Abstract
Previously, it was shown that an aconitase (citB) null mutation results in a vast overaccumulation of citrate in the culture fluid of growing Bacillus subtilis cells, a phenotype that causes secondary effects, including the hyperexpression of the citB promoter. B. subtilis aconitase is a bifunctional protein; to determine if either or both activities of aconitase were responsible for this phenotype, two strains producing different mutant forms of aconitase were constructed, one designed to be enzymatically inactive (C450S [citB2]) and the other designed to be defective in RNA binding (R741E [citB7]). The citB2 mutant was a glutamate auxotroph and accumulated citrate, while the citB7 mutant was a glutamate prototroph. Unexpectedly, the citB7 strain also accumulated citrate. Both mutant strains exhibited overexpression of the citB promoter and accumulated high levels of aconitase protein. These strains and the citB null mutant also exhibited increased levels of citrate synthase protein and enzyme activity in cell extracts, and the major citrate synthase (citZ) transcript was present at higher-than-normal levels in the citB null mutant, due at least in part to a >3-fold increase in the stability of the citZ transcript compared to the wild type. Purified B. subtilis aconitase bound to the citZ 5′ leader RNA in vitro, but the mutant proteins did not. Together, these data suggest that wild-type aconitase binds to and destabilizes the citZ transcript in order to maintain proper cell homeostasis by preventing the overaccumulation of citrate.
INTRODUCTION
Elimination of any one of the three enzymes of the tricarboxylic acid (TCA) branch of the Krebs cycle—citrate synthase (CS), aconitase (Acn), and isocitrate dehydrogenase—results in glutamate auxotrophy and a significant defect in spore formation in Bacillus subtilis (1–4). In particular, a null mutation in the aconitase (citB) gene causes a dramatic increase in the concentration of citrate in the culture fluid of growing cells (1). This accumulation of citrate prevents sporulation due to chelation by citrate of divalent cations required for proper functioning of the Spo0A-initiated phosphorelay (1, 4).
We had assumed that citB null cells accumulate citrate simply because of the lack of aconitase enzyme activity present in this strain. As B. subtilis lacks a citrate lyase enzyme, citB null cells have no way of enzymatically removing the citrate once it is formed; thus, the citrate accumulation phenotype has been attributed solely to this metabolic roadblock. However, in this report, we present data suggesting that the actual mechanism is more complex.
B. subtilis expresses one aconitase and two citrate synthase enzymes (3, 5). Aconitase is encoded by the citB gene in a single-gene transcription unit (5). The gene for the major citrate synthase, citZ, is the first gene in an operon that also includes the genes for isocitrate dehydrogenase (icd or citC) and malate dehydrogenase (mdh or citH). The gene for the minor citrate synthase, citA, is present at a separate locus and is expressed as a monocistronic RNA (3, 6). The coordinated expression of citZ, icd, and citB is controlled by three regulatory proteins (CcpA, CcpC, and CodY) that independently sense the nutritional state of the cell by interacting with specific metabolites (reviewed in reference 7). While CcpA and CodY are global regulatory proteins that respond to specific metabolites (fructose-1,6-bisphosphate for CcpA, and GTP and branched-chain amino acids for CodY), regulation by CcpC is specific to the TCA branch genes and responds to citrate. Antagonism of CcpC-dependent repression of the citB and citZ promoters by citrate has been described in detail (8–10). When citrate is absent, CcpC binds to sites in or near the citB and citZ promoters and blocks expression of these genes. When citrate is present, it causes a change in the interaction of CcpC with its binding sites, resulting in derepression of citB and citZ. When citrate is very abundant, CcpC activates citB expression, presumably reflecting a change in the interaction of CcpC with RNA polymerase (11).
In addition to having enzymatic activity, the B. subtilis aconitase protein has a second function as an RNA-binding regulatory protein (12, 13). Whether aconitase is active as an enzyme or an RNA-binding protein is determined by the status of an iron-sulfur (4Fe-4S) cluster that is essential for the catalytic activity of all aconitases in nature (14). The 4Fe-4S cluster interacts directly with the enzyme substrates and products (citrate, cis-aconitate, and isocitrate) and thus is exposed to solvent. This positioning makes the cluster vulnerable to oxidation by reactive oxygen species. At low levels of oxidation, one of the four iron atoms, Fea, is lost, resulting in a catalytically inactive 3Fe-4S cluster (15). The inactive form of the enzyme is also subject to more extensive cluster disassembly, resulting in an apoprotein that lacks the cluster entirely. The classic bifunctional aconitase, eukaryotic iron regulatory protein 1 (IRP1), is a cytosolic protein that responds to cellular iron levels by alternating between two functional states: the iron-sulfur cluster-containing aconitase enzyme and the RNA-binding apoprotein, which acts as a posttranscriptional regulator (16–18). IRP1 regulates mRNA translatability or stability by binding to iron regulatory elements (IREs), stem-loop structures present in the 5′ or 3′ untranslated regions (UTRs) of mRNA (14). IREs are found in the mRNAs of genes involved in the uptake, transport, and storage of iron (19). Since the discovery of IRP1, bifunctional aconitases have been described for prokaryotes including Escherichia coli (20), Mycobacterium tuberculosis (21), and B. subtilis (12, 13).
Recent work in our laboratory with an aconitase mutant (citB5) defective in RNA binding has provided an unexpected twist regarding the question of citrate accumulation in a citB null mutant. The citB5 mutant is a glutamate prototroph and exhibits high levels of aconitase activity in cell extracts (13), indicating that it has ample capacity to metabolize citrate. However, the citB5 mutant also accumulates citrate in the culture fluid at levels near those generated by the citB null mutant strain (22). This result suggests that there is a connection between aconitase RNA-binding activity and citrate accumulation and thus a more complex relationship between the activity of aconitase and the pool of citrate than originally assumed. In the current work, we sought to examine the roles of the individual functions of aconitase in the maintenance of citrate levels within the cell. Mutant citB alleles were created in an attempt to study the two functions of aconitase separately in vivo. Through work on these mutants, we established that mutations in aconitase lead to overproduction of both aconitase itself and citrate synthase. In addition, we present evidence that aconitase directly regulates citrate synthase production by interacting with the citZ transcript. The data presented here support a new model in which both functions of aconitase contribute to the maintenance of citrate levels in B. subtilis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains DH5α and JM107 were used as hosts for cloning; they were grown in L broth or on L agar plates supplemented with ampicillin (50 μg/ml) when necessary (23). All B. subtilis strains used in this work are listed in Table 1. Unless otherwise indicated, B. subtilis cells were grown at 37°C in liquid DS medium [0.8% nutrient broth, 0.1% KCl, 0.025% MgSO4 · 7H2O, 1 mM Ca(NO3)2, 10 μM MnCl2, 1 μM FeSO4] (24) or on DS agar plates. B. subtilis liquid cultures were prepared in Erlenmeyer flasks (culture-to-flask volume ratio of 1:10) and aerated by agitation at 200 rpm. When necessary, DS medium was supplemented with chloramphenicol (2.5 μg/ml), tetracycline (15 μg/ml), spectinomycin (50 μg/ml), or 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal) (80 μg/ml). TSS minimal medium (0.05 M Tris [pH 7.5], 40 μg each of FeCl3 · 6H2O and trisodium citrate dihydrate per ml, 2.5 mM K2HPO4, 0.02% MgSO4 · 7H2O, 0.5% glucose) (25), supplemented with tryptophan (0.004%) and phenylalanine (0.004%), was utilized for the determination of glutamate auxotrophy. When necessary, glutamine (0.2%) was added. CSE minimal medium [70 mM K2HPO4, 30 mM KH2PO4, 25 mM (NH4)2SO4, 0.5 mM MgSO4, 10 mM MnSO4] (26) was supplemented with ferric ammonium citrate (0.0022%), sodium succinate (0.6%), potassium glutamate (0.8%), and tryptophan (0.005%) and utilized in Northern blotting experiments.
Table 1.
B. subtilis strains
| Strain | Genotype | Reference(s) or source |
|---|---|---|
| 168 | trpC2 | 60 |
| JH642 | trpC2 pheA1 | 61, 62 |
| AF21 | ΔamyE::Φ(citBp21-lacZ cat) | 24 |
| CJB8 | ccpC::spc | 8 |
| HKB186 | ΔamyE::Φ(citBp21-lacZ cat) ΔccpC::ble | 50 |
| LS1003 | trpC2 pheA1 citZ::Φ(citZ′-lacZ cat) | 6 |
| MAB160 | trpC2 pheA1 ΩcitB::spc | 1 |
| SJB66 | trpC2 pheA1 ΔcitZ471 | 3 |
| SJB67 | trpC2 pheA1 ΔcitA::neo ΔcitZ471 | 3 |
| SJB231 | trpC2 pheA1 ΔcitZC::spc | 4 |
| AWS96 | trpC2 pheA1 | 13 |
| AWS173 | citA::neo citZ517 ΔamyE::Φ(citBp21-lacZ tet) | 22 |
| AWS174 | trpC2 pheA1 ΩcitB::spc ΔamyE::Φ(citBp21-lacZ tet) | 22 |
| AWS198 | trpC2 pheA1 His10-citB+::pAWS50(cat) | 13 |
| KBP17 | trpC2 pheA1 His10-citB2::pKP12(cat) | AWS96 × pKP12 |
| KBP22 | trpC2 pheA1 citB2 citZ340 | Serial passage of KBP17 |
| KBP26 | trpC2 pheA1 ΔamyE::Φ(citBp21-lacZ cat) | 11 |
| KBP44 | trpC2 pheA1 citZ::Φ(citZ′-lacZ cat) | AWS96 × LS1003 DNA |
| KBP45 | trpC2 pheA1 ΩcitB::spc citZ::Φ(citZ′-lacZ cat) | MAB160 × LS1003 DNA |
| KBP48 | trpC2 pheA1 citZ::Φ(citZ′-lacZ cat) ΔccpC::ble | KBP44 × HKB186 DNA |
| KBP49 | trpC2 pheA1 ΩcitB::spc citZ::Φ(citZ′-lacZ cat) ΔccpC::ble | KBP45 × HKB186 DNA |
| KBP51 | trpC2 pheA1 ΩcitB::spc ΔamyE::Φ(citBp21-lacZ cat) | 11 |
| KBP52 | trpC2 pheA1 ΔamyE::Φ(citBp21-lacZ cat) ΔccpC::ble | 11 |
| KBP54 | trpC2 pheA1 ΩcitB::spc ΔamyE::Φ(citBp21-lacZ cat) ΔccpC::ble | 11 |
| KBP72 | trpC2 pheA1 citB7 ΔamyE::Φ(citBp21-lacZ cat) | AWS96 × citB7PCR, AF21 DNA |
| KBP81 | trpC2 pheA1 citB7 ΔamyE::Φ(citBp21-lacZ tet) | KBP72 × AWS173 DNA |
| KBP85 | trpC2 pheA1 ΔamyE::Φ(citBp21-lacZ tet) | JH642 × AWS173 DNA |
| KBP86 | trpC2 pheA1 citZ340 ΔamyE::Φ(citBp23-lacZ cat) | SJB231 × pAF23, KBP22 DNA |
| KBP94 | trpC2 pheA1 ΔamyE::Φ(citBp21-lacZ tet) | AWS96 × AWS173 DNA |
| KBP96 | trpC2 pheA1 ccpC::spc ΔamyE::Φ(citBp21-lacZ tet) | KBP85 × CJB8 |
| KBP118 | trpC2 pheA1 citB2 ΔamyE::Φ(citBp21-lacZ tet) | AWS96 × KBP94, KBP22 DNA |
| KBP125 | trpC2 pheA1 citB+::pKP29(cat) | AWS96 × pKP29 |
| KBP126 | trpC2 pheA1 citB2::pKP29(cat) citZ340 | KBP22 × KBP125 DNA |
| KBP127 | trpC2 pheA1 citB+::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP94 × KBP125 DNA |
| KBP128 | trpC2 pheA1 citB2::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP94 × KBP126 DNA |
| KBP129 | trpC2 pheA1 citB7::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP81 × KBP125 DNA |
| KBP135 | trpC2 pheA1 citB+::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP85 × KBP127 DNA |
| KBP136 | trpC2 pheA1 citB2::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP85 × KBP128 DNA |
| KBP137 | trpC2 pheA1 citB7::pKP29(cat) ΔamyE::Φ(citBp21-lacZ tet) | KBP85 × KBP129 DNA |
| KBP138 | trpC2 pheA1 citB+::pKP29(cat) ccpC::spc ΔamyE::Φ(citBp21-lacZ tet) | KBP96 × KBP127 DNA |
| KBP139 | trpC2 pheA1 citB2::pKP29(cat) ccpC::spc ΔamyE::Φ(citBp21-lacZ tet) | KBP96 × KBP128 DNA |
| KBP140 | trpC2 pheA1 citB7::pKP29(cat) ccpC::spc ΔamyE::Φ(citBp21-lacZ tet) | KBP96 × KBP129 DNA |
| GP1441 | trpC2 ΩcitB::spc | 168 × KBP49 DNA |
Construction of the citB2 mutant.
To create a point mutant defective in aconitase enzyme activity, cysteine residue 450, a residue integral to the 4Fe-4S cluster, was mutated to serine. To do so, the promoter region and N-terminal portion of citB with a decahistidine tag were amplified and mutagenized by site-directed PCR mutagenesis. The template was genomic DNA from strain AWS198 [His10-citB+::pAWS50(cat)] (13). Primer citBF6 (5′-GGGCATGCGAGAACCTCCTTAAAAGAGTTCGGTGTTATT-3′ [the SphI restriction site is underlined]) and mutagenic primer OKP37 (5′-GTTTGATGTATTTGTAGAGCTTGTAATCGCAGCAATGGC-3′ [mutated residues are in boldface type]) were used to amplify the citB locus from 400 bp upstream to 1,365 bp downstream of the start codon. Mutagenic primer OKP36 (5′-GCTGCGATTACAAGCTCTACAAATACATCAAACCCATACGTG-3′ [mutated residues are in boldface type]) and primer OKP38 (5′-ATACCCGGGTTGACCATCCTTGCCCACACC-3′ [the XmaI restriction site is underlined]) were used to amplify citB from 1,333 bp to 1,785 bp downstream of the start codon. The products of the two reactions were annealed and amplified by using citBF6 and OKP38, yielding a final product of 2,203 bp. This product was purified, digested with restriction enzymes SphI and XmaI, and ligated to the B. subtilis integrative vector pJPM1 (27), creating pKP12. After transformation of E. coli and verification of its structure, pKP12 was used to transform B. subtilis strain AWS96. Chloramphenicol resistance was used to select for integration of pKP12 at the citB locus by homologous recombination, creating strain KBP17 [His10-citB2::pKP12(cat)]. The serial passage of KBP17 in L broth, without selection, to obtain a second crossover (and, thus, an unmarked citB2 strain) resulted in the isolation of KBP22, a strain that had retained the citB2 allele and lost the integrated plasmid but had acquired a suppressor mutation (citZ340). The citB2 mutation was separated from citZ340 by introducing genomic DNAs from strains KBP22 and KBP94 (amyE::citBp21-lacZ tet) simultaneously into wild-type strain AWS96. (In such experiments, 5 to 10% of transformants selected for one marker will have acquired a second, unlinked marker by congression.) Tetracycline-resistant transformants were isolated on DS medium containing X-Gal, and blue colonies were selected, indicative of derepressed expression of the citB-lacZ fusion. The resulting strain was named KBP118 (citB2 amyE::citBp21-lacZ tet).
Isolation of the citZ340 mutation.
Genomic DNA from strain KBP22 was introduced into strain SJB231 (citZC::spc) along with pAF23 plasmid DNA (amyE::citBp23-lacZ cat). Chloramphenicol-resistant transformants were isolated on DS medium and screened for spectinomycin sensitivity and glutamate auxotrophy. The resulting strain was named KBP86 (citZ340 amyE::citBp23-lacZ cat).
Construction of the citB7 mutant.
A single-amino-acid substitution expected to reduce RNA binding was introduced into the citB gene by transformation with a PCR product. The replacement of arginine-741 by glutamate was engineered by site-directed mutagenesis. Primer OKP71 (5′-ATAGCATGCAGCGGTTGAGTTAGGGCTTAAG-3′ [the SphI restriction site is underlined]) and mutagenic primer OKP73 (5′-GATTTGGTTTTTGATTTCAATGTTGGCAAATGTTCCTCTC-3′ [mutated residues are in boldface type]) were used to amplify the citB locus from 1,404 bp to 2,238 bp downstream of the start codon. Mutagenic primer OKP74 (5′-ACATTTGCCAACATTGAAATCAAAAACCAAATCGCACCG-3′ [mutated residues are in boldface type]) and primer OKP72 (5′-ATACCCGGGATTGATTCATCAGGACTGCTTCATTTTTTCACGAAGC-3′ [the XmaI site is underlined]) were used to amplify the citB locus from 2,206 bp downstream of the start codon to the stop codon. The products of the reactions were annealed and amplified with primers OKP71 and OKP72, yielding a final product of 1,341 bp (including restriction site overhangs on the outside primers). The final product was purified and introduced into B. subtilis strain AWS96 by transformation along with genomic DNA from strain AF21 (amyE::citBp21-lacZ cat). Chloramphenicol-resistant transformants were selected and screened for colonies that overexpressed the citBp21-lacZ fusion, a phenotype expected based on results obtained for the citB5 strain (A. W. Serio and K. B. Pechter, unpublished data). The citB locus was amplified by PCR and sequenced to confirm the presence of the citB R741E mutation. The resulting strain, KBP72 (citB7 amyE::citBp21-lacZ cat), also had an unplanned silent mutation near the 3′ end of the gene. To create an isogenic strain for analysis, genomic DNA containing the amyE::citBp21-lacZ tet fusion (AWS173) was introduced into KBP72 by transformation, and tetracycline-resistant colonies were selected, resulting in strain KBP81 (citB7 amyE::citBp21-lacZ tet).
Construction of a citB integrative vector and derivative strains.
To create a marked but untagged citB construct for genetic manipulations in B. subtilis, primers citMF1 (5′-GCGTCTAGAACCGTAACTTTGAAGGACGTATTCAC-3′ [the XbaI restriction site is underlined]) (13) and OKP11 (5′-AATAAGAGCTCGATTCATCAGGACTGCTTC-3′ [the 5′ SacI site is underlined]) were used to amplify the C-terminal 1.2 kb of the citB gene. The resulting PCR product was digested with XbaI and SacI and ligated into pJPM1 digested with the same enzymes, producing plasmid pKP29. This plasmid was introduced into E. coli by transformation, and the sequence was verified before introduction into B. subtilis strain AWS96 by a single crossover at the citB locus, producing strain KBP125 [citB+::pKP29(cat)]. Genomic DNA from KBP125 was introduced into strain KBP94, producing strain KBP127 [citB+::pKP29(cat) amyE::citBp21-lacZ tet). To create a marked version of the citB2 allele, genomic DNA from KBP125 was introduced into strain KBP22, producing strain KBP126 [citB2::pKP29(cat) citZ340]. Genomic DNA from KBP126 was then used to transform KBP94 to chloramphenicol resistance, and transformants that retained hyperexpression of the citB-lacZ reporter on DS medium containing X-Gal were selected. The resulting strain was named KBP128 [citB2::pKP29(cat) amyE::citBp21-lacZ tet]. To create a marked version of the citB7 allele, genomic DNA from KBP125 was introduced into strain KBP81, producing strain KBP129 [citB7::pKP29(cat) amyE::citBp21-lacZ tet]. To create isogenic strains for the analysis of the effect of a ccpC mutation on the phenotypes due to the citB2 and citB7 mutations, genomic DNA from strains KBP127, KBP128, and KBP129 was introduced into strains KBP85 (amyE::citBp21-lacZ tet) and KBP96 (ccpC amyE::citBp21-lacZ tet), producing strains KBP135, KBP136, KBP137, KBP138, KBP139, and KBP140 (Table 1).
Preparation of cell extracts for enzyme assays and Western blots.
Samples were removed from B. subtilis DS broth cultures at various times, collected by centrifugation, washed in TEG buffer (20 mM Tris [pH 8], 1 mM EDTA, 20% glycerol), and stored at −20°C. Thawed cells were resuspended in the same buffer supplemented with 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and incubated with 0.4 mg lysozyme per ml for 30 min at 37°C. If necessary, the resulting lysate was gently sonicated on ice to break up genomic DNA by using a Branson sonifier (30% duty, level 2, 30-s intervals with 20-s rests, 3 to 4 times). Sonication was avoided, if possible, due to negative effects on aconitase enzyme activity; importantly, samples were treated identically within a single experiment. Cell extracts were clarified by centrifugation at 16,000 × g for 10 to 15 min at 4°C. Clarified extracts were kept on ice and assayed immediately for aconitase and citrate synthase activities. Protein concentrations of the samples were determined by a Bradford assay using the Bio-Rad reagent and bovine serum albumin (BSA) as a standard.
Aconitase activity assay.
Aconitase enzyme activity was determined according to established methods (5, 28). One unit of activity is equivalent to 1 nmol cis-aconitate produced per minute, and units are expressed per milligram of protein.
Citrate synthase activity assay.
Citrate synthase activity was determined by using previously described methods, with slight modifications (29–31). Briefly, 10 μl of cell extract or purified protein was mixed with 0.1 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and 0.3 mM acetyl coenzyme A (CoA) in TEG buffer (see above). All steps were carried out at room temperature. After a 3-min preincubation step, the absorbance at 412 nm (A412) was measured to obtain a background reading before oxaloacetate was added to 1 mM. Samples were incubated for 10 min, and the A412 was measured to detect the formation of the thionitrobenzoate ion (TNB2−). The corrected value (32) for the TNB2− ion extinction coefficient (e = 14.15 ml cm−1 μmol−1) was utilized, and citrate synthase specific activity was expressed as micromoles of TNB2− produced (i.e., CoA released) per minute per milligram of protein.
Determination of intracellular and extracellular citrate concentrations.
B. subtilis cells were grown in DS broth (25- to 50-ml cultures). At various times, samples (1 to 5 ml) were removed and harvested by centrifugation. The culture fluid was removed, adjusted to pH 8.0 to 8.4, and analyzed by using a citric acid detection kit (R-Biopharm) according to the manufacturer's instructions. The cell pellet was washed with 20 mM Tris-Cl (pH 8.0)–1 mM EDTA, resuspended in perchloric acid (0.3 M HClO4), and incubated on ice for 10 min. Cell debris was removed by centrifugation, and the supernatant fluid was incubated with potassium carbonate (final concentration of 0.25 M K2CO3) for 15 min on ice. Any precipitate was removed by centrifugation, and the supernatant fluid was assayed for citrate content as described above.
β-Galactosidase activity assays.
For β-galactosidase activity assays, samples (0.5 to 1 ml) were removed from B. subtilis DS broth cultures at various times during growth after determining the A600 of the culture at that time point. Cells were pelleted by centrifugation, and pellets were frozen on dry ice. Cells were permeabilized and assayed as described previously (33). β-Galactosidase activity (Miller units) was calculated as described previously (23); however, a correction factor of 1.25 was used to account for the contribution of the sodium carbonate to the final reaction volume.
Western blot analysis of cell extracts.
Preparation of cell extracts for Western blot analysis of aconitase, citrate synthase, and isocitrate dehydrogenase levels is described above. When comparing extracts, equivalent amounts of protein (∼0.5 μg) were subjected to SDS–10% polyacrylamide gel electrophoresis (PAGE) before transfer onto an Immobilon polyvinylidene difluoride (PVDF) membrane (Millipore). Reconstituted nonfat dry milk (5%) was used as a blocking agent, and washes were performed with Tris-buffered saline (pH 8). Polyclonal antibodies raised in rabbits to B. subtilis aconitase (22), citrate synthase (34), isocitrate dehydrogenase (K. Matsuno and A. L. Sonenshein, unpublished data), and CodY (35) were used. Anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (Upstate Biotechnology, Inc.) were used, and blots were developed by using the ECL Plus Western blotting kit (GE Healthcare), respectively. Quantification of blots was performed by using ImageQuant TL software (GE Healthcare).
Northern blotting.
For all Northern blot experiments, cells were grown in CSE minimal medium (25-ml cultures in 250-ml flasks agitated at 200 rpm) and harvested in exponential phase (optical density at 600 nm [OD600] of 0.5). Total RNA and Northern blot analyses were carried out as described previously (36). All RNA samples were tested for integrity by agarose gel electrophoresis. In all cases, 16S and 23S rRNA bands were intact and in the same relative proportions from sample to sample. Digoxigenin (DIG)-labeled RNA probes were obtained by in vitro transcription with T7 RNA polymerase (Roche Diagnostics), using PCR-generated DNA fragments as templates. Primer pair FM167 (5′-GCGACACGCGGTCTTGAAGGG-3′) and FM168 (5′-CTAATACGACTCACTATAGGGAGAGGCGGATCAGACGGTTGTTGTC-3′ [the T7 promoter sequence is underlined]) was used to amplify the citZ open reading frame (ORF) from positions 7 to 1057. Primer pair FM174 (5′-CAGTCTCTAACGGAGTATTAAACGTACC-3′) and FM175 (5′-CTAATACGACTCACTATAGGGAGAGCCGCTTCGTTCCATCCTAAATGC-3′ [the T7 promoter sequence is underlined]) was used to amplify the icd ORF from positions 23 to 1137, and primer pair FM176 (5′-CGGAGCAGGTTTTACCGGAGCT) and FM177 (5′-CTAATACGACTCACTATAGGGAGAGATTCAACTGATTTATTCAGCTGCGCTC [the T7 promoter sequence is underlined]) was used to amplify the mdh ORF from positions 33 to 909. In vitro RNA labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions (Roche Diagnostics). The sizes of the transcripts were estimated based on transcripts of the gapA operon (36). RNA stability was analyzed as described previously (37). Briefly, rifampin (100 mg ml−1) was added to cultures growing logarithmically in CSE minimal medium, and samples were taken at time points after drug addition. Quantification was performed by using Image J software v1.42 (38).
Purification of wild-type, C450S, and R741E aconitase proteins.
Untagged wild-type, C450S, and R741E aconitase (Acn) proteins were purified from B. subtilis by using methods based on those described previously by Dingman and Sonenshein (5). For each strain (KBP94, KBP22, and KBP81), two independent cultures were grown in DS medium and harvested by centrifugation at 4°C (4,400 × g; JA-10 rotor) at the end of the exponential growth phase (OD600 of ∼0.8 to 1.0). While the volume of the cultures ranged from 500 ml to 2 liters, in each case, the amount of cell extract equivalent to a 500-ml culture was used as the input for a single preparation. Cell pellets were washed twice with ice-cold 20 mM Tris-citrate (TC) buffer (20 mM Tris plus 20 mM citrate; the pH was adjusted to 7.35 with NaOH) and stored at −80°C. Pellets were thawed and resuspended in ice-cold 20 mM TC buffer supplemented with 1 mM PMSF before being subjected to two passages through a French pressure cell (15,000 lb/in2). The resulting lysate was sonicated (50% duty, level 5, 30-s intervals with 20-s rests, 3 times) on ice to fragment genomic DNA. The sonicated lysate was clarified by centrifugation at 4°C (20,000 × g; JA-20 rotor); the soluble fraction was precipitated with ammonium sulfate on ice in a two-step process. First, ammonium sulfate was added to 40% saturation, and the sample was centrifuged as described above. The supernatant fluid was then adjusted with ammonium sulfate to give 85% saturation and subjected to centrifugation as described above. The pellet was resuspended in 20 mM TC buffer; concentrated to 1 ml, if necessary, by using a spin column (Millipore); and subjected to gel filtration chromatography on a Superose 12 column (10/300 GL, 24-ml bed volume; GE Healthcare) equilibrated with 20 mM TC buffer. Fractions containing aconitase were identified by an aconitase activity assay or by SDS-PAGE/Coomassie blue analysis (for the C450S mutant). The Superose 12 fractions containing aconitase were pooled (but not concentrated), giving a total volume of ∼1 ml, and subjected to anion-exchange chromatography using a MonoQ column (5/50 GL, 1-ml bed volume; GE Healthcare) prepared by sequential washing with 20 mM and 100 mM TC buffer (pH 7.35), followed by equilibration with 20 mM TC buffer. Protein was eluted with a linear gradient of 20 to 50 mM TC buffer (pH 7.35); aconitase eluted very early in the gradient. Fractions containing aconitase were identified by an aconitase enzyme activity assay (or SDS-PAGE analysis for the C450S mutant), pooled, and concentrated via a spin column to ∼0.5 ml (Millipore). The MonoQ column concentrate was loaded onto a Superdex 200 gel filtration column (10/300 GL, 24-ml bed volume; GE Healthcare) equilibrated with 2× storage buffer (20 mM Tris, 70 mM KCl). Protein was eluted with the same buffer; eluted fractions containing aconitase were identified by SDS-PAGE analysis with Coomassie blue. Fractions containing aconitase were pooled prior to concentration via a spin column (Millipore). Pure aconitase was diluted 2-fold with 100% glycerol (final concentrations of 10 mM Tris, 35 mM KCl, and 50% glycerol) and was stored at −20°C.
Alternatively, wild-type aconitase (Acnwt) for gel shift assays was purified with a cleavable C-terminal hexahistidine tag. To do so, the citB gene was amplified by PCR using primers OFM3 (5′-AAAGAGCTCTGATCTGAAGGGGGATTTTG-3′ [the SacI site is underlined]) and OFM4 (5′-AAATCTAGATCAGTGATGGTGATGGTGATGGCCCTGAAAATACAGGTTTTCGGACTGCTTCATTTTTTCACG-3′ [the XbaI site underlined]). The PCR product was digested with SacI and XbaI, and the resulting fragment was ligated to the expression vector pBAD30 (39); the resulting plasmid was named pFM1. E. coli R1279 (40) was used as the host for the overexpression of recombinant aconitase during growth in L broth. Expression was induced by the addition of 0.2% arabinose to exponentially growing, 500-ml cultures (OD600 = 0.6). Cells were harvested 2 h after the addition of arabinose and lysed by two passes at 18,000 lb/in2 through an HTU DIGI-F-Press (G. Heinemann, Germany). After lysis, the crude extract was centrifuged at 30,000 × g for 60 min and then passed over a Ni+-nitrilotriacetic acid (NTA) column (IBA, Göttingen, Germany). The protein was eluted with an imidazole gradient. After elution, the fractions were screened for the presence of His6-Acn by SDS-PAGE and subsequent staining with Coomassie blue. To remove the hexahistidine tag, the His6-Acn-containing fractions were adjusted to 1 mM dithiothreitol (DTT) and treated with AcTEV protease (Invitrogen) according to the manufacturer's instructions. After removal of the protease beads and free His6 tag by Ni+-NTA chromatography, the untagged, purified aconitase was concentrated by using a Vivaspin 500 concentrator (Sartorius Stedim, Göttingen, Germany). For all aconitase preparations, the protein concentration was determined by the Bradford method, as described above.
Gel mobility shift assays.
To create RNA targets for gel mobility shift assays, the citZ leader region and hag transcript were amplified by PCR to generate templates for in vitro runoff transcription. Primers FM169 (5′-CTAATACGACTCACTATAGGGAGATAGGCTTAAACTTAAATAAGCTTATAAAAATTTG-3′ [the T7 promoter is underlined, the T7 transcriptional start site is in italic boldface type, and the citZ transcriptional start site is in boldface type) and FM170 (5′-CATATATAACATCTCCTTTTCAATAAATTTCC-3′ [the complement of the citZ start codon is underlined]) were used to amplify an ∼200-bp region of the B. subtilis chromosome extending from just upstream of the citZ transcriptional start site to the start codon of citZ. In addition, primers CD53 (5′-CTAATACGACTCACTATAGGGAGATCCGATATTAATGATGTAGCCGGG-3′ [the T7 promoter is underlined]) and CD54 (5′-CTCCATGTTCTTTTGGCTCGC-3′) were used to amplify a 190-bp 5′ region of the hag locus from 106 bp upstream to 84 bp downstream of the start codon. In vitro transcription reactions were performed by using T7 RNA polymerase (Roche Diagnostics). The integrity of the RNA transcripts was analyzed by denaturing agarose gel electrophoresis (36). Binding of aconitase to the citZ and hag RNAs was assayed by gel retardation experiments as described previously, with minor modifications (41). Briefly, the RNA was incubated for 2 min at 95°C and placed on ice for 5 min. Purified aconitase was added to the RNA, and the reaction mixtures were incubated for 10 min on ice in Tris-acetate-EDTA (TAE) buffer in the presence of 300 mM NaCl. Glycerol was added to a final concentration of 10% (wt/vol); the reaction mixtures were then subjected to electrophoresis in 8% polyacrylamide gels in Tris-acetate buffer and visualized with ethidium bromide.
Filter binding assays.
To create an RNA target for filter binding assays, the citZ leader region was amplified by PCR to generate a template for in vitro runoff transcription. Primers OKP98 (5′-TAATACGACTCACTATAGGAGGCTTAAACTTAAATAAGCTT-3′ [the T7 promoter is underlined, the T7 transcriptional start site is in italic boldface type, and the citZ transcriptional start site is in boldface type]) and OKP99 (5′-CATATATAACATCTCCTTTTC-3′ [the complement of the citZ start codon is underlined]) were utilized to amplify an ∼200-bp region of the B. subtilis chromosome extending from the citZ transcriptional start site (position +1) to the start codon of citZ (i.e., the 5′ untranslated region [UTR]). Transcription reactions were performed by using T7 RNA polymerase (Stratagene) in the presence of [α-32P]UTP (Perkin-Elmer) to produce internally radiolabeled RNA. Radioactive RNA transcripts were purified by phenol-chloroform extraction, precipitated with isopropanol and sodium acetate to remove unincorporated nucleotides, and resuspended in diethyl pyrocarbonate (DEPC)-treated deionized water supplemented with RNaseOut (Invitrogen) prior to storage at −20°C in small aliquots to avoid freeze-thaw damage.
To assay aconitase binding to the citZ 5′ UTR, labeled RNA was briefly heated to 80°C and then allowed to cool slowly to ambient temperature. Cooled RNA was supplemented with RNaseOut and added to reaction mixtures containing different concentrations of aconitase in buffer (10 mM Tris [pH 7.5], 35 mM KCl, 20% glycerol, 0.5 mM dipyridyl, 5 mM β-mercaptoethanol). Reaction mixtures were allowed to equilibrate for 30 min at room temperature before filtration through nitrocellulose discs (0.45-μm HAWP; Millipore) (presoaked in reaction buffer), using a multisample filtration apparatus and the in-house vacuum line. Filters were washed twice with 0.5 ml reaction buffer, removed, and placed into scintillation fluid. A mock reaction mixture was added directly to scintillation fluid to obtain a measure of the total radioactivity of the RNA.
RESULTS
Construction of an enzymatically inactive mutant form of aconitase (C450S).
To determine the specific contribution of the enzymatic activity of aconitase to the maintenance of citrate levels within the cell, we sought to create an enzymatically inactive mutant of aconitase that retained RNA-binding activity. One of the cluster-ligating cysteine residues (C450) was mutated to serine by PCR mutagenesis; we refer to the C450S allele as citB2. To create the citB2 mutation, a 5′ region of the citB gene containing the C450S mutation and an N-terminal decahistidine (His10) tag sequence were amplified. This construct was ligated into the B. subtilis integrative vector pJPM1 (cat) (27), and the resulting plasmid, pKP12, was introduced by single crossover at the citB locus by selection for chloramphenicol resistance. The resulting transformants were a mixed population of glutamate auxotrophs and prototrophs. One of the glutamate auxotrophs was purified, producing a merodiploid strain [KBP17; His10-citB2::pKP12(cat)], in which the mutant allele was associated with the promoter. The glutamate auxotrophy presumably indicates that the C450S mutation abrogates aconitase enzymatic activity. Indeed, purified His10-AcnC450S protein is not an active enzyme (data not shown).
To allow a second crossover event and thereby obtain an unmarked citB2 strain, the His10-citB2::pKP12(cat) strain was passaged in rich medium without selection, plated for single colonies, and screened for glutamate auxotrophy and sensitivity to chloramphenicol. Such a segregant was isolated, but sequencing of the citZ locus from the resulting strain (KBP22) revealed an additional mutation in the major citrate synthase gene (citZ340). This mutation, H340Y, produces a stable CitZ protein (as determined by Western blotting) that is inactive in enzyme assays (data not shown). The appearance of citrate synthase mutations in aconitase null mutant populations has been reported for several organisms, including E. coli (42), Corynebacterium glutamicum (43), Sinorhizobium meliloti (44), and Vibrio fischeri (E. Stabb, personal communication). Overproduction of citrate is apparently deleterious to bacteria.
To separate the citB2 mutation from the citZ340 mutation, KBP22 (citB2 citZ340) genomic DNA and DNA containing a citBp-lacZ tet construct at the nonessential amyE locus (KBP94) were introduced together into a wild-type strain by transformation. Tetracycline-resistant colonies were screened for the acquisition by congression of the citB-lacZ hyperexpression phenotype (expected based on results from citB null cells [9]); the resulting strain was citB2 amyE::citBp21-lacZ tet (KBP118). KBP118 cannot grow on minimal medium without glutamate supplementation, indicating that it is a glutamate auxotroph. Sequencing of KBP118 genomic DNA confirmed the presence of the citB2 mutation. In addition, sequencing of the citZ and citA genes indicated that KBP118 carries the wild-type versions of both citrate synthase genes; therefore, the glutamate auxotrophy is due solely to the citB2 mutation.
Construction of an RNA-binding mutant of aconitase (R741E).
We previously described a B. subtilis citB mutant strain that produces an RNA-binding-defective aconitase protein (13). However, this mutant (citB5) has five amino acid substitutions, making it difficult to discern which of these residues is important for RNA binding. In addition, the need to retain all five mutations during strain passaging makes genetic manipulation cumbersome. To alleviate these concerns, we sought a single-point mutation that would mimic the citB5 phenotype. Given the great similarity between B. subtilis aconitase and mammalian IRP1, we consulted the crystal structure of IRP1 in complex with its IRE target (45). Based on this structure, only one of the five residues mutated in the citB5 strain, Arg-741, has an IRP1 counterpart (Arg-728) in close proximity to the RNA. To determine if this residue was solely responsible for the citB5 phenotype, we constructed an R741E point mutant (citB7) by site-directed PCR mutagenesis (see Materials and Methods). The resulting strain, KBP81 (citB7 amyE::citBp21-lacZ tet), was a glutamate prototroph, indicating that it possesses an active aconitase enzyme.
Aconitase activity in cell extracts.
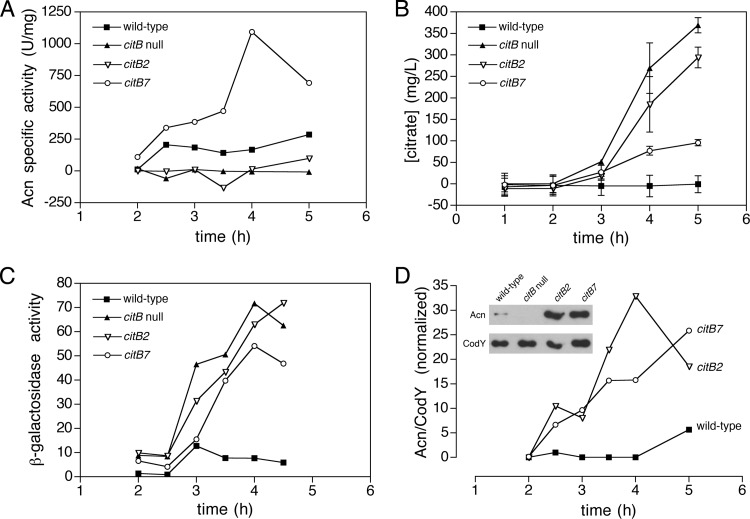
Aconitase activities in crude extracts of the wild-type (KBP94), citB null (AWS174), citB2 (KBP118), and citB7 (KBP81) strains were compared. As expected, the citB null strain had very low levels of activity throughout growth (Fig. 1A). The citB2 strain exhibited low levels of aconitase activity for the majority of the experiment; this was expected based on the strain's glutamate auxotrophy. There was, however, a small increase in aconitase activity in the citB2 strain above the background at the 5-h time point; we attribute this to the increased presence of suppressor and/or reversion mutations in the population (see below).
Fig 1.
Analysis of aconitase enzyme activity and expression levels in citB mutant strains. (A) Cell extracts were prepared from B. subtilis strains grown in DS medium and analyzed for aconitase specific activity. Data from a representative experiment of two are shown. (B) The concentration of citrate in the culture supernatant during growth in DS medium was determined. The means and standard deviations from two biological replicates are shown. (C) The expression level of a citB-lacZ fusion during growth in DS medium was determined. Samples were taken at the indicated time points, and β-galactosidase activity was measured. Data from a single experiment are shown; all strains were assayed in multiple experiments and gave similar results. (D) Cell extracts were prepared from strains grown in DS medium and analyzed by Western blotting using antibodies specific to Acn and CodY (a loading control). Bands were quantitated, and values are presented as a ratio of Acn to CodY, normalized to the ratio of the citB+ strain expression level at 2.5 h. Data from a representative experiment of two are shown; an immunoblot from a single time point (2.5 h) from that experiment is shown in the inset. The strains used were wild-type (KBP94), citB null (AWS174), citB2 (KBP118), and citB7 (KBP81) strains.
We were surprised to find that the citB7 strain showed very high levels of aconitase activity in cell extracts. In this strain, aconitase activity reached a peak of over 1,000 U/mg at the 4-h time point, a level more than 6-fold higher than that seen in the wild-type cell extract (167 U/mg).
citB2 and citB7 mutants accumulate citrate.
To determine how the aconitase activity levels in the citB2 and citB7 mutants affect the citrate levels in these cells, we examined citrate accumulation in the citB2 and citB7 mutant strains. Based on the existing metabolic roadblock model, we anticipated that any strain expressing an inactive aconitase enzyme (e.g., the citB2 mutant) would accumulate citrate to high levels, while a strain expressing a functional enzyme (e.g., the citB7 mutant) would not.
To assay the citB2 and citB7 strains for citrate accumulation in cells and in the culture fluid, the relevant strains were grown in broth cultures, samples were taken during growth, and the cell extracts and culture supernatants were analyzed by using a citric acid assay kit (R-Biopharm). As expected, the citB null and citB2 strains accumulated citrate both intracellularly and in the culture fluid; surprisingly, the citB7 mutant also accumulated more citrate than did wild-type cells albeit not to the same level as the citB2 mutant (Fig. 1B and Table 2). Because all of these strains are derivatives of JH642, the accumulation of citrate in the culture supernatant may be exacerbated by a mutation in that strain that reduces citrate import (46).
Table 2.
Intracellular and extracellular citrate concentrations in citB mutant strainsb
| Strain | Genotype | Samplea | Mean intracellular citrate concn (mg/liter) ± SD | Mean extracellular citrate concn (mg/liter) ± SD | pH of culture fluid |
|---|---|---|---|---|---|
| KBP94 | Wild type | 1 | 0.27 ± 0.09 | 3.3 ± 0.74 | 7.72 |
| 2 | 0.22 ± 0.16 | 4.1 ± 0.22 | 8.28 | ||
| 3 | 0.81 ± 0.57 | 5.7 ± 1.41 | 8.50 | ||
| AWS174 | citB null | 1 | 10.7 ± 1.4 | 223 ± 20 | 6.81 |
| 2 | 14.0 ± 1.4 | 323 ± 25 | 6.89 | ||
| 3 | 6.8 ± 0.3 | 365 ± 14 | 6.89 | ||
| KBP118 | citB2 | 1 | 23.4 ± 0.7 | 255 ± 50 | 6.87 |
| 2 | 9.0 ± 3.9 | 306 ± 39 | 6.94 | ||
| 3 | 7.9 ± 1.9 | 349 ± 38 | 6.88 | ||
| KBP81 | citB7 | 1 | 15.0 ± 1.9 | 97 ± 1.6 | 6.64 |
| 2 | 5.4 ± 2.0 | 108 ± 4.0 | 8.06 | ||
| 3 | 5.3 ± 1.1 | 121 ± 16 | 8.35 |
B. subtilis cultures were sampled at three time points during stationary phase in DS medium. Strains KBP94, KBP118, and KBP81 were sampled at 4.3, 5.45, and 6.88 h of growth, and strain AWS174 was sampled at 3.92, 5.35, and 7.22 h of growth; all cultures reached the end of exponential growth phase at ∼4 h.
For citrate samples, the means and standard deviations from 2 to 4 replicate assays are shown.
citB2 and citB7 mutants overexpress citB-lacZ and aconitase protein.
To further explore this citrate accumulation phenotype, we monitored an expected side effect of citrate accumulation, i.e., increased expression from the citB promoter (9). The β-galactosidase activities of wild-type, citB null, citB2, and citB7 strains carrying a citB-lacZ fusion at the amyE locus were assayed during growth in broth cultures. As shown in Fig. 1C, the wild-type strain induced citB expression at the 3-h time point, after which the expression level dropped but remained higher than the original basal level. As seen previously (9), the citB null strain induced citB-lacZ to levels far beyond those seen in the wild type; the citB2 strain exhibited a pattern of expression very similar to that of the null mutant. Surprisingly, the citB7 strain also overexpressed citB-lacZ. Although β-galactosidase activity in the citB7 strain did not reach the same level of Miller units as that in the citB null strain, the pattern of expression was very similar. In other experiments, the citB5 strain also showed increased expression levels of a citB-lacZ fusion (Serio and Pechter, unpublished).
To determine if hyperexpression from the citB promoter leads to elevated aconitase protein levels in the citB2 and citB7 strains, cell extracts of the relevant strains were prepared from samples taken during growth in broth cultures. Equivalent amounts of total protein were analyzed by immunoblotting with polyclonal antibodies raised against aconitase and CodY (35, 47). Protein bands were quantified by using ImageQuant TL software. For each sample, the amount of aconitase protein was normalized to that of CodY, which was used as a loading control. To allow comparison across samples, the ratio of Acn to CodY for each sample was normalized to the ratio at the 2.5-h time point. Both the citB2 and citB7 strains overexpressed aconitase protein (Fig. 1D). This result provides an explanation for the result presented in Fig. 1A; citB7 cells have very high levels of aconitase activity in cell extracts due to the hyperaccumulation of aconitase protein in this strain.
The accumulation of citrate in the citB7 strain was perplexing, however. Since the citB7 strain is a glutamate prototroph, it must have a TCA branch of the citric acid cycle that is functional enough to produce adequate 2-ketoglutarate to serve as a substrate for glutamate biosynthesis. In fact, given that glutamate is the cell's most abundant anion, with an in vivo concentration of ∼100 to 200 mM (48), the rate of 2-ketoglutarate synthesis must be high to maintain those levels. Indeed, we showed above that citB7 cells have increased aconitase activity in vivo. Why, then, would a strain overexpressing a functional aconitase protein accumulate citrate? To be certain that the R741E mutation does not have an effect on enzyme activity, we purified the protein and studied it in vitro. In addition, to confirm that the glutamate auxotrophy in the citB2 strain results from an inactive aconitase protein, we purified and studied the C450S aconitase protein as well.
Specific activities of C450S and R741E Acn proteins.
To determine how the C450S and R741E mutations affect the specific activity of aconitase, the wild-type and mutant aconitase proteins were purified from B. subtilis by using classical biochemical methods updated for use with fast pressure liquid chromatography (FPLC). (In our unpublished work, we found that polyhistidine tags on aconitase can lead to inappropriate phenotypes in vivo.) Strains KBP94 (citB+) and KBP81 (citB7) were used to purify the wild-type and R741E proteins, respectively. However, all attempts to obtain a large (500-ml) culture of strain KBP118 (citB2) resulted in a significant portion of the population acquiring suppressor or revertant mutations. (When KBP118 cultures were sampled and plated hourly between 2 and 5 h after inoculation, the proportion of suppressors/revertants increased from approximately 0.1% to 10% of the population over time.) To circumvent this issue, the C450S protein was purified from strain KBP22 (citB2 citZ340); the presence of the inactivating H340Y citrate synthase mutation (citZ340) prevents the accumulation of citrate, the condition that we assume is responsible for the selection of suppressor mutations.
The aconitase specific activities of two preparations of each of the three proteins (Acnwt, AcnC450S, and AcnR741E) were determined (Fig. 2). Since the proteins were purified aerobically, we also assayed their activities after incubation with reduced iron and DTT (28); activities were affected only minimally (data not shown). As expected, the C450S mutation abolished essentially all enzyme activity; a barely detectable level of activity was seen after incubation with Fe and S. Surprisingly, the R741E mutation also caused a defect in specific activity but not nearly as severe as the C450S mutation. This result raised the possibility that citrate accumulation in citB7 cells was due to a partial enzymatic defect. However, our assays of crude extracts indicated that the citB7 mutant exhibits higher-than-normal aconitase enzyme activity and protein levels in vivo (see above), suggesting that overexpression of aconitase in citB7 cells more than compensates for the reduced activity of individual enzyme molecules.
Fig 2.
Specific activity of purified aconitase proteins. Wild-type and C450S and R741E mutant aconitase proteins were purified from B. subtilis by a four-step purification scheme. Two preparations of each protein were purified from independent cultures, producing six total preparations. The concentrations of the purified proteins were determined by a Bradford assay (Bio-Rad). The individual preparations are presented separately. Values shown are the means and standard deviations from two technical replicates.
However, there was still no explanation for why citB7 cells, which contain a vast excess of aconitase activity (i.e., the capacity to convert citrate to isocitrate) compared to wild-type cells, would accumulate so much citrate. To examine this phenomenon further, we considered the possibility that the accumulation of citrate in citB mutant strains was not due solely to the lack of citrate catabolism but rather to hyperactive citrate synthesis as well.
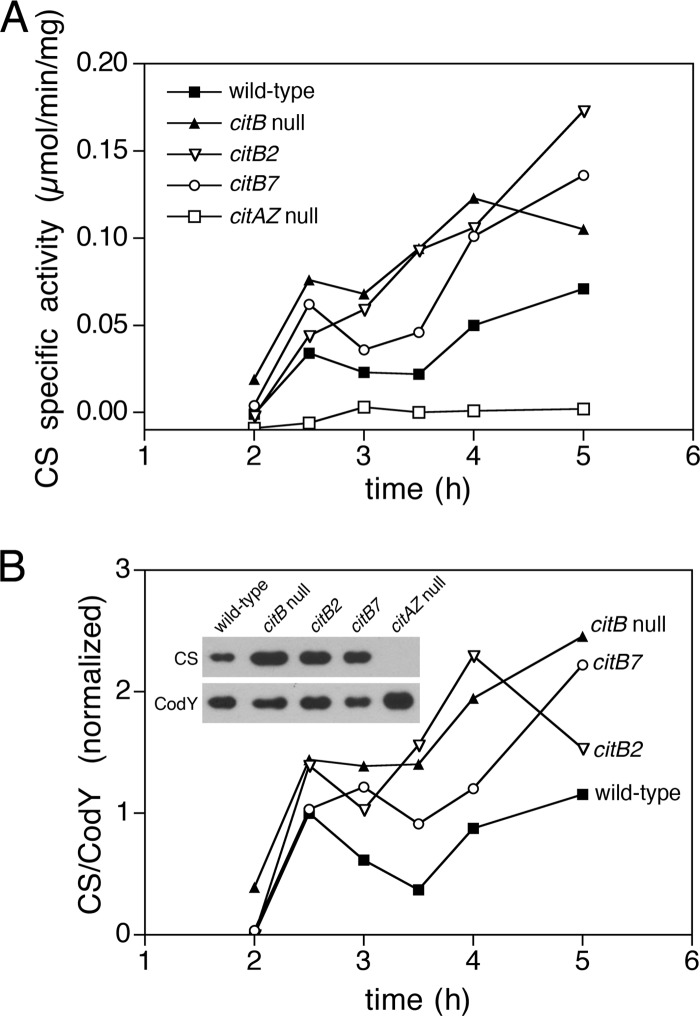
Increased CS activity in cell extracts of citB mutants.
Citrate synthase (CS) catalyzes the condensation of oxaloacetate and acetyl-CoA to citrate and is the only TCA branch enzyme that forms a carbon-carbon bond (49). The reaction is irreversible, and in B. subtilis, which lacks citrate lyase, aconitase is the only enzyme capable of citrate catabolism. Therefore, maintaining a balance between citrate synthase and aconitase enzyme activities is essential for preventing a buildup of citrate in vivo.
To determine if increased CS activity is involved in the overaccumulation of citrate in the citB mutant strains, extracts of wild-type (KBP94), citB null (AWS174), citB2 (KBP118), and citB7 (KBP81) strains were tested for CS enzyme activity. All of the strains exhibited an initial spike in CS activity at the 2.5- to 3.0-h time period (Fig. 3A). This is likely caused by an initial flux of carbon into the TCA cycle; leaky expression of citZ provides basal CS activity, and once the substrates become available, citrate is produced and CcpC is inactivated, resulting in derepression of citZ. In wild-type cells, citrate is quickly catabolized; when sources of the substrates oxaloacetate and acetyl-CoA are consumed, the citrate pool disappears, and repression by CcpC is reestablished. The three mutant strains began to diverge from the wild type after 2.5 h; the citB null and citB2 mutants did so the most dramatically. This is likely due to the lack of aconitase enzyme activity in these cells; the high citrate levels maintained in these mutants ensure that CcpC-dependent repression of citZ cannot be reinstated. The citB7 mutant, however, has ample aconitase activity in vivo; nonetheless, CS activity in the citB7 strain increased after 2.5 h to a level well above that seen for wild-type cells.
Fig 3.
citB mutant strains exhibit high levels of citrate synthase activity and protein levels in cell extracts. Strains were grown in DS medium, cells were harvested by centrifugation at the indicated time points, and crude cell extracts were generated. Separately, cell extracts were examined to determine the specific activity of citrate synthase (A) and the amount of CS protein (B). Cell extracts (0.25 to 0.5 μg) were analyzed by Western blotting using antibodies specific to CS and CodY (a loading control). Bands were quantitated, and values are presented as a ratio of CS to CodY, normalized to the ratio of the wild-type strain expression level at 2.5 h. Data from a representative experiment of two are shown. An immunoblot from a single time point (3 h) in a representative experiment is shown in the inset. The following strains were used: wild-type (KBP94), citB null (AWS174), citB2 (KBP118), citB7 (KBP81), and citAZ null (SJB67) strains.
Increased CS protein levels in cell extracts of citB mutants.
To determine if increased synthesis of CS protein was responsible for the increased CS enzymatic activity found in citB mutant cells, the cell extracts described above were analyzed by quantitative Western blotting using antibodies to CS and CodY. The immunoblots were quantified by using ImageQuant TL (GE Healthcare), CS levels were normalized to the CodY loading control, and the ratios of CS levels/CodY levels were normalized to values for the wild-type sample at 2.5 h. As with specific activity levels, CS protein levels of all strains increased at between 2 and 2.5 h of growth (Fig. 3B). However, the citB null, citB2, and citB7 strains had higher CS levels than the wild type at time points after 2.5 h.
citZ transcript levels are increased in the aconitase null mutant strain.
To determine if increases in CS protein levels and enzyme activity are due to higher-than-normal levels of citZ transcription, we used Northern blots to analyze the steady-state levels of citZ mRNA. Given the complex nature of the citZ locus, it was necessary to examine all transcripts that include the citZ coding sequence. The citZ gene is the first gene of an operon that includes the genes for isocitrate dehydrogenase (icd) and malate dehydrogenase (mdh). Three promoters (P1, P2, and P3) contribute to the synthesis of at least four different transcripts, as shown in Fig. 4A. The extent to which RNA processing also contributes to the diversity of citZ operon transcripts is not known. Total RNA from wild-type and citB null cells was analyzed by using probes specific to citZ, icd, and mdh (Fig. 4B). Transcripts detected by the citZ-specific probe, i.e., the citZ-icd-mdh, citZ-icd, and citZ transcripts, were much more abundant in the citB null mutant than in wild-type cells. The icd- and mdh-specific probes also detected a higher level of citZ-icd-mdh and citZ-icd transcripts in the citB null strain. In addition, we also saw increases in levels of icd-mdh, icd, and mdh transcripts, which do not contain citZ. It is possible that posttranscriptional RNA processing of the P1-driven message is responsible for this effect. Importantly, a control transcript, gapA mRNA, was unaffected by the citB null mutation (data not shown).
Fig 4.
citZ locus transcript levels in the citB null mutant. (A) Overview of the citZ locus. The citrate synthase II (citZ), isocitrate dehydrogenase (icd), and malate dehydrogenase (mdh) genes are transcribed by promoters P1, P2, and P3 (putative) to produce mono- and polycistronic messages. In addition, posttranscriptional processing of the full citZ-icd-mdh transcript likely contributes to the monocistronic transcript pools. Transcript lengths are indicated. (B) Northern blot analysis of citZ locus transcripts in the wild type and the citB null mutant. RNA was isolated from wild-type (168) and citB null (GP1441) strains grown in CSE minimal medium and analyzed by Northern blotting using probes specific for the citZ, icd, and mdh sense RNAs.
Overexpression of citZ in an aconitase null mutant is not suppressed by a ccpC mutation.
To clarify the basis for the higher citZ transcript level in the citB null mutant throughout growth, a previously described citZ-lacZ transcriptional fusion was utilized (6). This construct contains a 1.3-kb fragment that includes the citZ promoter, a 5′ leader region of 195 bp, and the first 30 codons of the citZ gene and is integrated by homologous recombination at the citZ locus. The fusion was introduced into wild-type (AWS96) and citB null (MAB160) strains, producing strains KBP44 and KBP45, respectively. The production of β-galactosidase was assayed during growth in broth cultures. The citB null mutation caused citZ-lacZ levels to increase 5-fold over wild-type levels after 5 h of growth (Fig. 5A); this result matches well with that obtained by Northern blotting, as described above. However, given that CcpC is a repressor of citZ, and increased citrate levels (such as those found in the citB null mutant) cause CcpC to dissociate from the citZ promoter (8), it was necessary to determine if the increase in citZ-lacZ levels in the citB null mutant was due merely to alleviation of CcpC repression. To do so, a previously described ccpC::ble null mutation (50) was introduced into KBP44 and KBP45, producing strains KBP48 and KBP49. Similar to results reported previously (8), the citZ-lacZ expression level increased 2-fold in the ccpC null mutant (Fig. 5B). However, the ccpC mutation did not suppress the citB phenotype; in fact, citZ-lacZ expression levels in the citB null mutant were higher than those in the ccpC null mutant strain. Moreover, the two mutations had an additive effect; in a citB ccpC double-null strain, citZ-lacZ expression rose to levels approximately 5-fold higher than that in the ccpC null mutant alone and approximately 10-fold higher than that in the wild type.
Fig 5.
Overexpression of citrate synthase in citB mutant strains is not suppressed by a ccpC null mutation. (A) Expression levels of citZ-lacZ in wild-type (KBP44), citB null (KBP45), ccpC null (KBP48), and ccpC citB null (KBP49) cells were determined, and β-galactosidase activity was measured at the indicated time points during growth in DS medium. (B) Levels of CS protein in cell extracts of wild-type (KBP26), citB null (KBP51), ccpC null (KBP52), and ccpC citB null (KBP54) strains were determined. Cell extracts were generated after 3 and 4 h of growth in DS medium. Extracts were analyzed by Western blotting using polyclonal antibodies raised to CS-II and CodY (loading control). The bands were quantitated, and values are presented as a ratio of CS to CodY, normalized to the ratio of the wild-type strain expression level at 3 h. Immunoblotting and quantification of a representative experiment of two are shown.
To determine how the increased expression level of citZ affects CS protein levels, cell extracts from wild-type (KBP26), citB null (KBP51), ccpC null (KBP52), and citB ccpC double mutant (KBP54) strains were analyzed by using quantitative Western blotting, as described above (Fig. 5B). Citrate synthase protein levels correlated well with the β-galactosidase activity results described here; overexpression could be seen in both citB null and ccpC null strains, and the citB ccpC double mutant increased the levels of CS protein even further. Similar results were seen for the citB2 and citB7 strains; introduction of citB2 or citB7 mutations into a ccpC null strain resulted in increased CS protein levels beyond those conferred by the ccpC null mutation alone (data not shown). These results indicate that aconitase exerts a regulatory effect on CS synthesis that is independent of CcpC.
In recent work (11), we described the conversion of CcpC from a repressor of citB gene expression to an activator when citrate accumulates. Due to the CcpC-binding-site architecture at the citZ locus, we hypothesized that CcpC could act only as a negative regulator of citZ; that is, one of the CcpC-binding sites overlaps the promoter −10 region, and the other is located downstream of the transcriptional start site (8). Importantly, if CcpC were able to act as a positive regulator of citZ in high concentrations of citrate, the increase in the citZ-lacZ expression level seen for the citB mutant would be suppressed by the addition of the ccpC mutation. Therefore, the results shown in Fig. 5A confirm that CcpC is not a positive regulator of citZ.
The stability of the citZ message is increased in an aconitase null mutant strain.
The canonical IRP1 bifunctional aconitase model holds that binding within the 5′ end of RNA targets leads to decreased transcript stability, caused by the blockage of ribosome loading (14). To test the stability of citZ mRNA, we utilized a Northern blot-based assay. Wild-type and citB null cells were treated with rifampin to block RNA synthesis, and total RNA was isolated from samples at specific time points following drug treatment. The RNA was then analyzed by Northern blotting using a probe specific to the citZ RNA, as described above. While in wild-type cells, the major citZ-icd-mdh transcript began decreasing in intensity after 2 min, with an estimated half-life of 3 min, the same transcript in the citB mutant exhibited the first drop in intensity after 6 min, with an estimated half-life of 11 min (Fig. 6). (Although the shorter transcript detected with the citZ probe was at a lower steady-state level in the wild-type strain than in the citB null mutant [Fig. 4], the relative stability of this transcript was unclear [Fig. 6]. Furthermore, the origin and coordinates of this transcript are uncertain.) The increased steady-state concentration of citZ mRNA in citB mutant cells may be explained by its increased stability, but we have not ruled out other potential contributing factors.
Fig 6.
The citZ transcript is stabilized by a citB null mutation. Wild-type (168) and citB null (GP1441) cells were grown in CSE minimal medium. Rifampin (final concentration, 100 mg/ml) was added to logarithmically growing cultures, and samples were taken at the indicated time points after treatment. RNA was isolated and analyzed by Northern blotting using a probe specific to the citZ sense strand. The citB null RNA blot was exposed for a shorter time period than the wild-type blot to accommodate the differences in RNA levels between the strains.
Aconitase interacts directly with the citZ 5′ leader RNA in vitro.
Given the greater abundance of citZ mRNA in citB mutant cells and the ability of B. subtilis aconitase to bind to certain mRNAs (12, 13), we considered the possibility that aconitase regulates citZ expression directly at the RNA level. We hypothesized that aconitase might regulate the translation or stability of the citZ message by binding to the 195-nucleotide (nt) 5′ untranslated leader region of citZ mRNA.
We first compared the binding of aconitase to the citZ 5′ leader region and to a 5′ region of the hag gene (a negative control) by a gel shift assay. The citZ leader RNA (195 nt) and the hag RNA (190 nt) were synthesized in vitro by using PCR products that included a phage T7 late promoter upstream of the template DNA. Increasing concentrations of purified Acn (0.25 to 4.5 μM) were incubated with a constant concentration of probe (250 nM) before analysis by polyacrylamide gel electrophoresis. A >50% shift of citZ 5′ leader RNA was evident at between 0.75 and 1.5 μM Acn; for the hag RNA, the highest concentration of Acn (4.5 μM) did not result in a complete shift (Fig. 7).
Fig 7.
Aconitase binds specifically to the citZ 5′ leader region in vitro. Wild-type aconitase purified from E. coli (His6-Acn, histidine tag cleaved with TEV protease) was mixed in increasing concentrations with in vitro-transcribed citZ 5′ leader RNA (250 nM). The hag RNA was used as a negative control. Reactions were analyzed by polyacrylamide gel electrophoresis and visualized by staining with ethidium bromide.
C450S and R741E proteins cannot bind to citZ RNA in vitro.
To discover whether the accumulation of CS in the citB2 and citB7 strains was explainable by differences in binding to the citZ mRNA, we tested binding of the wild-type and mutant aconitase proteins to the citZ mRNA 5′ leader region using a filter binding assay. Increasing concentrations of purified aconitase (in the nanomolar range) were incubated with a constant, low concentration of radiolabeled probe (83 pM) in buffer containing 0.5 mM dipyridyl, an iron chelator that is used to increase the fraction of aconitase molecules in the RNA-binding form (12). Whereas two independent preparations of wild-type aconitase both bound to the citZ leader RNA, they had different RNA-binding activities (Fig. 8). As described above, the enzyme activities of the two preparations were also different but inversely so (Fig. 2); that is, wild-type preparation 1 was a better RNA-binding protein but a less active enzyme than was wild-type preparation 2. The two preparations were presumably at different equilibria between the two forms of aconitase. Importantly, none of the C450S or R741E aconitase preparations bound to the citZ 5′ leader RNA.
Fig 8.
Differential binding of wild-type and mutant aconitase proteins to the citZ 5′ leader RNA in vitro. Each of six Acn protein preparations (described in the legend of Fig. 2) were mixed separately at the indicated concentrations with radiolabeled citZ 5′ leader RNA synthesized in vitro (83 pM). Reactions were allowed to equilibrate in buffer containing RNase inhibitor (RNaseOut; Invitrogen), dipyridyl (0.5 mM), and β-mercaptoethanol (5 mM) prior to passage through nitrocellulose membranes. RNA retained on each membrane was detected by scintillation counting, and the fraction of RNA retained was calculated, after background subtraction, as a percentage of the input RNA. The data shown are from a single experiment; wild-type preparation 2, C450S preparation 1, and R741E preparation 1 were assayed in other experiments and gave similar results.
DISCUSSION
We describe here the contributions of the two functions of B. subtilis aconitase to the accumulation of citrate in B. subtilis and, ultimately, to the regulation of the TCA branch enzymes. We found that citrate accumulates in two different aconitase mutants: a citB2 strain, which hyperexpresses enzymatically inactive aconitase, and a citB7 strain, which hyperexpresses enzymatically active aconitase. The unexpected and puzzling nature of the latter result prompted us to look for a novel explanation unrelated to aconitase enzyme activity. In fact, we found that the citZ transcript, CS protein, and CS activity levels were significantly increased in citB mutants. Our data indicate that wild-type aconitase, but neither the C450S nor the R741E mutant, binds to a region of citZ RNA containing the untranslated, 195-nt 5′ leader region in vitro. In addition, we found that the stability of the major citZ transcript is increased in the citB null mutant compared to the wild type, suggesting that aconitase regulates citZ at the posttranscriptional level. Analysis of the citZ 5′ UTR by using mfold (51) revealed several possible stem-loop structures that may form in vivo. None of these possible citZ stem-loops contains the IRE loop consensus sequence CAGUG (52) bound by eukaryotic IRP1, but this observation is not surprising. No consensus sequence has yet been determined for prokaryotic aconitase RNA binding, and it is possible that one of these stem-loops is, in fact, a target of Acn.
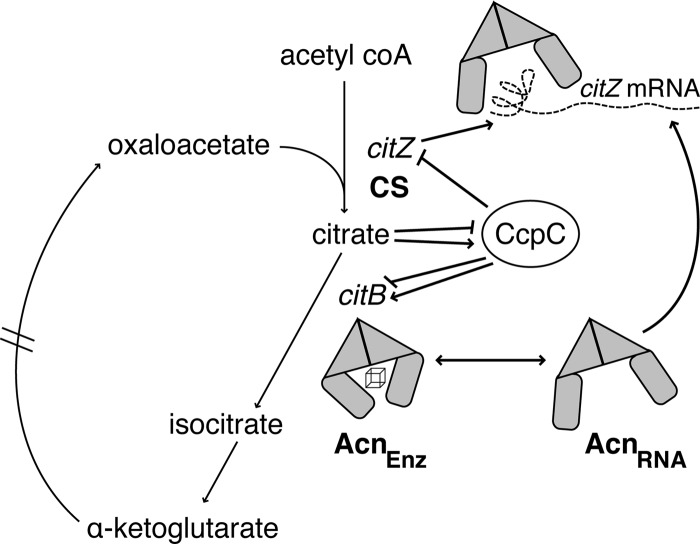
By integrating these new data with data from previous studies, a more complete model for regulation of the TCA branch of the Krebs cycle can be proposed (Fig. 9). When rapidly metabolizable carbon sources are available (e.g., glucose), the citZ gene is repressed by CcpA, and both the citZ and citB genes are repressed by CcpC. (Neither CcpA nor CcpC is by itself able to repress citZ completely, but the combined effects of the two repressors give very strong repression.) When glucose is exhausted, repression by CcpA is relieved, allowing partial derepression of citZ. Citrate begins to accumulate, leading to complete derepression of citZ and at least partial derepression of citB. The aconitase produced, in concert with isocitrate dehydrogenase, metabolizes citrate to 2-ketoglutarate. To prevent the accumulation of excessive levels of citrate, aconitase binds near the 5′ end of the citZ mRNA, decreasing its stability and thereby limiting the concentration of citrate synthase in the cell.
Fig 9.
Model for multilayered regulation of the TCA cycle in B. subtilis. At the transcriptional level, the citZ gene is repressed by CcpA (not pictured), which is activated by high glucose levels. In addition, both citZ and citB are repressed by CcpC, which is active in the absence of citrate. When glucose is exhausted, CcpA is inactivated, and citZ is partially depressed, resulting in the production of a small amount of citrate. This citrate causes CcpC repression of citZ to be alleviated; however, CcpC also becomes a positive regulator of citB (11), resulting in an increase in Acn levels. The aconitase protein is present in two pools in the cell; the enzymatic form (AcnEnz) and the RNA-binding form (AcnRNA). The RNA-binding form of Acn interacts with the citZ mRNA at the posttranscriptional level and decreases its stability (dashed line), leading to decreases in citrate synthase levels and citrate production within the cell. In this manner, the cell utilizes a citrate-metabolizing enzyme, aconitase, to tightly control the levels of citrate within the cell by regulating both the enzyme that produces citrate, citrate synthase, and aconitase itself, making this model a form of autoregulatory loop.
As a possible rationale for the latter mode of regulation, we propose that the key factor is the relationship of citrate to iron in the cell. Citrate is both cotransported with iron and a chelator of divalent cations, including iron. If the intracellular citrate level becomes excessive, iron will be sequestered away from iron-containing proteins, including aconitase. Since excess citrate greatly stimulates aconitase synthesis via the positive regulatory effect of CcpC (11), the cell will gain the ability to metabolize citrate at a higher rate. If so much iron has been sequestered that aconitase loses enzymatic activity, the cell will acquire a high concentration of enzymatically inactive but RNA-binding-competent aconitase molecules. These aconitase proteins can bind to the citZ mRNA and reduce the rate of citrate accumulation by restricting the synthesis of citrate synthase protein. As shown here, the lack of aconitase enzyme activity and the lack of aconitase RNA-binding activity both contribute to hyperaccumulation of citrate. These two roles of aconitase explain the fact that the citB7 mutant accumulates citrate to abnormal levels despite having higher-than-normal aconitase enzyme activity in vivo.
In constructing this model, we have ignored the potential contribution of the minor B. subtilis citrate synthase CitA (or CS-I) (3). CitA contributes only a small amount of CS activity to B. subtilis cells due to replacement of the active-site aspartate-307 by glutamate. Whereas citZ null strains are glutamate auxotrophs, citA null mutants have little observable phenotype (3). The regulation of citA remains poorly understood, but it is unlikely that CitA contributes a great deal to cellular physiology under the growth conditions used; cell extracts from citZ null mutants have very little residual citrate synthase signal when analyzed by immunoblotting with anti-CitZ antibodies that cross-react with CitA (34).
A recent paper suggested another potential level of complexity for the aconitase-CS regulation story. Schmalisch et al. (53) discovered a small RNA with strong complementarity to the citZ 5′ leader region. The small RNA, whose synthesis is regulated by the motility sigma factor, σD, is suspected to act as an antisense RNA for citZ, but the effect of the RNA and its mechanism of action have not been elucidated. Given the sequence complementarity between the citZ leader and the small RNA, it is possible that aconitase binds to this small RNA in addition to (or instead of) the citZ transcript.
Although we hoped to isolate aconitase mutants that were selectively deficient in either enzymatic activity or RNA binding, both of the mutants that we have reported here are at least partially defective in both activities. The C450S Acn protein is stable and produced in large amounts but is neither an enzyme nor an RNA-binding protein. The basis for the RNA-binding defect in the C450S mutant is unclear. The IRP2 protein, a homolog of IRP1 that lacks aconitase enzymatic activity, is subject to regulation via oxidation of cysteine residues that lie close to the IRE-binding site (54). It is possible that, in the absence of the iron-sulfur cluster, the two other cluster-ligating cysteine residues in the C450S mutant form a disulfide bond that prevents RNA binding. Similarly, the R741E Acn protein, designed to be defective in RNA binding, also exhibits a partial defect in enzymatic activity. While it is not as severe a defect as that of the C450S Acn protein, it is still surprising given the expectation that arginine-741 is not involved in enzyme activity. Several arginine residues contribute to enzyme activity in IRP1 (55), but the homolog of Arg-741 in IRP1 (Arg-728) is not one of them. However, a previous study of presumed nonenzymatic residues of IRP1 revealed that mutations of some residues resulted in decreased enzyme activity. Those authors reported altered Km and Vmax values for the enzyme activity of certain RNA-binding point mutants (56), although mutations of Arg-728 were not specifically tested. Interestingly, mutation of a neighboring residue, Arg-732, to Glu resulted in a 9-fold increase in the Km (56). These data, together with our results, suggest that the alteration of residues near the RNA-binding pocket of aconitase can have unexpected effects on enzymatic activity.
While this work was in progress, Gao et al. (57) reported that a strain with the aconitase substitutions R741E and Q745E exhibited higher-than-normal levels of citB-lacZ expression, aconitase protein, and aconitase activity in crude cell extracts. While those authors did not purify this protein to test its RNA-binding activity in vitro, their results are consistent with the results that we report here for the citB7 mutant (R741E) and with our work with the citB5 strain that carries R741E, Q745E, and three additional mutations (22; Serio and Pechter, unpublished).
While knowledge about the regulatory roles of bacterial aconitase proteins is limited, Tang and Guest (20) showed previously that the two aconitase proteins of E. coli, AcnA and AcnB, both bind to the 3′ UTRs of their own transcripts and that this binding increases levels of production of the AcnA and AcnB proteins in an in vitro transcription-translation assay. Aconitase levels were shown to increase in response to oxidative stress, despite a loss of aconitase activity, suggesting that the apo-Acn proteins activate this autoregulatory loop (20). In addition, the role of aconitase in two pathogenic bacteria has been explored. Deletion of the Staphylococcus aureus aconitase gene resulted in increased survival in stationary phase and reduced levels of virulence factors (58), while in Xanthomonas campestris pv. vesicatoria, mutations in acnB were found to result in decreased proliferation on pepper plant hosts (59). In each of these cases, the mechanism of regulation is unknown, but it is conceivable that aconitase is acting as an RNA-binding regulator of virulence.
In a broader sense, careful regulation of citrate synthase, aconitase, and citrate levels is likely to be vital in all organisms that lack a citrate lyase enzyme. For B. subtilis and other such organisms, once citrate is produced, aconitase is the only option for citrate removal. Other organisms that share this metabolic inflexibility are likely to have similarly layered regulatory mechanisms for preventing a potential citrate catastrophe.
ACKNOWLEDGMENTS
We thank C. Kumamoto, J. Mecsas, B. Schaffhausen, C. Squires, B. Belitsky, K. O'Day-Kerstein, and C. Majerczyk for helpful discussions; D. Dingman for sharing his aconitase purification protocol; R. Isberg, A. Camilli, and K. Heldwein for the use of their FPLC equipment; A. Hempstead, W. Amyot, and J. Pitts for sharing their FPLC expertise; and J. Busse and C. Diethmaier for their help with gel shift experiments.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM036718 to A.L.S. and by the German Federal Ministry of Education and Research SYSMO network (PtJ-BIO/0315784B) to J.S. F.M.M. was supported by Göttingen International.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 25 January 2013
REFERENCES
- 1. Craig JE, Ford MJ, Blaydon DC, Sonenshein AL. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179: 7351–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jin S, Levin P, Matsuno K, Grossman AD, Sonenshein AL. 1997. Deletion of the Bacillus subtilis isocitrate dehydrogenase gene causes a block at stage I of sporulation. J. Bacteriol. 179: 4725–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jin S, Sonenshein AL. 1994. Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 176: 4669–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsuno K, Blais T, Serio AW, Conway T, Henkin TM, Sonenshein AL. 1999. Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus subtilis. J. Bacteriol. 181: 3382–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dingman DW, Sonenshein AL. 1987. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citB gene. J. Bacteriol. 169: 3062–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin S, Sonenshein AL. 1994. Transcriptional regulation of Bacillus subtilis citrate synthase genes. J. Bacteriol. 176: 4680–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5: 917–927 [DOI] [PubMed] [Google Scholar]
- 8. Jourlin-Castelli C, Mani N, Nakano MM, Sonenshein AL. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295: 865–878 [DOI] [PubMed] [Google Scholar]
- 9. Kim HJ, Kim SI, Ratnayake-Lecamwasam M, Tachikawa K, Sonenshein AL, Strauch M. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim SI, Jourlin-Castelli C, Wellington SR, Sonenshein AL. 2003. Mechanism of repression by Bacillus subtilis CcpC, a LysR family regulator. J. Mol. Biol. 334: 609–624 [DOI] [PubMed] [Google Scholar]
- 11. Mittal M, Pechter KB, Picossi S, Kim HJ, Kerstein KO, Sonenshein AL. 8 November 2012. Dual role of CcpC protein in regulation of aconitase gene expression in Listeria monocytogenes and Bacillus subtilis. Microbiology doi:10.3891/acta.chem.scand.17s-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alén C, Sonenshein AL. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 96: 10412–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Serio AW, Pechter KB, Sonenshein AL. 2006. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188: 6396–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beinert H. 2000. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5: 2–15 [DOI] [PubMed] [Google Scholar]
- 15. Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59: 1073–1082 [DOI] [PubMed] [Google Scholar]
- 16. Emery-Goodman A, Hirling H, Scarpellino L, Henderson B, Kühn LC. 1993. Iron regulatory factor expressed from recombinant baculovirus: conversion between the RNA-binding apoprotein and Fe-S cluster containing aconitase. Nucleic Acids Res. 21: 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haile DJ, Rouault TA, Harford JB, Kennedy MC, Blondin GA, Beinert H, Klausner RD. 1992. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. U. S. A. 89: 11735–11739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H. 1992. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl. Acad. Sci. U. S. A. 89: 11730–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haile DJ. 1999. Regulation of genes of iron metabolism by the iron-response proteins. Am. J. Med. Sci. 318: 230–240 [DOI] [PubMed] [Google Scholar]
- 20. Tang Y, Guest JR. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145: 3069–3079 [DOI] [PubMed] [Google Scholar]
- 21. Banerjee S, Nandyala AK, Raviprasad P, Ahmed N, Hasnain SE. 2007. Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J. Bacteriol. 189: 4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serio AW. 2005. Ph.D. thesis. Tufts University, Boston, MA [Google Scholar]
- 23. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Fouet A, Sonenshein AL. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fisher SH, Magasanik B. 1984. Synthesis of oxaloacetate in Bacillus subtilis mutants lacking the 2-ketoglutarate dehydrogenase enzyme complex. J. Bacteriol. 158: 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wacker I, Ludwig H, Reif I, Blencke H-M, Detsch C, Stülke J. 2003. The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149: 3001–3009 [DOI] [PubMed] [Google Scholar]
- 27. Mueller J, Bukusoglu G, Sonenshein AL. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174: 4361–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kennedy MC, Emptage MH, Dreyer J-L, Beinert H. 1983. The role of iron in the activation-inactivation of aconitase. J. Biol. Chem. 258: 11098–11105 [PubMed] [Google Scholar]
- 29. Fortnagel P, Freese E. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 95: 1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srere P, Brazil H, Gonen L. 1963. The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem. Scand. 17: S129–S134 doi:10.3891/acta.chem.scand.17s-0129 [Google Scholar]
- 31. Jin S. 1995. Ph.D. thesis. Tufts University, Boston, MA [Google Scholar]
- 32. Riddles P, Blakeley R, Zerner B. 1983. Reassessment of Ellman's reagent. Methods Enzymol. 91: 49–60 [DOI] [PubMed] [Google Scholar]
- 33. Belitsky BR, Janssen PJ, Sonenshein AL. 1995. Sites required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J. Bacteriol. 177: 5686–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin S, Sonenshein AL. 1996. Characterization of the major citrate synthase of Bacillus subtilis. J. Bacteriol. 178: 3658–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ludwig H, Homuth G, Schmalisch M, Dyka FM, Hecker M, Stülke J. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41: 409–422 [DOI] [PubMed] [Google Scholar]
- 37. Meinken C, Blencke H-M, Ludwig H, Stülke J. 2003. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149: 751–761 [DOI] [PubMed] [Google Scholar]
- 38. Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- 39. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schnetz K, Stülke J, Gertz S, Krüger S, Krieg M, Hecker M, Rak B. 1996. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol. 178: 1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schilling O, Herzberg C, Hertrich T, Vörsmann H, Jessen D, Hübner S, Titgemeyer F, Stülke J. 2006. Keeping signals straight in transcription regulation: specificity determinants for the interaction of a family of conserved bacterial RNA-protein couples. Nucleic Acids Res. 34: 6102–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gruer MJ, Bradbury AJ, Guest JR. 1997. Construction and properties of aconitase mutants of Escherichia coli. Microbiology 143: 1837–1846 [DOI] [PubMed] [Google Scholar]
- 43. Baumgart M, Mustafi N, Krug A, Bott M. 2011. Deletion of the aconitase gene in Corynebacterium glutamicum causes strong selection pressure for secondary mutations inactivating citrate synthase. J. Bacteriol. 193: 6864–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koziol U, Hannibal L, Rodríguez MC, Fabiano E, Kahn ML, Noya F. 2009. Deletion of citrate synthase restores growth of Sinorhizobium meliloti 1021 aconitase mutants. J. Bacteriol. 191: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. 2006. Structure of dual function iron regulatory protein I complexed with ferritin IRE-RNA. Science 314: 1903–1908 [DOI] [PubMed] [Google Scholar]
- 46. Srivatsan A, Han Y, Peng J, Tehranchi A, Gibbs R, Wang J, Chen R. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4: e1000139 doi:10.1371/journal.pgen.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serio AW, Sonenshein AL. 2006. Expression of yeast mitochondrial aconitase in Bacillus subtilis. J. Bacteriol. 188: 6406–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fisher SH, Magasanik B. 1984. 2-Ketoglutarate and the regulation of aconitase and histidase formation in Bacillus subtilis. J. Bacteriol. 158: 379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wiegand G, Remington S. 1986. Citrate synthase: structure, control, and mechanism. Annu. Rev. Biophys. Biophys. Chem. 15: 97–117 [DOI] [PubMed] [Google Scholar]
- 50. Kim HJ, Mittal M, Sonenshein AL. 2006. CcpC-dependent regulation of citB and lmo0847 in Listeria monocytogenes. J. Bacteriol. 188: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klausner RD, Rouault TA, Harford JB. 1993. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72: 19–28 [DOI] [PubMed] [Google Scholar]
- 53. Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, Muffler A, Rodriguez S, Perkins J, Losick R. 2010. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 192: 5402–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zumbrennen KB, Wallander ML, Romney SJ, Leibold EA. 2009. Cysteine oxidation regulates the RNA-binding activity of iron regulatory protein 2. Mol. Cell. Biol. 29: 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Philpott C, Klausner RD, Rouault TA. 1994. The bifunctional iron-responsive element binding protein/cytosolic aconitase: the role of active-site residues in ligand binding and regulation. Proc. Natl. Acad. Sci. U. S. A. 91: 7321–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kaldy P, Menotti E, Moret R, Kühn LC. 1999. Identification of RNA-binding surfaces in iron regulatory protein-1. EMBO J. 18: 6073–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao W, Dai S, Liu Q, Xu Y, Bai Y, Qiao M. 2010. Effect of site-directed mutagenesis of citB on the expression and activity of Bacillus subtilis aconitase. Mikrobiologiia 79: 774–778 [PubMed] [Google Scholar]
- 58. Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70: 6373–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kirchberg J, Büttner D, Thiemer B, Sawers R. 2012. Aconitase B is required for optimal growth of Xanthomonas campestris pv. vesicatoria in pepper plants. PLoS One 7: e34941 doi:10.1371/journal.pone.0034941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Burkholder PR, Giles NH., Jr 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34: 345–348 [PubMed] [Google Scholar]
- 61. Dean DR, Hoch JA, Aronson AI. 1977. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J. Bacteriol. 131: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brehm SP, Staal SP, Hoch JA. 1973. Phenotypes of pleiotropic-negative sporulation mutants of Bacillus subtilis. J. Bacteriol. 115: 1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]