Abstract
Spores of Clostridium difficile play a key role in the dissemination of this important human pathogen, and until recently little has been known of their functional characteristics. Genes encoding six spore coat proteins (cotA, cotB, cotCB, cotD, cotE, and sodA) were disrupted by ClosTron insertional mutagenesis. Mutation of one gene, cotA, presented a major structural defect in spore assembly, with a clear misassembly of the outermost layers of the spore coat. The CotA protein is most probably subject to posttranslational modification and could play a key role in stabilizing the spore coat. Surprisingly, mutation of the other spore coat genes did not affect the integrity of the spore, although for the cotD, cotE, and sodA mutants, enzyme activity was reduced or abolished. This could imply that these enzymatic proteins are located in the exosporium or alternatively that they are structurally redundant. Of the spore coat proteins predicted to carry enzymatic activity, three were confirmed to be enzymes using both in vivo and in vitro methods, the latter using recombinant expressed proteins. These were a manganese catalase, encoded by cotD, a superoxide dismutase (SOD), encoded by sodA, and a bifunctional enzyme with peroxiredoxin and chitinase activity, encoded by cotE. These enzymes being exposed on the spore surface would play a role in coat polymerization and detoxification of H2O2. Two additional proteins, CotF (a tyrosine-rich protein and potential substrate for SodA) and CotG (a putative manganese catalase) were shown to be located at the spore surface.
INTRODUCTION
Spores of Clostridium difficile are integral to the infection and disease caused by this Gram-positive pathogen (1). As robust dormant entities, spores can survive indefinitely outside their host, and their persistence and resistance to many disinfectants have provided a serious problem for public health (2, 3). Antibiotic treatment which is used to treat C. difficile-associated diarrhea may itself promote the proliferation and dissemination of highly infectious spores of C. difficile, based on studies carried out in mice (4). Although this “supershedder” state ends once antibiotic treatment is discontinued, this finding demonstrates the potential threat of C. difficile spores within a hospital environment. Intriguingly, this work also suggested that C. difficile spores grown in the laboratory may be phenotypically different from those spores shed in feces (4).
Over the last decade, “hypervirulent” strains of C. difficile have emerged that have been responsible for a number of epidemics of C. difficile infection (CDI) (5–7). The best known of these hypervirulent strains is the PCR ribotype O27/restriction endonuclease analysis (REA) group B1 (6, 7). These strains are known to produce significantly higher levels of toxins and potentially higher numbers of spores (8–10), although the latter phenotype has been refuted by some (11).
The supershedder state and hypervirulent nature of some C. difficile isolates have focused attention on understanding spore formation in this bacterium. The spore proteome has been examined, with the identification of numerous genes related to sporulation genes found in Bacillus subtilis (12). Interestingly, however, substantial differences both in the regulation of spore formation and in spore structure exist. For example, in C. difficile, the sigma factor σK is encoded following excision of a 14.6-kbp prophage-like element, skinCd, which creates the structural gene sigK (13). In B. subtilis, following excision of a similar skin element, the sigK structural gene is subject to a further level of posttranslational control, where it is cleaved from an inactive form known as pro-σK to its mature form (σK) by a forespore-associated protease, SpoIVFB (14). So, while the key components of sporulation-specific gene expression appear similar to those of other spore formers, some notable differences exist (15). Bioinformatic studies (16) have identified only 18 orthologues of the 70 or so B. subtilis spore coat proteins known to exist (17), suggesting that the composition of the coat must be very different. In a recent study, we made an initial characterization of spore coat proteins and identified five distinct proteins that could be extracted from the coat using an SDS-borate-dithiothreitol (DTT) buffer (18). Based on sequence analysis, three of these proteins, CotCB, CotD, and CotE, could have enzyme activity. CotCB and CotD are putative manganese catalases, and CotE is a novel bifunctional protein with peroxiredoxin (amino-terminal) and chitinase (carboxy-terminal) activity, and the corresponding enzymatic activity was demonstrated in suspensions of pure spores (18).
In this work, we have used targeted mutagenesis to inactivate each of the spore coat genes, and for four genes, we have been able to assign a function. Three additional spore protein genes, cotF, cotG, and sodA, have also been investigated. Our studies show the following: first, that CotA plays a significant role in spore coat assembly; second, that catalase activity could be attributed to CotD and peroxiredoxin and chitinase activity to CotE; and finally, that the spore coat layers are not only enzyme rich but particularly fragile.
MATERIALS AND METHODS
Strains.
The wild-type strain of C. difficile used in this work, 630Δerm, is a spontaneously cured derivative of 630 (tcdA+ tcdB+) that is erythromycin sensitive (19). Strain 630 was isolated from a patient with pseudomembranous colitis during an outbreak of CDI (20).
Growth of C. difficile and preparation of spores.
C. difficile was routinely grown in vegetative culture by overnight growth in TGY vegetative medium (21). Spores of C. difficile were prepared by growth on SMC agar plates using an anaerobic incubator (Don Whitley, United Kingdom) as described previously (22). After growth for 7 days at 37°C, spores were either washed three times with water or subjected to a protocol of purification as follows. Initial incubation of harvested spore crops in phosphate-buffered saline (PBS) (containing 125 mM Tris, 200 mM EDTA, 0.3 mg/ml proteinase K, and 1% sarcosyl) for 3 h at 37°C. Next, resuspension of the pellet in sterile water was followed by 10 washes in water (40 ml). The purified spore suspension was heat treated (60°C, 20 min), and the number of CFU/ml was determined by serial dilution and colony counting.
ClosTron mutagenesis.
Insertional mutations in cot genes and sodA were made using the ClosTron system developed at the University of Nottingham (23–25). The Perutka algorithm (26), available at www.clostron.com, was used to design 45-bp retargeting sequences for each gene (Table 1). A derivative of plasmid pMTL007C-E2 carrying these retargeting sequences was obtained from DNA2.0 (Menlo Park, CA). Using the protocols provided by Heap et al. (23–25), plasmids were first introduced into E. coli and then conjugated with C. difficile 630Δerm. For each mutant, five erythromycin-resistant (Ermr) transconjugants were checked by PCR for ClosTron insertion. Genomic DNA was prepared as described previously (27), and then three PCR validations were performed (see Fig. S1 and Table S1 in the supplemental material). First, PCR using the ErmRAM primers resulted in a 900-bp product confirming that the Ermr phenotype was due to splicing of the group I intron from the group II intron following integration (see Fig. S1A in the supplemental material). Second, primers targeting the gene left and right ends of the insertion site were used to check the site of insertion (see Fig. S1B in the supplemental material). If insertion occurred, a PCR product 1,800 bp greater than that obtained for the wild-type strain would be found. Third, PCR was carried out using primers targeting the 5′ end of the gene and the intron (EBS-universal) to confirm again the right insertion site. No product would be expected for the wild-type strain (see Fig. S1C in the supplemental material).
Table 1.
C. difficile spore coat genes and mutations
| Gene | Locus taga | Encoded protein (mass [kDa])b | Mutant allelec | Retargeted sequenced |
|---|---|---|---|---|
| cotA | CD1613 | Coat protein (34) | cotA::CT555a | TCTACGTTTGCTAATGGAGTAGATGCAGCA<intron>AGTATAAAATCAATT |
| cotB | CD1511 | Coat protein (35) | cotB::CT329a | CTAAATCTTGACCAACTTGTGGGTTTACAC<intron>CACTTTGTAGATTTA |
| cotCB | CD0598 | Coat protein, manganese catalase (21) | cotCB::CT220s | GAGGAAATGGCTCATGTTGAGATAATTTCT<intron> TCAGTGTTATATCAA |
| cotD | CD2401 | Coat protein, manganese catalase (21) | cotD::CT302s | CAGGTCTTGGTTCAAACTATGCTCAAAATG<intron>GATATGGAATTTATC |
| cotE | CD1433 | Coat protein, peroxiredoxin-chitinase (81) | cotE::CT220s | AAAAAAAGAAATACAGAACTAATTGGTCTA<intron>AGTGTTGATAGTAAC |
| cotE::CT1203s | ACTAATGTAGATGCTCAACTATTAGATGCA<intron> GTTATATTTGCTTTT | |||
| cotF | CD0597 | Coat protein (11) | NDe | ND |
| cotG | CD1567 | Coat protein, manganese catalase (27) | ND | ND |
| sodA | CD1631 | Coat protein, superoxide dismutase (28) | sodA::CT394s | TTTCAAAAGAACCAAAGTCTCTATCAATAG<intron>CTTCTTTTAAAGATT |
As described in reference 16. Map positions are shown in Fig. S6 in the supplemental material.
Function where known; predicted mass of full-length protein in parentheses.
The mutant allele is shown, with “CT” designating ClosTron insertion, the number showing the base pairs within the ORF immediately preceding the ClosTron insertion, and either “a,” indicating insertion in the antisense strand, or “s,” in the sense strand.
The 45-bp targeting sequence produced using the aligorithm at www.clostron.com and used for mutant construction. The intron insertion site within the 45-mer target sequence is shown.
ND, not done.
Complementation analysis.
All cot mutants were complemented with wild-type copies of the respective genes using pRPF185 (28). Briefly, a DNA fragment including the entire coding sequence of each gene and the Shine-Dalgarno site was PCR amplified using KOD Hot Start polymerase (Merck) and cloned using SacI/BamHI into pRPF185 under the control of the inducible Ptet promoter. The resulting plasmids were transferred into the corresponding cot mutant strains via conjugation. Gene expression was induced when desired using anhydrous tetracycline (ATc) at 500 ng/ml. For the cotA mutant, which carried sensitivity to ethanol and lysozyme, full resistance was observed in the complemented strains (see Table S2 in the supplemental material), while for the other mutants, immunofluorescence was used to show restoration of the corresponding Cot protein on the spore surface (see Fig. S2 in the supplemental material).
Recombinant proteins and antibody production.
pET28b expression vectors that express the cotA, cotB, cotCB, cotD, and cotE open reading frames (ORFs) and polyclonal antibodies recognizing the encoded proteins have been described elsewhere (18). In the case of cotE, antibodies were raised against an amino-terminal fragment of CotE, rCotEN, carrying the entire peroxiredoxin domain (residues 1 to 214). For the cotF, cotG, and sodA genes, pET28b expression vectors carrying the entire ORF were constructed using primers shown in Table S1 in the supplemental material. For cotE, an additional carboxy-terminal clone, pCotEC, which carried the 3′ end of cotE starting at codon 390 (Pro) and ending with the stop codon, was made. High levels of expression were obtained upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction and purification of proteins by passage of the cell lysate through a HiTrap chelating HP column on a Pharmacia AKTA liquid chromatography system. Polyclonal antibodies were raised in BALB/c mice (female; age, 5 weeks) immunized by the intraperitoneal route with 2 μg of purified recombinant proteins on days 1, 14, and 28. Antibodies were first purified using a Protein G HP Spin-Trap column (GE Healthcare). Antibodies were successfully made to rCotF, rCotG, and rCotG but not to rCotEC. Work was performed under the UK Home Office license PPL 70/7025.
Spore coat extractions and Western blotting.
Spore coat proteins were extracted from approximately 2 × 109 purified spores as described previously (18). Twelve percent SDS-PAGE gels were used for protein analysis, using approximately 10 μg of protein per lane. For blotting, the gel was transferred to a nitrocellulose membrane (100 V, 70 min) and blocked with 5% skimmed milk for 1 h. Antibodies were used at the following dilutions: anti-CotA, 1:1,000; anti-CotB, 1:10,000; anti-CotC, 1:4,000; anti-CotD, 1:3,000; anti-CotE(C) and anti-CotEN, 1:500; anti-CotF, 1:4,000; anti-CotG, 1:4,000; anti-SodA, 1:4,000.
Resistance assays.
Spores (1 × 106) of wild-type 630Δerm or mutants were resuspended in a volume of 0.5 ml in PBS buffer and incubated for 20 min at 37°C with 70% ethanol, 1% Virkon, 3% H2O2, lysozyme (250 μg/ml), toluene, or chloroform, followed by serial dilution and plating on SMC agar to determine surviving CFU. For heat resistance, spores were heated at 65°C or 80°C for 20 min before enumeration of CFU.
TEM.
Spores were processed for ultramicrotomy according to standard procedures (29). Briefly, spore suspensions were diluted 10 times in H2O and washed twice by centrifugation at 10,000 × g for 10 min in order to eliminate spore debris. Spore pellets were fixed for 12 h at 4°C in a mixture of 2.5% glutaraldehyde and 4% paraformaldehyde in 0.2 M cacodylate buffer (pH 7.4) and then postfixed for 1 h at ambient temperature with 1% osmium tetroxide in the same buffer. Sample pellets were dehydrated with ethanol and embedded in Epon-Araldite resin. Thin sections were stained successively with 5% uranyl acetate and 1% lead citrate. Transmission electron microscopy (TEM) observation was performed using a FEI CM120 electron microscope operated at 120 kV.
Immunoelectron microscopy (IEM) methods.
Spores were washed as described above by two cycles of centrifugation at 10,000 × g for 10 min. Pellets were then resuspended in 200 μl PBS supplemented with 1% bovine serum albumin (BSA), incubated for 10 min, and then sedimented by centrifugation at 10,000 × g for 10 min. Pellets were resuspended as above and incubated with primary antibodies diluted 100× in PBS in the presence of 0.1% BSA for 1 h. The excess of antibody was eliminated by three cycles of centrifugation at 10,000 × g for 10 min and resuspension in PBS–0.1% BSA. Spore suspensions were then incubated for 30 min with 20×-diluted anti-mouse IgG antibody coupled to 10-nm gold particles (Beckman Coulter) in PBS–0.1% BSA. The excess antibody was eliminated as described above by a triple centrifugation at 10,000 × g for 10 min. The final pellets were fixed and processed for TEM as described above.
Spore enzyme assays.
For all assays, suspensions of purified wild-type 630Δerm or mutant spores were used.
(i) Catalase.
For spore-associated catalase activity, 1 × 107 spores were resuspended in 60 μl of 50 mM potassium phosphate buffer (pH 7.0) in an Eppendorf tube. The reaction was started with 1.94 ml of H2O2 solution (0.036%). After 15 min at room temperature, spores were pelleted and the optical density at 240 nm (OD240) of the supernatant was measured. Enzyme activity was calculated as the percentage decrease in OD.
(ii) SOD.
Superoxide dismutase (SOD) activity has been described elsewhere and relies upon the inhibitory action of SOD on the autooxidation of pyrogallol in alkaline buffer (30, 31). Spores (1 × 108) were incubated for 10 min at 37°C in 50 mM Tris-HCl buffer, pH 8.2, 1 mM EDTA, and 0.2 mM pyrogallol (P0381; Sigma). Spores were pelleted for 1 min at maximum speed, and the OD420 of the supernatant was measured.
(iii) Peroxiredoxin.
The peroxiredoxin assay was carried out as described previously (32, 33). Two hundred fifty micromolar β-NAD (NADH, N8129; Sigma), 500 μM H2O2, 4 μM thioredoxin (from E. coli, T0910; Sigma), and 0.1 μM thioredoxin reductase (from E. coli, T7915; Sigma) were mixed with 1 × 108 spores in 500 μl of buffer [containing 0.3 mM EDTA, 0.5 M KH2PO4, and 150 mM (NH4)2SO4]. Samples were incubated at 37°C for 15 min. Spores were pelleted for 1 min at maximum speed, and the OD340 of the supernatant was determined. Activity was calculated as the percentage difference from the starting OD.
(iv) Chitinase.
A chitinase assay kit (CS0980; Sigma) was used, with minor modifications. To perform the assay, 1 × 108 spores were suspended in 100 μl of buffer with 1 mg/ml of substrate (4-nitrophenyl N-acetyl-β-d-glucosaminide) and incubated for 3 h at 37°C. After 3 h, 200 μl of 0.4 M sodium carbonate was added to stop the reaction. Spores were pelleted for 1 min at maximum speed, and the OD405 was read. Activity was calculated as the percentage difference from the starting OD.
Recombinant protein enzyme assays. (i) Catalase.
For catalase, Akta-purified rCotC, rCotD, and rCotG were washed three times in a 10,000-molecular-weight-cutoff (MWCO) polyethersulfone (PES) concentrator (VS2001; Sartorius Stedim) using 50 mM Tris-H2SO4 (pH 7.2) and concentrated to 0.9 mg/ml. The protein concentration was measured using the Bradford assay. Catalase activity was determined as described by Mizobata et al. (34). Manganese chloride was added to the protein solution (to 50 mM), which was dialyzed for 16 h at 4°C against 50 mM Tris-H2SO4 (pH 7.2) containing 50 mM MnCl2. Enzyme reactions were started by mixing 0.5 mg/ml of dialyzed protein with 2 ml of 0.036% H2O2 (in 50 mM Tris-H2SO4). The decrease in the OD240 was followed for 1 min at room temperature. As a positive control, 5 U/ml of catalase from Aspergillus niger (C3515; Sigma) was used. Enzyme activity was calculated as the percentage difference from the starting OD. One unit of enzyme is defined as that which can decompose 1 μM H2O2 per minute (34).
(ii) SOD.
For SOD, Akta-purified rSodA was washed three times with 50 mM Tris-H2SO4 buffer (pH 7.2) in a 10,000-MWCO PES concentrator until a concentration of 0.9 mg/ml was achieved. The protein (0.9 mg/ml) was dialyzed at 4°C overnight in the presence of 50 mM manganese chloride (Sigma) in 50 mM Tris-H2SO4 buffer (pH 7.2). Enzyme activity was measured using inhibition of pyrogallol autooxidation as described previously (30, 31). The enzyme assay was performed in 50 mM Tris-HCl buffer, pH 8.2, 1 mM EDTA, 0.2 mM pyrogallol (Sigma), and 20 μg/ml catalase (from Aspergillus niger; Sigma). The increase in absorbance was measured at 420 nm for 5 min at 37°C. Four μg/ml superoxide dismutase from bovine erythrocytes (S5395; Sigma) was used as a positive control. Enzyme activity was calculated as the percentage of inhibition of pyrogallol autooxidation. One unit of SodA activity is the amount required to inhibit autooxidation of pyrogallol by 50% (31).
(iii) Peroxiredoxin.
For peroxiredoxin, the Akta-purified rCotEN protein was dialyzed overnight against PBS (pH 7.4). Peroxiredoxin activity was determined as described previously (32) with a reaction volume of 0.5 ml consisting of rCotEN (0.3 mg/ml), 250 μM NAD, 500 μM H2O2, 4 μM thioredoxin (E. coli; Sigma), and 0.1 μM thioredoxin reductase (E. coli; Sigma) in EKN buffer [0.3 mM EDTA, 0.5 M KH2PO4, and 150 mM (NH4)2SO4]. The OD340 was measured continuously for 3 min at 37°C. As a positive control 4 μg/ml of human peroxiredoxin 1 (P8986; Sigma) was used. Enzyme activity was calculated as a percentage difference from the starting OD. One unit of peroxiredoxin activity is defined as the amount required to oxidize 1 nmol NADPH/min (35).
(iv) Chitinase.
For chitinase, Akta-purified rCotEC was dialyzed overnight against PBS (pH 7.4). The protein concentration was measured, and 0.5 mg/ml (in PBS; pH 7.4) was incubated at 37°C in the presence of 1 mg/ml 4-nitrophenyl N-acetyl-β-d-glucosamide in a volume of 0.1 ml. The OD405 was measured every 30 s for 30 min. As a positive control, 0.05 mg/ml chitinase from Trichoderma viridae (C6242; Sigma) was used. Enzyme activity was calculated as the percentage difference from the starting OD. One unit of chitinase activity is defined in the Sigma assay kit as the amount necessary to release 1 μmol of p-nitrophenol/min.
Immunofluorescence microscopy.
For immunofluorescence microscopy, the procedure followed was as described by Duc et al. (36) with minor changes. Microscope coverslips were first treated with 0.01% poly-l-lysine overnight. Spores (1 × 107) were added to the slide and dried for 1 h at room temperature. After 3 washes with PBS (pH 7.4) and blocking in PBS plus 2% BSA plus 0.05% Tween 20 for 1.5 h, the first antibody was added (1:1,000). Spores were incubated for 30 min at room temperature. After 3 washes with PBS plus 0.05% Tween 20, anti-mouse IgG serum (1:1,000) was added, followed by incubation for 30 min at room temperature. After six more washes, the slide was viewed under a Nikon Eclipse Ti-S fluorescence microscope.
Detection of surface display using ELISA.
The whole-spore enzyme-linked immunosorbent assay (ELISA) protocol was followed as described by Duc et al. (37) with small changes. Purified spores (1 × 107/ml) were suspended in CO3 buffer (50 mM sodium carbonate-sodium bicarbonate buffer, pH 9.6) with 4% formaldehyde. ELISA plates were coated with 50 μl of the spore suspension and left at 4°C overnight and an additional 2 h at room temperature. Plates were washed three times with PBS (pH 7.4) and blocked with 2% BSA in PBS for 1 h at 37°C. Antibodies were added in a 2-fold dilution series starting with a 1/40 dilution in buffer (0.1 M Tris-HCl [pH 7.4], 3% NaCl, 0.5% BSA, 10% sheep serum [Sigma], 0.1% Triton X-100, and 0.05% Tween 20). The plates were incubated at 37°C for 2 h before washing three times with PBS plus 0.05% Tween 20, with 2-min intervals between washes. Anti-mouse IgG-horseradish peroxidase conjugate was added at 1:3,000 in PBS plus 1% BSA plus 0.05% Tween 20. The plates were incubated for an additional 1 h at 37°C and washed three times again, with 5-min intervals between washes. Color was developed with 1 mg/ml 3,3′,5,5′-tetramethylbenzidine (TMB) (Sigma) in 9 ml water, 1 ml sodium acetate buffer (1 M, pH 5.5), and 2 μl of H2O2. Reactions were stopped with 1 volume H2SO4/4 volumes H2O, and the OD450 was read.
RESULTS
ClosTron mutagenesis of spore coat genes.
Five spore coat genes, cotA, cotB, cotCB, cotD, and cotE, from the spore coats of C. difficile were identified in a recent study (18). In addition, a putative superoxide dismutase (CD1631; see Fig. S3 in the supplemental material) and a tyrosine-rich spore protein (CD0596; see Fig. S4) were implicated as potential spore coat proteins, and we have named the corresponding genes sodA and cotF, respectively. Finally, in addition to cotCB and cotD, a further gene (CD1567) that could encode a spore-associated manganese catalase was identified from bioinformatic analysis and named cotG (see Fig. S5). We used the ClosTron system for targeted mutagenesis of these genes (with the exception of cotF and cotG) as described in Materials and Methods and summarized in Table 1. In the case of cotE, which carries two enzymatic domains (peroxiredoxin and chitinase), we made two mutations, inactivating the amino-terminal (cotE::CT220s) and carboxy-terminal (cotE::CT1203s) domains, respectively.
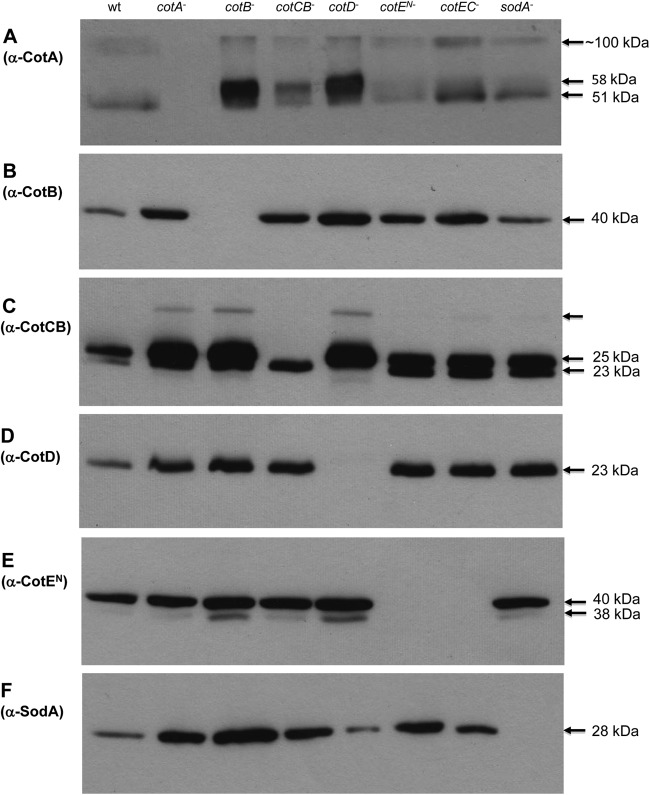
Spores from all of the mutants were prepared, coat proteins were extracted and fractionated, and gels were probed with antibodies raised against each of the individual spore coat proteins (Fig. 1). For CotE, we used antisera raised against the amino-terminal peroxiredoxin (27-kDa) domain. As a control, we also probed coat extracts from the isogenic wild-type strain 630Δerm. For cotA (Fig. 1A) mutant spores, no CotA species was detectable, but its presence was found in the other mutants. Interestingly, we observed that CotA was present as more than one polypeptide species. In wild-type spores, we could detect a band of about 51 kDa and a higher-molecular-mass species of ∼100 kDa. In previous work, we had observed only one species running at about 47 kDa (18). In the five cot mutants, in addition to the 51-and 100-kDa species, a further band of about 58 kDa was present. cotB or cotD mutant spores lacked the corresponding coat protein of 40 kDa (CotB) or 23 kDa (CotD), respectively, but it was present in the other mutants (Fig. 1B and D).
Fig 1.
Western blot analysis of spore coat mutants. Spores of wild-type 630Δerm and spore coat mutants were purified, and spore coat proteins were extracted and fractionated on 12% SDS-PAGE gels (A to F). For cotE, two mutants are shown, as cotEC− (cotE::CT1203s mutant) and cotEN− (cotE::CT220s mutant), representing insertions at the carboxy and amino termini, respectively. Gels were transferred to membranes and probed with polyclonal antibodies to each of the recombinant proteins shown. For CotE, antibodies to the amino terminus (CotEN) were used. In panel C, the two bands that are recognized by CotCB antibodies are shown, with the upper band corresponding to CotCB and the lower to CotD, which is cross-reactive; the unlabeled arrow indicates a high-molecular-mass potential multimer.
For the cotCB mutant, a 23-kDa band of lower intensity was observed (Fig. 1C). This species can be explained by the fact that antibodies to CotCB cross-react with CotD, as shown previously (18), and closer examination of the bands recognized by anti-CotCB revealed two distinct bands, for CotCB (25 kDa) and CotD (23 kDa). However, despite the similarity of CotCB and CotD as manganese catalases, CotD antibodies do not recognize CotC. It is also apparent that in the cotA, cotB, and cotD mutants, an additional band of about 30 to 32 kDa was observed. This was not apparent for wild-type spores and presumably must represent a multimeric form of CotCB which is revealed in some mutants.
Antibodies to the amino terminus of CotE recognized a 40-kDa species and a minor band of about 38 kDa, both of which were absent in the cotE::CT220s and cotE::CT1203s mutants (Fig. 1E). The intact size of CotE is 81 kDa, and in previous work we have shown that this protein band is labile and is lost during spore purification, so it was not present in these purified preparations of spores (18). For the amino-terminal mutant, we would expect no recognition since the mutation would be polar, but for the carboxy-terminal mutation we might have expected labeling by antibodies to the amino terminus. This observation suggests that despite being able to express the amino-terminal domain in E. coli, when presented on the spore, it is unstable and presumably degraded. Finally, for labeling with anti-SodA, the 28-kDa SodA protein was absent in the sodA mutant and present in the other mutants, albeit at lower levels in the cotD mutant (Fig. 1F).
We first examined the resistances of spores compared to that of the isogenic wild-type strain 630Δerm using a selection of treatments, including heat, solvents, disinfectants (Virkon), H2O2, lysozyme, and alcohol (Table 2). We observed that all strains (wild type and mutants) were sensitive to Virkon, hydrogen peroxide, and a temperature of 80°C, in agreement with previous work (38). Spores of all strains were essentially resistant to heat exposure at 65°C, but in some cases (630Δerm and the cotE::CT1203s and sodA::CT394s strains), counts of survivors were greater than the starting CFU. We attribute this to superdormancy and the ability of spores to enter a state where they fail to completely germinate and outgrow unless heat activated (39). Only the cotA::CT555a mutant showed a clear sensitivity to any of the treatments compared to wild-type spores, specifically a 74% or 71% reduction in spore CFU after treatment with lysozyme or ethanol, respectively.
Table 2.
Resistance of mutant and wild-type sporesa
| Strain or genotype | % CFU after treatment |
|||||||
|---|---|---|---|---|---|---|---|---|
| Heat (°C) |
Virkon | H2O2 | Chloroform | Toluene | Lysozyme | Ethanol | ||
| 65 | 80 | |||||||
| WT 630Δerm | 113 | 4 | <1 | <1 | 82 | 87 | 91 | 90 |
| cotA::CT555a | 83 | 2 | <1 | <1 | 82 | 83 | 26 | 29 |
| cotB::CT329a | 72 | 3 | <1 | <1 | 80 | 83 | 91 | 91 |
| cotCB::CT220s | 73 | 5 | <1 | <1 | 79 | 89 | 92 | 91 |
| cotD::CT302s | 73 | 4 | <1 | <1 | 79 | 93 | 91 | 93 |
| cotE::CT220s | 77 | 3 | <1 | <1 | 78 | 95 | 92 | 90 |
| cotE::CT1203s | 104 | 3 | <1 | <1 | 87 | 94 | 90 | 88 |
| sodA::CT394s | 112 | 6 | <1 | <1 | 82 | 80 | 95 | 79 |
Suspensions of spores were treated with a selection of noxious chemicals or heat, and the numbers of CFU before and after treatment are shown as average percentages for three independent experiments. CFU data and statistical analysis are given in Table S3 in the supplemental material. WT, wild type. Boldface indicates values representing sensitivity.
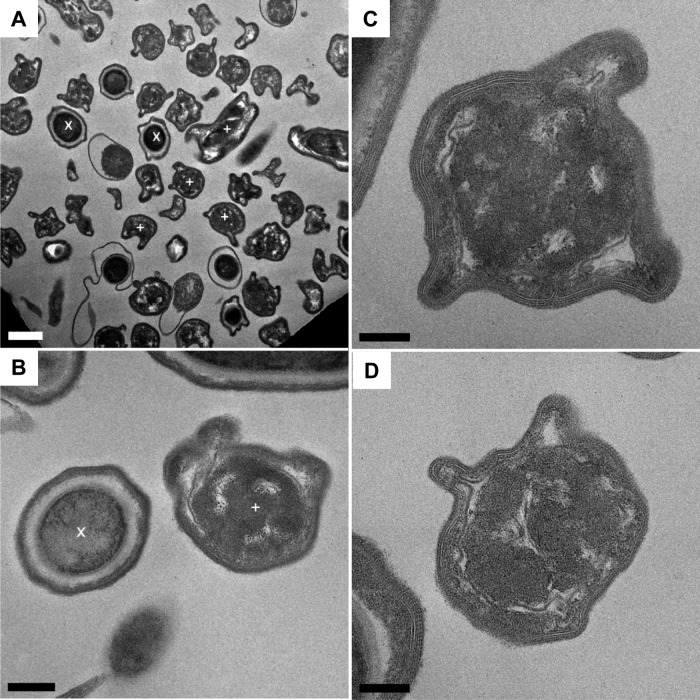
Mutants were examined by TEM in comparison to the wild-type strain (630Δerm), and with the exception of the cotA::CT555a mutant, no obvious differences were observed. The cotA::CT555a mutant, however, demonstrated an unusual phenotype (Fig. 2). In samples of purified spores, two morphologies were apparent. Approximately half of the spores examined were similar to wild-type spores (marked “x” in Fig. 2A and B), while the other half presented structural defects (marked “+” in Fig. 2A and B; see also panels C and D). With the mutant spores, the electron-dense outer layer of the coat appeared absent, leaving only a lamellated inner coat apparent (Fig. 2B). In some cases, what appeared to be ribbons of coat material (possibly an outer coat layer) were partially attached to the spore (Fig. 2A). TEM analysis was conducted in two experiments using independently produced spores with exactly the same phenotype. The images suggest that ill-formed spores present a defect in the formation of the inner content of the spore, which is responsible of the deformed morphology of the spores.
Fig 2.
Ultrastructure of cotA mutant spores. (A) Low-magnification image showing ill-formed purified cotA::CT555a spores (three of them are labeled with a white “+” symbol), together with spores presenting a normal morphology (two of them are labeled with a white “x” symbol). Bar, 1 μm. (B) An ill-formed cotA::CT555a mutant spore (+) located next to a spore with normal morphology (x). Bar, 200 nm. (C and D) Examples of ill-formed cotA::CT555a mutant spores. Bars, 100 nm.
Analysis of surface location.
Formaldehyde-fixed spores (purified) of 630Δerm and mutants were used in a whole-spore ELISA using antibodies to each of the spore coat proteins (Fig. 3). In each case, significant levels of detection were found, and with the exception of the cotCB mutant, no antibody-antigen interactions were found using the mutant spores. This showed that each protein was surface exposed. In the case of CotCB, as mentioned earlier, anti-CotCB antibodies are able to cross-react with CotD, which we assume accounts for the labeling observed in the mutant.
Fig 3.
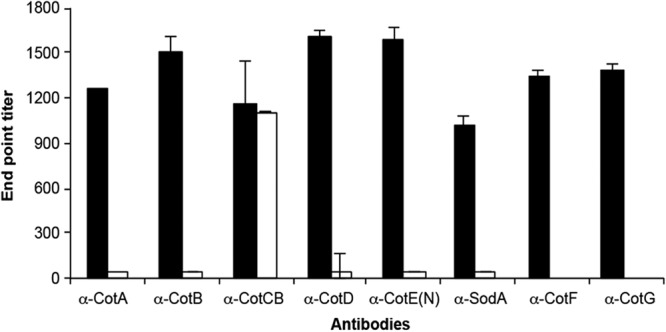
Surface location determined by “whole-spore ELISA.” Microtiter plates were coated with spores, which were probed with antibodies (α) to each of the spore coat proteins. Spores were either wild-type 630Δerm (black bar) or the corresponding isogenic Clostron mutant (unfilled bar), with the exception of anti-CotF and anti-CotG, where mutants were not available. Naive serum was used also for comparison, and basal levels were subtracted.
For CotF, CotG, and SodA, surface expression in 630Δerm spores was also confirmed using immunofluorescence, as was the absence of expression in the sodA::CT394s mutant (Fig. 4A to D). Since none of these proteins were identified in our first analysis of spores, where we had identified CotA-CotE (18), we used antibodies to CotF, CotG, and SodA to probe extractions of spore coat proteins extracted in an SDS-borate-DTT buffer as done previously (18). Only SodA was identified in these extracts (lane 3 of Fig. 4E). However, using sonication of purified spores, we were able to identify all three proteins by Western blotting (lane 2 of Fig. 4E). This demonstrates that additional coat proteins exist and that these are refractory to solubilization in SDS-borate-DTT buffer.
Fig 4.
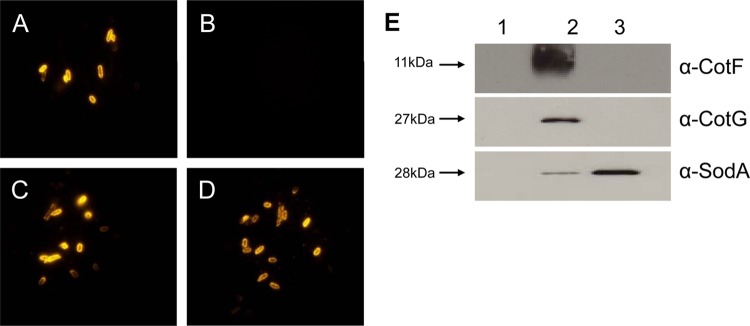
CotF, CotG, and SodA. Based on bioinformatics analysis, three additional spore coat proteins were identified: CotF (tyrosine rich), CotG (putative catalase), and SodA (superoxide dismutase). Polyclonal antibodies to each of these recombinant proteins were used to examine surface location on the 630Δerm spore coat. Panels A to D show immunofluorescent labeling of spores: 630 spores labeled with anti-SodA antibodies (A), spores of the sodA::CT394s mutant labeled with anti-SodA (B), 630 spores labeled with anti-CotF (C), or 630 spores labeled with anti-CotG (D). Spores showed no labeling with preimmune serum (data not shown). Panel E shows Western blots probed with anti-CotF, anti-CotG, and anti-SodA. Lane 1, extracts of proteins found in the supernatant of vegetative cells that had been sonicated (10 bursts of 30 s with cooling); lane 2, extracts from the supernatant of pelleted spores that had been sonicated (10 bursts of 30 s with cooling); lane 3, proteins extracted from spores using a protein extraction buffer (sodium borate-SDS-DTT) described previously (18). Every lane carried 10 μg of protein, determined using the Bradford assay.
IEM analysis.
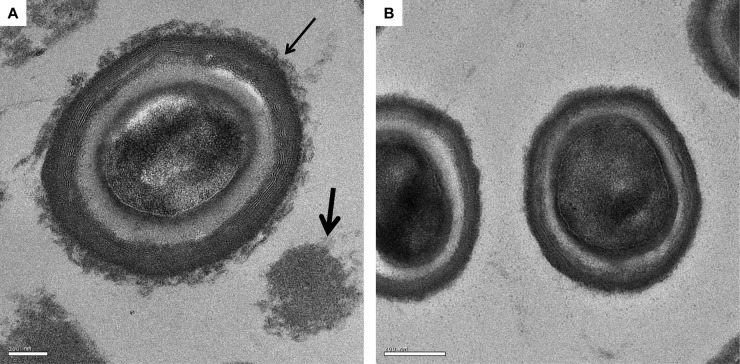
For more detailed analysis of the spore coats, we first examined spores of wild-type spores (630Δerm) prepared with or without purification (Fig. 5). Clear differences were apparent, with unpurified spores showing an abundance of material (>90%) detached from the spore. In unpurified preparations, a diffuse material associated with the outer layer of the spore coat (thin arrow in Fig. 5A) was nearly always seen. We believe the diffuse surface material is most likely the exosporium, the sac-like structure that encases the mature spore (17) and which has been described for C. difficile (12, 40). Our image however, does not agree well with that shown by Lawley et al. (12), where the exosporium is clearly detached from the spore. Either the exosporium has partially degraded, or its appearance somehow relates to the method of spore preparation.
Fig 5.
Ultrastructure of unpurified and purified C. difficile spores. (A) Unpurified spores; (B) purified spores. Unpurified spores present some material either associated with the spore coat (thin arrow) or present in the bulk between and detached from spores (thick arrow).
The material detached from the spore consisted of two types. The first was clumps of uniform density, most probably unstructured proteins that have not assembled, and we refer to this as the bulk material (thick arrow in Fig. 5A). The second was a sheet-like material resembling layers of coat material (shown in Fig. 6D). Purified spores, on the other hand, resembled those shown in Fig. 5B, where the diffuse surface layer was absent. However, our preparations did show some evidence of detached material, mostly sheet-like material (not shown), but this represented far less of the total material observed. Whatever the nature of this material, it is clear that the diffuse surface (“exosporial”) material associated with mature C. difficile spores is apparently easily removed by the purification methods employed here, and even in preparations of spores that had been washed only with water they still carried a considerable amount of detached material.
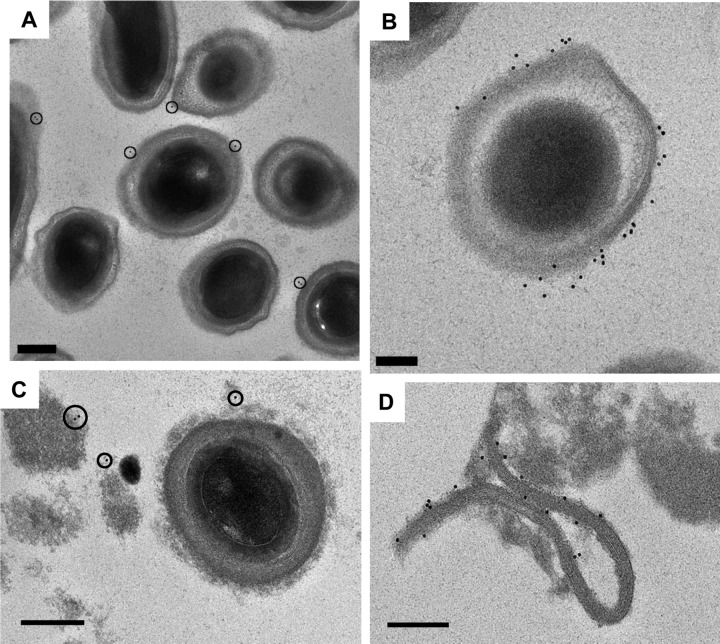
Fig 6.
Immunogold labeling of C. difficile spores. (A) Weak labeling of the surface of purified spores with anti-CotD antibodies. less than 50% of the spores are labeled, with 0 to 1 particle per spore section. Bar, 200 nm. (B) Strong labeling of the surface of purified spores with anti-CotD antibodies. More than 75% spores are labeled, with as many of 10 particles per spore section. Bar, 100 nm. (C) Labeling of material associated with the outer coat of unpurified spores with anti-CotEN antibodies. Bar, 200 nm. (D) Labeling of sheet-like material of unpurified spores with anti-CotEN antibodies. Bar, 200 nm.
Suspensions of unpurified and purified spores were labeled with a series of antibodies raised against spore coat proteins, namely, the CotA, CotB, CotCB, CotD, and CotEN proteins. Immunolabeling was revealed by means of gold particles. A summary of the results is presented in Table 3. Basically, most antibodies recognized epitopes present either at the level of the spore surface or associated with detached material present in the suspension (Fig. 6). In most cases, labeling was weak, meaning that on the electron microscopy (EM) micrographs, only half of the spores were labeled with a few gold particles. It must be noted, however, that the EM micrographs presented in Fig. 6 represent thin sections, about 70 nm thick, of spores that have a width of at least 700 nm. Therefore, if half of the spores are labeled in a thin section, this means that all of the spores are indeed labeled, yet with a limited number of gold particles. Remarkably, in some cases spores were significantly more strongly labeled than the other spores in the same suspension, such as, for example, the spore presented in Fig. 6B. Since spores were 7 days old, we reason that all of the spores must be suitably mature, so these differences in labeling most likely indicate that some proteins have been removed. If we broadly define the spore surface as two components, the diffuse, exosporial layer and the outer layer of the spore coat proper, then we can state that CotB is most likely associated with the exosporial layer, CotA with the spore coat, and CotCB, CotD, and CotE with both layers.
Table 3.
IEM analysisa
| Antibody | Labeling of spore suspension |
|||
|---|---|---|---|---|
| Unpurifiedb |
Purified, spore surface | |||
| Diffuse surface material (exosporium?) | Bulk | SLM | ||
| Anti-CotA | No labeling | No labeling | No labeling | Weak |
| Anti-CotB | Weak | Strong | Strong | No labeling |
| Anti-CotCB | Weak | Weak | Weak | Weak |
| Anti-CotD | Weak | No labeling | No labeling | Strong |
| Anti-CotE(N) | Strong | Weak | Strong | Strong |
Summary of IEM labeling of wild-type 630 spores; weak, less than 50% spores labeled, 0 to 1 particles/spore section; strong, more than 75% spores labeled, up to 10 particles/spore section.
Enzyme activities.
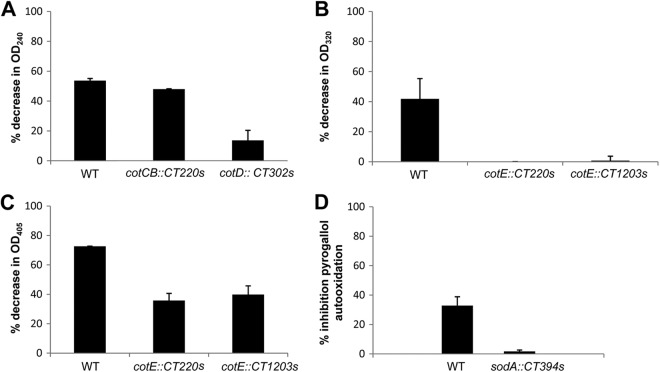
Using purified wild-type (630Δerm) and mutant spores, we measured enzyme activity (Fig. 7). For catalase activity, the cotCB::CT220s mutant showed a small reduction in activity compared to that of 630Δerm, but for the cotD::CT302s mutant, the reduction was more significant (Fig. 7A). Since there are three putative manganese catalases (CotCB, CotD, and CotG) on the spore coat, we would not necessarily expect either the CotCB or CotD mutant to show a full reduction in enzyme activity due to functional redundancy. Moreover, each protein may be displayed differently (e.g., partially hidden), but clearly the cotD mutant demonstrated a noticeable reduction in activity. For CotE, there are two enzyme activities, a peroxiredoxin activity and a chitinase activity. As shown in Fig. 7B, both cotE mutants abolished peroxiredoxin activity, and for chitinase activity, a 50% reduction was observed (Fig. 7C). Finally, for the sodA::CT394s mutant, SOD activity was abolished (Fig. 7D).
Fig 7.
Spore-associated enzyme activity. Enzyme assays were made using purified spores of wild-type 630Δerm or isogenic ClosTron mutants carrying insertions in cotCB (cotCB::CT220s), cotD (cotD::CT302s), sodA (sodA::CT394s), or the cotE amino terminus (cotE::CT220s) or carboxy terminus (cotE::CT1203s). (A) Catalase activity; (B) peroxiredoxin activity; (C) chitinase activity; (D) superoxide dismutase activity.
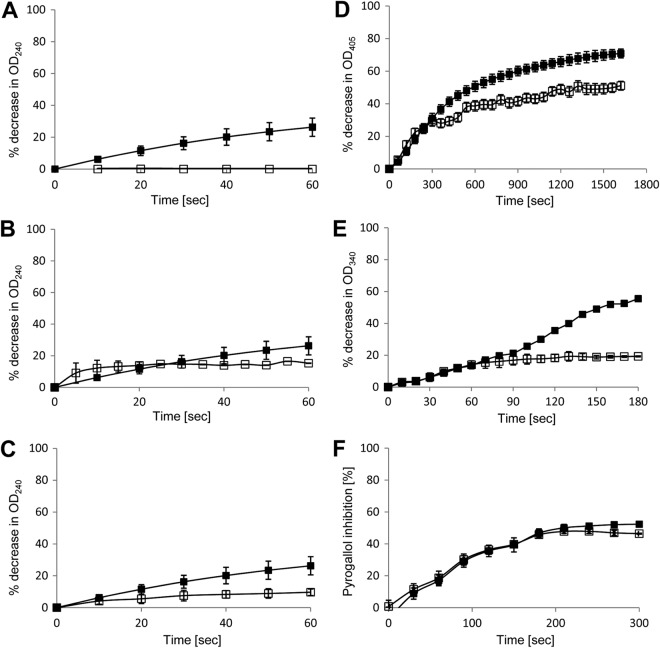
We expressed and purified the enzymatic spore coat proteins as His-tagged recombinant proteins in E. coli. After purification, these proteins were used in enzyme assays (Fig. 8). Using positive-control enzymes, we were able to demonstrate that rCotD carried catalase activity (Fig. 8B; specific activity [SA], 796 U/mg) while rCotG displayed a small amount of activity (Fig. 8C; SA, 431 U/mg). No activity was found for rCotCB (Fig. 8A). The carboxy-terminal domain (rCotEC) carried substantial levels of chitinase activity (Fig. 8D; SA, 10 U/mg), and the amino-terminal domain of CotE (rCotEN) carried a low level of peroxiredoxin activity (Fig. 8E; SA, 146 U/mg). Finally, rSodA carried high levels of SodA activity (Fig. 8F; SA, 12 U/mg).
Fig 8.
Enzyme activities of recombinant proteins. Purified recombinant proteins (□) were assessed for enzyme activity in parallel with positive-control enzymes (■). Experiments were repeated two times with examples shown. Panels A to C show catalase activity for rCotCB (A), rCotD (B), or rCotG (C). The control enzyme used was catalase from Aspergillus niger. Panel D shows chitinase activity of rCotEC and the control chitinase from Trichoderma viridae. Panel E shows peroxiredoxin activity of rCotEN, and the control enzyme is peroxiredoxin 1. Panel F shows superoxide dismutase activity of rSodA, and the control enzyme is superoxide dismutase from bovine erythrocytes. All enzyme assays were repeated at least two times, and standard deviations are shown.
The CotE chitinase is most similar to family 18 chitinases (41), which can be inhibited by pentoxyfilline (an anti-inflammatory drug derived from methylxanthine [42]). We showed that pentoxyfilline could inhibit the activity of rCotEC in a dose-dependent manner and, for comparison, that of a family 18 chitinase purified from Trichoderma viridae (Fig. 9).
Fig 9.
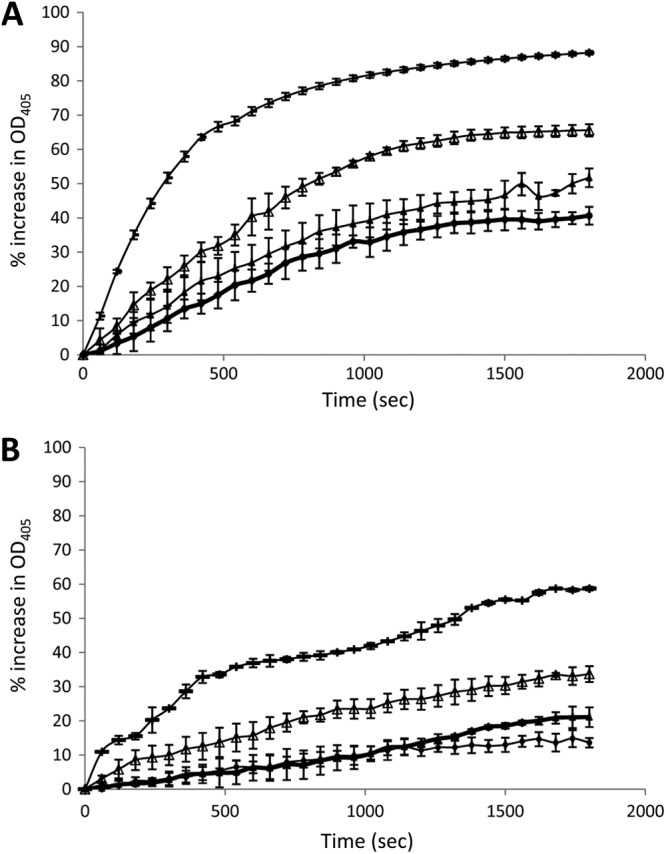
Inhibition of rCotEC chitinase activity. Trichoderma viridae chitinase (0.05 mg/ml) (A) or rCotEC (0.5 mg/ml) (B) was mixed with a 0 (-), 2 (△), 20 (▲), or 200 (●) mM concentration of the family 8 chitinase inhibitor pentoxifylline (P1784; Sigma) in PBS (pH 7.4), and chitinase activity was assessed as described in Materials and Methods.
DISCUSSION
The purpose of this study was to determine the functions of proteins found on the surface of C. difficile spores. Our rationale was that spores are particularly relevant to CDI for three reasons: first, their ability to disseminate and survive in the environment; second, the emergence of hypervirulent strains that produce higher levels of toxins; and finally, the interesting possibility that spores might exist in different infectious states. Using ClosTron mutagenesis, we have examined six genes (cotA, cotB, cotCB, cotD, and cotE) which encode proteins found on the spore coat and one, sodA, which we had previously hypothesized could encode a spore coat protein. Of the corresponding mutants, only the cotA mutant exhibited a clear functional defect in spore coat assembly. In this mutant, spores were clearly sensitive to ethanol and lysozyme, which is a clear functional indicator that the spore coat has been disturbed. TEM analysis revealed an unusual phenotype where half of the spores examined were clearly defective, with spore coats apparently lacking the electron-dense outer coat layer. This implies that CotA may play an important role in assembly of the outer layer of the spore coat. Parallels exist in B. subtilis, for which spore coat assembly is better understood (17). Here, the C. difficile cotA mutant resembles the cotE mutant of B. subtilis, which also fails to assemble the outer electron-dense layer and has a lysozyme-sensitive phenotype (43). With B. subtilis CotE, at least 24 proteins are assembled on the spore in a CotE-dependent manner (44), so it is possible that CotA may also play a key role in outer coat assembly. We found here that CotA and CotCB appear to exists in multimeric forms, which was previously unnoticed. For CotA, the predicted molecular mass of this protein is 34 kDa, but it runs in SDS-PAGE at ∼51 kDa. In our analysis, we observed a high-molecular-mass species of ∼100 kDa, and in mutants lacking the CotB, CotCB, CotD, and CotE proteins, an additional polypeptide species of about 58 kDa was apparent. Multimeric spore coat proteins have been observed before, and in B. subtilis the 12-kDa CotC protein is present as multimers of 21 and 30 kDa (45). With B. subtilis CotC, oligomerization may be linked to posttranslational modification of CotC monomers due to their unusually high tyrosine content of CotC (∼30%). Neither CotA nor CotCB appear to carry any obvious feature within the amino acid sequence that might indicate how these proteins may be posttranslationally modified, but in the case of CotA, due to its functional phenotype, we believe it probably plays an important role in assembly of the outer layers of the spore coat. One important question, though, is why half of the cotA mutant spores we examined do not exhibit a phenotype. One possibility is oligosporogeny, a phenomenon where, despite a defective gene product, sporulation can still proceed to completion in a proportion of cells (46). Oligosporogenous mutants usually produce very low levels of wild-type spores and rarely above 1 to 5%, so we are surprised if this is the case here. A second possibility is that what we observed as wild-type spores are in fact still defective but appear under TEM as wild type. Using IEM analysis, we were also able to show that at least for CotA, this protein was located in the outer layer of pure spores and not in the diffuse surface layer. CotB, on the other hand, appeared to be located only in the diffuse surface layer, while CotB, CotCB, and CotD could not be suitably resolved to either layer.
EM analysis also revealed that the state of spore purification markedly affected the spore coats. Spores that had been harvested from solid agar and washed three times with water retained what appeared to be a diffuse surface layer, as well as considerable material detached from the spore. This layer was completely lost upon further purification using a proteinase K buffer. This shows that the spore surface layers are particularly fragile and can easily be lost. We believe the diffuse surface material may be the exosporium that has been described for C. difficile. Images of 630 spores grown in liquid show a classical, electron-dense, sac-like structure, referred to as an exosporium (12). In our previous study, spores grown on solid agar showed an electron-dense outer layer that was partially detached from the spore, but as noted, this was rarely observed (18). In both of these studies, a thin, electron-dense layer can be discerned between the inner lamellar layer and the putative exosporium. In the images we present here (Fig. 5), we can identify a thick lamellar layer, a more electron-dense outer coat layer, and then the diffuse surface material, which is removed by further purification. Alternatively, we believe this material may comprise the exosporium, which is either not completely formed or alternatively partially degraded and thus consisting of remnants. Whatever the explanation, it is clear that considerable work needs to be done to define the precise nature of the exosporium, and we suspect that the appearance of the exosporium is very much dependent on the growth conditions, as well as the purification procedure.
The most interesting aspect of the spore surface layers is the presence and confirmation of enzymatic activity. These assays carry a number of potential caveats: for example, do the recombinant proteins possess full activity, and are the reaction conditions optimal, i.e., have we identified the correct enzyme substrate? For example, some chitinases can also recognize cellulose, so it is formally possible that we have yet to identify the true substrate for CotE (47). Despite this, it is clear that CotD and CotG carry catalase activity, SodA carries SOD activity, and CotE carries both peroxiredoxin and chitinase activities. SodA most likely plays a role in polymerizing spore coat protein monomers (by oxidative cross-linking) in the presence of H2O2 (48), and we have shown that being located in the spore coat of C. difficile spores, SodA would be well placed to perform this function. In B. subtilis, SodA is thought to cross-link tyrosine-rich coat protein monomers (48), and C. difficile CotF, which we have shown here to be located in the outer spore coat, is a likely candidate.
In summary, we have confirmed the surface location of eight spore coat proteins from C. difficile and have shown for three (CotD, CotE, and SodA) that they carry enzymatic activity. We have also identified CotA as one protein that is subject to posttranslational modification and is likely to play a major role in stabilizing the outer spore coat. The most immediate question now is to determine which of these proteins are located in the exosporium, with only CotA being most likely to have a spore coat, rather than an exosporial, location. The enzymatic proteins, CotD, SodA, and CotE may also be located in the exosporium. In support of this, exosporia in other spore-forming species have been shown to carry a number of enzymatic proteins (49), and deletion of the C. difficile proteins showed no apparent spore structural defect, unless of course these proteins were structurally redundant.
Supplementary Material
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02104-12.
REFERENCES
- 1. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 2. Gerding DN, Muto CA, Owens RC., Jr 2008. Measures to control and prevent Clostridium difficile infection. Clin. Infect. Dis. 46(Suppl 1:S43–S49 [DOI] [PubMed] [Google Scholar]
- 3. Vonberg RP, Kuijper EJ, Wilcox MH, Barbut F, Tull P, Gastmeier P, van den Broek PJ, Colville A, Coignard B, Daha T, Debast S, Duerden BI, van den Hof S, van der Kooi T, Maarleveld HJ, Nagy E, Notermans DW, O'Driscoll J, Patel B, Stone S, Wiuff C. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin. Microbiol. Infect. 14(Suppl 5):2–20 [DOI] [PubMed] [Google Scholar]
- 4. Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eggertson L, Sibbald B. 2004. Hospitals battling outbreaks of C. difficile. CMAJ 171:19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 7. McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 8. Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192:4904–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuijper EJ, van Dissel JT, Wilcox MH. 2007. Clostridium difficile: changing epidemiology and new treatment options. Curr. Opin. Infect. Dis. 20:376–383 [DOI] [PubMed] [Google Scholar]
- 10. Vohra P, Poxton IR. 2011. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology 157:1343–1353 [DOI] [PubMed] [Google Scholar]
- 11. Burns DA, Heeg D, Cartman ST, Minton NP. 2011. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One 6:e24894 doi:10.1371/journal.pone.0024894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191:5377–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haraldsen JD, Sonenshein AL. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48:811–821 [DOI] [PubMed] [Google Scholar]
- 14. Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. 1990. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell 62:239–250 [DOI] [PubMed] [Google Scholar]
- 15. Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 16. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 17. Henriques AO, Moran CP., Jr 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588 [DOI] [PubMed] [Google Scholar]
- 18. Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM. 2011. Surface layers of Clostridium difficile endospores. J. Bacteriol. 193:6461–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Derm) and demonstration that the conjugative transposon Tn916DE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137–141 [DOI] [PubMed] [Google Scholar]
- 20. Wust J, Sullivan NM, Hardegger U, Wilkins TD. 1982. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol. 16:1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paredes-Sabja D, Bond C, Carman RJ, Setlow P, Sarker MR. 2008. Germination of spores of Clostridium difficile strains, including isolates from a hospital outbreak of Clostridium difficile-associated disease (CDAD). Microbiology 154:2241–2250 [DOI] [PubMed] [Google Scholar]
- 22. Permpoonpattana P, Hong HA, Phetcharaburanin J, Huang JM, Cook J, Fairweather NF, Cutting SM. 2011. Immunization with Bacillus spores expressing toxin A peptide repeats protects against infection with Clostridium difficile strains producing toxins A and B. Infect. Immun. 79:2295–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 24. Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85 [DOI] [PubMed] [Google Scholar]
- 25. Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80:49–55 [DOI] [PubMed] [Google Scholar]
- 26. Perutka J, Wang W, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 336:421–439 [DOI] [PubMed] [Google Scholar]
- 27. Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899 [DOI] [PubMed] [Google Scholar]
- 28. Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286:27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong HA, Khaneja R, Tam NM, Cazzato A, Tan S, Urdaci M, Brisson A, Gasbarrini A, Barnes I, Cutting SM. 2009. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 160:134–143 [DOI] [PubMed] [Google Scholar]
- 30. Marklund S, Marklund G. 1974. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 47:469–474 [DOI] [PubMed] [Google Scholar]
- 31. Attar F, Keyhani E, Keyhani J. 2006. A comparative study of superoxide dismutase activity assays in Crocus sativus L. Corm. Appl. Biochem. Microbiol. 42:101–106 [PubMed] [Google Scholar]
- 32. Greetham D, Grant CM. 2009. Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol. Cell. Biol. 29:3229–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Logan C, Mayhew SG. 2000. Cloning, overexpression, and characterization of peroxiredoxin and NADH peroxiredoxin reductase from Thermus aquaticus. J. Biol. Chem. 275:30019–30028 [DOI] [PubMed] [Google Scholar]
- 34. Mizobata T, Kagawa M, Murakoshi N, Kusaka E, Kameo K, Kawata Y, Nagai J. 2000. Overproduction of Thermus sp. YS 8-13 manganese catalase in Escherichia coli production of soluble apoenzyme and in vitro formation of active holoenzyme. Eur. J. Biochem. 267:4264–4271 [DOI] [PubMed] [Google Scholar]
- 35. Sekiya M, Mulcahy G, Irwin JA, Stack CM, Donnelly SM, Xu W, Collins P, Dalton JP. 2006. Biochemical characterisation of the recombinant peroxiredoxin (FhePrx) of the liver fluke, Fasciola hepatica. FEBS Lett. 580:5016–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duc LH, Hong HA, Fairweather N, Ricca E, Cutting SM. 2003. Bacterial spores as vaccine vehicles. Infect. Immun. 71:2810–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duc LH, Hong HA, Uyen NQ, Cutting SM. 2004. Intracellular fate and immunogenicity of B. subtilis spores. Vaccine 22:1873–1885 [DOI] [PubMed] [Google Scholar]
- 38. Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, Dougan G. 2010. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl. Environ. Microbiol. 76:6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei J, Shah IM, Ghosh S, Dworkin J, Hoover DG, Setlow P. 2010. Superdormant spores of Bacillus species germinate normally with high pressure, peptidoglycan fragments, and bryostatin. J. Bacteriol. 192:1455–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panessa-Warren BJ, Tortora GT, Warren JB. 1997. Exosporial membrane plasticity of Clostridium sporogenes and Clostridium difficile. Tissue Cell 29:449–461 [DOI] [PubMed] [Google Scholar]
- 41. Henrissat B, Davies GJ. 2000. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124:1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rao FV, Andersen OA, Vora KA, Demartino JA, van Aalten DM. 2005. Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes. Chem. Biol. 12:973–980 [DOI] [PubMed] [Google Scholar]
- 43. Zheng L, Donovan WP, Fitz-James PC, Losick R. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047–1054 [DOI] [PubMed] [Google Scholar]
- 44. Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487–502 [DOI] [PubMed] [Google Scholar]
- 45. Isticato R, Esposito G, Zilhao R, Nolasco S, Cangiano G, De Felice M, Henriques AO, Ricca E. 2004. Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J. Bacteriol. 186:1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piggot PJ, Coote JG. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanaka T, Fujiwara S, Nishikori S, Fukui T, Takagi M, Imanaka T. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microbiol. 65:5338–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henriques AO, Melsen LR, Moran CP., Jr 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henriques AO, Moran CP. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95–110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.