations of oxygen. As oxygen concentrations increased, the colonies spread less and formed fewer wrinkles. In addition, wrinkle thickness increased with increasing oxygen concentration (Fig. 2H and I). To further probe the link between oxidant availability and colony structure, we took advantage of P. aeruginosa's ability to use nitrate instead of oxygen for respiration and redox homeostasis. Two P. aeruginosa nitrate reductase complexes, Nar and Nap, might be expected to affect the intracellular redox state under hypoxic conditions. While Nar is a cytoplasmic, membrane-associated complex that contributes to the production of a proton motive force and ATP generation, the Nap complex is periplasmic and is thought to balance the intracellular redox state without directly contributing to the generation of a transmembrane electrochemical gradient (33, 34). Both Nap and Nar catalyze the reduction of nitrate to nitrite. Three additional enzyme complexes, known as Nir (nitrite reductase), Nor (nitric oxide reductase), and Nos (nitrous oxide reductase), allow P. aeruginosa to perform denitrification, the full reduction of nitrate to nitrogen gas (N2).
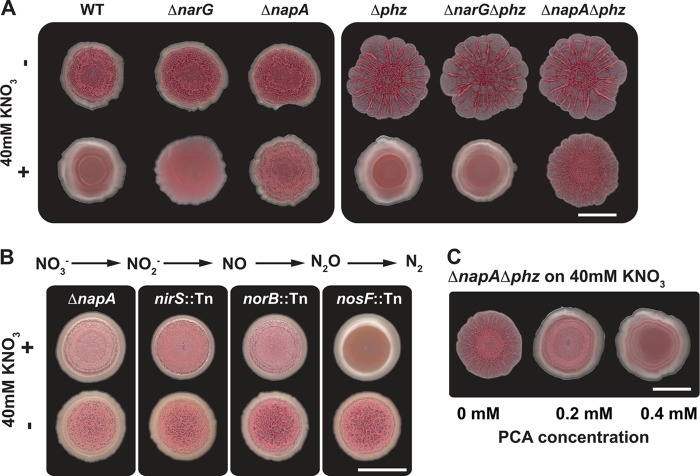
We grew a Δphz colony under atmospheric oxygen on medium amended with 40 mM potassium nitrate and found that these conditions rendered the colony smooth (Fig. 3A). To determine whether this was due to nitrate reduction, we generated mutants lacking the nitrate reductase subunits NarG and NapA. The ΔnarG mutant formed a smooth colony when grown on nitrate, suggesting that nitrate respiration is not required for the nitrate-dependent smooth-colony phenotype. Strikingly, the ΔnapA mutant wrinkled when grown on medium amended with nitrate (Fig. 3A) but reverted to smooth when complemented by the nap operon on a plasmid (see Fig. S2 in the supplemental material). We then tested whether the downstream enzymes in the denitrification pathway were required for Nap-mediated colony smoothness. Mutants deficient in Nir and Nor also formed wrinkled colonies on 40 mM nitrate, while the mutant lacking functional Nos remained smooth, suggesting that reduction to nitrous oxide is required for NapA-dependent nitrate reduction (Fig. 3B). Furthermore, these results suggest that P. aeruginosa community structure is determined, at least in part, by the intracellular redox state.
Fig 3.
Nap-mediated nitrate reduction supports redox homeostasis for cells in colonies. (A) Nap-mediated nitrate reduction prevents colony wrinkling. Shown is the effect of medium amendment with 40 mM potassium nitrate on colony morphology. The images are representative of 3 independent experiments. The scale bar represents 1 cm. (B) Colony morphology of denitrification mutants. Specific steps in the canonical denitrification pathway catalyzed by Nap, Nir, and Nor contribute to the nitrate-induced smoothness observed for wild-type colonies. The images shown are representative of 3 independent experiments. The scale bar represents 1 cm. (C) PCA rescues the ΔnapA phenotype on nitrate; addition of exogenous PCA promotes smoothness in a ΔnapA Δphz mutant. The images are representative of 3 independent experiments. The scale bar represents 1 cm.
If both phenazine reduction and nitrate reduction contribute to oxidizing the intracellular redox state, one would predict that these two activities could complement each other to promote colony smoothness. Nitrate addition to a Δphz mutant decreased wrinkling (Fig. 3A), and phenazine-1-carboxylic acid (PCA) addition to a ΔnapA Δphz mutant (grown in the presence of nitrate) also had this effect (Fig. 3C).
To further test the hypothesis that community structure and the intracellular redox state are linked, we set out to measure and manipulate the NADH-to-NAD+ ratio in colony biofilms under different growth conditions. Building on previous results showing that phenazines and nitrate decreased NADH/NAD+ ratios in liquid cultures (26), we first asked whether nitrate-dependent redox balancing required napA. We extracted NADH and NAD+ from planktonically grown cells and measured their levels using an enzyme-based cycling assay (24, 25). We found that the ΔnapA mutant showed a partial but significant defect in nitrate-dependent oxidation of the intracellular redox state relative to Δphz and wild-type P. aeruginosa (Fig. 4A). We next adapted the NADH/NAD+ extraction and quantification protocol for use with colony samples. This method revealed that the NADH/NAD+ ratio of phenazine-null colonies reached a maximum that coincided with the induction of wrinkling, while the NADH/NAD+ ratio of wild-type colonies remained relatively consistent throughout the time course (Fig. 4B). Absolute quantitation of the NADH and NAD+ levels (see Fig. S3 in the supplemental material) confirmed that the total NAD(H) pools of cells in wild-type and Δphz colonies were approximately equivalent.
Fig 4.
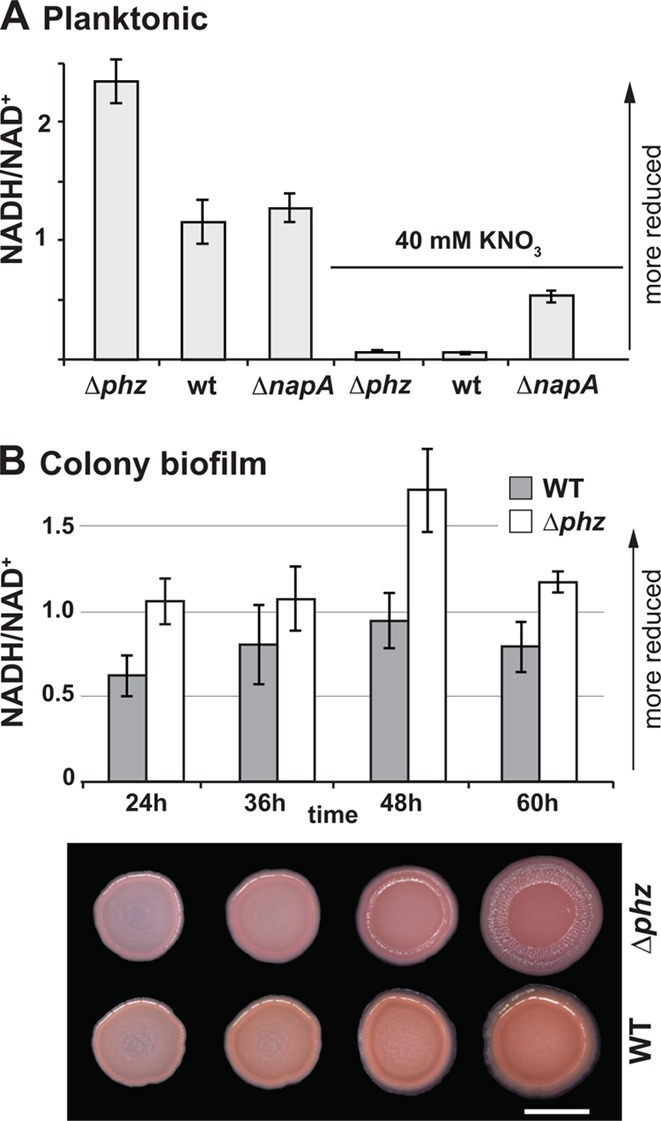
(A) NADH/NAD+ ratios for liquid cultures grown for 16 h. For 40 mM KNO3 cultures, potassium nitrate was added to the medium at the same time that blue pigmentation (pyocyanin production) was apparent in the wild-type cultures. The error bars represent the standard deviations of triplicate cultures. (B) (Top) NADH/NAD+ for wild-type and Δphz colonies. The error bars represent the standard deviations of measurements from triplicate colonies. (Bottom) Representative images of Δphz and wild-type colonies at the time points for collection. The scale bar represents 0.5 cm. The images are representative of 3 independent experiments.
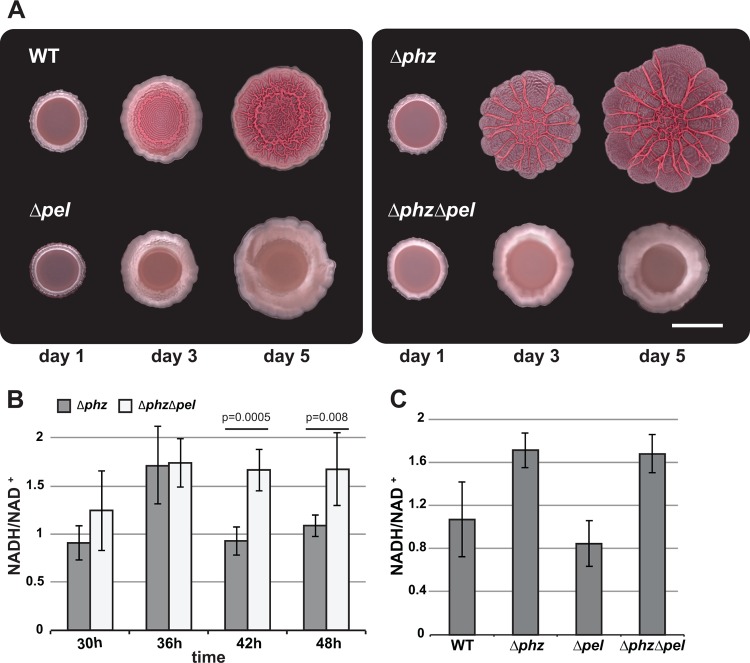
Rugosity appears to be induced when the cytoplasmic reducing potential reaches a threshold value, indicating that increased colony surface area is an adaptive response to oxidant limitation that promotes rebalancing of the intracellular redox state. To test this idea, we used a mutant defective in wrinkle formation. Previously, we observed a correlation between wrinkle formation and colony staining with Congo red, suggesting that production of the PEL polysaccharide is critical for rugose morphology (7). We therefore obtained Δpel mutants lacking genes required for PEL biosynthesis to test in our colony biofilm assay (35). As expected, colony wrinkle formation was abolished when this mutation was made in both wild-type and Δphz backgrounds (Fig. 5A). We then measured NADH/NAD+ ratios during colony maturation and found that the transient increased ratio that coincided with the onset of wrinkling in the Δphz mutant persisted in the Δphz Δpel mutant (Fig. 5B). In contrast, the pel deletions had no effect on the NADH/NAD+ ratios of planktonically grown cells (Fig. 5C). This supports the hypothesis that wrinkling is a strategy for balancing the intracellular redox state of cells within a community. These results suggest that colony morphological development is an active process in which a critical redox state is sensed, leading to a biological response. We are in the process of identifying the circuitry responsible for this phenomenon and the extent to which wrinkling is an emergent property. We note that the specific time when the NADH/NAD+ ratio peaks can vary from experiment to experiment as a function of slight differences in plate thickness, density of the initial inoculum, etc. However, occurrence of the peak just prior to colony wrinkling is highly reproducible.
Fig 5.
(A) PEL production is required for wrinkle formation. pelB-pelG were deleted in the wild-type and Δphz backgrounds. Colonies were grown for 5 days on 1% tryptone, 1% agar supplemented with 40 μg/ml Congo red and 20 μg/ml Coomassie blue. (B) NADH/NAD+ ratios in colony biofilms for Δphz and Δphz Δpel mutants. The error bars represent the standard deviations of measurements from triplicate colonies. P values were determined using the one-tailed heteroscedastic Student's t test comparing Δphz and Δphz Δpel colonies at 42 and 48 h. (C) NADH/NAD+ ratios in liquid culture (early stationary phase) for wild-type, Δphz, Δpel, and Δphz Δpel colonies grown in 1% tryptone to early stationary phase. The error bars represent the standard deviations of measurements from biological triplicates.
All cells catalyze a repertoire of catabolic and anabolic reactions that must be balanced so that the cytoplasm remains a hospitable environment for protein function. In bacterial physiology, there has traditionally been a focus on the individual cell level, and redox reactions have been identified that appear to serve the sole purpose of modulating the intracellular redox state, i.e., they do not contribute directly to energy generation or the production of biomass. We have shown that P. aeruginosa can use endogenous phenazines and/or exogenous nitrate to balance the intracellular redox state when oxygen is limiting. The observation that mutants unable to produce phenazines form structurally more complex communities with increased surface area led us to propose that this morphogenetic switch is a response to a reduced cytoplasm. Similarly, we have observed that electron acceptor limitation elicits a community-wide response that maximizes oxygen accessibility—and thereby redox balance—in a bacterial system (Fig. 6). Furthermore, inhibiting this response disrupts redox balancing (Fig. 5B). Whether rugosity has a similar effect in other microbial species remains to be investigated. It would not be surprising if bacteria with different metabolic properties (e.g., with or without the ability to produce electron shuttles) have different mechanisms for growing and surviving in multicellular communities. Intriguingly, a recent study of patterning in Bacillus subtilis biofilms (36) suggested that localized cell death promotes wrinkle formation. Redox homeostasis may also play a role in this system, but to our knowledge, experiments have not yet been performed that enable a direct comparison. Understanding how active processes and passive physical effects interweave to achieve multicellular patterning in Pseudomonas is our long-term goal; it will be interesting to learn whether the mechanisms that underpin these patterns are generalizable. As has been well articulated by others (37), the complex interplay between physical, chemical, and biological processes in microbial communities provides a rich subject for future research.
Fig 6.
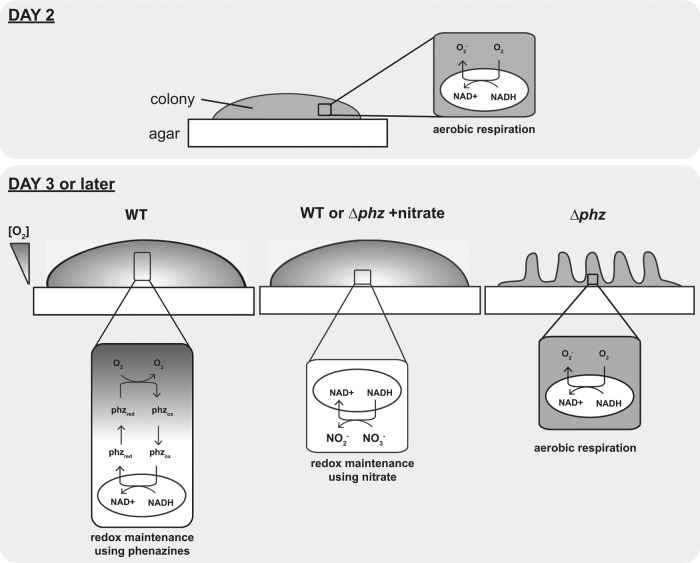
P. aeruginosa communities employ various strategies that enable access to electron acceptors. From inoculation to day 2, oxygen is detectable throughout the structure. By day 3, wild-type colonies and Δphz colonies grown on nitrate-containing medium exceed the critical height that allows full oxygenation, and the lower portion of the colony becomes anoxic. Cells in the anoxic zone can maintain redox homeostasis by reducing phenazines or nitrate. In the absence of phenazines or nitrate, redox homeostasis can be mediated by a community-wide response: colony wrinkling increases the colony surface area and oxygen accessibility.
In conclusion, as Friedheim recognized nearly a century ago (15), cellular aggregation leads to gradient formation due to limited diffusion and consumption of substrates by individual cells within a community. Whether they subsist in bacterial colonies or eukaryotic tissues, cells in multicellular environments likely employ differing strategies to ensure substrate acquisition and survival, depending on the specific microenvironment they inhabit. This concept is well recognized in the biofilm field, and numerous reports have discussed the fact that oxygen defines metabolic zones in biofilms (19, 20, 38). Here, for the first time, we have demonstrated that it is not oxygen per se but rather changes in the intracellular redox state that correlate with biofilm morphological development. Mechanisms that aid in redox homeostasis at the cellular level have been characterized in diverse organisms. In metazoans, redox-balancing mechanisms that function at the multicellular level are also well known; for example, the development of the vascular system prevents oxygen starvation of the growing embryo (39). In multicellular aerobes, cells must cope with limited oxygen availability that leads to the formation of aerobic, microaerobic, and anaerobic zones. During processes such as tumor angiogenesis, relative oxygen concentrations act as cues that determine adaptive morphological features, facilitating oxygen delivery to cells within the macroscopic structure (40). Our findings suggest that, like metazoans, bacteria can also respond to electron acceptor limitation and balance intracellular redox levels through morphological changes at the community level. Morphological adaptation to redox imbalance thus appears to be a conserved biological strategy.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Ziebis, P. Girguis, G. Squyres, and D. Chapman for technical assistance.
This research was supported by funding from the Howard Hughes Medical Institute (HHMI) (L.E.P.D., A.P.-W., R.C.H., and D.K.N.), a Gilliam fellowship from the HHMI (C.O.), an IGERT fellowship from the NSF (H.S.), and a startup fund from Columbia University to L.E.P.D. D.K.N. is an HHMI Investigator.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02273-12.
REFERENCES
- 1. Grosberg RK, Strathmann RR. 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Systematics 38:621–654 [Google Scholar]
- 2. Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U. S. A. 99:7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398 doi:10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 5. Booth SC, Workentine ML, Wen J, Shaykhutdinov R, Vogel HJ, Ceri H, Turner RJ, Weljie AM. 2011. Differences in metabolism between the biofilm and planktonic response to metal stress. J. Proteome Res. 10:3190–3199 [DOI] [PubMed] [Google Scholar]
- 6. Hoiby N, Frederiksen B, Pressler T. 2005. Eradication of early Pseudomonas aeruginosa infection. J. Cyst. Fibros. 4(Suppl. 2):49–54 [DOI] [PubMed] [Google Scholar]
- 7. Dietrich LE, Teal TK, Price-Whelan A, Newman DK. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramos I, Dietrich LE, Price-Whelan A, Newman DK. 2010. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Price-Whelan A, Dietrich LE, Newman DK. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71–78 [DOI] [PubMed] [Google Scholar]
- 10. Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, Mazurier S, Heide L, Blankenfeldt W, Weller DM, Thomashow LS. 2010. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76:866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42:2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedheim EA. 1931. Pyocyanine, an accessory respiratory enzyme. J. Exp. Med. 54:207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrop GA, Barron ES. 1928. Studies on blood cell metabolism. I. The effect of methylene blue and other dyes upon the oxygen consumption of mammalian and avian erythrocytes. J. Exp. Med. 48:207–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barron ES, Hoffman LA. 1930. The catalytic effect of dyes on the oxygen consumption of living cells. J. Gen. Physiol. 13:483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedheim EA. 1934. The effect of pyocyanine on the respiration of some normal tissues and tumours. Biochem. J. 28:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barron ES. 1930. The catalytic effect of methylene blue on the oxygen consumption of tumors and normal tissues. J. Exp. Med. 52:447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kerr JR. 2000. Phenazine pigments: antibiotics and virulence factors. Infect. Dis. Rev. 2:184–194 [Google Scholar]
- 18. Hernandez ME, Newman DK. 2001. Extracellular electron transfer. Cell. Mol. Life Sci. 58:1562–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peters AC, Wimpenny JW, Coombs JP. 1987. Oxygen profiles in, and in the agar beneath, colonies of Bacillus cereus, Staphylococcus albus and Escherichia coli. J. Gen. Microbiol. 133:1257–1263 [DOI] [PubMed] [Google Scholar]
- 20. Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y, Kern SE, Newman DK. 2010. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192:365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. San KY, Bennett GN, Berrios-Rivera SJ, Vadali RV, Yang YT, Horton E, Rudolph FB, Sariyar B, Blackwood K. 2002. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4:182–192 [DOI] [PubMed] [Google Scholar]
- 25. Bernofsky C, Swan M. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 53:452–458 [DOI] [PubMed] [Google Scholar]
- 26. Price-Whelan A, Dietrich LE, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 28. Sullivan NL, Tzeranis DS, Wang Y, So PT, Newman D. 2011. Quantifying the dynamics of bacterial secondary metabolites by spectral multiphoton microscopy. ACS Chem. Biol. 6:893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vander Wauven C, Pierard A, Kley-Raymann M, Haas D. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heim R, Prasher DC, Tsien RY. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 91:12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C, Xing X-H, Lou K. 2005. Rapid detection of a GFP-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol. Lett. 249:211–218 [DOI] [PubMed] [Google Scholar]
- 33. Williams HD, Zlosnik JE, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52:1–71 [DOI] [PubMed] [Google Scholar]
- 34. Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 36. Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. U. S. A. 109:18891–18896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klapper I, Dockery J. 2010. Mathematical description of microbial biofilms. Siam Rev. 52:221–265 [Google Scholar]
- 38. Xavier JB, Foster KR. 2007. Cooperation and conflict in microbial biofilms. Proc. Natl. Acad. Sci. U. S. A. 104:876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simon MC, Keith B. 2008. The role of oxygen availability in embryonic development and stem cell function. Nat. Rev. Mol. Cell Biol. 9:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giaccia AJ, Simon MC, Johnson R. 2004. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 18:2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lambertsen L, Sternberg C, Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726–732 [DOI] [PubMed] [Google Scholar]
- 42. Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 43. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.