Abstract
The perspective of the cytoskeleton as a feature unique to eukaryotic organisms was overturned when homologs of the eukaryotic cytoskeletal elements were identified in prokaryotes and implicated in major cell functions, including growth, morphogenesis, cell division, DNA partitioning, and cell motility. FtsZ and MreB were the first identified homologs of tubulin and actin, respectively, followed by the discovery of crescentin as an intermediate filament-like protein. In addition, new elements were identified which have no apparent eukaryotic counterparts, such as the deviant Walker A-type ATPases, bactofilins, and several novel elements recently identified in streptomycetes, highlighting the unsuspected complexity of cytostructural components in bacteria. In vivo multidimensional fluorescence microscopy has demonstrated the dynamics of the bacterial intracellular world, and yet we are only starting to understand the role of cytoskeletal elements. Elucidating structure-function relationships remains challenging, because core cytoskeletal protein motifs show remarkable plasticity, with one element often performing various functions and one function being performed by several types of elements. Structural imaging techniques, such as cryo-electron tomography in combination with advanced light microscopy, are providing the missing links and enabling scientists to answer many outstanding questions regarding prokaryotic cellular architecture. Here we review the recent advances made toward understanding the different roles of cytoskeletal proteins in bacteria, with particular emphasis on modern imaging approaches.
INTRODUCTION
When the term “cytoskeleton” was first coined in 1931 (1), cytoskeletons were thought to consist of fibrous structural elements within a cell which, like the bones in our body, exist to provide reinforcement. It gradually became clear, however, that the cytoskeleton is not so much a static structural system like spokes in a wheel but is rather a highly dynamic system responsible for major processes in the cell, including muscle contraction (2), the beating of cilia (3), chromosome segregation (4), cell division (5), phagocytosis (6), and organelle transport (7, 8), besides providing cell structure.
Still, it was a widely held notion that the cytoskeleton, consisting of microtubuli, microfilaments, and intermediate filaments (IFs), with cross-linking and other associating proteins providing additional levels of complexity (9), is a feature unique to eukaryotic cells. The existence of a multifunctional cytoskeleton in bacteria became generally accepted only in the last decade, when the concept of bacterial cells as sacculi of freely diffusible proteins was overturned, and it was established that they, in fact, contain homologs of all known eukaryotic cytoskeletal elements (10–12). FtsZ (a tubulin homolog [13]) and MreB (an actin homolog [14]) were the first to be characterized; later, crescentin, the first intermediate filament (IF)-like protein, was discovered in Caulobacter crescentus (15). Currently, there are also newly identified elements with no eukaryotic counterparts, namely, the deviant Walker A-motif ATPases (16) and bactofilins (17), clear evidence of the complexity of the bacterial cytoskeleton, while many elements are likely still to be discovered.
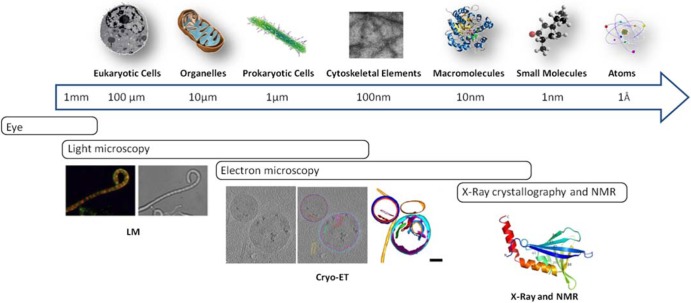
On the cellular scale, much has been learned about the cytoskeleton based on fluorescence light microscopy (fLM) studies and, in recent years, also via atomic force microscopy (AFM), which has been applied for the study of live cells as well as of isolated membrane proteins or microtubules (18), by measurement of surface properties. On the molecular scale, X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy are providing valuable structural information. In fact, rather than sequence similarity analyses, the main methods used for identification of prokaryotic cytoskeletal elements have been based on a combination of crystal structures, in vitro properties, and in vivo functional behavior (19). Bridging the gap between cellular and molecular structural studies (Fig. 1), cryo-electron tomography (cryo-ET) is taking its place as an important part of the imaging arsenal, providing structural information about protein complexes under conditions directly relevant to the native state of the cell (20–24). Combining tomography with the aforementioned imaging methods provides the multiscale and multidisciplinary approach needed to understand how cytoskeletal proteins function within the context of the cell.
Fig 1.
Resolution ladder demonstrating imaging techniques which can be used at different scales. Light microscopy (LM) can be used to image the live localization of proteins tagged with fluorescent reporters to obtain dynamic information; at higher resolution, cryo-electron tomography (cryo-ET) can provide structural information about the localization of, for example, cytoskeletal elements within the cell. Crystal structures of the proteins of interest can be obtained via X-ray crystallography or nuclear magnetic resonance spectroscopy. LM images adapted from reference 104 with permission from Cold Spring Harbor Laboratory Press; cryo-ET images adapted from reference 21 with permission from Elsevier; X-ray and NMR images adapted from reference 109 with permission from the American Society for Biochemistry and Molecular Biology.
In this review, we focus on the different roles of the major cytoskeletal proteins and demonstrate ways in which multiscale imaging techniques have provided insight into the organization and spatial arrangement of cytoskeletal filaments. In addition, we highlight their function in the morphologically unusually complex mycelial Streptomyces, where many of the elements take on new and distinct roles. We believe that streptomycetes are a good illustration of the flexibility of core cytoskeletal protein motifs, where one type of element can perform various functions and one function can be performed by several types of elements (10, 19). The review is nonexhaustive, and for detailed information on specific filament-forming proteins in bacteria we refer the reader to excellent reviews elsewhere (10, 19, 25–27).
DETERMINING (AND MAINTAINING) BACTERIAL CELL SHAPE
Bacteria have a wide variety of shapes, from the more common spherical and rod shapes to spiral, square, and filamentous shapes, and the molecular basis underlying cell shape is complex (11, 12, 28). Maintaining shape throughout cell growth and division is important if cells are to proliferate. In most bacteria, the external peptidoglycan (PG) wall, or murein sacculus, is responsible for maintaining cell shape by providing a rigid protection against osmotic pressure. Some bacteria, however, lack a cell wall and still maintain clearly defined shapes, ranging from the cocci of Acholeplasma to the tapered flask-like shape of some Mycoplasma species and the spiral shape of Spiroplasma species (28). It is little wonder that a wide variety of shape-defining and -maintaining cytoskeletal elements, forming a variety of superstructures (29), must exist to enable this diversity.
MreB AND MreB-LIKE PROTEINS
In rod-shaped bacteria, mutation of rodA, rodZ, and mreBCD resulted in loss of shape, with formation of round cells that eventually die (14, 30–32). Their structural analysis identified MreB as an actin homolog (33). Gram-negative bacteria apparently have a single mreB gene, typically in an operon with mreC and mreD, while genomes of Gram-positive bacteria encode up to three MreB-like proteins (MreB, Mbl, and MreBH in Bacillus subtilis). Imaging of MreB-green fluorescent protein (MreB-GFP) revealed patches localized in a spiral-like fashion along the long axis of cells, which was explained by MreB aiding in peptidolycan deposition (14, 34–36) (Fig. 2). Recent experiments, however, contradicted the notion of the existence of continuous helices and suggested rather a model whereby MreB moves circumferentially around the cell, perpendicular to its length, with synthesis complexes moving independently of each other in both directions (37). Inhibition of peptidoglycan synthesis blocks filament motion within 10 to 30 s, suggesting that PG synthesis drives the motion of MreB—and not vice versa. In fact, though previous studies have suggested that MreB dynamics are driven by its own polymerization, MreB rotation around the long axis of the cell requires the assembly of the peptidoglycan cell wall (38). Total internal-reflection fluorescence microscopy visualized the dynamic relationship between MreB paralogs and the cell-wall synthesis machinery in Bacillus (39, 40), and similar patch-like localizations of MreB and dynamics were found in Escherichia coli and Caulobacter crescentus, suggesting that the behavior is widely conserved.
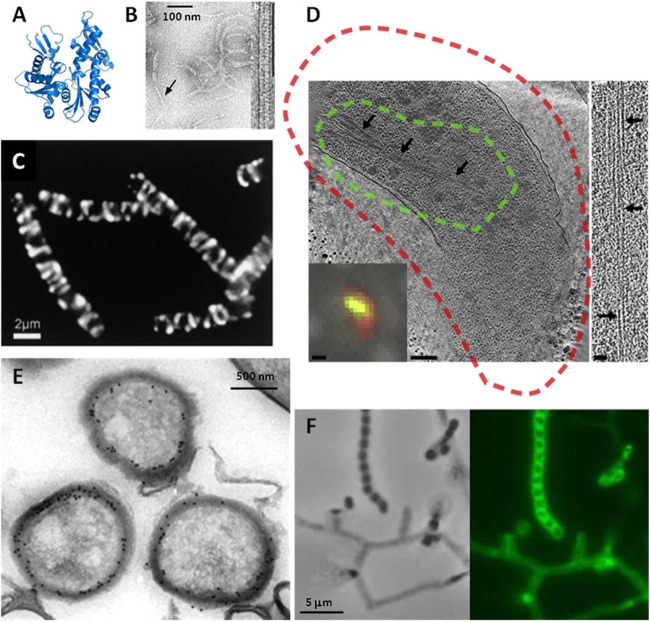
Fig 2.
Structure and microscopy of MreB. (A and B) The fold of prokaryotic MreB is similar to that its eukaryotic counterpart, actin (A), and MreB assembles into actin-like filaments (B). (C) The seemingly helical localization of an (MreB-like) Mbl-GFP fusion protein in cells in rod-shaped B. subtilis is now called into question (41–43). (D) In fact, though cytoplasmic MreB filaments are visible in tomograms, no helical filaments were seen in several rod-shaped bacteria, and helical filaments seen in E. coli were shown to be an artifact of the N-terminal YFP tag. In panel D, a tomographic slice through a Vibrio cholerae cell overexpressing GFP-MreB is shown. The cryo-fLM inset shows the cell stained with membrane dye FM 4-64 (red) and expressing GFP-MreB (green); dashed lines in the main image represent the fluorescent signal boundaries. On the right side, a 15-nm-thick tomographic slice through an MreB bundle not fused to GFP is shown. The scale bars represent 1 μm in the fLM inset, 200 nm in the cryo-electron tomography slice, and 50 nm in the higher-magnification inset. In Streptomyces coelicolor, MreB is not essential for vegetative growth but is essential for spore integrity and has been shown by immunoelectron microscopy (E) and fluorescence microscopy (F) to localize to the spore envelope. Panel B adapted from reference 33 with permission from the Nature Publishing Group; panel C from reference 14 with permission from Elsevier Ltd.; panel D from reference 42 with permission from Elsevier Ltd.; and panels E and F from reference 59 with permission from John Wiley and Sons.
These studies sparked a large debate. Is the helical organization the natural configuration that a filamentous structure on the inner surface of a cylinder will assume? Or are the localization patterns an artifact of images taken with long exposure times? Or is helical localization perhaps due to the effect of fluorescent fusions on tagged proteins? It is now becoming clear that different imaging techniques are required to corroborate the observed localization patterns (41). Cryo-electron tomography corroborated the absence of long (>80-nm) MreB filaments near or along the inner membrane of six different rod-shaped bacteria, although use of correlative cryo-light and electron tomography of GFP-MreB allowed the identification of cytoplasmic MreB bundles, showing that MreB is indeed detectable by cryo-ET (42). The same researchers later demonstrated that helical localization may be induced by fluorescent tagging: cryo-ET on E. coli cells producing native and yellow fluorescent protein (YFP)-tagged MreB demonstrated that MreB localizes in a helix when it is N-terminally tagged with YFP, while, when tagged with mCherry within an internal loop, it localizes in the same manner as native MreB (43).
COILED-COIL PROTEINS: INTERMEDIATE FILAMENTS IN BACTERIA?
If MreB is mainly involved in maintaining the rod shape, some intermediate filament-like proteins act to control other bacterial shapes. Crescentin (CreS), bearing remarkable architectural and biochemical relatedness to eukaryotic IF proteins, was the first to be identified in this class; other IF-like proteins include CfpA (44, 45), Scc (46), AglZ (47), and the four Ccrp proteins of Helicobacter (48, 49), as well as FilP (51) and Scy (54) in Streptomyces. Because, unlike crescentin, the other proteins do not actually have a high degree of structural similarity to IF proteins and may in fact present a case of convergent evolution (50), they have also been termed coiled-coil rich proteins, or Ccrps.
Crescentin (CreS) forms a filamentous structure at the short axis of the curved bacterium C. crescentus (15). Deletion of the creS gene turns curved C. crescentus cells into straight rods, demonstrating that crescentin is required for the curved (crescent) cell shape. Cryo-electron tomography of C. crescentus revealed multiple filament bundles, which fall into four major classes based on their shape and location (inner curvature, cytoplasmic, polar, and ring like [15]). Bundles, however, persisted in crescentinless mutants and in cells treated with A22, a compound with causes depolymerization of MreB filaments (53), suggesting that they are composed of as-yet-unidentified cytoskeletal elements. This led to the identification of the metabolic enzyme CtpS as a novel filament-forming protein in C. crescentus, interacting dynamically with CreS to regulate cell curvature (Fig. 3). The bifunctionality of CtpS filaments, which have a metabolic as well as a morphogenic role, demonstrates that protein polymerization may serve different functions within different cell contexts. Polymerization may have initially occurred for nonstructural, regulatory reasons—and the development of cytoskeletal and structural function of some proteins occurred later, during diversification and evolution.
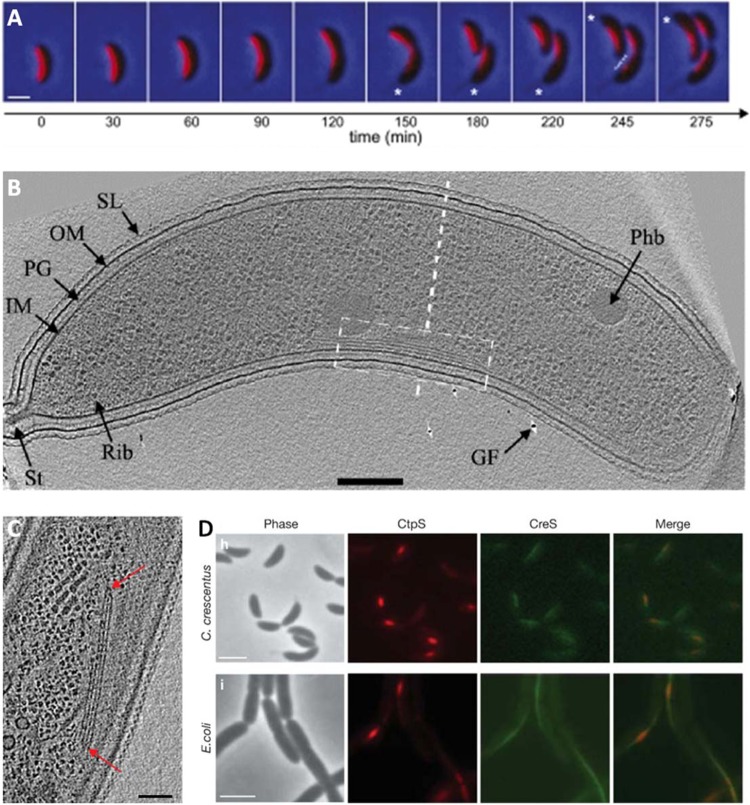
Fig 3.
Discovery of metabolic enzyme CtpS as a novel filament-forming protein in Caulobacter crescentus. (A) A time-lapse series of images of the GFP-labeled crescentin structure (red; laid over phase-contrast images) during the course of the cell cycle shows crescentin localizing to the short axis of curved C. crescentus cells. (B and C) Cryo-ET confirmed the presence of filament bundles (B), though bundles were still present in a crescentin deletion strain (C). This led to the discovery of metabolic enzyme CtpS as a novel filament-forming protein in C. crescentus. (D) CtpS and CreS colocalize. Bars, 1 μm (A), 200 nm (B), 50 nm (C), and 2 μm (D). Structural features in panel B: SL, surface layer; OM, outer membrane; PG, peptidoglycan layer; IM, inner membrane; St, stalk; Rib, probable ribosome; GF, gold fiducial used to align images; Phb, putative poly-β-hydroxybutyrate granule. Panel A adapted from reference 133 with permission from Cold Spring Harbor Laboratory Press; panel B adapted from reference 52 with permission from John Wiley and Sons; panels C and D adapted from reference 134 with permission from the Nature Publishing Group.
The four coiled-coil proteins of Helicobacter pylori (Ccrp48, Ccrp59, Ccrp1142, and Ccrp1143) have all been shown to be essential for the maintenance of proper (spiral) cell shape (48, 49). Deletion of these genes results in almost straight chained cells; in addition, though flagella are not affected, motility is reduced. All four Ccrps have different multimerization and filamentation properties and different types of smallest subunits and do not copurify, suggesting that the filaments have different though complementary roles (48).
The Streptomyces cytoskeletal element Scy was recently proposed to control polarized growth in existing hyphal tips, as well as to promote new tip formation during branching (54). Deletion of scy affects polarized growth and, as a consequence, also hyphal geometry, resulting in irregular hyphal width, short hyphal length, and aberrant branching. FilP, encoded from a gene immediately downstream of scy, is important for the stability of filamentous hyphae and for correct DNA segregation (51), again underlining the versatile architecture of IF-like proteins, offering different solutions for a variety of cytoskeletal tasks. Characterization of these coiled-coil proteins was performed by analyzing protein architecture and sequence conservation and, in the case of FilP, by AFM to analyze the rigidity of wild-type hyphae and deletion mutants; cryo-ET should allow visualization of such filaments in situ. In fact, Streptomyces hyphae are ideally suited for study with whole-cell cryo-electron tomography, as they are thinner than 500 nm—almost half the width of unicellular bacteria such as E. coli and B. subtilis—and therefore within the range of permissible thickness for ET. Moreover, other than those already identified, streptomycetes may have many more cytostructural elements, and preliminary mutational analysis suggests specific functions for a number of the large coiled-coil proteins found in these organisms in the control of cell integrity, growth, development, protein secretion, and DNA segregation (our unpublished data). It will be interesting to see if cryo-ET can reveal these elements within the hyphae. Indeed, despite their expected significant width (eukaryotic IFs are around 10 nm wide), to date, no confirmed intermediate filament-like structures (of CreS or others) have been unequivocally identified in tomograms of bacteria. Confirmation of the existence of prokaryotic IFs by, for example, high-resolution imaging combined with fluorescence microscopy is therefore eagerly awaited.
OTHER ROLES FOR MreB AND THE IF-LIKE PROTEINS
Difficult as it may be to group the intermediate filament-like proteins based on their structural similarity, it is even more difficult to classify them based on their function. Though several have an important role in shape control in non-rod-shaped bacteria, others have diverse roles in, for example, cell division (CfpA) or motility (AglZ). In a similar fashion, MreB and its homologs not only function to dictate rod shape but also play a role in motility (discussed below) (55, 56), chromosome segregation (53, 57), establishing cell polarity (58), and providing stability to the spore coat wall in Streptomyces (59).
In Myxococcus xanthus, filament-forming proteins AglZ and, interestingly, MreB are involved in (adventurous) A-type motility, a type of gliding for which the mechanism is not yet well understood. Many models were proposed (60, 61), but the latest results indicate that A-motility involves distributed motors and focal adhesion complexes, involving up to 40 proteins in a large multiprotein structural complex (62, 63). Coiled-coil protein AglZ localizes in clusters at the leading cell pole and, as the cell moves, is transported toward the lagging cell pole, where clusters are disassembled (63). These clusters are associated with A-motility motors that are hypothesized to power motility by coupling movement on a rigid cytoskeletal filament with adhesion complexes on the surface (64). Recently, MreB, in cooperation with MglA, a Ras-like GTPase, was shown to be critical for the proper positioning and stabilization of polar motility proteins and the focal adhesion complexes (55). A-type gliding motion of M. xanthus is thus actually remarkably similar to eukaryotic cell migration.
In the mollicute Spiroplasma melliferum, propulsion is also assisted by the action of MreB filaments: two types of filaments were found arranged in three parallel ribbons underneath the cell membrane, with the two outer ribbons built of fibril protein and the inner ribbon suggested to be composed of MreB (56). The structural data suggest a model explaining propulsion of helical mollicutes by means of coordinated length changes of the ribbons.
In the filamentous actinomycetes, which grow by apical extension, MreB homologs occur only in species that differentiate by forming an aerial mycelium and spores, with a less pronounced and nonessential role during vegetative growth (59). The absence in other apically growing actinomycete genera such as corynebacteria and mycobacteria suggests that the function of MreB is directly related to the way bacteria elongate and divide (reviewed in reference 65). In Streptomyces, the main function of MreB is to provide stability to the spore wall, which is corroborated by the dense crowding of the inner spore wall by MreB, as shown by immunoelectron microscopy (immuno-EM), suggesting that a large part of the spore wall is associated with MreB molecules (59). Deletion of the paralogous mbl also compromises spore-wall integrity (66). Detailed biochemical analysis of MreB and Mbl in streptomycetes should teach us more about the diverse roles these proteins can play in non-rod-shaped bacteria.
CELL DIVISION AND THE TUBULIN ANCESTOR FtsZ
Although some of the IF-like proteins assist in cell division processes, the major cell players in binary cell division are the Fts proteins, originally identified through analysis of temperature-sensitive mutants that fail to divide (the term fts, for filamentous temperature sensitive, was coined by Van de Putte and colleagues [135]). Upon completion of chromosome segregation (influenced by MreB in E. coli, Caulobacter, and B. subtilis [31, 53, 57]), the bacterial protein FtsZ directs the formation of the cytokinetic ring. A guanosine triphosphatase (GTPase) that is widely conserved and located within a cluster of genes involved in division and cell wall (dcw) synthesis, FtsZ polymerizes to form a scaffold of cell division proteins (the Z-ring) at the midplane of dividing cells. Interestingly, while in eukaryotes the cytokinetic ring is formed by actin, FtsZ is a structural homolog (and ancestor) of tubulin (13, 67, 68). The highly conserved FtsZ protein is found in virtually all bacteria and archaea (26), with only a few exceptions (69).
CLUES FROM IMAGING OF CELL DIVISION
Cryo-ET imaging of bacterial cell division recently provided new insight into FtsZ localization. Cryo-ET images of dividing C. crescentus cells showed short, separated, arc-like filaments of FtsZ and not a complete ring or spiral (70). In fact, the formation of FtsZ arcs was reported previously (71) as a stage in ring maturation. Cryo-ET revealed irregularly spaced protofilaments of FtsZ, seemingly connected to the inner membrane by other electron-dense protein complexes. Some were curved and others were straight, suggesting that, as speculated (72–74), FtsZ generates the constricting force for cell division itself through the nucleotide-hydrolysis-driven conformational change from straight to curved protofilaments.
Cross-wall formation in Streptomyces may, however, prove to be an exception to this model. Indeed, cell division in Streptomyces is remarkable. Not only is its division controlled in an entirely different manner, but it is also the only organism known to grow without cell division; the creation of a knockout mutant of ftsZ in S. coelicolor is an important event in cell biology (75). Availability of null mutants for the canonical cell division genes such as ftsEX, ftsI, ftsL, ftsQ, ftsW, and ftsZ makes Streptomyces an important object for cell division studies (76, 77). Additionally, in Streptomyces two types of cell division occur: in aerial hyphae, septation results in formation of spores which can separate to disperse, and in vegetative hyphae, cross-walls form at irregular intervals, do not constrict, and do not result in cell fission (Fig. 4). Amazingly, most canonical cell division proteins such as FtsI and FtsW are not even required for cross-wall formation. This suggests an entirely different cell division mechanism, whereby another pair consisting of a SEDS (shape, elongation, division, and sporulation) protein and a cognate class B penicillin-binding protein (PBP) may carry out septum synthesis (77, 78). This is something that has so far gained very little attention. One explanation is that because cross walls do not constrict but, rather, form semipermeable barriers that separate connecting compartments, the main function of the divisome is to mediate the activation of Z-ring contraction and that FtsZ does not generate a constricting force if other divisome components are absent. It will be interesting to see what role FtsZ plays in cross-wall formation.
Fig 4.
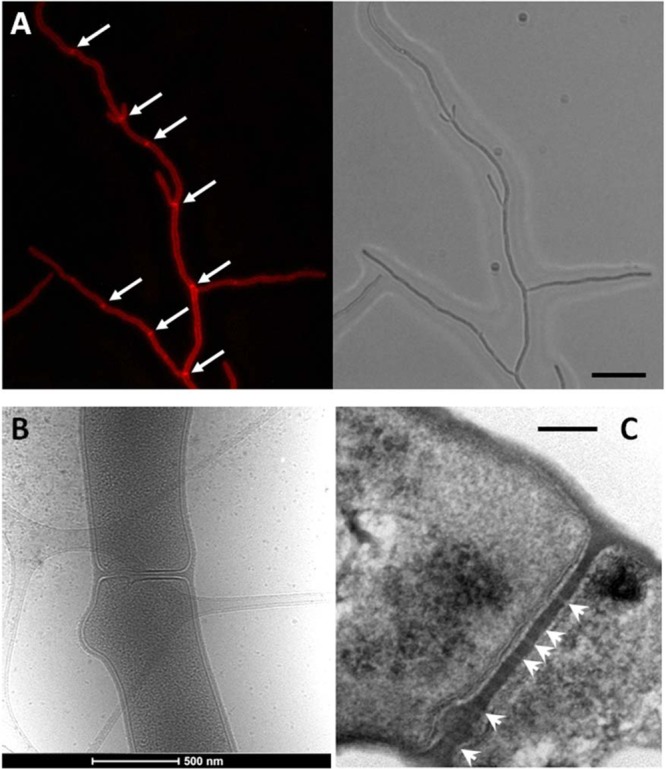
Cross-wall formation in Streptomyces coelicolor. (A) Cross walls (arrows) are formed at irregular intervals in vegetative hyphae of Streptomyces, and their structure and control of their localization are still poorly understood. Left, fluorescence micrograph after staining with the membrane dye FM5-95; right, corresponding light image. Bar, 5 μm. (B) Electron micrograph of a cross wall at higher resolution. (C) Transmission electron micrograph of a complete cross-wall with likely channels (arrowheads; apparent as lighter sections) and a bulge. Bar, 100 nm. It should be noted that cross walls may not all have channels. Figure 4C adapted from reference 76 with permission.
OTHER TUBULIN HOMOLOGS
Although FtsZ is clearly the most common tubulin homolog, other bacterial tubulin homologs exist, including TubZ and RepX in Bacillus (79, 80) and BtubA and BtubB in Prosthecobacter (81, 82). TubZ and RepX are plasmid-encoded proteins that play an important role in maintaining the stability of the plasmids that encode them and are therefore discussed in the following section. In contrast to TubZ, BtubA and BtubB are more closely related to eukaryotic α- and β-tubulin than to any other bacterial protein, forming heterodimers which polymerize into protofilaments in vitro. Based on comparative modeling data, and because microtubules were not found in thin EM sections, researchers initially predicted that BtubA and -B protofilaments are unlikely to form microtubule-like structures (83). When the ultrastructure of BtubA and BtubB was recently revisited using cryo-ET, however, it was shown that these proteins indeed assemble to form microtubules—consisting of five protofilaments instead of the 13 found in eukaryotes (84)—but with the same basic architecture. Their existence suggests that microtubule organization may have originated in bacteria, although horizontal transfer of the eukaryotic tubulins cannot be ruled out.
POSITIVE CONTROL OF CELL DIVISION AND THE SsgA-LIKE PROTEINS
In bacteria that divide by binary fission, FtsZ is the first protein to localize at the midcell position at the onset of cell division, followed by the subsequent recruitment of the other cell division components (85). For details on prokaryotic cell division and the cell division machinery, we refer the reader to excellent reviews published elsewhere (for instance, references 11, 12, 85, 86, and 87). In E. coli and Bacillus, septum-site localization and stabilization of the Z-ring require, among others, FtsA and ZipA (88–90), ZapA (91), and SepF (92), and the positioning and timing of septum formation involve the action of negative-control systems such as Min, which prevents Z-ring assembly at the cell poles (93, 94), and nucleoid occlusion, which prevents formation of the Z-ring over nonsegregated chromosomes (95–98).
Remarkably, division site selection during sporulation in Streptomyces, where up to a hundred septa are constructed almost simultaneously in the long aerial hyphae (76, 77), appears to be positively controlled. Similar positive control of cell division was also recently described in another multicellular bacterium, namely, the fruiting body-forming Myxococcus xanthus, where FtsZ is recruited by the ParA-like protein PomZ (99). In Streptomyces, division is mediated via the SsgA-like proteins, a family of small proteins that occur exclusively in morphologically complex actinomycetes and play a role in the control of morphogenesis (100, 101), with SsgA and SsgB required for sporulation (102, 103). During sporulation-specific cell division, FtsZ is actively recruited by the membrane-associated divisome component SsgB, which also stimulates FtsZ polymerization in vitro (104). The technique of Förster fluorescence resonance energy transfer combined with fluorescence lifetime imaging (FRET-FLIM), a powerful tool for the in vivo imaging and calculation of distances between proteins or between a protein and another cellular component such as the membrane, cell wall, or DNA (105, 106), revealed that SsgB indeed interacts closely with FtsZ and with the membrane (104). In turn, SsgB is guided to future septum sites by its paralog SsgA, a multifunctional protein that directly activates cell division (107) but also other events relating to cell-wall remodeling such as germination and branching (108). SsgB (and probably also SsgA) forms multimeric complexes, with the crystal structure of SsgB revealing a bell-shaped trimer (109). Whether or not SsgA-like proteins should be regarded as cytoskeletal elements themselves is yet unclear. Another interesting aspect of the control of division in Streptomyces is that reaching a threshold level of FtsZ expression appears to be the decisive step in the onset of division (110), and enhanced expression of FtsZ indeed overrules many sporulation (whi) mutants (111). This again points at a different way of decision making toward the initiation of division.
The concept of positive control of division apparently violates the general idea that in nature all major checkpoints are negatively regulated (9). However, positive division control is probably less expensive in terms of ATP (e.g., not requiring the energy-consuming oscillation of the Min proteins). In the case of Streptomyces, occasional defective spores in a long spore chain are less consequential than mistakes during binary fission, which could be considered an advantage of a multicellular lifestyle (76). Having said that, PomZ is required for binary fission in M. xanthus (99), while FtsZ can also localize (though inefficiently) to division sites in the absence of Min and Noc in B. subtilis (112). It remains to be seen how widespread active FtsZ recruitment in unicellular bacteria actually is (113).
ON PAR: THE CYTOSKELETON AND CHROMOSOME SEGREGATION
As we have seen, components of the cytoskeleton play an important role in guiding the spatiotemporal dynamics that govern the assembly of cellular components into higher-order structures. Chromosome and plasmid segregation is a good case in point. Segregation is mediated by tripartite partitioning systems (114, 115), which consist of a cytoskeletal nucleotide triphosphatase that provides the energy (ParA, ParM, or TubZ), a DNA-binding protein that forms higher-order nucleoprotein complexes with the DNA (ParB, ParG, or ParR), and a centromere site (parC, parS, or parH) close to the origin of replication (ori) that is recognized by dimers of the respective DNA binding proteins (114, 115).
Interestingly, the cytoskeletal partitioning NTPases all have different structural folds, suggesting that convergent evolution resulted in these different elements (and solutions) for the general problem of DNA separation. ParM is an actin family ATPase (116, 117), TubZ is a tubulin homolog (thus hydrolyzing GTP) (79), and ParA is a deviant Walker A-type Cytoskeletal ATPase (WACA) protein (16), a bacterial cytoskeletal element that has no eukaryotic counterparts. ParM forms dynamic, actin-like filaments that segregate plasmids in a mitosis-like process. In E. coli, cryo-ET was used to identify small bundles of three to five intracellular ParM filaments located close to the nucleoid, confirming that plasmid-segregating ParM filaments are associated with the nucleoid (118). A recent model suggests that antiparallel ParM filaments work together to drive plasmid segregation (119). TubZ assembles into highly dynamic, linear polymers with directional polymerization that are involved in plasmid segregation and move by a process called treadmilling. This treadmilling has so far been observed only in eukaryotes and involves assembly at one end of the filament and disassembly at the other, with, as a result, the net movement of the filament. Yet, unlike the hollow cylinders formed by tubulin, TubZ forms a two-stranded doubly helical filament which much more resembles actin-like ParM, which is also doubly helical. ParA functions by fuelling ParB, which in turn forms higher-order nucleoprotein complexes at partitioning (parS) sites near the chromosomal origin of replication, or oriC (120, 121). In rod-shaped bacteria, ParB complexes actively transfer the oriC proximal chromosomal region to the cell poles after completion of DNA replication. ParA most likely attaches to a chromosome with bound ParB and then pulls the chromosome across the cell by depolymerizing (114, 122, 123). It may also play a role in the control of chromosome replication, since B. subtilis ParA directly affects the function of replication initiator DnaA (124, 125). However, the cytoskeletal role of ParA is as yet controversial, and the filaments produced in vitro (126) have not yet been unequivocally established in vivo.
FUTURE PERSPECTIVES
In the span of a few years, we have made leaps and bounds toward understanding the mechanisms behind bacterial shape and structure. Twenty years ago, bacterial cytoskeletal elements were unknown; today, actin, tubulin, and intermediate-filament homologs, as well as novel cytoskeletal elements with no apparent eukaryotic counterparts, have all been identified in bacteria. The examples provided in this review demonstrate the vast plasticity and wide variety of roles taken on by prokaryotic cytoskeletal proteins and illustrate how multiscale imaging techniques are leading to new insights and improving our understanding of how bacterial cells function.
To come even closer to an understanding of the complex interactions that occur within the molecular landscape of the cell, static structural information must be coupled with in vivo dynamic studies. For this, correlative approaches are necessary. In correlative light and electron microscopy, proteins tagged with a fluorescent reporter, such as enhanced GFP (eGFP), or cell components stained with a selective dye can be directly identified on an EM grid and a tilt series acquired at the location of interest. This should enable mapping of cytoskeletal proteins onto high-resolution images created by electron microscopy, preferably in three dimensions (127–131). In addition, to catch dynamic structural changes, using a rapid-transfer system, samples can be cryoimmobilized once a physiological state has been observed in the cell (132). In this way, the missing links needed to resolve physical models for bacterial growth, division, or propulsion can be determined.
Adding the insight provided by correlative methods to the multiscale data of other techniques, we can get even further toward understanding the relationship between the structure of cytoskeletal elements and their position within the cell and function. Given the centrality of the cytoskeleton in regulating and executing key cellular processes, this would mark a great milestone in the field of cell biology.
ACKNOWLEDGMENTS
We are very grateful to Grant Jensen, William Margolin, and the anonymous referees for their valuable comments on the manuscript.
G.P.V.W. acknowledges support from the Netherlands Technology Foundation STW (VICI grant 10379). A.J.K. and R.I.K. acknowledge funding from the Dutch funding sources Cyttron II 20559 and NIMIC SSM06002 supporting the development of correlative imaging tools.
Footnotes
Published ahead of print 15 February 2013
REFERENCES
- 1. Wintrebert P. 1931. La rotation immédiate de l'oeuf pondu et la rotation d'activation chez Discoglossus pictus Otth. C. R. Soc. Biol. 106:439–442 [Google Scholar]
- 2. Banga I, Szent-Györgyi A. 1942. Preparation and properties of myosin A and B. Stud. Inst. Med. Chem. Univ. Szeged. I:5–15 [Google Scholar]
- 3. Gibbons IR, Rowe AJ. 1965. Dynein: a protein with adenosine triphosphatase activity from cilia. Science 149:424–426 [DOI] [PubMed] [Google Scholar]
- 4. Inoué S, Sato H. 1967. Cell motility by labile association of molecules—nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 50(Suppl):259–292 [PMC free article] [PubMed] [Google Scholar]
- 5. Schroeder TE. 1972. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J. Cell Biol. 53:419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaplan G. 1977. Differences in the mode of phagocytosis with Fc and C3 receptors in macrophages. Scand. J. Immunol. 6:797–807 [DOI] [PubMed] [Google Scholar]
- 7. Brady ST. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature 317:73–75 [DOI] [PubMed] [Google Scholar]
- 8. Vale RD, Reese TS, Sheetz MP. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42:39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular biology of the cell, 4 ed. Garland Science, New York, NY [Google Scholar]
- 10. Graumann PL. 2007. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61:589–618 [DOI] [PubMed] [Google Scholar]
- 11. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young KD. 2010. Bacterial shape: two-dimensional questions and possibilities. Annu. Rev. Microbiol. 64:223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- 14. Jones LJ, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 15. Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 16. Koonin EV. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165–1174 [DOI] [PubMed] [Google Scholar]
- 17. Kühn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 29:327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamon L, Curmi PA, Pastre D. 2010. High-resolution imaging of microtubules and cytoskeleton structures by atomic force microscopy. Methods Cell Biol. 95:157–174 [DOI] [PubMed] [Google Scholar]
- 19. Cabeen MT, Jacobs-Wagner C. 2010. The bacterial cytoskeleton. Annu. Rev. Genet. 44:365–392 [DOI] [PubMed] [Google Scholar]
- 20. Subramaniam S. 2005. Bridging the imaging gap: visualizing subcellular architecture with electron tomography. Curr. Opin. Microbiol. 8:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koning RI, Koster AJ. 2009. Cryo-electron tomography in biology and medicine. Ann. Anat. 191:427–445 [DOI] [PubMed] [Google Scholar]
- 22. Tocheva EI, Li Z, Jensen GJ. 2010. Electron cryotomography. Cold Spring Harb. Perspect. Biol. 2:a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McIntosh R, Nicastro D, Mastronarde D. 2005. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 15:43–51 [DOI] [PubMed] [Google Scholar]
- 24. Lucić V, Forster F, Baumeister W. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833–865 [DOI] [PubMed] [Google Scholar]
- 25. Shih YL, Rothfield L. 2006. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70:729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michie KA, Lowe J. 2006. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 75:467–492 [DOI] [PubMed] [Google Scholar]
- 27. Ingerson-Mahar M, Gitai Z. 2012. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 36:256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabeen MT, Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 29. Pilhofer M, Jensen GJ. 2013. The bacterial cytoskeleton: more than twisted filaments. Curr. Opin. Cell Biol. 25:125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 31. Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soufo HJ, Graumann PL. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916–1920 [DOI] [PubMed] [Google Scholar]
- 33. van den Ent F, Amos LA, Lowe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44 [DOI] [PubMed] [Google Scholar]
- 34. Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 102:18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Srivastava P, Demarre G, Karpova TS, McNally J, Chattoraj DK. 2007. Changes in nucleoid morphology and origin localization upon inhibition or alteration of the actin homolog, MreB, of Vibrio cholerae. J. Bacteriol. 189:7450–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vats P, Rothfield L. 2007. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U. S. A. 108:15822–15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228 [DOI] [PubMed] [Google Scholar]
- 40. Vats P, Yu J, Rothfield L. 2009. The dynamic nature of the bacterial cytoskeleton. Cell. Mol. Life Sci. 66:3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Margolin W. 2012. The price of tags in protein localization studies. J. Bacteriol. 194:6369–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swulius MT, Chen S, Jane Ding H, Li Z, Briegel A, Pilhofer M, Tocheva EI, Lybarger SR, Johnson TL, Sandkvist M, Jensen GJ. 2011. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem. Biophys. Res. Commun. 407:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swulius MT, Jensen GJ. 2012. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J. Bacteriol. 194:6382–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Izard J, McEwen BF, Barnard RM, Portuese T, Samsonoff WA, Limberger RJ. 2004. Tomographic reconstruction of treponemal cytoplasmic filaments reveals novel bridging and anchoring components. Mol. Microbiol. 51:609–618 [DOI] [PubMed] [Google Scholar]
- 45. You Y, Elmore S, Colton LL, Mackenzie C, Stoops JK, Weinstock GM, Norris SJ. 1996. Characterization of the cytoplasmic filament protein gene (cfpA) of Treponema pallidum subsp. pallidum. J. Bacteriol. 178:3177–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mazouni K, Pehau-Arnaudet G, England P, Bourhy P, Saint Girons I, Picardeau M. 2006. The scc spirochetal coiled-coil protein forms helix-like filaments and binds to nucleic acids generating nucleoprotein structures. J. Bacteriol. 188:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang RF, Bartle S, Otto R, Stassinopoulos A, Rogers M, Plamann L, Hartzell P. 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 186:6168–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Specht M, Schatzle S, Graumann PL, Waidner B. 2011. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J. Bacteriol. 193:4523–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. 2009. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 5:e1000669 doi:10.1371/journal.ppat.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Graumann PL. 2009. Dynamics of bacterial cytoskeletal elements. Cell Motil. Cytoskeleton 66:909–914 [DOI] [PubMed] [Google Scholar]
- 51. Bagchi S, Tomenius H, Belova LM, Ausmees N. 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 70:1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol. 62:5–14 [DOI] [PubMed] [Google Scholar]
- 53. Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- 54. Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. 2013. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 110:E397–E406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. 2010. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 29:315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kürner J, Frangakis AS, Baumeister W. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436–438 [DOI] [PubMed] [Google Scholar]
- 57. Defeu Soufo HJD, Graumann PL. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mazza P, Noens EE, Schirner K, Grantcharova N, Mommaas AM, Koerten HK, Muth G, Flardh K, van Wezel GP, Wohlleben W. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60:838–852 [DOI] [PubMed] [Google Scholar]
- 60. Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. 2002. How myxobacteria glide. Curr. Biol. 12:369–377 [DOI] [PubMed] [Google Scholar]
- 61. Mignot T. 2007. The elusive engine in Myxococcus xanthus gliding motility. Cell. Mol. Life Sci. 64:2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45:21–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. 2011. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. U. S. A. 108:7559–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Letek M, Fiuza M, Villadangos AF, Mateos LM, Gil JA. 2012. Cytoskeletal proteins of actinobacteria. Int. J. Cell Biol. 2012:905832 doi:10.1155/2012/905832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Heichlinger A, Ammelburg M, Kleinschnitz EM, Latus A, Maldener I, Flärdh K, Wohlleben W, Muth G. 2011. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J. Bacteriol. 193:1533–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Erickson HP. 1995. FtsZ, a prokaryotic homolog of tubulin? Cell 80:367–370 [DOI] [PubMed] [Google Scholar]
- 68. Löwe J, Amos LA. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206 [DOI] [PubMed] [Google Scholar]
- 69. Bernander R, Ettema TJ. 2010. FtsZ-less cell division in archaea and bacteria. Curr. Opin. Microbiol. 13:747–752 [DOI] [PubMed] [Google Scholar]
- 70. Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Addinall SG, Lutkenhaus J. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231–237 [DOI] [PubMed] [Google Scholar]
- 72. Erickson HP, Taylor DW, Taylor KA, Bramhill D. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U. S. A. 93:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lutkenhaus J, Addinall SG. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93–116 [DOI] [PubMed] [Google Scholar]
- 74. Erickson HP. 1997. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 7:362–367 [DOI] [PubMed] [Google Scholar]
- 75. McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14:243–254 [DOI] [PubMed] [Google Scholar]
- 76. Jakimowicz D, van Wezel GP. 2012. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85:393–404 [DOI] [PubMed] [Google Scholar]
- 77. McCormick JR. 2009. Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Biotechnol. 12:689–698 [DOI] [PubMed] [Google Scholar]
- 78. Bennett JA, Yarnall J, Cadwallader AB, Kuennen R, Bidey P, Stadelmaier B, McCormick JR. 2009. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J. Bacteriol. 191:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. 2007. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 21:1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tinsley E, Khan SA. 2006. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 188:2829–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sontag CA, Staley JT, Erickson HP. 2005. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J. Cell Biol. 169:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schlieper D, Oliva MA, Andreu JM, Lowe J. 2005. Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc. Natl. Acad. Sci. U. S. A. 102:9170–9175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jenkins C, Samudrala R, Anderson I, Hedlund BP, Petroni G, Michailova N, Pinel N, Overbeek R, Rosati G, Staley JT. 2002. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl. Acad. Sci. U. S. A. 99:17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. 2011. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 9:e1001213 doi:10.1371/journal.pbio.1001213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. de Boer PA. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 87. Dajkovic A, Lutkenhaus J. 2006. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11:140–151 [DOI] [PubMed] [Google Scholar]
- 88. Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185 [DOI] [PubMed] [Google Scholar]
- 89. Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. RayChaudhuri D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59:989–999 [DOI] [PubMed] [Google Scholar]
- 93. Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Raskin DM, de Boer PA. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685–694 [DOI] [PubMed] [Google Scholar]
- 95. Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Woldringh CL, Mulder E, Huls PG, Vischer N. 1991. Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol. 142:309–320 [DOI] [PubMed] [Google Scholar]
- 97. Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925 [DOI] [PubMed] [Google Scholar]
- 98. Wu LJ, Errington J. 2012. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 10:8–12 [DOI] [PubMed] [Google Scholar]
- 99. Treuner-Lange A, Aguiluz K, van der Does C, Gomez-Santos N, Harms A, Schumacher D, Lenz P, Hoppert M, Kahnt J, Munoz-Dorado J, Sogaard-Andersen L. 2013. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol. Microbiol. 87:235–253 [DOI] [PubMed] [Google Scholar]
- 100. Traag BA, van Wezel GP. 2008. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Noens EE, Mersinias V, Traag BA, Smith CP, Koerten HK, van Wezel GP. 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58:929–944 [DOI] [PubMed] [Google Scholar]
- 102. Keijser BJ, Noens EE, Kraal B, Koerten HK, van Wezel GP. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225:59–67 [DOI] [PubMed] [Google Scholar]
- 103. van Wezel GP, van der Meulen J, Kawamoto S, Luiten RG, Koerten HK, Kraal B. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182:5653–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25:89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Alexeeva S, Gadella TW, Jr, Verheul J, Verhoeven GS, den Blaauwen T. 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol. 77:384–398 [DOI] [PubMed] [Google Scholar]
- 106. Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887 [DOI] [PubMed] [Google Scholar]
- 107. van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B. 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 72:5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 64:1244–1259 [DOI] [PubMed] [Google Scholar]
- 109. Xu Q, Traag BA, Willemse J, McMullan D, Miller MD, Elsliger MA, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Carlton D, Chen C, Chiu HJ, Chruszcz M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Ernst D, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Grzechnik SK, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Marciano D, Minor W, Mommaas AM, Morse AT, Nigoghossian E, Nopakun A, Okach L, Oommachen S, Paulsen J, Puckett C, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Wang S, Weekes D, et al. 2009. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J. Biol. Chem. 284:25268–25279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Flärdh K, Leibovitz E, Buttner MJ, Chater KF. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38:737–749 [DOI] [PubMed] [Google Scholar]
- 111. Willemse J, Mommaas AM, van Wezel GP. 2012. Constitutive expression of ftsZ overrides the whi developmental genes to initiate sporulation of Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rodrigues CD, Harry EJ. 2012. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 8:e1002561 doi:10.1371/journal.pgen.1002561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Monahan LG, Harry EJ. 2013. Identifying how bacterial cells find their middle: a new perspective. Mol. Microbiol. 87:231–234 [DOI] [PubMed] [Google Scholar]
- 114. Gerdes K, Howard M, Szardenings F. 2010. Pushing and pulling in prokaryotic DNA segregation. Cell 141:927–942 [DOI] [PubMed] [Google Scholar]
- 115. Leonard TA, Moller-Jensen J, Lowe J. 2005. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21:6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Møller-Jensen J, Jensen RB, Lowe J, Gerdes K. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Salje J, Zuber B, Lowe J. 2009. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science 323:509–512 [DOI] [PubMed] [Google Scholar]
- 119. Gayathri P, Fujii T, Moller-Jensen J, van den Ent F, Namba K, Lowe J. 2012. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science 338:1334–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hayes F, Barilla D. 2006. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4:133–143 [DOI] [PubMed] [Google Scholar]
- 121. Leonard TA, Butler PJ, Lowe J. 2004. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53:419–432 [DOI] [PubMed] [Google Scholar]
- 122. Banigan EJ, Gelbart MA, Gitai Z, Wingreen NS, Liu AJ. 2011. Filament depolymerization can explain chromosome pulling during bacterial mitosis. PLoS Comput. Biol. 7:e1002145 doi:10.1371/journal.pcbi.1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84 [DOI] [PubMed] [Google Scholar]
- 125. Scholefield G, Errington J, Murray H. 2012. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31:1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hui MP, Galkin VE, Yu X, Stasiak AZ, Stasiak A, Waldor MK, Egelman EH. 2010. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc. Natl. Acad. Sci. U. S. A. 107:4590–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Plitzko JM, Rigort A, Leis A. 2009. Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Curr. Opin. Biotechnol. 20:83–89 [DOI] [PubMed] [Google Scholar]
- 128. Briegel A, Chen S, Koster AJ, Plitzko JM, Schwartz CL, Jensen GJ. 2010. Correlated light and electron cryo-microscopy. Methods Enzymol. 481:317–341 [DOI] [PubMed] [Google Scholar]
- 129. Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. 2007. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160:135–145 [DOI] [PubMed] [Google Scholar]
- 130. van Driel LF, Valentijn JA, Valentijn KM, Koning RI, Koster AJ. 2009. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur. J. Cell Biol. 88:669–684 [DOI] [PubMed] [Google Scholar]
- 131. Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. 2007. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J. Microsc. 227:98–109 [DOI] [PubMed] [Google Scholar]
- 132. Müller-Reichert T, Srayko M, Hyman A, O'Toole ET, McDonald K. 2007. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 79:101–119 [DOI] [PubMed] [Google Scholar]
- 133. Charbon G, Cabeen MT, Jacobs-Wagner C. 2009. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 23:1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Van de Putte P, Van Dillewijn J, Rorsch A. 1964. The selection of mutants of Escherichia coli with impaired cell division at elevated temperature. Mutat Res. 1:121–128 [DOI] [PubMed] [Google Scholar]