Abstract
Enterotoxigenic Escherichia coli (ETEC) is a bacterial pathogen that causes diarrhea in children and travelers in developing countries. ETEC adheres to host epithelial cells in the small intestine via a variety of different pili. The CS1 pilus is a prototype for a family of related pili, including the CFA/I pili, present on ETEC and other Gram-negative bacterial pathogens. These pili are assembled by an outer membrane usher protein that catalyzes subunit polymerization via donor strand complementation, in which the N terminus of each incoming pilin subunit fits into a hydrophobic groove in the terminal subunit, completing a β-sheet in the Ig fold. Here we determined a crystal structure of the CS1 major pilin subunit, CooA, to a 1.6-Å resolution. CooA is a globular protein with an Ig fold and is similar in structure to the CFA/I major pilin CfaB. We determined three distinct negative-stain electron microscopic reconstructions of the CS1 pilus and generated pseudoatomic-resolution pilus structures using the CooA crystal structure. CS1 pili adopt multiple structural states with differences in subunit orientations and packing. We propose that the structural perturbations are accommodated by flexibility in the N-terminal donor strand of CooA and by plasticity in interactions between exposed flexible loops on adjacent subunits. Our results suggest that CS1 and other pili of this class are extensible filaments that can be stretched in response to mechanical stress encountered during colonization.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) bacteria cause travelers' diarrhea and are important agents of diarrhea in children in regions with poor sanitation and limited access to clean water (1). ETEC colonizes the small intestine, where it produces heat-labile and/or heat-stable toxins that lead to severe diarrhea that can be fatal in infants and small children. ETEC adheres to receptors on epithelial cells using filamentous surface appendages called pili or fimbriae. ETEC possesses multiple serologically distinct pili, called coli surface (CS) antigens or colonization factor antigens (CFAs), which differ from strain to strain (2). The class 5 family includes CS1, CS2, CS14, CS17, CS19, and CFA/I of ETEC and the cable pili of Burkholderia cepacia (3). These pili require only four specific proteins to assemble: the major pilin subunit that forms the pilus rod, a tip adhesin, a periplasmic chaperone, and an outer membrane usher. All four proteins are encoded on a single operon (4). Individual components share amino acid sequence homology within this class of pili. Class 5 pilus assembly is referred to as the “alternate” chaperone pathway to distinguish it from the classical chaperone-usher assembly pathway used for the related class 1 pili (5, 6), which include the P pili and type I pili of uropathogenic E. coli (UPEC). Like the class 5 pili, class 1 pili assemble via a periplasmic chaperone and an outer membrane usher, but there is little to no sequence similarity between components of the two classes. Furthermore, the class 1 pili are more complex, having a fibrillar structure at their distal tip as well as adaptor proteins between the tip adhesin, fibril, and pilus rod. P pili have an additional termination subunit. However, strong structural similarities between the two pilus classes suggest that the distinction between alternate chaperone and chaperone-usher assembly pathways may not be meaningful (7).
Both class 1 and class 5 pilin subunits have an incomplete immunoglobulin (Ig) protein fold that lacks an internal β-strand in the second β-sheet. This missing strand results in a groove along the length of the cylinder-shaped protein that is lined with hydrophobic residues. In the absence of the periplasmic chaperone, pilin subunits are unstable and are degraded (8–10). The chaperone donates a β-strand from one of its two Ig domains, stabilizing the pilin subunit and allowing it to fold in the periplasm (11–14). This process is called donor strand complementation. Four alternating hydrophobic residues on the donated chaperone strand, termed P1 to P4, insert into 4 hydrophobic pockets in the pilin groove. The chaperone P3 residue is either a glycine or an alanine. Only these small side chains fit into the P3 site, which is obstructed by bulky aromatic residues, thus providing a specific register for the donated strand. The chaperone-pilin complex is targeted to the pore-forming usher complex in the outer membrane (15). The chaperone β-strand is displaced by the N-terminal extension (Nte) of a pilin on an incoming pilin-chaperone complex. All pilin subunits in the chaperone-usher pilus family have an exposed and flexible 10- to 20-residue Nte with four alternating hydrophobic residues, P2 to P5, that insert into the hydrophobic groove of the terminal subunit in the growing pilus filament, displacing the chaperone strand. This “donor strand exchange” process occurs when the P5 side chain of the Nte in the incoming pilin inserts into the unoccupied P5 pocket of the terminal chaperone-complemented pilin and “unzips” the chaperone strand, replacing its P4, P3, and P2 residues with the corresponding residues on the pilin Nte (14–17). In this way, pilin subunits are added iteratively to the base of the pilus filament, which grows out through the usher pore into the extracellular space.
Electron microscopy and three-dimensional (3D) image processing of P pili and type I pili from UPEC and CFA/I pili from ETEC, combined with X-ray crystallography of the pilin subunits and subunit complexes, reveal a conserved filament architecture, where subunits are associated in a head-to-tail manner, with the Nte on one subunit, n+1, inserted into the hydrophobic groove of the adjacent subunit, n (7, 18–20). Subunits align with their cylindrical axes almost perpendicular to the filament axis and wrap around the filament with a rotation of ∼110o and a rise of ∼8 Å per pilin monomer for a right-handed 1-start helix. The filament diameter is ∼8 nm, and an ∼25 Å channel runs through the filament. These data together indicate that although the class 1 and 5 pili are dissimilar in amino acid sequence, their subunit structures, filament architectures, and assembly mechanisms are very similar. CFA/I filaments often possess a thin fibrillar structure at their tips, which appears to represent extended CFA/I filaments. The extended form would allow passage through the narrow channel of the usher complex and then collapse into the more compact and rigid helical rod that is displayed on the bacterial surface (7).
The ETEC CS1 pili are closely related to CFA/I. The major pilin subunit of CS1, CooA, shares 53% identity with the CFA/I pilin CfaB, with the Nte of these pilins being particularly well conserved (see Fig. 1A). CS1 is composed of hundreds of CooA subunits plus a single tip-associated minor pilin, CooD, which is necessary for initiating CS1 assembly and for adherence of ETEC to host cells (21–23). CooB is the CS1 periplasmic chaperone, and CooC is the outer membrane usher (4). CS1-expressing ETEC strains use the CooD adhesin to bind to intestinal cells and also to bovine erythrocytes in an in vitro hemagglutination assay (21). Here we present the crystal structure of recombinantly expressed ETEC CooA together with a 3D reconstruction of the CS1 pilus filament. CooA, like CfaB and the class 1 pilins, undergoes donor strand complementation. The CS1 filaments exhibit structural polymorphism that likely relates to pilus extensibility.
Fig 1.
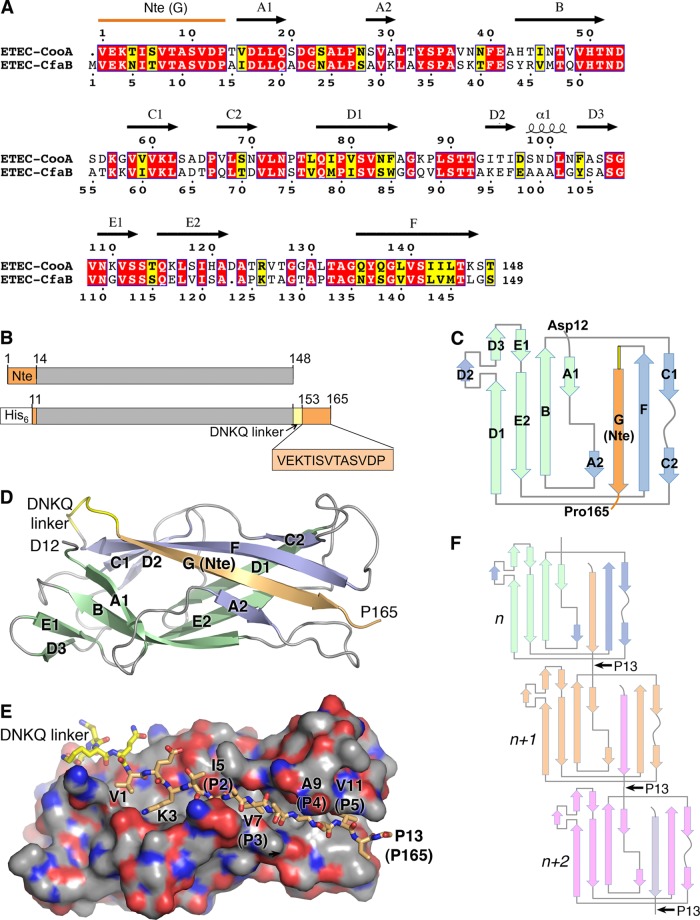
Crystal structure of ETEC CooAdsc. (A) Amino acid sequence alignment of CooA and CfaB. Secondary structure elements of CooA are indicated above its sequence. Red background, identical residues; yellow background, similar residues. (B) Schematic representation of wild-type CooA (top) and the CooAdsc construct (bottom). The sequence shown with orange background represents the Nte, which has been removed from the N terminus and attached to the C terminus. (C) Schematic of the secondary structure of CooAdsc. Green β-strands belong to the first β-sheet and blue and orange strands belong to the second β-sheet in the Ig fold. The orange G strand is the Nte, which has been fused to the C terminus. (D) Ribbon representation of the 1.6-Å-resolution CooAdsc crystal structure. (E) Space-filling representation of CooA with the Nte shown in stick representation. Carbon atoms are colored orange in the Nte, yellow in the DNKQ linker, and gray in the remainder of the protein. Oxygen atoms are red, and nitrogens are blue. Residues in the Nte are numbered as in native mature CooA, where the Nte of subunit n+1 (not shown) is donated to subunit n (shown). The Nte fits into the hydrophobic groove with hydrophobic side chains at positions P2 to P5, indicated in parentheses, filling corresponding pockets in the groove. (F) CS1 assembly scheme showing donor strand complementation between adjacent CooA subunits. Each subunit has a different color to illustrate donor strand complementation with an adjacent subunit in the filament.
MATERIALS AND METHODS
Bacterial strains and plasmids.
CS1-producing E. coli strains were a gift from June Scott, Emory University. E. coli MC4100/pEU494 (24) was used to PCR amplify the cooA gene for recombinant expression. ETEC strain LMC10/pEU2030 was used to express CS1 pili for electron microscopy (EM) analysis. All bacterial strains, plasmids, and oligonucleotides are listed in Table 1. Antibiotic concentrations were as follows: kanamycin (Kan), 45 μg/ml; ampicillin (Amp), 100 μg/ml.
Table 1.
Bacterial strains, plasmids, and primers
| Reagent | Description | Source/reference(s) |
|---|---|---|
| Bacterial strains | ||
| E. coli MC4100/pEU494 | F− araD139 Δ(argF-lac)169 λe14− flhD5301 Δ(fruK-yeiR)725 (fruA25) relA1 rpsL150 (Strr) fibB5301 deoC1 ptsF25 rbsR22 Δ(fimB-fimE)632(::IS1) pEU494 | 24, 51 |
| ETEC LMC10/pEU2030 | ETEC lacking the plasmid encoding the toxins and the positive regulator of coo; pEU2030 | 35 |
| E. coli RosettaBlue | endA1 hsdR17(rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proA+B+ lacIqZΔM15::Tn10] pRARE2 (Camr Tetr) | Novagen |
| Plasmids | ||
| pET-15b | T7 promoter, Ampr | Novagen |
| pEU494 | pHGS576, coo genes under control of lac promoter | 52 |
| pEU2030 | pUC18 encoding rns under its own promoter, Ampr | This study |
| pET-15b-CooBA | pET-15b, cooB-cooA gene segment in NdeI/BamHI sites | This study |
| pETDuet-CooB1 | pETDuet, cooB inserted into BamHI and SalI sites | This study |
| pETDuet-CooB1-Ntd11CooA2 | pETDuet-CooB1 with cooA gene encoding residues 12–148 (cooAΔN11) inserted into NdeI and XhoI sites | This study |
| pET-28b-Ntd11CooA-dsc13 | pET-28b with Ntd11CooA-dsc13 inserted into NdeI and XhoI sites | This study |
| Primers | ||
| ETEC–CooBA-fPCR | GGAATTCATATGCGAAAATTATTTTTAAGTTTGCTTATGATTC | This study |
| ETEC–CooBA-rPCR | CGGGATCCTTACGTTGACTTAGTCAGGATAATTGATACG | This study |
| Ntd11CooA-dsc13-F | GGAATTCATATGGACCCGACTGTTGACCTTC | This study |
| CooA-R | CGCTCGAGTTACGTTGACTTAGTCAGGATA | This study |
| Ntd11CooA-dsc13-R | CGCTCGAGTTACGGGTCAACACTCGCCGTAACGCTAATGGTCTTCTCGACTTGTTTATTGTCCGTTGACTTAGTCAGGATAATTGATACG | This study |
Cloning and expression of CooAdsc.
The cooB-cooA genes were PCR amplified from pEU494 (24) using primers ETEC-CooBA-fPCR and ETEC-CooBA-rPCR (Table 1). The PCR product was ligated into the NdeI and BamHI restriction sites of the pET-15b vector (Novagen) to produce pET-15b-CooBA. The cooA gene fragment encoding residues 12 to 148 of the mature CooA protein (cooAΔN11) was subcloned from this vector using primers Ntd11CooA-dsc13-F and CooA-R and ligated into the pETDuet-CooB1 vector at the NdeI and XhoI sites, which had the cooB gene inserted into the BamHI and SalI sites, to produce pETDuet-CooB1-Ntd11CooA2. Primers Ntd11CooA-dsc13-F and Ntd11CooA-dsc13-R were used to PCR amplify cooAΔN11 from pETDuet-CooB1-Ntd11CooA2 and add a gene segment to its 3′ end encoding a flexible linker, DNKQ, followed by the 13 N-terminal amino acids of mature CooA. This new gene fragment, Ntd11CooA-dsc13, was inserted into the NdeI and XhoI restriction sites of pET-28b to produce pET-28b-Ntd11CooA-dsc13 for expression of a soluble CooA protein lacking its N-terminal 11 residues, with its 13 N-terminal residues fused to its C terminus via a DNKQ linker (donor-strand-complemented CooA [CooAdsc]). CooAdsc was expressed with an N-terminal hexahistidine tag for the purposes of protein purification.
E. coli RosettaBlue (Novagen) was transformed with pET-28b-Ntd11CooA-dsc13. Cells were grown overnight in Luria broth (LB)-Kan, and 100 ml of the overnight cultures was used to inoculate 1 liter LB-Kan. Cells were grown at 37°C to an optical density of 0.75 at a 600-nm wavelength (OD600), at which time isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce CooAdsc expression. Cells were grown for an additional 3.5 h at 30°C with vigorous shaking and harvested by centrifugation at 5,000 × g for 30 min at 4°C. Cells were lysed by incubation in lysis buffer (500 mM NaCl, 50 mM phosphate buffer [pH 7.4], and 40 mM imidazole with 0.2 mg/ml lysozyme) followed by sonication 3 times (2 min each) at 45% amplitude. The lysed cells were centrifuged at 25,000 × g for 40 min at 4°C to remove cell debris, and CooAdsc was isolated from the supernatant using a nickel-nitrilotriacetic acid (NTA) column (GE Healthcare). CooAdsc was eluted using an imidazole gradient. Fractions containing CooAdsc were pooled and concentrated on an Amicon stirred-cell concentrator (Millipore) and exchanged into CooA buffer (100 mM NaCl and 20 mM Tris-HCl, pH 7.5). CooAdsc was further purified on a Sephacryl S-100 HR size exclusion column (GE Healthcare) and concentrated to 15 mg/ml for crystallization.
CooAdsc crystallization, data collection, and structure determination.
CooAdsc was crystallized by the hanging drop vapor diffusion method in 1.0 M sodium citrate and 0.1 M imidazole, pH 8.0, using 2 μl protein solution and 2 μl reservoir buffer. Crystals were flash-frozen and stored in liquid nitrogen in reservoir buffer with 25% glycerol as a cryoprotectant. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) Beamline 11-1. The raw data were processed for Lorentz and polarization corrections and integrated by using the XDS software program, followed by scaling with the program XSCALE (25). The crystal belonged to the triclinic space group P1, with two molecules in the unit cell. The solvent content was 35% as determined from the Matthews coefficient (26). The structure was solved by molecular replacement using the CfaB crystal structure as a search model (PDB code 3F83) (7). Nonidentical residues in CfaB were changed to Ser, Ala, or Gly in the software program COOT. An initial search by using the program PHASER (27) yielded two unambiguous solutions with fairly high Z scores (100 and 14.9). Several cycles of rigid-body refinement with tight NCS (noncrystallographic symmetry) restraint and density modification procedures were performed using the software programs REFMAC5 (28) and DM (29), respectively. The electron density map was interpretable and clearly showed density for many missing side chains, which were built using COOT (30). Many iterative cycles of model building and restrained refinement by the software program ARP/wARP (31) improved the model and revealed density for most of the side chains as well as the missing portions of the main chain. Water oxygens were located using ARP/wARP, and their positions were manually checked in COOT with difference map output by REFMAC5 and the composite annealed omit map calculated using the software program CNS (32). Further refinement using TLS (thermal libration screw-motion) gave an Rcryst value of 0.222 and an Rfree value of 0.251. No electron density was observed for the His tag or linker. The model was validated by the software programs PROCHECK (33) and MOLPROBITY (34). Data collection and refinement statistics are shown in Table 2.
Table 2.
Crystallographic data collection and refinement statistics
| Parameter | Name or value(s) for CooAdsc |
|---|---|
| Data collection | |
| Beamline | SSRL 11-1 |
| Wavelength (Å) | 1.07 |
| Space group | P1 |
| Cell a, b, c (Å) | 29.99, 46.87, 51.27 |
| Cell α, β, γ (o) | 83.72, 89.95, 74.58 |
| Resolution (Å) | 1.6 |
| Completeness (%)a | 94.9 (92.5) |
| No. of observed reflections | 131,428 |
| No. of unique reflections | 33,503 |
| Rsym (%)a,b | 0.025 (0.117) |
| I/σ(I)a | 34.1 (10.8) |
| Redundancy | 3.9 (4.0) |
| Wilson B value (Å2) | 24.5 |
| Mosaicity (o) | 0.2 |
| Refinement statistics | |
| Resolution limits (Å) | 19.7–1.6 |
| Molecules/unit cell | 2 |
| No. of reflections used | 31,826 |
| Rcryst (%)c | 22.2 |
| Rfree (%)d | 25.1 |
| No. of atoms | |
| Protein | 2,295 |
| Ligands | 6 |
| Water | 156 |
| Avg B factor (Å2) | |
| Protein | |
| Chain A | 21.6 |
| Chain B | 22.0 |
| Ligands | |
| Imidazole | 29.7 |
| Sodium ion | 21.1 |
| Water oxygen | 29.0 |
| RMSD bond angles (o) | 1.37 |
| RMSD bond lengths (Å) | 0.008 |
| Ramachandron plot | |
| Favored (%) | 99.0 |
| Allowed (%) | 1.0 |
Overall/last shell.
Rsym is the unweighted R value on I between symmetry mates.
Rcryst = Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖/Σhkl|Fobs(hkl)|.
Rfree is the cross-validation R factor for 5% of reflections against which the model was not refined.
Purification of CS1 filaments.
CS1 pili were purified from ETEC LMC10 cells carrying the pEU2030 plasmid (35). ETEC LMC10 has lost the plasmid carrying rns, which encodes the positive transcriptional regulator of coo operon expression. pEU2030 encodes rns under its own promoter. Cells were grown for 16 h in LB-Amp with shaking at 37°C. Cells were harvested by centrifugation at 8,000 × g for 30 min at 4°C. The cell pellet was resuspended in phosphate-buffered saline (PBS) (pH 7.4: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) with 10 mM EDTA. CS1 pili were sheared from the cells by passing the mixture through a 25-gauge needle four times followed by vigorous vortexing for 1 min. Cell debris was removed by centrifugation at 8,000 × g for 30 min at 4°C. Ammonium sulfate (20% [wt/vol]) was added to the supernatant and incubated at 4°C for 2 h to precipitate CS1 filaments, which were isolated by centrifugation. The CS1 pellet was resuspended in PBS-EDTA and dialyzed exhaustively in the same buffer to remove residual ammonium sulfate. Purity was assessed by SDS-PAGE analysis (see Fig. S1 in the supplemental material), and the major 15-kDa CooA band was excised from the gel and analyzed by tandem mass spectrometry at the University of Victoria (UVic) Genome BC Proteomics Centre to confirm its identity.
Electron microscopy and image processing.
CS1 pili in PBS buffer were diluted 20× with distilled water. Five microliters of sample was applied to glow-discharged continuous carbon-coated grids, negatively stained by 2% uranyl acetate, and imaged on an FEI Tecnai T12 microscope at 80 keV with a nominal magnification of ×30,000. Images were recorded on Kodak SO163 film and digitized using a Nikon Coolpix 8000 scanner at a raster of 4.16 Å per pixel. The EMAN software package (36) was used to extract filament images from micrographs, and the SPIDER software package (37) was used for most image processing.
Segments (100 × 100 pixels each; n = 10,443) of the CS1 filaments were extracted from the micrographs, and their power spectra were added together to generate an average power spectrum (see Fig. 2B). Layer lines were indexed and used along with the metal shadowing data (see below) to determine the mean helical symmetry. The data set was reconstructed using the iterative helical real-space reconstruction (IHRSR) method (38) using three starting points: 114°/7.7 Å, 113°/8.7 Å, and 112°/9.7 Å. The two stable solutions were used for modeling. University of California, San Francisco (UCSF), Chimera software (39) was used to fit the high-resolution CooAdsc structure into the experimental density maps after converting from the SPIDER format to BRIX. Atomic coordinates from the crystal structure were converted into a density map, filtered to a 20-Å resolution, and docked into each map manually. The orientation of the subunits in the two preliminary pseudoatomic models differed by a ∼30° tilt and a ∼25° counterclockwise rotation. We used the CooA crystal structure to generate four model volumes. In the first model, the orientation of the protomers was the same as that in the preliminary atomic model derived from the 113°/8.7-Å stable solution; in the second model, protomers were rotated counterclockwise by 25°; in the third model, protomers were tilted by 25°; finally, in the fourth model, subunits were rotated counterclockwise by 25° and tilted by 25°. The first two models had a symmetry of 113°/8.7 Å, while a symmetry of 112°/10 Å was imposed on the third and fourth models. It was not possible to apply the symmetry of 113°/8.7 Å on tilted subunits because of the severe clashes between the protomers in such a model. The four model volumes were scaled to 4.16 Å per pixel and projected into 100 × 100-pixel images with an azimuthal rotational increment of 4°, generating 360 reference projections (4 × 90). The CS1 segments were cross-correlated with these 360 reference projections. Less than 6% of segments yielded the best correlation with model 3, and thus that model was eliminated in the final run. Finally, 10,443 CS1 segments were cross-correlated with 270 model projections, and each of the three resultant groups was independently reconstructed by the IHRSR algorithm. The first group (n = 6,012) converged to a symmetry of 113.2°/8.7 Å, and the second group (n = 2,052) yielded a symmetry of 113.1°/8.6 Å, while the third group (n = 2,379) returned a 111.9°/10-Å value. Reference-free 2D averages were calculated for each group using the AP SR SPIDER algorithm. The conservative Fourier shell correlation (FSC) 0.5 criterion was used for resolution determination. Each of the three data sets was split in two, and the IHRSR procedure was used on these half sets, starting each at different helical symmetries. The correlation between the corresponding Fourier shells of the two resultant volumes was calculated for each data set, all of which yielded a resolution of ∼20 Å. The UCSF Chimera software program (39) was used to manually fit the high-resolution structures of CooAdsc (chain A) into the experimental density maps for the three structural groups to generate pseudoatomic-resolution CS1 models, with N-terminal residues 13 and 14 removed due to redundancy of these residues in the C terminus.
Fig 2.
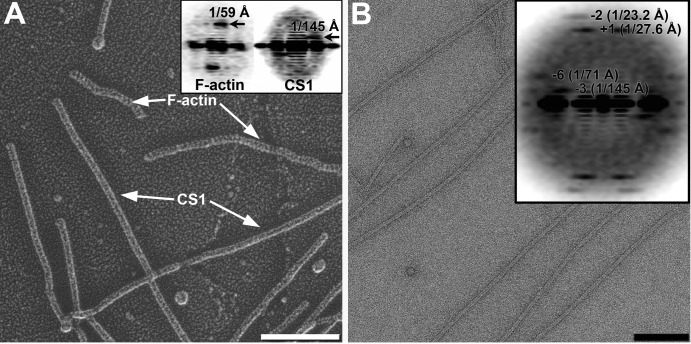
Determination of the helical parameters of the CS1 pili. (A) Quick-freeze/deep-etch metal shadowed micrograph of CS1 pili and actin filaments. Scale bar, 1,000 Å. (Inset) Diffraction patterns obtained from F-actin (left) and CS1 pili (right). (B) Micrograph of the negatively stained CS1 filaments. Scale bar, 1,000 Å. (Inset) Diffraction pattern of negatively stained CS1 filaments reveals four major layer lines. Bessel orders and positions of the peaks are indicated.
Scanning transmission electron microscopy of CS1.
To establish the mass per unit length of CS1, the filaments were examined by scanning transmission electron microscopy (STEM). CS1 filaments were freeze-dried at the Brookhaven National Laboratory (BNL) STEM facility. Digital dark-field images were collected, and the data were analyzed using the PCMass software program. CS1 data were normalized to data for tobacco mosaic virus (using a mass per unit length of 13.1 kDa Å−1).
Metal shadowing.
To determine the helical handedness of CS1, samples were first adsorbed to small glass coverslips at 37°C for 5 min in preparation for freeze-drying and platinum replication. (In some cases, F-actin was added to the samples to provide an internal standard to determine the orientation of the final replica and thus to determine the correct handedness of CS1.) The glass was then rinsed vigorously with 10 ml of buffer solution to remove unattached proteins, passed very briefly through distilled water to remove buffer and salts, and “quick-frozen” by contact with a copper block cooled to ∼4 K with liquid helium. Frozen samples were then stored in liquid nitrogen until mounting in a Balzers 301 vacuum evaporator, wherein they were freeze-dried for 20 min at −80°C and then rotary replicated with 2 nm of platinum deposited from an electron-beam gun mounted at 15° above the horizontal. Replicas were then stabilized with a carbon film 10 nm thick, rotary deposited from a 70° angle. Replicas were floated off the glass onto concentrated hydrofluoric acid and then transferred through several rinses of distilled water and picked up on Formvar-coated copper grids. Finally, replicas were examined with a Jeol 100CX electron microscope operating at 100 keV and photographed at magnification ×150,000 with an AMT digital camera.
Figure preparation.
Molecular graphic images were produced using the UCSF Chimera software package (39) and the PyMOL molecular graphics system, Schrödinger, LLC.
Accession numbers.
The atomic coordinates for CooAdsc have been deposited in the Protein Data Bank under accession number 4HJI. The CS1 electron density maps have been deposited in the Electron Microscopy Data Bank under accession numbers 1951 (reconstruction 1), 1952 (reconstruction 2), and 1953 (reconstruction 3).
RESULTS AND DISCUSSION
X-ray crystal structure of CooAdsc.
The CooA pilin subunit was expressed as a soluble recombinant protein in which the N-terminal 11 residues were removed and the Nte (residues 1 to 13) was appended to the CooA C terminus (Fig. 1A) with an intervening 4-residue Asp-Asn-Lys-Gln linker sequence (the DNKQ linker) (Fig. 1B). This strategy has been used to produce soluble subunits and complexes for CfaB and pilins in the chaperone-usher pilus family to prevent misfolding and spontaneous subunit polymerization (7, 8, 40, 41). The crystal structure of recombinant donor-strand-complemented CooA, CooAdsc, containing an N-terminal hexahistidine tag was solved to a 1.6-Å resolution by molecular replacement using CfaBdsc (3F84 [7]) as a model (Table 2).
CooAdsc is a single-domain protein that forms a 7-stranded β-sandwich with an Ig fold (Fig. 1C to E). CooAdsc crystallized as a dimer with the two monomers packed together by hydrogen bonding between the second sheet of the β-sandwich in each subunit. The monomers cross over each other at an ∼45o angle and are essentially identical in structure (see Fig. S2A in the supplemental material), but the N- and C-terminal residues Asp12 and Pro165, respectively, are resolved only for chain A. The first β-sheet of the β-sandwich is antiparallel, comprising strands A1, B, E, and D (Fig. 1C). The second β-sheet comprises strands A2, G, F, and C. Strands G, F, and C are antiparallel, whereas A2, which lies at the edge of the second β-sheet, runs parallel to G. Several of the β-strands span both β-sheets and are interrupted by loops that protrude from the surface of the protein. The N- and C-terminal halves of strand A, A1 and A2, are part of β-sheets 1 and 2, respectively, and are connected by an 8-residue loop. A similar arrangement is observed for strand D on the opposite side of the protein, with D1 being part of β-sheet 1 and D2 being part of β-sheet 2, connected by a 9-residue loop. D2 is followed by a single helical turn and then D3, which rejoins the first β-sheet. Strand C also has a short loop interruption, with its N- and C-terminal halves being part of β-sheet 2 but C2 also forming canonical backbone hydrogen bonds with strand D1 of β-sheet 1. The connectivity and orientation of the β-strands are typical of a c-type Ig fold (42), but the protein has an overall cylindrical shape and resembles a β-barrel due to the curvature of the β-sheets and their continuity at strand C2. The loops within the β-strands, as well as those at the ends of the β-sheets, dominate the surface of the protein. These loops are considerably more flexible than the β-strands, as shown by their higher B factors (see Fig. S2A), although they are constrained by extensive interactions with adjacent molecules in the closely packed crystal lattice, which has a solvent content of only 35%. The conformation of the loop between strand A2 and B differs slightly between the two molecules in the crystallographic dimer, with the Cα atoms of Ala36 at the tip of this loop being ∼2.2 Å apart. The CooA structure is very similar to that of the CFA/I major pilin, CfaB, as expected from the high sequence identity (53%) (Fig. 1A; see also Fig. S2B in the supplemental material). The only significant differences in backbone structure occur at the ends of the proteins: the D3-E1 loop, which is a β-hairpin in CooA and an irregular loop in CfaB, and divergent conformations for the E2-F loop.
Strand G of the second β-sheet (residues 153 to 165) is the Nte in native mature CooA that has been fused to the C terminus in CooAdsc (Fig. 1C to E). Strand G fills a hydrophobic groove between strands A2 and F to complete the second β-sheet. Strand G forms canonical backbone hydrogen bonds with strands A2 and F and interacts with A1 in sheet 1 via its side chains. Although strand G is the C-terminal strand of CooAdsc, its residues are numbered in Fig. 1E according to their position at the N terminus of the protein, since this strand would originate with an adjacent subunit (n+1) in the pilus filament via donor strand complementation (Fig. 1F). As is seen for pilins in the chaperone-usher family, four alternating hydrophobic side chains at positions P2 to P5 fit into their respective P2 to P5 pockets in the hydrophobic groove. In addition, Lys3 forms a salt bridge with Asp16 in strand A1. Clear electron density is seen for residues 1 to 12 of the Nte (residues 153 to 164 in CooAdsc) in chain A, but only the backbone atoms are apparent for Pro13 (Pro165). Pro13 is conserved among the class 5 fimbrial family and was proposed to form a flexible hinge between subunits in CFA/I due to cis-trans isomerization (7). Pro13 is also present at the N terminus in our recombinant CooAdsc; however, neither the electron density at the N terminus nor that at the C terminus is sufficient to distinguish between the cis and trans proline conformations. The next residue in native CooA, Thr14, is the first residue in β-strand A1 in the body of the pilin subunit. Since residues 1 to 11 of the Nte for subunit n+1 form the donor G strand for subunit n in the pilus filament and residues 14 to 19 form the first β-strand of subunit n+1, the linker between the subunits is comprised of only two residues, Asp12 and Pro13. Thus, the donor strand complementation interaction requires the ends of the CooA subunits to be very close together in a head-to-tail interaction in the CS1 filament (Fig. 1F), as was seen in the crystal structure of the ternary complex between two CfaB subunits and the CfaE chaperone (7).
3D reconstruction of the CS1 filament.
We used quick-freeze/deep-etch metal shadowing EM to visualize the surface features of the CS1 pili (Fig. 2A). F-actin filaments were used as a control because they possess a left-handed 1-start helix that gives rise to a sharp layer line in the power spectrum at 1/(59 Å) (Fig. 2A, inset, left). An averaged power spectrum generated from 174 nonoverlapping segments of the CS1 filaments (Fig. 2A, inset, right) revealed a layer line at 1/(145 Å) arising from a left-handed helix. To further determine the helical symmetry of the CS1 pili, we calculated power spectra for 10,443 overlapping segments from negatively stained CS1 filaments and added these together (Fig. 2B). This method is independent of any alignment needed to average together images and is thus unbiased. The resulting power spectrum shows four layer lines. The layer line at ∼1/(27.6 Å) arose from a 1-start helix, based upon the diameter of the filaments (∼75 Å) and the distance of the peak from the meridian (Fig. 2B, inset). The mass per unit length was independently determined to be 1.76 kDa/Å by STEM (see Fig. S3 in the supplemental material). Taking into account the molecular mass of CooA (∼15.2 kDa), the average rise per subunit was determined to be ∼8.6 Å. This gives a twist of ∼3.2 subunits per turn of the 1-start helix. Indexing of the remaining layer lines in the power spectrum shown in Fig. 2B suggested that the reflection at 1/(145 Å) arose from the 3-start helix, while the layer line at 1/(23.2 Å) originated with the 2-start helix. Since metal shadowing unambiguously showed that the 3-start helix is left-handed, the 1-start helix must be right-handed. Thus, the average symmetry of the CS1 pili was determined to be a 113.5° right-handed rotation around the helical axis with an 8.7-Å rise per subunit. These helical parameters are close to those of CFA/I pili (113.6°/8.3 Å [19]).
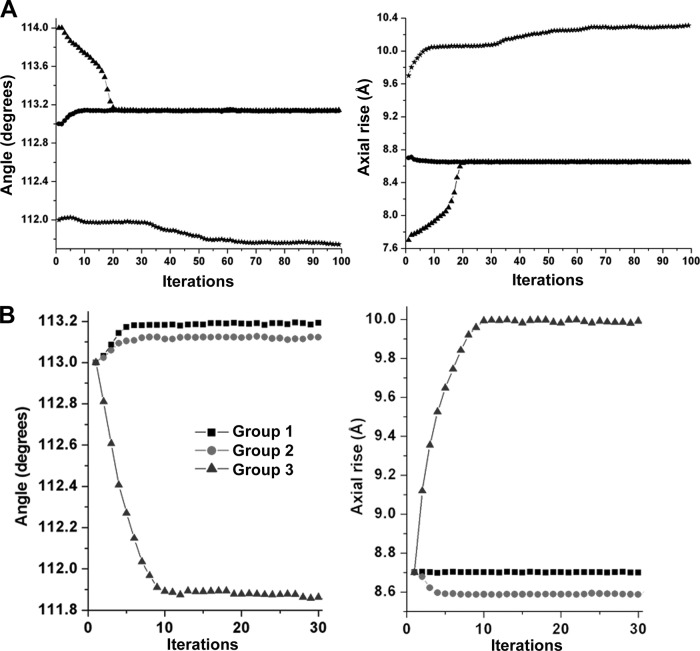
We used IHRSR (38) to determine the CS1 pilus structure. IHRSR was run using 10,443 overlapping pilus segments with three starting symmetries distributed about the mean helical parameters determined from the power spectra and the STEM data (Fig. 2B; see also Fig. S3 in the supplemental material). The IHRSR run converged to the same solution of 113.1° and 8.7 Å from the two starting values of 113°/8.7 Å and 114°/7.7 Å but failed to converge to a solution from a starting symmetry of 112°/9.7 Å (Fig. 3A). In our experience, when two or more closely related starting symmetries do not converge to the same solution, it indicates structural heterogeneity for the filaments. Thus, a single-particle approach was used to sort CS1 pilus segments into three more homogeneous groups (see Materials and Methods). IHRSR was then run for each group using a starting symmetry of 113o and 8.7 Å. Each group converged to a stable solution: the symmetries determined for group 1, the largest group, which accounts for ∼58% of all segments, and for group 2 (∼20%) are very similar, 113.2°/8.7 Å and 113.1°/8.6 Å, respectively, whereas group 3 (∼22%) has a symmetry of 111.9°/10.0 Å (Fig. 3B). Importantly, all three symmetries were randomly distributed among the CS1 filament particles extracted for image analysis, indicating that the heterogeneity exists within the filaments rather than between them.
Fig 3.
IHRSR to determine CS1 symmetry. (A) IHRSR convergence of the overall set of CS1 segments from the three starting points yields two stable solutions. (B) IHRSR convergence of the three groups to their stable solutions starting from the same helical parameters.
Pseudoatomic-resolution CS1 filament structures.
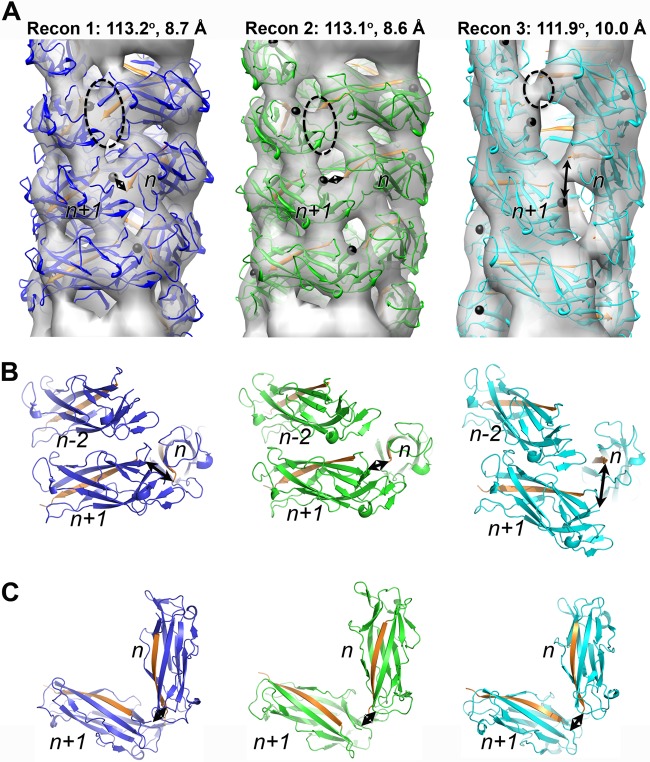
The three independent CS1 reconstructions generated using IHRSR are shown in Fig. 4A. Each reconstruction was estimated to be at ∼20-Å resolution using an FSC of 0.5 for two completely independent reconstructions generated from different starting symmetries (see Materials and Methods). Since the best measure of resolution is the quality of the map itself, we filtered our high-resolution structure of the CooAdsc pilin to 20 Å, docked it into each of the three 3D reconstructions, and generated pseudoatomic-resolution filament models by imposing the symmetry parameters onto the subunit (Fig. 4A). An important fitting constraint was the requirement for a head-to-tail interaction between each subunit such that donor strand complementation could occur. To test the validity of the CS1 filament models, each model was filtered to a 20-Å resolution and superimposed on the corresponding 3D reconstruction (Fig. 4B). The FSC for the models and reconstructions (see Fig. S4 in the supplemental material) confirm the resolution estimates and provide confidence in the 3D reconstructions of the three CS1 structural classes. Since each reconstruction was generated starting from a featureless solid cylinder in the IHRSR procedure, all the details in each reconstruction that emerge during the iterative process arose from the information contained in the pilus images. To exclude any bias from the projections used for the sorting of the CS1 segments into the three groups (see Materials and Methods), we calculated reference-free 2D averages for each group and compared those with the corresponding projections from the 3D reconstructions (Fig. 4C). An excellent match between the 2D averages and the projections of the actual reconstructions excluded the possibility that the 3D reconstructions are model biased.
Fig 4.
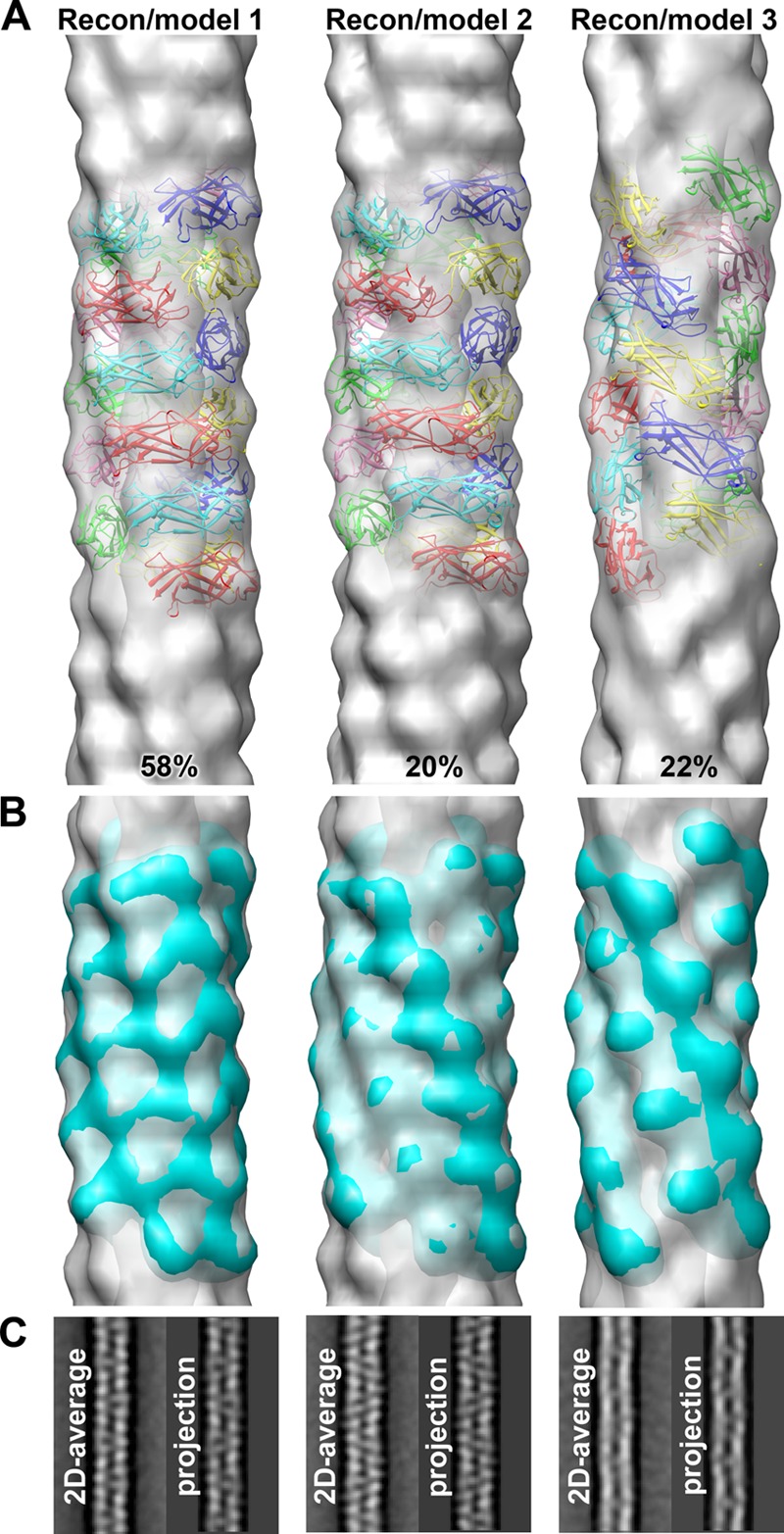
Characterization of three CS1 structural groups. (A) 3D reconstructions of the three structural groups (gray transparent surfaces) with the corresponding pseudoatomic-resolution models (colored ribbons). (B) Pseudoatomic models filtered to 20-Å resolution (cyan surfaces) superimposed on the 3D reconstructions (gray transparent surfaces). (C) Reference-free 2D averages of each structural group are compared with the projections of the corresponding 3D reconstructions.
An important feature of the class 1 and class 5 pili from Gram-negative bacteria is the head-to-tail connectivity between pilin subunits n+1 and n along the 1-start helix of the filament as a result of donor strand complementation (6). Bullitt and colleagues found that the head-to-tail interactions between the CfaB subunits of the 1-start helix in CFA/I were more extensive than those between subunits on consecutive turns or “layers” of that helix (19). Our structural analysis of the CS1 pili shows that the extent of the head-to-tail interactions depends upon the structural state of the filament. To compare the connectivity along the 1-start helix, we contoured the EM reconstructions of the three structural groups at a higher threshold (Fig. 5A) than that required to accommodate the crystal structure of the CooA pilin (as shown in Fig. 4B). The group 1 reconstruction, which represents the majority of CS1 segments, possesses a prominent bridge of density between subunits along the 1-start helix (Fig. 5A, dashed oval). This connectivity is also apparent in the group 2 reconstruction, though the density is weaker. Only a narrow bridge of density connects subunits in the 1-start helix in the group 3 reconstruction. The extent of the end-to-end connectivity correlates with the distance between the C terminus of the Nte of CooAdsc subunit n (Pro13/Pro165, end of orange strand, Fig. 5) and the N terminus of subunit n+1 (Thr14, black sphere), which are joined by a peptide bond in native CooA. In the CS1 models 1 and 2, these distances are ∼8 Å and ∼5 Å, respectively, which would allow the N terminus of subunit n+1 to complement the β-sheet of subunit n without major conformational changes in the CooA structure. However, in filament model 3, these residues are ∼19 Å apart. Head-to-tail connectivity could be accommodated in this filament structure if a small conformational change occurs at the CooA N terminus such that β-strand A1 is released from hydrogen bonding with strand B and side chain interactions with strand G (Fig. 1C and D; see also Fig. S5 in the supplemental material), allowing it to extend far enough for the Nte to reach its neighboring subunit in the 1-start helix. Support for such a mechanism is provided by the crystal structure of EcpA, the major pilin from the E. coli common pilus (ECP) (43). EcpA is similar in structure to CooA and crystallized as a domain-swapped dimer, with the A1 β-strands crossing over to hydrogen bond with strand B of their partner monomer. The exposed loop connecting strands A1 and A2 is present in CfaB and other pilins belonging to the chaperone-usher family, and thus flexibility in the N terminus may be a common feature of these pili.
Fig 5.
Comparison of the three CS1 structural states. (A) 3D reconstructions (Recon 1, 2, and 3) of the three structural states of the CS1 pili (transparent gray surfaces) shown with corresponding pseudoatomic models (ribbons). The reconstructions are contoured to show the head-to-tail connectivity between CooA subunits n and n+1 in the right-handed 1-start helix (dashed ovals). The Nte is shown in orange, and the N terminus of each CooAdsc subunit, Thr14, is shown as a black sphere. In the native CS1 filament, the Nte shown for subunit n is in fact the N-terminal strand of subunit n+1, linked via a peptide bond to Thr14 (indicated by arrows). (B) Orientation of and interactions between three subunits in the CS1 filament models, colored as in panel A. (C) Top view of subunits n and n+1 in the 1-start helix. The double-headed arrows indicate connectivity that would exist between Pro13 on the Nte bound to subunit n and Thr14 at the N terminus of subunit n+1.
Despite the relatively low resolution of the reconstructions, the fitting of the CooAdsc structure into the EM maps was unambiguous. Surprisingly, small differences in the helical symmetries of the three reconstructions are accompanied by significant structural perturbations. Reconstructions 1 and 2 have similar helical symmetries but differ in orientations of the subunits, where CooA is rotated ∼25o about its long axis in model 2 relative to its orientation in model 1 (Fig. 5B). Reconstruction 3, with its larger rise (10.0 Å), is more extended than reconstructions 1 and 2 (8.7 Å and 8.6 Å, respectively). CooA subunits are held together in the CS1 filament by head-to-tail donor strand complementation (n to n+1) and by interactions between the subunit bodies, both head-to-tail (n to n+1) and along the filament axis between each successive turn of the 1-start helix (i.e., between n+1 and n−2 in Fig. 5B). Only minor clashes occur between subunits in each of the three models, mainly between side chains in loops. This is not surprising for models derived from rigid body fits, and these clashes could be eliminated with minor adjustments in side- and main-chain torsion angles. In support of this idea, the A2-B loop at the “head” of CooA subunit n is closely associated with the tail of subunit n+1 in models 1 and 2; this loop has relatively high temperature factors, and its conformation differs slightly between the A and B chains of the CooA crystallographic dimer (see Fig. S2A in the supplemental material), suggesting flexibility in this region. Interestingly, despite the fact that many of the surface loops in neighboring subunits contact each other in both the filament models and the crystallographic lattice, these contacts are not the same. This is not surprising given that there is no head-to-tail constraint for subunits in the crystal lattice.
The filament symmetries determined here for CS1, 112o-113° rotation and ∼9-Å rise, are comparable to those of CFA/I class 5 pili (113.6°/8.3 Å [19]) and those of UPEC P pili and type I pili from the class 1 family (109.6°/7.6 Å for P pili [18]; 106.7/7.2 Å for type I pili [20]). The orientation of the CooA subunits in filament models 1 and 2 is similar to that of CfaB in the CFA/I model (7), while model 3 differs significantly.
The origin of the filament polymorphism is not clear, but it is not introduced by negative staining for EM, which may actually limit the observed heterogeneity. In an effort to obtain a higher-resolution structure, we imaged and processed frozen hydrated CS1 pili in order to preserve the filaments in a near-native state. Surprisingly, the averaged power spectra for CS1 pili in vitrified ice showed a single layer line representing the left-handed 3-start helix, with other layer lines being weakened or absent altogether (see Fig. S6 in the supplemental material). The loss of higher-resolution data is indicative of structural heterogeneity in the frozen hydrated sample. It is possible that structural perturbations were introduced into the filaments during purification, where they were mechanically sheared from E. coli cells. Yet Vibrio cholerae type IV pili are heterogeneous even without being subjected to mechanical shearing (44), and Neisseria gonorrhoeae type IV pili are not polymorphic although they were prepared by shearing (45). Interestingly, the N. gonorrhoeae type IV pili were shown to undergo a conformational change that exposed new epitopes when force was applied (46). Filament polymorphism implies structural plasticity, which would allow the pili to maintain adherence to the host epithelium when subjected to high flow forces. The ability of the filaments to alter their subunit orientations and contacts may be explained by having (i) most contacts mediated by flexible surface-exposed loops and (ii) an extensible N terminus.
In addition to the filament polymorphism, we observed a more dramatic structural perturbation of the CS1 pili that may provide further flexibility for the filaments. A fraction of the pili in the quick-freeze and deep-etch images appear to be partially unraveled at their ends, as well as within the filaments (Fig. 6). These thin fibrillar segments likely represent sections where the axial interactions between CooA subunits (i.e., between n+1 and n−2 in Fig. 5B) are disrupted but subunits remain associated via donor strand complementation. Pilus unraveling has also been reported for CFA/I, P pili, and type I pili (18, 20, 47, 48) and may thus represent an important adaptation to stress for pili in the chaperone-usher family. Force measurements of CFA/I pili using optical tweezers showed that unraveling occurs at relatively low forces (49) compared to the forces required to unravel P pili and type 1 pili (50). These differences were explained by fewer layer-to-layer interactions between subunits in consecutive turns of the CFA/I filaments than was the case for those for P pili and type 1 pili (49). The close similarity between the CS1 and CFA/I structures suggests that CS1 pili may also unravel at low forces, consistent with our observations of structural polymorphism. Importantly, unraveling of CFA/I, P pili, and type I pili is reversible, implying that the pili can behave in a spring-like manner (18, 20, 47–49). Together, pilus extensibility and reversible unraveling would provide considerable pilus length variability while maintaining the structural integrity of the filament. These features are likely to be important to pathogenic E. coli in countering flow forces in the intestines and the urethra.
Fig 6.
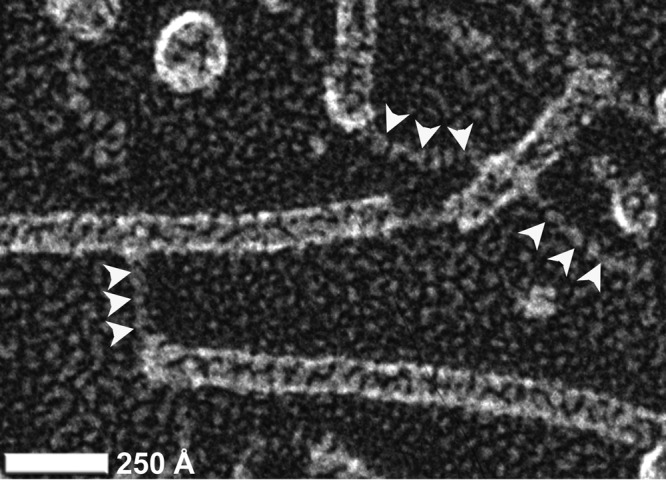
Unraveling of CS1 pili. Quick-freeze/deep-etch metal shadowed micrograph of CS1 pili reveals an unraveling of helical filaments into fibrils at the ends and within CS1 filaments.
To our knowledge, this is the first report of structural heterogeneity for a pilus belonging to the chaperone-usher family. Most likely, structural polymorphism and filament extensibility exist in all members of this pilus family, where the filament structure is determined by donor strand complementation as well as flexible axial interactions between subunits. Such structural plasticity would represent an important pathogenic adaptation for these adhesive filaments and may also contribute to antigenic variability and immune evasion.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to June Scott at Emory University, who initially proposed structural studies for CS1 and provided the expression strains. We thank Joseph Wall at the BNL STEM facility. We also thank John Heuser and Robyn Roth at the Washington University School of Medicine for metal shadowing analysis, Darryl Hardie and Derek Smith at the UVic Genome BC Proteomics Center for mass spectrometry analysis, and the staff at SSRL Beamline 11-1 for their assistance with remote collection of X-ray diffraction data. SSRL is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University.
This work was supported by grants from the NIH (EB001567 to E.H.E.) and the Natural Sciences and Engineering Research Council (to L.C.). L.C. is supported by salary awards from the Canadian Institutes of Health Research (CIHR) and the Michael Smith Foundation for Health Research. J.L. is the recipient of a Frederick Banting and Charles Best Canada Graduate Scholarship from CIHR.
Footnotes
Published ahead of print 23 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01989-12.
REFERENCES
- 1. Qadri F, Svennerholm AM, Faruque AS, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444–452 [DOI] [PubMed] [Google Scholar]
- 3. Low D, Braaten B, Van de Woude M. 1996. Fimbriae, p 146–157 In Neidhardt RCIFC, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 1 ASM Press, Washington, DC [Google Scholar]
- 4. Sakellaris H, Scott JR. 1998. New tools in an old trade: CS1 pilus morphogenesis. Mol. Microbiol. 30:681–687 [DOI] [PubMed] [Google Scholar]
- 5. Poole ST, McVeigh AL, Anantha RP, Lee LH, Akay YM, Pontzer EA, Scott DA, Bullitt E, Savarino SJ. 2007. Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol. Microbiol. 63:1372–1384 [DOI] [PubMed] [Google Scholar]
- 6. Waksman G, Hultgren SJ. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7:765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li YF, Poole S, Nishio K, Jang K, Rasulova F, McVeigh A, Savarino SJ, Xia D, Bullitt E. 2009. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ. 2000. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl. Acad. Sci. U. S. A. 97:7709–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vetsch M, Puorger C, Spirig T, Grauschopf U, Weber-Ban EU, Glockshuber R. 2004. Pilus chaperones represent a new type of protein-folding catalyst. Nature 431:329–333 [DOI] [PubMed] [Google Scholar]
- 10. Voegele K, Sakellaris H, Scott JR. 1997. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94:13257–13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061–1066 [DOI] [PubMed] [Google Scholar]
- 12. Holmgren A, Branden CI. 1989. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature 342:248–251 [DOI] [PubMed] [Google Scholar]
- 13. Kuehn MJ, Ogg DJ, Kihlberg J, Slonim LN, Flemmer K, Bergfors T, Hultgren SJ. 1993. Structural basis of pilus subunit recognition by the PapD chaperone. Science 262:1234–1241 [DOI] [PubMed] [Google Scholar]
- 14. Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058–1061 [DOI] [PubMed] [Google Scholar]
- 15. Phan G, Remaut H, Wang T, Allen WJ, Pirker KF, Lebedev A, Henderson NS, Geibel S, Volkan E, Yan J, Kunze MB, Pinkner JS, Ford B, Kay CW, Li H, Hultgren SJ, Thanassi DG, Waksman G. 2011. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474:49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. 2006. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell 22:831–842 [DOI] [PubMed] [Google Scholar]
- 17. Sauer FG, Barnhart M, Choudhury D, Knight SD, Waksman G, Hultgren SJ. 2000. Chaperone-assisted pilus assembly and bacterial attachment. Curr. Opin. Struct. Biol. 10:548–556 [DOI] [PubMed] [Google Scholar]
- 18. Mu XQ, Bullitt E. 2006. Structure and assembly of P-pili: a protruding hinge region used for assembly of a bacterial adhesion filament. Proc. Natl. Acad. Sci. U. S. A. 103:9861–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mu XQ, Savarino SJ, Bullitt E. 2008. The three-dimensional structure of CFA/I adhesion pili: traveler's diarrhea bacteria hang on by a spring. J. Mol. Biol. 376:614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller S. 2002. Exploring the 3D molecular architecture of Escherichia coli type I pili. J. Mol. Biol. 323:845–857 [DOI] [PubMed] [Google Scholar]
- 21. Sakellaris H, Balding DP, Scott JR. 1996. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol. Microbiol. 21:529–541 [DOI] [PubMed] [Google Scholar]
- 22. Sakellaris H, Munson GP, Scott JR. 1999. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc. Natl. Acad. Sci. U. S. A. 96:12828–12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sakellaris H, Penumalli VR, Scott JR. 1999. The level of expression of the minor pilin subunit, CooD, determines the number of CS1 pili assembled on the cell surface of Escherichia coli. J. Bacteriol. 181:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Froehlich BJ, Karakashian A, Melsen LR, Wakefield JC, Scott JR. 1994. CooC and CooD are required for assembly of CS1 pili. Mol. Microbiol. 12:387–401 [DOI] [PubMed] [Google Scholar]
- 25. Kabsch H. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800 [Google Scholar]
- 26. Matthews BW. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491–497 [DOI] [PubMed] [Google Scholar]
- 27. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240–255 [DOI] [PubMed] [Google Scholar]
- 29. Cowtan K. 1994. DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34–38 [Google Scholar]
- 30. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 32. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54(Part 5):905–921 [DOI] [PubMed] [Google Scholar]
- 33. Laskowski RA, McArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 34. Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caron J, Coffield LM, Scott JR. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 37. Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 [DOI] [PubMed] [Google Scholar]
- 38. Egelman EH. 2000. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85:225–234 [DOI] [PubMed] [Google Scholar]
- 39. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 40. Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543–551 [DOI] [PubMed] [Google Scholar]
- 41. Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. 2003. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587–596 [DOI] [PubMed] [Google Scholar]
- 42. Bork P, Holm L, Sander C. 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242:309–320 [DOI] [PubMed] [Google Scholar]
- 43. Garnett JA, Martinez-Santos VI, Saldana Z, Pape T, Hawthorne W, Chan J, Simpson PJ, Cota E, Puente JL, Giron JA, Matthews S. 2012. Structural insights into the biogenesis and biofilm formation by the Escherichia coli common pilus. Proc. Natl. Acad. Sci. U. S. A. 109:3950–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J, Egelman E, Craig L. 2012. Electron microscopy reconstruction of the Vibrio cholerae toxin coregulated pilus and comparative analysis with the Neisseria gonorrhoeae GC pilus. J. Mol. Biol. 418:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662 [DOI] [PubMed] [Google Scholar]
- 46. Biais N, Higashi DL, Brujic J, So M, Sheetz MP. 2010. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. U. S. A. 107:11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miller E, Garcia T, Hultgren S, Oberhauser AF. 2006. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 91:3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saulino ET, Bullitt E, Hultgren SJ. 2000. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. U. S. A. 97:9240–9245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andersson M, Bjornham O, Svantesson M, Badahdah A, Uhlin BE, Bullitt E. 2012. A structural basis for sustained bacterial adhesion: biomechanical properties of CFA/I pili. J. Mol. Biol. 415:918–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andersson M, Uhlin BE, Fallman E. 2007. The biomechanical properties of E. coli pili for urinary tract attachment reflect the host environment. Biophys. J. 93:3008–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peters JE, Thate TE, Craig NL. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perez-Casal J, Price JA, Maguin E, Scott JR. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809–819 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.