Abstract
Bacteria use a number of small basic proteins for organization and compaction of their genomes. By their interaction with DNA, these nucleoid-associated proteins (NAPs) also influence gene expression. Rv3852, a NAP of Mycobacterium tuberculosis, is conserved among the pathogenic and slow-growing species of mycobacteria. Here, we show that the protein predominantly localizes in the cell membrane and that the carboxy-terminal region with the propensity to form a transmembrane helix is necessary for its membrane localization. The protein is involved in genome organization, and its ectopic expression in Mycobacterium smegmatis resulted in altered nucleoid morphology, defects in biofilm formation, sliding motility, and change in apolar lipid profile. We demonstrate its crucial role in regulating the expression of KasA, KasB, and GroEL1 proteins, which are in turn involved in controlling the surface phenotypes in mycobacteria.
INTRODUCTION
Tuberculosis is a major infectious disease with an ever-expanding global importance. Current estimates indicate that about one-third of the human population is latently infected with Mycobacterium tuberculosis, and an estimated 2.5 million people die of tuberculosis annually (1). M. tuberculosis is one of the most successful human pathogens, well adapted to long-term residence inside the host macrophages. The secret of M. tuberculosis as a successful pathogen can be ascribed to its extraordinary stealth and ability to adapt within the host cell during the course of infection (2). Genes that are involved in the adaptation to environmental changes and intracellular survival are controlled by specific transcription factors, as well as by global regulators, such as the nucleoid-associated proteins (NAPs) (3, 4).
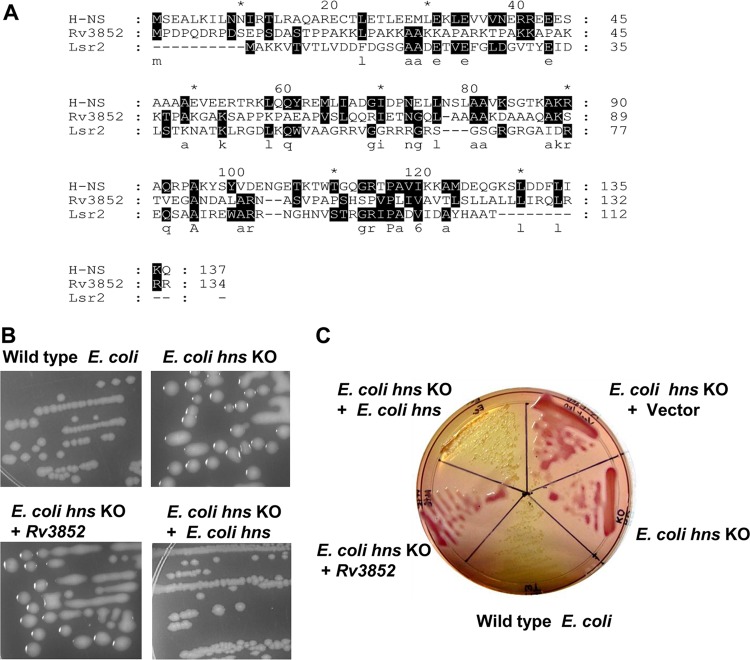
NAPs, also referred to as histone-like proteins, are low-molecular-weight, basic proteins primarily responsible for the compaction of the bacterial genome. Apart from their role in the organization and condensation of the bacterial chromosome, they influence different DNA transaction processes, viz., replication, regulation of gene expression, and recombination (5). H-NS is one of the abundant NAPs in the Enterobacteriaceae and has been well characterized from Escherichia coli and Salmonella enterica serovar Typhimurium (4). In these organisms, H-NS regulates the expression of a large number of genes by cooperative binding to high-affinity sites in the vicinity of the promoters (6, 7). In accordance with its global regulatory role in E. coli and Salmonella, mutations in hns are highly pleiotropic. H-NS homologues from other Gram-negative bacteria share a high degree of sequence similarity with the E. coli protein (8). Lsr2, a functional homologue of H-NS with remote sequence conservation (Fig. 1A), has been identified in various actinomycetes (9). The M. tuberculosis Lsr2 protein exhibits H-NS-like DNA binding properties and has also been shown to complement E. coli H-NS knockout (KO) cells (9, 10). Lsr2 regulates various cellular processes, including antibiotic resistance, biofilm formation, and cell wall biosynthesis (11–13), in mycobacteria, thus exhibiting pleiotropic effects similar to those of H-NS.
Fig 1.
Rv3852 does not complement the E. coli hns knockout strain. (A) Sequence alignment of E. coli H-NS, M. tuberculosis Rv3852, and Lsr2 proteins. The shaded regions denote the sequence conservation, and the letters below indicate the identical amino acids. Every 10th residue in the sequence is marked either by asterisks or by the exact number. (B) The mucoidy phenotype of the hns KO strain was complemented by E. coli hns, but not by Rv3852. (C) Fermentation of salicin on a MacConkey agar plate indicated that Rv3852 fails to complement E. coli H-NS.
Although M. tuberculosis seems to have an underrepresentation of NAPs compared to E. coli (14), one of the NAPs, Rv3852, in the genome sequence was annotated as a possible H-NS (15). Notably, E. coli H-NS showed only ∼10% sequence similarity to Rv3852 (Fig. 1A). Thus, neither Lsr2 nor Rv3852 shows significant sequence similarity to the canonical H-NS, although functionally Lsr2 resembled it in many respects (9, 10). These differences warranted a thorough investigation of the properties of Rv3852. Here, we demonstrate that the properties of Rv3852 are markedly different from those of H-NS. The carboxy-terminal (C-terminal) region of the protein contains a transmembrane helix that anchors the protein to the cell membrane. Its expression in M. smegmatis resulted in pleiotropic phenotypic changes. In addition to the change in nucleoid structure, the alterations include compromised ability of the bacterium to form biofilm, reduction in sliding motility, and altered colony morphology and lipid profile.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Mycobacterium smegmatis mc2155 cells were grown in Middlebrook 7H9 broth or 7H9 agar. The E. coli K-12 strain MC4100 was used as the wild-type strain. PD32, the hns knockout strain of MC4100 (E. coli K-12 MC4100 hns-260::Apr), and PD32/pAF201 (strain PD32 complemented with E. coli hns) were kind gifts from C. J. Dorman (Trinity College, Dublin, Ireland). To analyze biofilm formation, M63 minimal medium supplemented with 0.2% glycerol, 0.5% Bacto-Casitone (BD), 1 mM MgSO4, and 0.7 mM CaCl2 was used. Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; and chloramphenicol, 25 μg/ml.
Sequence alignment and analysis.
Sequence searches of the nonredundant (nr) database were carried out with the BLASTP program (http://www.ncbi.nlm.nih.gov), using the sequence of Rv3852 as a query. Multiple-sequence alignment was carried out using ClustalW2 and edited with the Genedoc software. The transmembrane helix prediction was carried out using the TMHMM Server, v. 2.0 (http://www.cbs.dtu.dk). α-Helical regions were identified with the secondary-structure prediction program TMpred. (http://www.ch.embnet.org/software/TMPRED_form.html). The helical-wheel arrangement of the transmembrane region of Rv3852 was derived from http://cti.itc.virginia.edu/∼cmg/Demo/wheel/wheelApp.html.
Cloning, expression, and purification of Rv3852.
The Rv3852 gene (GenBank CAB06214.1) was PCR amplified from M. tuberculosis H37Rv genomic DNA using the forward primer 5′-ATGGATCCATGCATCATCATCATCATCACGCAGACCCGCAGG-3′, containing the six histidine codons, and the reverse primer 5′-GCTCTAGATCAGCGGCGGCGCAG-3′. The PCR amplicon was cloned into the BamHI and XbaI sites of expression vectors pJAM2 (16) and pBAD33 to generate pJ-Rv3852 and pB-Rv3852, respectively. The C-terminal deletion construct (pJ-Rv3852ΔCTD) was created by introducing a stop codon after 330 nucleotides. Plasmid pJ-Rv3852 or pJ-Rv3852ΔCTD was electroporated into M. smegmatis, and pB-Rv3852 was used to transform the E. coli PD32 strain. The M. smegmatis transformants were grown on Middlebrook 7H9 broth. The recombinant protein was expressed by inducing the cells with 2% acetamide for 8 h at 37°C. Cells were harvested; resuspended in buffer containing 10 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM imidazole; and lysed using a French press. The lysate was centrifuged at 13,000 rpm for 20 min at 4°C. The pellet was subjected to extraction in the presence of 0.5% Triton X-100 and 800 mM KCl. The supernatant was mixed with Ni-nitrilotriacetic acid metal affinity chromatography matrix (Qiagen) and purified according to the manufacturer's instructions. The protein-containing fractions were pooled and dialyzed against buffer B (10 mM Tris-HCl, 100 mM KCl, and 1 mM EDTA), applied to an SP Sepharose HiTrap column, and eluted with 0.1 to 1 M linear KCl gradient.
Glucoside fermentation assay.
An aryl-β-d-glucoside fermentation assay was carried out in MacConkey agar (Difco) supplemented with 0.5% salicin. E. coli strains were streaked on the plates and incubated at 37°C for 18 h. MacConkey agar is an indicator medium used to differentiate Gram-negative bacteria on the basis of their sugar utilization abilities. Those bacteria that ferment the given sugar produce acidic by-products, causing a drop in pH and giving rise to pink colonies, while the others that fail to ferment the sugar grow using the peptone and produce ammonia, leading to an increase in pH, resulting in yellow/white colonies.
Electrophoretic mobility shift assays (EMSA).
End-labeled 30-bp double-stranded DNA (5′CCAACTTCCCTAACCCAGCTGCGATCCGTT3′) (1 nM) was incubated with increasing concentrations of protein on ice for 20 min in the binding buffer (10 mM HEPES, pH 7.4, and 50 mM KCl) in a 20-μl reaction volume. DNA-protein complexes were electrophoresed on 7% (wt/vol) polyacrylamide gels (39:1 acrylamide-bisacrylamide) in TBE (45 mM Tris-borate, pH 8.3, 1 mM EDTA) at 100 V in 4°C for 2.5 h. The DNA-protein complexes were visualized by phosphorimaging.
Topoisomerase assays.
The M. smegmatis DNA gyrase and topoisomerase I (TopoI) were purified as described previously (17, 18). For supercoiling, 400 ng of relaxed circular pUC18 DNA was incubated with Rv3852 in a buffer (35 nM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM potassium glutamate, 2 mM spermidine, 1.4 mM ATP, 50 μg/ml bovine serum albumin [BSA], 90 μg/ml yeast tRNA, and 5% glycerol) and M. smegmatis DNA gyrase at 37°C for 30 min. After heat inactivation of the assay mixture at 68°C and Proteinase K treatment (5 μg/20-μl reaction mixture) at 37°C for 15 min, the products were resolved on a 1% agarose gel. To monitor the DNA relaxation, negatively supercoiled pUC18 DNA (400 ng) was incubated with Rv3852 (10 to 500 nM) in reaction buffer (40 mM Tris-HCl, pH 8.0, 20 mM NaCl, 5 mM MgCl2, and 1 mM EDTA) at room temperature for 15 min, followed by the addition of 5 nM M. smegmatis TopoI and further incubation at 37°C for 30 min. The reactions were terminated and analyzed as described above.
Western blot analysis.
The cellular level of Rv3852 was determined by semiquantitative Western blotting, as described previously (19). Briefly, the M. tuberculosis H37Ra cells were grown to an optical density at 600 nm (OD600) of 0.6, corresponding to the mid-log phase (day 5) of growth, and cells were suspended in lysis buffer and boiled for 45 min. The total cell lysate was subjected to SDS-PAGE, along with different amounts of purified Rv3852 (40, 30, 20, and 15 ng), and detected by immunoblotting with anti-Rv3852 antibody. Quantitation of band intensities was carried out by scanning the bands and analyzing the image with Image Gauge V2.54 software. The number of molecules per cell was obtained from the total number of cells used for the preparation of cell lysates. To compare the levels of Rv3852 expression in different cells, M. tuberculosis, E. coli hns KO harboring pB-Rv3852, and M. smegmatis cells harboring pJ-Rv3852, were grown to mid-log phase. Equal amounts of total cell lysate were resolved on 12% SDS-PAGE, followed by immunoblotting, as described above. The experiments were repeated three times.
Fluorescence microscopy and transmission electron microscopy.
M. smegmatis cells harboring pJ-Rv3852, pJ-Rv3852ΔCTD, or pJAM2 vector alone (control) were grown in Middlebrook 7H9 in the presence of 2% acetamide. Cells were fixed overnight with phosphate-buffered saline (PBS) containing 1% toluene and 2% Triton X-100 and stained with 0.1 mg/ml 4′-6-diamidino-2-phenylindole (DAPI). The cells were visualized in a Zeiss Axio Imager fluorescence microscope under a 100× objective. To analyze the ultrastructure of the cells by transmission electron microscopy, samples were prepared following the previously published protocols (20). In brief, the cells were washed, fixed with 4% paraformaldehyde and 2.5% glutaraldehyde, postfixed with 1% OsO4, dehydrated in a graded series of acetone, and embedded in Spurr's resin. Thin sections (70 nM) were cut using a Reichert Ultracut E microtome, placed on a copper grid, and stained with 2% uranyl acetate and Reynold's lead citrate. A JEOL TEM 1200 EX electron microscope operating at 80 kV was used for imaging.
Immunoelectron microscopy.
The bacteria were first embedded in agar and fixed in a mixture of 4% freshly prepared paraformaldehyde and 0.5% glutaraldehyde in PBS (pH 7.4). The samples were then treated with 0.05 M NH4Cl buffer for 15 min and washed with PBS. The cells were dehydrated in graded ethanol and embedded in London Resin white. Ultrathin sections were mounted on nickel grids and treated with blocking solution (2% BSA in PBS). The sections were incubated overnight with affinity-purified mouse anti-Rv3852 polyclonal antibody, followed by anti-mouse IgG–gold for 30 min. After five washings, the sections were poststained with 2% uranyl acetate in methanol, followed by staining with lead citrate.
Subcellular fractionation.
Subcellular fractionation was carried out as described previously (21) with minor modifications. M. smegmatis cells were resuspended in buffer containing 10 mM Tris HCl, pH 7.5, 100 mM NaCl, and 0.1 mM EDTA and lysed by sonication. The lysates were treated with DNase I and RNase for 30 min, and the resulting lysate was subjected to differential centrifugation as described previously (21). The pellet from centrifugation at 27,000 × g was resubmitted to lysis and recentrifuged at 1,000 × g, and the resulting supernatant was centrifuged at 27,000 × g. The supernatants from the two rounds of centrifugation at 27,000 × g were pooled and centrifuged at 100,000 × g. The resulting pellet contained the cell membranes, whereas the supernatant corresponded to the cytosol. The factions were loaded onto 12% SDS-PAGE, and the presence of the recombinant proteins was detected by Western blotting with anti-His antibody.
To determine the distribution of DNA in the membrane fraction, equal amounts of M. smegmatis cell pellets were resuspended in lysis buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA) and lysed by sonication. The lysate was treated with 5 μg/ml RNase A for 1 h, and 5 mg of the total protein from the 27,000 × g supernatants of the respective lysates were subjected to fractionation as described above. The membrane fractions were solubilized with 8 M urea and diluted in lysis buffer. The DNA content was estimated by DAPI-based fluorescence assay (22).
Pellicle and biofilm formation assay.
Pellicle formation by different strains was monitored by growing the cultures of M. smegmatis without shaking at 37°C for 24 h in Middlebrook 7H9 medium devoid of Tween 80. An assay for biofilm formation was carried out as described previously (12) with minor modifications. M63 medium containing 2% acetamide (2 ml) was added to 24-well polyvinyl chloride (PVC) plates and inoculated with cells to an optical density at 600 nm of 0.03. The plates were incubated at 37°C for 48 h, and free-floating cells were washed with deionized water, stained with 1% crystal violet, and assayed by spectrophotometric reading of the ethanol extract at 570 nm.
Colony morphology analysis and sliding motility assay.
M. smegmatis cells transformed with pJAM2, pJ-Rv3852, or pJ-Rv3852ΔCTD were grown on Middlebrook 7H9 agar containing 2% acetamide to analyze the colony morphology. Motility assays were carried out as described previously (23). Briefly, cells were cultured in 7H9 broth to mid-logarithmic phase (OD600, 0.8 to 1.0) before 2-μl aliquots were spotted onto motility medium consisting of M63 salts supplemented with 0.5% Casamino Acids, 0.2% glycerol, 1 mM MgCl2, and 10 μM FeCl2 and solidified with 0.3% (wt/vol) agarose. The plates were incubated for 24 h at 37°C under humidified conditions.
Cell wall lipid analysis.
M. smegmatis cells were grown on Middlebrook 7H9 medium, and equal cell mass (50 mg) was taken from control and Rv3852- and Rv3852ΔCTD-expressing cells for lipid analysis. Lipid analysis was carried out using thin-layer chromatography (TLC) on silica gel 60 F254 plates (Merck). The glycopeptidolipids (GPLs) were isolated from cells as described previously (23). Deacetylation of lipid extracts was carried out by alkaline methanolysis. TLC plates were developed in the solvent mixture, chloroform-methanol-water (90:10:1), after application of the GPL samples. The GPL spots were visualized by spraying 10% H2SO4 in ethanol and heating at 110°C. The apolar and polar lipids were prepared from M. smegmatis cells according to the published procedures (24). Apolar lipids were developed with chloroform-methanol (96:4; three times) in the first dimension and toluene-acetone (80:20) in the second dimension. Lipids were detected by charring with 5% phosphomolybdic acid.
Proteomics.
M. smegmatis control and Rv3852- and Rv3852ΔCTD-expressing cells were grown in 7H9 medium with 2% acetamide at 37°C to an OD600 of ∼1.5. The cells were harvested, washed twice with PBS, and resuspended in 2 ml of lysis buffer (10 mM HEPES, pH 8.0, 7% urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2 M thiourea, 10 mM dithiothreitol [DTT]), followed by sonication. The lysate was cleared with a ReadyPrep 2-D Cleanup Kit (Bio-Rad). Immobilized pH gradient strips, pH 4 to 7 (Bio-Rad; 17 cm), were rehydrated with 300 μg of total protein. Isoelectric focusing (IEF) was carried out on a Protean i12 IEF Cell (Bio-Rad) using the following steps: (i) 0 to 500 V linear for 2 h, (ii) 250 V rapid for 2 h, (iii) 250 to 6,000 V linear for 3 h, and (iv) 6,000 V constant to 50 kVh. After equilibration, the strips were loaded and resolved on 10% SDS-PAGE. The gels were stained with a ProteoSilver Kit (Sigma-Aldrich), and the spots that differed in intensity between the samples were identified by mass spectrometry.
RNA extraction and qPCR.
RNA was extracted from E. coli, M. smegmatis, and M. tuberculosis cells using a Qiagen RNeasy kit according to the manufacturer's protocol. From the total RNA, cDNAs were synthesized using a Fermentas first-strand synthesis kit. cDNA generated with random primers was used for quantitative real-time PCR (qPCR), with SYBR green as the indicator dye. The expression of the groEL1, kasA, and kasB genes was quantified after normalization of RNA levels to the expression of the rpoB genes, as previously described (19). The qPCR cycling conditions were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 20 s.
RESULTS AND DISCUSSION
Rv3852 does not complement an E. coli hns knockout strain.
From the sequence alignment of E. coli H-NS and Rv3852 depicted in Fig. 1A, it is evident that the two proteins have little similarity; although they are annotated as H-NS, only 12 residues are conserved between them. Initially, to determine whether Rv3852 could functionally mimic H-NS and complement its deficiency in an E. coli hns KO strain, the gene was cloned into the E. coli expression vector pBAD33 and transformed into an E. coli hns KO strain. Western blot analysis using anti-Rv3852 antibody indicated that the expression of the protein is ∼1.7-fold higher than the endogenous level in M. tuberculosis (see Fig. S1A in the supplemental material). Deficiency of H-NS in E. coli is associated with several phenotypes, including mucoidy (25), glucoside utilization (26), and hemolytic activity (27). The E. coli hns KO strain harboring the pBAD33 vector alone or a plasmid expressing Rv3852 remained mucoid, while the strain complemented with E. coli hns became nonmucoid and resembled the wild-type colonies (Fig. 1B). Next, whether Rv3852 is functionally similar to H-NS in the regulation of β-glucoside metabolism was investigated. Genes necessary for uptake and fermentation of aryl-β-d-glucosides are included in the bgl operon, which is predominantly cryptic due to its repression by H-NS (26). The fermentation of aryl-β-d-glucosides was examined on MacConkey agar plates supplemented with 0.4% salicin. The wild-type E. coli strain, unable to ferment salicin, grew as yellow colonies, and the hns KO strain appeared red, indicating the fermentation of salicin. The hns KO strain harboring Rv3852 also remained red, while the positive-control E. coli hns-complemented strain gave rise to yellow colonies (Fig. 1C). Thus, in spite of expression ∼1.7-fold-higher than the endogenous level in M. tuberculosis (see Fig. S1A in the supplemental material), Rv3852 did not functionally complement the E. coli hns KO phenotypes. On the other hand, Lsr2, another nucleoid-associated protein found in mycobacteria and considered yet another homologue of H-NS, complements the E. coli hns KO phenotypes (9). Further, Lsr2 has been shown to possess H-NS-like DNA binding properties in vitro, indicating that there is a functional relatedness between Lsr2 and H-NS (10).
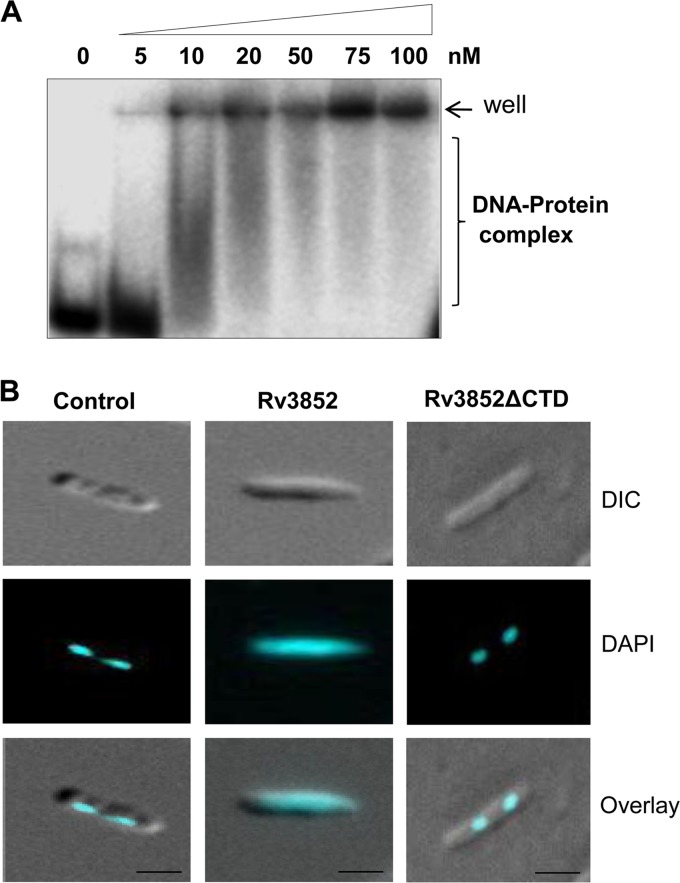
Next, to study the biochemical properties of the protein, the Rv3852 gene was cloned and expressed in M. smegmatis using the pJAM2 inducible expression system (16). The ectopic expression of the protein caused marginal reduction in the growth of M. smegmatis cells (see Fig. S1A and B in the supplemental material). The recombinant protein showed an anomalous mobility of ∼21 kDa in 15% SDS-PAGE and was identified as Rv3852 by peptide mass fingerprinting (data not shown). The purified protein formed DNA-protein complexes upon incubation with 30-bp double-stranded DNA substrate (Fig. 2A). The protein bound to DNA in a sequence-independent manner, and with increasing protein concentrations, the DNA-protein complexes were increased. At higher concentrations (75 to 100 nM) of the protein, the DNA-protein complexes failed to enter the gel, suggesting the formation of higher-order oligomeric DNA-protein structures, as reported previously (28). These structures presumably formed due to the cooperative binding of the protein on DNA. Self-association, DNA-dependent oligomerization, and sequence-independent DNA binding are the characteristic features of many NAPs (29, 30). In addition to the DNA binding properties, the abundance of the NAPs in the cells contributes to the compact organization of the genome. Semiquantitative Western blotting using ani-Rv3852 antibody (see Fig. S2 in the supplemental material) indicated that the endogenous Rv3852 level in M. tuberculosis H37Ra was ∼80,000 molecules/cell at exponential phase, a level comparable to those of major NAPs of E. coli (14).
Fig 2.
Rv3852 binds to DNA and changes the nucleoid morphology. (A) The DNA binding property of Rv3852 was analyzed by EMSA using 30 bp double-stranded DNA (dsDNA). (B) Fluorescent micrographs of the DAPI-stained nucleoid of M. smegmatis cells. (Left) Condensed nucleoid in control cells. (Middle) Decompaction of nucleoid in Rv3852-expressing cells. (Right) Compact nucleoid in Rv3852ΔCTD-expressing cells. The cells shown are representative of ∼85% of the population. Bars, 1 μM. DIC, differential interference contrast.
Alteration of nucleoid morphology.
When the nucleoid morphology was examined using fluorescence microscopy, Rv3852-expressing cells were of normal size and shape, but the DAPI-stained nucleoids were more diffused and spread throughout the cell than the control cells, which had compact nucleoids (Fig. 2B). The decompaction of the nucleoid seen with the overproduction of Rv3852 is in contrast to the overexpression of E. coli H-NS (31). The NAPs are the key players in the organization and compaction of bacterial chromosomes (32–34). Most of the NAPs alter the trajectory of the DNA by bridging, bending, or wrapping it (32, 33, 35). Genome-wide studies in E. coli indicated that they are directly involved in the formation or maintenance of topological domains in the chromosome, in addition to topoisomerases, which also influence the nucleoid structure (5). The topological status of the genome is maintained by the combined actions of DNA gyrase, which catalyzes negative supercoiling, and topoisomerase I, which relaxes DNA and prevents hypernegative supercoiling (36). Genetic studies in E. coli showed that some of the NAPs, such as HU, cross talk with topoisomerases and influence their function (37). To examine whether Rv3852 has any influence on topoisomerase I and DNA gyrase activities, coupled topoisomerase assays were carried out as described in Materials and Methods. There was no significant effect of Rv3852 on the supercoiling activity of gyrase or on the DNA relaxation activity of topoisomerase I (see Fig. S3A and B in the supplemental material), indicating that the change in nucleoid morphology seen with the protein is most likely due to the direct DNA-protein interaction and not to any effect through topoisomerases.
Ultrastructure analysis.
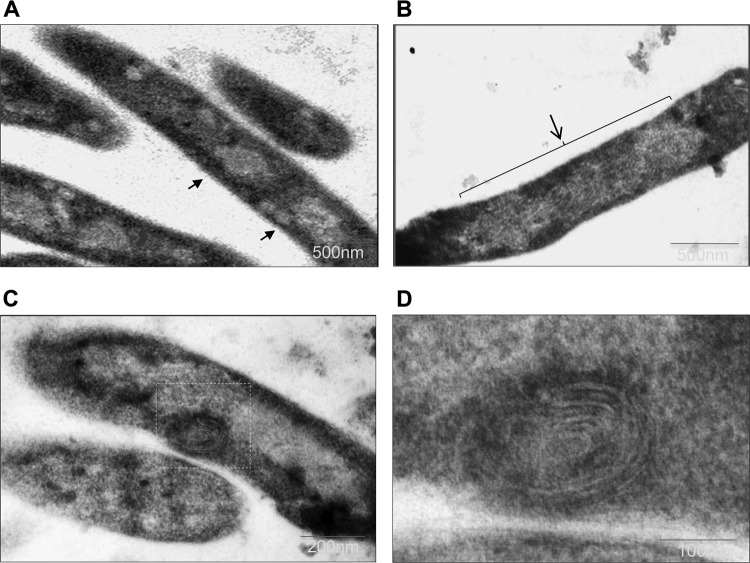
To investigate the finer details of the nucleoid organization, ultrastructure analysis was carried out using transmission electron microscopy. The electron micrograph of Rv3852-expressing cells showed an altered nucleoid structure, characterized by less condensed and decompacted nucleoid (Fig. 3B and C), compared to that of control cells (Fig. 3A). These results correlated well with fluorescence microscopic studies. In addition to the difference in nucleoid morphology, unusual membrane structures were also observed in Rv3852-expressing cells (Fig. 3C). Higher-magnification imaging revealed the presence of spiral membrane invaginations in the cytoplasm of the cells (Fig. 3D) resembling membrane structures associated with the overproduction of the b subunit of F1Fo ATP synthase (38) or chemotaxis receptor Tsr protein (39) in E. coli. In this context, it is noteworthy that electron tomography analysis had demonstrated that the overproduction of integral membrane proteins led to the formation of large membrane networks that extended into the cytoplasm as zippered and rounded assemblies, probably to accommodate large amounts of membrane proteins within a defined space (39). There are also examples of naturally occurring membrane clusters in certain cyanobacteria (40). The formation of the extended membrane structures in Rv3852-overexpressing cells thus indicated that the protein may be associated with the membrane.
Fig 3.
Ultrastructure analysis of Rv3852-expressing M. smegmatis cells. (A and B) The compact nucleoid regions of control cells are indicated by arrows (A), and the decompacted nucleoid of an Rv3852-expressing cell is indicated by an arrow and bracket (B). (C) Cells expressing Rv3852 showing the presence of a spiral membrane structure (boxed). (D) Higher-magnification image of the spiral structure.
Rv3852 has a transmembrane helix.
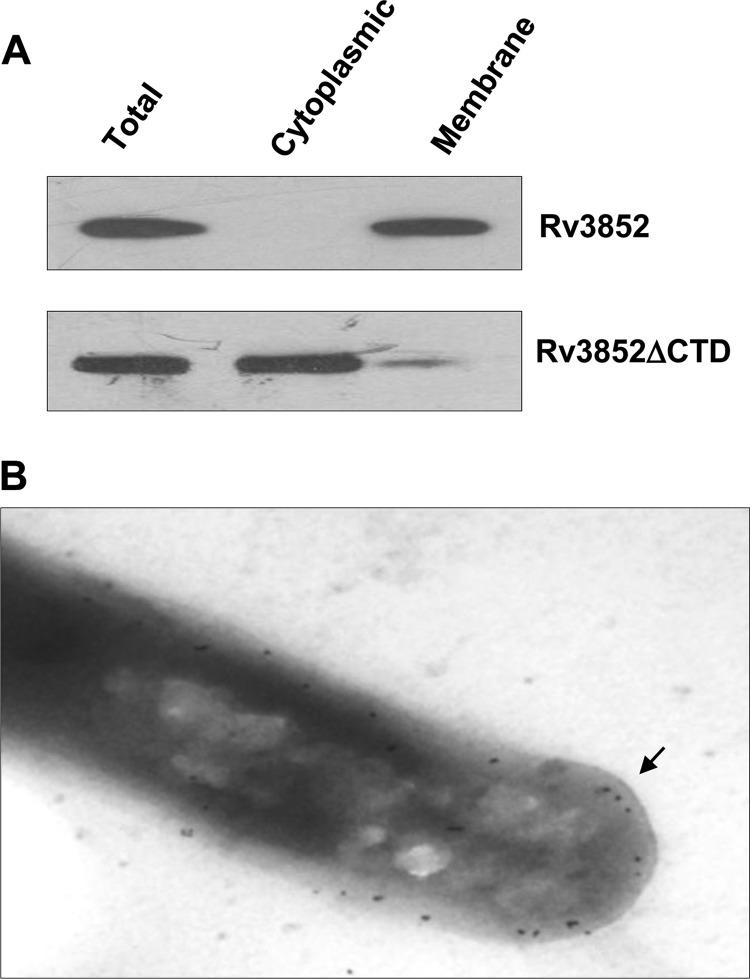
To investigate the possibility that Rv3852 is indeed a membrane-bound protein, first, a detailed bioinformatics analysis was carried out. The multiple-sequence alignment of Rv3852 homologues among the pathogenic mycobacterial species showed a highly conserved sequence at the C-terminal end (see Fig. S4A in the supplemental material), and the hydropathy index analysis revealed it to be hydrophobic (see Fig. S4B in the supplemental material). The TMHMM and TMpred tools predicted a transmembrane helix spanning from 111 to 132 amino acids (see Fig. S5A and B in the supplemental material). Analysis of the predicted helix by a helical-wheel algorithm indicated the helical arrangement of the hydrophobic amino acids in the helix (see Fig. S5C and D in the supplemental material). The subcellular fractionation of cells expressing Rv3852 and Western blot analysis with anti-Rv3852 antibody demonstrated the localization of the protein in the membrane fraction (Fig. 4A). To investigate further the role of the predicted C-terminal transmembrane helix in membrane localization, a truncated Rv3852 construct (Rv3852ΔCTD) that lacked the C-terminal 24 amino acids was generated. Unlike the full-length protein, the truncated protein was retained predominantly in a cytosolic fraction (Fig. 4A), suggesting that the C-terminal helix is responsible for membrane localization of the protein. To analyze the localization of endogenous Rv3852 in M. tuberculosis H37Ra cells, immunoelectron microscopy was carried out with anti-Rv3852 polyclonal antibody. Similar to the above observations, the protein was found localized in the membrane (Fig. 4B). A recent study on the membrane proteome of M. tuberculosis also showed that Rv3852 is present in the membrane (41). Thus, in addition to its role in nucleoid organization, Rv3852 may function as an anchorage to tether the DNA to the membrane. To test whether Rv3852 indeed has the DNA anchorage property, the DNA was isolated from membrane fractions of Rv3852- and Rv3852ΔCTD-expressing cells, as well as from control cells. The ∼3-fold increase of the DNA content in the membrane fraction of Rv3852-expressing cells compared to the control and Rv3852ΔCTD cells (Table 1) indicated that Rv3852 can tether DNA to the membrane. To further assess the contribution of Rv3852 in anchoring DNA to the membrane, the nucleoid morphology of the cells expressing the Rv3852ΔCTD protein was analyzed. Unlike the full-length protein, the nucleoids of Rv3852ΔCTD-expressing cells appeared compact (Fig. 2B, right). Thus, Rv3852 could be decompacting the nucleoid by tethering DNA to the membrane.
Fig 4.
Rv3852 is localized in the cell membrane. (A) Subcellular fractionation of Rv3852-containing cells indicating the presence of the protein in membrane fractions. Truncated Rv3852 lacking the transmembrane helix (Rv3852ΔCTD) localizes in the cytosolic fraction. (B) Immunogold staining of M. tuberculosis cells with anti-Rv3852 antibody. The gold particles (arrow) indicate the presence of Rv3852 in the membrane.
Table 1.
DNA contents of membrane fractions derived from different M. smegmatis strains
| M. smegmatis strain | DNA content (ng/20 µg of membrane protein)a |
|---|---|
| Control | 3.02 ± 0.15 |
| Rv3852 | 9.84 ± 0.22 |
| Rv3852ΔCTD | 3.68 ± 0.11 |
Mean ± standard deviation; n = 3.
Phenotypic changes: pellicle and biofilm formation, colony morphology, and sliding motility.
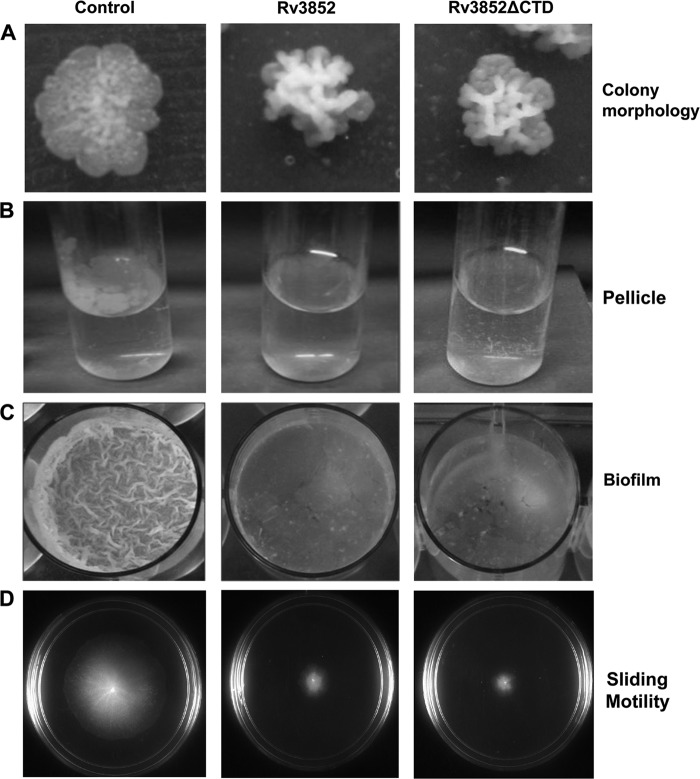
To understand the physiological role of Rv3852, the phenotypic changes associated with the ectopic expression of the protein in M. smegmatis were examined. Cells showed increased clumping (data not shown) upon expression of the protein. The strain expressing Rv3852 showed wrinkled, rough, and dry colonies with an uneven colony surface in contrast to control colonies with more even edges (Fig. 5A). Defects in cell wall components can modify the surface properties of the individual cells, alter cell-to-cell interactions, and give rise to colonies with visibly different morphologies (42). Mycobacteria grown in the absence of detergent form a surface pellicle in standing liquid media, and this property is attributed to the high lipid content of the cells (43). However, in contrast to the control cells, which formed pellicles, pellicles were not detected in the Rv3852-expressing cells (Fig. 5B). Pellicle growth and the formation of biofilm are well correlated in different bacterial species (44, 45). Biofilms are considered to be surface-associated bacterial communities comprising microcolonies surrounded by extracellular matrix (46). Mycobacteria form biofilms either attached to hydrophobic solid surfaces or floating as pellicles on the surfaces of liquid culture media. Therefore, the ability of the Rv3852-expressing strain to form biofilms on PVC plastic surfaces was tested using an M63-based liquid medium. Biofilm formation was significantly reduced in the strain compared to that of the control cells (Fig. 5C). Quantification by crystal violet staining assay revealed an ∼5-fold reduction in the biofilm upon Rv3852 expression (data not shown).
Fig 5.
Phenotypic changes of M. smegmatis cells. The Rv3852- and Rv3852ΔCTD-expressing cells show altered colony morphology (A), reduced pellicle (B) and biofilm formation (C), and decreased sliding motility (D) compared to control cells.
Biofilm formation and sliding motility are well correlated in mycobacteria (47). The members of the genus Mycobacterium were originally considered to be nonmotile until sliding motility in M. smegmatis and Mycobacterium avium was demonstrated on solid surfaces (23). As with biofilm formation, the sliding motility of the Rv3852-expressing strain on a motility plate showed an altered phenotype. The diameter of the sliding halo showed ∼4-fold reduction in size (Fig. 5D), indicating reduced sliding motility.
To investigate whether the changes in surface phenotypes were due to the membrane localization of Rv3852, the truncated protein, Rv3852ΔCTD, which exhibited cytosolic localization (Fig. 4A), was expressed in M. smegmatis and tested for the phenotypic changes. The expression of the truncated protein in M. smegmatis cells showed phenotypes similar to that of expression of the full-length protein (Fig. 5). These data indicated that the change in surface properttes is not solely correlated with the membrane localization, and there may be a regulatory role for Rv3852 in modulation of biofilm formation and different surface phenotypes. Several key regulatory proteins of biofilm formation have been identified in different bacteria. It has been shown that CsrA (a carbon storage regulator), an RNA-binding protein, and Fis, a NAP, act as regulators of biofilm formation in E. coli and Dickeya dadantii, respectively (48, 49). Overexpression of a diguanylate cyclase or phosphodiesterase resulted in modulating the intracellular cyclic di-GMP (c-di-GMP) pool and consequently induced the dispersion of Shewanella oneidensis biofilms (50). A relationship between a defect in biofilm formation and inability to slide has been shown in M. smegmatis GPL biosynthesis gene (atf1) mutants (51). Biofilm formation in M. smegmatis is associated with changes in the profile of mycolic acids in the mycobacterial cell wall, and GroEL1, a dedicated chaperone, has been shown to play a regulatory role in this process (52). Lsr2 has also been shown to be a regulator of mycobacterial biofilm formation; cells lacking Lsr2 were unable to form biofilm (12). In contrast to these observations, expression of Rv3852 affected biofilm formation. Thus, it appears that the regulatory role of Rv3852 in biofilm formation is different from and opposite to that of Lsr2.
Cell wall lipid analysis.
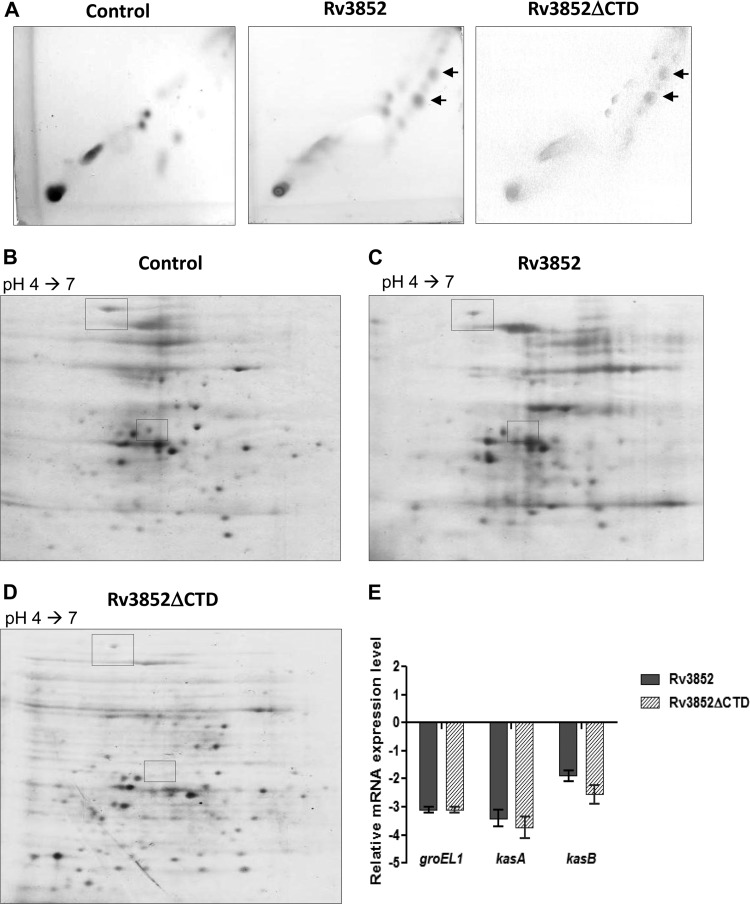
The changes in colony morphology and biofilm formation suggested that Rv3852 expression affected some component of the mycobacterial cell wall. Since the mutation in the GPL biosynthetic pathway affects biofilm formation (51), we first tested whether the GPL composition was changed due to Rv3852 expression. GPLs extracted from the strains were analyzed by TLC. The Rv3852- and Rv3852ΔCTD-expressing cells exhibited normal GPLs with no detectable change in the profile (data not shown), and hence, we examined the profiles of other cell wall lipids. TLC analysis of polar lipids also did not reveal any differences (data not shown), but apolar lipids had distinct TLC profiles. Two lipid spots present in Rv3852- and truncated-protein-expressing cells (Fig. 6A) were detected in relatively small amounts in the control cells, indicating that the protein alters the cell wall lipid composition by modulating the lipid biosynthesis pathway. Contributions by lipids, in addition to outer membrane proteins and extracellular polysaccharides, in biofilm matrix formation have been reported previously (46, 52, 53). The alteration in the extracellular-matrix content can probably explain both the reduced sliding motility and the defective biofilm formation observed in cells expressing Rv3852.
Fig 6.
Effect of Rv3852 expression on the M. smegmatis lipid profile and proteome. (A) 2D-TLC profile of apolar lipids developed with chloroform-methanol (96:4; three times) in the first dimension and toluene-acetone (80:20) in the second dimension. The arrow indicates the lipid spots, which are more intense in Rv3852- and Rv3852ΔCTD-expressing cells. (B to D) Analysis of the total proteome profile of M. smegmatis cells. The boxes indicate the protein species present at reduced levels in the Rv3852- and truncated-protein-expressing cells relative to the control. (E) Relative expression profile of groEL1, kasA, and kasB in Rv3852- and Rv3852ΔCTD-expressing cells as determined by qPCR. The cycle threshold (CT) value of each gene was normalized with the housekeeping gene rpoB. The error bars indicate standard deviations.
All the results described so far indicate that Rv3852 differs from E. coli H-NS in its properties, whether it is the primary sequence, the role in nucleoid organization, or the phenotypic characteristics. On the other hand Lsr2 of M. tuberculosis, although structurally distinct, exhibited characteristics similar to those of E. coli H-NS (9). Hence, we suggest that Rv3852 could be renamed mycobacterial DNA binding protein 2 (MDP2), since HU (Rv2986c), another mycobacterial nucleoid-associated protein, is already named MDP1 (54).
MDP2 regulates the expression of lipid biosynthetic genes.
To analyze the effect of MDP2 expression on the total proteome, two-dimensional (2D) PAGE analysis was carried out as described in Materials and Methods. The comparative profiling of control cells versus cells expressing MDP2 or the truncated protein resulted in identification of two unique spots that show reduction upon expression of the proteins (Fig. 6B to D). Peptide mass fingerprinting analysis identified the spots as GroEL1 chaperone and KasA. GroEL1 is a dedicated chaperone involved in heat shock response in mycobacteria. The lack of GroEL1 results in reduction in biofilm maturation and accumulation of free mycolic acids in M. smegmatis cells (52). GroEL is also known to stabilize the KasA protein, a key component of the mycolic acid biosynthetic pathway (52). Biosynthesis of mycolic acid requires two types of fatty acid synthase (FAS), FAS I, a multifunctional enzyme capable of de novo synthesis of medium-chain fatty acids, and the ACP-dependent FAS II system, responsible for their elongation (55). The FAS II pathway is composed of two main transcriptional units, fabD-acpM-kasA-kasB-accD6 (the fasII operon) and mabA-inhA (15). The KasA and KasB proteins expressed from the fasII operon are involved in elongation of mycolic acid chains (55) and deficiency or reduction in their levels is associated with reduction in biofilm formation (52), altered colony morphology (56), and accumulation of precursors of mycolic acid biosynthesis (55, 56). To investigate the regulatory role of MDP2 in the expression of these genes, the transcript level was estimated by qPCR. The steady-state groEL1 mRNA level was reduced by ∼3-fold, while kasA and kasB expression was downregulated by ∼3.5- and ∼2-fold, respectively, in MDP2- and truncated-protein-expressing cells compared to control cells (Fig. 6E). The repression of groEL1 and the fasII operon genes seen when MDP2 was expressed indicated that the protein might be acting as a negative regulator of their transcription.
In conclusion, MDP2 is a distinct NAP, different in all its properties from E. coli H-NS and M. tuberculosis Lsr2. The protein apparently binds to DNA in a nonspecific fashion and alters nucleoid compaction. Unlike the action of Lsr2, its expression leads to reduced biofilm formation and sliding motility and a change in colony morphology. These changes could be attributed to alteration in the lipid profile due to transcriptional downregulation of groEL1 and fasII operon genes by MDP2, suggesting a major role for the protein in regulation of lipid biosynthesis in mycobacteria. Thus, it appears that in M. tuberculosis each of the NAPs characterized (MDP1, MDP2, and Lsr2) has a designated physiological role important to the cellular function. Such designated roles may be necessary when there is an underrepresentation of NAPs in the genome.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. J. Dorman for E. coli strains MC4100, PD32, and PD32/pAF201 and discussions. S. Rimsky is acknowledged for suggestions on purification of the protein.
This work is financially supported by grants from the Department of Biotechnology, Government of India. S.G. is a Senior Research Fellow of the Council of Scientific and Industrial Research. V.N. is a J. C. Bose Fellow of the Department of Science and Technology, Government of India.
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02081-12.
REFERENCES
- 1. Dye C, Watt CJ, Bleed DM, Hosseini SM, Raviglione MC. 2005. Evolution of tuberculosis control and prospects for reducing tuberculosis incidence, prevalence, and deaths globally. JAMA 293:2767–2775 [DOI] [PubMed] [Google Scholar]
- 2. Karakousis PC, Bishai WR, Dorman SE. 2004. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol. 6:105–116 [DOI] [PubMed] [Google Scholar]
- 3. Atlung T, Ingmer H. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7–17 [DOI] [PubMed] [Google Scholar]
- 4. Dorman CJ. 2009. Nucleoid-associated proteins and bacterial physiology. Adv. Appl. Microbiol. 67:47–64 [DOI] [PubMed] [Google Scholar]
- 5. Johnson RC, Johnson MJ, Schmidt JW, Gardner JF. 2005. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome, p 65–132. In Higgins NP. (ed), The bacterial chromosome. ASM Press, Washington, DC [Google Scholar]
- 6. Hommais F, Krin K, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20–36 [DOI] [PubMed] [Google Scholar]
- 7. Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14:441–448 [DOI] [PubMed] [Google Scholar]
- 8. Tendeng C, Bertin PN. 2003. H-NS in Gram negative bacteria: a family of multifaceted proteins. Trends. Microbiol. 11:511–518 [DOI] [PubMed] [Google Scholar]
- 9. Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. 2008. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 190:7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. 2010. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 107:5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arora K, Whiteford DC, Lau-Bonilla D, Davitt CM, Dahl JL. 2008. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J. Bacteriol. 190:4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen JM, German GJ, Alexander DC, Ren H, Tan T, Liu J. 2006. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 188:633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Colangeli R, Helb D, Vilcheze C, Hazbon MH, Lee CG, Safi H, Sayers B, Sardone I, Jones MB, Fleischmann RD, Peterson SN, Jacobs WR, Jr, Alland D. 2007. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3:e87 doi:10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 16. Triccas JA, Parish T, Britton WJ, Gicquel B. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:151–156 [DOI] [PubMed] [Google Scholar]
- 17. Manjunatha UH, Dalal M, Chatterji M, Radha DR, Visweswariah SS, Nagaraja V. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 30:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain P, Nagaraja V. 2006. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J. Mol. Biol. 357:1409–1421 [DOI] [PubMed] [Google Scholar]
- 19. Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8:e1002621 doi:10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paul TR, Beveridge TJ. 1992. Reevaluation of envelope profiles and cytoplasmic ultrastructure of mycobacteria processed by conventional embedding and freeze-substitution protocols. J. Bacteriol. 174:6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rezwan M, Laneelle MA, Sander P, Daffe M. 2007. Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68:32–39 [DOI] [PubMed] [Google Scholar]
- 22. Kapuscinski J, Skoczylas B. 1977. Simple and rapid fluorimetric method for DNA microassay. Anal. Biochem. 83:252–257 [DOI] [PubMed] [Google Scholar]
- 23. Martinez A, Torello S, Kolter R. 1999. Sliding motility in mycobacteria. J. Bacteriol. 181:7331–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Besra GS. 1998. Mycobacteria protocals. Humana Press, Totowa, NJ [Google Scholar]
- 25. Harrison JA, Pickard D, Higgins CF, Khan A, Chatfield SN, Ali T, Dorman CJ, Hormaeche CE, Dougan G. 1994. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol. Microbiol. 13:133–140 [DOI] [PubMed] [Google Scholar]
- 26. Dole S, Nagarajavel V, Schnetz K. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589–600 [DOI] [PubMed] [Google Scholar]
- 27. Gomez-Gomez JM, Blazquez J, Baquero F, Martinez JL. 1996. Hns mutant unveils the presence of a latent haemolytic activity in Escherichia coli K-12. Mol. Microbiol. 19:909–910 [DOI] [PubMed] [Google Scholar]
- 28. Werlang IC, Schneider CZ, Mendonca JD, Palma MS, Basso LA, Santos DS. 2009. Identification of Rv3852 as a nucleoid-associated protein in Mycobacterium tuberculosis. Microbiology 155:2652–2663 [DOI] [PubMed] [Google Scholar]
- 29. Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. 2006. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156:262–272 [DOI] [PubMed] [Google Scholar]
- 30. Krylov AS, Zasedateleva OA, Prokopenko DV, Rouviere-Yaniv J, Mirzabekov AD. 2001. Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips. Nucleic Acids Res. 29:2654–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spurio R, Dürrenberger M, Falconi M, Teana A, Pon LC, Gualerzi CO. 1992. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 231:201–211 [DOI] [PubMed] [Google Scholar]
- 32. Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56:858–870 [DOI] [PubMed] [Google Scholar]
- 33. Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391–400 [DOI] [PubMed] [Google Scholar]
- 34. Macvanin M, Adhya S. 2012. Architectural organization in E. coli nucleoid. Biochim. Biophys. Acta 1819:830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8:185–195 [DOI] [PubMed] [Google Scholar]
- 36. Champoux JJ. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369–413 [DOI] [PubMed] [Google Scholar]
- 37. Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. 1996. Cross-talk between topoisomerase I and HU in Escherichia coli. J. Mol. Biol. 256:292–300 [DOI] [PubMed] [Google Scholar]
- 38. Arechaga I, Miroux B, Karrasch S, Huijbregts R, de Kruijff B, Runswick MJ, Walker JE. 2000. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett. 482:215–219 [DOI] [PubMed] [Google Scholar]
- 39. Lefman J, Zhang P, Hirai T, Weis RM, Juliani J, Bliss D, Kessel M, Bos E, Peters PJ, Subramaniam S. 2004. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 186:5052–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Porta D, Rippka R, Hernandez-Marine M. 2000. Unusual ultrastructural features in three strains of Cyanothece (cyanobacteria). Arch. Microbiol. 173:154–163 [DOI] [PubMed] [Google Scholar]
- 41. Bell C, Smith GT, Sweredoski MJ, Hess S. 2012. Characterization of the Mycobacterium tuberculosis proteome by liquid chromatography mass spectrometry-based proteomics techniques: a comprehensive resource for tuberculosis research. J. Proteome Res. 11:119–130 [DOI] [PubMed] [Google Scholar]
- 42. Belisle JT, McNeil MR, Chatterjee D, Inamine JM, Brennan PJ. 1993. Expression of the core lipopeptide of the glycopeptidolipid surface antigens in rough mutants of Mycobacterium avium. J. Biol. Chem. 268:10510–10516 [PubMed] [Google Scholar]
- 43. Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont MA, Daffe M. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089–3100 [DOI] [PubMed] [Google Scholar]
- 44. Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793–808 [DOI] [PubMed] [Google Scholar]
- 45. O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 46. Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends. Microbiol. 13:20–26 [DOI] [PubMed] [Google Scholar]
- 47. Agusti G, Astola O, Rodriguez-Guell E, Julian E, Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J. Bacteriol. 190:6894–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prigent-Combaret C, Zghidi-Abouzid O, Effantin G, Lejeune P, Reverchon S, Nasser W. 2012. The nucleoid-associated protein Fis directly modulates the synthesis of cellulose, an essential component of pellicle-biofilms in the phytopathogenic bacterium Dickeya dadantii. Mol. Microbiol. 86:172–186 [DOI] [PubMed] [Google Scholar]
- 49. Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Recht J, Kolter R. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183:5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Jr, Hatfull GF. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861–873 [DOI] [PubMed] [Google Scholar]
- 53. Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs W, Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69:164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsumoto S, Yukitake H, Furugen M, Matsuo T, Mineta T, Yamada T. 1999. Identification of a novel DNA-binding protein from Mycobacterium bovis bacillus Calmette-Guerin. Microbiol. Immunol. 43:1027–1036 [DOI] [PubMed] [Google Scholar]
- 55. Bhatt A, Molle V, Besra GS, Jacobs WR, Jr, Kremer L. 2007. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol. Microbiol. 64:1442–1454 [DOI] [PubMed] [Google Scholar]
- 56. Bhatt A, Fujiwara N, Bhatt K, Gurcha SS, Kremer L, Chen B, Chan J, Porcelli SA, Kobayashi K, Besra GS, Jacobs WR., Jr 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104:5157–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.