Abstract
Hfq is a global small RNA (sRNA) chaperone that interacts with Hfq-regulated sRNAs and functions in the posttranscriptional regulation of gene expression. In this work, we identified Hfq to be a virulence regulator in the Gram-negative fire blight pathogen Erwinia amylovora. Deletion of hfq in E. amylovora Ea1189 significantly reduced bacterial virulence in both immature pear fruits and apple shoots. Analysis of virulence determinants in strain Ea1189Δhfq showed that Hfq exerts pleiotropic regulation of amylovoran exopolysaccharide production, biofilm formation, motility, and the type III secretion system (T3SS). Further characterization of biofilm regulation by Hfq demonstrated that Hfq limits bacterial attachment to solid surfaces while promoting biofilm maturation. Characterization of T3SS regulation by Hfq revealed that Hfq positively regulates the translocation and secretion of the major type III effector DspE and negatively controls the secretion of the putative translocator HrpK and the type III effector Eop1. Lastly, 10 Hfq-regulated sRNAs were identified using a computational method, and two of these sRNAs, RprA and RyhA, were found to be required for the full virulence of E. amylovora.
INTRODUCTION
Erwinia amylovora is a Gram-negative bacterial plant pathogen and the causal agent of fire blight, a disease that occurs on rosaceous species, such as apples and pears. During infection, E. amylovora enters host plants through natural openings in flowers or shoot tips and is able to rapidly move within plant hosts in the vascular tissue and establish systemic infections. To date, many virulence factors of E. amylovora have been characterized, with the major determinants including the type III secretion system (T3SS), amylovoran exopolysaccharide production, biofilm formation, and motility (1). E. amylovora pathogenesis on apple trees is manifested through several distinct stages and interactions with living and nonliving host cells. The stigma surface of flowers is the primary site of multiplication of E. amylovora prior to infection of flowers through nectarthodes and internal invasion of the host (2). Infection of flowers by E. amylovora requires a functional T3SS, and motility is an important virulence factor affecting migration of cells downward from the stigma to the nectarthodes (3–5). The T3SS of E. amylovora is mainly required for the translocation of the effector protein DspE into plant cells; DspE is required for the pathogenicity of E. amylovora, multiplication in planta, and disease promotion by the alteration of host cell defenses (6–8). Genes associated with production of the T3SS and type III effector genes, including the alternate sigma factor hrpL, type III pilus hrpA, translocator hrpN, and effector dspE, are expressed at between 6 and 48 h of inoculation to flower stigmas (9), which correlates well with regulatory studies of the HrpL regulon performed in vitro (10). In addition to the T3SS and DspE, a third pathogenicity factor produced by E. amylovora is the exopolysaccharide amylovoran, which is an important component of biofilms (11, 12). One role of amylovoran is apparently to protect cells from exposure to antimicrobial compounds produced by the host in response to pathogen infection (11).
Infection of apple leaves and shoots by E. amylovora is a second common mode of infection that is initiated following cell entry through wounds. Leaf infection also requires the T3SS, as the initial interaction is with living parenchymal cells in leaf tissue. Systemic infection of apple and other hosts by E. amylovora is accomplished following invasion of xylem (the water-conducting tubes of plants), phloem, and the cortical parenchyma of stems. Xylem is composed of nonliving cells, and plant pathogens such as Xanthomonas albilineans that exclusively infect xylem do not possess a T3SS (13), suggesting that this determinant is of less importance for systemic infection by E. amylovora. In contrast, biofilm formation, which is not required for leaf infection, is critically important to the establishment of large cell populations in xylem tubes and to systemic movement of E. amylovora out of leaves and into apple stems (12, 14).
In order to successfully establish infections, E. amylovora utilizes a complicated regulatory network involving two-component signal transduction systems, alternate sigma factors, quorum sensing, and the second messenger cyclic di-GMP to collectively control the expression of virulence genes at different stages of infection (1, 4, 10, 15–18). In addition, the requirement at different times of infection for the T3SS and biofilm formation and the importance of cellular motility suggest that E. amylovora cells must be able to rapidly alter the production of distinct virulence factors in response to specific host cues.
One method utilized by bacteria to facilitate rapid responses to environmental changes is through the use of regulatory small RNAs (sRNAs). These noncoding RNAs range from 50 to 400 nucleotides (nt) and target specific mRNA transcripts in cells, effecting posttranscriptional regulation of the target mRNAs by either altering their translational efficiency or affecting mRNA stability, or both (19–21). The stability and functional activation of sRNAs are controlled by the RNA chaperone protein Hfq (22).
Hfq forms a hexameric ring structure and preferentially binds to U-rich sequences of sRNAs on the proximal side of its central core and to A-rich sequences on the distal face (23, 24). The sRNAs bound by Hfq target specific mRNAs in the bacterial cell and exert posttranscriptional regulatory effects. Two roles of Hfq in sRNA-mRNA interactions have been implicated: first, Hfq-sRNA binding enhances the stability of sRNAs; second, Hfq-sRNA binding also facilitates the imperfect base pairings of sRNAs to the 5′ untranslated regions (5′ UTRs) of their target mRNAs (20, 21, 25). The binding of sRNAs to the 5′ UTRs would either lead to translational repression of the target mRNAs, when the binding of sRNAs blocks the ribosomal binding site (RBS), or lead to translational activation, when the binding of sRNAs competes with inhibitory intramolecular base-pairing interactions (21). In addition, RsmB/CsrB-type sRNAs also play critical roles in the posttranscriptional regulation by binding to and sequestering the RsmA/CsrA (26, 27).
Because Hfq is a global sRNA chaperone and binds to sRNAs with diverse functions, its regulation in bacteria is often pleiotropic. Genes controlled by Hfq encode diverse traits, including cell membrane protein composition, cell surface structures, stress tolerance, motility, and sugar, nitrogen, and fatty acid metabolism (28). Recent reports have also shown that Hfq and sRNAs play important roles in virulence regulation in animal pathogens (28, 29). For example, in Vibrio cholerae, Hfq is required for intestinal colonization of suckling mice (30), and in enterohemorrhagic Escherichia coli (EHEC), Hfq negatively controls T3SS-encoding genes in the locus of enterocyte effacement (LEE) (31). A reduced-virulence phenotype in hfq mutants was also commonly observed in other Gram-negative pathogens, including Brucella abortus, Francisella tularensis, Neisseria meningitidis, Pseudomonas aeruginosa, and Yersinia pestis, and in the Gram-positive pathogen Listeria monocytogenes (28). Although Hfq has been implicated in controlling virulence in many bacterial pathogens, the regulatory targets of Hfq vary among species, from regulation of the T3SS (31, 32) and stress tolerance (33, 34) to biofilm formation (35). In some cases, the regulatory function of Hfq is still not clear (28).
Although the contribution of Hfq and Hfq-regulated sRNAs to virulence has been described in detail in bacterial pathogens of animals, the role of Hfq and Hfq-regulated sRNAs in virulence and host colonization has been reported in only one bacterial plant pathogen, Agrobacterium tumefaciens (36). We hypothesized that Hfq and Hfq-regulated sRNAs would regulate the critical components of pathogenesis in E. amylovora, including the T3SS and amylovoran exopolysaccharide biosynthesis. In this study, we constructed an hfq deletion mutant in E. amylovora Ea1189 and demonstrated that Hfq is an important virulence regulator. By combining virulence assays, electron microscopy analyses, and effector translocation and secretion assays, we showed that the virulence regulation of Hfq is exerted through its control of amylovoran biosynthesis, biofilm formation, motility, and the T3SS. Finally, 10 potential Hfq-regulated sRNAs were identified in the E. amylovora genome, and 2 of them were shown to be important for virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and culture conditions.
The bacterial strains and plasmids used in this study and their relevant characteristics are listed in Table 1. The sequences of the oligonucleotide primers used for cloning and mutations are listed in Table S1 in the supplemental material. All strains were stored at −80°C in 15% glycerol and cultured in Luria-Bertani (LB) medium at 28°C. For biofilm assays, strains were cultured in 0.5× LB broth. For Northern blot assays and protein secretion assays, strains were cultured in LB broth at 28°C overnight and then cultured in Hrp-inducing minimal medium (41), which induces the expression of the T3SS regulon. When required, antibiotics were added to the media at the following concentrations: gentamicin, 15 μg ml−1; chloramphenicol, 30 μg ml−1; kanamycin, 50 μg ml−1; and ampicillin, 100 μg ml−1.
Table 1.
Bacterial strains and plasmids used in this study and their relevant characteristics
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 λ− | Invitrogen |
| Erwinia amylovora | ||
| Ea1189 | Wild type | 37 |
| Ea1189Δhfq | hfq deletion mutant, Cmr | This study |
| Ea1189ΔhrpL | hrpL deletion mutant, Cmr | 10 |
| Ea1189Δams | Deletion of 12-gene ams operon, Cmr | 37 |
| Ea1189ΔT3SS | Deletion of 24-gene T3SS pathogenicity island, Kmr | 37 |
| Ea1189ΔryhA | ryhA sRNA deletion mutant, Cmr | This study |
| Ea1189ΔrprA | rprA sRNA deletion mutant, Cmr | This study |
| Ea1189Δspf | spf sRNA deletion mutant, Cmr | This study |
| Ea1189ΔmicA | micA sRNA deletion mutant, Cmr | This study |
| Ea1189ΔomrAB | omrAB sRNA deletion mutant, Cmr | This study |
| Ea1189ΔryhB | ryhB sRNA deletion mutant, Cmr | This study |
| Ea1189ΔsroB | sroB sRNA deletion mutant, Cmr | This study |
| Ea1189ΔryeA | ryeA sRNA deletion mutant, Cmr | This study |
| Ea1189ΔglmZ | glmZ sRNA deletion mutant, Cmr | This study |
| Plasmids | ||
| pKD4 | Apr Kmr, mutagenesis cassette template | 38 |
| pKD46 | Apr, expresses bacteriophage λ red recombinase | 38 |
| pML123 | RSF1010-derived expression and lac fusion broad-host-range vector, Gmr | 39 |
| pMLhfq | 530-bp fragment containing hfq with its native promoter cloned at XbaI/SacI in pML123, Gmr | This study |
| pLRT201 | pMJH20 expressing DspE(1-737)-CyaA | 40 |
| pLRT8 | pMJH20 expressing DspE(1-15)-CyaA | 40 |
Cmr, Kmr, Gmr, and Apr, chloramphenicol, kanamycin, gentamicin, and ampicillin resistance, respectively.
Deletion mutagenesis of hfq and sRNA-encoding genes.
E. amylovora chromosomal deletion mutants were constructed using the red recombinase method (38). Briefly, recombination fragments consisting of 50-nucleotide homology arms of flanking regions of hfq or sRNA-encoding genes flanking a chloramphenicol resistance cassette were amplified from the plasmid pKD4. PCR products were purified by gel purification and electroporated into E. amylovora Ea1189 expressing recombinase genes from the helper plasmid pKD46. Mutants were selected on LB medium amended with kanamycin. Mutations of target genes were confirmed by PCR and sequencing. Deletion of sRNA-encoding genes in E. amylovora was based on their sequence homologies to corresponding sRNA-encoding genes in E. coli.
Virulence assays.
The virulence of wild-type strain Ea1189 and mutant strains was tested using an immature pear fruit assay and an apple shoot assay as previously described (17, 42). Briefly, for the immature pear fruit assay, bacteria were inoculated on wounded immature pears at a concentration of 1 × 104 CFU ml−1, and the pears were incubated at 25°C under high-relative-humidity conditions. Lesion diameters were measured at 3, 5, and 7 days postinoculation. Bacterial populations within immature pear fruits were quantified at 3, 27, and 51 h postinoculation. For the apple shoot assay, the youngest apple leaves were inoculated by cutting with scissors dipped in a bacterial suspension of 2 × 108 CFU ml−1. The progression of symptoms was observed at 3, 8, and 14 days postinoculation. All assays were repeated three times, with five biological replicates in each experiment. Statistical analyses of treatment means was done by one-way analysis of variance, and mean separation (P < 0.05) was accomplished using Fisher's protected-least-significant-difference test.
Amylovoran production assay and motility assays.
The amylovoran concentration in supernatants of bacterial cultures was quantified using a turbidity assay with cetylpyrimidinium chloride (CPC) as previously described (42). Briefly, cells from overnight LB medium cultures were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and inoculated into MBMA medium (43) with 1% sorbitol. The supernatant of the MBMA culture was tested for the amylovoran concentration by adding 50 μl of CPC (50 mg ml−1) per ml of supernatant sample, followed by measuring the optical density at 600 nm (OD600). The experiments were repeated three times with four biological replicates in each experiment.
To measure bacterial swarming motility, cells from overnight cultures were collected by centrifugation, resuspended in PBS, and diluted in sterile water. The diluted bacterial suspension was plated onto the center of swarming agar plates (10 g tryptone, 5 g NaCl, 3 g agar per liter). Swarming diameters were measured at 18 h postinoculation. The experiments were repeated three times with four biological replicates in each experiment.
Biofilm quantification by crystal violet staining and analysis using SEM.
To quantify the amount of biofilm by crystal violet staining, bacterial strains were cultured in 0.5× LB broth in a 24-well plate with a glass coverslip placed in each well at a 30° angle. After 48 h of incubation at 28°C, the bacterial culture was removed from the wells, 10% crystal violet was added to the wells, and the plate was incubated at room temperature for 1 h. Glass coverslips were rinsed with water, air dried for 2 h, and eluted with 200 μl of elusion solution (40% methanol, 10% glacial acetic acid). The solubilized crystal violet in the elution solution was quantified by measuring the light absorbance at OD600 using a Safire microplate reader (Tecan, Research Triangle Park, NC). The experiment was repeated three times with 12 replicates in each experiment.
For the observation of biofilm formation using scanning electron microscopy (SEM), strains to be tested were cultured in 100 μl of 0.5× LB broth in a 96-well plate with a 300-mesh transmission electron microscopy (TEM) gold grid in each well (G300-Au; Electron Microscopy Sciences, Hatfield, PA). The plates were incubated at 28°C for 48 h, and 100 μl of paraformaldehyde-glutaraldehyde (2.5% of each compound in 0.1 M sodium cacodylate buffer; Electron Microscopy Sciences) was added to each well. The mixture was incubated at room temperature for 1 h, and grids were successively dehydrated in 25, 50, 75, and 90% ethanol for 30 min each and in 100% ethanol three times for 15 min each. Grids were then dried to the critical point using a critical point drier (Balzers CPD, Lichtenstein) and mounted on aluminum mounting stubs (Electron Microscopy Sciences). Samples were then coated with osmium using a pure osmium coater (Neoc-an; Meiwa Shoji Co. Ltd., Japan). Images were taken on a JEOL 6400V scanning electron microscope (Japan Electron Optics Laboratories) equipped with an LaB6 emitter (Noran EDS) using analySIS software (Soft Imaging System, GmbH).
Protein purification and analyses.
Strains were cultured in 50 ml LB broth overnight at 28°C. Cells were harvested by centrifugation, washed with 20 ml of PBS, and resuspended in 100 ml Hrp-inducing minimal medium. After 48 h of induction in Hrp-inducing minimal medium, culture supernatant was collected by centrifugation, phenylmethylsulfonyl fluoride (PMSF) was added to a concentration of 0.5 mM, and the resulting solution was filtered through a 0.22-μm-pore-size filter (Stericup; Millipore, Billerica, MA) to obtain a cell-free supernatant. The cell-free supernatant was concentrated to 1 ml using an Amicon 15-ml centrifugal filter unit (10-kDa-molecular-mass cutoff). Proteins was extracted from the cell-free supernatant using a previously described method (44) with modifications. Proteins from the concentrated supernatant were extracted twice by mixing with 0.5 volume of water-saturated phenol at 4°C with agitation for 30 min. Phases of the mixture were separated by centrifugation; the lower phases of the phenol fractions from each extraction were combined, and the proteins in the phenol fraction were precipitated by adding 5 volumes of 100 mM ammonium acetate in methanol. Samples were incubated overnight at −20°C, followed by centrifugation at 13,000 × g at 4°C for 30 min. The protein pellets were resuspended in 50 μl of water and reprecipitated by adding 500 μl of cold acetone. Samples were incubated overnight at −20°C. Protein pellets were collected by centrifugation at 13,000 × g at 4°C for 30 min and resuspended in 50 μl of water with 0.5 mM PMSF. Protein concentrations were measured with a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL) and were adjusted to 1 μg μl−1. For the SDS-PAGE analysis of proteins from the E. amylovora secretome, proteins were separated using a Mini-PROTEAN 3 system (Bio-Rad, Hercules, CA), and gels were stained with Coomassie blue. Protein bands of interest were excised from the Coomassie blue-stained polyacrylamide gels and identified by mass spectrometry (MS) using a Thermo Scientific LTQ linear ion trap mass spectrometer. For Western blot analysis, 8 μg of proteins of each sample was analyzed using anti-CyaA antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Anti-DnaK antibody was used as a lysis control. The band intensities of proteins of interest following Western blot analysis were quantified using ImageJ software (http://rsbweb.nih.gov.ij/).
Hypersensitive response and DspE-CyaA translocation assay.
For hypersensitive response (HR) assays, strains were cultured in LB broth overnight, harvested by centrifugation, and washed with 0.5× PBS twice. Cells were resuspended in 0.5× PBS and adjusted to a concentration of 1 × 107 CFU ml−1. Approximately 100 μl of cell suspension was infiltrated into 9-week-old Nicotiana benthamiana leaves using a needleless syringe, and the HR was observed at 16 h after infiltration.
The DspE-CyaA translocation assay was performed as previously described (40). Briefly, bacterial strains carrying DspE-CyaA fusion plasmids pRLT201 and pRLT8 were cultured in LB medium overnight, washed with 0.5× PBS, and resuspended in 0.5× PBS. Cells were adjusted to a concentration of 6 × 108 CFU ml−1 and were infiltrated into the youngest three fully expanded leaves of an 8-week-old Nicotiana tabacum plant. After 20 h postinoculation, leaf disks were collected using a 1-cm hole puncher and were immediately frozen in liquid nitrogen. Cyclic AMP (cAMP) was extracted from the leaf disks by grinding leaf disks in liquid nitrogen and resuspending them in 325 μl 1.1 M HCl. cAMP levels in the supernatants were then quantified using a cyclic AMP enzyme immunoassay kit (Cayman Chemical Co., Ann Arbor, MI). Protein levels in leaf pellets were measured using the Bradford method. The final cAMP concentration of each sample was the measured cAMP level adjusted by the amount of proteins (pg cAMP/μg protein).
RNA isolation, qRT-PCR, and Northern blot analysis.
Total bacterial RNA from cultures grown in Hrp-inducing medium was isolated by using the RNeasy minikit method (Qiagen, Valencia, CA) and treated with Turbo DNA-free DNase (Ambion, Austin, TX). cDNA was synthesized from 1 μg of DNase-treated total RNA using TaqMan reverse transcription (RT) reagents (Applied Biosystems, Foster City, CA). SYBR green PCR master mix (Applied Biosystems) was used for real-time PCRs to quantify the cDNA levels of target genes. The oligonucleotide primer sequences used in quantitative RT-PCRs (qRT-PCRs) are listed in Table S1 in the supplemental material. recA was used as an endogenous control for data analysis (45). Data were collected using a StepOne Plus real-time PCR system (Applied Biosystems) and analyzed using the Relative Expression software tool as described previously (36).
5′ rapid amplification of cDNA ends (RACE) assay.
Twelve micrograms of total bacterial RNA from E. amylovora Ea1189 was treated with tobacco acid pyrophosphatase (Epicentre, Madison, WI) at 37°C for 0.5 h, following which 300 pmol of RNA oligonucleotide linker was added. An extraction with a 25:24:1 (vol/vol) solution of water-saturated phenol-chloroform-isoamyl alcohol (P-C-I) was added to the tobacco acid pyrophosphatase-treated sample in a 2:1 (vol/vol) ratio, followed by vigorous shaking for 30 s and centrifugation at 13,000 × g for 15 min. RNA was pelleted from the aqueous phase by adding 3 volumes of ethanol containing 0.3 M sodium acetate, followed by incubation on ice for 1 h and centrifugation at 13,000 × g at 4°C for 40 min. The RNA pellet was then dissolved in 14 μl of RNase-free H2O. Purified RNA-linker mix was denatured at 90°C for 2 min and was ligated by T4 RNA ligase (New England BioLabs, Ipswich, MA). The ligated RNA-linker mix was purified by the P-C-I extraction again and was dissolved in 10 μl of RNase-free H2O. cDNA was synthesized by SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) using random hexamers following the instructions of the kit. The cDNA of ryhA and rprA was amplified by PCR using the total cDNA as the template and RNA linker primer and primers specific for the ryhA and rprA genes. cDNA of ryhA and rprA was gel purified and sequenced to map the 5′ end of the transcript.
Nucleotide sequence accession numbers.
The ryhA and rprA sequences from E. amylovora Ea1189 were deposited in GenBank with accession numbers KC357251 and KC357252, respectively.
RESULTS
Hfq is an important virulence regulator in Erwinia amylovora.
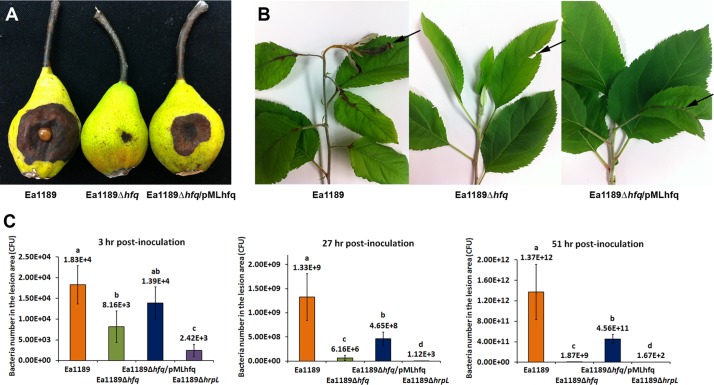
To determine whether Hfq plays a role in virulence regulation of E. amylovora, an hfq deletion mutant (EAM0436) was constructed and the levels of virulence of wild-type strain E. amylovora Ea1189 and E. amylovora mutant strain Ea1189Δhfq were compared in immature pears and in apple shoots (Fig. 1). In the immature pear fruit assay, Ea1189 caused dramatic necrosis symptoms with associated ooze production at 5 days postinoculation (dpi), while Ea1189Δhfq caused significantly reduced necrosis symptoms that were restricted to the inoculation site (Fig. 1A). In apple shoots at 8 dpi, Ea1189 caused complete necrosis of the inoculated leaf and surrounding leaves and exhibited systemic spreading within the shoot, resulting in an overall wilt symptom (Fig. 1B). In contrast, Ea1189Δhfq caused only slight necrosis at the inoculation site (denoted by the arrow) and showed no systemic movement to other parts of the plant (Fig. 1B). Enumeration of bacterial populations at the early stage of infection indicated that Ea1189Δhfq failed to rapidly multiply following inoculation and exhibited significantly reduced populations compared to wild-type Ea1189 at 3, 27, and 51 h postinoculation (Fig. 1C). The reduced-virulence phenotype and reduced growth following inoculation into pears of Ea1189Δhfq were partially complemented by plasmid pMLhfq, which encoded the hfq gene from Ea1189 (Fig. 1A and C).
Fig 1.
Effect of Hfq on E. amylovora virulence. (A) Virulence of E. amylovora Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq in immature pears at 5 dpi. (B) Virulence of Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq in apple shoots, at 8 dpi. Arrows denote the disease symptoms. (C) Populations of Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189ΔhrpL bacteria in immature pear fruits measured at 3 h, 27 h, and 51 h postinoculation. An identical amount of inoculum (1 × 104 CFU) was inoculated into each wounded pear. Sample means were compared by an analysis of variance and separated using the Student t test. The presence of different letters indicates that the means were significantly different (P < 0.05).
Hfq regulates amylovoran production, motility, and biofilm formation.
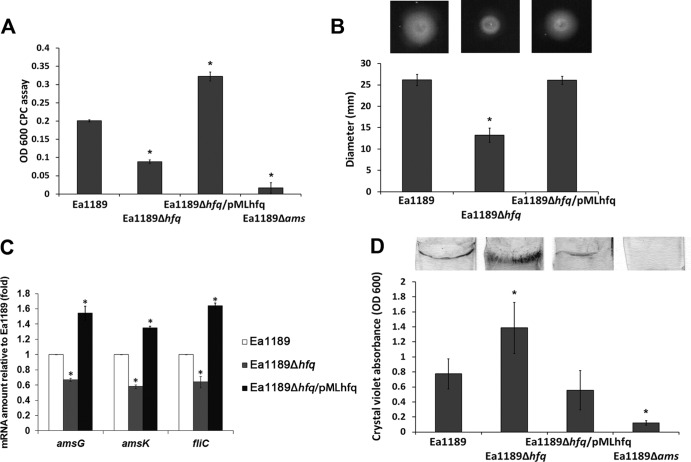
We next conducted experiments to assess the involvement of Hfq in regulating critical virulence factors in E. amylovora. We first compared the amylovoran production, motility, and biofilm formation in strains Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq. Production of amylovoran was detected in Ea1189 but not in Ea1189Δams, a deletion mutant of the amylovoran biosynthesis operon (Fig. 2A). Compared to Ea1189, Ea1189Δhfq exhibited a significant reduction in amylovoran production that was complemented by pMLhfq (Fig. 2A). Similar to the observation of amylovoran production, bacterial swarming motility was also significantly reduced in Ea1189Δhfq to about 0.5 that in Ea1189 (Fig. 2B). The reduced-motility phenotype in Ea1189Δhfq was also restored by pMLhfq (Fig. 2B).
Fig 2.
Effect of Hfq on virulence-related functions in E. amylovora. (A) Amylovoran production of E. amylovora Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189Δams. Bacterial strains were cultured in MBMA medium for 2 days, and the amount of amylovoran produced was quantified using the cetylpyrimidinium chloride (CPC) assay. (B) Swarming motility of Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq. Bacterial strains were inoculated at the center of swarming agar plates (0.3% agar), and the swarming diameters were measured at 18 h postinoculation. Asterisks indicate significant differences (P < 0.05) compared to Ea1189. (C) Relative amount of amsG, amsK, and fliC mRNA in Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq compared to Ea1189, measured by qRT-PCR. (D) Biofilm formation of Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189Δams. Bacterial strains were incubated with glass coverslips in static cultures of 0.5× LB broth. The biofilm formed on the coverslips was stained with crystal violet and quantified by measuring light absorbance at OD600. Asterisks indicate significant differences (P < 0.05) compared to Ea1189.
To confirm the regulation of amylovoran biosynthesis and motility by Hfq, the expression of two genes in the amylovoran biosynthesis operon, amsG and amsK, along with one gene encoding flagellin, fliC, was compared in strains Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq. Consistent with the phenotypic observations, the mRNA levels of amsG, amsK, and fliC were all reduced in Ea1189Δhfq and restored in Ea1189Δhfq/pMLhfq (Fig. 2C). Interestingly, although amylovoran is critical for biofilm formation and Ea1189Δhfq produced significantly less amylovoran than Ea1189, a significant increase in biofilm formation was observed in Ea1189Δhfq compared to Ea1189, detected by crystal violet staining of biofilm formation on glass coverslips (Fig. 2D). This increased biofilm phenotype of Ea1189Δhfq was restored to wild-type levels in Ea1189Δhfq/pMLhfq. Together, these results indicate that Hfq controls different virulence determinants in E. amylovora.
Hfq limits cell attachment to solid surfaces while promoting cell aggregation and biofilm maturation.
The crystal violet staining method used as described above measures the amount of cells attaching to and presumably forming biofilms on glass coverslips but does not provide additional information regarding the biofilm structure and complexity. To test whether the increased biofilm formed by strain Ea1189Δhfq was similar in structure to that formed by the wild type, we examined the biofilm structures of Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq produced in vitro on 300-mesh gold grids using SEM. After 52 h, the majority of Ea1189 cells formed highly structured cell aggregates that attached to the grid rim and expanded into the center space of the mesh (Fig. 3A). In addition to the aggregated cells, single, nonaggregated cells individually attached to the grid surfaces were observed. Compared to Ea1189, the majority of cells of Ea1189Δhfq observed were nonaggregated individual cells that formed a lawn evenly covering the grid surfaces. Fewer cell aggregates with less complex structures were observed for Ea1189Δhfq than for Ea1189 (Fig. 3B). The complemented strain Ea1189Δhfq/pMLhfq showed fewer cells attached to the grid surfaces and formed fewer cell aggregates than Ea1189 (Fig. 3C). Greatly reduced cell attachment and aggregation on the TEM grids were observed in the Δams strain (unpublished data). Detailed structures of cell aggregates of Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq were further characterized by SEM under higher magnification (×14,000). In the biofilm formed by Ea1189, large amounts of extracellular fibrillar material were observed among cell aggregates (Fig. 3D, indicated by the arrow). The amount of this material was greatly reduced in Ea1189Δhfq (Fig. 3E) and was restored in Ea1189Δhfq/pMLhfq (Fig. 3F, denoted by the arrow).
Fig 3.
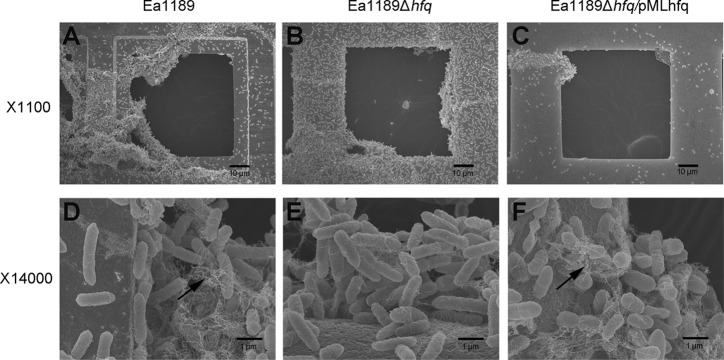
Effect of Hfq on biofilm formation in E. amylovora. SEM observation of biofilm structures formed on 300-mesh grids by E. amylovora Ea1189 (A, D), Ea1189Δhfq (B, E), and Ea1189Δhfq/pMLhfq (C, F). Bacterial strains were cultured in 0.5× LB broth with 300-mesh TEM grids for 52 h. Samples were fixed with paraformaldehyde-glutaraldehyde, and each grid was examined for biofilm formation by SEM at ×1,100 and ×14,000 magnifications. Arrows in the micrographs at ×14,000 magnification denote fibrillar material associated with biofilm formation.
In order to quantify the density of individual, nonaggregated cells of strains Ea1189 and Ea1189Δhfq attaching to the grid surfaces, the cell density of nonaggregated cells attached to the grid surfaces was determined. Compared to Ea1189, a significant increase in density of individually attached cells was observed in Ea1189Δhfq (Fig. 4A). This phenotype could be complemented by pMLhfq. We also quantified the percentage of the mesh space covered by the aggregated cells expanding from grid rims. Aggregated cells of Ea1189Δhfq covered only 15% of the mesh spaces, whereas cell aggregation in Ea1189 covered 26% of the mesh space, on average (Fig. 4B). Although the average percent mesh space covered by Ea1189Δhfq/pMLhfq was not restored (5%), the majority of cells attached to the grid surfaces were observed to be in the form of aggregated cells, similar to the wild type. These results indicate that Hfq negatively controls bacterial attachment while positively controlling cell aggregation in E. amylovora. Taken together, our results suggest that the regulation of the biofilm by Hfq occurs on two different levels: negative control over initial attachment and positive regulation of cell aggregation and of biofilm maturation.
Fig 4.
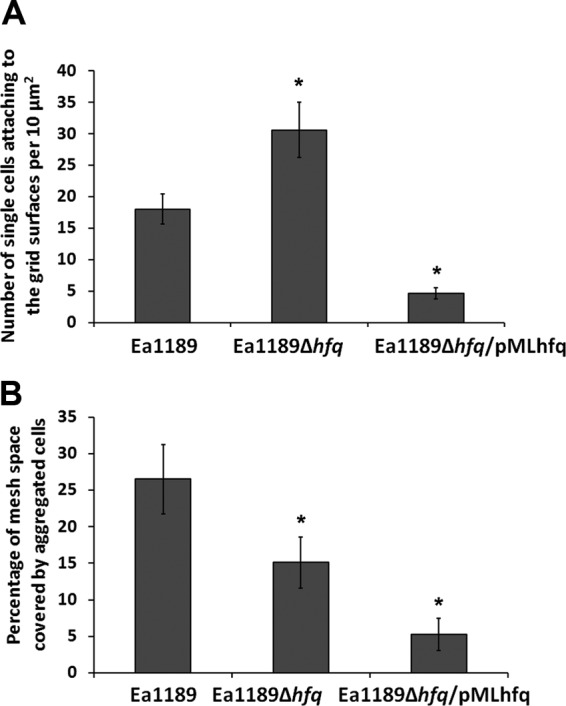
Effect of Hfq on the density of cellular attachment in E. amylovora. (A) Density of individual, nonaggregated cells attached to the surface of 300-mesh grids of E. amylovora Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq. Cells present in randomly selected 10-μm2 areas were quantified. (B) Percentage of center spaces of grids covered by aggregated cells of Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq. Bacterial strains were cultured in 0.5× LB broth with 300-mesh TEM grids for 52 h. Samples were fixed with paraformaldehyde-glutaraldehyde, and each grid was examined for biofilm formation by SEM at ×1,100 magnification. Asterisks indicate significant differences (P < 0.05) compared to Ea1189.
Hfq controls T3SS by upregulating the secretion and translocation of DspE and downregulating the secretion of HrpK and Eop1.
To determine if Hfq regulated the normal functioning of T3SS, strains Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189ΔT3SS were infiltrated into leaves of the non-host plant Nicotiana benthamiana and compared for their abilities to elicit the HR. The HR is a phenotype of programmed cell death exhibited by non-host plants in response to bacterial pathogens (46). At 16 h postinfiltration, a typical HR was observed at the site of infiltration of Ea1189 but not Ea1189ΔT3SS (Fig. 5A). A reduction in the intensity of the HR was observed in the leaf area infiltrated with Ea1189Δhfq; this reduction was restored to wild-type levels in the complemented strain Ea1189Δhfq/pMLhfq (Fig. 5A). These results suggest that Hfq is important for the normal function of the T3SS in E. amylovora.
Fig 5.
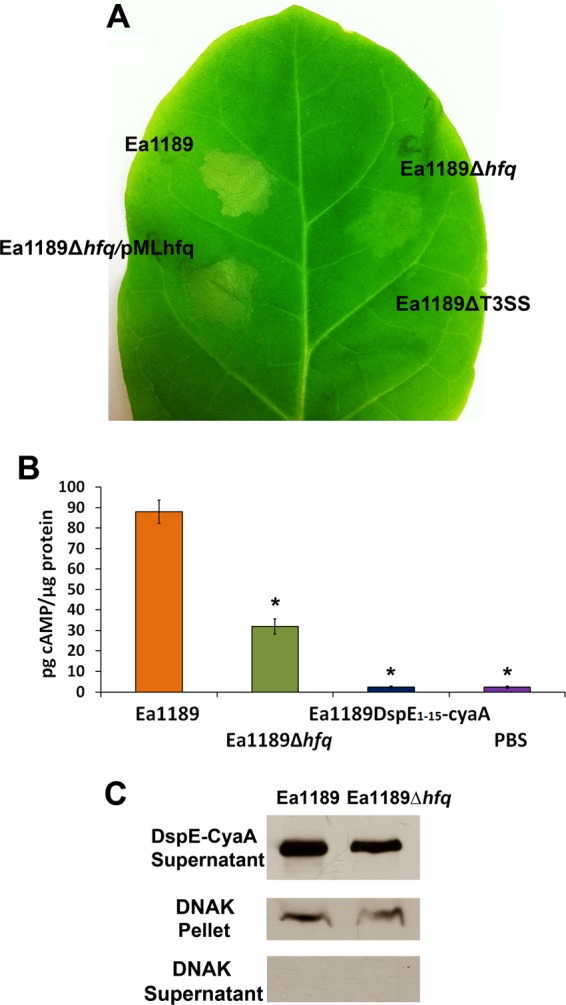
Effect of Hfq on type III effector translocation in E. amylovora. (A) HR elicited by E. amylovora Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189ΔT3SS. Bacterial strains (1 × 107 CFU ml−1) were infiltrated into Nicotiana benthamiana leaves, and the HR was observed at 16 h postinfiltration. (B) Levels of DspE1-737–CyaA translocation into N. tabacum by Ea1189 and Ea1189Δhfq. The translocation of DspE1-15–CyaA by Ea1189, which contains only the first 15 amino acid of DspE and lacks the translocation signal, was included as a negative control. The cAMP accumulation in tobacco leaves was measured 16 h after infiltrating 1 × 108 CFU ml−1 of bacterial cells expressing DspE-CyaA. Asterisks indicate significant differences (P < 0.05) compared to Ea1189. (C) Secretion of DspE-CyaA protein into the culture supernatant by Ea1189 and Ea1189Δhfq. DspE-CyaA detection was assessed by Western blotting using anti-CyaA antibody. DnaK, a nonsecreted protein that is constitutively expressed in E. amylovora, was used as a cell lysis control.
DspE is the most important type III effector protein and is required for the pathogenicity of E. amylovora. To examine whether the translocation of DspE is affected by the deletion of hfq, the N-terminal portion of DspE from positions 1 to 737 (DspE1-737), which is required for translocation by the T3SS (40), was fused with the reporter domain CyaA. The translocation of DspE1-737–CyaA into N. tabacum was compared in strains Ea1189 and Ea1189Δhfq by measuring the cAMP levels in plants (Fig. 5B). The translocation of a previously reported nontranslocatable construct (DspE1-15–CyaA) (40) by Ea1189 was measured as a negative control. At 20 h postinoculation, cAMP was detected in plant tissues inoculated with Ea1189 (88.0 ± 5.7 pg cAMP/μg protein) but was not detected following inoculation with Ea1189 producing DspE1-15–CyaA (2.3 ± 0.4 pg cAMP/μg protein) (Fig. 5B). Consistent with the HR assay result, the cAMP level in plant tissues inoculated with Ea1189Δhfq containing DspE1-737–CyaA was reduced to about one-third of the level in plant tissues inoculated with Ea1189 (32.0 ± 3.7 pg cAMP/μg protein) (Fig. 5B). The results from the HR assay and translocation assay both indicate that Hfq positively controls the translocation of the T3SS effector DspE into plant cells and is important for the normal function of the T3SS.
To test whether the reduced translocation of DspE1-737–CyaA in Ea1189Δhfq was because of a reduced secretion of DspE, Ea1189 and Ea1189Δhfq producing DspE1-737–CyaA were cultured in Hrp-inducing minimal medium, and the culture supernatant was examined for the presence of DspE1-737–CyaA by Western blotting using anti-CyaA antibody. A reduction in secretion of DspE1-737–CyaA (band intensity, 0.77, the level for Ea1189) was observed in Ea1189Δhfq (Fig. 5C).
To test whether the secretion of other type III-secreted proteins was affected by Hfq, we conducted a secretome analysis of strains Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189ΔhrpL. Strains were cultured in Hrp-inducing minimal medium, and the profile of proteins from the culture supernatant was examined by SDS-PAGE. DspE (band 1, at 198 kDa; protein identity was confirmed by MS) was detected in both Ea1189 and Ea1189Δhfq but not in Ea1189ΔhrpL (Fig. 6A). Two proteins, identified in Fig. 6A as band 2 and band 3, showed enhanced secretion in Ea1189Δhfq but were not secreted by Ea1189ΔhrpL; these proteins were selected as potential T3SS-secreted proteins regulated by Hfq and were further characterized using MS. Protein 2, an 80-kDa protein, and protein 3, a 44-kDa protein, were identified as HrpK, a putative type III translocator protein, and Eop1, a type III secreted effector protein, respectively. Using a qRT-PCR assay, significantly enhanced levels of mRNA of hrpK and the Eop1 gene compared to the levels for the wild-type strain were also detected (Fig. 6B). The enhanced mRNA levels could be restored in the hfq-complemented strain (Fig. 6B). These results indicate that Hfq controls the production and secretion of T3SS-secreted proteins, including HrpK and Eop1.
Fig 6.
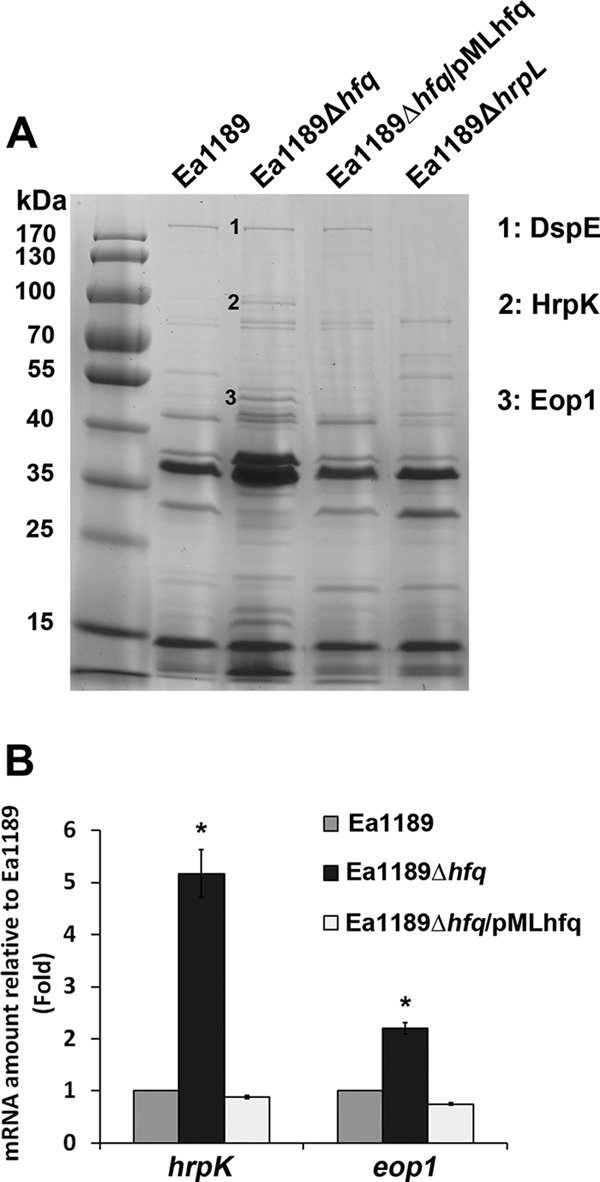
Effect of Hfq on protein secretion in E. amylovora. (A) Proteins in the secretomes of E. amylovora Ea1189, Ea1189Δhfq, Ea1189Δhfq/pMLhfq, and Ea1189ΔhrpL. Bacterial strains were cultured in Hrp-inducing minimal medium for 48 h, and proteins from the culture supernatant were purified, concentrated, and quantified by BCA assay. Ten micrograms of protein from the supernatant of each strain was analyzed by SDS-PAGE. Proteins of interest were purified from the gel and identified using mass spectrometry. (B) Relative amounts of mRNA of hrpK and the Eop1 gene in Ea1189, Ea1189Δhfq, and Ea1189Δhfq/pMLhfq compared to those in Ea1189, measured by qRT-PCR. Asterisks indicate significant differences from Ea1189 (P < 0.05).
Identification of 10 Hfq-regulated sRNAs in E. amylovora by computational analysis.
To identify Hfq-regulated sRNAs in E. amylovora, sequences of 27 previously characterized Hfq-regulated sRNAs in Escherichia coli, another member of the Enterobacteriaceae family, were used in a BLAST search for homologs in the genome of E. amylovora ATCC 49946 (GenBank accession number FN666575). Candidate sRNA-encoding genes were selected on the basis of the following criteria: 50% or greater sequence homology, the presence of an sRNA sequence immediately upstream of a Rho-independent terminator, and location in the intergenic regions of the chromosome. Homologs of 10 of the total of 27 sRNA genes from E. coli were found in E. amylovora (Table 2). To test whether the sRNA-encoding genes identified by the computational methods were actually expressed in E. amylovora, strains Ea1189 and Ea1189Δhfq were cultured in Hrp-inducing minimal medium for 6 and 12 h, and the expression of the sRNAs was examined by Northern blotting. Among all 10 sRNAs tested, RprA, RyhA (ArcZ), OmrA-OmrB, and RyhB were detected in Ea1189 at both 6 and 12 h (Fig. 7), GcvB was detected only at 12 h (Fig. 7), and Spf, MicA, GlmZ, RyeA, and MicM were not detected at either time point (data not shown). Similar to previous observations in E. coli (47), two RyhA transcripts, at approximately 50 nt and 200 nt, were detected in E. amylovora (Fig. 7). None of the sRNAs was detected in Ea1189Δhfq, indicating that Hfq might be important for the expression and stability of these sRNAs and further suggesting that the sRNAs identified are Hfq-regulated sRNAs.
Table 2.
sRNA-encoding genes identified in E. amylovora by sequence homology search of known sRNAs in E. coli
| sRNA (or alternate name) | % sequence identity to homolog in E. coli | Predicted size (nt) | Location in Erwinia genomea | Presence of Rho-independent terminator | Putative function |
|---|---|---|---|---|---|
| spf (spot42) | 99.1 | 108 | 52709–52816 | Yes | Regulator of DNA polymerase I activity, gal operon |
| rprA | 73.0 | 111 | 1771945–1771835 (c) | Yes | Translational activator of RpoS, biofilm formation |
| omrA-omrB | 57.5 | 79 | 3009425–3009347 (c) | Yes | Outer membrane protein regulator |
| micA | 79.8 | 84 | 2872079–2872162 | Yes | Outer membrane protein regulator |
| gcvB | 73.2 | 209 | 2962739–2962947 | Yes | Regulator of periplasmic ABC transporter |
| glmZ | 70.4 | 198 | 218841–219038 | Yes | Translational activator of GlmS (aminotransferase) |
| ryeA (sraC) | 58.6 | 251 | 2132789–2133039 | Nob | Unknown function |
| ryhB | 73.3 | 93 | 1981747–1981655 (c) | Yes | Regulator of iron uptake |
| ryhA (arcZ, sraH) | 62.3 | 121 | 3399430–3399550 | Yes | Translational activator of RpoS |
| micM (sroB) | 62.2 | 85 | 1149212–1149296 | Yes | Negative regulator of DpiA/DpiB (TCSTS) |
(c), the sequence is present on the complementary strand of the chromosomal sequence.
This observation is consistent with the observation of ryeA in E. coli.
Fig 7.
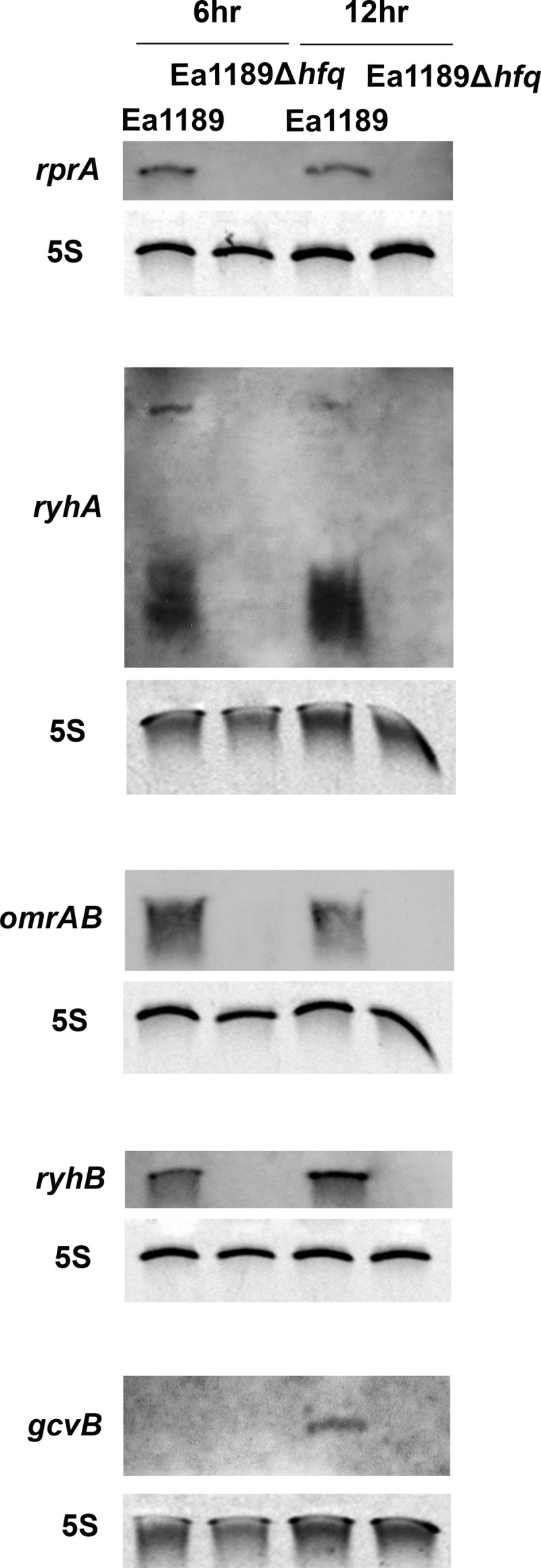
Examination of expression of sRNAs in E. amylovora. Detection of the sRNAs rprA, ryhA, omrAB, ryhB, and gcvB in Ea1189 and Ea1189Δhfq at 6 and 12 h postinoculation. Bacterial strains were cultured in Hrp-inducing minimal medium for 6 and 12 h prior to total RNA isolation. Each specific sRNA (and 5S rRNA control) in the total RNA sample was detected by Northern hybridization.
Mutational analysis of sRNA-encoding genes revealed two Hfq-regulated sRNAs that are important for virulence in E. amylovora.
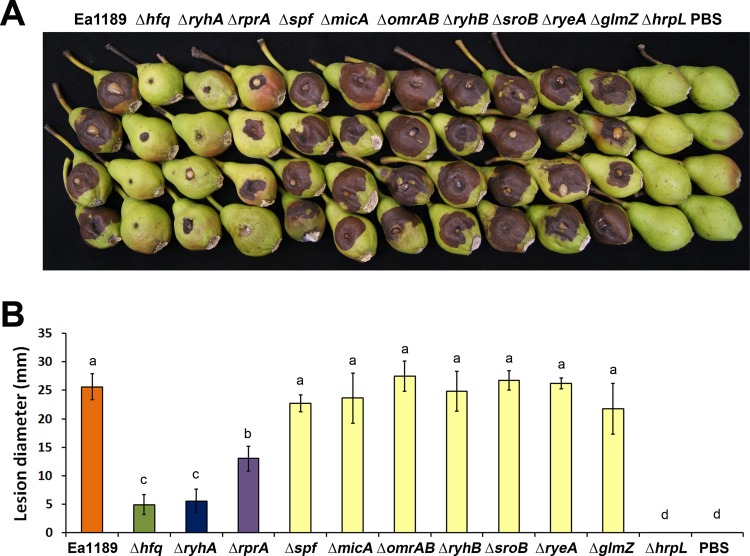
Since Hfq controls virulence in E. amylovora and Hfq exerts its regulation mainly by interacting with sRNAs, we next wanted to determine whether any sRNAs identified in the computational analysis play a role in regulating virulence in E. amylovora. Seven out of the nine sRNA deletion mutants constructed showed a level of virulence similar to that of strain Ea1189 in an immature pear assay (Fig. 8A and B). However, two sRNA mutants, those with the ΔryhA and ΔrprA mutations, showed significantly reduced virulence compared to Ea1189 (Fig. 8A and B).
Fig 8.
Analysis and quantification of virulence of E. amylovora deletion mutants of hfq and sRNAs. (A) Virulence of E. amylovora Ea1189, Ea1189Δhfq, and Ea1189ΔhrpL and sRNA mutants Ea1189ΔryhA, Ea1189ΔrprA, Ea1189Δspf, Ea1189ΔmicA, Ea1189ΔomrAB, Ea1189ΔryhB, Ea1189ΔsroB, Ea1189ΔryeA, and Ea1189ΔglmZ in immature pears at 5 dpi. (B) Average lesion diameters of immature pears (± standard error) inoculated with each E. amylovora deletion mutant. Sample means were compared by an analysis of variance and separated using the Student t test. The presence of different letters indicates that the means were significantly different (P < 0.05).
The transcriptional start sites of ryhA and rprA were determined using a 5′RACE assay (see Fig. S1 in the supplemental material). Based on our identification of the ryhA and rprA transcriptional start sites, we determined that the deletions constructed in strains Ea1189ΔrprA and Ea1189ΔryhA encompassed 80% and 100% of the sequences of these sRNAs (see Fig. S2 in the supplemental material). In addition, an examination of the expression of flanking genes mtgA and arcB for ryhA and ppsA, respectively, and EAM1647 for rprA indicated that the deletions of ryhA and rprA did not affect the expression levels of these genes (see Fig. S3 in the supplemental material).
At 5 dpi, the mean lesion diameter of Ea1189ΔryhA was reduced to a level similar to that of Ea1189Δhfq. The mean lesion diameter of Ea1189ΔrprA was also reduced to about half of that of Ea1189 but was still significantly higher than the lesion diameters of Ea1189Δhfq and Ea1189ΔryhA (Fig. 8B). The Ea1189ΔhrpL mutant, with deletion of the alternate sigma factor that regulates T3SS-encoding genes, was nonpathogenic (Fig. 8B). The reduced-virulence phenotype observed in Ea1189ΔrprA and in Ea1189ΔryhA was complemented by plasmids carrying rprA and ryhA, respectively (see Fig. S4 in the supplemental material). These results indicate that the Hfq-regulated sRNAs RyhA and RprA may work together with the sRNA chaperone Hfq and collaboratively control virulence in E. amylovora.
DISCUSSION
In this study, we showed that Hfq regulated all of the known essential pathogenicity determinants in E. amylovora, including type III secretion, translocation of the major effector DspE, and production of amylovoran EPS. Hfq is a known regulator of virulence in many bacterial pathogens and is also important in the response to various environmental stress factors in pathogens and other bacteria (28). However, the role of Hfq as a virulence regulator is not universal among pathogens. For example, in Staphylococcus aureus, an hfq mutant, virulence for the worm Caenorhabditis elegans was not affected (48), and in Neisseria gonorrhoeae, the virulence of an hfq mutant was not altered in cell culture models (49).
Biofilm formation is critical for E. amylovora to develop large populations in apple xylem and to move systemically through the host (12, 14, 42). The Ea1189Δhfq mutant appears to be unique among E. amylovora biofilm mutants in that although the mutant produced a decreased amount of amylovoran, it exhibited an altered enhanced biofilm phenotype. However, further examination of biofilm formation by this mutant indicated that Ea1189Δhfq exhibited a hyperattachment phenotype that presumably led to an artifact result suggesting an increase in biofilm formation on a polystyrene surface. In addition, although Ea1189Δhfq cells were capable of aggregation, the fibrillar matrix material characteristic of wild-type E. amylovora Ea1189 biofilms was not present. Thus, it is likely that Ea1189Δhfq cells are not capable of formation of mature biofilms. Since biofilm formation is important for the establishment of large populations of E. amylovora in apple xylem (12, 14, 42), we hypothesize that the lack of biofilm formation by Ea1189Δhfq resulted in an inability of these cells to establish in apple xylem and move systemically through the host. Furthermore, our results suggest that multiple in vitro and in vivo experiments are necessary for the characterization of mutants affecting biofilm formation in bacterial pathogens.
The regulatory effect of Hfq on biofilm formation had been studied in a few bacterial species prior to this study. In Moraxella catarrhalis, when the Δhfq and wild-type M. catarrhalis strains were mixed and inoculated in a continuous-flow biofilm system, the number of Δhfq cells clearly predominated over the number of wild-type cells in the population recovered from the biofilm after an overnight incubation (50). This observation is consistent with our finding that Ea1189Δhfq exhibited a hyperattaching phenotype on polystyrene solid surfaces compared to the wild-type strain Ea1189. Similarly, increased biofilm formation was observed in the absence of Hfq in Yersinia pestis (35). However, in uropathogenic Escherichia coli (UPEC), a mutation of hfq caused reduced biofilm formation when examined by a crystal violet staining method, which suggests a general activating effect of Hfq on biofilm formation (51). These observations indicate that biofilm regulation by Hfq may vary in different pathogens.
Biofilm formation is a complex developmental process that typically involves four phases: planktonic phase, attachment phase, maturation phase, and detachment phase (52). The transition between different phases is tightly regulated by multiple mechanisms, including the second messenger molecule cyclic di-GMP, and by quorum sensing (53, 54). Our observation that Hfq negatively controls bacterial attachment while positively controlling the production of amylovoran and biofilm maturation may provide insight into the transition of biofilm development processes. The attachment or adhesion to solid surfaces is the initial and prerequisite step of biofilm formation. This could be seen in our observation that much reduced attachment and biofilm formation were observed in Ea1189Δhfq/pMLhfq compared to Ea1189 when a multicopy plasmid, pMLhfq, was used for complementation. However, although attachment is an important step of biofilm formation, it has been proposed in animal pathogens that bacterial adhesion to host epithelial cells may also come at a cost due to the possibility of inducing host immunity (55). To minimize host recognition and possible host immune responses while establishing colonization, bacterial animal pathogens likely regulate the transition from initial attachment to biofilm maturation. Our work indicates that Hfq and potentially Hfq-regulated sRNAs may play important roles in this transition.
The delivery of effector proteins from bacteria into plant cells requires the processes of secretion and translocation. The fact that the translocation of DspE-CyaA in Ea1189Δhfq was reduced to ∼0.3-fold of that of wild-type Ea1189 while its secretion was reduced to only ∼0.77-fold of that of Ea1189 suggests that Hfq may also affect the translocation of DspE, in addition to regulating its secretion. Increased secretion of HrpK, a type III translocator protein (56), was observed in Ea1189Δhfq. The disruption of the production of HrpK, which is important for effector translocation, may be the reason for the reduced DspE translocation observed in Ea1189Δhfq. Similar to the downregulation of the expression of hrpK and the Eop1 gene observed in this work, Hfq was also reported to repress the production of type III effectors encoded on the LEE pathogenicity island in EHEC strains (31). Further study is needed to characterize the detailed mechanism of control of the T3SS by Hfq and small RNAs.
We observed pleiotropic regulation of amylovoran production, biofilm formation, motility, and the T3SS in E. amylovora by Hfq. These observations imply that Hfq-regulated sRNAs may likewise be important virulence regulators in E. amylovora. To determine the role of Hfq-regulated sRNAs in virulence regulation, we first identified 10 sRNAs using computational analysis and found 2 of them (RprA and RyhA) that contribute to the virulence of E. amylovora. Both RprA and RyhA were previously demonstrated to positively regulate translation of the stationary-phase sigma factor RpoS in E. coli (57). To test whether the reduced virulence in strains Ea1189ΔrprA and Ea1189ΔryhA was due to a potential downregulation of rpoS, a ΔrpoS mutant was constructed and its virulence phenotype was compared with that of Ea1189 in an immature pear fruit assay (data not shown). A similar level of virulence was observed in Ea1189ΔrpoS and Ea1189, which is consistent with a previous observation that RpoS is not involved in the induction of fire blight disease symptoms by E. amylovora (58). This suggests that the regulation of virulence by RprA and RyhA is likely mediated via targets different from those in E. coli. We are currently examining the regulatory mechanisms of these sRNAs. Also of note, prior to our study, Schmidtke et al. identified one virulence-related sRNA, sX12, in the plant-pathogenic bacterium Xanthomonas campestris (59). Neither RprA nor RyhA in E. amylovora shares any sequence homology with sX12.
In conclusion, we provide evidence that, similar to its role in animal pathogens, the RNA chaperone Hfq also plays important roles in controlling virulence in the plant-pathogenic bacterium E. amylovora. Compared to previously characterized regulators such as HrpL, the master regulator for T3SS (10), and RcsBCD, the key regulator of amylovoran production (42), Hfq appears to more broadly regulate many virulence determinants, including amylovoran production, biofilm formation, T3SS translocation and secretion, and bacterial motility. Thus, Hfq and the sRNAs that it regulates likely play a central role in the fine-tuning of virulence gene expression in E. amylovora. During fire blight pathogenesis, E. amylovora cells colonize and grow on the relatively nutrient-rich stigma surface, on the high-osmotic flower nectarthode, within the leaf apoplast, and in the potentially low-nutrient vascular system (1). Transitions between these various host environments may occur over relatively short time scales, thus necessitating the ability to rapidly alter the production of virulence factors via posttranscriptional regulation. Our identification of pleiotropic virulence effects in Ea1189Δhfq and of individual sRNAs with strong effects on virulence provides a promising start for characterization of the detailed regulatory mechanisms of Hfq and Hfq-regulated sRNAs in the virulence of E. amylovora.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a special grant from the United States Department of Agriculture CSREES, Project GREEEN, a Michigan plant agriculture initiative at Michigan State University, and Michigan AgBioResearch.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02056-12.
REFERENCES
- 1. Malnoy M, Martens S, Norelli JL, Barny MA, Sundin GW, Smits TH, Duffy B. 2012. Fire blight: applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 50:475–494 [DOI] [PubMed] [Google Scholar]
- 2. Thomson SV. 1986. The role of the stigma in fire blight infections. Phytopathology 76:476–482 [Google Scholar]
- 3. Bayot RG, Ries SM. 1986. Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology 76:441–445 [Google Scholar]
- 4. Oh CS, Kim JF, Beer SV. 2005. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6:125–138 [DOI] [PubMed] [Google Scholar]
- 5. Venisse JS, Malnoy M, Faize M, Paulin JP, Brisset MN. 2002. Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol. Plant Microbe Interact. 15:1204–1212 [DOI] [PubMed] [Google Scholar]
- 6. Boureau T, ElMaarouf-Bouteau H, Garnier A, Brisset MN, Perino C, Pucheu I, Barny MA. 2006. DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol. Plant Microbe Interact. 19:16–24 [DOI] [PubMed] [Google Scholar]
- 7. DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. 2004. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. U. S. A. 101:9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaudriault S, Malandrin L, Paulin JP, Barny MA. 1997. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol. Microbiol. 26:1057–1069 [DOI] [PubMed] [Google Scholar]
- 9. Pester D, Milcevicova R, Schaffer J, Wilhelm E, Blumel S. 2012. Erwinia amylovora expresses fast and simultaneously hrp/dsp virulence genes during flower infection on apple trees. PLoS One 7:e32583 doi:10.1371/journal.pone.0032583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McNally RR, Toth IK, Cock PJ, Pritchard L, Hedley PE, Morris JA, Zhao Y, Sundin GW. 2012. Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Mol. Plant Pathol. 13:160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geider K. 2006. Twenty years of molecular genetics with Erwinia amylovora: answers and new questions about EPS-synthesis and other virulence factors. Acta Hortic. 704:397–402 [Google Scholar]
- 12. Koczan JM, McGrath MJ, Zhao Y, Sundin GW. 2009. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99:1237–1244 [DOI] [PubMed] [Google Scholar]
- 13. Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, Cociancich S, Couloux A, Darrasse A, Gouzy J, Jacques MA, Lauber E, Manceau C, Mangenot S, Poussier S, Segurens B, Szurek B, Verdier V, Arlat M, Rott P. 2009. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616 doi:10.1186/1471-2164-10-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koczan JM, Lenneman BR, McGrath MJ, Sundin GW. 2011. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol. 77:7031–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castiblanco L, Edmunds A, Waters CM, Sundin GW. 2011. Characterization of quorum sensing and cyclic-di-GMP signaling systems in Erwinia amylovora. Phytopathology 101:S2.2 doi:10.1094/PHYTO-101-10-S2.1 [Google Scholar]
- 16. Wang D, Qi M, Calla B, Korban SS, Clough SJ, Cock PJ, Sundin GW, Toth I, Zhao Y. 2012. Genome-wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora. Mol. Plant Microbe Interact. 25:6–17 [DOI] [PubMed] [Google Scholar]
- 17. Zhao Y, Blumer SE, Sundin GW. 2005. Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187:8088–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y, Wang D, Nakka S, Sundin GW, Korban SS. 2009. Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics 10:245 doi:10.1186/1471-2164-10-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frohlich KS, Vogel J. 2009. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12:674–682 [DOI] [PubMed] [Google Scholar]
- 20. Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:pii=a003798 doi10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Link TM, Valentin-Hansen P, Brennan RG. 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U. S. A. 106:19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sauer E, Weichenrieder O. 2011. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U. S. A. 108:13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeng Q, Ibekwe AM, Biddle E, Yang CH. 2010. Regulatory mechanisms of exoribonuclease PNPase and regulatory small RNA on T3SS of Dickeya dadantii. Mol. Plant Microbe Interact. 23:1345–1355 [DOI] [PubMed] [Google Scholar]
- 28. Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13:24–33 [DOI] [PubMed] [Google Scholar]
- 29. Toledo-Arana A, Repoila F, Cossart P. 2007. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10:182–188 [DOI] [PubMed] [Google Scholar]
- 30. Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345–354 [DOI] [PubMed] [Google Scholar]
- 31. Shakhnovich EA, Davis BM, Waldor MK. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 74:347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 66:1174–1191 [DOI] [PubMed] [Google Scholar]
- 33. Chambers JR, Bender KS. 2011. The RNA chaperone Hfq is important for growth and stress tolerance in Francisella novicida. PLoS One 6:e19797 doi:10.1371/journal.pone.0019797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186:3355–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bellows L, Koestler BJ, Karaba SM, Walters CM, Lanthem WW. 2012. Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilms I, Moller P, Stock AM, Gurski R, Lai EM, Narberhaus F. 2012. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J. Bacteriol. 194:5209–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao Y, Sundin GW, Wang D. 2009. Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora. Can. J. Microbiol. 55:457–464 [DOI] [PubMed] [Google Scholar]
- 38. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Labes M, Puhler A, Simon R. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 89:37–46 [DOI] [PubMed] [Google Scholar]
- 40. Triplett LR, Melotto M, Sundin GW. 2009. Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Mol. Plant Microbe Interact. 22:1282–1292 [DOI] [PubMed] [Google Scholar]
- 41. Huynh TV, Dahlbeck D, Staskawicz BJ. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377 [DOI] [PubMed] [Google Scholar]
- 42. Wang D, Korban SS, Pusey PL, Zhao Y. 2011. Characterization of the RcsC sensor kinase from Erwinia amylovora and other enterobacteria. Phytopathology 101:710–717 [DOI] [PubMed] [Google Scholar]
- 43. Bellemann P, Bereswill S, Berger S, Geider K. 1994. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 16:290–296 [DOI] [PubMed] [Google Scholar]
- 44. Nissinen RM, Ytterberg AJ, Bogdanove AJ, Van Wijk KJ, Beer SV. 2007. Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular hairpin levels. Mol. Plant Pathol. 8:55–67 [DOI] [PubMed] [Google Scholar]
- 45. Takle GW, Toth IK, Brurberg MB. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 7:50 doi:10.1186/1471-2229-7-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dangl JL, Jones JD. 2001. Plant pathogens and integrated defence responses to infection. Nature 411:826–833 [DOI] [PubMed] [Google Scholar]
- 47. Mandin P, Gottesman S. 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29:3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bohn C, Rigoulay C, Bouloc P. 2007. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 7:10 doi:10.1186/1471-2180-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dietrich M, Munke R, Gottschald M, Ziska E, Boettcher JP, Mollenkopf H, Friedrich A. 2009. The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS J. 276:5507–5520 [DOI] [PubMed] [Google Scholar]
- 50. Attia AS, Sedillo JL, Wang W, Liu W, Brautigam CA, Winkler W, Hansen EJ. 2008. Moraxella catarrhalis expresses an unusual Hfq protein. Infect. Immun. 76:2520–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76:3019–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Monroe D. 2007. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 5:e307 doi:10.1371/journal.pbio.0050307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 56. Petnicki-Ocwieja T, van Dijk K, Alfano JR. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. 2010. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U. S. A. 107:9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anderson M, Pollitt CE, Roberts IS, Eastgate JA. 1998. Identification and characterization of the Erwinia amylovora rpoS gene: RpoS is not involved in induction of fireblight disease symptoms. J. Bacteriol. 180:6789–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. 2012. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 40:2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.