Abstract
In streptococci, ComX is the alternative sigma factor controlling the transcription of the genes encoding the genetic transformation machinery. In Streptococcus thermophilus, comX transcription is controlled by a complex consisting of a transcriptional regulator of the Rgg family, ComR, and a signaling peptide, ComS, which controls ComR activity. Following its initial production, ComS is processed, secreted, and imported back into the cell by the Ami oligopeptide transporter. We characterized these steps and the partners interacting with ComS during its extracellular circuit in more detail. We identified the mature form of ComS and demonstrated the involvement of the membrane protease Eep in ComS processing. We found that ComS was secreted but probably not released into the extracellular medium. Natural competence was first discovered in a chemically defined medium without peptides. We show here that the presence of a high concentration of nutritional peptides in the medium prevents the triggering of competence. In milk, the ecological niche of S. thermophilus, competence was found to be functional, suggesting that the concentration of nutritional peptides was too low to interfere with ComR activation. The kinetics of expression of the comS, comR, and comX genes and of a late competence gene, dprA, in cultures inoculated at different initial densities revealed that the activation mechanism of ComR by ComS is more a timing device than a quorum-sensing mechanism sensu stricto. We concluded that the ComS extracellular circuit facilitates tight control over the triggering of competence in S. thermophilus.
INTRODUCTION
Natural competence for transformation is a physiological state allowing bacteria to bind, take up, and integrate exogenous DNA in their genomes by homologous recombination. The triggering of competence is tightly controlled in Gram-positive bacteria and has been studied extensively in Streptococcus pneumoniae (for reviews, see references 1 to 3). In this bacterium, a signaling heptadecapeptide, the competence-stimulating peptide (CSP), encoded by the comC gene, is secreted and processed by an ABC transporter, ComAB. Extracellular CSP is detected by a two-component system. The membrane-embedded histidine kinase, ComD, is thought to undergo autophosphorylation in response to CSP, subsequently phosphorylating its cognate response regulator, ComE, which then activates the transcription of the early CSP-induced genes, including comCDE, comAB, and comX. ComX, which is also known as SigX, is an alternative competence-specific sigma factor required for expression of the late CSP-induced genes, which encode components of the DNA uptake machinery. This regulation of the early steps of the cascade, up to the transcription of comX, occurs in streptococci from the mitis and anginosus groups. Sequencing studies have highlighted an absence of early competence genes in other streptococci, despite the presence of late competence genes and the comX gene in their genomes (4). Recent discoveries that S. thermophilus is competent in a chemical medium lacking peptides and that the Ami oligopeptide transporter is essential for the expression of comX have constituted the first step toward the deciphering of an alternative mechanism (5). This mechanism is based on a signaling peptide, ComS, and a transcriptional regulator of the Rgg family, ComR. ComS is thought to be processed, secreted, and imported back into the cell by Ami. There, it activates ComR, which then binds to the comX promoter, thereby controlling the transcription of this gene, in both S. thermophilus and S. salivarius (6). This mechanism has also been shown to occur in S. mutans, whose genome also encodes a ComCDE system, and probably also operates in the streptococci of the pyogenic and bovis groups, which have orthologs of comR and comS in their genomes. Based on the sequences of ComS peptides, the ComS-ComR systems of streptococci of the salivarius group were named type I, and those of the mutans, pyogenic, and bovis groups were named type II. Mature ComS peptides are also known as sigX-inducing peptides (XIP) (7).
Two type II ComS-ComR systems, those of S. mutans and S. pyogenes, were recently studied in more detail. The amino acid sequence of the XIP of S. mutans has been determined. This heptapeptide (ComS11–17) is released by C-terminal cleavage of the ComS propeptide, in a process that does not involve the Eep membrane protease (8). The development of competence in S. mutans was studied in a medium lacking nutritional peptides, and this state was found to be triggered at a high cell density. XIP is released into the extracellular medium at concentrations equivalent to 1 μM ComS11–17 (9). For S. pyogenes, despite the presence of the genes required for transformation in the genome and the control of comX transcription by ComR and ComS, the bacterium has been found to remain noncompetent under the many laboratory conditions tested (10).
We focused on the initial steps—production of the mature type I ComS, its extracellular circuit, and its uptake via the Ami oligopeptide transport system—to validate the proposed model in S. thermophilus and to decipher its workings further. We found that the type I and II ComS-ComR mechanisms are similar but not identical. The slight differences observed resulted in significant discrepancies, particularly in terms of the kinetics of competence in laboratory media.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used are listed in Table 1. S. thermophilus strains were grown at 28 or 37°C in M17 medium (Difco) supplemented with 10 g liter−1 lactose (M17lac), a chemically defined medium (CDM) (11), or reconstituted Nilac low-heat skim milk (Nizo, Ede, Netherlands), without shaking, under a normal air atmosphere. Escherichia coli strains were grown at 37°C in Luria-Bertani broth, with shaking (12). Agar (1.5%) was added to the media as appropriate. When required, antibiotics were added to the media at the following final concentrations: erythromycin, 200 μg ml−1 for E. coli and 5 μg ml−1 for S. thermophilus; kanamycin, 1 mg ml−1 for S. thermophilus; and spectinomycin, 100 μg ml−1 for S. thermophilus. The optical density at 600 nm (OD600) of each culture was measured with a Uvikon 931 (Kontron) spectrophotometer or a microplate reader (Infinite M200 spectroluminometer; Tecan).
Table 1.
Bacterial strains used in this study
| Strain | Relevant propertya | Descriptionb | Reference or source |
|---|---|---|---|
| S. thermophilus strains | |||
| LMD-9 | Wild type | 32 | |
| CB001 | blp::PcomX-luxAB | 6 | |
| LF121 | blp::PcomS-luxAB | 35 | |
| LF123 | blp::PcomR-luxAB | 35 | |
| TIL773 | Δeep | 14 | |
| TIL1179 | pBV5030::P32 | pBV5030::P32 → LMD-9 | This study |
| TIL1199 | ΔamiA1 amiA3::erm | 5 | |
| TIL1202 | ΔamiCDE::spec | 14 | |
| TIL1209 | comS::spec | comS::spec PCR fragment → LMD-9 | This study |
| TIL1307 | pBV5030::P32-comS | pBV5030::P32-comS → LMD-9 | This study |
| TIL1311 | blp::PcomX-luxAB aphA3 | pGICB004a::PcomX → LMD-9 | This study |
| TIL1314 | blp::PcomX-luxAB aphA3 comS::spec | TIL1209 DNA → TIL1311 | This study |
| TIL1329 | pBV5030::P32-comSΔamiCDE::spec | TIL1202 DNA → TIL1307 | This study |
| TIL1349 | blp::PdprA-luxAB aphA3 | pGICB004a::PdprA → LMD-9 | This study |
| TIL1371 | blp::comX-luxAB aphA3 Δeep | TIL1311 DNA → TIL773 | This study |
| TIL1391 | blp::PcomX-luxAB comS::spec | TIL 1209 DNA → CB001 | This study |
| TIL1392 | blp::PcomX-luxAB comS::spec amiA3::erm | TIL1199 DNA → TIL1391 | This study |
| TIL1393 | blp::PcomX-luxAB comS::spec ΔamiA1 amiA3::erm | TIL1199 DNA → TIL1391 | This study |
| TIL1394 | blp::PcomX-luxAB comS::spec ΔamiA1 | pG+host9::updown.amiA1→ TIL1391 | This study |
| TIL1404 | comS::erm | PCR fragment comS::erm → LMD-9 | This study |
| TIL1405 | blp::PcomX-luxAB comS::erm | TIL1404 DNA → CB001 | This study |
| E. coli strain | |||
| TG1 repA+ | TG1 derivative with repA gene integrated into the chromosome | P. Renault, INRA, Jouy-en-Josas, France |
erm, aphA3, and spec indicate resistance to erythromycin, kanamycin, and spectinomycin, respectively.
Arrows indicate construction by transformation with chromosomal DNA or plasmid.
Natural transformation of S. thermophilus LMD-9 in milk.
Cells of S. thermophilus strain LMD-9 were grown overnight at 42°C in Nilac milk. The OD480 of the culture was measured after clarification of milk by 10-fold dilution in 5 mM EDTA (pH 12). The culture was then diluted in milk to an OD480 of 0.05. One hour after dilution, 100 μl of the culture was mixed with 1 μg of plasmid DNA (pG+host9), with or without 1 μM ComS17–24, and incubated for 2 h at 28°C. The culture was then serially diluted and spread on M17lac plates with erythromycin. For calculation of the transformation rate (i.e., the number of antibiotic-resistant transformants per ml divided by the number of viable CFU per ml), we also spread cells on M17lac plates without antibiotic. All these experiments were carried out at least three times.
DNA manipulation and sequencing.
Standard methods were used for DNA purification, restriction digestion, PCR, ligation, and sequencing. The E. coli TG1 repA+ strain was used as the host for cloning experiments. The plasmids used are listed in Table 2, and the oligonucleotides used for PCR are listed in Table S1 in the supplemental material. Naturally competent (5) or electrocompetent (13) S. thermophilus cells were used for transformation.
Table 2.
Plasmids used in this study
| Plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| pG+host9 | Erm Ts | 33 |
| pG+host9::updown.amiA1 | Erm; derivative of pG+host9 used for amiA1 gene replacement by double-crossover integration | 5 |
| pGICB004 | Erm Ts; allows the integration of transcriptional fusions to the luxAB reporter genes at the blp locus | 14 |
| pGICB004a | Erm; derivative of pGICB004 containing the aphA3 gene downstream from the luxB gene | B. Fleuchot, INRA, Jouy-en-Josas, France, unpublished data |
| pGICB004a::PcomX | Erm; derivative of pGICB004a used to introduce a PcomX-luxAB transcriptional fusion at the blp locus | This study |
| pGICB004a::PdprA | Erm; derivative of pGICB004a used to introduce a PdprA-luxAB transcriptional fusion at the blp locus | This study |
| pBV5030 | Erm; replicative plasmid in E. coli and Gram-positive bacteria | 34 |
| pBV5030::P32 | Erm; derivative of pBV5030 containing the constitutive promoter P32 of Lactococcus lactis | This study |
| pBV5030::P32-ster1357 | Erm; derivative of pBV5030 allowing overexpression of the ster1357 gene of S. thermophilus LMD-9 | 14 |
| pBV5030::P32-comS | Erm; derivative of pBV5030 allowing overexpression of the comS gene of S. thermophilus LMD-9 | This study |
Erm, resistance to erythromycin; Ts, the plasmid encodes a thermosensitive RepA protein.
Construction of strains containing luxAB reporters.
We used derivatives of the plasmid pGICB004a (B. Fleuchot, unpublished data) to easily integrate, by natural transformation and double-crossover events, transcriptional fusions between the promoter of comX or dprA and the luxAB reporter genes at the blp locus in S. thermophilus LMD-9. We constructed pGICB004a::PcomX as follows. The comX promoter was amplified by PCR with oligonucleotides PcomX-EcoRI and PcomX-SpeI, and the resulting fragment was digested with the SpeI and EcoRI restriction enzymes and ligated between the corresponding restriction sites of pGICB004a. Strain TIL1311 (blp::PcomX-luxAB aphA3) was obtained by natural transformation of strain LMD-9 with pGICB004a::PcomX linearized with the restriction enzyme ScaI. The same strategy was used to obtain strain TIL1349 (blp::PdprA-luxAB aphA3), with plasmid pGICB004a::PdprA constructed with oligonucleotides PdprA-EcoRI and PdprA-SpeI. Strain TIL1371 (blp::PcomX-luxAB aphA3 Δeep) was obtained by the natural transformation, in the presence of ComS17–24, of strain TIL773 (Δeep) with chromosomal DNA from strain TIL1311.
Construction of ComS-overproducing strains.
We used pBV5030::P32-comS, a derivative of the plasmid pBV5030::P32-ster1357, to overproduce a ComS peptide containing a glycine residue between the methionine at position 1 and the lysine at position 2 in the wild-type sequence. The comS gene was amplified by PCR with oligonucleotides ComS-NcoI and ComS-PstI. The resulting fragment was digested with the NcoI and PstI restriction enzymes and inserted into pBV5030::P32-ster1357 digested with the same enzymes. The resulting plasmid was then introduced by transformation into strain LMD-9, generating strain TIL1307. Strain TIL1307 was transformed with chromosomal DNA of strain TIL1202 (ΔamiCDE::spec), generating strain TIL1329 (pBV5030::P32-comS ΔamiCDE::spec). We constructed pBV5030::P32 from pBV5030::P32-ster1357 by digestion with the NcoI and PstI restriction enzymes, treatment with T4 DNA polymerase, and self-ligation. Strain TIL1179 was constructed by transforming strain LMD-9 with pBV5030::P32 and was used as a negative control in the experiments.
Construction of mutant strains.
Deletion of genes was performed by using either the overlapping PCR method or the thermosensitive plasmid pG+host9, as follows. We used the overlapping PCR method to replace the comS gene with the spectinomycin (spec) or erythromycin (erm) resistance cassette. Briefly, the spectinomycin resistance cassette (spec) was amplified by PCR (14) and fused, by PCR, to fragments located upstream and downstream of the comS gene. The oligonucleotides used for amplification of the upstream fragment were comS-uF and comS-uRspec, and those used for the downstream fragment were comS-dFspec and comS-dR. The resulting PCR fragment was used to transform naturally competent LMD-9 cells to generate strain TIL1209 (comS::spec). The erythromycin resistance cassette was amplified by PCR (5), and the oligonucleotides used for amplification of fragments upstream and downstream of comS were comS-uF plus comS-uRerm and comS-dFerm plus comS-dR, respectively. The resulting PCR fragment was used to transform naturally competent LMD-9 cells to generate strain TIL1404 (comS::erm). Both comS deletions were then introduced into different strains by natural transformation. Chromosomal DNA from strain TIL1209 was used to transform strain TIL1311 to generate strain TIL1314 (blp::PcomX-luxAB aphA3 comS::spec). Chromosomal DNA from strain TIL1404 was used to transform strain CB001 to generate strain TIL1405 (blp::PcomX-luxAB comS::erm). Chromosomal DNA from strain TIL1209 (comS::spec) was used to transform naturally competent cells of strain CB001 (blp::PcomX-luxAB) to generate strain TIL1391 (blp::PcomX-luxAB comS::spec). We used a pG+host9 plasmid containing flanking regions of the amiA1 gene (pG+host9::updown.amiA1) to delete the amiA1 gene from strain TIL1391 by integration and excision of the plasmid as previously described (5), generating strain TIL1394 (blp::PcomX-luxAB comS::spec ΔamiA1). Finally, chromosomal DNA from strain TIL1199 (ΔamiA1 amiA3::erm) was used to transform naturally competent cells of strain TIL1391 in the presence of the synthetic peptide ComS17-24, generating strains TIL1392 (blp::PcomX-luxAB comS::spec amiA3::erm) and TIL1393 (blp::PcomX-luxAB comS::spec ΔamiA1 amiA3::erm), which could be distinguished by PCR. All constructs were checked by PCR and sequencing.
Luciferase assays.
Cells were grown in CDM overnight at 37°C. The monocultures were then diluted to a final OD600 of 0.05 in all experiments except those reported in Fig. 7, in which case the cultures were diluted to various initial OD600 values. In coculture experiments, each individual culture was diluted in CDM to an OD600 of 0.025. Cell supernatant or CDM was used as the dilution medium, sometimes with the addition of other compounds (tryptone or Casamino Acids). Unless otherwise specified, ComS14–24 or ComS17–24 was added 50 min after the dilution of the cultures, to a final concentration of 0.05 or 1 μM, depending on the experiment. We transferred 250-μl aliquots of these diluted cultures to the wells of a sterile covered white microplate with a transparent bottom (Greiner). The OD600 and luminescence (expressed in relative light units [RLU]) of the cultures were monitored at 37°C as previously described (6).
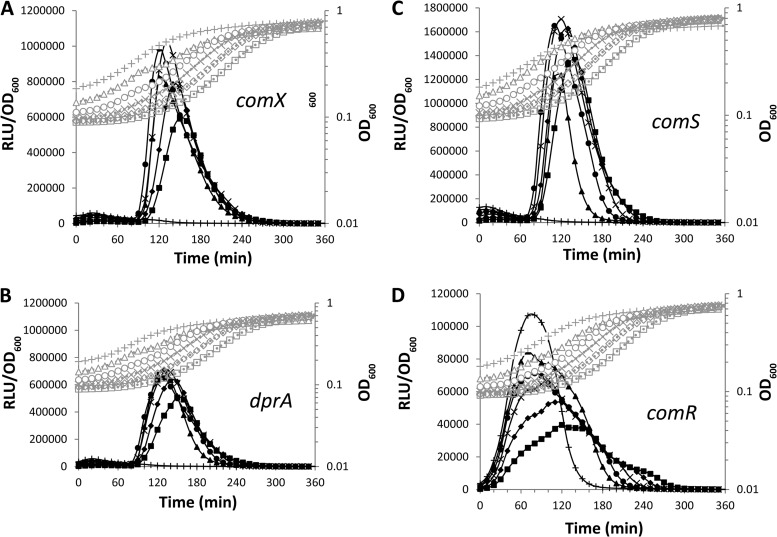
Fig 7.
Effect of initial cell density on the expression of com genes. The graphs show the growth (OD600; gray lines and symbols) and relative luciferase activities (RLU/OD600; black lines and symbols) in CDM of strains containing a PcomX-luxAB fusion (CB001) (A), a PcomS-luxAB fusion (LF121) (B), a PdprA-luxAB fusion (TIL1349) (C), or a PcomR-luxAB fusion (LF123) (D), with inoculation at various concentrations, i.e., OD600 values of 0.012 (squares), 0.025 (diamonds), 0.05 (×'s), 0.1 (circles), 0.2 (triangles), and 0.4 (+ symbols). The data shown are representative of three independent experiments.
Liquid chromatography-tandem MS (LC-MS/MS).
The supernatants of strains TIL1329 (pBV5030::P32-comS ΔamiCDE) and TIL1209 (comS::spec) were recovered by centrifugation from 80-ml cultures in CDM when the OD600 reached 1. They were subjected to ultrafiltration through Amicon devices (Centriplus YM-10 [10-kDa cutoff]; Millipore), and the peptides present in the ultrafiltrates were concentrated by solid-phase extraction (Sep-Pak Plus C18 cartridges; Waters) after washing with 4 ml of 0.1% trifluoroacetic acid and 30% acetonitrile and elution in 2 ml of 40% acetonitrile. The eluted fractions were dried further and resuspended in 50 μl of 0.1% trifluoroacetic acid and 2% acetonitrile. We then loaded 5-μl aliquots of the fractions onto a Pepmap C18 column (150-mm length by 75-μm internal diameter [ID] by 100 Å; Dionex, Voisins-le-Bretonneux, France) for analysis by online mass spectrometry (MS) on an LTQ-Orbitrap Discovery apparatus (Thermo Fischer, San Jose, CA). We initially analyzed only the m/z ion current corresponding to all possible C-terminal amino acid sequences produced by the cleavage of the ComS precursor in the supernatant from strain TIL1329. We checked that these masses were not present in the supernatant from strain TIL1209 and that they were eluted from the Pepmap C18 column with a retention time compatible with the hydrophobicity of their putative amino acid sequences. We then fragmented the ions with a mass satisfying these conditions and analyzed them on the Orbitrap mass analyzer. However, peptide ion fragmentation was found to be impossible due to the weakness of the mass spectrometry signals. We therefore alkylated the culture supernatants with iodoacetamide to strengthen the signal. NaOH and iodoacetamide were added to the supernatant at final concentrations of 25 mM and 20 mM, respectively, and the mixture was stored in the dark for 30 min. Samples were then enriched in peptides and run on a Pepmap C18 column as described above, but with the following modification: 50 μl of formic acid was added to adjust the pH to 6.5 after the ultrafiltration step. The masses of the alkylated form of the ions previously identified were determined, and these ions were fragmented and analyzed on an Orbitrap mass analyzer with a PAPPSO platform (INRA, Jouy-en-Josas, France).
RESULTS
Location and identification of the secreted form of ComS.
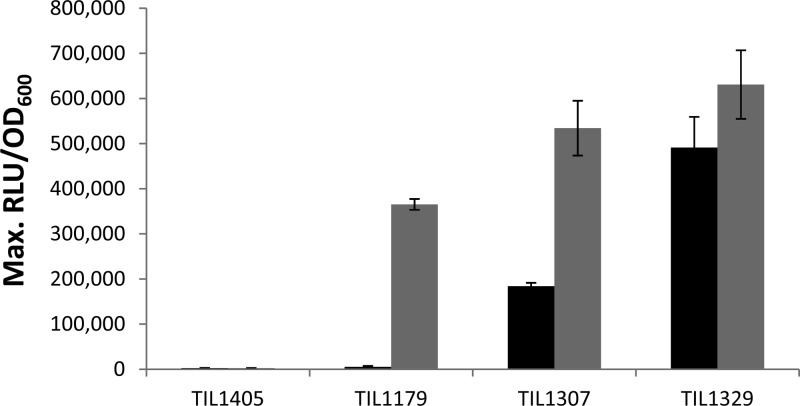
The addition of the ComS17–24 synthetic peptide (LPYFAGCL) to CDM induces the expression of comX in a strain lacking the comS gene, demonstrating that ComS is the precursor of a competence-stimulating peptide in S. thermophilus LMD-9 (6). However, the natural form(s) of the secreted peptide has yet to be identified. To check if the competence-stimulating peptide was released into the external medium or not, we collected the supernatant from strain LMD-9 at various time points and mixed it with cells of a reporter strain containing a PcomX-luxAB fusion but lacking the comS gene (TIL1405). Unexpectedly, we detected no luciferase activity with supernatant samples collected 30, 60, 90, or 120 min after the start of LMD-9 strain growth. This observation suggested that the mature form of ComS did not accumulate in the supernatant in sufficient amounts to induce the activity of the comX promoter of the reporter strain. We thus constructed a replicative plasmid containing the comS gene under the control of a strong constitutive promoter, which we then introduced into strain LMD-9 and a ΔamiCDE mutant impaired in peptide import, generating strains TIL1307 and TIL1329, respectively. The supernatants of these two strains and that of the control strain TIL1179, which naturally produces ComS, were collected after 90 min of growth. The level of expression of comX was 35 and 90 times higher than that observed in the presence of the control strain supernatant when the cells of the reporter strain (TIL1405) were grown in the presence of supernatants from strains TIL1307 and TIL1329, respectively (Fig. 1). These results indicate that in a ComS-overproducing context, ComS becomes detectable in supernatants, and we learned from this experiment that the Ami system plays a significant role in the disappearance of ComS from the medium. These results also suggest that under natural conditions of expression, ComS is either (i) secreted and rapidly reimported or (ii) secreted but kept at the bacterial surface before its reimport. In both cases, we cannot exclude the possibility that part of ComS was released into the supernatant but in too small amounts to trigger competence. We tested these hypotheses with cocultures of the reporter strain TIL1405 and strain TIL1179, which naturally produces ComS, or one of the two overproducing strains, i.e., TIL1307 and TIL1329. Under these conditions, contact between the cells of the reporter strain and each of the ComS-producing strains tested was possible, and ComS sensing occurred. Coculture with strain TIL1179 resulted in levels of comX expression that were 135 times higher than those in a monoculture of strain TIL1405, and induction reached levels of 200- and 230-fold with strains TIL1307 and TIL1329, respectively (Fig. 1), confirming the overproduction of ComS in these two strains and the role of the transporter Ami in the rapid import of ComS. These results demonstrate that even under conditions of natural production of ComS, the peptide is produced in amounts sufficient to induce competence but is not released freely into the external medium. Our results also indicate that a ComS peptide produced by one bacterium can be sensed by another one, indicating that all pheromones are not immediately reimported by the producer bacteria. This pheromone exchange needs bacterial proximity and does not occur via supernatant.
Fig 1.
Detection of ComS in the extracellular space. The graph shows the maximum relative luciferase activities of strain TIL1405 (blp::PcomX-luxAB comS::erm) cultured with the supernatants of strains TIL1405, TIL1179 (pBV5030::P32), TIL1307 (pBV5030::P32-comS), and TIL1329 (pBV5030::P32-comS ΔamiCDE::spec) (black bars) or cocultured with these four strains (gray bars). All cultures were performed in CDM. The means for three independent experiments are presented, and error bars indicate standard deviations.
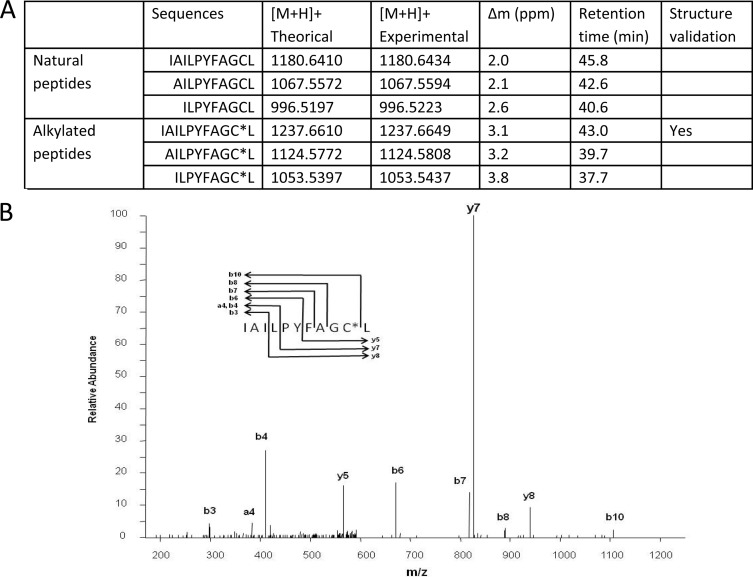
We determined the amino acid sequence of the natural secreted form(s) of ComS by analyzing the supernatant of the ComS-overproducing strain TIL1329 by LC-MS/MS and comparing the results obtained with those for the ComS-nonproducing strain TIL1209. The C-terminal Leu residue of ComS is known to be essential for ComS activity (6). We therefore looked for unmodified peptides derived from the C-terminal cleavage of ComS that were present in the TIL1329 supernatant but absent from the TIL1209 supernatant. Three masses, corresponding to peptides ComS14–24, ComS15–24, and ComS16–24, met these criteria (Fig. 2A). At this stage, peptide ion fragmentation was not possible because the mass spectrometry signals were too weak. These signals were strengthened by alkylating the supernatants of both strains with iodoacetamide. As expected, masses corresponding to alkylated forms of the three peptides were detected only in the supernatant from the overproducing strain, with the expected shift in high-pressure liquid chromatography (HPLC) retention times (Fig. 2A). Alkylation strengthened the ComS14–24 peptide sufficiently for its fragmentation and the validation of its sequence (Fig. 2B). It was not possible to validate the sequences of ComS15–24 and ComS16–24 because their mass spectrometry signals were too weak. We therefore validated the undecapeptide ComS14–24 (IAILPYFAGCL) as a natural mature competence pheromone in S. thermophilus LMD-9 and confirmed that this fragment contained a free cysteine residue (Fig. 2A). As cysteine residues are known to be highly reactive, we checked the capacity of a synthetic ComS14–24 peptide that had been alkylated with iodoacetamide at the cysteine to complement a comS deletion mutant and observed that the mutant was not transformable and that such a peptide was not active any more (data not shown). We compared the activities of the synthetic peptides ComS14–24 and ComS17–24 by using the reporter strain TIL1314 (blp::PcomX-luxAB aphA3 comS::spec). In CDM, no significant difference was observed between ComS14–24 and ComS17–24 when these two peptides were added at a nonsaturating concentration of 0.05 μM (6; also data not shown).
Fig 2.
Identification of the mature forms of ComS in the supernatant of a ComS-overproducing strain with impaired peptide import (TIL1329). (A) Features of peptides corresponding to fragments of the ComS precursor detected with an Orbitrap mass analyzer. (B) Fragmentation spectrum of the alkylated ion m/z 1,237.66 analyzed in a linear ion trap. The stars indicate the alkylation of the cysteine residue.
The membrane protease Eep is involved in the processing of ComS.
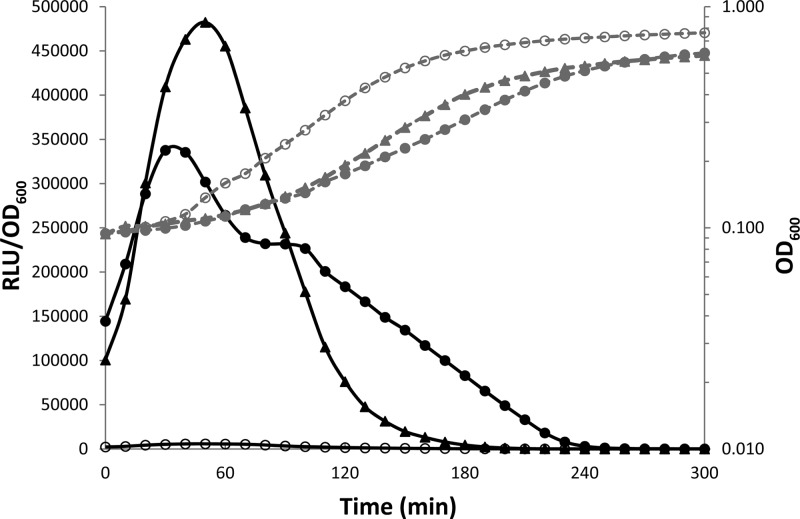
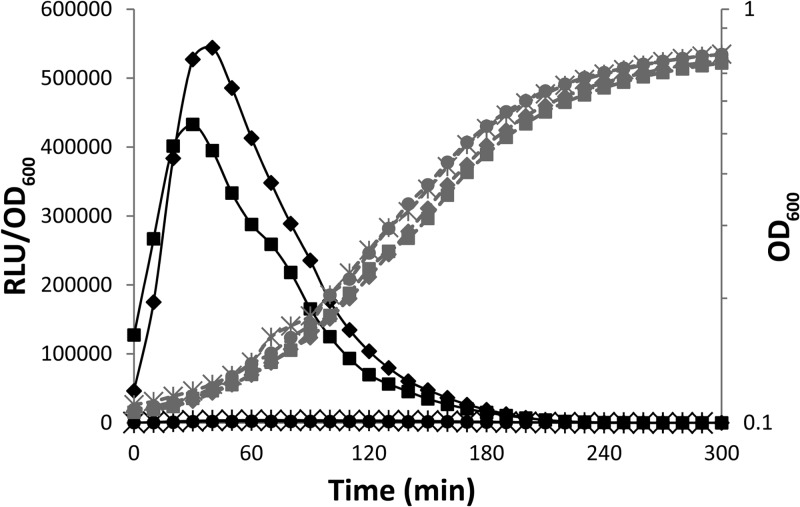
The Eep (enhanced expression of pheromone) protease is involved in the processing of several pheromones in Enterococcus faecalis (15), S. thermophilus (14), and S. pyogenes (16). We investigated its possible involvement in the processing of proComS by constructing a strain containing a PcomX-luxAB reporter fusion from which the eep gene was deleted (TIL1371). No significant luciferase activity was detected with this strain in CDM. However, the addition of the ComS17–24 peptide to the growth medium restored luciferase activity to levels slightly lower than that obtained with the PcomX-luxAB ΔcomS strain (TIL1314) grown with ComS17–24 in CDM (Fig. 3), and for an extended period. We therefore concluded that the endoprotease Eep is involved at least in the processing of the ComS propeptide.
Fig 3.
Role of the Eep membrane protease in ComS processing. The graph shows the growth (OD600; gray) and relative luciferase activities (RLU/OD600; black) in CDM of a strain containing a PcomX-luxAB fusion in a Δeep background (TIL1371), without (open circles) or with (closed circles) 1 μM ComS17–24, and of a strain containing a comX-luxAB fusion in a ΔcomS background (TIL1314), with 1 μM ComS17–24 (closed triangles). The data shown are representative of three independent experiments.
The AmiA3 OBP is more important than AmiA1 in the import of ComS.
The last step of the extracellular circuit of ComS is its capture by the oligopeptide binding proteins (OBPs) of the Ami oligopeptide transporter. There are two OBPs in LMD-9: AmiA1 and AmiA3. We have already shown that an amiA3 deletion mutation results in a 98% loss of transformability, whereas an amiA1 deletion mutation results in the loss of only 50% transformability, indicating that AmiA3 predominates in the control of competence during culture in CDM (5). However, in this experiment, ComS was produced naturally, and the rate of transformability was controlled by pheromone import and the positive-feedback loop controlling the transcription of the corresponding gene. We assessed the role of each OBP in the import of ComS in more detail, by deleting either amiA1 or amiA3 from a ComS-nonproducing strain containing a PcomX-luxAB reporter fusion and adding synthetic ComS17–24, at a nonsaturating concentration of 0.05 μM, to the culture medium. Relative luciferase activity was much weaker in the amiA3 deletion mutant (TIL1392) than in the amiA1 deletion mutant (TIL1394), confirming the greater importance of AmiA3 than that of AmiA1 in the import of ComS17–24 during growth in CDM (Fig. 4). In the presence of both OBPs (TIL1391), luciferase activity levels were similar to those obtained with strain TIL1394, and as expected, no luciferase activity was detected if both OBPs were deleted (TIL1393).
Fig 4.
Roles of the AmiA1 and AmiA3 oligopeptide binding proteins in the import of ComS17–24. The graph shows the growth (OD600; gray symbols) and relative luciferase activities (RLU/OD600; black symbols) in CDM supplemented with 0.05 μM ComS17–24 of strains containing a comX-luxAB fusion in a ΔcomS background (TIL1391; squares) or a deletion of amiA1 (TIL1394; diamonds), amiA3 (TIL1392; stars), or both amiA1 and amiA3 (TIL1393; circles). The data shown are representative of three independent experiments.
Peptides in the growth medium prevent the triggering of competence.
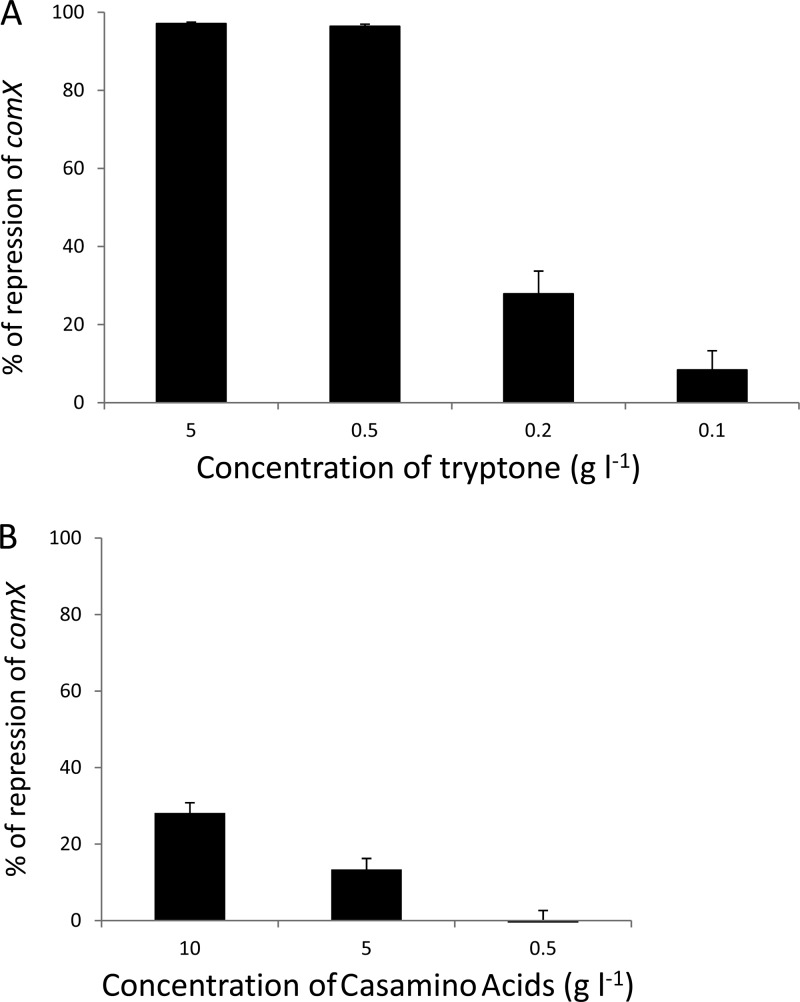
We have shown that comX expression is lower in M17lac than in CDM, by a factor of 200, resulting in an absence of transformants during growth in M17lac (5). M17 is a peptide-rich medium. We therefore hypothesized that the presence of high concentrations of peptides might prevent the triggering of competence. Tryptone, a tryptic digestion product of caseins, is one of the sources of the peptides in M17, in which its concentration is 5 g liter−1. We cultured a strain containing the PcomX-luxAB reporter fusion (CB001) in CDM supplemented with tryptone at concentrations of 0.1 to 5 g liter−1 and measured the maximum levels of relative luciferase activity. These levels were compared with those obtained in CDM to determine repression levels. We observed 97% repression of comX expression in CDM supplemented with 5 g liter−1 tryptone (Fig. 5A). A similar level of repression was observed in CDM supplemented with 0.5 g liter−1 tryptone, and the concentration of tryptone had to be lowered to 0.2 g liter−1 to achieve a significant decrease in the inhibitory effect. We performed a similar experiment with Casamino Acids, an acid hydrolysate of caseins consisting mostly of amino acids. In the presence of 10 g liter−1 Casamino Acids, the expression of comX was repressed by only 28%, with no repression detected at a Casamino Acids concentration of 0.5 g liter−1 (Fig. 5B). These findings confirm that the presence of tryptone and, more precisely, of peptides in M17 is sufficient to account for the inhibition of transformation observed in this medium.
Fig 5.
Inhibitory effect of tryptone or Casamino Acids in CDM on the expression of comX. The maximum level of relative luciferase activity of strain CB001 (blp::PcomX-luxAB) was measured in CDM supplemented with either tryptone or Casamino Acids and compared with that obtained in unsupplemented CDM to determine the percent repression of comX expression by tryptone (A) or Casamino Acids (B). The means for three independent experiments are presented, and error bars indicate standard deviations.
The competence of S. thermophilus has been studied only in laboratory media such as M17 and CDM. However, S. thermophilus is a streptococcus adapted to milk, which is believed to be its ecological niche (17). The peptide concentration of milk has been estimated to be about 0.05 g liter−1 (18). The results obtained with tryptone-complemented CDM suggest that this concentration is compatible with the triggering of competence in S. thermophilus. We thus assessed the rate of transformation with plasmid DNA of the strain LMD-9 in milk. We found it to be 6.6 × 10−4 μg−1 of DNA, increasing to 1.4 × 10−1 μg−1 of DNA when ComS was added to the milk at a final concentration of 1 μM. This observation confirms that the peptide content of milk is not high enough to prevent the triggering of competence.
The ComS-ComR complex is more a timing device than a quorum-sensing (QS) system.
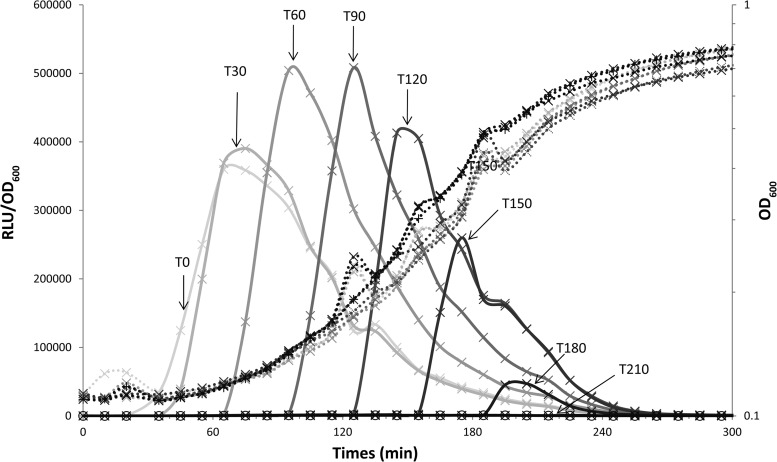
The triggering of comX expression occurs 90 min after the start of the culture of strain CB001 (blp::PcomX-luxAB), a naturally ComS-producing strain (6). To check if the cells were resistant to the presence of ComS during the beginning of the exponential growth phase, we performed a set of experiments in which ComS17–24 was added at different time points to cultures of the same strain, TIL1391 (blp::PcomX-luxAB comS::spec), inoculated at the same time and from the same preculture. ComS17–24 was added at the beginning of the culture (T0) and then every 30 min during the exponential growth phase, to a nonsaturating concentration of 0.05 μM. The addition of the ComS17–24 peptide at T0 triggered the expression of comX 30 min later (Fig. 6). The addition of ComS17–24 between 30 and 180 min after the start of the culture (between T30 and T180) was followed by comX expression, which was first detected about 10 min after addition of the peptide. At the end of the exponential growth phase (T210), no comX expression was detected, probably due to the limitation of luciferase measurement at low pH and/or when the cofactor FMNH2 is limiting (19). Levels of comX expression were maximal after the addition of ComS17–24 at T60 and T90. Thus, cells can respond to ComS early or at any point during growth, indicating that bacteria are ready to become competent at any time. The addition of ComS at the beginning of growth triggered the expression of comX approximately 50 min sooner than that under natural conditions of ComS synthesis (Fig. 7A).
Fig 6.
Expression of comX in a ComS-nonproducing strain is triggered by the presence of the ComS17–24 pheromone in the culture medium during the exponential growth phase. The graph shows the growth (OD600; dotted lines) and relative luciferase activities (RLU/OD600; solid lines) of the LMD-9 strain containing a PcomX-luxAB fusion in a ΔcomS background (TIL1391) after (×) or in the absence of (○) the addition of the synthetic ComS17–24 pheromone 0, 30, 90, 120, 150, 180, and 210 min after culture inoculation at an OD600 of 0.05. The time at which ComS17–24 was added (from T0 to T210) is represented on a gray scale. The data shown are representative of three independent experiments.
QS mechanisms combine cell-cell signaling and the measurement of population density through the detection of a signal following its passive accumulation during growth (2). We investigated whether the timing of competence depended on quorum cell density by measuring comX expression in cultures of the CB001 strain (blp::PcomX-luxAB) inoculated at various initial concentrations. Because no peptide was added to the cultures, PcomX activation was controlled by the production of ComS by strain CB001. We observed that the expression of comX was triggered independently of the initial OD600 of the culture and always occurred 80 to 100 min after inoculation, except in the culture inoculated with the highest initial density of cells, in which no comX expression was detected (Fig. 7A). Similar results were obtained with strain TIL1349 (blp::PdprA-luxAB) and strain LF121 (blp::PcomS-luxAB), in which we measured expression of the late competence gene dprA and the comS gene, respectively (Fig. 7B and C). In all cases, the maximum level of gene expression was not correlated with initial inoculums, as similar values were obtained with all the initial densities tested. We also performed similar experiments to investigate comR expression with strain LF123. In this case, expression was triggered immediately. We also observed a clear correlation between the level of inoculums and gene expression: the larger the inoculum, the higher was the level of comR expression (Fig. 7D). Conversely, the inoculum level and time course of expression were inversely correlated, with larger inoculums being associated with shorter durations of comR expression.
DISCUSSION
The ComS-associated ComR regulator belongs to an ever-expanding family of regulators involved in cell-cell communication, such as quorum-sensing mechanisms, and in interacting with intracellular signaling peptides. This family seems to be restricted to Gram-positive bacteria and includes the SHP-associated Rgg proteins in streptococci and the regulators of the RNPP family: the PlcR and NprR activators of bacilli, the PrgX repressor of Enterococcus faecalis, and the Rap phosphatases of bacilli (20). To date, only a few mature forms of signaling peptides from this family—cCF10, PhrC, PapR, SHP1358, and the XIP of S. mutans, a type II ComS pheromone—have been found in the extracellular medium (8, 14, 21–23) (see Table S2 in the supplemental material). We describe here the early steps of the extracellular circuit of another member of this family of signaling peptides, ComS from S. thermophilus, a type I ComS pheromone.
We first identified the active form of ComS in the extracellular medium of a ComS-overproducing strain. Mature ComS, i.e., ComS14–24, is an undecapeptide and is thus longer than the synthetic peptides identified as active in a previous study (6). Natural mature ComS is probably unmodified, like previously identified signaling peptides, which bind directly to their effectors (PrgX, PlcR, and Rap phosphatases) (see Table S2 in the supplemental material), although we cannot rule out the possibility of intracellular proteolysis or modifications. Indeed, it is difficult to determine whether ComS14–24 is the peptide preferentially interacting with ComR, a precursor of shorter active peptides generated by cleavage with intracellular aminopeptidases, or both. Six intracellular aminopeptidases with general or more specific cleavage specificities are encoded in the S. thermophilus genome and could potentially cleave ComS after its import (24). Two shorter forms of ComS14–24, ComS15–24 and ComS16–24, which are probably products of ComS14–24 degradation, have been detected in the extracellular medium. This observation suggests that ComS14–24 is degraded extracellularly by an aminopeptidase that remains to be identified. We also showed that synthetic ComS17–24 was as efficient as synthetic ComS14–24. The extracellular mature form of ComS is slightly longer than the other internalized signaling peptides identified in supernatants from Gram-positive bacteria (see Table S2), which are between five and nine amino acids long. However, ComS14–24 could clearly be transported by the Ami oligopeptide transporter, which can import peptides of 3 to 23 residues (25). Furthermore, we demonstrated that the thiol of the ComS cysteine residue must remain unmodified for ComS transport or activity. This observation indicates a possible role of the oxydo-reduction status of the environment in the triggering and regulation of competence.
ComS14–24 is generated by C-terminal cleavage of the peptide precursor. The Eep membrane protease plays a direct or indirect role in the processing of proComS and of several pheromones and antipheromones of E. faecalis (26, 27), the SHP1358 pheromone of S. thermophilus (14), and SHP3 from S. pyogenes (16). If Eep cleaves the pheromone precursors, as seems likely, in all cases it would recognize hydrophobic peptides of about 20 amino acids in length, signal peptides, or peptides resembling signal peptides. These characteristics must be sufficient for Eep recognition, because no obvious amino acid signature could be identified by sequence alignment analysis. These features are also probably necessary for peptide secretion. We do not know whether these peptides are secreted via the Sec machinery or via another transporter or whether their hydrophobicity allows them to cross the membrane directly. The processing and secretion steps are probably tightly linked and occur simultaneously in the membrane predicted to contain Eep. Interestingly, Eep is not involved in the processing of ComS from S. mutans (8). The ComS peptides of other streptococci, also named XIP, vary in length between 15 and 32 amino acids and are less hydrophobic than SHP signaling peptides. It will be interesting to identify the ComS species processed by Eep for the identification of additional features potentially accounting for the specificity of this protease. It would also be very interesting to identify proteases that can substitute for Eep. The situation seems to be different in bacilli, in which PapR and Phr peptides, working with PlcR and Rap phosphatases, respectively, have a peptide signal toward the N terminus, in front of the active part of the peptide, suggesting probable secretion via the Sec machinery. It remains unclear how PapR is processed, but PhrC (CSF) in Bacillus subtilis is processed by at least three proteases: subtilisin, Epr, and Vpr (28).
We have confirmed that the AmiA3 oligopeptide plays a more important role than that of AmiA1 in the import of ComS. However, it is difficult to determine whether this is due to a higher abundance of AmiA3 than of AmiA1 or to a higher specificity of AmiA3 for ComS. Previous proteomic results obtained by our group (5) indicated that AmiA3 has a higher abundance factor (3.93 ± 0.37) than that of AmiA1 (2.76 ± 0.35) (data not shown), suggesting that AmiA3 is probably more abundant than AmiA1 in CDM. Indeed, the abundance factor is the ratio between the total number of spectra obtained during the protein identification process and the theoretical number of peptides detectable for a given protein by mass spectrometry. In the case of AmiA1 and AmiA3, homologous proteins of the same size, these factors can be compared.
We were unable to detect ComS in the extracellular medium of a strain naturally producing this molecule by mass spectrometry or complementation experiments with supernatants. ComS does not seem to diffuse freely or to accumulate in the medium. However, a nonproducing strain can obtain this molecule from a strain that produces it naturally, as shown by our coculture experiments. In addition, ComS was detected in the supernatant from a ComS-overproducing strain in which Ami-mediated import was impaired. Several hypotheses can be put forward to explain these findings: ComS may remain stuck on the membrane after secretion, or it may rapidly disappear from the cell surface, being captured rapidly by the OBPs of the Ami transporter. These situations allow sensing of ComS by the producer bacteria as well as by bacteria located in the close vicinity of the producers, which is probably relevant in bacterial chains, colonies, or biofilms. Different findings have been reported for S. mutans, as type II ComS is released into the medium, in which it accumulates (9). The behavior of type I ComS from S. thermophilus is very similar to that reported for the PhrA signaling peptide involved in controlling sporulation in B. subtilis. PhrA was detected only with a mutant with impaired oligopeptide transport (Δopp), and complementation experiments were successful with cocultures but not with supernatants. As pointed out by M. Perego, this behavior suggests that PhrA acts in cell-to-cell communication between interacting cells and over a short distance or that it is a more fundamental component of a timing device (29). We addressed this question in our system, by following the triggering of comX expression in a ComS-nonproducing strain in response to the addition of synthetic ComS17–24 during the exponential phase of growth. Our results suggest that cells can respond immediately to the presence of the signal peptide, except at the beginning of the culture (when the cells are used to inoculate fresh medium), probably because some of the components of the cell-to-cell communication machinery must first be synthesized in sufficiently large amounts. The response of the cells to the presence of ComS is transient. Following the dilution of a culture naturally producing ComS and containing a comX-luxAB fusion to various initial OD600 values, the expression of comX was always first detected 90 min after dilution and reached high levels regardless of the OD600, except in the least-diluted culture, in which no comX expression was detected. Even if we cannot completely exclude that, whatever the dilution, the ComS concentration remained above the threshold for quorum sensing detection, our findings suggest that the mechanism triggering comX expression resembles a timing device much more than a quorum-sensing system. The component that is limiting during the first 90 min remains to be identified. Similar results were obtained with a late competence gene, dprA, and comS, whose expression seems to be triggered slightly earlier, possibly because the ComR regulator is synthesized in the vicinity of the comS promoter. As observed with comX, no expression was detected for the three genes in the least-diluted culture (OD600 = 0.4). More experiments will be needed to explain this surprising result, but we can hypothesize that at an OD600 of 0.4, as the cultures are less diluted, a competence inhibitor (protein or signal) is present, or that the physiological state of the cells is different than in less-diluted cultures. The comR gene was expressed immediately from the start of culture, even in the least-diluted culture. In S. mutans, the kinetics of competence triggering is completely different, with this state occurring at the end of the exponential phase, at high cell density. Moreover, the addition of synthetic ComS to the medium leads to a linear increase in comX expression that continues for up to 4 h (9). In this case, the type II ComS-ComR mechanism appears to match the criteria required for a QS mechanism more closely.
We have demonstrated that the presence of nutritional peptides in CDM at the concentration found in M17 medium inhibits the triggering of competence. A very recent work described similar results for S. mutans, in which small peptides from brain heart infusion (BHI) medium or tryptone suppress the XIP response (30). The simplest explanation for this is that the Ami transporter is saturated with the nutritional peptides, which are present at a much higher concentration than that of ComS. This is not the only possible explanation for the complete lack of competence development of S. thermophilus in M17lac medium. Indeed, Boutry et al. recently suggested that in this rich medium, MecA sequesters ComX, allowing it to be degraded by the ClpCP complex (31). As milk is thought to be the ecological niche of S. thermophilus, we assessed the rate of transformation of strain LMD-9 in this medium. Levels of competence in milk were lower than those in CDM by a factor of only 10. Interestingly, the addition of synthetic ComS17–24 increased the transformation rate by a factor of only 3 in CDM but by a factor of 200 in milk. Thus, the LMD-9 strain is more competent in the presence of ComS17–24 in milk than in CDM, suggesting that one of the steps of the development of competence (occurring after the processing of proComS) is limiting in CDM but not in milk. For example, the Ami level may be higher in milk, in which this transporter is essential for growth (25), than in CDM. It will be interesting to study the regulation of competence triggering in this complex medium from both fundamental and applied research viewpoints.
In conclusion, the type I ComS-ComR mechanism of S. thermophilus is clearly different from the type II ComS-ComR mechanism of S. mutans and the SHP-Rgg1358 QS mechanism of S. thermophilus strain LMD-9, although it has several features in common with these other mechanisms. The pheromones differ: the type II ComS of S. mutans and SHP1358 are released into the extracellular medium, although SHP1358 is more hydrophobic than type I and type II ComS. The SHP-Rgg1358 mechanism and probably also the type II ComS-ComR mechanism of S. mutans act as QS mechanisms, whereas the type I ComS-ComR mechanism of S. thermophilus is more likely to correspond to a timing device. SHP1358 and type I ComS are processed by an Eep-dependent process, whereas type II ComS is not. Thus, the type II ComS-ComR mechanism of S. mutans seems to resemble the SHP1358 mechanism of S. thermophilus more closely than the type I ComS-ComR mechanism of S. thermophilus. This highlights the importance of not extending the conclusions drawn from a single system to all SHP-Rgg mechanisms.
If we compare the mechanisms controlling the expression of comX in S. pneumoniae and S. thermophilus, it is clear that there are differences in the extracellular step. In S. pneumoniae, the ComC (or CSP) signal peptide, which is very different from ComS, is detected in the extracellular medium by the ComDE two-component system. However, the kinetics of comX expression in both species are strikingly similar, with transient triggering early in the exponential growth phase. The ComCDE and ComS-ComR mechanisms seem to control the timing of comX expression rather than acting as a quorum-sensing mechanism for this gene. These similarities are probably linked to a physiological constraint on competence in both streptococci that has yet to be identified.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Institut National de la Recherche Agronomique (INRA). The Plateforme d'Analyse Protéomique Paris Sud-Ouest (PAPPSO; specifically Alain Guillot) received financial support from the Ile-de-France regional council and from ISTREA. The work carried out in the group of P. Hols was financially supported by FNRS.
L. Fontaine is a postdoctoral worker and P. Hols is a research associate at FNRS. We thank V. Juillard and B. Fleuchot for critical readings of the manuscript and M. Coté for technical assistance.
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02196-12.
REFERENCES
- 1. Johnsborg O, Håvarstein LS. 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 33:627–642 [DOI] [PubMed] [Google Scholar]
- 2. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451–475 [DOI] [PubMed] [Google Scholar]
- 3. Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767–778 [DOI] [PubMed] [Google Scholar]
- 4. Martin B, Quentin Y, Fichant G, Claverys JP. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339–345 [DOI] [PubMed] [Google Scholar]
- 5. Gardan R, Besset C, Guillot A, Gitton C, Monnet V. 2009. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191:4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B, Petersen FC. 2012. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194:3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation by Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194:3774–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mashburn-Warren L, Morrison DA, Federle MJ. 2012. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194:4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Letort C, Juillard V. 2001. Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J. Appl. Microbiol. 91:1023–1029 [DOI] [PubMed] [Google Scholar]
- 12. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Ibrahim M, Guillot A, Wessner F, Algaron F, Besset C, Courtin P, Gardan R, Monnet V. 2007. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189:8844–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80:1102–1119 [DOI] [PubMed] [Google Scholar]
- 15. An FY, Sulavik MC, Clewell DB. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 7:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delorme C, Bartholini C, Bolotine A, Ehrlich SD, Renault P. 2010. Emergence of a cell wall protease in the Streptococcus thermophilus population. Appl. Environ. Microbiol. 76:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Juillard V, Guillot A, Le Bars D, Gripon J-C. 1998. Specificity of milk peptide utilization by Lactococcus lactis. Appl. Environ. Microbiol. 64:1230–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waterfield NR, Le Page RW, Wilson PW, Wells JM. 1995. The isolation of lactococcal promoters and their use in investigating bacterial luciferase synthesis in Lactococcus lactis. Gene 165:9–15 [DOI] [PubMed] [Google Scholar]
- 20. Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:18490–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, Adsit JC, Dunny GM, Suzuki A. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263:14574–14578 [PubMed] [Google Scholar]
- 22. Solomon JM, Lazazzera BA, Grossman AD. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 10:2014–2024 [DOI] [PubMed] [Google Scholar]
- 23. Bouillaut L, Perchat S, Arold S, Zorrilla S, Slamti L, Henry C, Gohar M, Declerck N, Lereclus D. 2008. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 36:3791–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rul F, Monnet V. 1997. Presence of additional peptidases in Streptococcus thermophilus CNRZ 302 compared to Lactococcus lactis. J. Appl. Microbiol. 82:695–704 [DOI] [PubMed] [Google Scholar]
- 25. Garault P, Le Bars D, Besset C, Monnet V. 2002. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J. Biol. Chem. 277:32–39 [DOI] [PubMed] [Google Scholar]
- 26. An FY, Clewell DB. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 190:1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. 2007. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol. Microbiol. 65:1321–1333 [DOI] [PubMed] [Google Scholar]
- 29. Perego M. 1999. Self-signaling by Phr peptides modulates Bacillus subtilis development, p 243–258 In Dunny GM, Winans SC. (ed), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, DC. [Google Scholar]
- 30. Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86:258–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boutry C, Wahl A, Delplace B, Clippe A, Fontaine L, Hols P. 2012. Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J. Bacteriol. 194:1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bojovic B, Djordjevic G, Topisirovic L. 1991. Improved vector for promoter screening in lactococci. Appl. Environ. Microbiol. 57:385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 4 February 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. [Epub ahead of print.] doi:10.1111/mmi.12157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.