Abstract
Bacillus subtilis cells were exposed to decoyinine to trigger stringent transcription control through inhibition of GMP synthase; amino acid starvation results in the same control through inhibition of GMP kinase by 5′-diphosphate 3′-diphosphate guanosine. The positive and negative transcription control of the stringent genes involves adenine and guanine at the transcription initiation sites, whereby they sense an increase and a decrease in the in vivo ATP and GTP pools, respectively. Decoyinine also induces sporulation in minimum medium. DNA microarray analysis revealed that decoyinine induced two major sensor kinase genes, kinA and kinB, involved in the phosphorelay leading to spore formation. lacZ fusion experiments involving the core promoter regions of kinA and kinB, whose transcription initiation bases are adenines, indicated that decoyinine induced their expression. This induction was independent of CodY and AbrB. When the adenines were replaced with guanines or cytosines, the induction by decoyinine decreased. The in situ replacement of the adenines with guanines actually affected this decoyinine-induced sporulation as well as massive sporulation in nutrient medium. These results imply that operation of the positive stringent transcription control of kinA and kinB, which is mediated by an increase in the ATP pool, is likely a prerequisite for the phosphorelay to transfer the phosphoryl group to Spo0A to initiate sporulation.
INTRODUCTION
Entry into the sporulation pathway is governed by a member of the response regulator family of transcription factors known as Spo0A (1) (Fig. 1). Most response regulators are phosphorylated directly by the respective cognate sensor kinases that carry out autophosphorylation at a histidine residue and then transfer a phosphoryl group to an aspartyl residue in the response regulator. Spo0A, in contrast, is indirectly phosphorylated by a multicomponent phosphorelay system involving at least two kinases called KinA and KinB (9). The kinases phosphorylate Spo0F, and the resulting Spo0F∼P, in turn, transfers the phosphoryl group to Spo0B. Finally, Spo0B∼P transfers the phosphoryl group to, and thereby activates, Spo0A (Fig. 1) (10). An increased level of Spo0A∼P results in repression of transcription of the abrB gene for AbrB (11), leading to derepression of transcription of the sigH (spo0H) gene, encoding σH, an alternative σ subunit of RNA polymerase (RNAP), as well as of kinB (12–14). Accordingly, elevation of the concentration of σH RNAP leads to triggering of the transcription of kinA, spo0F, and spo0A (Fig. 1) (1, 15). Additionally, Spo0A∼P is required for the induced transcription of spo0F and spo0A (12, 14, 16), thereby setting up a self-reinforcing closed cycle.
Fig 1.
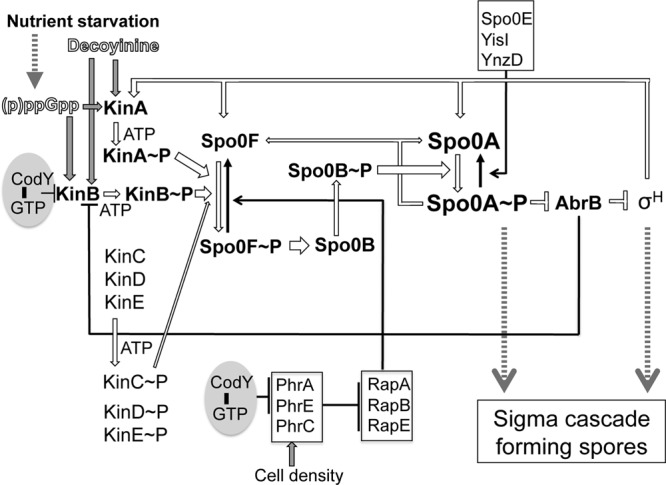
Phosphorelay regulatory network leading to the formation of spores in B. subtilis. Upon nutrient starvation or decoyinine addition, two major sensor kinases (KinA and KinB) undergo autophosphorylation. In addition, three minor sensor kinases (KinC, KinD, and KinE) are considered to be involved in the phosphorylation of Spo0F (2). However, it was recently reported that not only KinC but also KinD and KinE are unlikely to be involved in this phosphorylation (3, 4). KinA∼P and KinB∼P provide phosphate input to the master transcriptional regulator, Spo0A, yielding Spo0A∼P via two additional regulators, i.e., the phosphorylated forms of Spo0F and Spo0B (Spo0F∼P and Spo0B∼P). Spo0A∼P becomes a positive or negative regulator for sporulation genes, including those for Spo0A itself, Spo0F, and the transition state transcription regulator AbrB. AbrB represses the transcription of the gene of σH, which is also essential for sporulation, as well as that of kinB. Thus, Spo0A∼P represses abrB, thereby stimulating σH synthesis. kinA is transcribed with RNAP possessing σH. As a result, the transcription of the genes for KinA, Spo0F, and Spo0A is triggered in a closed-loop system. The accumulation of Spo0A∼P and σH leads to the sigma cascade to form spores. The kinB gene is likely a target of CodY (5). The cell density is sensed by Phr peptides that are secreted, processed, and imported as pentapeptides back into the cell, where they inhibit the Rap proteins (RapA, RapB, and RapE) that cause dephosphorylation of Spo0F∼P. The phrA and phrE genes are CodY candidate targets (5). Spo0A∼P is susceptible to dephosphorylation through the action of Spo0E (6, 7) and two homologues, YisI and YnzD (8); expression of the last two proteins increases under nonsporulation conditions (8). Open and gray arrows and black arrows indicate forward and backward sporulation, respectively.
However, the key unresolved issue regarding the feedback regulation of the phosphorelay is identification of the first component to be activated upon starvation triggering phosphorelay. Currently, GTP is known as a metabolite whose intracellular level is monitored by a GTP-sensing repressor, CodY (17, 18). When cells are grown under nutrient-rich conditions, the cellular GTP level is elevated, and the genes under the control of CodY are repressed. Conversely, when cells have limited nutrients, the GTP level is low, resulting in the derepression of CodY-regulated genes. Actually, the kinB gene is a candidate target of CodY, as revealed by genome-wide transcript analysis (5). However, CodY itself cannot be the primary factor initiating sporulation because a mutant lacking it does not exhibit massive sporulation during growth under usual medium conditions (17).
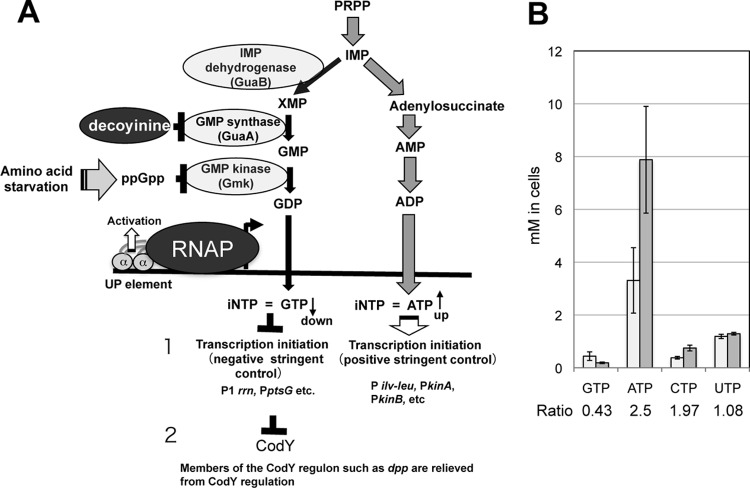
More than a quarter of a century ago, decoyinine was found to induce sporulation of Bacillus subtilis cells exponentially growing in the presence of rapidly metabolizable carbon, nitrogen, and phosphate sources (19). Decoyinine causes the reciprocal concentration changes of a decrease in GTP and an increase in ATP (Fig. 2A), both being substrates of RNAP, by inhibiting GMP synthase (21, 22, 24, 26). (The changes of the in vivo NTP concentrations after decoyinine addition are shown in Fig. 2B [24]; the changes of the CTP and UTP concentrations were less than 2-fold, which are relatively tolerant to decoyinine addition [24].) These reciprocal concentration changes of GTP and ATP were supposed to be also evoked by inhibition of IMP dehydrogenase by 5′-diphosphate 3′-diphosphate guanosine (ppGpp), whose synthesis is triggered by a stringent response (27, 28). However, it was very recently shown that ppGpp does not inhibit IMP dehydrogenase efficiently, but it severely inhibits GMP kinase to convert GMP to GDP, resulting in the decrease of the in vivo GTP concentration (20). It is known that the stringent response also induces sporulation (28, 29). These reciprocal changes can be sensed through increases and decreases in the rate of transcription initiation from numerous positive and negative stringent promoters depending on their first and/or second initiation bases at nucleotide + 1 and +2, i.e., adenine and guanine, respectively (Fig. 2). Therefore, this stringent transcription control involves no transcriptional regulators. Our DNA microarray analysis involving the ΔcodY strain with and without decoyinine (22) revealed that the kinA and kinB promoters are likely included in numerous positive stringent promoters that are inducible upon decoyinine addition and possess adenines at their transcription initiation nucleotides (30, 31).
Fig 2.
Molecular mechanism underlying stringent transcription control in B. subtilis. (A) Illustration of its molecular mechanism. ppGpp is synthesized upon amino acid starvation, which is involved in inhibition of GMP kinase rather than IMP dehydrogenase (20). This inhibition results in the reciprocal changes of a GTP decrease and an ATP increase, which result in negative and positive regulation of numerous stringent promoters. Negative stringent promoters such as P1rrn (21) and PptsGHI (22) have guanines at their transcription initiation sites (nucleotides +1 and/or +2), whereas positive stringent promoters such as Pilv-leu (23, 24), PkinA, and PkinB have adenines at these sites. So, the iNTPs are GTP and ATP for the negative and positive stringent promoters. Thus, the GTP decrease and ATP increase upon the stringent response result in downregulation of negative stringent promoters and upregulation of positive ones, respectively. The addition of decoyinine, a GMP synthase inhibitor, also evokes these reciprocal concentration changes of GTP and ATP. In addition, members of the CodY regulon such as dpp (25) are relieved from CodY regulation due to the decrease in GTP, a corepressor of CodY. Phosphoribosyl pyrophosphate (PRPP), IMP, and XMP are intermediates of purine biosynthesis. (B) In vivo concentration changes of NTP upon the stringent response in B. subtilis. In vivo NTP concentrations changed from the initial values (light gray bars) to the ones shown 30 min after decoyinine addition (darker gray bars); these values determined, by use of capillary electrophoresis mass spectrometry, as well as their standard deviations were taken from Table 3 in our previous publication (24). Ratios of values obtained with and without decoyinine are shown.
We show in this communication that the replacement of these adenines of kinA and kinB with guanines or cytosines decreased their induction by decoyinine. Furthermore, when the adenine of either kinA or kinB was replaced with a guanine in situ, decoyinine-induced sporulation as well as massive sporulation in nutrient medium was well affected. These findings imply that the operation of the positive stringent control of transcription of kinA and kinB, which is enhanced by an increase in an RNAP substrate of ATP, is likely a prerequsite for the phosphorelay to transfer the phosphate group to Spo0A that eventually leads to the formation of spores.
MATERIALS AND METHODS
Bacterial strains and their construction.
The B. subtilis strains used in this work are listed in Table 1. The ΔabrB::erm strain FU1106 was constructed by means of recombinant PCR as follows. The regions upstream and downstream of the abrB gene were first amplified by PCR using DNA of the wild-type strain 168 as the template and primer pairs ABu-F/ABu-R and ABd-F/ABd-R (see Table S1 in the supplemental material), respectively. The erm cassette was amplified by PCR using DNA of plasmid pMUTIN2 (37) as the template and primer pair EM-F/EM-R. Second, recombinant PCR using primer pair ABu-F/ABd-R and three PCR fragments resulted in a PCR product covering the region upstream of abrB, the erm gene, and the region downstream of abrB. The resultant recombinant PCR product was used to transform strain 168 to erythromycin resistance (0.3 μg/ml) on tryptose blood agar base (Difco) plus 10 mM glucose (TBABG) plates to produce the ΔabrB::erm strain FU1106. The ΔkinA::erm and ΔkinB::kan strains FU1095 and FU1096 were provided by T. Sato (Hosei University) and K. Kobayashi (Nara Institute of Science and Technology), respectively. The ΔkinA::erm strain FU1095 was transformed to kanamycin resistance (5 μg/ml) with DNA of the ΔkinB::kan strain FU1096 to produce the ΔkinA::erm ΔkinB::kan strain FU1098. The gid::spc ΔcodY strain PS37t was obtained by transformation of wild-type strain 168 with DNA of the gid::spc ΔcodY strain PS37 to spectinomycin resistance (60 μg/ml). The gid::spc ΔcodY ΔkinA and gid::spc ΔcodY ΔkinB strains FU1063 and FU1064 were obtained by transformation of the gid::spc ΔcodY strain PS37t with DNAs of the ΔkinA and ΔkinB strains FU1095 and FU1096 to erythromycin and kanamycin resistance, respectively. The presence of ΔcodY in strains PS37t, FU1063, and FU1064 was confirmed by the appearance of the PCR product that was 250 bp shorter than that obtained for the CodY+ strain, as described previously (38). Disruption of the gid gene, present in the ΔcodY strain, did not affect the expression of the target genes in this work or sporulation efficiency.
Table 1.
B. subtilis strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| 168 | trpC2 | 32 |
| PS37 | trpC2 gid::spc ΔcodY | 33 |
| PS37t | trpC2 gid::spc ΔcodY | This work |
| FU1095 | trpC2 ΔkinA::erm | T. Sato, this work |
| FU1096 | trpC2 ΔkinB::kan | K. Kobayashi, this work |
| FU1063 | trpC2 gid::spc ΔcodY ΔkinA::erm | This work |
| FU1064 | trpC2 gid::spc ΔcodY ΔkinB::kan | This work |
| FU1098 | trpC2 ΔkinA::erm ΔkinB::kan | This work |
| ASK201 | trpC2 Δspo0H::erm | 34 |
| FU1121 | trpC2 Δspo0A::spc | 35 |
| TM0310 | trpC2 aprE::[spc lacI Pspac-mazF] | 36 |
| FU1106 | trpC2 ΔabrB::erm | This work |
| FU1087 | trpC2 amyE::[cat PkinA (−55/+10)-lacZ] | This work |
| FU1088 | trpC2 amyE::[cat PkinA (−55/+10)(A + 1G)-lacZ] | This work |
| FU1115 | trpC2 amyE::[cat PkinB (−55/+10)-lacZ] | This work |
| FU1116 | trpC2 amyE::[cat PkinB (−55/+10)(A + 1G)-lacZ] | This work |
| FU1107 | trpC2 Δspo0H::erm amyE::[cat PkinA (−55/+10)-lacZ] | This work |
| FU1108 | trpC2 Δspo0H::erm amyE::[cat PkinA (−55/+10)(A + 1G)-lacZ] | This work |
| FU1117 | trpC2 Δspo0H::erm amyE::[cat PkinB (−55/+10)-lacZ] | This work |
| FU1118 | trpC2 Δspo0H::erm amyE::[cat PkinB (−55/+10)(A + 1G)-lacZ] | This work |
| FU1109 | trpC2 gid::spc ΔcodY amyE::[cat PkinA (−55/+10)-lacZ] | This work |
| FU1110 | trpC2 gid::spc ΔcodY amyE::[cat PkinA (−55/+10)(A + 1G)-lacZ] | This work |
| FU1119 | trpC2 gid::spc ΔcodY amyE::[cat PkinB (−55/+10) -lacZ] | This work |
| FU1120 | trpC2 gid::spc ΔcodY amyE::[cat PkinB (−55/+10)(A + 1G)-lacZ] | This work |
| FU1111 | trpC2 ΔabrB::erm amyE::[cat PkinA (−55/+10)-lacZ] | This work |
| FU1112 | trpC2 ΔabrB::erm amyE::[cat PkinA (−55/+10)(A + 1G)-lacZ] | This work |
| FU1122 | trpC2 ΔabrB::erm amyE::[cat PkinB (−55/+10)-lacZ] | This work |
| FU1123 | trpC2 ΔabrB::erm amyE::[cat PkinB (−55/+10)(A + 1G)-lacZ] | This work |
| FU1124 | trpC2 Δspo0A::spc amyE::[cat PkinA (−55/+10)-lacZ] | This work |
| FU1125 | trpC2 Δspo0A::spc amyE::[cat PkinA (−55/+10)(A + 1G)-lacZ] | This work |
| FU1126 | trpC2 Δspo0A::spc amyE::[cat PkinB (−55/+10)-lacZ] | This work |
| FU1127 | trpC2 Δspo0A::spc amyE::[cat PkinB (−55/+10)(A + 1G)-lacZ] | This work |
| FU1102 | trpC2 kinA (A + 1G) | This work |
| FU1103 | trpC2 kinA (A + 1G) ΔkinB::kan | This work |
| FU1113 | trpC2 kinB (A + 1G) | This work |
| FU1114 | trpC2 kinB (A + 1G) ΔkinA::erm | This work |
| FU1153 | trpC2 amyE::[cat PkinA (−55/+10)(A + 1C)-lacZ] | This work |
| FU1154 | trpC2 amyE::[cat PkinB (−55/+10)(A + 1C)-lacZ] | This work |
| FU1155 | trpC2 kinA (A + 1G) kinB (A + 1G) | This work |
| FU1156 | trpC2 kinA (A + 1C) | This work |
| FU1170 | trpC2 kinA (A + 1C) ΔkinB::kan | This work |
To construct transcriptional promoter-lacZ fusion strains with the kinA and kinB promoters, their core regions comprising nucleotides −55 to +10 of kinA (30) and kinB (31) (nucleotide +1 is the transcription initiation nucleotide) were amplified using primer pairs KA-F1/KA-R1 and KB-F1/KB-R1 (Table S1) and DNA of wild-type strain 168 as the template, respectively. The PCR products were trimmed with XbaI and BamHI and then ligated with the XbaI-BamHI arm of plasmid pCRE-test2 (39). The ligated DNAs were used for transformation of Escherichia coli strain DH5α to ampicillin resistance (50 μg/ml) on Luria-Bertani medium plates (40). Correct construction of the fusions in the resulting plasmids was confirmed by DNA sequencing. The plasmids carrying the promoter regions of kinA and kinB were linearized with PstI and then used for double-crossover transformation of strain 168 to chloramphenicol resistance (5 μg/ml) on TBABG plates, which produced the PkinA-lacZ and PkinB-lacZ strains FU1087 and FU1115, respectively.
To construct strains carrying the kinA and kinB promoter-lacZ fusions with guanine or cytosine substitutions at nucleotide +1, the core promoter regions (nucleotides −55 to +10) were amplified using primer pairs KA-F1/KA-R1g and KB-F1/KB-R1g or KA-F1/KA-R1c and KB-F1/KB-R1c (Table S1), respectively, and chromosomal DNA of wild-type strain 168 as the template. The PCR products trimmed with XbaI and BamHI were cloned into plasmid pCRE-test2 in E. coli strain DH5α, as described above. Correct construction of the fusions in the resulting plasmids was confirmed by DNA sequencing. The plasmids that had a guanine or cytosine substitution at nucleotide +1 were each linearized with PstI and then used for transformation of strain 168, resulting in the PkinA(A + 1G)-lacZ and PkinB(A + 1G)-lacZ strains FU1088 and FU1116 or the PkinA(A + 1C)-lacZ and PkinB (A + 1C)-lacZ strains FU1153 and FU1154, respectively.
To introduce Δspo0H, ΔabrB, ΔcodY, and Δspo0A into the PkinA-lacZ, PkinB-lacZ, PkinA (A + 1G)-lacZ, and PkinB(A + 1G)-lacZ strains FU1087, FU1115, FU1088, and FU1116, they were transformed with DNAs of the Δspo0H::erm, ΔabrB::erm, gid::spc ΔcodY, and Δspo0A::spc strains ASK201, FU1106, PS37, and FU1121 to erythromycin and spectinomycin resistance, yielding the Δspo0H strains FU1107, FU1117, FU1108, and FU1118; the ΔabrB strains FU1111, FU1122, FU1112, and FU1123; the ΔcodY strains FU1109, FU1119, FU1110, and FU1120; and the Δspo0A strains FU1124, FU1126, FU1125, and FU1127, respectively. The presence of ΔcodY in strains FU1109, FU1119, FU1110, and FU1120 was confirmed as described above.
In situ replacement of adenines at nucleotide +1 of the kinA and kinB promoters with guanines or cytosines was carried out by means of positive selection using the mazF gene (36) (see Fig. S1 in the supplemental material). First, three PCR fragments carrying the spectinomycin resistance gene (spc), lacI, and mazF under the control of Pspac, and the respective kinA regions upstream and downstream of its promoter containing guanine instead of adenine at the kinA transcription initiation nucleotide, were prepared by means of PCRs using DNA of the aprE::(spc lacI Pspac-mazF) strain TM0310 as the template and primer pairs MF-F/MF-R, KA-F2/KA-R2g or KA-F2/KA-R2c, and KA-F3g/KA-R3 or KA-F3c/KA-R3 (Table S1), respectively. Second, recombinant PCR to combine the three fragments with themselves as templates and primer pair KA-F2/KA-R3 was carried out. Then, wild-type strain 168 was transformed to spectinomycin resistance (60 μg/ml) with the recombinant PCR fragment comprising the three fragments. The resulting spectinomycin-resistant cells were treated with isopropyl-β-d-thiogalactopyranoside (IPTG), and the IPTG-resistant and spectinomycin-sensitive transformants, which had resulted from the self-recombination between the short repeated regions including guanine and cytosine at the kinA transcription initiation site, were the kinA (A + 1G) strain FU1102 and the kinA (A + 1C) strain FU1156. Correct construction of the kinA (A + 1G) strain FU1102 and the kinA (A + 1C) strain FU1156 was confirmed by DNA sequencing. Similarly, the kinB (A + 1G) strain FU1113 was obtained by means of the same positive selection as described above, the first PCR resulting in the three fragments carrying the spc, lacI, and Pspac-mazF and the kinB regions upstream and downstream of its promoter containing guanine instead of adenine at the kinB transcription initiation site, using primer pairs MF-F/MF-R, KB-F2/KB-R2g, and KB-F3g/KB-R3 (Table S1), respectively, and the second recombinant PCR involving them as the templates and primer pair KB-F2/KB-R3. Although we attempted to similarly isolate the kinB (A + 1C) strain using primer pairs KB-F2/KB-R2c and KB-F3c/KB-R3 (Table S1) for the first PCR, we could not obtain the right kinB (A + 1C) strain due to the occurrence of various unexpected base substitutions and deletions in the kinB promoter region during the self-recombination to result in the IPTG-resistant and spectinomycin-sensitive transformants. Moreover, the kinA (A + 1G) strain FU1102, kinA (A + 1C) strain FU1156, and kinB (A + 1G) strain FU1113 were transformed with DNAs of the ΔkinB::kan and ΔkinA::erm strains FU1096 and FU1095 to kanamycin resistance (5 μg/ml) and erythromycin resistance (0.3 μg/ml) to yield the kinA (A + 1G) ΔkinB strain FU1103, kinA (A + 1C) ΔkinB strain FU1170, and kinB (A + 1G) ΔkinA strain FU1114, respectively.
The kinA (A + 1G) kinB (A + 1G) strain FU1155 was constructed as follows. The kinA (A + 1G) strain was transformed with the PCR product amplified by use of primer pair KB-F2/KB-R3 and a template DNA of an intermediate strain during the construction of the kinB (A + 1G) strain FU1102, which carries the mazF cassette (spc lacI Pspac-mazF) between the PkinB (A + 1G) repeated regions to result in spectinomycin resistance (60 μg/ml). The spectinomycin-resistant cells were treated with IPTG, and one of the IPTG-resistant and spectinomycin-sensitive transformants was the kinA (A + 1G) kinB (A + 1G) strain FU1155. Correct construction of strain FU1155 was confirmed by DNA sequencing.
Cell cultivation, β-Gal assay, and spore titer.
The lacZ fusion strains were grown at 30°C overnight on TBABG plates containing the appropriate antibiotic(s), chloramphenicol (5 μg/ml), spectinomycin (60 μg/ml), and/or erythromycin (0.3 μg/ml). The cells were inoculated and grown in 50 ml of S6 medium (41) containing 25 mM glucose and 50 μg of tryptophan per ml. When the cells reached to an optical density at 600 nm (OD600) of 0.5, 10-ml aliquots were distributed into two flasks, and decoyinine was added to one culture to obtain a final concentration of 500 μg/ml (18 mM). The cultures with and without decoyinine were further incubated. During incubation before and after decoyinine addition, 1-ml aliquots of the culture were withdrawn at 30-min intervals, and the β-galactosidase (β-Gal) activity in crude cell extracts was measured spectrophotometrically as described previously (37). At 10 h after decoyinine addition (T10), the titers of viable cells (V) and spores (S) that were heat resistant (75°C for 20 min) were measured to obtain the sporulation percentage (S/V × 100). The sporulation percentage at T20 (20 h after the entry into the stationary cell phase) was also determined using nutrient sporulation medium (NSMP) (41).
RESULTS
DNA microarray analysis of stringent transcription control of the genes involved in phosphorelay that initiates sporulation.
Decoyinine has been known for a long time to induce the sporulation of cells exponentially growing in the presence of rapidly metabolizable carbon, nitrogen, and phosphate sources (19). However, CodY cannot be the primary factor initiating the massive sporulation, as discussed above. So, we examined the levels of induction by decoyinine of the genes involved in the sporulation phosphorelay (Fig. 1) by using the data from the previous DNA microarray analysis involving the ΔcodY strain with and without decoyinine to eliminate expression disturbance by decoyinine induction of members of the CodY regulon (22) (see the DNA microarray data deposited in the KEGG Expression Database [http://www.genome.jp/kegg/expression]). This examination revealed that out of the sporulation sensor kinase genes (kinA to kinE) (Fig. 1), kinA, kinB, kinD, and kinE are likely included in numerous positive stringent promoters that are inducible upon decoyinine addition (Table 2). Among the five sensor kinases, KinA and KinB are likely major sensor kinases involved in the phosphorelay that initiates sporulation (31, 42). Recently, KinC and KinD were found to be involved in biofilm formation rather than sporulation (4, 43). Moreover, KinD and KinE are unlikely to be involved in the phosphorelay leading to the formation of spores (3). Therefore, kinA and kinB were targets of the current study on metabolic regulation of sporulation initiation, because KinA and KinB are major sensor kinases to have the phosphoryl group in sporulation phosphorelay and their genes are likely under positive stringent transcription control involving the adenines at their transcription initiation nucleotides (30, 31).
Table 2.
DNA microarray analysis of induction of the spo genes involved in sporulation phosphorelay upon addition of decoyininea
| spo gene | Fold expression (with decoyinine/without decoyinine) |
|---|---|
| kinA | 3.33 |
| kinB | 1.96 |
| kinC | 0.657 |
| kinD (ykvD) | 1.53 |
| kinE (ykrQ) | 4.25 |
| spo0F | 8.09 |
| spo0B | 0.854 |
| spo0A | 4.31 |
| abrB | 0.546 |
| sigH | 1.92 |
| rapA | 14.4 |
| rapB | 4.84 |
| rapE | 2.06 |
| spo0E | 2.28 |
| ynzD | 0.560 |
| yisI | 0.752 |
DNA microarray analysis was performed previously (22). Also, the data were deposited in the KEGG Expression Database (http://www-genome.jp/kegg/expression).
Expectedly, the DNA microarray analysis had also revealed that spo0F, spo0A, and sigH, besides kinA and kinB, were induced upon decoyinine addition (Table 2). The increased level of Spo0A∼P upon the onset of the phosphorelay results in repression of transcription of the abrB gene for AbrB (11), leading to the derepression of transcription of the sigH (spo0H) gene, encoding σH, as well as of kinB (12–14), and RNAP containing σH transcribes kinA, spo0A, and spo0F (Fig. 1) (1, 15, 30). Also, Spo0A∼P is required for the induced transcription of spo0F and spo0A (12, 14, 16).
lacZ expression under the control of the promoters of kinA and kinB upon decoyinine addition.
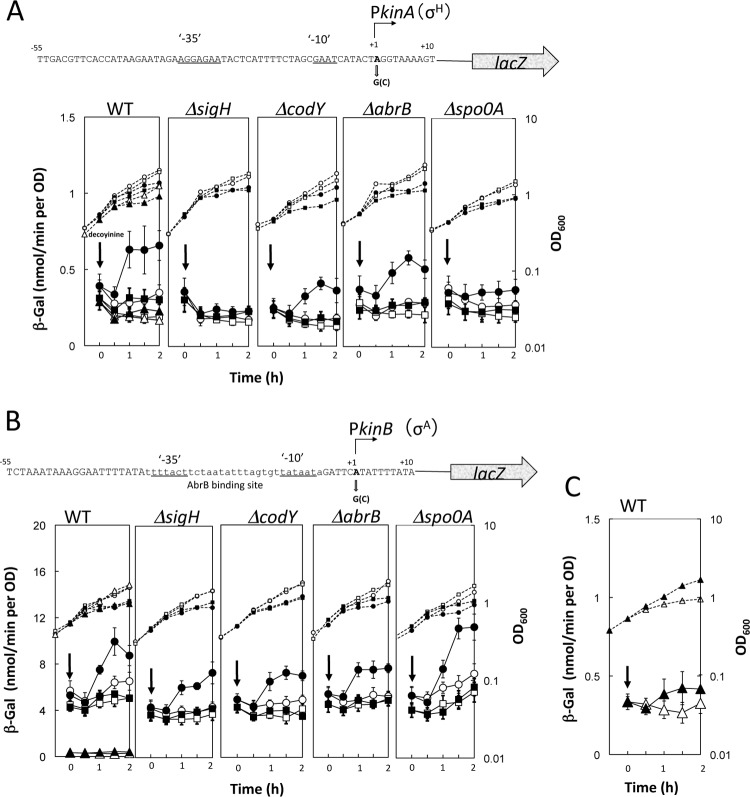
To confirm the DNA microarray data showing that the kinA and kinB promoters are under positive stringent control in response to decoyinine addition (Table 2), the kinA and kinB core promoter regions (nucleotides −55 to +10) were placed upstream of lacZ in the amyE locus to yield the PkinA-lacZ and PkinB-lacZ strains FU1087 and FU1115, respectively. (The B. subtilis strains are listed in Table 1.) As shown in Fig. 3 (refer to WT), synthesis of β-Gal in the cells growing in a synthetic medium (S6 medium) (41), which is encoded by lacZ under the control of the kinA and kinB promoters, significantly increased after addition of decoyinine in the wild-type background. To demonstrate that each adenine at the initiation nucleotide for transcription of kinA and kinB is involved in this positive stringent control, they were replaced with guanines to yield the PkinA (A + 1G)-lacZ and PkinB (A + 1G)-lacZ strains FU1088 and FU1116. (Adenine replacement with guanine at nucleotide +1 of the positive stringent promoter always diminishes its positive regulation, but the extent of the diminution [sometimes entering negative regulation] depends on the delicate nucleotide sequence close to nucleotide +1 of the positive stringent promoter [22].) As shown in Fig. 3 (refer to WT), this replacement did not significantly reduce the kinA and kinB promoter activities, but the positive stringent control of these promoters was diminished [PkinA (A + 1G)] or abolished [PkinB (A + 1G)]. To confirm the results of the adenine replacement with guanine, the adenines were also replaced with cytosines to yield the PkinA (A + 1C)-lacZ and PkinB (A + 1C)-lacZ strains FU1153 and FU1154, as the change of the CTP concentration is relatively tolerant to decoyinine addition (Fig. 2B) (24). As shown in Fig. 3 (WT), this replacement did not reduce the kinA promoter strength, but it reduced the kinB promoter strength largely. The lacZ expression from the recombinant promoter [PkinA(A + 1C)] was slightly induced, although it was less induced than that from the recombinant promoter [PkinA (A + 1G)] (Fig. 3A; refer to WT). The positive transcription control of PkinB (A + 1C) promoter was not significantly detected, that is, within the standard deviations (Fig. 3C). As cytosine at nucleotide + 1 is a rare phenomenon in B. subtilis (44), substituting +1 A/G for C leads to a decrease in the promoter activity as seen with the kinB promoter. In the kinA promoter, nucleotide +2 is a G, and it is possible that by replacing the +1 A with C moves the transcription start site 1 bp downstream so that it now starts with G. This would explain the still relatively high activity of this promoter version as well as its inability to increase transcription after decoyinine treatment. These overall results of the adenine replacements with guanine and cytosine indicated that the adenines are actually involved in the positive stringent transcription control of the kinA and kinB promoters.
Fig 3.
Activation of the kinA and kinB core promoters upon addition of decoyinine. (A) The nucleotide sequence of the core kinA promoter region (nucleotides −55 to +10) is shown in the upper part, along with the transcription initiation adenine (boldfaced A at nucleotide +1) and the −10 and −35 regions that are recognized by RNAP possessing σH (30). This promoter region was transcriptionally fused with lacZ. The adenine at nucleotide +1 was replaced with a guanine. The synthesis of β-Gal encoded by lacZ under the control of the wild-type and guanine substitution kinA promoters was monitored after the addition of decoyinine in the wild-type (WT), ΔsigH, ΔcodY, ΔabrB, and Δspo0A backgrounds. Closed and open symbols indicate the presence and absence of decoyinine addition; circles, squares, and triangles represent the kinA promoters possessing adenine, guanine, and cytosine at nucleotide +1, respectively. Large and small symbols denote β-Gal activity and OD600, respectively. (B) The nucleotide sequence of the core kinB promoter region (nucleotides −55 to +10) is shown, along with the transcription initiation adenine (boldfaced A at nucleotide +1) and the −10 and −35 regions (underlined) that are most likely recognized by RNAP possessing σA (31). This promoter region containing the AbrB binding site (lowercase letters) (13) was fused with lacZ, whose transcription initiation adenine was replaced with guanine. β-Gal synthesis under the control of the WT and guanine substitution kinA promoters was monitored after the addition of decoyinine in the WT, ΔsigH, ΔcodY, ΔabrB, and Δspo0A backgrounds. The symbol assignment is the same as in panel A. (C) β-Gal synthesis under the control of the WT and cytosine substitution kinB promoter was monitored after the addition of decoyinine in the WT, where a scale of the vertical axis (WT in panel B) for the β-Gal synthesis is enlarged to the maximal value of 1.5. In all panels, the standard deviations of the average β-Gal activity values from the duplicated or triplicate experiments are shown by error bars. Thick and thin error bars are used for the adenine and guanine at nucleotide +1 of kinA and kinB (A and B), respectively. The error bars for the cytosine at nucleotide +1 of kinA and kinB are not shown in panel A or B (WT) to avoid unnecessary complication, but they are shown in panel C.
kinA and kinB are known to be transcribed by RNAP possessing σH and σA (30, 31), so the PkinA-lacZand PkinB-lacZ fusions were placed in the ΔsigH genetic background to yield the PkinA-lacZ ΔsigH and PkinB-lacZ ΔsigH strains FU1107 and FU1117, respectively. As expected, the induction of lacZ under the control of PkinA after addition of decoyinine was not observed in the ΔsigH genetic background (Fig. 3A), but lacZ under the control of PkinB was expressed in the ΔsigH genetic background as in the wild-type background (Fig. 3B). The results indicate that lacZ transcription from PkinA, which increased upon decoyinine addition, is actually conducted by RNAP containing σH, whereas that from PkinB is most likely by RNAP containing σA. The kinB gene is a candidate of the CodY targets, as revealed by genome-wide transcript analysis (5). So, the observed positive stringent regulation of the kinA and kinB promoters might be explained by derepression due to CodY inactivation through a decrease in the in vivo GTP pool upon decoyinine addition. To determine if the lacZ expression from the kinA or kinB promoter is affected by codY deletion, the PkinA-lacZ and PkinB-lacZ fusions were placed in the ΔcodY background to yield the PkinA-lacZ ΔcodY and PkinB-lacZ ΔcodY strains FU1109 and FU1119, respectively. As shown in Fig. 3, lacZ expression from neither PkinA nor PkinB was affected by ΔcodY. Hence, it is considered that CodY is not involved in the positive stringent control of kinA and kinB. Even if kinB is a direct target of CodY, which is inactivated upon decoyinine addition, the binding site of CodY in the kinB promoter region is supposed to be located outside the core kinB promoter region (nucleotides −55 to +10). It is known that kinB is repressed by AbrB and that its binding site is located between nucleotides −34 and −6 (Fig. 3B) (13). To determine if the lacZ expression from the kinA or kinB promoter is affected by abrB deletion, the PkinA-lacZ and PkinB-lacZ fusions were placed in the ΔabrB background to yield the PkinA-lacZ ΔabrB and PkinB-lacZ ΔabrB strains FU1111 and FU1122, respectively. As shown in Fig. 3, lacZ expression from neither PkinA nor PkinB was affected by ΔabrB. Hence, it is considered that AbrB is not involved in the positive stringent control of kinA and kinB. Although the fact that the transcription from the kinB promoter is not affected by ΔabrB cannot be properly explained, it is possible to consider that AbrB is too rapidly repressed after decoyinine addition by Spo0A∼P, which increased upon the onset of the phosphorelay, to be detected even in the abrB+ genetic background.
Furthermore, kinA is reported to be negatively regulated by Spo0A∼P (12), but its Spo0A box is located outside the core kinA promoter (nucleotides −55 to +10). However, kinA is transcribed by RNAP possessing σH, and the expression of sigH encoding σH is activated by Spo0A∼P through repression of AbrB (Fig. 1), so it is expected that kinA is not transcribed in the Δspo0A genetic background. To determine if the lacZ expression from the kinA or kinB promoter is affected by the spo0A deletion, the PkinA-lacZ and PkinB-lacZ fusions were placed in the Δspo0A background to yield the PkinA-lacZ Δspo0A and PkinB-lacZ Δspo0A strains FU1124 and FU1126, respectively. As expected, lacZ expression from PkinA was not induced by decoyinine addition in the spo0A background, but that from PkinB was not affected by the spo0A deletion (Fig. 3).
Even when PkinA (A + 1G)-lacZ and PkinB (A + 1G)-lacZ were introduced into the ΔsigH, ΔcodY, ΔabrB, and Δspo0A backgrounds, no significant induction of lacZ expression was observed, as in the wild-type background. The overall results indicate that the positive stringent control of σH- and σA-dependent transcription from the respective kinA and kinB promoters, which is evoked by decoyinine addition, involves the adenines at their transcription initiation nucleotides and that this positive stringent control is most likely independent of either CodY or AbrB.
Sporulation efficiency of kinA and kinB mutants with replacement of adenine with guanine (or cytosine) at the transcription initiation nucleotide.
lacZ expression under the control of either the PkinA or PkinB promoter carrying adenine at the transcription initiation nucleotide was induced by the addition of decoyinine, but it was not significantly induced when the adenine was replaced with guanine (Fig. 3). To examine the effect of this replacement on decoyinine-induced sporulation as well as on the massive sporulation using NSMP (41) as a control experiment, the transcription initiation bases, adenines, of the kinA and kinB promoters were replaced with guanines in situ by means of positive selection using the mazF gene (36), as described in Materials and Methods and illustrated in Fig. S1, to yield the kinA (A + 1G) and kinB (A + 1G) strains FU1102 and FU1113, respectively.
Furthermore, the replacement of the adenine with cytosine at the transcription initiation nucleotide rendered only the kinB promoter inefficient (Fig. 3B and C), but decoyinine did not induce β-Gal synthesis under the control of PkinA (A + 1C) and PkinB (A + 1C) significantly (Fig. 3). We also attempted to replace the adenine with cytosine in situ as described above. We successfully introduced the adenine replacement with cytosine at the transcription initiation nucleotide of the kinA promoter to obtain the kinA (A + 1C) strain FU1156. However, we were unable to introduce this replacement at nucleotide +1 of the kinB promoter in spite of our numerous trials due to the occurrence of various unexpected base substitutions and deletions in the kinB promoter region at the final stage of the nucleotide replacement protocol (see Fig. S1 in the supplemental material). This might possibly be related to the fact that the replacement of the adenine with cytosine severely affected this kinB promoter strength (Fig. 3B and C).
We examined the sporulation efficiencies of various wild-type and mutant strains under decoyinine-induced sporulation conditions using S6 medium as well as under the nutrient sporulation conditions using NSMP (Table 3). When the cells of wild-type strain 168 exponentially growing in the S6 medium were exposed to decoyinine and further incubated for 10 h, they sporulated at the sporulation percentage of 19%. As a control, the wild-type cells were not exposed to decoyinine and were further incubated for 10 h, their sporulation percentage was 0.37%, indicating that decoyinine induced sporulation 51-fold. This decoyinine-induced sporulation was completely blocked in the Δspo0A, Δspo0H, or ΔkinA ΔkinB genetic background (less than 5 × 10−6%, the experimental limit to count heat-resistant spores), whereas it was only partially affected by ΔkinA (0.26%) or ΔkinB (0.67%) (Table 3). Interestingly, ΔcodY enhanced this sporulation (36%). Such enhancement with ΔcodY was also observed in the genetic background of ΔkinA (7.3%) or ΔkinB (1.5%) (Table 3). In contrast, ΔabrB well inhibited this sporulation (0.40%) (Table 3). Thus, ΔcodY or ΔabrB well affected the metabolic network leading to the sporulation in S6 medium. It is reasonable that ΔcodY allows the decoyinine-induced sporulation with S6 medium to proceed, because kinB, phrA, and phrE are probable targets of CodY (5), whose CodY derepression is favorable for sporulation initiation. However, it cannot be properly explained at present why ΔabrB affected sporulation in S6 medium in spite of possibly enhancing sporulation due to derepression of sigH.
Table 3.
Effects of variable mutations, including the substitution mutations of the respective adenines at nucleotide +1 of PkinA and PkinB with guanines, on sporulation efficiency
| Strain | Sporulation percentage (%)a |
|
|---|---|---|
| Decoyinine induced, S6 medium, T10 | Nutrient medium (NSMP), T20 | |
| Wild-type 168 | 19 | 71 |
| PS37t (ΔcodY) | 36 | 29 |
| FU1106 (ΔabrB) | 0.40 | 34 |
| FU1121 (Δspo0A) | <5 × 10−6 | <5 × 10−6 |
| ASK201 (Δspo0H) | <5 × 10−6 | <5 × 10−6 |
| FU1095 (ΔkinA) | 0.26 | 7.9 |
| FU1063 (ΔkinA ΔcodY) | 7.3 | 8.5 |
| FU1096 (ΔkinB) | 0.67 | 22 |
| FU1064 (ΔkinB ΔcodY) | 1.5 | 12 |
| FU1098 (ΔkinA ΔkinB) | <5 × 10−6 | <5 × 10−6 |
| FU1102 [kinA (A + 1G)] | 3.5 | 36 |
| FU1103 [kinA (A + 1G) ΔkinB] | 6.2 × 10−4 | 0.055 |
| FU1113 [kinB (A + 1G)] | 1.7 | 25 |
| FU1114 [ΔkinA kinB (A + 1G)] | 2.1 × 10−5 | 0.0019 |
| FU1155 [kinA(A + 1G) kinB (A + 1G)] | 0.0011 | 2.3 |
| FU1156 [kinA (A + 1C)] | 4.2 | 27 |
| FU1170 [kinA (A + 1C) ΔkinB] | 7.6 × 10−5 | 0.0052 |
The sporulation experiments were repeated at least three times. Representative values are presented. The standard deviations were less than 15% of the values shown.
Sporulation of wild-type strain 168 in NSMP (71%), which was perfomed as a control experiment for decoyinine-induced sporulation, was completely blocked in the Δspo0A, Δspo0H, or ΔkinA ΔkinB genetic background (less than 5 × 10−6 %), whereas it was only partially affected by ΔkinA (7.9%) or ΔkinB (22%) (Table 3). These results indicate that the sporulation phosphorelay involving kinA and kinB is similarly operated in the massive sporulation in NSMP as in the decoynine-induced sporulation. However, ΔcodY and ΔabrB only slightly reduced the wild-type sporulation percentage of 71% to 29 and 34%, respectively, in contrast to their considerable effect on the decoyinine-induced sporulation as described above.
The kinA (A + 1G) and kinB (A + 1G) strains FU1102 and FU1113 with adenine replacement with guanine at the transcription initiation nucleotide affected decoyinine-induced sporulation, reducing the wild-type sporulation of 19% to 3.5% and 1.7%, respectively (Table 3). Also, the kinA (A + 1C) strain FU1155 with adenine replacement with cytosine at nucleotide +1 affected decoyinine-induced sporulation, reducing the wild-type sporulation of 19% to 4.2%, which is essentially indistinguishable from the effect of the adenine replacement with guanine at nucleotide +1 in spite of slightly lower induction of β-Gal synthesis by decoyinine than in the kinA (A + 1G) strain (Fig. 3A). The kinA (A + 1G) ΔkinB, kinA (A + 1C) ΔkinB, and ΔkinA kinB (A + 1G) strains FU1103, FU1170, and FU1114 were able to sporulate only at very low frequencies (6.2 × 10−4, 7.6 × 10−5, and 2.1 × 10−5%), whereas the ΔkinB and ΔkinA strains FU1096 and FU1095 formed considerable numbers of spores (0.67 and 0.26%, respectively). Also, the kinA (A + 1G) kinB (A + 1G) strain FU1155 sporulated at a comparatively low frequency, 0.0011%.
Similarly, these replacements, kinA (A + 1G), kinA (A + 1C), and kinB (A + 1G), affected the massive sporulation in NSMP moderately, reducing the wild-type sporulation percentage of 71% to 36%, 27%, and 25%, respectively (Table 3). The kinA (A + 1G) ΔkinB, kinA (A + 1C) ΔkinB, and ΔkinA kinB (A + 1G) strains FU1103, FU1170, and FU1114 sporulated at low frequencies (0.055, 0.0052, and 0.0019%, respectively), whereas the ΔkinB and ΔkinA strains FU1096 and FU1095 formed considerable numbers of spores (22 and 7.9%, respectively). Also, the kinA (A + 1G) kinB (A + 1G) strain FU1155 formed a low but considerable number of spores (2.3%). These results clearly indicate that the respective adenine replacements with guanine at the transcription initiation nucleotides of kinA and kinB as well as the adenine replacement with cytosine at nucleotide +1 of kinA actually affect the decoyinine-induced sporulation in S6 medium as well as the massive sporulation in NSMP.
DISCUSSION
Decoyinine, a GMP synthase inhibitor, induces the sporulation of exponentially growing wild-type cells in the presence of rapidly metabolizable carbon, nitrogen, and phosphate sources (Table 3) (19) and also causes the reciprocal concentration changes of an increase in ATP and a decrease in GTP, which can be sensed through increases and decreases in the rate of transcription initiation from positive and negative stringent promoters depending on their transcription initiation bases, i.e., adenine and guanine, respectively (22–24). DNA microarray analysis indicated that out of the genes involved in sporulation phosphorelay, decoyinine induced kinA, kinB, kinD, kinE, spo0F, spo0A, sigH, rapA, rapB, rapE, and spo0E more than 1.5-fold and repressed kinC, abrB, and ynzD the same amount (Table 2). The former genes, except rap and spo0E, were expected to be induced according to the sporulation phosphorelay (Fig. 1). KinC and KinD are involved in biofilm formation rather than sporulation (4, 43). The repression of ynzD and yisI (1.3-fold) coincided with the fact that YnzD and YisI increased under nonsporulation conditions (8).
KinA and KinB, whose synthesis was found to be under positive stringent transcription control in this study, are likely major sensor kinases that initiate sporulation (31, 42). Recently, it was reported that using an IPTG-inducible promoter, the induction of the synthesis of KinA beyond a certain level leads to the entry of the irreversible process of sporulation irrespective of nutrient availability (45) and that the primary role of the N-terminal domain of KinA is to form a functional tetramer that is necessary for the kinase activity catalyzed by the C-terminal domain (46). These facts imply that KinA as well as KinB is synthesized in an active form, suggesting that the threshold autophosphorylation level of the KinA (plus KinB) protein governs entry into sporulation under usual sporulation conditions.
The experiments involving the lacZ fusions of the promoter regions of kinA and kinB confirmed their induction by decoyinine on DNA microarray analysis. As shown in Fig. 3, the positive stringent transcription control of σH- and σA-dependent transcription from the respective kinA and kinB promoters involves the adenine at their transcription initiation nucleotides, and this positive control is most likely independent of either CodY or AbrB. Either Δspo0A or ΔsigH only affected kinA expression, because kinA is mainly transcribed by RNAP possessing σH, and sigH is indirectly activated by Spo0A through repression of AbrB (Fig. 1). Transcription of kinA and kinB is initiated at an adenine, so the initiation nucleotide triphosphate (iNTP) of kinA and kinB transcription is ATP. The increase in in vivo ATP concentration, which is evoked by decoyinine addition, enhances transcription initiation through RNAP, causing kinA and kinB activation. As far as we know, the positive stringent transcription control is only one device to activate the transcription of kinA and kinB by the modulation of the medium conditions, except that kinA transcription is driven by RNAP possessing σH inducible upon the onset of the phosphorelay.
It is possible to speculate that the kinA induction by decoyinine might reflect the derepression of sigH through AbrB repression by Spo0A∼P. However, this is unlikely, because the kinA gene was induced by decoyinine in an abrB+ genetic background essentially in the same fashion as in a ΔabrB background (Fig. 3B). This observation suggests that AbrB could be rapidly repressed by Spo0A∼P, which is formed through the phosphorelay triggered by KinA and KinB, leading to the immediate formation of σH.
Our recent study suggested that the presence of a guanine(s) and an adenine(s) at nucleotide +1 and nucleotide +2 might be indispensable for negative and positive stringent control, respectively (22). However, nucleotide +2 of kinA is guanine, which likely represses transcription initiation. It is possible to assume that the purine base at nucleotide +1 might be mainly involved in the formation of the transcription initiation complex in the case of the transcription conducted by RNAP possessing σH.
Spo0A, AbrB, CodY, and σH are all transcriptional regulators, and σH is the only sigma factor, involved in sporulation phosphorelay (Fig. 1). Expectedly, Δspo0A and ΔsigH blocked sporulation completely under the two sporulation conditions (decoyinine-induced sporulation in S6 medium and massive sporulation in NSMP) (Table 3). ΔcodY did not affect the sporulation in NSMP largely, but it enhanced the decoyinine-induced sporulation in S6 medium (Table 3). CodY negatively regulates kinB and phrAE (21); PhrA and PhrE inhibit RapA and RapE, respectively (Fig. 1), so ΔcodY was likely to suppress this negative effect on the sporulation. However, it is conceivable that ΔcodY affects the metabolic network leading to the sporulation in S6 medium more than in NSMP.
kinA (A + 1G), kinA (A + 1C), and kinB (A + 1G) mutants with adenine replacement with guanine or cytosine at the transcription initiation nucleotides of kinA and kinB were constructed (see Fig. S1 in the supplemental material); the construction of kinB (A + 1C) failed in spite of numerous trials. With respect to iNTP, the reciprocal changes of ATP increase and GTP decrease were provoked upon decoyinine addition, but CTP remained relatively unchanged (Fig. 2B) (24). The kinA (A + 1G) and kinA (A + 1C) mutants exhibited sporulation percentages between those of wild-type strain 168 and ΔkinA strains for the decoyinine-induced sporulation in S6 medium as well as for the massive sporulation in NSMP (Table 3). Also, the kinB (A + 1G) mutant exhibited sporulation percentages between those of the wild-type and ΔkinB strains under these two sporulation conditions (Table 3). Moreover, the kinA (A + 1G) kinB (A + 1G) strain sporulated at a lower frequency than that of either the kinA (A + 1G) or kinB (A + 1G) strain. Consequently, almost the same sporulation initiation mechanism, involving kinA and kinB, most likely operates for the decoyinine-induced sporulation in S6 medium and for the massive sporulation in NSMP. Although the ΔkinA and ΔkinB strains formed considerable numbers of spores under these two sporulation conditions, the ΔkinA kinB (A + 1G), ΔkinA kinB (A + 1C), and ΔkinB kinA (A + 1G) strains were scarcely able to form spores. These findings strongly indicated that the positive stringent transcription control of kinA and kinB without the involvement of any other transcription regulators is involved in enhancement of the phosphorelay that eventually leads to the formation of spores. In other words, operation of the positive stringent transcription control of kinA and kinB is a prerequisite to enter sporulation, whereby it can be gauged whether the energy charge of the cells, nearly parallel to in vivo ATP concentration, would be enough for sporulation to proceed. The ATP increase is sensed by putting the RNAP reaction with ATP as an iNTP of kinA and kinB transcription into the threshold kinetic range, leading to activation of the phosphorelay. In addition, an increase in the in vivo ATP pool likely facilitates the autophosphorylation of KinA and KinB for sporulation phosphorelay to proceed.
Conversely, the phosphoryl group is drained from the phosphorelay system through the actions of dedicated phosphatases; RapA, RapE, and RapB dephosphorylate Spo0F∼P by sensing a low cell density, and Spo0E, YisI, and YnzD dephosphorylate Spo0A∼P (Fig. 1) (2, 6). The opposing actions of the kinases and the phosphatases are believed to integrate environmental and physiological signals for the decision to sporulate by governing the flux through the relay system and hence the level of Spo0A∼P, which must reach a threshold concentration to trigger sporulation (47). Therefore, the negative stringent transcription control and the CodY-mediated repression of the stringent genes, both of which are evoked by stringent conditions such as decoyinine addition, contribute to the inhibition of dephosphorylation of Spo0A∼P and Spo0F∼P through decreases in and inhibition of the phosphatases such as YisI and YnzD and RapA and RapE, respectively (Fig. 1 and Table 2). It is notable that the efficiency of the induction of sporulation, which is induced by the stringent response, likely depends on the degree of the downregulation of the phosphatases that interfere with the phosphorelay system leading to the formation of spores, which is affected by the medium constituents; for example, decoyinine addition did not cause the efficient sporulation in the presence of good nitrogen sources such as ammonium and glutamate or glutamine in the medium in spite of sufficient induction of kinA and kinB (S.Tojo and Y. Fujita, unpublished observations).
In conclusion, we suggest that operation of the positive stringent transcription control is a prerequisite for phosphorelay to transfer a phosphate group to Spo0A, leading to spore formation. However, this does not necessarily mean that the operation of the positive stringent transcription regulation triggers the sporulation to proceed. The delicate balance of the input of the phosphate group into the phosphorelay by KinA and KinB and its drain from it by the Rap phosphatase determines whether the cell can enter sporulation. Thus, it would be possible that a decrease of the Rap phosphatase and kinB derepression, which might be evoked by CodY inactivation probably through a GTP decrease, could trigger sporulation during the low but constant operation of the positive stringent control under certain sporulation conditions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for the Strategic Support Project of Research Infrastructure Formation for Private University (S1001056) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Also, this work was supported by JSPS KAKENHI grant number 23380053.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02131-12.
REFERENCES
- 1. Hoch JA. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441–465 [DOI] [PubMed] [Google Scholar]
- 2. Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579–586 [DOI] [PubMed] [Google Scholar]
- 3. Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genomewide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perego M, Hoch JA. 2002. Two-component systems, phosphorelays, and regulation of their regulation by phosphatases, p 473–482 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 7. Sonenshein AL. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566 [DOI] [PubMed] [Google Scholar]
- 8. Perego M. 2001. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol. Microbiol. 42:133–143 [DOI] [PubMed] [Google Scholar]
- 9. Stephenson K, Hoch JA. 2002. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46:297–304 [DOI] [PubMed] [Google Scholar]
- 10. Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 11. Fürbass R, Gocht M, Zuber P, Marahiel MA. 1991. Interaction of AbrB, a transcriptional regulator from Bacillus subtilis with the promoters of the transition state-activated genes tycA and spoVG. Mol. Gen. Genet. 225:347–354 [DOI] [PubMed] [Google Scholar]
- 12. Fujita M, Sadaie Y. 1998. Promoter selectivity of the Bacillus subtilis RNA polymerase σA and σH holoenzymes. J. Biochem. 124:89–97 [DOI] [PubMed] [Google Scholar]
- 13. Strauch MA. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strauch MA, Trach KA, Day J, Hoch JA. 1992. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 74:619–626 [DOI] [PubMed] [Google Scholar]
- 15. Strauch MA, Hoch JA. 1993. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 7:337–342 [DOI] [PubMed] [Google Scholar]
- 16. Strauch MA, Wu JJ, Jonas RH, Hoch JA. 1993. A positive feedback loop controls transcription of the spo0F gene, a component of the sporulation phosphorelay in Bacillus subtilis. Mol. Microbiol. 7:967–974 [DOI] [PubMed] [Google Scholar]
- 17. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 19. Mitani T, Heinze JE, Freese E. 1977. Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem. Biophys. Res. Commun. 77:1118–1125 [DOI] [PubMed] [Google Scholar]
- 20. Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krásný L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23:4473–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tojo S, Kumamoto K, Hirooka K, Fujita Y. 2010. Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis. J. Bacteriol. 192:1573–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krásný L, Tišerová H, Jonák J, Rejman D, Sanderová H. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 69:42–54 [DOI] [PubMed] [Google Scholar]
- 24. Tojo S, Satomura T, Kumamoto K, Hirooka K, Fujita Y. 2008. Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J. Bacteriol. 190:6134–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Slack FJ, Serror P, Joyce E, Sonenshein AL. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689–702 [DOI] [PubMed] [Google Scholar]
- 26. Lopez JM, Marks CL, Freese E. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587:238–252 [DOI] [PubMed] [Google Scholar]
- 27. Lopez JM, Dromerick A, Freese E. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ochi K, Kandala JC, Freese E. 1981. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 256:6866–6875 [PubMed] [Google Scholar]
- 29. Ochi K, Kandala JC, Freese E. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J. Bacteriol. 151:1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Predich M, Nair G, Smith I. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH. J. Bacteriol. 174:2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trach KA, Hoch JA. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 8:69–79 [DOI] [PubMed] [Google Scholar]
- 32. Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serror P, Sonenshein AL. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshida K, Fujita Y, Ehrlich SD. 1999. Three asparagine synthetase genes of Bacillus subtilis. J. Bacteriol. 181:6081–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Isezaki M, Hosoya S, Takeuchi M, Sato T. 2001. A putative ATP-binding cassette transporter YbdA involved in sporulation of Bacillus subtilis. FEMS Microbiol. Lett. 204:239–245 [DOI] [PubMed] [Google Scholar]
- 36. Morimoto T, Ara K, Ozaki K, Ogasawara N. 2009. A new simple method to introduce marker-free deletions in the Bacillus subtilis genome. Genes Genet. Syst. 84:315–318 [DOI] [PubMed] [Google Scholar]
- 37. Yoshida K, Ishio I, Nagakawa E, Yamamoto Y, Yamamoto M, Fujita Y. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573–579 [DOI] [PubMed] [Google Scholar]
- 38. Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56:1560–1573 [DOI] [PubMed] [Google Scholar]
- 39. Miwa Y, Fujita Y. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 183:5877–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Fujita Y, Freese E. 1981. Isolation and properties of a Bacillus subtilis mutant unable to produce fructose-bisphosphatase. J. Bacteriol. 145:760–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayashi Y. 1995. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that KinC encodes a histidine protein kinase. J. Bacteriol. 177:176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shemesh M, Kolter R, Losick R. 2010. The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J. Bacteriol. 192:6352–6356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sierro N, Makita Y, de Hoon M, Nakai K. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36(Suppl 1):D93–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. 2010. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J. Bacteriol. 192:3870–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eswaramoorthy P, Dravis A, Devi SN, Vishnoi M, Dao HA, Fujita M. 2011. Expression level of a chimeric kinase governs entry into sporulation in Bacillus subtilis. J. Bacteriol. 193:6113–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fujita M, González-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.