Abstract
Germination of Bacillus subtilis spores can be triggered by the binding of specific nutrients, called germinants, to germinant receptors (GRs) in the spore's inner membrane. This interaction eventually initiates, with variable time delays, the release of dipicolinic acid and cations from the spore core—a key step in spore germination. The kinetics of this process are highly heterogeneous for individual spores. In this work, we sought to investigate how the germination heterogeneity was controlled. In particular, we tested whether the rates of germination were determined by GR levels, which vary from spore to spore due to stochastic gene expression. Both the expression levels of GRs and the germination rate were measured in single spores, and the experimental results were compared to theoretical predictions. Our results indicated that the variation in the expression levels of GRs was not the primary factor that controls spore germination heterogeneity. Two alternative hypotheses are discussed in light of this experimental discovery.
INTRODUCTION
The bacterium Bacillus subtilis forms spores under starvation in order to survive, and these spores resume vegetative growth through germination when suitable nutrients become available (1). Each spore contains a core region containing nucleic acid and protein that is, in essence, the spore protoplast, and the core is surrounded by an inner membrane and then a nascent germ cell wall, followed by a cortex, with the latter two layers composed largely of peptidoglycan (2). The cortex is, in turn, surrounded by an outer membrane that may not be intact in the dormant spore and then by the several layers of the proteinaceous spore coats (1).
A key step in the spore germination process is the release of the core's large depot (∼25% of the core [dry weight]) of dipicolinic acid (DPA) chelated to divalent cations, largely Ca2+ (Ca-DPA), and the uptake of water molecules. For nutrient-induced germination, Ca-DPA release requires the presence of the germinant receptors (GRs)–trimeric protein complexes (with A, B, and C subunits) situated in the inner spore membrane (3). Three functional GRs (GerA, GerB, and GerK) (4–6) have been identified in B. subtilis spores, and their orthologs have been found in other related spore-forming bacterial species (2, 3), but how GRs induce Ca-DPA release is not fully understood. Interestingly, a recent study showed that all GRs in B. subtilis spores colocalize within a nanometer-sized germinosome structure (7). Another inner membrane protein involved in germination, GerD, was also found to localize within this germinosome. Deletion of the gerD gene results in a dispersed distribution of GRs as well as a significantly lower rate of spore germination (8), suggesting that the structure of the germinosome may be important for the spore germination process.
Studies on individual spores of B. subtilis have shown that nutrient-induced germination is a two-step process. The release of most Ca-DPA follows sigmoidal kinetics and takes only a few minutes (9–13). However, this fast Ca-DPA release step is preceded by a time delay, termed Tlag, between the addition of a nutrient germinant and the rapid Ca-DPA release. Tlag varies significantly (from less than 1 min to many hours) among individuals in a population of isogenic spores under identical environmental conditions, and thus, variations in Tlag represent a form of pure phenotypic heterogeneity. Little is known about the root causes of this large heterogeneity in spore germination time (3), even though a mechanistic understanding would have practical significance to the food and medical product industries (14), because the elimination of spore-forming bacteria often can be carried out readily only after the spores germinate. One hypothesis, first proposed by Woese et al. in 1968 (15), is that the heterogeneity in germination times arises from stochastic variations in the steady-state expression levels of proteins required for the germination. This model assumes that the release of Ca-DPA during spore germination is triggered by the buildup of a molecule p to a threshold level with the pattern p = 0 → p = 1 → … . → p = n.
Furthermore, the rate of the buildup of molecule p is determined by the amount of a key protein that is essential for the process. Thus, if the expression of the key protein is at a low number, the stochastic variations in such levels may be large enough to lead to significant heterogeneity in spore germination. Among known proteins that are important for the germination process, the GRs are thought to be present in spores at relatively low levels (4, 5, 16). Therefore, we suspected that spore-to-spore variations in GR expression would be large enough to account for the broad heterogeneous distribution in the germination times. The purpose of the current study was to test this hypothesis by experimentally evaluating the correlation between single-spore germination times and their GR levels and by measuring the germination time heterogeneity among subpopulations of spores with relatively uniform levels of GRs.
MATERIALS AND METHODS
Strain and spore preparation.
The construction of the B. subtilis strains used for this work was described previously (7). These are all isogenic derivatives of strain PS4150, a mutant of wild-type strain PS832 lacking two genes (cotE and gerE) in spore coat assembly, which leads to a significantly decreased autofluorescence background in these spores. Each strain has a GR protein translationally fused to a green fluorescent protein (GFP) derivative (7) expressed from its native promoter. These mutant strains are (i) strain KGB174, which lacks the wild-type gerA operon and has a gerAA-mCherry gerAB gerAC operon downstream of the native gerA promoter at the amyE locus, (ii) strain KGB202, which lacks the wild-type gerB operon and has a gerBA-gfp gerBB gerBC operon downstream of the native gerB promoter at the amyE locus, and (iii) strain KGB08, obtained by inserting the GFP coding sequence at the end of the wild-type gerKB gene through a double crossover. Spores from all three strains germinate relatively normally with appropriate nutrients, indicating that the fusion proteins are functional. All GRs were expressed from their respective native promoters; therefore, the expression levels are expected to be comparable to those in the wild-type strain.
Spores of B. subtilis strains were prepared on 2 SG agar plates without antibiotics at 37°C, and spores were incubated, harvested, and cleaned as described previously (17). All spores used in this work were free (>98%) of growing or sporulating cells, germinated spores, and cell debris, and spores were stored at 4°C protected from light.
Microscopy and spore germination.
For microscopy experiments, spores were first heat activated for 30 min at 75°C and then cooled on ice for at least 15 min. The heat-activated spores were spread on 0.1% polylysine-coated glass dishes (18). Levels of GRs in spores on the polylysine-coated glass dishes were measured with epifluorescence microscopy as described previously (7). To quantify the steady-state expression levels of GRs, we took 500 frames of fluorescent images. The image stacks were examined, and the initial segments of the images, for which no significant photobleaching had taken place (<10% drop of fluorescent signal, typically ∼80 to 200 images depending on the strain), were averaged to give a final fluorescence image. The relative levels of each GR were quantified by fitting the fluorescent foci from each spore with the following two-dimensional (2D) Gaussian function:
where I is the image intensity, H is proportional to the level of GR, B is the background signal, and σ is the width of the point spread function of the microscope.
To obtain the germination times of individual spores, germinants were added after the fluorescence images described above were taken. Differential interference contrast (DIC) time-lapse images were recorded until most spores underwent germination. Germination times (Tlag), defined as the time delay between the addition of germinant and the time the DIC signal drops by 50% in pixel intensity, were obtained for each spore using an automated image analysis algorithm as previously described (13). Unless otherwise specified, spores of strain KGB174 were germinated at 25°C in 50 mM Tris-HCl buffer (pH 8.6) plus 10 mM l-valine, and spores of strains KGB202 and KGB08 were germinated at 25°C in AGFK (10 mM [each] l-asparagine, d-glucose, and d-fructose and 10 mM K-HEPES buffer, pH 7.4). These germinant concentrations are all saturating for the appropriate GRs. DIC images were taken with a time lapse using an Olympus IX81 microscope equipped with a ×60 microscope objective (NA = 1.45; Olympus), with frame rates of 1 to 2 frames/min. Image analysis was carried out in ImageJ and Matlab. For strains KGB08, KGB174, and KGB202, the numbers of spores analyzed are 3,769, 4,316, and 2,237, respectively.
RESULTS
Stochastic expression of GRs in B. subtilis spores.
The levels of specific GRs in individual dormant spores were measured using GRs fused to fluorescent proteins. For GerA and GerB GRs, we used strains in which the A subunit of the receptor was fused with mCherry (for GerA) or GFP (for GerB). For GerK expression, the B subunit was labeled with GFP (see Materials and Methods) (7).
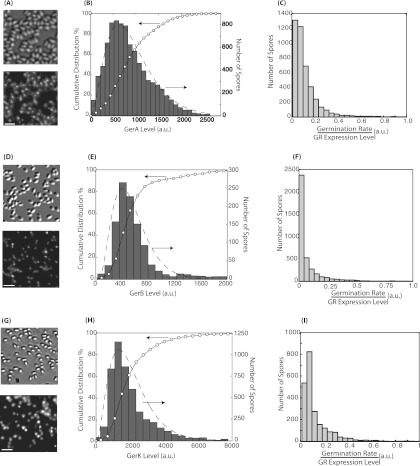
Figure 1A, D, and G shows the fluorescence images of spores of the various strains prior to the addition of germinants and the corresponding bright-field images. As reported previously (7), the fluorescence signal from each spore was confined within a diffraction-limited spot. The intensity of each diffraction-limited spot was quantified for individual spores (see Materials and Methods), and histograms were plotted (Fig. 1B, E, and H). In all cases, isogenic spores with >10-fold differences in GR expression were able to be easily identified, consistent with the hypothesis of a large expression heterogeneity for these likely low-abundance proteins. To quantify the exact amplitudes of the heterogeneity in GR expression, we used the coefficient of variation (CV) with the equation CV = standard deviation/mean, where CV is a unitless parameter that varies from 0 to infinity. We obtained CVs of 0.58 for GerAA-mCherry expression, 0.64 for GerBA-GFP expression, and 0.66 for GerKB-enhanced GFP (EGFP) expression. These values are comparable to CVs for germination times measured from the same spores (Table 1), a finding which suggests that, at least in principle, the stochastic variations in GR expression are large enough to account for the heterogeneity in spore germination.
Fig 1.
Quantification of the stochastic expression of GerA (A to C), GerB (D to F), and GerK (G to I). Bright-field (top) and fluorescence (bottom) images of KGB174 (A), KGB202 (D), and KGB08 (G) spores show the clustered localization of the GRs and variations in GR expression levels among spores. Bar, 3 μm. The distributions of GerA (B), GerB (E), and GerK (H) expression levels in individual spores were quantified from the analysis of the fluorescence intensities of individual spores. The histograms of the GR levels are plotted with gray bars (right axes). The open circles represent the corresponding cumulative distributions (left axes). The dashed lines are the maximum likelihood fit to a Gamma distribution function. The distributions of the ratio of the germination rate and the GR expression level measured for individual spores are shown in panels C (l-valine germination rate/GerA expression level), F (AGFK germination rate/GerB expression level), and I (AGFK germination rate/GerK expression level). The broad distributions in panels C, F, and I indicate that the relationship between the germination rate and GR levels cannot be modeled by a simple linear function.
Table 1.
Stochastic expression of GRs and CVs of spore germination times
| GR | CV | k | θ | CV of the germination time | Strain used |
|---|---|---|---|---|---|
| GerA | 0.58 | 3.07 ± 0.13 | 11.2 | 0.72a | KGB174 |
| GerB | 0.64 | 3.63 ± 0.14 | 9.5 | 0.53b | KGB202 |
| GerK | 0.68 | 2.94 ± 0.50 | 18.8 | 0.91b | KGB08 |
Germinated with l-valine.
Germinated with AGFK.
The probability distributions of low-level protein expression in prokaryotic systems can often be modeled with the following Gamma distribution function (19) based on the assumption that proteins are produced in random bursts:
in which g denotes the expression level, k is the shape parameter which is an estimator for the rate of bursts, and θ is the scale parameter and an estimator for burst sizes. It has been experimentally shown in Escherichia coli, for example, that the expression of many proteins can be described with this model (20). Therefore, we fitted the distributions of the expression of GerA, GerB, and GerK to a Gamma distribution function using the maximum likelihood method (Fig. 1B, E, and H, dashed line). We found that the data from all three GRs can be well fitted with the Gamma distribution (Fig. 1), suggesting that the variations in expression levels are likely also due to the burst protein production mechanism. The resulting fitting parameters are summarized in Table 1.
Germination rates of individual spores.
The spores of all three fluorescently labeled strains used in this study germinated with the appropriate nutrients, indicating that the chimeric GR subunits produced were functional, as found previously (7). However, the germination kinetics were somewhat altered from those of spores with the wild-type GRs. For KGB174 spores (expressing GerAA-mCherry), germination with l-alanine, which requires the GerA receptor, was 2-fold faster than that for the parent strain (PS4150) (7). For KGB202 spores (expressing GerBA-GFP) and KGB08 spores (expressing GerKB-GFP), germination was 2- to 3-fold slower than that of the parent strain (PS4150) using AGFK, which requires both the GerK and GerB GRs (7).
After the fluorescence levels of individual spores were measured, germinants were added and the germination times of the individual spores were measured using DIC microscopy. After a variable delay time (Tlag), the DIC intensity of an individual spore underwent a sudden drop, corresponding to the change in refractive index upon Ca-DPA release (8). However, in ∼50% of spores, there was a second, slower drop in DIC intensity that was able to be resolved from the first with a few minutes separation, most likely corresponding to the hydrolysis of the spore cortex and the accompanying swelling of the spore core (8). We note that due to the deletion of spore coat protein (cotE and gerE mutations), the cortex hydrolysis rate in these strains may be lower than that in wild-type spores (7, 21). However, we do not expect the Ca-DPA release time to be significantly altered, because previous research has shown that the Ca-DPA release and cortex hydrolysis were controlled by independent mechanisms (22).
For the spores that germinated in the measurement period, we defined the germination rate as the inverse of the Tlag with the equation R = 1/Tlag.
According to the model of Woese et al. (15), R = α × g, where α is a constant and g is the expression level of the protein. To test if such a linear relationship exists, we computed for each spore the ratio of R to g and examined the distribution of these values (Fig. 1C, F, and I). With l-valine as the germinant, we used the level of GerA as g (Fig. 1C). Instead of obtaining a narrow distribution around a constant, α, we found that the ratio is asymmetrically distributed, showing a long tail, indicating that the germination rate cannot be described by a simple linear function of GerA expression. Using AGFK as the germinant, we examined effects of both GerB expression (Fig. 1F) and GerK expression (Fig. 1I) on spore germination rates and obtained similarly broad and asymmetric distributions, indicating that spores' germination rate with AGFK is not a simple linear function of either the GerB or GerK level. However, it remains possible that the spore germination rate with AGFK is determined by a complex combinatorial function of the levels of both GerB and GerK.
Effect of GR expression level on average germination rate.
To test if the expression level of GRs had any effect on spore germination rates at all, we divided the total spore population into four percentile subgroups according to the expression levels of the specific GRs: 0 to 25th, 25th to 50th, 50th to 75th, and 75th to 100th percentiles. If GR expression level determines the germination rate, we would expect the spore subgroups with lower expression to have a lower germination rate. The median germination rate for each subpopulation was then plotted against the median GR expression level (Fig. 2). The results clearly show that, for l-valine-induced germination, spores belonging to the highest GerA expression quantile germinated significantly faster than spores that belong to the lowest quantile (P < 0.01). Similarly, for AGFK-induced germination, spores with the highest GerK levels germinated faster (P < 0.01). In contrast, for GerB expression, the germination rates were not significantly different between different quantiles (Fig. 2). Both GerB and GerK are needed for AGFK-induced germination. However, our results suggest that GerK is the rate-limiting one. In all cases, the correlation seems to approach a saturation level at higher expression, a finding which is consistent with effects of GR overexpression on spore germination rates (23) that showed that, for example, overexpression of either GerB or GerK does not increase rates of AGFK germination. Combined with these previous findings, we concluded that increasing GR (especially GerA and GerK) numbers increase the rate of nutrient-induced germination, but only to a point, because the effects saturate at high expression levels.
Fig 2.
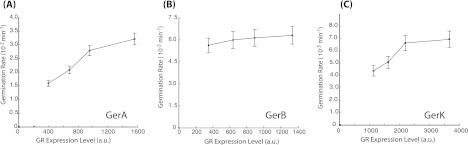
Correlation between germination rates and GR expression levels. The median germination rate with l-valine (A) or AGFK (B and C) for subsets of spores sorted by their expression levels of GerA (A), GerB (B), or GerK (C) GR was measured and plotted against the median expression level of the corresponding GR. Error bars indicate the standard errors of the mean (SEM).
Effect of GR expression level on germination heterogeneity.
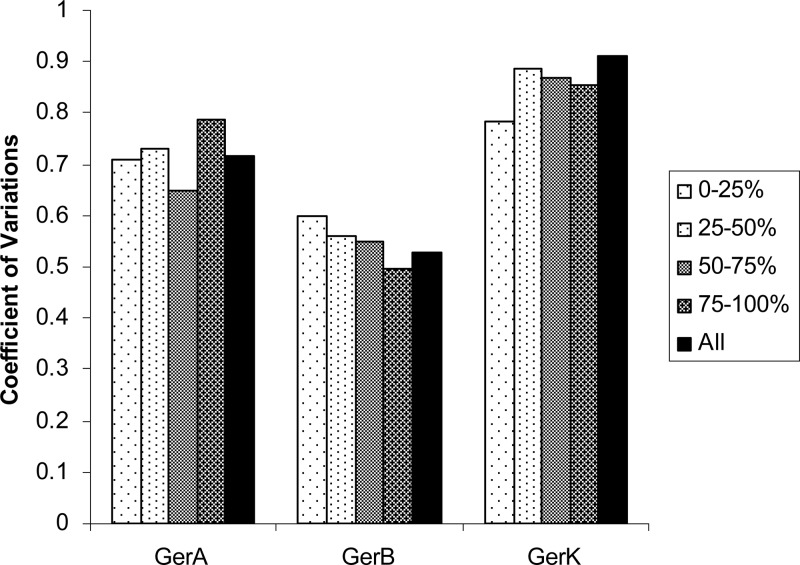
The heterogeneity of a kinetic process can be quantitatively evaluated using the coefficient of variation (CV) of the distribution. It is important to note that the heterogeneity, which is the spread of a distribution, can be completely independent of the average value of the distribution. Therefore, the fact that the average germination rate is dependent on GerA and GerK expression levels does not necessarily indicate that the heterogeneity has a similar dependence. To test if the stochastic expression of GRs is a major cause of germination heterogeneity, we again divided the total spore population into four percentile subgroups according to the expression levels of the specific GRs, 0 to 25th, 25th to 50th, 50th to 75th, and 75th to 100th percentiles, and computed the coefficient of variation (CV) of germination times for each of the four subgroups of spores. Since the GR expression of each subgroup is significantly narrower than that of the overall population, we expected the CV of germination times of subgroups to be smaller than that of the whole population if heterogeneity of germination is due mainly to the variations in the GR expression level. Interestingly, as shown in Fig. 3, we found no significant reduction of germination time heterogeneity when examining individual subgroups for all three strains we measured. Dividing the spores into even finer subpopulations (e.g., eight subpopulations) yielded no improvement in germination heterogeneity (data not shown). Therefore, we concluded that the stochastic expression of GRs is not a significant factor in the heterogeneity of spore germination rates.
Fig 3.
Germination heterogeneity is not correlated with GR expression levels. Total spore populations were divided into four subpopulations (0 to 25th percentiles, 25th to 50th percentiles, 50th to 75th percentiles, and 75th to 100th percentiles) based on their GerA (KGB174), GerB (KGB202), or GerK (KGB08) expression level. The CV (coefficient of variation) of each subpopulation was computed and compared with the overall CV. The results showed that populations of spores with a narrower distribution of GR expression did not show reduced heterogeneity of germination times.
Non-Gaussian germination time distribution.
Next we examined the exact distribution of germination times for each subgroup of spores. If heterogeneity is controlled mainly by the GR expression level, reducing the GR expression variations (as in each subgroup) would result in a narrow distribution of germination times that approaches a Gaussian function. Furthermore, the peak of the distribution should be inversely dependent on the average GR expression level of each subgroup, at least for GRs that affect the germination rate (GerA and GerK). The results shown in Fig. 4 instead show that these distributions are highly non-Gaussian. The dominant feature of the distributions is a decay that follows roughly an exponential function (note the semilog scale). For strains KGB174 and KGB08, the distributions peaked very early in time (∼30 min for KGB174 and <45 min for KGB08) and exhibited no significant dependence on GR expression levels. Interestingly, for strain KGB202, the distributions peaked at a relatively later time (60 to 120 min) and the peak positions seem to be dependent on the expression level of GerB.
Fig 4.
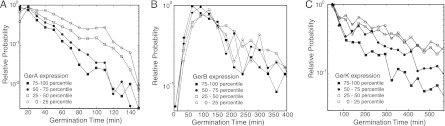
Non-Gaussian distribution of germination time in spore populations with narrow distributions of GR expression. Germination times were plotted for subpopulations of spores (0 to 25th percentiles, 25th to 50th percentiles, 50th to 75th percentiles, and 75th to 100th percentiles) based on their GerA (A), GerB (B), or GerK (C) expression level. Each subpopulation has a significantly more narrow distribution of corresponding GR expression than the overall population, and the germination time distributions show significant non-Gaussian characteristics.
The shape of these distributions is consistent with the observation that the average germination rate is dependent on GR expression but the germination heterogeneity is not. In the case in which l-valine is used as a germinant, for example, the distribution of germination times from spores with higher GerA expression decays faster, resulting in a smaller average value as well as a narrower overall distribution. Since the heterogeneity measure, CV, is the ratio between the width of the distribution and the average value of the distribution, it remains relatively constant. In fact, if the distribution is truly exponential, the CV would be constant (CV = 1) and independent of the average value of the distribution.
The non-Gaussian shape of the distribution is inconsistent with our original hypothesis that the germination time is the time it takes for an n-step buildup process, in which the number of a hypothetical substance p increases during germination until it reaches a threshold value n. Assuming that the rate of an individual reaction step is relatively constant for each subpopulation and that spores within each subgroup have similar expression levels, the central-limit theorem would predict that the overall reaction time should follow a Gaussian distribution as long as n is a large number. Thus, our results indicated that either reaction rates were not constant among spores (i.e., reaction rates are controlled by other unknown proteins) or n is a very small number (i.e., not a “buildup” process).
DISCUSSION
The primary goal of this study was to test if the stochastic variation in the GR expression levels is sufficient to explain the heterogeneity in rates of spore germination. We found that the spore-to-spore variation in GR expression was indeed large and the variation indeed contributes to the heterogeneity in germination time, at least partly, but does not seem to be the determining factor in spore germination heterogeneity. On average, spores with higher levels of GerA expression tend to germinate faster under l-valine and spores with higher levels of GerK expression tend to germinate faster under AGFK. These results indicate that GRs are involved in the rate-limiting step of the overall germination process. On the other hand, an important observation from our experiment is that even the spores with relatively uniform GR levels still germinated with very heterogeneous rates.
What, then, controls the single-spore germination time? Based on our experimental observations, we suggest two possibilities. First, it is entirely possible that the stochastic expression of another unknown protein molecule, presumably one also expressed at low levels, causes the variations in germination time. However, if such a protein exists, it is unlikely to be one of the known proteins involved in the germination. The GerD protein, for example, is important for fast nutrient-triggered spore germination (1, 3, 8). However, the level of GerD in spores is believed to be ∼10- to 100-fold higher than those of the GRs (24); thus, the heterogeneity in GerD expression should be much smaller. Furthermore, overexpression of GerD has little effect on the average germination time, indicating that the expression level of GerD is not rate limiting in the germination process. A set of lytic enzymes, SleB and CwlJ (1), is also important for germination by degrading the cortex layer. However, this degradation step happens after Ca-DPA release during germination and thus is not likely to be the rate-limiting step. Finally, the various SpoVA proteins thought to comprise the channel for Ca-DPA release in spore germination (1, 25) are present in spores at ∼103× higher levels than the GRs (26), and fluorescence microscopy of spores carrying SpoVA-GFP showed relatively constant fluorescence intensity in spores (data not shown), consistent with a relatively uniform expression level of SpoVA proteins. Unfortunately, the overall molecular mechanism of the nutrient-induced germination process is still not completely clear. Therefore, it is currently not possible to completely rule out that stochastic gene expression is indeed a key factor in spore germination time heterogeneity.
Another possible mechanism for the spore germination time heterogeneity is that the rate-limiting step of germination is a single-step chemical process as opposed to a multistep buildup process. Single-step chemical processes produce particularly broad distributions of waiting times that follow a single-exponential distribution, and thus, a large heterogeneity may arise independent of the fluctuations in protein expression levels. A classic example of such heterogeneity is the decay of radioactive elements. While a radioactive element has a well-defined average lifetime, the waiting times of the reaction for individual atoms vary greatly, with a CV value of 1. Furthermore, our experimental data showed that the distribution of germination times for spores with similar GR expression can be approximated by a single-exponential distribution, consistent with such a mechanism. The single-step process may be related to an activation of the germinosome, such as an allosteric structural rearrangement of the cluster. Such a highly cooperative transition step is conceivable in light of the finding that the molecules important for germination colocalize within a germinosome structure and may be in direct contact with each other. Additional information on the molecular arrangements within the germinosome may shed light on whether such an activation event is the cause of the observed heterogeneity in spore germination.
Footnotes
Published ahead of print 8 February 2013
REFERENCES
- 1. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 2. Setlow P. 1988. Small acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 42:319–338 [DOI] [PubMed] [Google Scholar]
- 3. Paidhungat M, Setlow P. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hudson KD, Corfe BM, Kemp EH, Feavers IM, Coote PJ, Moir A. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paredes-Sabja D, Setlow P, Sarker MR. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl. Environ. Microbiol. 75:3813–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81(4):1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang G, Yi X, Li YQ, Setlow P. 2011. Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins, which are important in spore germination. J. Bacteriol. 193:2301–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen D, Huang SS, Li YQ. 2006. Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal. Chem. 78:6936–6941 [DOI] [PubMed] [Google Scholar]
- 10. Huang SS, Chen D, Pelczar PL, Vepachedu VR, Setlow P, Li YQ. 2007. Levels of Ca2+-dipicolinic acid in individual Bacillus spores determined using microfluidic Raman tweezers. J. Bacteriol. 189:4681–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kong L, Zhang P, Setlwo P, Li YQ. 2010. Characterization of bacterial spore germination using integrated phase contrast microscopy, Raman spectroscopy, and optical tweezers. Anal. Chem. 82:3840–3847 [DOI] [PubMed] [Google Scholar]
- 12. Peng L, Chen D, Setlow Li YQ. 2009. Elastic and inelastic light scattering from single bacterial spores in an optical trap allows the monitoring of spore germination dynamics. Anal. Chem. 81:4035–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang P, Kong L, Wang G, Setlow P, Li YQ. 2010. Combination of Raman tweezers and quantitative differential interference contrast microscopy for measurement of dynamics and heterogeneity during the germination of individual bacterial spores. J. Biomed. Opt. 15:056010 doi:10.1117/1.3494567 [DOI] [PubMed] [Google Scholar]
- 14. Gould GW. 1969. Spore germination, p 397–444 In Gould GW, Hurst A. (ed), The bacterial spore. Academic Press, New York, NY [Google Scholar]
- 15. Woese CR, Vary JC, Halvorson HO. 1968. A kinetic model for bacterial spore germination. Proc. Natl. Acad. Sci. U. S. A. 59:869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moir A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526–530 [DOI] [PubMed] [Google Scholar]
- 17. Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England [Google Scholar]
- 18. Zhang J, Garner W, Setlow P, Yu J. 2011. Quantitative analysis of spatial-temporal correlations during germination of spores of Bacillus species. J. Bacteriol. 193:3765–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedman N, Cai L, Xie XS. 2006. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys. Rev. Lett. 97:168302. [DOI] [PubMed] [Google Scholar]
- 20. Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bagyan I, Setlow P. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Popham DL, Meador-Parton J, Costello CE, Setlow P. 1999. Spore peptidoglycan structure in a cwlD dacB double mutant of Bacillus subtilis. J. Bacteriol. 181:6205–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pelczar PL, Setlow P. 2008. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J. Bacteriol. 190:5635–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vepachedu VR, Setlow P. 2007. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol. Lett. 274:42–47 [DOI] [PubMed] [Google Scholar]
- 26. Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187:5677–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]