Abstract
d-Cycloserine (DCS) is a broad-spectrum antibiotic that inhibits d-alanine ligase and alanine racemase activity. When Escherichia coli K-12 or CFT073 is grown in minimal glucose or glycerol medium, CycA transports DCS into the cell. E. coli K-12 cycA and CFT073 cycA mutant strains display increased DCS resistance when grown in minimal medium. However, the cycA mutants exhibit no change in DCS sensitivity compared to their parental strains when grown in LB (CFT073 and K-12) or human urine (CFT073 only). These data suggest that cycA does not participate in DCS sensitivity when strains are grown in a non-minimal medium. The small RNA GvcB acts as a negative regulator of E. coli K-12 cycA expression when grown in LB. Three E. coli K-12 gcvB mutant strains failed to demonstrate a change in DCS sensitivity when grown in LB. This further suggests a limited role for cycA in DCS sensitivity. To aid in the identification of E. coli genes involved in DCS sensitivity when grown on complex media, the Keio K-12 mutant collection was screened for DCS-resistant strains. dadA, pnp, ubiE, ubiF, ubiG, ubiH, and ubiX mutant strains showed elevated DCS resistance. The phenotypes associated with these mutants were used to further define three previously characterized E. coli DCS-resistant strains (χ316, χ444, and χ453) isolated by Curtiss and colleagues (R. Curtiss, III, L. J. Charamella, C. M. Berg, and P. E. Harris, J. Bacteriol. 90:1238–1250, 1965). A dadA mutation was identified in both χ444 and χ453. In addition, results are presented that indicate for the first time that DCS can antagonize d-amino acid dehydrogenase (DadA) activity.
INTRODUCTION
d-Cycloserine (DCS) is a broad-spectrum antibiotic produced by Streptomyces garyphalus, Streptomyces orchidaceus, and Streptomyces lavendulae. DCS is a cyclic, structural analog of d-alanine that inhibits alanine racemase and d-alanine ligase activity (1, 2). Inactivation of these enzymes results in a failure to produce mature peptidoglycan and an increased susceptibility to osmotic lysis. DCS is occasionally used as a second-line drug in the treatment of multidrug-resistant Mycobacterium tuberculosis infections (3, 4).
DCS is not commonly used in chemotherapy regimens due to its adverse neurological side effects when administered at an effective dose (5). Unfortunately, physicians are increasingly being forced to use drugs like DCS to combat antibiotic-resistant bacterial infections. The development of new drugs is in high demand. The continued characterization of drugs like DCS could play a pivotal role in the development of novel drugs. The identification of additional DCS targets and the characterization of additional DCS resistance mechanisms could contribute to the development of new drugs with novel targets that possess less adverse effects.
The Escherichia coli cycA gene codes for a permease that transports the antibiotic d-cycloserine and the amino acids β-/l-/d-alanine, glycine, and d-serine when grown in minimal glucose or glycerol media (6–10). A mutation in the cycA gene in the E. coli K-12 and the uropathogenic E. coli (UPEC) CFT073 strains results in increased DCS resistance when grown in a minimal medium (6, 11, 12). The resistance to and transport of DCS in a complex medium, like Luria Bertani (LB), or a biologically relevant medium, such as human urine, has not been reported for E. coli. As a result, the role cycA plays in the resistance to and transport of DCS in media other than a minimal medium is not defined. The goal of our research was to characterize DCS resistance in E. coli when grown in a non-minimal medium and to determine cycA's role in resistance and transport.
In E. coli K-12, the expression of cycA is regulated by the 206-base small RNA carried by gcvB. GcvB, in conjunction with the RNA-binding protein, Hfq, negatively regulates the expression of genes involved in the transport of amino acids, dipeptides, and oligopeptides (13–17). Recently, Pulvermacher et al. determined in E. coli K-12 that GcvB negatively regulates cycA in an Hfq-dependent manner when grown in LB (18). When grown in minimal glucose medium containing glycine, Hfq, in a GcvB-independent manner, negatively regulates cycA expression. Furthermore, they showed that when growth is monitored by optical density changes, an E. coli K-12 gcvB mutant strain lyses more frequently than the wild-type strain when grown in LB containing DCS. Their data suggest that GcvB interacts with the cycA 5′ untranslated region (5′ UTR) to decrease cycA expression.
Curtiss and colleagues were the first to characterize DCS resistance in E. coli (11). They isolated three DCS-resistant E. coli strains, originating from the wild-type χ289, via successive isolation (χ289→χ316→χ444→χ453) by growing each strain in minimal glucose agar medium containing successively higher concentrations of DCS. They determined the DCS MIC for each strain in minimal glucose medium and the amino acids that antagonized DCS's negative effects, and they created a preliminary genetic map of the mutations in these strains. Russell further refined the location of the mutation in the χ316 strain to the 83-min region of the K-12 chromosome and designated it cycA (12).
Wargel et al. utilized these χ strains in the characterization of the E. coli transport systems involved in the uptake of l-/d-alanine, glycine, and DCS (9, 10). They performed DCS uptake experiments with χ289 and generated a Lineweaver-Burk plot that displays a high-affinity (Km = 4.2 × 10−5) and a low-affinity (Km = 2.0 × 10−4) line segment (10). These data suggest that E. coli K-12 possesses a high- and low-affinity DCS transport system. Similar biphasic uptake kinetics were seen in their Lineweaver-Burk plots generated from transport experiments in χ289 when glycine and d-alanine are used as substrates (9, 10). These data suggest that d-alanine, glycine, and DCS share the same transport systems in E. coli. Wargel et al. also performed DCS uptake experiments with χ316 that show a loss of the high-affinity transport system (9). These data suggest that cycA is the high-affinity transport system suggested in the uptake experiments with χ289. From these data, they determined that the increase in resistance to DCS displayed by χ316 when grown in minimal glucose medium is due to a decrease in DCS uptake. Their kinetic analyses of glycine and d-alanine uptake experiments performed with χ444 and χ453 show a loss of both the apparent high- and low-affinity transport systems in these strains. Since d-alanine, glycine, and DCS uptake experiments suggest that these molecules are transported through the same systems, these data indicate that the χ444 and χ453 strains have lost both DCS transport systems. The DCS low-affinity transport system remains unidentified.
In this study, the loss of cycA is shown to not affect E. coli DCS sensitivity when grown in LB (CFT073 and K-12) or human urine (CFT073 only). To further characterize DCS resistance in E. coli, the Keio K-12 mutant collection was screened to identify strains with an increased DCS resistance when grown in LB. Seven Keio mutant strains (mutated in dadA, pnp, ubiE to ubiH, and ubiX) were identified with increased DCS resistance. The further characterization of three K-12 DCS-resistant mutants (χ316, χ444, and χ453) isolated by Curtiss et al. was also performed in an attempt to identify genes involved in DCS transport (11). Finally, due to the association of a dadA mutation with increased DCS resistance, a competition assay was performed between DCS and d-alanine for DadA activity.
MATERIALS AND METHODS
Strains and plasmids.
Table 1 contains a description of the plasmids and strains used in this study.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| WAM2267 | UPEC strain CFT073 | 21 |
| WAM2850 | CFT073 + pKD46 | 22 |
| WAM4282 | BW25113 + pKD46 | This study |
| WAM4249 | CFT073 ΔcycA | This study |
| WAM4290 | CFT073 ΔcycA (WAM4249) + pACYC177 | This study |
| WAM4291 | CFT073 ΔcycA (WAM4249) + pcycA | This study |
| WAM4326 | CFT073 gcvB::kan | This study |
| WAM4258 | K-12 strain BW25113 | 19 |
| WAM4372 | BW25113 gcvB::kan | This study |
| GS162 | K-12 thi pheA905 ΔlacU169 araD129 rpsL150 | 23 |
| GS1144 | GS162 ΔgcvB::Ωcm | 13 |
| WAM4420 | BW25113 ΔcycA | This study |
| WAM4418 | WAM4420 + pACYC177 | This study |
| WAM4419 | WAM4420 + pcycA | This study |
| WAM4324 | BW25113 cycA::kan | 20 |
| WAM4316 | BW25113 dadA::kan | 20 |
| WAM4375 | BW25113 pnp::kan | 20 |
| WAM4376 | BW25113 ubiE::kan | 20 |
| WAM4379 | BW25113 ubiF::kan | 20 |
| WAM4388 | BW25113 ubiG::kan | 20 |
| WAM4389 | BW25113 ubiH::kan | 20 |
| WAM4390 | BW25113 ubiX::kan | 20 |
| χ289 | K-12 W1485F− | 11 |
| χ316 | χ289 cycR1 | 11 |
| χ444 | χ316 cycR1 cycR2 | 11 |
| χ453 | χ444 cycR1 cycR2 cycR3 | 11 |
| Plasmids | ||
| pcycA | CFT073 cycA gene cloned into pACYC177 (pWAM3479) | This study |
| pACYC177 | Cloning vector | 24 |
| pKD4 | Template for λ-red kanamycin cassette | 19 |
| pKD13 | Template for λ-red kanamycin cassette | 19 |
| pCP20 | λ-Red Flp recombinase for removal of resistance cassette | 19 |
| pKD46 | λ-Red recombinase expression plasmid | 19 |
Media and antibiotics.
LB broth and LB agar mix (Difco) were used as complex media. MOPS medium [3-(N-morpholino)propanesulphonic acid] was used as a minimal medium with various carbon sources added as described below (25). Glycerol (43.4 mM) was used to grow overnight minimal medium cultures and perform DCS sensitivity assays. Sodium succinate (20 mM) was used to assay for growth with succinate as a carbon source. l-Alanine was added, to a final concentration of 5.6 mM, to MOPs glycerol minimal medium for d-amino acid dehydrogenase induction. Agar was added to the minimal medium to make plates to a final concentration of 1.5%. Pooled human urine was mixed, filter sterilized, and stored at 4°C. To make human urine agar plates, 3 parts heated (50°C) human urine was added to 1 part sterilized 6% agar water solution, mixed thoroughly, and poured immediately. Each plate used to assay for antibiotic sensitivity was poured to contain exactly 23 ml of media. Working stocks of DCS were made fresh each day by dissolving the antibiotic in double-distilled water (ddH2O) that was then filter sterilized with a 0.45-μm filter. Erythromycin was dissolved in 100% ethanol to a final concentration of 40 mg/ml and then diluted in sterile ddH2O to make a working stock at a final concentration of 2.0 mg/ml. Other antibiotics used as needed were kanamycin (50 μg/ml) and chloramphenicol (20 μg/ml).
Genetic techniques.
WAM4249, WAM4326, and WAM4372 strains were constructed using the λ-red method developed by Datsenko and Wanner (19). To replace gcvB in BW25113 and CFT073, p0 primer (5′-CCGTTGAGCTTCTACCAGCAAATACCTATAGTGGCGGCTGTGTAGGCTGGAGCTGCTTCG-3′) and p4 primer (5′-GAGATGGTCGAACTGGATCAGTAATTCGCGATCGCAAGGTATTCCGGGGATCCGTCGACC-3′), with pKD13 as a template, were used to generate the λ-red PCR fragment. To replace cycA in CFT073, p0 primer (5′-CCTGAACAACACAGACAGGTACAGGAAGAAAAAAAACTGTGTAGGCTGGAGCTGCTTC-3′) and p2 primer (5′-AAAGCTGGATGGCATTGCGCCATCCAGCATGATAATGCGACATATGAATATCCTCCTTA-3′), with pKD4 as a template, were used to generate the λ-red PCR fragment. The kanamycin cassette was excised from WAM4249 by the Flp recombinase encoded by pCP20 (19). Confirmation of a gene replacement with the kanamycin cassette and subsequent cassette removal were performed by PCR with primers flanking the gene of interest.
Antibiotic sensitivity assays.
For the antibiotic disk diffusion assays, each strain was grown overnight at 37°C with shaking in the various liquid media. The next day, the cultures were diluted to approximately 107 CFU/ml. A cotton swab was used to inoculate each agar plate. Sterile 6-mm blank Whatman paper disks were placed on each plate. For DCS sensitivity assays, a 10-μl aliquot of a 2.5 mg/ml (24.5 mM) solution of DCS was placed on each disk. For the erythromycin sensitivity assay, a 10-μl aliquot of a 2.0 mg/ml solution of erythromycin was placed on each disk. Plates were inverted and incubated at 37°C for 16 to 18 h for assays performed on LB and human urine plates and 24 h for assays performed on MOPS glycerol minimal medium plates. The zones of growth inhibition were measured with a digital caliper in two directions and averaged. MICs were determined by utilizing a broth microdilution MIC protocol (26).
Glycine accumulation assay.
A modified version of the Anfora and Welch transport assay was used in this glycine uptake study (6). Ten ml of the appropriate broth was inoculated with an overnight culture to a starting optical density at 600 nm (OD600) of 0.03 and incubated at 37°C with shaking. When each culture reached an OD600 of 0.3 to 0.4, 25.0 μl of chloramphenicol (20 mg/ml) was added to each culture and placed on ice for 10 to 15 min. The cells grown in MOPs glucose media were washed twice in 1 volume of ice-cold survival buffer containing 50 μg/ml of chloramphenicol. Cells grown in LB were washed twice in 1 volume of ice-cold LB containing 50 μg/ml of chloramphenicol. The cells were then resuspended into 1 ml of their respective wash buffers containing 50 μg/ml of chloramphenicol. A 50-μl cell aliquot of each strain was removed and placed on ice to be used for a total protein assay (Bio-Rad, CA). A 350-μl cell aliquot was then incubated at 37°C for 5 min. A 9.0-μl aliquot of [14C]glycine (0.1 Ci/mol, 1 mM) was then added to the cells and vortexed briefly. A 50-μl aliquot was taken at designated times and added to 5 ml of MOPS-Tris buffer and vortexed briefly. The suspension was then filtered through a 0.45-μm nitrocellulose membrane filter. The filters were then allowed to dry and were placed in a scintillation vial containing 3 ml of Biosafe counting cocktail. The radioactivity was assayed for each vial using a Beckman scintillation counter.
Quantification of bacterial growth.
Measurements of bacterial growth kinetics were performed in a 24-well flat-bottom tissue culture plate (Costar) using a BioTek Synergy HT plate reader (BioTek, Vermont). Approximately 107 CFU (OD600 of 0.03) was inoculated into 1 ml of LB broth. Cultures were incubated at 37°C with continuous fast shaking. The OD600 was measured every 30 min for 8 h.
d-Alanine dehydrogenase assays.
A modified version of the d-amino acid dehydrogenase assay described by Wild et al. was used (27). A brief description of the cell preparation and assay procedure are included below.
To induce dadA expression, each strain was grown overnight at 37°C with shaking in MOPS glycerol broth containing 5.6 mM l-alanine. The next day, 1 ml of culture was washed in an equal volume of 0.01 M phosphate buffer (PB), pH 7.4. The cells were resuspended in 0.1 M PB, pH 7.4, and placed on ice for 5 min. A 10-μl aliquot of cells was taken for a total protein assay and 20 μl of toluene was added to the remaining cells, which were then mixed for 30 s and placed on ice for 20 min. A 40-μl aliquot of toluenized cells was added to 960 μl of prewarmed 0.1 M PB, pH 7.4, containing 20 mM d-alanine and incubated for 30 min at 37°C. After 30 min of incubation, 0.25 ml of 2,4-dinitrophenylhyrazine (0.17 mg/ml in 1.2 N HCl) was added and the mixture was incubated at room temperature for 15 min. One ml of a 2.5 N NaOH solution then was added, and the mixture was incubated for 10 min. The OD520 was then measured.
The Bio-Rad protein assay was used to determine the total amount of protein in each DadA assay. To a 10-μl cell aliquot, 1 μl of popculture reagent (Novagen) was added, and the mixture was incubated at room temperature for 10 min to lyse the cells. A standard curve was generated using bovine serum albumin. DadA units of activity were represented as OD520/h/mg of total protein.
Competition assays for DadA activity were performed with toluenized cells. The reactions were set up as outlined above, but 1 mM instead of 20 mM d-alanine was used. Also, DCS at various concentrations was added to the prewarmed PB/d-alanine solution prior to the addition of the toluenized cells. Care was taken to make sure each reaction was allowed to progress for the same amount of time.
Succinate utilization.
Strains were grown overnight at 37°C with shaking in MOPS glycerol. The next day, each strain was streaked onto MOPS succinate minimal medium agar plates and incubated for 24 h at 37°C. The ability of a strain to utilize succinate was considered positive if it could produce colonies after overnight incubation.
RESULTS
A comparison of the DCS sensitivity phenotypes of E. coli cycA strains when grown in minimal versus complex media.
CycA was originally identified by selection for DCS-resistant mutants when grown on a minimal glucose medium (11). However, the DCS sensitivity of E. coli when grown in a non-minimal medium and the possible role of cycA in that sensitivity was not investigated. Therefore, to further characterize this phenotype, DCS sensitivity was tested by antibiotic disk diffusion when grown in minimal glycerol and non-minimal media. This was done by comparing the sensitivity of the laboratory E. coli K-12 (BW25113) and UPEC CFT073 wild-type strains to their respective cycA mutants when grown in a MOPS minimal glycerol medium, LB medium, and human urine (CFT073 only). As expected, compared to the UPEC wild-type strain CFT073, a CFT073 cycA mutant (WAM4249) possessed an increased DCS resistance phenotype similar to that of the E. coli K-12 cycA mutant (WAM4324) when grown in a MOPS minimal glycerol medium (Table 2). Compared to their respective wild-type strains, when grown in MOPs glycerol agar plates WAM4249 and WAM4324 displayed a 4- and 5-fold increase in the diameter of growth inhibition to DCS. The cycA trans-complementation of the cycA mutation in the CFT073 cycA (WAM4249) and BW2513 cycA (WAM4324) mutant strains restored DCS sensitivity to wild-type levels while the strains carrying the vector only (WAM4290 and WAM4418) did not.
Table 2.
DCS sensitivity of E. coli CFT073 and K-12 grown on MOPS glycerol, LB, or human urine agar plates
| Strain | Description | Avg zone of inhibition (mm) ona: |
||
|---|---|---|---|---|
| MOPS glycerol | LB | Human urineb | ||
| CFT073 | Wild type | 51.1 (±1.2) | 14.0 (±1.2) | 22.2 (±0.8) |
| WAM4249 | CFT073 cycA | 13.4 (±0.4) | 13.8 (±0.8) | 21.4 (±0.9) |
| WAM4290 | CFT073 cycA/pACYC177 | 12.9 (±1.0) | 13.7 (±0.7) | 22.6 (±0.8) |
| WAM4291 | CFT073 cycA/pcycA | 53.2 (±1.4) | 13.7 (±0.7) | 22.4 (±0.7) |
| BW25113 | Wild type | 67.1 (±1.3) | 13.1 (±0.3) | ND |
| WAM4420 | BW25113 cycA | 15.2 (±1.5) | 12.9 (±0.2) | ND |
| WAM4418 | BW25113 cycA/pACYC177 | 13.7 (±1.6) | 12.4 (±0.4) | ND |
| WAM4419 | BW25113 cycA/pcycA | 66.1 (±2.2) | 12.4 (±0.2) | ND |
DCS sensitivity was determined by antibiotic disk diffusion assay. A 10-μl aliquot from a 24.5 mM solution of DCS was added to each disk. Each data point represents the average zone of growth inhibition and the resulting standard deviations from three experimental replicates.
ND, not determined.
Surprisingly, the DCS sensitivity of CFT073 and BW25113 in LB medium was unaltered compared to those of their respective cycA mutant strains (Table 2 and 3). WAM4291 and WAM4420 or CFT073 cycA and BW25113 cycA, respectively, each contained a plasmid copy of cycA (pcycA) constitutively expressed from the plasmid's kanamycin cassette promoter. The constitutive expression of the cycA gene in CFT073 cycA/pcycA (WAM4291) and BW25113 cycA/pcycA (WAM4419) mutant strains did not increase the DCS sensitivity levels above the levels observed for the cycA mutant strains containing the vector only (WAM4490 and WAM4418) or their respective wild-type strains (Table 2 and 3). In addition, the wild-type K-12 and CFT073 strains were transformed with pcycA and assayed for DCS sensitivity when grown in LB. These strains also did not display an increase in DCS sensitivity compared to their respective strain carrying the vector only (data not shown).
Table 3.
DCS sensitivity of E. coli CFT073 and K-12 gcvB mutant strains grown on LB agar
| Strain | Description | Avg zone of inhibitiona (mm) | MICb (μg/ml) |
|---|---|---|---|
| CFT073 | Wild type | 14.0 (±1.2) | 20 (±5) |
| WAM4326 | CFT073 gcvB | 13.6 (±0.7) | 40 (±5) |
| WAM4249 | CFT073 cycA | 13.8 (±0.8) | 20 (±5) |
| WAM4290 | CFT073 cycA/pACYC177 | 13.7 (±0.7) | 20 (±5) |
| WAM4291 | CFT073 cycA/pcycA | 13.7 (±0.7) | 20 (±5) |
| BW25113 | Wild type | 13.0 (±0.5) | 30 (±5) |
| WAM4372 | BW25113 gcvB | 12.5 (±0.5) | 40 (±5) |
| WAM4420 | BW25113 cycA | 12.9 (±0.2) | 30 (±5) |
| WAM4418 | BW25113 cycA/pACYC177 | 12.4 (±0.4) | 25 (±5) |
| WAM4419 | BW25113 cycA/pcycA | 12.4 (±0.2) | 30 (±5) |
| GS162 | Wild type | 13.8 (±0.4) | 15 (±5) |
| GS1148 | GS162 gcvB | 14.3 (±0.4) | 15 (±5) |
DCS sensitivity was determined by antibiotic disk diffusion assay. A 10-μl aliquot from a 24.5 mM solution of DCS was added to each disk. Each data point represents the average zone of growth inhibition and the resulting standard deviations from three experimental replicates.
Andrew's microdilution method was used to determine the DCS MIC for each strain (26). The MIC for each strain was determined in LB broth. The MIC was reported as the median from three experimental replicates performed for each strain.
To determine if cycA was involved in DCS sensitivity in a biologically relevant growth medium for UPEC, CFT073 and the CFT073 cycA (WAM4249) mutant strain were assayed for DCS sensitivity when grown in human urine agar plates (Table 2). CFT073 and the CFT073 cycA mutant strain displayed the same levels of sensitivity to DCS. Also, the CFT073 cycA/pcycA (WAM4291) strain did not display an increase in sensitivity to DCS compared to the CFT073 cycA/pACYC177 (WAM4290) strain.
These results suggest that cycA does not play a role in DCS transport when E. coli is grown in a complex or biologically relevant medium but does so only when the bacteria are grown in a minimal medium where a simple carbon source like glycerol is available. These data also suggest that cycA does not play a role in DCS sensitivity when uropathogenic E. coli resides in the urinary tract.
We hypothesize that cycA does not affect E. coli's sensitivity to DCS when grown in a non-minimal medium because of either negative regulation of cycA expression or competition from other CycA substrates present in LB and human urine for transport through CycA. To test this hypothesis, an investigation into whether the negative regulation of cycA affects E. coli's DCS sensitivity was performed.
E. coli K-12 and CFT073 gcvB mutants do not display an increase in sensitivity to DCS.
Pulvermacher et al. recently determined in E. coli K-12 that GcvB, in conjunction with Hfq, negatively regulates cycA expression when grown in LB broth (18). By optical density measurements, they showed that a K-12 gcvB strain (GS1144) lyses earlier and more completely than the parent strain (GS162) when grown in LB containing DCS.
K-12 strains GS162 and GS1144 (GS162 mutated in gcvB) were kindly provided by George Stauffer and tested for DCS sensitivity by antibiotic disk diffusion assay when grown in LB plates (Table 3). The gcvB mutants in both the CFT073 (WAM4326) and BW25113 (WAM4372) background strains were also examined for DCS sensitivity when grown in LB. None of these gcvB mutant strains showed an increased sensitivity to DCS compared to their respective parent strain (Table 3). Compared to CFT073, the CFT073 gcvB mutant strain also did not show an increase in DCS sensitivity when assayed on human urine plates (data not shown).
The MICs for each of the gcvB mutants and their respective wild-type strains were also determined. The BW25113 gcvB and CFT073 gcvB mutant strains displayed higher DCS MICs than their parent strains, while the GC162 gcvB mutant strain displayed the same MIC as its parent strain.
These data suggest that the increased expression of CycA, due to the lack of repression by GcvB, did not result in increased DCS sensitivity. These data suggest that CycA did not transport DCS due to competition from other CycA substrates present in LB and human urine. To test this hypothesis, we determined K-12's ability to accumulate a radiolabeled analog of DCS, glycine, in complex media.
The accumulation of DCS in a K-12 cycA mutant and wild-type strain are similar when assayed in complex media.
The acquisition of radiolabeled DCS was not cost-effective at the time of this study. As a result, the characterization of DCS accumulation in E. coli when grown in complex media was not possible. Fortunately, previous studies have shown that the affinities of CycA for glycine, d-alanine, and DCS were similar when assayed in a minimal medium (9, 10). As a result, the accumulation of [14C]glycine in a BW25113 cycA mutant strain and its respective wild-type strain when grown in complex and minimal medium was determined (Fig. 1). The 30-s initial velocities of wild-type and cycA mutant strains in MOPs glucose medium were 240 (±48.7) × 10−6 and 35 (±20.8) × 10−6 μmol/mg, respectively. This ∼7-fold difference in glycine accumulation supports previously published observations of CycA's role in glycine accumulation (9, 10). The 30-s initial velocities of the wild-type and cycA mutant strains when assayed in LB medium were 4.7 (±0.8) × 10−6 and 3.1 (±0.2) × 10−6 μmol/mg. Although the glycine accumulation rates are similar when assayed in LB medium, the wild type displays a 1.5-fold higher accumulation rate compared to that of the cycA mutant. The wild-type strain displayed a 51-fold decrease in glycine accumulation when assayed in LB versus MOPS glucose medium, showing the negative effect a rich medium has on glycine accumulation. When comparing the cycA mutant's glycine accumulation in MOPS glucose medium to that of the wild-type and mutant strains assayed in LB medium, the mutant displayed a 7- and 11-fold increase, respectively, in glycine accumulation. The increase in glycine accumulation of the cycA mutant when assayed in a minimal medium versus complex media supports previously published observations of an additional glycine accumulation mechanism in E. coli K-12 (9, 10).
Fig 1.
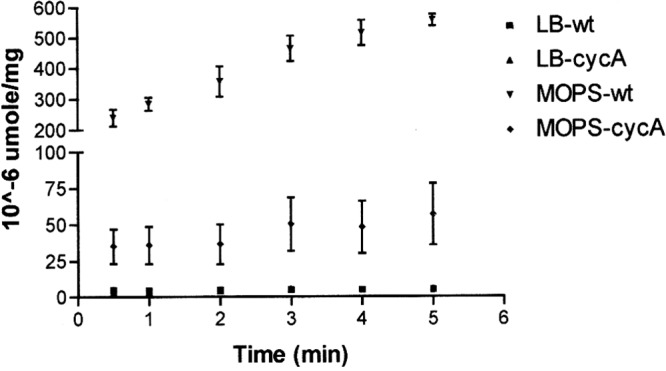
[14C]Glycine accumulation of an E. coli K-12 wild-type strain and cycA mutant strain. [14C]Glycine (2.5 ×10−5 M final concentration) was added to an aliquot of the wild type (■, LB; ▲, MOPs) and mutant cycA (▼, LB; ◆, MOPs) strains grown and resuspended in LB or MOPs glucose medium and assayed over a 5-min period. Each data point represents the average from three experimental replicates for each condition tested. The error bars represent the standard deviations from the 3 experimental replicates of each condition.
These glycine accumulation data suggest that DCS is not transported in complex medium due to either the negative regulation of CycA in complex media or as the result of additional CycA substrates (i.e., d/β/l-alanine, glycine and d-serine) present in the complex media outcompeting DCS for transport through CycA. To determine whether additional CycA substrates are outcompeting DCS for transport through CycA, competition assays were performed.
The presence of glycine and d-alanine decreases E. coli CFT073's and K-12's sensitivity to DCS.
Free glycine and d-alanine are found in significant quantities in human urine (28–32). Both of these amino acids are CycA substrates and are potential competitors for DCS transport through CycA. As a result, the capability of glycine or d-alanine to decrease CFT073's and K-12's DCS sensitivity when grown in MOPS glycerol agar plates was determined. DCS sensitivity was tested by antibiotic disk diffusion assay with a series of disks impregnated with the same amount of DCS but with an increasing amount of glycine or d-alanine (Table 4). For CFT073, a 10:1 ratio of glycine or d-alanine to DCS was necessary for the elimination of DCS sensitivity. For K-12, a 100:1 ratio of glycine or d-alanine to DCS was necessary to eliminate sensitivity to DCS. These data show that glycine and d-alanine can successfully antagonize DCS's effects on CFT073 and K-12 in MOPS glycerol agar plates.
Table 4.
E. coli CFT073 and K-12 DCS sensitivity when grown on MOPs glycerol medium in the presence of increasing concentrations of d-alanine or glycine
| A.A./DCS ratioa | Avg. zone of inhibitionb (mm) for: |
|||
|---|---|---|---|---|
| CFT073 |
K-12 |
|||
| d-Alanine | Glycine | d-Alanine | Glycine | |
| 0:1 | 18.9 (±0.4) | 21.5 (±1.6) | 32.7 (±1.1) | 32.4 (±1.1) |
| 1:1 | 15.3 (±0.3) | 19.8 (±1.9) | 31.4 (±0.3) | 32.9 (±1.7) |
| 10:1 | Resistant | Resistant | 29.1 (±0.4) | 33.5 (±0.4) |
| 100:1 | Resistant | Resistant | Resistant | Resistant |
A.A./DCS ratio, amino acid/d-cycloserine ratio. A 10-μl aliquot from a 24.5 mM solution of DCS was added to each disk. For the amino acid competition assays, d-alanine or glycine solutions of the appropriate concentrations were added to each disk in a 10-μl aliquot. For the 0:1 A.A./DCS experiment, in addition to DCS a 10-μl aliquot of H2O was added to the disk.
DCS sensitivity was determined by antibiotic disk diffusion assay on a MOPS glycerol plate containing no antibiotics or amino acids. Each data point represents the average zone of growth inhibition and the resulting standard deviations from three experimental replicates.
These data show that cycA does not affect E. coli's sensitivity to DCS in LB or human urine. This does not appear to be due to negative regulation by GcvB but is the result of competition from other CycA substrates present in human urine and LB. Our data suggest that cycA does not play a role in DCS transport or resistance when E. coli was grown in a non-minimal medium. As a result, the Keio E. coli mutant strain collection was screened to determine the genes involved in DCS transport and resistance under complex medium growth conditions.
The identification of Keio strains that have increased resistance to DCS.
The Keio collection contains approximately 4,000 E. coli strains, each with a kanamycin gene cassette replacement of a different nonessential gene (20, 33). This collection of strains was screened for growth on LB agar plates containing 25 μg/ml of DCS. Seven mutants with increased resistance to DCS were identified (Table 5). To minimize the complications of potential secondary-site mutations, each mutation was moved into a clean K-12 background via P1 transduction and retested for DCS sensitivity by antibiotic disk diffusion assay on LB plates (data not shown). All seven of the Keio strains displayed a significant increase in resistance to DCS compared to the wild-type strain. Complementation of the pnp, dadA, ubiF, ubiG, and ubiH mutant strains to their respective wild-type allele in trans resulted in an increase in DCS sensitivity similar to that of the wild type (data not shown).
Table 5.
Keio strains identified in DCS resistance screen
| Straina | Gene description | Function | MICb (μg/ml) | Gen timec (min) |
|---|---|---|---|---|
| BW25113 dadA | d-Amino acid dehydrogenase | Catalyzes the oxidative deamination of most d-amino acids; involved in alanine metabolism | ∼37.5 | ∼33 |
| BW25113 pnp | RNA polynucleotide phosphorylase/polymerase | 3′ to 5′ RNA exonuclease and 3′-terminal RNA oligonucleotide polymerase; involved in RNA maturation and degradation | ∼50 | ∼47 |
| BW25113 ubiE | C-methyltransferase | 2-Octaprenyl-6-methoxy-1,4-benzoquinone methylase and S-adenosylmethionine:2-DMK methyltransferase; involved in ubiquinone and menaquinone metabolism | ∼75 | ∼56 |
| BW25113 ubiF | Hydroxylase | 2-Octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone hydroxylase; involved in ubiquinone metabolism | ∼62.5 | ∼47 |
| BW25113 ubiG | O-methyltransferase | 3-Demethylubiquinone-8 3-O-methyltransferase and 2-octaprenyl-6-hydroxyphenol methylase; involved in ubiquinone metabolism | ∼112.5 | ∼103 |
| BW25113 ubiH | Putative hydroxylase | 2-Octaprenyl-6-methoxyphenol hydroxylase; involved in ubiquinone metabolism | ∼112.5 | ∼69 |
| BW25113 ubiX | Decarboxylase | Together with UbiG forms a 3-octaprenyl-4-hydroxybenzoate decarboxylase; involved in ubiquinone metabolism | ∼87.5 | ∼135 |
| BW25113 | K-12 wt strain | Parent strain of the Keio collection | ∼15 | ∼35 |
The gene name represents the Keio strain. BW25113 is the parent strain (wild type) of the Keio collection.
The MIC for each strain was determined in LB broth as described in the text. Three experimental replicates were performed for each strain.
Generation (Gen) times determined during growth in LB broth. Two experimental replicates were performed for each strain and averaged. Generation times for each strain were extrapolated from the log phase of each strain's growth curve.
The MIC for DCS was determined in LB broth for each of the seven strains (Table 5). The different ubi mutant strains showed the highest level of resistance to DCS compared to the other mutants. While testing the MICs for each of these strains, some of the Keio mutants were observed to have different degrees of growth attenuation in LB broth. To determine if the DCS resistance was due to the different growth rates, the generation time for each Keio mutant was determined and compared to their MICs (Table 5). Although some strains (mutated in dadA, pnp, ubiE, ubiF, or ubiG) showed a correlation between resistance and generation times, some strains (mutated in ubiH or ubiX) strains did not, which suggest that the resistance is not due to growth rate differences.
This is the first study to show that a mutation in genes involved in alanine catabolism, ubiquinone and menaquinone synthesis, and RNA processing results in an increase in E. coli DCS resistance. The E. coli DCS-resistant strains from the Curtiss et al. study (11) were characterized to determine if these strains contained a mutation in one of the genes identified in the Keio screen. Since DCS transport was characterized in these strains, the identification of a mutation in one of the genes identified in the Keio screen would aid in our understanding of DCS resistance and transport in E. coli.
Characterization of two E. coli DCS-resistant mutants defective in d-cycloserine transport (χ316 and χ444).
In 1965, Curtiss et al. were the first to report DCS resistance in E. coli K-12 (11). They isolated three DCS-resistant E. coli strains, originating from the wild-type χ289, via successive isolation (χ289→χ316→χ444→χ453) by growing each strain on minimal glucose plates containing successively higher concentrations of DCS. Wargel et al. determined that χ316's increase in DCS resistance when grown in a minimal glucose medium is due to a decrease in DSC transport resulting from a mutation in cycA (9). They also showed, with additional transport experiments performed with χ444 and χ453, that these strains do not transport DCS through CycA or through a second uncharacterized transport system (9). The χ strains show an association between DCS resistance and transport such that a decrease in DCS transport results in an increase in DCS resistance. The further characterization of DCS resistance in these strains will give a better understanding of DCS transport in E. coli.
The χ strains were assayed by antibiotic disk diffusion when grown in MOPS glycerol and LB medium to determine each strain's DCS-resistant phenotype. The following phenotypes are associated with the DCS-resistant Keio strains: inability to utilize succinate as a carbon and energy source (ubi mutants), increased erythromycin sensitivity and generation time (pnp mutants), and the inability to degrade d-alanine (dadA mutants). Each χ strain was assayed for these phenotypes to determine if they contained a mutation in one of the seven genes identified in the Keio screen. The results of these assays are presented in Table 6.
Table 6.
Phenotypes of the χ289, χ316, χ444, and χ453 strains
| Straina | DadA activityb | Succinate utilizationc | Erm sensitivity (mm)d | DCS sensitivity (mm)d on: |
|
|---|---|---|---|---|---|
| MOPS glycerol | LB | ||||
| χ289 | 45.6 (±3.5) | Yes | 10.4 (±0.6) | 64.4 (±2.7) | 12.6 (±0.6) |
| χ316 | 47.0 (±3.3) | Yes | 11.0 (±0.6 | 16.3 (±0.7) | 12.6 (±0.5) |
| χ444 | 2.4 (±1.1) | Yes | 10.7 (±0.4) | Resistant | Resistant |
| χ453 | 2.0 (±0.8) | Yes | 10.5 (±0.7) | Resistant | Resistant |
| BW25113 | 61.9 (±10.9) | Yes | 12.0 (±1.5) | 67.1 (±1.3) | 13.1 (±0.3) |
| BW25113 dadA | 3.0 (±2.4) | Yes | 11.4 (±0.3) | 46.2 (±2.6) | Resistant |
| BW25113 pnp | 79.9 (±12.3) | Yes | 23.6 (±1.6) | 63.4 (±2.2) | Resistant |
| BW25113 ubiF | ND | No | ND | ND | Resistant |
χ Strains are from Curtiss et al.'s study of DCS resistance in E. coli (11). The gene name represents the Keio strain identified in the DCS resistance screen. BW25113 is the parent of the Keio strain collection.
d-Alanine dehydrogenase (DadA) activity is reported as OD520/h/mg of total protein. Each data point represents the average and the resulting standard deviations from three experimental replicates.
From an overnight culture, each strain was streaked onto a MOPS succinate agar plate and incubated at 37°C for 24 h. Growth was determined by visual inspection.
Antibiotic sensitivity was determined by antibiotic disk diffusion assay on LB plates. For erythromycin sensitivity assays, 20 μg of erythromycin was added to each disk. For DCS sensitivity assays, a 10-μl aliquot from a 24.5 mM solution of DCS was added to each disk. Each data point represents the average zone of growth inhibition and the resulting standard deviations from three experimental replicates.
The magnitude of DCS sensitivity for χ289 and χ316 showed levels of sensitivity similar to those of the BW25113 strain and BW25113 cycA mutant strain when assayed by antibiotic disk diffusion in MOPS glycerol and LB plates (Tables 2 and 6). These data support Wargel et al.'s conclusion that cycA is mutated in χ316 (9). Because χ444 and χ453 were derived from χ316, these strains should also contain the same cycA mutation. The cycA genes from χ289, χ316, χ444, and χ453 were PCR amplified and sequenced, and their nucleotide sequences were compared to the K-12 (MG1655) cycA sequence. The cycA gene from χ289 was identical to the MG1655 sequence, while the cycA genes from χ316, χ444, and χ453 all contained an identical 19-bp deletion (bp 930 to 949). This mutation was predicted, via in silico translation, to cause a nonsense mutation at amino acid 342 of the 470-amino-acid protein. Complementation of the χ316 cycA allele with a wild-type cycA allele in trans resulted in an increase in DCS sensitivity similar to that of the wild type (data not shown).
Ubiquinone mutants are unable to utilize nonfermentable substrates as a carbon source and thus are unable to grow in a MOPS minimal succinate medium (34–37). All three χ mutant strains and the parent strain were capable of utilizing succinate as a sole carbon source, as shown by their ability to grow in a MOPS succinate plate. The Keio pnp and dadA mutants were also capable of utilizing succinate as a sole carbon source; the ubiF mutant, however, was not (Table 6). These data suggest that the χ strains do not contain a mutation in the genes responsible for ubiquinone biosynthesis.
E. coli pnp mutant strains display longer generation times (45 to 60 min) than their parents when grown in LB and possess increased sensitivity to the antibiotics nitrofurantoin, rifampin, spectinomycin, chloramphenicol, and erythromycin (38, 39). The generation times for all of the χ strains were calculated to be ∼33 min when grown in LB broth. These generation times are more similar to that of BW25113 (∼35 min) than the Keio pnp mutant (∼47.3 min). Compared to the pnp mutant, all four χ strains and BW25113 were less sensitive to erythromycin when assayed by disk diffusion on LB plates (Table 6). These data suggest that the χ strains, particularly χ444 and χ453, contain a functional pnp gene. This was also confirmed by DNA sequence analysis of the pnp gene in each strain (data not shown).
E. coli dadA strains are unable to catabolize d-alanine into ammonia and pyruvate (40, 41). DadA activity was assayed in the selected Keio mutants and χ strains (Table 6). BW25113, the BW25113 pnp mutant, χ289, and χ316 strains all possessed similar DadA activity levels, while χ444, χ453, and the Keio dadA mutant strain did not. The dadA genes from χ289, χ316, χ444, and χ453 were PCR amplified, sequenced, and compared to the MG1655 dadA gene sequence. The MG1655, χ289, and χ316 dadA nucleotide sequences were 100% identical, while the dadA nucleotide sequences from χ444 and χ453 contained a 57-bp insertion after bp 1138 of the 1,299-bp gene. When translated in silico, the 57-bp insertion is predicted to result in an in-frame insertion of 19 amino acids after amino acid 371 of the 432-amino-acid DadA protein. Complementation of this dadA allele in χ444 and χ453 with a wild-type dadA allele in trans resulted in an increase in DCS sensitivity similar to that of the wild type (data not shown). These data indicate that dadA was one of the additional mutations in χ444 and χ453.
An attempt was made to identify the third mutation in χ453 that was responsible for this strain's higher DCS resistance when grown in a minimal glycerol medium. The genes that encode known DCS-sensitive enzyme targets, the alanine racemases (alr and dadX) and the d-alanine ligases (ddlA and ddlB), were PCR amplified from the χ453 chromosome, sequenced, and compared to the respective MG1655 gene sequences. In addition, 600 to 800 bp upstream of each of these genes in χ453 was analyzed in the same manner. The sequence comparison of all four genes and their promoter regions from χ453 were identical to the MG1655 gene sequences. These data suggest that the increase in χ453 DCS resistance is not due to a mutation in the alanine racemase or the d-alanine ligase genes or their putative promoter regions (data not shown).
The χ444 and χ453 strains are the second instance in this study in which a mutation in dadA has been associated with DCS resistance in E. coli. As a result, the mechanism behind this phenotype was further characterized.
d-Cycloserine competes with d-alanine for DadA activity.
As mentioned previously, DCS is a structural analog of d-alanine that inhibits alanine racemase and d-alanine ligase activity (1, 2). To determine if DCS can antagonize E. coli DadA activity, a series of competition assays were performed with BW25113 toluenized cells to determine if DCS can compete with d-alanine for DadA activity. As the concentration of DCS is increased, the rate of d-alanine degradation decreased (Fig. 2). These data suggest that DCS can antagonize the degradation of d-alanine by DadA.
Fig 2.
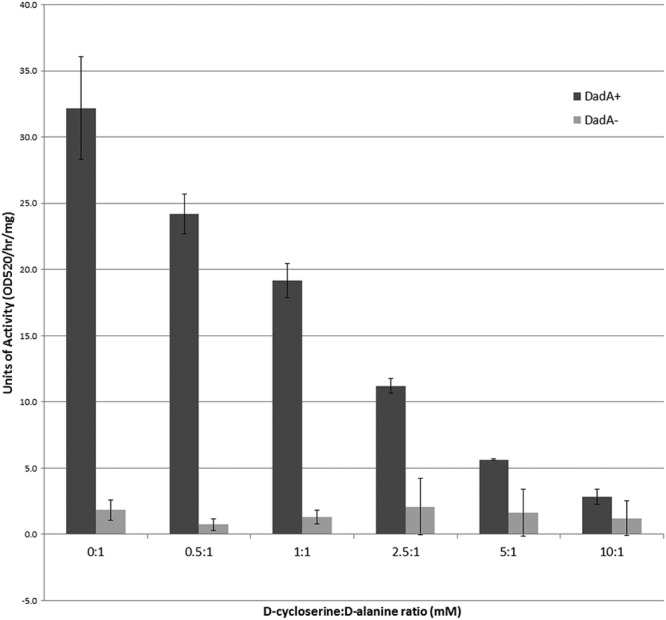
d-Amino acid dehydrogenase (DadA) competition assays performed with toluenized BW25113 and BW25113 dadA mutant cells. Six DadA reactions were performed with the same amount of d-alanine (1 mM) but increasing concentrations of DCS (0 to 10 mM) for each strain. Each data point represents the average from three experimental replicates for each condition tested. The error bars represent the standard deviations from the 3 experimental replicates of each condition.
DISCUSSION
The DCS resistance and transport studies performed by Curtiss et al. and Wargel et al. identified and characterized the role of CycA in the resistance and transport of DCS in E. coli when grown in a simple defined minimal glucose medium (9, 11). Neither of these studies characterized the effect of CycA on DCS resistance or transport in a complex medium. In this study, we confirmed observations that showed that an E. coli cycA mutant is more resistant to DCS than the wild-type parent when grown in a minimal medium (Table 2). These data support Wargel et al.'s conclusion that E. coli transports DCS through CycA when grown in a minimal glucose or glycerol medium (9). In this study, we also showed that compared to a wild-type strain, a cycA mutant strain did not display an increase in sensitivity when assayed by antibiotic disk diffusion and grown in complex media (i.e., LB) or a biologically relevant medium (i.e., human urine). These data suggest that E. coli does not transport DCS through CycA when grown in a complex or biologically relevant medium.
The lack of DCS transport through CycA could be the result of negative regulation of cycA when grown in LB or human urine. The results of Pulvermacher et al. suggest that when the E. coli K-12 strain GS162 is grown in LB, the small RNA (sRNA) encoded by gcvB, in conjunction with Hfq, interacts with the cycA 5′ UTR, which results in the negative regulation of cycA (18). The GS162 gcvB mutant strain lysed at an earlier time point than the parent when growth in LB broth containing DCS was measured by changes in optical density (18). Our results indicate that the loss of gcvB did not affect DCS sensitivity when an E. coli mutant and its respective parent were assayed by disk diffusion on LB agar medium (Table 3). Our data suggest that the increase in cycA expression due to the lack of gcvB repression does not result in increased DCS sensitivity.
The DCS sensitivity of the gcvB mutants tested in this study varied (Table 3). The CFT073 and BW25113 gcvB mutant strains displayed similar sensitivities to DCS relative to their respective wild-type parents when tested via disk diffusion, but these mutants displayed an increased MIC compared to the wild type. The GS162 gcvB mutant strain, on the other hand, displayed the same MIC and similar DCS sensitivities when assayed by disk diffusion. In addition, the GS162 gcvB mutant strain did not show an increased sensitivity to DCS as observed by Pulvermacher et al. These variable results may be due to the different techniques used to assay DCS sensitivity and pleiotropic effects of a gcvB mutation in different E. coli strains.
The recombinant plasmid pcycA was used to complement the K-12 and CFT073 cycA mutant strains. This plasmid has the CFT073 cycA gene cloned downstream of the kanamycin resistance gene in the vector pACYC177. The location of the cycA gene in pACYC177 allows for the constitutive expression of cycA from the kanamycin resistance gene (APH promoter). In addition, this vector construct lacks the cycA 5′ UTR, which is the region regulated by GcvB. Expression constructs lacking the cycA 5′ UTR result in increased cycA expression when grown in LB (18). The presence of pcycA in the different cycA mutant backgrounds did not confer increased sensitivity to DCS compared to those strains with the pACYC177 vector when grown in LB or human urine (Table 2). In addition, when grown in LB, the presence of pcycA in wild-type CFT073 and BW25113 backgrounds also did not confer an increase in sensitivity to DCS compared to that of these strains with the vector only (data not shown). These data suggest that the overexpression of cycA does not affect E. coli's sensitivity to DCS when grown in LB medium.
Pulvermacher et al. studies of cycA regulation show that even though GcvB negatively regulates CycA expression 2.5-fold when E. coli K-12 is grown in LB, CycA is still expressed when this strain is grown in complex media (18). Along with the results of our DCS sensitivity assays, these data suggest that the lack of DCS transport in E. coli through CycA is not due to the negative regulation of cycA. We hypothesize instead that the lack of DCS transport through CycA most likely is due to competition from other CycA substrates present in LB and human urine. CycA is capable of transporting d/l/β-alanine, d-serine, glycine, and DCS (6–10). Uptake experiments performed on E. coli CFT073 show that glycine and d-alanine are capable of outcompeting d-serine for transport through CycA (6). Our data showed that glycine and d-alanine are capable of antagonizing DCS's antibiotic effect on E. coli when grown in MOPS glycerol medium (Table 4). In addition, the [14C]glycine accumulation experiments performed in this study showed antagonism of glycine uptake when an E. coli K-12 wild-type strain was assayed in complex media (Fig. 1). LB is rich in oligopeptides derived from the proteolytic digest of casein (tryptone) and yeast extract (Saccharomyces cerevisiae) (42). Estimates on the amounts of l-alanine and glycine in LB are 0.5 and 0.3 to 1.35 mg/ml, respectively (43). The amount of these two amino acids present as free amino acids is unknown. The major organic components of human urine include urea, uric acid, and creatinine (44). Free amino acids are also present in urine, including all of the known amino acid substrates of CycA in the following concentration ranges: l-alanine (0.26 to 9.1 μg/ml) (28–30), β-alanine (0.2 to 1.0 μg/ml) (45), d-serine (1 to 39 μg/ml) (28, 30, 46), d-alanine (0.004 to 7.5 μg/ml) (28–30), and glycine (0.34 to 7.5 μg/ml) (31, 32). The presence of CycA substrates in human urine, and possibly LB, suggests E. coli most likely does not transport DCS through CycA due to substrate competition.
To further characterize DCS resistance and transport in complex media, the Keio mutant collection was screened to identify gene deletions that allow for growth in increased concentrations of DCS. Seven Keio strains (dadA, pnp, ubiE, ubiF, ubiG, ubiH, and ubiX mutant strains) were identified that could form colonies on LB plates after 16 to 18 h at 37°C containing 25 μg/ml of DCS.
The pnp gene encodes the phosphorylase Pnpase, which is involved in the processing and degradation of RNA. In vivo, Pnpase functions primarily as a 3′ to 5′ exonuclease responsible for the processive degradation of single-stranded RNA. Pnpase can also act as a polymerase in vivo in an E. coli mutant lacking the major RNA polyadenylating enzyme, PAP I. This activity involves the addition of heteropolymeric tails to the 5′ end of single-stranded RNA (47–49). While Pnpase consists of a homotrimer primarily found unassociated with other proteins, approximately 10 to 20% of the Pnpase molecules do associate with the RhlB RNA helicase, the glycolytic enzyme enolase, and the endoribonuclease RNase E in a complex designated the RNA degradosome (50–53). Pnpase is also involved in the processing and degradation of mRNA, rRNA, tRNA, and sRNA (54, 55).
Pnpase is a pleiotropic regulator, and a K-12 pnp mutant strain displays multiple phenotypes compared to the wild-type strain. Previously reported phenotypes of K-12 pnp mutants are longer lag and generation times when grown in LB (56), increased sensitivity to a broad spectrum of antibiotics (38, 57), and growth attenuation at low temperatures (58). In addition, median mRNA steady-state levels and half-lives of an E. coli pnp strain are different from those of its parent strain (39, 56). These differences lead to a general fitness defect and are responsible for the range of phenotypes associated with a pnp mutant. We confirmed an increased sensitivity to the antibiotics chloramphenicol (data not shown) and erythromycin (Table 6) (38). In addition, we also determined that the Keio pnp strain is more sensitive to carbenicillin, nalidixic acid, rifampin, and colistin (data not shown).
We observed for the first time that the Keio pnp mutant strain is more resistant to DCS. How increased DCS resistance is conferred by the Keio pnp strain is unknown at this time. Due to the observed alterations in mRNA abundance and stability in an E. coli K-12 pnp strain, we hypothesize that the altered mRNA abundance and stability results in an increase in the expression of one or more of the DCS-sensitive enzymes (i.e., alanine racemase and d-alanine ligase). This hypothesis is supported by multiple studies that show that the overexpression of alanine racemase and/or d-alanine ligase in E. coli (59), Streptococcus challis (3), and Mycobacterium smegmatis (59, 60) results in an increased DCS resistance. Alternatively, the altered mRNA abundance and stability may lead to a decrease in expression of DCS transporters, thus increasing resistance.
Ubiquinone, also called coenzyme Q, is an isoprenoid quinone that acts as an electron carrier in the aerobic respiratory chain. We identified several ubiquinone synthetic mutant strains (mutated in ubiE, ubiF, ubiG, ubiH, or ubiX) in our screening of the Keio collection for DCS-resistant strains (Table 5). The ubiE gene also participates in menaquinone synthesis. It should be noted that ubiA, ubiB, and ubiD mutant strains were not present in the version of the Keio collection used in our screen. These genes are also involved in the synthesis of ubiquinone, and whether ubiA, ubiB, and ubiD mutant strains would display an increased resistance to DCS remains to be determined.
E. coli ubi mutant strains display resistance to multiple antibiotics. K-12 ubiA and ubiD mutants were identified in the characterization of resistance to the antibiotics phleomycin and bleomycin (61). An E. coli K-12 ubiF mutant was isolated in a screen for resistance to the aminoglycoside streptomycin (62). This mutant strain displayed an increased resistance not only to streptomycin but also to other aminoglycosides, including gentamicin, kanamycin, and neomycin. Aminoglycoside transport experiments determined that resistance is partially dependent upon proton-motive force (PMF). A decrease in the PMF potential results in a decrease in aminoglycoside uptake and an increase in resistance (62–65). The identification of multiple ubi mutant strains in this study suggests that a fully functioning aerobic respiratory chain is necessary for DCS uptake and sensitivity.
In addition to the primary mode of action displayed by bactericidal antibiotics, they also contribute to cell death through a common free radical-based mechanism that results in damage to proteins, membrane lipids, and DNA (66). Bactericidal antibiotics hyperactivate the electron transport chain, which results in a transient NADH depletion and the formation of superoxide. The increased superoxide damages iron-sulfur clusters, generating ferrous iron, which participates in hydroxyl radical formation via the Fenton reaction (66). Since DCS is a bactericidal antibiotic and a fully functioning respiratory chain is necessary for increased DCS sensitivity, hydroxyl radical damage induced by DCS may also contribute to cell death.
The dadA gene encodes a membrane-bound d-amino acid dehydrogenase capable of the oxidative deamination of a broad spectrum of d-amino acids with its highest affinity toward d-alanine (40, 41, 67). DadA is an iron-sulfur flavoprotein capable of donating electrons directly to the electron transport chain via ubiquinone (68). DadA gives E. coli strains the ability to utilize d-alanine as a sole carbon, nitrogen, and energy source (69).
The DCS–d-alanine competition experiments, performed via the d-alanine dehydrogenase assay (Fig. 2), showed that an increased DCS concentration resulted in a decrease in d-alanine degradation by DadA. Two sources of intracellular d-alanine are the conversion of l-alanine to d-alanine by the alanine racemases and the recycling of the d-alanine-containing peptidoglycan stem peptide during peptidoglycan metabolism (70, 71). If DCS truly inhibits DadA activity, these two sources of d-alanine would lead to an increase in the intracellular d-alanine concentration. This increased intracellular d-alanine concentration should make it more difficult for DCS to compete for not only DadA activity but also its enzyme targets (alanine racemases and d-alanine ligases), negating DCS's inhibitory properties. These data suggest that DCS does not inhibit DadA activity but instead is a DadA substrate that can antagonize DadA for d-alanine degradation.
The DCS resistance displayed by a dadA mutant may be the result of its inability to modify DCS into a more potent form via DadA. DadA has been shown to convert d-chloroalanine to β-chloropyruvate, which inhibits alanine racemase activity (72, 73). In addition, β-chloropyruvate inhibits solute transport in E. coli by a process that has not been fully characterized, but the strong alkylating properties of this molecule are believed to be responsible for this effect (73–75). Whether DadA modifies DCS into a more potent antibiotic remains to be determined.
Interestingly, there is a potential metabolic connection between the DCS resistance displayed by the ubi and dadA mutants. DadA donates electrons to ubiquinone in the process of metabolizing its substrates (68). The absence of a fully functional electron transport chain results in a decrease in DadA activity (68, 69). If DCS is modified by DadA, resulting in a more potent antibiotic, the decrease in DadA activity would result in less DCS conversion and an increase in resistance. These data support the hypothesis that DCS is modified to a more potent form via DadA.
We were unable to identify a third mutation in χ453 responsible for the higher DCS resistance of this strain when grown in a minimal glycerol medium. Screening of the χ453 strain for phenotypes associated with the seven Keio DCS-resistant mutants failed to identify an additional mutation in this strain. Sequence analysis of the genes (ddlA, ddlB, dadX, and alr) and their promoter regions that encode known DCS targets also failed to identify a mutation. Recently, Pathania et al. determined that E. coli strains that contain ftsA or yadR on a high-copy-number plasmid possess a 16-fold increased DCS resistance (76). Their study was the first study to associate these genes with an increase in DCS resistance in E. coli. DNA sequence analysis of the ftsA and yadR genes and their promoter regions in χ453 were not performed in our study.
The data presented in this study suggest that DCS has additional enzyme targets and a previously unknown mechanism of transport. Even though DCS is toxic to humans, the identification of new enzyme targets could encourage the development of less toxic d-cycloserine derivatives or entirely new antibiotics and aid in the treatment of antibiotic-resistant pathogens.
ACKNOWLEDGMENTS
We thank Sid Withers and Miguel Dominguez from the Great Lake Bioenergy Research Center for access to their Keio collection and reagents and also for suggestions on experimental design. We also thank Roy Curtiss III and George Stauffer for providing us with the χ and GS strains, respectively. Furthermore, we thank Eric Battaglioli, Andrew Hryckowian, and Erica Raterman for their critical reading of the manuscript.
This research was funded by NIH grants RO1 DK063250 and R21 AI074592.
Footnotes
Published ahead of print 11 January 2013
REFERENCES
- 1. Lambert MP, Neuhaus FC. 1972. Mechanism of D-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zawadzke LE, Bugg TD, Walsh CT. 1991. Existence of two D-alanine:D-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry 30:1673–1682 [DOI] [PubMed] [Google Scholar]
- 3. Reitz RH, Slade HD, Neuhaus FC. 1967. The biochemical mechanisms of resistance by Streptococci to the antibiotics D-cycloserine and O-carbamyl-D-serine. Biochemistry 6:2561–2570 [DOI] [PubMed] [Google Scholar]
- 4. Riccardi G, Pasca MR, Buroni S. 2009. Mycobacterium tuberculosis: drug resistance and future perspectives. Future Microbiol. 4:597–614 [DOI] [PubMed] [Google Scholar]
- 5. Cohen AC. 1969. Pyridoxine in the prevention and treatment of convulsions and neurotoxicity due to cycloserine. Ann. N. Y. Acad. Sci. 166:346–349 [DOI] [PubMed] [Google Scholar]
- 6. Anfora AT, Welch RA. 2006. DsdX is the second D-serine transporter in uropathogenic Escherichia coli clinical isolate CFT073. J. Bacteriol. 188:6622–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cosloy SD. 1973. D-serine transport system in Escherichia coli K-12. J. Bacteriol. 114:679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider F, Kramer R, Burkovski A. 2004. Identification and characterization of the main beta-alanine uptake system in Escherichia coli. Appl. Microbiol. Biotechnol. 65:576–582 [DOI] [PubMed] [Google Scholar]
- 9. Wargel RJ, Hadur CA, Neuhaus FC. 1971. Mechanism of D-cycloserine action: transport mutants for D-alanine, D-cycloserine, and glycine. J. Bacteriol. 105:1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wargel RJ, Shadur CA, Neuhaus FC. 1970. Mechanism of D-cycloserine action: transport systems for D-alanine, D-cycloserine, L-alanine, and glycine. J. Bacteriol. 103:778–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curtiss R, III, Charamella LJ, Berg CM, Harris PE. 1965. Kinetic and genetic analyses of D-cycloserine inhibition and resistance in Escherichia coli. J. Bacteriol. 90:1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell RR. 1972. Mapping of a D-cycloserine resistance locus in Escherichia coli K-12. J. Bacteriol. 111:622–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pulvermacher SC, Stauffer LT, Stauffer GV. 2008. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol. Lett. 281:42–50 [DOI] [PubMed] [Google Scholar]
- 14. Pulvermacher SC, Stauffer LT, Stauffer GV. 2009. The small RNA GcvB regulates sstT mRNA expression in Escherichia coli. J. Bacteriol. 191:238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma CM, Darfeuille F, Plantinga TH, Vogel J. 2007. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 21:2804–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urbanowski ML, Stauffer LT, Stauffer GV. 2000. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 37:856–868 [DOI] [PubMed] [Google Scholar]
- 17. Wonderling LD, Urbanowski ML, Stauffer GV. 2000. GcvA binding site 1 in the gcvTHP promoter of Escherichia coli is required for GcvA-mediated repression but not for GcvA-mediated activation. Microbiology 146(Pt 11):2909–2918 [DOI] [PubMed] [Google Scholar]
- 18. Pulvermacher SC, Stauffer LT, Stauffer GV. 2009. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology 155:106–114 [DOI] [PubMed] [Google Scholar]
- 19. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22. Reference deleted.
- 23. Reference deleted.
- 24. Reference deleted.
- 25. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for Enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrews JM. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1):5–16 [DOI] [PubMed] [Google Scholar]
- 27. Wild J, Walczak W, Krajewska-Grynkiewicz K, Klopotowski T. 1974. D-amino acid dehydrogenase: the enzyme of the first step of D-histidine and D-methionine racemization in Salmonella typhimurium. Mol. Gen. Genet. 128:131–146 [DOI] [PubMed] [Google Scholar]
- 28. Bruckner H, Haasmann S, Friedrich A. 1994. Quantification of D-amino acids in human urine using GC-MS and HPLC. Amino Acids 6:205–211 [DOI] [PubMed] [Google Scholar]
- 29. Bruckner H, Schieber A. 2001. Determination of amino acid enantiomers in human urine and blood serum by gas chromatography-mass spectrometry. Biomed. Chromatogr. 15:166–172 [DOI] [PubMed] [Google Scholar]
- 30. Huang Y, Nishikawa T, Satoh K, Iwata T, Fukushima T, Santa T, Homma H, Imai K. 1998. Urinary excretion of D-serine in human: comparison of different ages and species. Biol. Pharm. Bull. 21:156–162 [DOI] [PubMed] [Google Scholar]
- 31. Kaneta T, Maeda H, Miyazaki M, Miyake R, Izaki H, Sakoda Y, Kinoshita S, Imasaka T. 2008. Determination of amino acids in urine by cyclodextrin-modified capillary electrophoresis-laser-induced fluorescence detection. J. Chromatogr Sci. 46:712–716 [DOI] [PubMed] [Google Scholar]
- 32. Petty F, Tucker HN, Molinary SV, Flynn NW, Wander JD. 1976. Quantitation of glycine in plasma and urine by chemical ionization mass fragmentography. Clin. Chim. Acta 66:111–117 [DOI] [PubMed] [Google Scholar]
- 33. Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, Mori H. 2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335 doi:10.1038/msb.2009.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gulmezian M, Hyman KR, Marbois BN, Clarke CF, Javor GT. 2007. The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta S, Mat-Jan F, Latifi M, Clark DP. 2000. Acetaldehyde dehydrogenase activity of the AdhE protein of Escherichia coli is inhibited by intermediates in ubiquinone synthesis. FEMS Microbiol. Lett. 182:51–55 [DOI] [PubMed] [Google Scholar]
- 36. Soballe B, Poole RK. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145(Pt 8):1817–1830 [DOI] [PubMed] [Google Scholar]
- 37. Wallace BJ, Young IG. 1977. Aerobic respiration in mutants of Escherichia coli accumulating quinone analogues of ubiquinone. Biochim. Biophys. Acta 461:75–83 [DOI] [PubMed] [Google Scholar]
- 38. Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic barcode. Antimicrob. Agents Chemother. 54:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohanty BK, Kushner SR. 2003. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 50:645–658 [DOI] [PubMed] [Google Scholar]
- 40. Franklin FC, Venables WA, Wijsman HJ. 1981. Genetic studies of D-alanine-dehydrogenase-less mutants of Escherichia coli K-12. Genet. Res. 38:197–208 [DOI] [PubMed] [Google Scholar]
- 41. Wild J, Klopotowski T. 1981. d-Amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol. Gen. Genet. 181:373–378 [DOI] [PubMed] [Google Scholar]
- 42. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brunzel NA. 2004. Fundamentals of urine & body fluid analysis, 2nd ed. Saunders, Philadelphia, PA [Google Scholar]
- 45. Kuo KC, Cole TF, Gehrke CW, Waalkes TP, Borek E. 1978. Dual-column cation-exchange chromatographic method for beta-aminoisobutyric acid and beta-alanine in biological samples. Clin. Chem. 24:1373–1380 [PubMed] [Google Scholar]
- 46. Jin D, Miyahara T, Oe T, Toyo'oka T. 1999. Determination of D-amino acids labeled with fluorescent chiral reagents, R(−)- and S(+)-4-(3-isothiocyanatopyrrolidin-1-yl)-7-(N, N-dimethylaminosulfonyl)-2,1,3-benzoxadiazoles, in biological and food samples by liquid chromatography. Anal. Biochem. 269:124–132 [DOI] [PubMed] [Google Scholar]
- 47. Mohanty BK, Kushner SR. 1999. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol. Microbiol. 34:1109–1119 [DOI] [PubMed] [Google Scholar]
- 48. Mohanty BK, Kushner SR. 2000. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:1807–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172 [DOI] [PubMed] [Google Scholar]
- 51. Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889–900 [DOI] [PubMed] [Google Scholar]
- 52. Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93:3865–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. 2009. The role of 3′-5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 85:187–229 [DOI] [PubMed] [Google Scholar]
- 55. Deutscher MP, Li Z. 2001. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 66:67–105 [DOI] [PubMed] [Google Scholar]
- 56. Bernstein JA, Lin PH, Cohen SN, Lin-Chao S. 2004. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 101:2758–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang W, Bechhofer DH. 1996. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178:2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luttinger A, Hahn J, Dubnau D. 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19:343–356 [DOI] [PubMed] [Google Scholar]
- 59. Caceres NE, Harris NB, Wellehan JF, Feng Z, Kapur V, Barletta RG. 1997. Overexpression of the D-slanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis. J. Bacteriol. 179:5046–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Feng Z, Barletta RG. 2003. Roles of Mycobacterium smegmatis D-alanine:D-alanine peptidoglycan inhibitor D-cycloserine. Antimicrob. Agents Chemother. 47:283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Collis CM, Grigg GW. 1989. An Escherichia coli mutant resistant to phleomycin, bleomycin, and heat inactivation is defective in ubiquinone synthesis. J. Bacteriol. 171:4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muir ME, Hanwell DR, Wallace BJ. 1981. Characterization of a respiratory mutant of Escherichia coli with reduced uptake of aminoglycoside antibiotics. Biochim. Biophys. Acta 638:234–241 [DOI] [PubMed] [Google Scholar]
- 63. Bryan LE, Kwan S. 1983. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 23:835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bryan LE, Van Den Elzen HM. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob. Agents Chemother. 12:163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Damper PD, Epstein W. 1981. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob. Agents Chemother. 20:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 67. Raunio RP, Jenkins WT. 1973. D-alanine oxidase form Escherichia coli: localization and induction by L-alanine. J. Bacteriol. 115:560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Olsiewski PJ, Kaczorowski GJ, Walsh C. 1980. Purification and properties of D-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J. Biol. Chem. 255:4487–4494 [PubMed] [Google Scholar]
- 69. Franklin FC, Venables WA. 1976. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coli K-12. Mol. Gen. Genet. 149:229–237 [DOI] [PubMed] [Google Scholar]
- 70. Wild J, Hennig J, Lobocka M, Walczak W, Klopotowski T. 1985. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol. Gen. Genet. 198:315–322 [DOI] [PubMed] [Google Scholar]
- 71. Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Manning JM, Merrifield NE, Jones WM, Gotschlich EC. 1974. Inhibition of bacterial growth by beta-chloro-D-alanine. Proc. Natl. Acad. Sci. U. S. A. 71:417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kaczorowski G, Shaw L, Laura R, Walsh C. 1975. Active transport in Escherichia coli B membrane vesicles. Differential inactivating effects from the enzymatic oxidation of beta-chloro-L-alanine and beta-chloro-D-alanine. J. Biol. Chem. 250:8921–8930 [PubMed] [Google Scholar]
- 74. Kaczorowski G, Shaw L, Fe M, Walsh C. 1975. Coupling of alanine racemase and D-alanine dehydrogenase to active transport of amino acids in Escherichia coli B membrane vesicles. J. Biol. Chem. 250:2855–2865 [PubMed] [Google Scholar]
- 75. Kaczorowski G, Walsh C. 1975. Active transport in Escherichia coli B membrane vesicles. Irreversible uncoupling by chloropyruvate. J. Biol. Chem. 250:8931–8937 [PubMed] [Google Scholar]
- 76. Pathania R, Zlitni S, Barker C, Das R, Gerritsma DA, Lebert J, Awuah E, Melacini G, Capretta FA, Brown ED. 2009. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 5:849–856 [DOI] [PubMed] [Google Scholar]


