Abstract
The facultative human pathogen Vibrio cholerae transits between the gastrointestinal tract of its host and aquatic reservoirs. V. cholerae adapts to different situations by the timely coordinated expression of genes during its life cycle. We recently identified a subclass of genes that are induced at late stages of infection. Initial characterization demonstrated that some of these genes facilitate the transition of V. cholerae from host to environmental conditions. Among these genes are uptake systems lacking detailed characterization or correct annotation. In this study, we comprehensively investigated the function of the VCA0682-to-VCA0687 gene cluster, which was previously identified as in vivo induced. The results presented here demonstrate that the operon encompassing open reading frames VCA0685 to VCA0687 encodes an ABC transport system for hexose-6-phosphates with Km values ranging from 0.275 to 1.273 μM for glucose-6P and fructose-6P, respectively. Expression of the operon is induced by the presence of hexose-6P controlled by the transcriptional activator VCA0682, representing a UhpA homolog. Finally, we provide evidence that the operon is essential for the utilization of hexose-6P as a C and P source. Thereby, a physiological role can be assigned to hexose-6P uptake, which correlates with increased fitness of V. cholerae after a transition from the host into phosphate-limiting environments.
INTRODUCTION
The life cycle of the facultative human pathogen Vibrio cholerae is marked by repetitive transitions between aquatic environments and the host gastrointestinal tract. Besides many other variable conditions, V. cholerae has to adjust to different qualities and quantities of nutrient sources. This variability is emphasized by the fact that utilization of nutrients under laboratory conditions, such as in full broth or a chemically defined minimal medium, represents no growth limitation for clinical V. cholerae isolates (1).
In the open sea, bacteria such as V. cholerae face C, N, and P limitation and are restricted to limited nutrients on a picomolar or nanomolar scale, whereby substrates become available and accessible only in specific habitats (2). Therefore, marine bacteria are found close to organic particles ranging from micrometer- to millimeter-sized aggregates. These organic particles derive from different sources, such as lysed or dead phytoplankton, zooplankton, or fecal pellets. They deliver organic and inorganic substrates in concentrations that are up to 4 orders of magnitude greater than those found in particle-free open seawater (for a recent review, see reference 2). Therefore, many plants and animals in the ocean serve as microbial niches for V. cholerae (3–6). For example, copepods or other crustaceans contain or are covered with chitin, a β,1-4-linked polymer of 2-acetamido-2-deoxy-β-d-glucopyranoside (GlcNAc)n and its deacetylated form, chitosan. These substrates are utilized by Vibrio sp. as C and N sources (7–9). In addition, biofilm formation on chitinous surfaces plays a crucial role in V. cholerae persistence in the aquatic environment (5, 10, 11). Therefore, biofilms play a more vital role, as thought for V. cholerae. For example, as recently shown in our laboratory, extracellular DNA represents a component of the V. cholerae-produced biofilm matrix that can further be utilized as a P source (12). Whether or not hexose-6-phosphate (hexose-6P) can be found in the environment is not known to us, since we found no study addressing the environmental monitoring of these substrates.
After oral infection of its human host, V. cholerae passes the stomach and enters the gastrointestinal tract. V. cholerae colonizes primarily the small intestine while penetrating the intestinal mucin close to the epithelial barrier, where nutrient-rich cells are located and bacterial propagation takes place (13). As shown by scanning electron microscopy in the infant mouse model (14), V. cholerae cells rapidly penetrate the intervillous space and associate with the mucosal surface close to crypts. Besides penetrating the mucus lining, products of mucin or mucous surfaces can serve as a C source. This was shown recently (15), whereby the uptake system for sialic acids was identified in V. cholerae and found to be encoded on the Vibrio pathogenicity island. Within the host, major changes in V. cholerae gene expression take place, whereby virulence gene regulation plays a central role in controlling several regulatory cycles and numerous genes (16). Examples are the well-characterized ToxRS and TcpPH regulons that represent the virulence gene expression system that results in colonization and toxin production (17). Once V. cholerae has colonized its human host, its growth ability is exceptional. It was reported that cell numbers reach up to 108 cells/g stool excreted by cholera patients (18). Thereby, a highly active metabolism is necessary to support V. cholerae cells to such high growth rates within the host. In vivo expression technology (IVET) and recombination-based IVET (RIVET) were used to identify in vivo-induced genes in V. cholerae (19–25). Using such techniques, it was possible to identify and characterize several virulence genes under infection conditions, such as genes encoding cholera toxin and toxin-coregulated pili. Additionally, in these studies, many other genes encoding so-far-uncharacterized substrate metabolism and uptake systems were found to be induced in vivo. Recently, Schild et al. used RIVET in combination with the infant mouse model to identify late in vivo-induced genes (26). They showed that the induction of these genes facilitates the transition fitness of V. cholerae cells when they depart from the infection site of the host and enter the aquatic environment (27). For example, genes involved in chitin degradation or synthesis of the second messenger c-di-GMP have been characterized as being induced late under infection conditions and enhance survival in pond water after in vivo passage (16, 26). Interestingly, a considerable number of late in vivo-induced genes encode different metabolic enzymes and substrate uptake systems. For most of them, the exact function and transported substrates have not been comprehensively investigated. One example is the uncharacterized late in vivo-induced gene cluster encoded by hypothetical genes VCA0685 to VCA0687, which have been demonstrated to play a role in pond water survival after release from an in vivo colonization model, e.g., the infant mouse model (16, 26). In the present study, we have characterized VCA0685 to VCA0687 as an ABC transport system for hexose-6P, including its regulation and utilization of hexose-6P as a carbon and phosphate source. The results indicate that the utilization of hexose-6P as a phosphate source by V. cholerae is advantageous for subsequent survival under phosphate-limiting conditions.
MATERIALS AND METHODS
Strain construction and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1, and the oligonucleotides used are listed in Table 2. Unless noted otherwise, strains were grown with aeration in Luria-Bertani (LB) broth at 37°C. V. cholerae AC53, a spontaneous streptomycin-resistant mutant of V. cholerae O1 El Tor clinical isolate E7946 (34), was used as the wild-type (WT) strain in all experiments. For genetic manipulations, Escherichia coli strains DH5αλpir and SM10λpir were used (28, 29, 35). Antibiotics and other supplements were used at the following final concentrations: streptomycin (Sm), 100 μg/ml; ampicillin (Ap), 50 μg/ml in combination with other antibiotics, 100 μg/ml otherwise; glucose, 0.2%; glycerol, 0.4%; sucrose, 10%.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| DH5αλpir | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | 28 |
| SM10λpir | thi recA thr leu tonA lacY supE RP4-2-Tc::Mu λ::pir | 29 |
| V. cholerae strains | ||
| E7946 | O1 El Tor, Ogawa; Smr (WT) | 30 |
| ΔVCA0685-7 | Deletion of VCA0685, VCA0686, and VCA0687 in E7946; Smr | This work |
| ΔuhpA | Deletion of VCA0682 in E7946; Smr | This work |
| VCA0687::phoA | Insertion of pGPphoA downstream of VCA0687 in E7946; Smr Apr | This work |
| ΔuhpA VCA0687::phoA | Insertion of pGPphoA downstream of VCA0687 in ΔuhpA; Smr Apr | This work |
| Plasmids | ||
| pGP704 | ori6K mobRP4, Apr | 29 |
| pCVD442 | ori6K mobRP4 sacB, Apr | 30 |
| pTRC99A | pBR322 origin, Apr | 31 |
| pGPphoA | pGP704 with promoterless phoA of SM10λpir, Apr | 32 |
| pGPphoA-VCA0687 | pGPphoA with VCA0687 gene fragment, Apr | This work |
| pCVDΔVCA0685-7 | pCVD442 with upstream fragment of VCA0685 and downstream fragment of VCA0687, Apr | This work |
| pCVDΔuhpA | pCVD442 with up- and downstream fragments of VC0682, Apr | This work |
| pTRCuhpA | pTRC99A with reading frame VCA0682, Apr | This work |
| pTRC0685-7 | pTRC99A with reading frames VCA0685 to VCA0687, Apr | This work |
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| VCA0685-NcoI-2 | TTTCCATGGGTTTTGCATTAACAATCATTGGGTT |
| VCA0685-SacI-1 | TTTGAGCTCATTCACGTAGCGAACGGGGCT |
| VCA0687-XbaI-4 | TTTTCTAGACTCAATTTTGAAGACTATGTGCTC |
| VCA0687-NcoI-3 | TTTCCATGGCAGAATAGTGATAATACCCTGAG |
| VCA0687-KpnI-phoA-3′ | TTTGGTACCTATTATCACTATTCTGCGTAGGG |
| VCA0687-SacI-phoA-5′ | TATGAGCTCTTAGATGCCAACTTGCGCCG |
| UhpA-XbaI-1 | AAATCTAGAGCATCTCCACGTTCAGCCCTAG |
| UhpA-KpnI-2 | AAAGGTACCATGATGTTTATCACCTAGCAAAATC |
| UhpA-KpnI-3 | AAAGGTACCATGTGATCCGCTTTGCTTTGCAG |
| UhpA-SacI-4 | ATTGAGCTCAACGGATTTGCGAATTTGCTCTT |
| UhpA-EcoRI-5′ | TTTGAATTCTGATTTTGCTAGGTGATAAACATCA |
| UhpA-XbaI-3′ | TTTTCTAGAGTTAACCTTCCACGAGCAGTTG |
| VCA0685-EcoRI-5′ | AAAGAATTCAAGGACACAAGGGAGCATCAA |
| VCA0687-XbaI-3′ | TTTTCTAGAGGCTTTACTCAGGGTATTATCAC |
Restriction sites are underlined.
DNA manipulation and construction of suicide plasmids, reporter fusions, and mutant strains.
Chromosomal DNA was isolated as described by Grimberg et al. (36), whereas PCR products and digested plasmid DNA were purified with the QIAquick gel extraction and QIAquick PCR purification kits (Qiagen). PCRs for sequencing and subcloning were carried out with the Phusion High-Fidelity polymerase (NEB). For all other reactions, Taq DNA polymerase (NEB) was used.
Deletion mutants were constructed as described by Donnenberg and Kaper (31). PCR fragments of approximately 500 bp upstream and downstream of the gene of interest were amplified with oligonucleotides x-y-1 and x-y-2 or x-y-3 and x-y-4, where x represents the gene and y represents the restriction site/enzyme used (Table 2). After enzymatic digestion, PCR fragments were ligated into similarly digested pCVD442, resulting in the corresponding knockout plasmids (Table 2).
Derivatives of pGPphoA (33) were constructed to obtain chromosomal transcriptional fusions of phoA to respective genes. As the phoA gene acts as a useful genetic marker in V. cholerae, promoterless phoA was used to generate the VCA0687::phoA operon fusion. The gene fragment corresponding to VCA0687 containing the translational stop codon was amplified by PCR with oligonucleotides VCA0687-SacI-phoA-5′ and VCA0687-KpnI-phoA-3′ (Table 2). The PCR product was digested with SacI and KpnI and ligated into pGPphoA digested with the corresponding enzymes, resulting in plasmid pGPphoA-VCA0687 (Table 1).
In the case of the suicide plasmid derivatives used to generate deletion knockout mutants and transcriptional fusions, generated plasmids (Table 1) were first transformed into E. coli Sm10λpir and subsequently conjugated into V. cholerae. The resulting conjugants were purified via selection for Smr and Apr colonies. Insertion of pGPphoA-VCA0687 into the VCA0687 locus on the V. cholerae chromosome by homologues recombination resulted in transcriptional fusions to the VCA0687-encoding gene. For generation of chromosomal deletions in the case of pCVD442 derivatives (Table 1), sucrose selection was used to obtain Aps colonies (31). Correct chromosomal insertions or deletions were confirmed by PCR (data not shown).
Construction of expression plasmids.
All expression and complementing plasmids were constructed in a similar manner. PCR fragments of the respective genes, containing their own ribosomal binding sites, were generated with oligonucleotides x-y-5′ and x-y-3′, where x represents the gene and y represents the restriction site/enzyme used (Table 2). PCR fragments digested with the respective restriction enzymes were ligated into similarly digested plasmid pTRC99A as indicated (Tables 1 and 2) and shown in Results and Discussion.
Growth kinetics.
Growth kinetics were determined in transparent 24-well plates (Greiner) with a 1-ml culture volume, providing good aeration with a well-to-culture volume of 3:1, with M9 minimal medium supplemented with carbon sources such as glucose (11 mM), fructose-6P (10 mM, disodium salt hydrate; Sigma-Aldrich), and glucose-6P (10 mM, disodium salt hydrate; Sigma-Aldrich). The optical density at 600 nm (OD600) was monitored every 30 min in a FLUOstar OMEGA plate reader (BMG Labtech) under shaking conditions at 37°C. For presentation of data, at least three independent growth curves were monitored for each strain tested. The mean values were calculated and plotted. Standard deviations are not shown, since only growth and nongrowth phenotypes were evaluated.
Alkaline phosphatase assays.
To determine the enzymatic activities for the transcriptional phoA fusions, alkaline phosphatase assays were performed as described previously (33, 37). The activities were expressed in Miller units, given by A405 × 1.000/(A600 × ml × min). Note that no intrinsic alkaline phosphatase background activity was measured for V. cholerae strain E7946 in the presence of glucose or glucose-6P (data not shown).
14C-labeled compounds.
For substrate uptake and kinetics, labeled substrates were used as follows, [U-14C]d-fructose-6P and [U-14C]d-glucose-6P in disodium salt with specific activities of 248 and 245 mCi/mmol (Hartmann Analytic), respectively.
Uptake studies.
For uptake studies, the WT and ΔVCA0685-7 mutant strains transformed with the pTRC99A control plasmid and the ΔVCA0685-7 mutant strain transformed with complementing plasmid pTRC0685-7 and induced with 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were used. To demonstrate the uptake of [14C]labeled hexose-6P, cells were inoculated to an OD600 of 0.1 in M9 supplemented with glycerol (0.4%) and hexose-6P (10 mM) and grown to an OD600 of 0.5, representing the mid-log growth phase. M9 medium supplemented with glycerol and hexose-6P allows growth on both substrates and thereby induction of the hexose-6P uptake system. Note that glycerol was added as a C source to allow hexose-6P uptake-deficient mutants to grow. This growth strategy was used to generate energized cells in the mid-log growth phase by allowing induction of VCA0685-to-VCA0687 expression. The cultures were washed twice and resuspended in M9 to an OD600 of 0.5. Uptake studies were carried out at room temperature. The reaction was started with the addition of either 2.5 μl of [U-14C]fructose-6P (0.4 mM) and 7.5 μl of fructose-6P (0.4 mM) or 2.5 μl of [U-14C]glucose-6P (0.4 mM) and 7.5 μl of glucose-6P (0.4 mM) per ml of culture. After 30, 60, 90, 210, and 330 s, samples of 500 μl were taken, filtered through membranes (0.45-μm pore size; Schleicher & Schuell MicroScience), and washed under suction with 10 ml NaCl (100 mM). The membranes were then placed in vials containing 4 ml of scintillation fluid (Ultima Gold; PerkinElmer). To correlate the total added radioactivity, 500 μl of the whole reaction mixture was added directly to separate vials containing 4 ml of scintillation fluid. All samples were counted in a liquid scintillation analyzer (Tri-Carb 2300TR; Packard).
For substrate-dependent uptake kinetics, only the WT strain was used and cultures were prepared as previously described. The uptake reactions were each started with the addition of 25, 18.75, 12.5, 7.5, 5, or 2.5 μl of [U-14C]d-fructose-6P (0.04 mM) per ml of culture and 37.5, 31.25, 25, 18.75, or 12.5 μl of [U-14C]d-glucose-6P (0.04 mM) per ml of culture, respectively. After 10, 20, and 30 s, samples of 500 μl were taken, filtered through membrane filters, washed under suction with 100 mM NaCl, and measured as previously described. To detect the entire added radioactivity per volume tested, 500 μl of the whole reaction mixture was added directly to vials containing 4 ml scintillation fluid. Rates of substrate transport were determined from the number of counts obtained after 20 s of incubation with the labeled substrate, as described by Xavier et al. (38). It was verified that a linear correlation of the number of counts versus time was observed for at least 30 s for all of the substrate concentrations tested.
RESULTS AND DISCUSSION
VCA0685 to VCA0687 represent a hexose-6P uptake system.
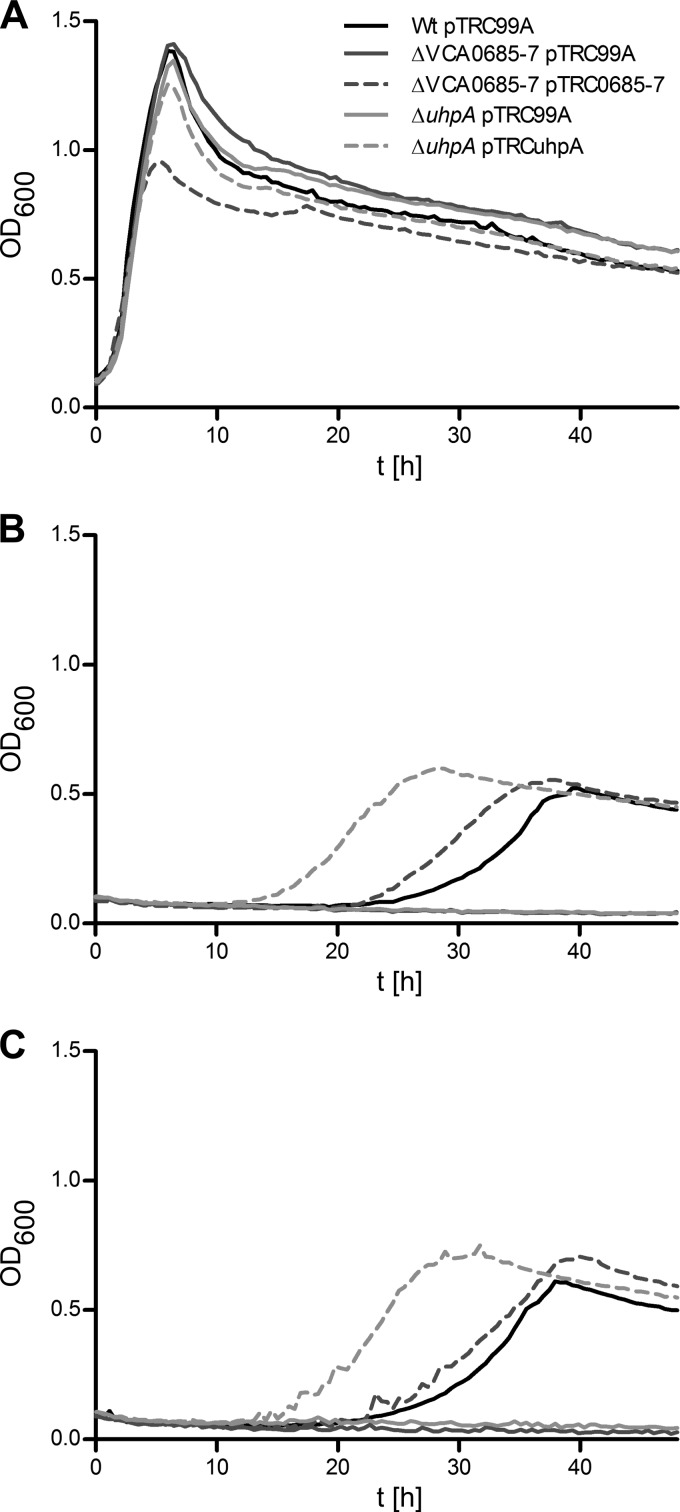
The gene cluster containing open reading frames (ORFs) VCA0685 to VCA0687 was found to be induced under late infection conditions (26, 39). Furthermore, it has been shown that its function contributes to survival after in vivo passage and subsequent transition into pond water (26). VCA0685 to VCA0687, originally annotated as ferric iron transporter system components (39), were recently investigated as a putative ABC iron uptake transport system showing 69, 65, and 60% amino acid identity to the AfuABC system of Actinobacillus pleuropneumoniae (40). Functional complementation analysis in iron transport-deficient Shigella flexneri, Fur-dependent reporter gene induction in E. coli, and mutant analysis in V. cholerae showed no evidence of iron uptake activity of VCA0685 to VCA0687 (40). However, in V. cholerae, VCA0682 to VCA0687 represent a putative gene cluster (Fig. 1) that corresponds in part to homologous genes of the uhp operon in E. coli (41, 42). As demonstrated in E. coli, UhpT is an antiporter for hexose-6P uptake (43) and UhpABC represent the sensor kinase-regulating components (44). To characterize cellular growth ability on hexose-6P substrates, two mutant strains were tested. In one mutant strain (ΔVCA0682), the transcriptional activator homologue VCA0682 (uhpA) was deleted and a second mutation comprises the deletion of three reading frames, constituting the putative hexose-6P uptake system, i.e., VCA0685, VCA0686, and VCA0687 (39), referred to here as VCA0685-7. The growth kinetics of these deletion mutants and the parental WT were monitored in M9 minimal medium supplemented with different sole C sources such as glucose (Fig. 2A), glucose-6P (Fig. 2B), or fructose-6P (Fig. 2C). All of the strains showed similar growth on M9 supplemented with glucose, except the VCA0685-7 complementation strain, which reached only 70% of the maximum cell density (OD600) of the WT. This effect is probably due to a reduced fitness of these cells bearing the burden of expressing the VCA0685-7 gene products from plasmid pTRC99A, thus reaching only partial maximum growth ability in M9 glucose. Such an effect was not observed in M9 hexose-6P-supplemented medium, where physiologically relevant complementation activity was observed. The deletion mutant strains were unable to grow in hexose-6P-supplemented medium. Complementation of the growth phenotype of both mutant strains was observed by the expression of UhpA or VCA0685-7 from pTRC99A even without the addition of IPTG, because of leaky promoter activity (Fig. 2B and C). Notably, growth in M9 glucose medium started immediately, whereas prolonged lag phases of up to 25 h were observed in the WT strain on hexose-6P substrates. Interestingly, expression of the putative response regulator UhpA from pTRC99A substantially shortened the lag phase (Fig. 2A and B).
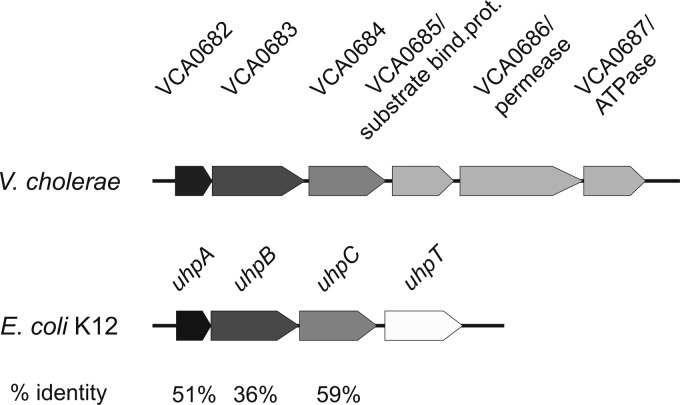
Fig 1.
Overview of the genomic organization of hexose-6P uptake systems in V. cholerae and E. coli. The genomic region of E. coli responsible for hexose-6P uptake consists of three regulatory genes, uhpA, uhpB, and uhpC, that control the expression of the corresponding transport protein UhpT (42). In V. cholerae, homologous genes are located on the small chromosome and are similarly arranged. They are regulatory genes uhpA/VCA0682, uhpB/VCA0683, and uhpC/VCA0684. While no uhpT homologue is found in V. cholerae, three additional genes are present: VCA0685 (substrate-binding protein [bind. prot.]), VCA0686 (permease), and VCA0687 (ATPase). According to NCBI BLASTP (57), V. cholerae ORFs VCA0682 to VCA0684 show amino acid identities to corresponding E. coli K-12 proteins, as indicated at the bottom.
Fig 2.
Growth curves of V. cholerae WT and mutant strains in M9 minimal medium supplemented with hexoses as carbon sources. Shown are the growth kinetics of the WT, the ΔVCA0685-7 and ΔuhpA deletion mutants, and the ΔVCA0685-7/pTRC0685-7 and ΔuhpA/pTRCuhpA complemented mutants in M9 minimal medium supplemented with glucose (A), fructose-6P (B), or glucose-6P (C). Data represent mean values of at least three independent measurements.
Additionally, we observed a decline in cell density during the late exponential growth phase (e.g., Fig. 2A). Most likely, this is an effect of stationary-phase resting cells, which may undergo lysis, change their shape, or form aggregates. It was also noted that the maximum OD600 reached by cells grown in M9 glucose was 2 to 3 times as high as that of cells grown in M9 hexose-6P. This represents a substrate concentration effect, since if we increased the glucose-6P concentration to 25 mM, then the OD600 increased to 1.4, which is similar to the observed density of cells grown in M9 glucose (data not shown). Finally, by reviewing published and deposited genome information on sequenced Vibrio sp. (KEGG, J. Craig Venter Institute, or the BROAD Institute), it seems that the VCA0682-to-VCA0687 gene cluster characterized here is represented inconsistently. Sequenced O1, O139, or non-O1/non-O139 V. cholerae, as well as V. mimicus, V. farnesi, V. metschnikovii, and V. anguillarum, isolates showed the presence of this gene cluster, whereas no such gene cluster was found in V. fischeri, V. parahaemolyticus, or V. vulnificus. Future bioinformatic analysis will address the genomic view of this gene region. Taken together, these results demonstrate that the components of VCA0685-7 and the putative activator UhpA are essential for the utilization of hexose-6P substrates.
Hexose-6P uptake and kinetics.
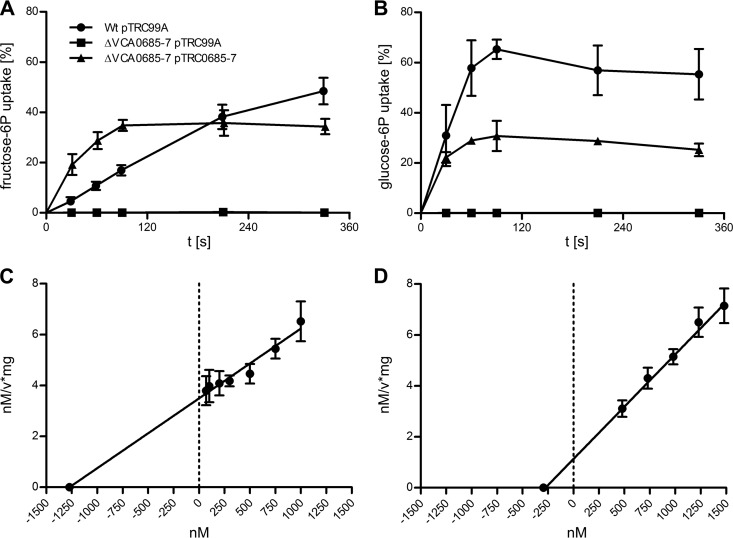
On the basis of the growth phenotypes shown in Fig. 2, we further characterized the specific transport kinetics of the VCA0685-7 deletion mutant compared to those of WT cells and complemented mutant strains. For example, the uptake of [14C]fructose-6P and [14C]glucose-6P was monitored (Fig. 3A and B). Substrate accumulations of 49.5% (fructose-6P) and 55% (glucose-6P) after 6 min of incubation were observed for the WT, and 33% (fructose-6P) and 25% (glucose-6P) accumulations were observed for the ΔVCA0685-7 mutant complemented with plasmid pTRC0685-7. In contrast, less than 0.1% of [14C]fructose-6P or [14C]glucose-6P accumulated in the ΔVCA0685-7 mutant strain containing control plasmid pTRC99A. For complementation, IPTG induction was necessary to enhance the Ptrc promoter activity of pTRC99A (32), allowing the expression of VCA0685-7 and monitoring of hexose-6P uptake, whereas no IPTG induction was necessary to complement the growth phenotypes (Fig. 2). As further shown in Fig. 3A and B, uptake of [14C]glucose-6P was quickly saturated, in <2 min, indicating that the extracellular [14C]glucose-6P concentration becomes limited for further uptake after 55% substrate accumulation. In contrast, [14C]fructose-6P uptake showed a lower velocity and did not reach saturation before the 6-min time point. Differences were also observed for uptake activities of complementing strains, where neither fructose-6P nor glucose-6P uptake reached WT levels, suggesting that expression of VCA0685-7 from pTRC99A is insufficient, resulting in partial complementation. According to Michaelis-Menten kinetics, the Vmax and Km values for the [14C]fructose-6P and [14C]glucose-6P uptake of the V. cholerae WT strain were determined (Fig. 3C and D). At least five different substrate concentrations were analyzed to determine the velocity of the uptake dynamics, according to the analysis method of Hanes (45). For [14C]glucose-6P and [14C]fructose-6P, Vmax values of 243.4 and 364.5 nM/min/mg were determined, and the apparent Km values deduced were 0.275 and 1.273 μM, respectively. Interestingly, the two substrates show similar Vmax values but the Km values differ by ∼5-fold. This finding correlates with glucose-6P transport saturation observed already after 2 min (Fig. 3B) and not observed for fructose-6P (Fig. 3A). The most plausible explanation is that for glucose-6P, the Vmax of the Uhp transporter is maintained, while decreasing substrate concentrations are well above the Km value in the medium. In contrast, for fructose-6P, the decreasing substrate concentration in the medium is approaching the Km value and therefore causing lower uptake activity. According to the literature, Km values for the glucose-6P uptake system were determined between 20 and 500 μM (46). It was suggested that these values were influenced by various Pi concentrations in the buffer or medium used, which may act as a competitive inhibitor (46). It is noteworthy that the Uhp transport components characterized here differ significantly from the known system components of E. coli or Salmonella enterica serovar Typhimurium (Fig. 1). According to bioinformatic analyses, the V. cholerae uhp-encoded transport components do not belong to the electrochemical gradient-coupled permease family, also termed secondary active transporters. Instead, the system characterized here constitutes an ATP-binding cassette (ABC) transporter system that resembles a primary active transporter family (47). As shown in V. cholerae (Fig. 1), three additional ORFs are present that replace the otherwise occurring single permease-encoding uhpT gene in E. coli. As bioinformatic analyses revealed, the three genes observed in V. cholerae encode sequences homologous to primary active transporters. These products are a substrate-binding protein, a transmembrane protein, and a nucleotide-binding domain-containing protein resembling an ABC-type transporter (48). The apparent Km values of a well-characterized prototype ABC transporter, the disaccharide maltose transport system of E. coli (49), were determined to be 0.8 to 1.4 μM (50), depending on the absence or presence of the maltose-binding protein, respectively. Another example is the binding protein-dependent ABC iron transporter FbpABC of Haemophilus influenzae. Its Fe3+ uptake was determined to have apparent Km of 0.9 to 1.2 μM, depending on the presence or absence of the iron-binding protein (51). Hence, this identified ABC hexose-6P transport system has a Km value that is similar to those of other ABC transporters and indicates higher hexose-6P substrate sensitivity, as known for the Uhp system in E. coli. This raises an interesting view, whereby V. cholerae cells may have an advantage over members of the Enterobacteriaceae flora by utilizing such substrates in the human gut. Whether this provides a competitive growth advantage needs to be proven in vitro or in vivo. The uhp system was found to be induced late in vivo, and in vivo colonization showed no effect on competition between WT V. cholerae and a uhp knockout strain (26). Therefore, the uhp system should play only a minor role in the colonization phase and this indicates that utilization of hexose-6P per se is not required for successful colonization. However, WT cells did show higher survival fitness after being released into the environment (26), for reasons addressed in more detail below.
Fig 3.
Hexose-6P uptake by V. cholerae WT and mutant strains. The time-dependent uptake of [U-14C]d-fructose-6P (A) and [U-14C]d-glucose-6P (B) by the WT/pTRC99A strain and the ΔVCA0685-7/pTRC99A and ΔVCA0685-7/pTRC0685-7 mutant strains was determined as a percentage of the total added radioactivity. Shown are mean values of at least four independent measurements. The error bars indicate standard deviations. (C, D) The uptake velocities at specific substrate concentrations were determined by measurements at three consecutive time points. Shown are mean values of at least two independent experiments. The error bars indicate standard deviations. From Hanes-Woolf plots, the calculated apparent Km was 1.273 μM with a Vmax of 364.5 nM/min/mg protein for fructose-6P (C). As shown in plot D, the apparent Km was 0.275 μM with a Vmax of 243.4 nM/min/mg protein for glucose-6P.
Regulation of the hexose-6P uptake system encoding homologues of uhp genes in V. cholerae.
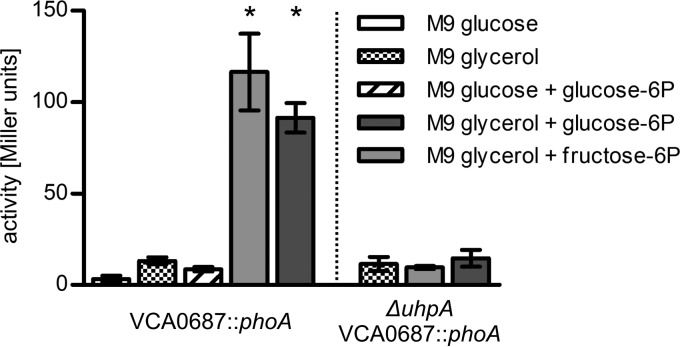
To characterize the transcriptional regulation of the VCA0685-7 operon in V. cholerae, we generated chromosomal transcriptional fusion strains consisting of a promoterless phoA reporter gene fused to VCA0687 (Fig. 1) (33). The resulting PhoA activity represents the transcriptional activity of the last gene of the putative uhp cluster, VCA0687. To test whether hexose-6P substrates such as fructose-6P or glucose-6P induce VCA0687 gene transcription, V. cholerae reporter strains were grown in M9 minimal medium supplemented with glycerol or glucose in the presence or absence of hexose-6P (Fig. 4). V. cholerae cultures grown in M9 glycerol reached mid-log phase (OD600 of 0.5) after 8 h and late exponential growth phase (OD600 of 1.5) after 16 h (data not shown). It was necessary to use glycerol to determine gene induction in a ΔuhpA background, since this strain would not grow on hexose-6P sources. In the presence of hexose-6P, 10- and 12-fold induction of VCA0687::phoA expression was observed with fructose-6P and glucose-6P, respectively (Fig. 4). This is consistent with data from E. coli showing 50-fold induction of uhp genes (44). In contrast, cultures grown in M9 glucose and glucose-6P showed no induction, indicating that catabolite repression might control uhp gene expression. However, neither cyclic AMP (cAMP; 3 mM) addition to cultures growing in such a medium nor cAMP addition to M9 glucose-6P cultures grown for 8 h showed induction of uhp genes (data not shown). Therefore, we exclude an influence of cAMP-cAMP receptor protein on uhp gene transcription at this point. Further characterization is necessary to elucidate any participation of catabolite repression in uhp gene regulation. So far in E. coli, only glucose-6P has been found to bind to a membrane receptor compound (52), which most likely constitutes the sensor kinase complex UhpBC (53), whereas fructose-6P binding could not be demonstrated. This might indicate that fructose-6P has to be converted to glucose-6P before induction can take place (for an overview, see reference 46). Additionally, a VCA0682 deletion mutant missing a uhpA homologue was tested. VCA0682 (39), showing 51% identity to response regulator UhpA (44), may represent the transcriptional activator of the uhp genes in V. cholerae. As shown in Fig. 4, in a ΔuhpA mutant strain, VCA0687::phoA expression does not respond to the inducing substrate fructose-6P or glucose-6P. Concordant with this observation, ΔuhpA prevents growth on hexose-6P, while expression of uhpA in trans from the complementation plasmid restores growth (Fig. 2). Thus, the results indicate that deletion of the transcriptional activator VCA0682 (UhpA) leads to loss of uhp gene expression. It is important to note that no uhp gene induction was observed when cells were grown in the presence of glycerol, glucose, or glucose and glucose-6P in combination (Fig. 4).
Fig 4.
PhoA activities of V. cholerae VCA0687::phoA and ΔuhpA VCA0687::phoA strains grown in M9 supplemented with various carbon sources. Strains were grown in M9 medium as indicated. Alkaline phosphatase activities (in Miller Units) of cultures of the VCA0687::phoA and ΔuhpA VCA0687::phoA transcriptional fusion strains grown for 8 h were measured. Shown are the means of at least four independent measurements. The error bars indicate standard deviations. *, significant differences between the respective data sets in comparison to the M9 glycerol control condition (Kruskal-Wallis test followed by posthoc Dunn's multiple comparisons; P < 0.05).
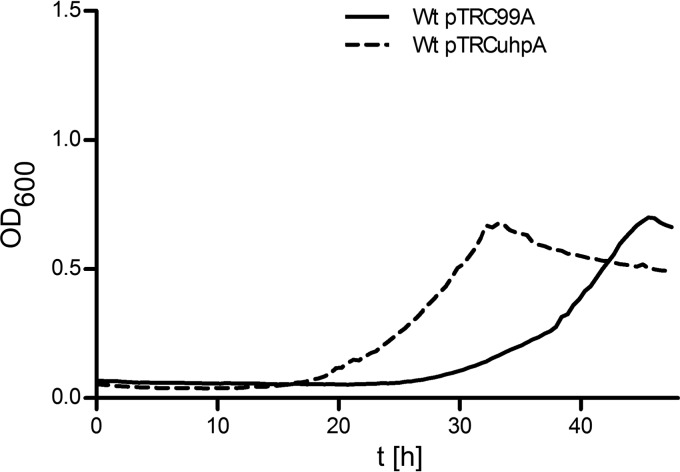
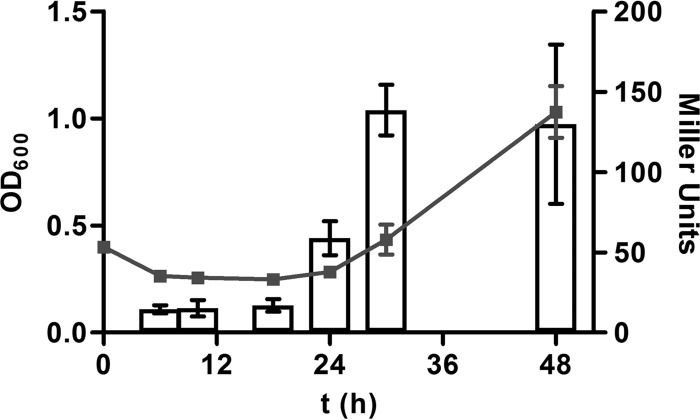
To correlate the observed prolonged lag phase of cell growth (Fig. 2) with uhp gene induction, additional experiments were performed. First we found that UhpA expression from pTRCuhpA shortened the lag phase when it was transformed into the WT strain (Fig. 5). Next, VCA0687::phoA fusion activity was monitored during the entire growth phase at 6, 10, 18, 24, 30, and 48 h (Fig. 6). The results show that at time points prior to 18 h, no significant induction could be observed. However, PhoA activity increased at 24 h, reaching a maximum at 30 h, and stayed high until 48 h. Thus, evidence is provided that uhp gene induction correlates with a prolonged observed lag phase and consequently an inhibition of uhp gene induction is responsible for the prolonged lag phase.
Fig 5.
Growth curves of WT V. cholerae and in trans expression of UhpA. Shown are the growth kinetics in M9 glucose-6P of the WT strain transformed with plasmids pTRC99A (control) and pTRCuhpA. Data represent mean values of at least three independent OD600 measurements.
Fig 6.
uhp gene expression of VCA0687::phoA fusion activity along the growth curve. Shown are the mean values and standard deviations of four independent growth curves (OD600, left y axis) of VCA0687::phoA in M9 glucose-6P measured at 6, 10, 18, 24, 30, and 48 h. At the same time points and from these cultures, PhoA activities (in Miller Units, right y axis) of VCA0687::phoA were determined; mean values and standard deviations are shown.
Survival under phosphate-limiting conditions.
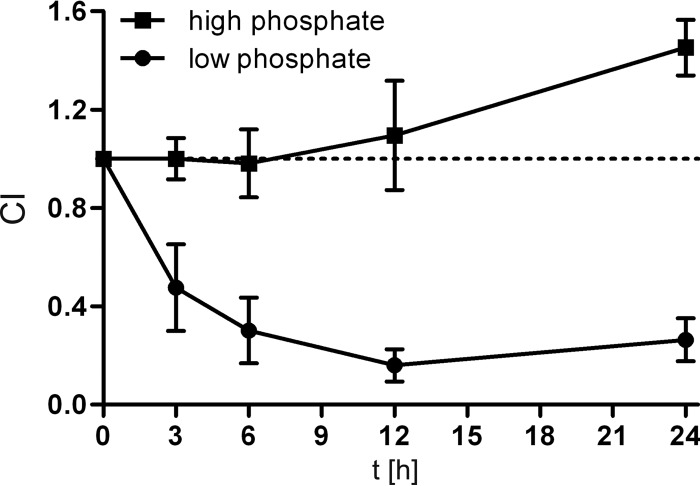
It was previously shown (26) that the hexose-6P uptake system components characterized here are induced late in vivo, suggesting the presence of the inducing substrate hexose-6P in the gut. Furthermore, the ΔVCA0686 mutant strain showed 5-fold less fitness than the WT in pond water survival after in vivo passage (26). Interestingly, this defect was compensated when the pond water was supplemented with M9 minimal medium salts. Thus, we speculated that the lack of at least one component of M9 minimal medium salts was the cause for the observed fitness disadvantage. Inorganic phosphate is especially limited in aquatic environments, and V. cholerae faces a 3-fold drop in the concentration of phosphate when transiting from cholera patient stool to pond water (54). Interestingly, E. coli is not able to grow on glucose-6P as the sole P source (55), since UhpT is an antiporter that imports hexose-6P by exporting cellular Pi (43). In contrast, the V. cholerae transporter described here should allow the utilization of hexose-6P as a C and P source. Thus, we investigated whether hexose-6P utilization accounts for increased cell survival under phosphate-limiting conditions. It is known that polyphosphate storage enhances the ability of V. cholerae to survive stress in a low-phosphate environment (56). Therefore, our experiments were carried out under such conditions, whereby the WT and ΔVCA0685-7 mutant strains were cultured for 40 h in low-Pi (150 μM) M9-Tris (pH 7, 100 mM) glycerol medium supplemented with hexose-6P (10 mM). After growth in this medium, the WT and mutant strains were washed and mixed at a ratio of 1:1 to achieve equal cell densities (OD600 of 0.5). Subsequently, the cells were incubated in either low-phosphate M9-Tris or standard M9-Tris medium, both lacking any C source. Next, the time-dependent number of CFU/ml and competitive index (CI) were determined. We observed an ∼5-fold lower CI of cells deficient in hexose-6P utilization after 12 to 24 h of incubation in low-phosphate M9-Tris, whereas in standard M9-Tris medium, no survival defect was observed (Fig. 7). This result is consistent with the previous observation that a ΔVCA0686 mutant exhibits a fitness disadvantage compared to the WT in pond water after in vivo passage. Thus, the data indicate that previous exposure to hexose-6P contributes to enhanced cell survival under phosphate-limiting culture conditions. The transition from the host to an aquatic reservoir represents such a scenario, providing an explanation for a physiological role for the hexose-6P uptake system in the life cycle of V. cholerae. Since the Uhp system in V. cholerae most likely includes an ABC transporter without Pi antiport, acquisition of phosphate should be considered. Therefore, it was tested whether this hexose-6P uptake system is already induced by phosphate limitation. Using the transcriptional phoA fusion to VCA0687, we tested whether phosphate limitation alone results in induction of the VCA0685-7 operon. The mean PhoA activities of the VCA0687::phoA strain were determined to be 5.5 Miller units in standard M9-Tris medium and 11.9 Miller units in low-phosphate M9-Tris. Thus, phosphate starvation does not result in significant induction of the hexose-6P uptake system. Also, on the basis of the finding that a relatively high phosphate concentration is present in cholera patient stool (54), it seems to be very unlikely that phosphate starvation triggers hexose-6P uptake at late stages of infection.
Fig 7.
VCA0685-7-dependent survival under phosphate limitation. Shown is the CI, which reflects the survival ratio of the WT and ΔVCA0685-7 mutant strains. The CI is expressed as a ratio of ΔVCA0685-7 CFU to WT CFU at 3 to 24 h normalized for the input ratio at time zero. Closed circles show CI values derived from cultures incubated in low-phosphate M9-Tris, and square symbols indicate CI values of cultures incubated in standard phosphate M9-Tris medium. Prior to M9-Tris incubation, cultures were grown in M9 supplemented with glycerol and glucose-6P (for details, see the text). Shown are the means of at least four independent measurements. Error bars indicate standard deviations.
In summary, this study characterized the hexose-6P substrate uptake system of V. cholerae. The reported induction of this system under infection conditions indicates that the corresponding substrate in the host is available to V. cholerae. In general, the Uhp regulatory system comprising UhpABC is highly conserved in E. coli and V. cholerae. However, in this report, distinct differences are revealed. For example, the hexose-6P permease-encoding uhpT gene of E. coli is absent from V. cholerae. Instead, a primary transport system showing lower Km values for glucose-6P uptake than UhpT in E. coli is present. Initially, we got interested in characterizing this late in vivo-induced operon because it was reported to facilitate the fitness of V. cholerae in the transition phase when the bacteria are released from the host back into the environment. By unraveling the function of the VCA0685-7 components as a hexose-6P uptake system, this work provides a first clue to how this uptake system contributes to enhanced survival fitness. As shown here, glucose-6P utilization serves as a suitable substrate to prepare V. cholerae cells for better survival in phosphate-limiting environments. Hence, these data suggest that hexose-6P substrates serve not only as valuable carbon sources but also as substrates from which cells benefit in the transition phase, most likely by building up their endogenous phosphate storage.
ACKNOWLEDGMENTS
This work was supported by Austrian Science Fund (FWF) grants W901 (DK Molecular Enzymology) to M.M., A.S., J.R., and S.S. and P22986 to S.S.
We thank C. Schweinzer for carefully reading the manuscript.
Footnotes
Published ahead of print 15 February 2013
REFERENCES
- 1. Kay BA, Cheryl AB, Wells JG. 1994. Isolation and identification of Vibrio cholerae O1 from fecal specimens, p 3–25 In Wachsmuth K, Blake PA, Olsvik O. (ed), Vibrio cholerae and cholera. ASM Press, Washington, DC [Google Scholar]
- 2. Stocker R, Seymour JR. 2012. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76:792–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huq A, Colwell RR, Rahmann R, Ali A, Chowdhury MAR, Parveen S, Sack DA, Russek-Chohen R. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56:1977–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Islam MS, Drasar BS, Sack RB. 1993. The aquatic environment as a reservoir of Vibrio cholerae: a review. J. Diarrhoeal Dis. Res. 11:197–206 [PubMed] [Google Scholar]
- 6. Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2011. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front. Microbiol. 2:260 doi:10.3389/fmicb.2011.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect. Immun. 25:768–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berg T, Schild S, Reidl J. 2007. Regulation of the chitobiose-phosphotransferase system in Vibrio cholerae. Arch. Microbiol. 187:433–439 [DOI] [PubMed] [Google Scholar]
- 10. Colwell RR. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025–2031 [DOI] [PubMed] [Google Scholar]
- 11. Huq A, Whitehouse CA, Grim CJ, Alam M, Colwell RR. 2008. Biofilms in water, its role and impact in human disease transmission. Curr. Opin. Biotechnol. 19:244–247 [DOI] [PubMed] [Google Scholar]
- 12. Seper A, Fengler VH, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. 2011. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82:1015–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125–139 [DOI] [PubMed] [Google Scholar]
- 14. Guentzel MN, Amerine D, Guerrero D, Gay TV. 1981. Association of Vibrio cholerae mutants with the intestinal mucosa of infant mice. Scan Electron Microsc. 4:115–124 [PubMed] [Google Scholar]
- 15. Chowdhury N, Norris J, McAlister E, Lau SY, Thomas GH, Boyd EF. 2012. The VC1777-VC1779 proteins are members of a sialic acid-specific subfamily of TRAP transporters (SiaPQM) and constitute the sole route of sialic acid uptake in the human pathogen Vibrio cholerae. Microbiology 158:2158–2167 [DOI] [PubMed] [Google Scholar]
- 16. Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matson JS, Withey JH, DiRita VJ. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233 [DOI] [PubMed] [Google Scholar]
- 19. Merrell DS, Camilli A. 2000. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J. Bacteriol. 182:5342–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lombardo MJ, Michalski J, Martinez-Wilson H, Morin C, Hilton T, Osorio CG, Nataro JP, Tacket CO, Camilli A, Kaper JB. 2007. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. U. S. A. 104:18229–18234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Osorio CG, Crawford JA, Michalski J, Martinez-Wilson H, Kaper JB, Camilli A. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Camilli A, Beattie DT, Mekalanos JJ. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. U. S. A. 91:2634–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camilli A, Mekalanos JJ. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634 [DOI] [PubMed] [Google Scholar]
- 25. Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 98:6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schild S, Bishop AL, Camilli A. 2008. Ins and outs of Vibrio cholerae: Vibrio cholerae transitions between the human gut and the aquatic environment are aided by specific shifts in gene expression. Microbe 3:131–136 [Google Scholar]
- 28. Kolter R, Inuzuka M, Helinski DR. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199–1208 [DOI] [PubMed] [Google Scholar]
- 29. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mekalanos JJ. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253–263 [DOI] [PubMed] [Google Scholar]
- 31. Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
- 33. Moisi M, Jenul C, Butler SM, New A, Tutz S, Reidl J, Klose KE, Camilli A, Schild S. 2009. A novel regulatory protein involved in motility of Vibrio cholerae. J. Bacteriol. 191:7027–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pearson GDN, Woods A, Chiang SL, Mekalanos JJ. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. U. S. A. 90:3750–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 36. Grimberg J, Maguire S, Belluscio L. 1989. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 17:8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schild S, Lamprecht AK, Fourestier C, Lauriano CM, Klose KE, Reidl J. 2005. Characterizing LPS and core lipid A mutant O1 and O139 Vibrio cholerae strains for adherence properties on mucus producing cell line HT-29-Rev MTX and virulence in mice. Int. J. Med. Microbiol. 295:243–251 [DOI] [PubMed] [Google Scholar]
- 38. Xavier KB, Martins LO, Peist R, Kossmann M, Boos W, Santos H. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill S, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishman RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188:6515–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Friedrich MJ, Kadner RJ. 1987. Nucleotide sequence of the uhp region of Escherichia coli. J. Bacteriol. 169:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weston LA, Kadner RJ. 1987. Identification of uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli K-12. J. Bacteriol. 169:3546–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonna LA, Ambudkar SV, Maloney PC. 1988. The mechanism of glucose-6-phosphate transport by Escherichia coli. J. Biol. Chem. 263:6625–6630 [PubMed] [Google Scholar]
- 44. Weston LA, Kadner RJ. 1988. Role of uhp genes in expression of the Escherichia coli sugar-phosphate transport system. J. Bacteriol. 170:3375–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanes CS. 1932. Studies on plant amylases: the effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem. J. 26:1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin ECC. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p 307–342 In Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger H. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 47. Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Locher KP. 2009. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ehrmann M, Ehrle R, Hofmann E, Boos W, Schloesser A. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685–694 [DOI] [PubMed] [Google Scholar]
- 50. Manson MD, Boos W. 1985. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J. Biol. Chem. 260:9727–9733 [PubMed] [Google Scholar]
- 51. Anderson DS, Adhikari P, Nowalk AJ, Chen CY, Mietzner TA. 2004. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 186:6220–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goldenbaum PE, Farmer KS. 1980. uhp-directed, glucose 6-phosphate membrane receptor in Escherichia coli. J. Bacteriol. 142:347–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Island MD, Kadner RJ. 1993. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J. Bacteriol. 175:5028–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nelson EJ, Chowdhury A, Flynn J, Schild S, Bourassa L, Shao Y, LaRocque RC, Calderwood SB, Qadri F, Camilli A. 2008. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 4(10):e1000187 sdoi:10.1371/journal.ppat.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoffer SM, van Uden N, Tommassen J. 2001. Expression of the pho regulon interferes with induction of the uhpT gene in Escherichia coli K-12. Arch. Microbiol. 176:370–376 [DOI] [PubMed] [Google Scholar]
- 56. Jahid IK, Silva AJ, Benitez JA. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]