Abstract
The central carbon metabolism genes in Corynebacterium glutamicum are under the control of a transcriptional regulatory network composed of several global regulators. It is known that the promoter region of ramA, encoding one of these regulators, interacts with its gene product, RamA, as well as with the two other regulators, GlxR and SugR, in vitro and/or in vivo. Although RamA has been confirmed to repress its own expression, the roles of GlxR and SugR in ramA expression have remained unclear. In this study, we examined the effects of GlxR binding site inactivation on expression of the ramA promoter-lacZ fusion in the genetic background of single and double deletion mutants of sugR and ramA. In the wild-type background, the ramA promoter activity was reduced to undetectable levels by the introduction of mutations into the GlxR binding site but increased by sugR deletion, indicating that GlxR and SugR function as the transcriptional activator and repressor, respectively. The marked repression of ramA promoter activity by the GlxR binding site mutations was largely compensated for by deletions of sugR and/or ramA. Furthermore, ramA promoter activity in the ramA-sugR double mutant was comparable to that in the ramA mutant but was significantly higher than that in the sugR mutant. Taken together, it is likely that the level of ramA expression is dynamically balanced by GlxR-dependent activation and repression by RamA along with SugR in response to perturbation of extracellular and/or intracellular conditions. These findings add multiple regulatory loops to the transcriptional regulatory network model in C. glutamicum.
INTRODUCTION
Corynebacterium glutamicum is a high-G+C-content Gram-positive soil bacterium which is used in biotechnological production of amino acids, organic acids, and alcohols (1–6). Since the complete genome sequence of C. glutamicum became available (7–9), a number of transcriptional regulators controlling genes involved in central carbon metabolism have been characterized (10, 11). Genome-wide studies reveal that a global transcription regulatory system for carbon source-dependent regulation in C. glutamicum is quite different from the well-established systems in Escherichia coli and Bacillus subtilis. A major control system for utilization of carbon sources, namely, carbon catabolite repression, is mediated by the cyclic AMP (cAMP) receptor protein (CRP) via intracellular cAMP levels in E. coli, while in B. subtilis, it is mediated by carbon control protein A (CcpA) via phosphorylation states of HPr, a component of a phosphotransferase system. In contrast to the situation in E. coli, where the phosphotransferase system for glucose uptake modifies adenylate cyclase activity to decrease the intracellular cAMP levels in the presence of glucose (12, 13), the C. glutamicum intracellular cAMP levels are increased in the presence of glucose by an unknown mechanism(s) (14–16). Besides, no HPr kinase/phosphatase system is found in the C. glutamicum genome. These differences in the molecular characteristics are likely reflected in the capacity of C. glutamicum to cometabolize glucose and a variety of carbon sources, including sugars, organic acids, and aromatic compounds without catabolite repression (17–24), expect for a few cases (25–27).
Carbon source-dependent regulation of gene expression in C. glutamicum has been investigated by comparing the expression profile in glucose-grown cells to that in acetate-grown cells (28–30). These studies reveal that genes for sugar uptake and metabolism, including the phosphotransferase system and glycolysis, are upregulated in the presence of glucose. In contrast, genes for enzymes involved in the tricarboxylic acid cycle, glyoxylate shunt, and gluconeogenesis are upregulated in the presence of acetate. Multiple transcription regulators are involved in the expression of these genes. For example, expression of the gapA-pgk-tpi operon, encoding the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and triose phosphate isomerase, respectively, is coordinately regulated by SugR, RamA, and GlxR (15, 31, 32). SugR is a global repressor of genes for sugar uptake and metabolism, including phosphotransferase systems, glycolysis, and fermentative lactate dehydrogenase (32–37). As sugar phosphates, e.g., fructose-1-phosphate and fructose-1,6-bisphosphate, act as negative effectors of SugR, the SugR regulon is derepressed in the presence of sugar. GlxR, a cAMP-responsive regulator, and RamA, a LuxR-type regulator, were first identified as the transcriptional repressor and activator, respectively, of the aceB gene, encoding malate synthase of the glyoxylate cycle (14, 38). Both regulators not only activate gapA expression (15, 31) but are also involved in expression of a number of genes for various physiological functions, including carbon and nitrogen metabolism, respiration, SOS and stress responses, and cell division (15, 39–41). However, in contrast to the mechanism by which SugR switches gene expression, how GlxR and RamA switch expression of genes involved in glucose and acetate metabolism in response to a carbon source provided is not fully understood. This is because the identity of an effector molecule that controls RamA activity and understanding how intracellular cAMP levels are controlled remain elusive.
Furthermore, carbon source-dependent regulation is complicated by a hierarchical and/or interactive transcriptional control of the transcription regulators. SugR, RamA, and GlxR act as transcription repressor of their own genes, sugR, ramA, and glxR, respectively (42–44). Expression of the sugR gene is directly activated by RamA (31). Expression of the ramA gene is upregulated by an unknown factor in the presence of acetate (43). Thus, the carbon metabolism genes in C. glutamicum are under the control of a complex hierarchical regulatory network consisting of transcription factors responding to various environmental and/or physiological signals.
In this study, to unveil a new hierarchical interaction in the regulatory network, we focus on regulation of the ramA gene. It has been reported that four transcription regulators, RamA, SugR, GlxR, and RamB, bind to the ramA promoter in vitro and/or in vivo (15, 35, 43). RamA is subject to negative autoregulation, as described above. RamB is involved in repression of the acetate metabolism genes in the presence of glucose (45), but disruption of its gene, ramB, has no effect on expression of ramA (43). However, the involvement of SugR and GlxR in ramA expression has not been investigated so far. Here, the effects of mutations in the GlxR binding site in combination with deletion of ramA, sugR, and ramB on ramA promoter activity are described.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. The oligonucleotide primers used are listed in Table S1 in the supplemental material. For genetic manipulation, E. coli strains were grown at 37°C in Luria-Bertani (LB) medium. C. glutamicum strains were grown at 33°C in nutrient-rich A medium (50), which contains 0.2% yeast extract and 0.7% Casamino Acids with 4% glucose. When appropriate, the media were supplemented with antibiotics. The final antibiotic concentrations for E. coli were 50 μg of ampicillin ml−1 and 50 μg of kanamycin ml−1; for C. glutamicum, kanamycin was used at 50 μg ml−1. For promoter-reporter assays, C. glutamicum strains chromosomally carrying a promoter-lacZ fusion were grown in A medium containing 1% glucose or acetate.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB+ lacIq lacZΔM15] | TaKaRa |
| JM110 | dam dcm supE44 hsdR17 tih leu rpsL lacy galK galT ara tonA thr tsx Δ(lac-proAB) [F′ traD36 proAB+ lacIq lacZΔM15] | 46 |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) | 47 |
| C. glutamicum | ||
| R (JCM 18229) | Wild-type strain | 9 |
| KT27 | R with mutations in the GlxR binding site in the ramA promoter region on the chromosome | This study |
| Plasmids | ||
| pCold | Apr; cold-inducible expression vector | TaKaRa |
| pGEM-T Easy | Apr; TA cloning vector | Promega |
| pCRA725 | Kmr; suicide vector containing the B. subtilis sacB gene | 2 |
| pCRA741 | Kmr; pCRA725 with a 2.0-kb PCR fragment from SSI7 and a 3.1-kb PCR fragment containing the E. coli lacZ gene | 48 |
| pCRC621 | Kmr; pCRA725 with a 2.65-kb fragment containing a mutated ramA promoter | This study |
| pCG1 | 3,069-bp plasmid from C. glutamicum | 49 |
| pKK223-3 | Apr; expression vector under the control of the tac promoter | Pharmacia |
| pDW363 | Apr; source of lacIq | NBRP (NIG, Japan) |
| pCRB12iP | Kmr lacIq Ptac, IPTG-inducible vector based on pCG1 | This study |
| pCRC622 | Kmr; pCRB12iP with a 909-bp PCR fragment containing the-ramA coding region | This study |
EMSA.
GlxR was expressed with an N-terminal His tag and purified by affinity chromatography as described previously (15). DNA fragments containing the ramA promoter region with a native or mutated GlxR binding site were amplified by PCR using primers PramAFW and PramARV and promoter-lacZ fusion plasmids, which were constructed as described below, as the templates and cloned into the pGEM-T Easy vector (Promega). The sequence and direction of the cloned fragments were confirmed, and the cloned fragments were labeled with Cy3 by PCR amplification using primers SP6Cy3 and T7 (see Table S1 in the supplemental material). The resulting fragments were purified with a QIAquick PCR purification kit (Qiagen). Electrophoretic mobility shift assay (EMSA) was performed as described previously (51). DNA and DNA-protein complexes were visualized by a Typhoon TRIO variable-mode imager (GE Healthcare Bioscience).
Construction of promoter-lacZ fusions.
The ramA promoter region from positions −474 to +15 and the cysK promoter region from positions −595 to +15 with respect to the translational start point were amplified from C. glutamicum R chromosomal DNA by PCR using primers PramAFW-PramARV and PcysKFW-PcysKRV, respectively (see Table S1 in the supplemental material). Mutations in the GlxR binding site were introduced by overlapping PCR using primers PramAmutFW and PramAmutRV, as described below. The fragments amplified were phosphorylated and cloned upstream of the lacZ gene in pCRA741 (48). The direction and sequence of the inserted fragment were confirmed by DNA sequencing. The plasmids were isolated as nonmethylated DNA from E. coli JM110, introduced into C. glutamicum, and subsequently integrated into a strain-specific island 7 (SSI7) on the chromosome of C. glutamicum R by markerless gene insertion methods, as described previously (2). The integration was confirmed by PCR using primers specific for the promoter region and the SSI7 region.
Construction of a genetically modified strain.
To modify the GlxR binding site on the chromosome, the GlxR binding site and both flanking regions were amplified by PCR using primer pairs ramA_integ_FW/PramAmutRV and ramA_integ_RVxba/PramAmutFW. The two fragments were used as a template for PCR using primers ramA_integ_FW and ramA_integ_RV. The resulting fragment with mutations in the GlxR binding site was digested with SalI-XbaI and cloned into pCRA725, a suicide vector for markerless gene disruption (2), yielding pCRC621. C. glutamicum R (wild type [WT]) was transformed with pCRC621, which was isolated as nonmethylated DNA from E. coli JM110 for efficient gene introduction into C. glutamicum (52), by electroporation. Screening for the mutants was performed as described previously (2). Introduction of the mutations into the site on the chromosome was confirmed by direct sequencing of a PCR product, which was amplified using primers PramAFW and PramRV and genomic DNA extracted from the strains obtained as the template.
Construction of an IPTG-inducible expression vector and overexpression of ramA.
To construct a ramA-overexpressing strain, we constructed an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector, pCRB12iP. This plasmid carries the LacI repressor gene lacIq and the tac promoter, which is followed by the rrnB1 and rrnB2 terminators. The DNA fragment containing lacIq was amplified by PCR using pDW363 as the template, while the DNA fragments containing the tac promoter and the terminators were amplified by PCR using pKK223-3 as the template. The coding region of ramA was amplified by PCR using primers ramAFWKpn and ramARVKpn and the genomic DNA as the template and cloned into pCRB12iP, yielding pCRC622. Overexpression of ramA in strains carrying pCRC622 was induced by supplementation of 0.5 mM IPTG.
β-Galactosidase assay.
C. glutamicum cells carrying the promoter-lacZ fusion were harvested, washed once with Z buffer (53), resuspended in the same buffer, and treated with toluene. The permeabilized cells were then incubated with o-nitrophenyl-β-galactoside, and activity was measured in Miller units, as previously described (53).
qRT-PCR.
Total RNA was extracted from C. glutamicum cells using an RNeasy minikit (Qiagen) as described previously (32). Isolated RNA samples were checked for purity using an Agilent RNA 6000 nanokit on an Agilent 2100 bioanalyzer (Agilent Technologies, CA) and stored at −80°C. Quantitative reverse transcription-PCR (qRT-PCR) was performed using an Applied Biosystems 7500 Fast real-time PCR system (Life Technologies, CA) and Power SYBR green PCR master mix with murine leukemia virus reverse transcriptase and the RNase inhibitor of the GeneAmp RNA PCR kit (Life Technologies) as described previously (32). Specific primers (see Table S1 in the supplemental material) were designed using Primer Express software (version 3.0; Life Technologies). The specificity of the amplicons was checked by DNA dissociation curve analysis. The comparative threshold cycle method (Life Technologies) was used to quantify relative expression.
5′ RACE-PCR.
For the identification of the transcriptional start points (TSPs) of cysK, 5′ rapid amplification of cDNA ends (RACE)-PCR analyses were carried out. Total RNA was extracted as described above. The RNA extracted was poly(A) tailed using a poly(A) tailing kit (Life Technologies) according to the manufacturer's instructions. After phenol-chloroform extraction, the RNA with the poly(A) tail was purified by ethanol precipitation. cDNA was synthesized using a SMARTer RACE cDNA amplification kit (Clontech, CA) with a supplied (oligo)dT-anchored primer and 1 μg of the tailed RNA prepared as described above. The cDNA was amplified with Universal Primer A (supplied with kit) and gene-specific primers (see Table S1 in the supplemental material). The resulting PCR products were cloned into the pGEM-T Easy vector (Promega). At least 10 clones of the 5′ RACE-PCR product were sequenced.
RESULTS
GlxR positively regulates ramA expression.
Our previous chromatin immunoprecipitation (ChIP) chip analysis identified the GlxR binding site at position −290 with respect to the transcriptional start point (TSP) of the ramA gene (15) (Fig. 1). To examine GlxR binding to the site, EMSA using purified His-tagged GlxR was performed as described previously (15). The results of EMSA demonstrated that GlxR actually binds to the 489-bp ramA promoter region in the presence of cAMP in vitro (Fig. 2). The GlxR binding was abolished by mutations (underlined) in the GlxR binding site (5′-AGTGTTCTATGACACT-3′ → 5′-ACACTTCTATGAGTGT-3′) (Fig. 2), confirming that the site identified is the sole binding site in this region. The binding was also abolished by removing cAMP from the reaction, demonstrating that the binding is cAMP dependent.
Fig 1.
Binding sites for transcriptional regulators in the ramA promoter region. While the intergenic region between ramA and cysK is indicated with lowercase letters, coding regions of the genes are indicated with uppercase letters. Primers used for amplification of the ramA promoter region are indicated with arrows. The transcriptional start points of ramA and cysK are indicated with +1. The binding sites for GlxR and RamB are boxed and indicated with a dotted line, respectively. The putative binding sites for RamA and SugR are indicated with bold italic letters and underlines, respectively.
Fig 2.
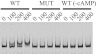
Binding of GlxR to the ramA promoter region. DNA fragments covering the ramA promoter region with the wild-type (WT) or mutated (MUT) GlxR site were incubated with the purified GlxR at the indicated concentrations (nM) in the presence of 0.5 mM cAMP or in the absence of cAMP (−cAMP) and analyzed by nondenaturing PAGE. Each well contained 10 nM DNA fragment.
Because a glxR mutant shows severe growth defects (31, 54), a role of GlxR in expression of ramA was assessed by promoter-reporter assays. The same DNA fragments as those examined in EMSA were fused to the promoter-less lacZ gene, and the fusions (PramA-lacZ) yielded were integrated into the wild-type chromosome. The β-galactosidase activity driven by the native and mutated promoters during growth in nutrient-rich medium (A medium) containing either 1% glucose or 1% acetate was determined. We used the medium because the ramA deletion mutant strains described in the following section are incapable of growing on acetate as the sole carbon source, as has been reported (38). The activities were largely unchanged during growth, although they slightly increased in some of these strains used in the later growth phase. The promoter activities in the cells cultured for 8 h (in the stationary phase) are summarized in Table 2. The activity of the wild-type promoter was higher in the cells grown on acetate than those grown on glucose (Table 2), which is consistent with previous results (43). The promoter carrying the above-described mutations in the GlxR binding site showed no detectable activity during growth under these conditions (Table 2), suggesting that GlxR plays an essential role in ramA expression.
Table 2.
Activity of the ramA promoter with or without mutations in the GlxR binding site in the genetic background of the wild type and ramA, sugR, ramA-sugR, and ramB mutantsa
| Genotype | GlxR siteb | β-Galactosidase activity (Miller units)c |
|
|---|---|---|---|
| Glucose | Acetate | ||
| WT | WT | 25.16 ± 3.75 | 61.32 ± 6.14 |
| Mut | BLD | BLD | |
| ΔsugR | WT | 64.18 ± 8.77 | 123.8 ± 20.6 |
| Mut | 39.84 ± 7.42 | 75.97 ± 7.06 | |
| ΔramA | WT | 474.2 ± 34.4 | 637.8 ± 68.1 |
| Mut | 245.7 ± 6.5 | 330.9 ± 33.3 | |
| ΔramA-ΔsugR | WT | 393.0 ± 1.5 | 634.8 ± 10.0 |
| Mut | 215.6 ± 20.4 | 349.8 ± 10.7 | |
| ΔramB | WT | 18.34 ± 1.30 | 52.08 ± 1.55 |
| Mut | BLD | BLD | |
The strains carrying the ramA promoter-lacZ fusion in the chromosome were cultured on either glucose or acetate for 8 h (in the stationary phase). The activities are the mean values from at least three independent cultivations with standard deviations.
The GlxR binding site was mutated (Mut) or not mutated (WT).
BLD, below the level of detection.
The GlxR binding site is not implicated in expression of cysK transcribed divergently from ramA.
The GlxR binding site in the ramA promoter region is located 336 bp upstream of the translational start codon of cysK, which is transcribed divergently from ramA. The cysK gene encodes cysteine synthase. The TSP of cysK was determined by 5′ RACE and sequencing to be an adenine located 99 bp upstream of the translational start codon; the GlxR binding site was centered at position −237 with respect to the TSP (Fig. 1). To examine whether GlxR regulates cysK expression, we investigated the effect of the mutations in the GlxR binding site on the promoter activity of the 595-bp upstream region of the cysK gene using the promoter-reporter assay as described above. The results obtained from the analyses showed that the mutations in the GlxR binding site had no effect on cysK expression under the conditions used (see Fig. S1 in the supplemental material), indicating that GlxR bound to the site regulates only ramA expression.
ramA expression is repressed by SugR.
SugR interacts with the ramA promoter region in vivo and in vitro (35). As one of the two deduced binding sites is found between positions −8 and −21 with respect to the TSP, SugR is predicted to act as a repressor of ramA (Fig. 1) (35). The other site is located between positions −55 and −68. To experimentally demonstrate a role of SugR in expression of ramA, the ramA promoter-lacZ fusion described above was integrated into the chromosome of a sugR mutant. The activity of the ramA promoter in the sugR mutant was 2-fold higher than that in the wild type under the same conditions described earlier (Table 2). These results confirmed that SugR represses ramA expression.
Repression by SugR is relieved in the ramA mutant.
Next, we examined whether RamA has a role in expression of the ramA gene, using a ramA mutant carrying the ramA promoter-lacZ fusion in the chromosome. The ramA promoter activity in the ramA mutant background was almost 10- to 20-fold higher than that in the wild-type background (Table 2). It is consistent with a previous finding that RamA represses its own expression (43), although the extent of derepression observed in our experiment was higher than that reported in the study (up to a 5-fold increase by ramA deletion). The promoter activity in the ramA mutant background was higher than that in the sugR mutant background in both glucose- and acetate-grown cells, indicating that the degree of repression by RamA is greater than that by SugR. Furthermore, the effects of disruption of both sugR and ramA on the ramA promoter activity were investigated. The activity of the ramA promoter in the sugR-ramA double mutant background was comparable to that in the ramA single mutant background (Table 2). These results indicate that SugR has no influence on the ramA promoter activity in the ramA mutant background, influence that it has in the wild-type background. As RamA directly activates sugR expression (31), it is conceivable that RamA is required for SugR to repress ramA expression.
GlxR activates ramA expression in ramA and sugR mutants.
The ramA promoter with the mutations in the GlxR binding site showed no activity in the wild type, as described earlier (Table 2). We examined the effects of the mutations on the promoter activity derepressed in the three genetic backgrounds, i.e., sugR, ramA, and sugR-ramA deletion mutants. The GlxR binding site mutations reduced 2-fold the ramA promoter activity in all the deletion mutants grown under the conditions used (Table 2), indicating that GlxR positively regulates ramA expression. It should be noted that the apparent essential role of GlxR in the ramA promoter activity observed in the wild-type background was compensated for to a large extent by inactivation of RamA and/or SugR.
The level of ramA mRNA expression is not affected by GlxR binding site mutation.
To examine the effects of the negative autoregulation of ramA on the positive regulation by GlxR, the ramA promoter at the original locus in the wild type was modified with the aforementioned GlxR binding site mutations, yielding strain KT27. Expression of ramA driven by the mutated promoter in strain KT27 grown under the conditions described earlier was compared to that in the wild type by qRT-PCR analysis. The ramA mRNA levels in these two strains were comparable (see Fig. S2 in the supplemental material). Moreover, expression of the representative RamA regulon members sugR and aceA in strain KT27 was also comparable to that in the wild type (data not shown). As RamA is essential for expression of aceA encoding isocitrate lyase in the glyoxylate shunt, the ramA mutant shows a growth defect on acetate (38). In contrast, the growth of strain KT27 on acetate was comparable to that of the wild type (data not shown). Taken altogether, these findings suggest that the level of ramA expression is maintained without the GlxR-dependent activation in strain KT27. This is probably due to the strong negative autoregulation of ramA.
We also compared the cysK mRNA levels between strain KT27 and the wild type, showing that the cysK mRNA level was not affected by the mutations in the GlxR binding site (data not shown). This is consistent with the results of the promoter-reporter assay that GlxR is not involved in cysK expression (see Fig. S1 in the supplemental material).
Overexpression of ramA shuts off ramA promoter activity.
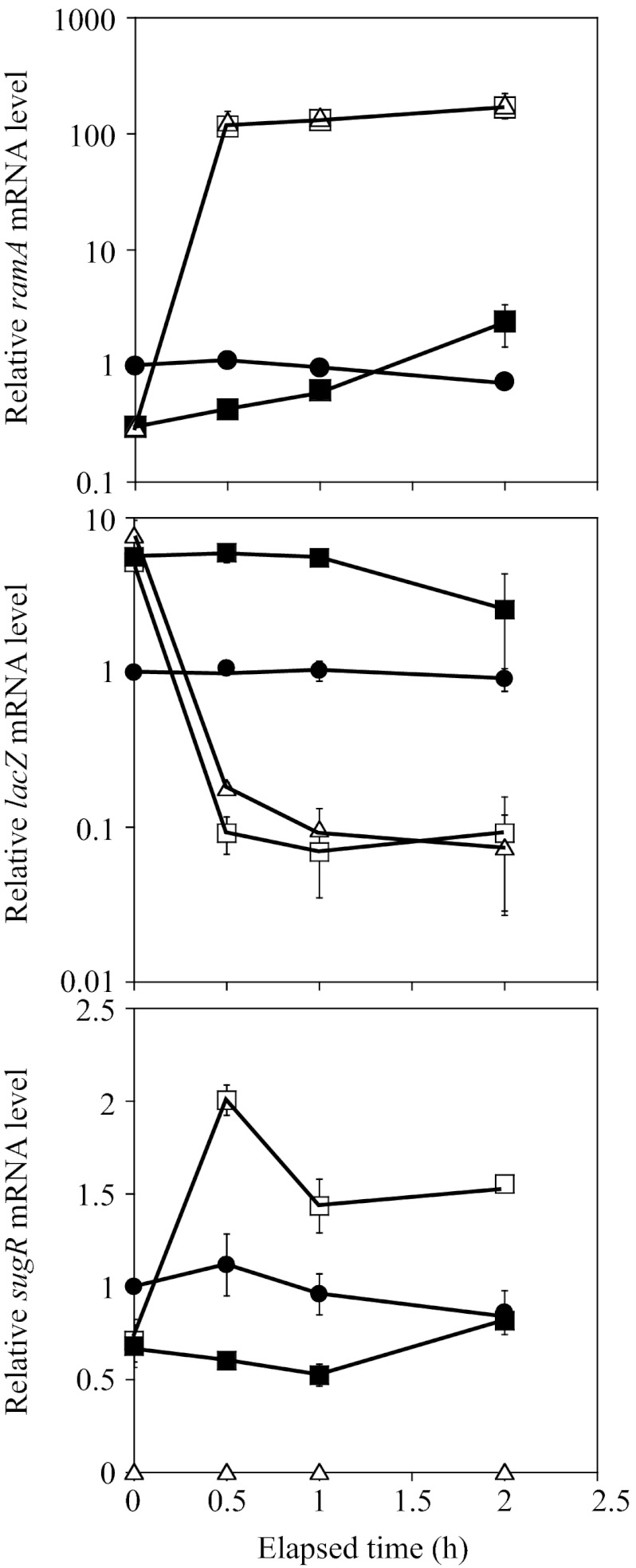
To assess the functional significance of the negative autoregulation of ramA, we examined the effects of overexpression of ramA on the activity of the PramA-lacZ fusion. A plasmid carrying the ramA gene under the control of the tac promoter and LacI repressor (pCRC622) was introduced into the ramA and ramA-sugR mutants that chromosomally carry the native PramA-lacZ fusion. Since the expression of the ramA gene in the resulting strain (the ΔramA PramA-lacZ/pCRC622 and ΔramA-sugR PramA-lacZ/pCRC622 strains) was derived only from the IPTG-inducible promoter on the plasmid, the autoregulation of ramA expression was eliminated. The cells exponentially growing on glucose (4 h after the start of culture) were supplemented with IPTG. Alterations in the levels of ramA and lacZ mRNA after the IPTG supplementation were analyzed by qRT-PCR. The mRNA levels relative to the value obtained from the wild type carrying the native PramA-lacZ fusion (the WT PramA-lacZ strain) before IPTG supplementation (0 h) are shown in Fig. 3. After the addition of IPTG, the level of ramA mRNA in the WT PramA-lacZ strain was nearly unchanged for 2 h, although a slight decrease was observed (Fig. 3, top, circles). Before the addition of IPTG, the level of ramA expression in the plasmid-carrying strains was 4-fold lower than that in the WT PramA-lacZ strain (Fig. 3, top, squares and triangles). The level of ramA mRNA in these strains increased 400-fold within 0.5 h of IPTG supplementation and slightly increased in the subsequent 1.5 h (Fig. 3, top, open symbols). Even in the absence of IPTG, the level of ramA mRNA in the ΔramA PramA-lacZ/pCRC622 strain gradually increased (Fig. 3, top, filled squares), indicating leaky expression of ramA from the plasmid. At the end of the sampling period (2 h), the level of ramA mRNA in this strain was about 2-fold higher than that in the WT PramA-lacZ strain but 80-fold lower than the IPTG-induced level (Fig. 3, top, filled and open squares). These results confirmed the IPTG-inducible overexpression of ramA in the ΔramA PramA-lacZ/pCRC622 strain.
Fig 3.
Overexpression of ramA represses the ramA promoter activity. The exponentially growing cells were supplemented with IPTG (0 h). The strains used carried the PramA-lacZ fusion in the chromosome in the genetic background of the wild type (filled circles), ramA mutant transformed with pCRC622 for IPTG-inducible ramA expression (open squares), and ramA-sugR mutant transformed with pCRC622 (open triangles). Changes in the levels of ramA (top), lacZ (middle), and sugR (bottom) mRNAs were analyzed using qRT-PCR. The mRNA levels in the ramA mutant carrying pCRC622 cultured in the absence of IPTG throughout are also shown (filled squares). The mRNA levels are presented relative to the value obtained from the wild type carrying PramA-lacZ before IPTG supplementation (0 h). Mean values obtained from at least three independent cultivations are shown with standard deviations.
qRT-PCR analysis revealed that the level of lacZ expression in the WT PramA-lacZ strain was unchanged during the period (Fig. 3, middle, circles). Before the addition of IPTG, the levels of lacZ expression in the ΔramA PramA-lacZ/pCRC622 and ΔramA-sugR PramA-lacZ/pCRC622 strains were 6- and 8-fold higher, respectively, than the level in the WT PramA-lacZ strain (Fig. 3, middle, squares and triangles). The level of lacZ expression in both the plasmid-carrying strains decreased about 60-fold within 0.5 h of IPTG supplementation (Fig. 3, middle, open symbols). When the ΔramA PramA-lacZ/pCRC622 strain was cultured in the absence of IPTG throughout, the level of lacZ expression remained 6-fold higher than that in the WT PramA-lacZ strain during the first 1 h and then decreased 2-fold in the subsequent 1 h (Fig. 3, middle, filled squares). This downregulation of PramA-lacZ expression in the absence of IPTG may be due to the leaky expression of ramA from the plasmid. These expression profiles of ramA and lacZ in the ramA mutant background in the absence of IPTG were comparable to those in the ramA-sugR mutant background (data not shown). These results indicated that RamA overexpressed from pCRC622 strongly repressed the ramA promoter activity.
We also examined the effects of overexpression of ramA on the expression profile of sugR under the same conditions. After the addition of IPTG, the level of sugR mRNA in the WT PramA-lacZ strain was nearly unchanged for 2 h (Fig. 3, bottom, circles). Before the addition of IPTG, the level of sugR expression in the ΔramA PramA-lacZ/pCRC622 strain was lower than that in the WT PramA-lacZ strain (Fig. 3, bottom, squares). The level of sugR mRNA in the ΔramA PramA-lacZ/pCRC622 strain increased 2.5-fold within 0.5 h of IPTG supplementation, and the level remained higher than that in the WT PramA-lacZ strain in the subsequent 1.5 h (Fig. 3, bottom, open squares). When the ΔramA PramA-lacZ/pCRC622 strain was cultured in the absence of IPTG during this period, the level of sugR mRNA remained lower than that in the WT PramA-lacZ strain in the first 1 h (Fig. 3A, bottom, filled squares). No difference in the level of sugR expression between these two strains was observed at the end of the period, probably due to the gradual increase in the level of ramA mRNA in the ΔramA PramA-lacZ/pCRC622 strain under the noninducing conditions. Thus, the changes in the level of sugR expression in the pCRC622-carrying strain seemed to be correlated with those in the level of ramA expression. This is consistent with the finding that RamA directly activates sugR expression (31).
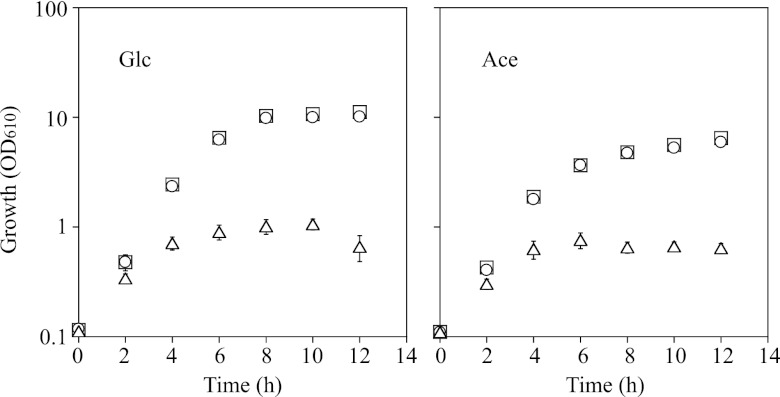
Addition of IPTG to the cell cultures in the exponential growth phase had no effect on the growth of these strains tested. However, when IPTG was supplemented at the start of culture, growth of the ramA mutant carrying pCRC622 was significantly retarded, and the final cell density was markedly lower than the density of cells cultured in the absence of IPTG (Fig. 4). The sugR-ramA mutant carrying pCRC622 showed the same growth profiles as the ramA mutant carrying pCRC622 (data not shown). These results indicate that overexpression of ramA is detrimental to the growth of C. glutamicum cells.
Fig 4.
Growth profiles of the ramA mutant carrying pCRC622 in A medium containing either 1% glucose (Glc) or 1% acetate (Ace) without (squares) or with IPTG. IPTG was added either 0 h (triangles) or 4 h (circles) after the start of culture. Mean values obtained from at least three independent cultivations are shown with standard deviations. OD610, optical density at 610 nm.
GlxR is important for the activation of ramA expression against repression by both SugR and RamA.
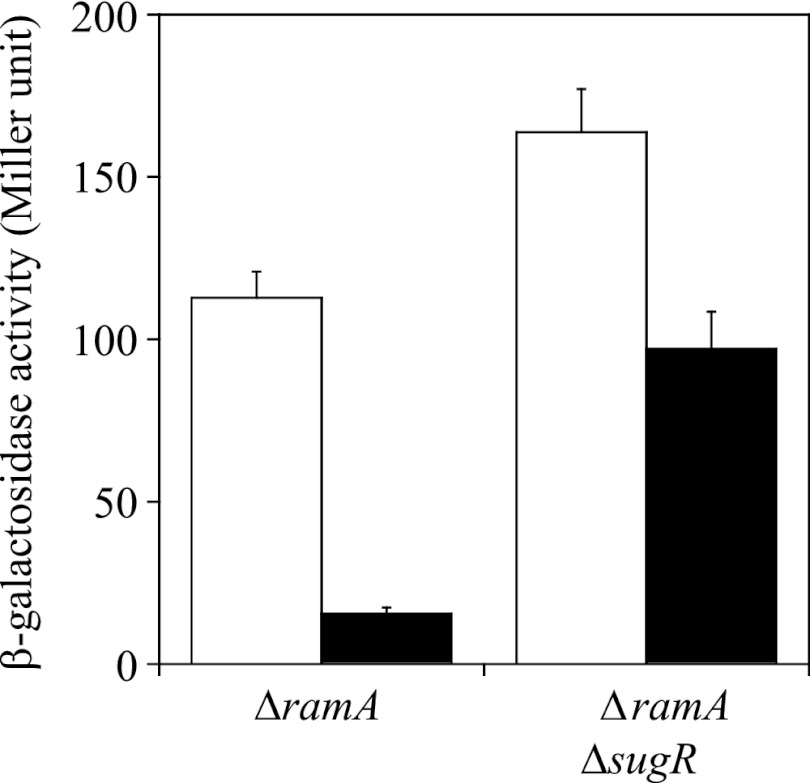
The level of expression of lacZ fused to PramA in the ramA and ramA-sugR mutants carrying pCRC622 in the absence of IPTG was much higher than that in the wild type, probably due to the low level of expression of ramA (Fig. 3). Therefore, we expected that the effect of the mutations in the GlxR binding site on ramA promoter activity could be evaluated quantitatively using these pCRC622-carrying strains, in contrast to the observation for the wild type, in which the activity of the mutated ramA promoter was undetectable. pCRC622 was introduced into the ramA and ramA-sugR mutants that chromosomally carry the PramA-lacZ fusion with the mutated GlxR binding site. The β-galactosidase activity of the resulting strains during growth on glucose in the absence of IPTG was determined as described earlier. The activity of the mutated ramA promoter in the ramA mutant carrying pCRC622 was more than 7-fold lower than that of the native one (Fig. 5). Meanwhile, the activity of the mutated ramA promoter in the ramA-sugR mutant carrying pCRC622 was less than 2-fold lower than that of the native one (Fig. 5). It was confirmed by qRT-PCR that there was no difference in the level of ramA mRNA between the strains carrying the lacZ gene under the control of the native and the mutated ramA promoters in the same genetic backgrounds (data not shown). These findings suggested that the GlxR-mediated activation has less of a contribution to the derepressed ramA expression. This is consistent with the observation that the same mutations reduced the ramA promoter activity to undetectable levels in the wild-type background but only by 2-fold in the genetic background of single and double deletion mutants of sugR and ramA (Table 2).
Fig 5.
Activity of the ramA promoter with (black bars) or without (white bars) mutations in the GlxR binding site in the ramA and ramA-sugR mutants carrying pCRC622 during growth on glucose without IPTG. The β-galactosidase activity of these strains carrying the PramA-lacZ fusion cultured for 8 h (in the stationary phase) was measured. Mean values obtained from at least three independent cultivations are shown with standard deviations.
RamB is not implicated in ramA expression.
It has been reported that the ramA promoter activity in the ramB mutant is comparable to that in the wild type (43). However, as the RamB binding site flanks the GlxR binding site on the ramA promoter region (Fig. 1), we posited that GlxR bound to the ramA promoter prevents RamB from binding to the adjacent site, which masks the effects of ramB deletion on ramA expression. However, this hypothesis was excluded because the activity of the ramA promoter with or without mutations in the GlxR binding site in the ramB mutant was comparable to that in the wild type (Table 2).
DISCUSSION
The current study shows that expression of ramA encoding a LuxR-type transcription regulator of various genes involved in central carbon metabolism is negatively and positively regulated by SugR and GlxR, respectively, in addition to negative autoregulation (43). Since both regulators' activity is modulated with biomolecules whose concentrations varied with the carbon sources used, it is conceivable that SugR and GlxR are involved in the carbon source-dependent regulation of ramA. However, it is interesting to note that the effects of inactivation of these regulators in any combinations tested on the ramA promoter activity appeared to be unaffected by the carbon source used. Because SugR represses genes involved in sugar uptake and metabolism in the absence of sugar and its repressor activity is inhibited by sugar metabolites (32, 35, 36, 55), the degree of derepression of most SugR target genes in the sugR mutant is higher in the absence of sugar than in its presence. However, the degree of derepression of ramA promoter activity in the sugR mutant in the presence of glucose was comparable to that in the presence of acetate. The discrepancy in the derepression levels in the sugR mutant between ramA and other SugR targets may be due to the negative autoregulation of ramA; the derepressed RamA in the sugR mutant in turn represses its own gene, which may lead to underestimation of the degree of derepression of ramA by relief of SugR repression, especially in the presence of acetate. It is also conceivable that relief of the RamA-mediated repression of the ramA promoter-lacZ fusion irrespective of the carbon source masks the effects of SugR inactivation on the ramA promoter activity in the ramA-sugR double mutant.
GlxR binds to its target sites in a cAMP-dependent manner in vitro. The intracellular cAMP levels of C. glutamicum are higher on glucose than on acetate (14–16). Although GlxR bound to the ramA promoter in a cAMP-dependent manner (Fig. 2), the ramA promoter activity in any of the strains examined in this study was decreased by the mutations in the GlxR binding site irrespective of the carbon source (Table 2). We previously showed that GlxR mediates not only glucose-dependent upregulation of the glycolytic genes gapA and pfk, encoding glyceraldehyde-3-phosphate dehydrogenase and phosphofructokinase, respectively, but also the carbon source-independent upregulation of genes for ATP synthase and cytochrome c oxidase (15). It has also been reported that the glyoxylate pathway genes aceA and aceB, which encode isocitrate lyase and malate synthase, respectively, are negatively regulated by GlxR in the presence of either glucose or acetate (54). These findings indicate that the intracellular cAMP levels and the regulatory roles of GlxR are not necessarily correlated. An unidentified regulator other than SugR and GlxR may be responsible for the upregulation of ramA observed in the presence of acetate compared to the level of regulation observed in the presence of glucose. Although previous DNA affinity purification with the ramA promoter region as a ligand identified neither SugR nor GlxR, it identified, in addition to RamA and RamB, a GntR-type transcriptional regulator (cg0764), which has not yet been characterized (43).
Deletion of ramA had the most prominent effects on the ramA promoter activity (Table 2), indicating that of the four global regulators investigated, expression of ramA is primarily regulated by RamA itself. The large increase in the ramA promoter activity in the ramA mutant background is due to relief of repression mediated not only by RamA but also by SugR. This is supported by the findings that the activity of the ramA promoter in the ramA mutant background was comparable to that in the sugR-ramA mutant background (Table 2). In this context, it should be noted that RamA acts as a transcription activator of sugR and sugR mRNA is markedly downregulated by ramA deletion (31). We further confirmed that β-galactosidase activity of the sugR promoter-lacZ fusion in the ramA mutant background was undetectable (data not shown). In this context, it is noteworthy that the overexpression of ramA markedly repressed the expression of the ramA promoter-lacZ fusion and enhanced sugR expression to some extent (Fig. 3). These findings suggest that C. glutamicum cells need to maintain ramA expression levels below a certain threshold under the strict negative control of RamA along with SugR. This idea may be supported by our observation that overexpression of ramA had a negative effect on growth (Fig. 4).
The strong direct and indirect negative autoregulation may explain the discrepancy between the effects of the GlxR binding site mutations on the level of ramA mRNA expression in strain KT27 and their effects on expression of the ramA promoter-lacZ fusion. Given that GlxR activates ramA, the decrease in ramA expression by the loss of GlxR binding in strain KT27 was possibly compensated for by alleviation of the repression by RamA itself, resulting in the wild-type level of ramA expression. On the other hand, in the promoter-reporter assays, the loss of GlxR binding to the ramA promoter that is fused to lacZ had no effect on expression of ramA at the original chromosomal locus, resulting in the undetectable level of β-galactosidase activity by RamA-mediated repression. Taken together, it is likely that the level of ramA expression is dynamically balanced by the GlxR-dependent activation and the RamA- and/or SugR-dependent repression in response to perturbation of extracellular and/or intracellular conditions. It is interesting to note that the GlxR-mediated activation appears to have a relatively minor role in ramA expression when the repression by either SugR or RamA is relieved, as shown in this study (Table 2 and Fig. 5). The detailed molecular mechanism of this regulation mediated by the three regulators is a subject for future studies.
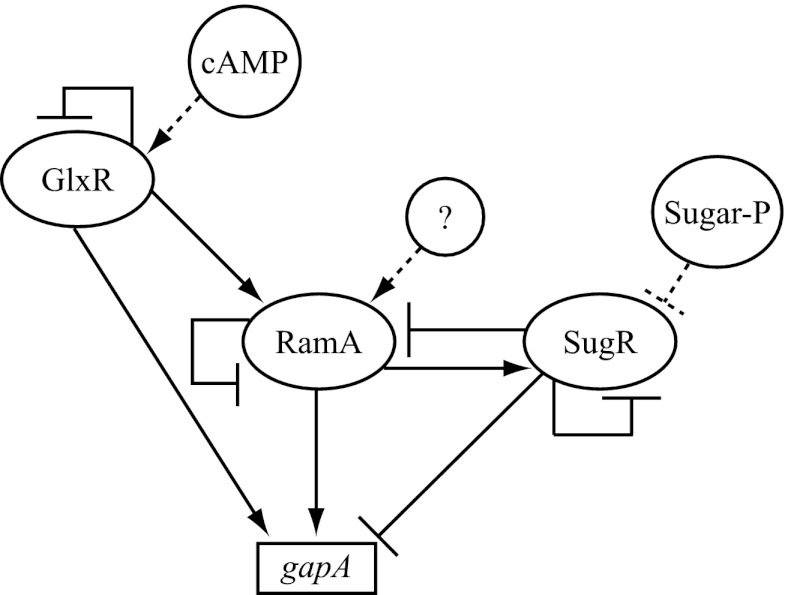
The current study demonstrated a new regulatory connection between GlxR and RamA, which has a great impact on the C. glutamicum genome-wide transcriptional regulatory network structure (56), because the connection not only expands the GlxR regulon by including the RamA regulon but also newly creates multiple feed-forward loops (FFLs) in the regulatory cascades. The FFL, which is one of the most significant network motifs found in a transcriptional regulatory network in both E. coli (57) and Saccharomyces cerevisiae (58), comprises two cascaded transcription factors that jointly regulate a common gene (59). The coherent FFL, in which the direct effect of the upper regulator on target gene expression is consistent with the indirect effect of that via regulation of the lower regulator, has been reported to contribute to sense signal persistency, thereby filtering noise or fluctuations in the environmental input signal (60). In contrast, the incoherent FFL, in which the direct and indirect effects of the upper regulator are opposite, accelerates the response of the regulatory system (61). In E. coli, genes for nonglucose sugar metabolism are under the control of the FFL composed of CRP and the carbon-specific transcriptional regulator to respond to a combination of the respective signals (59, 61). In the case of C. glutamicum gapA for a key glycolytic enzyme, one incoherent FFL composed of RamA and SugR and two coherent FFLs based on the newly identified regulatory interactions among GlxR, SugR, and RamA are involved in gene regulation (Fig. 6). On the basis of the presumed physiological function of each type of FFL, the coherent FFLs formed with GlxR-RamA and SugR-RamA are predicted to play a role in maintaining gapA expression levels in response to fluctuations in cAMP and sugar phosphate levels, whereas the incoherent FFL composed of RamA-SugR may be involved in a rapid response to physiological and/or environmental changes, which are sensed by RamA. Moreover, the FFLs formed by GlxR and RamA are possibly involved in regulation of other central carbon metabolism genes, i.e., ptsF, gltA, acn, sdhCAB, aceA, and aceB, encoding the fructose uptake phosphotransferase system, citrate synthase, aconitase, succinate dehydrogenase, isocitrate lyase, and malate synthase, respectively (35, 38, 45, 54, 62–65). Further insights into the biological roles of these complex regulatory connections will be provided by elucidation of the environmental signals to which RamA and RamB respond and how the intracellular cAMP levels are controlled. These important issues need to be addressed in future studies.
Fig 6.
Model of the transcriptional regulatory network involved in expression of gapA in C. glutamicum. Arrows indicate transcriptional activation, while T bars indicate repression. Effector molecules controlling the activity of transcriptional regulators are shown with circles and dotted lines. An unknown effector for RamA is indicated with a question mark. Sugar-P, sugar phosphate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript.
This work was partially supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00016-13.
REFERENCES
- 1. Wendisch VF, Bott M, Eikmanns BJ. 2006. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9:268–274 [DOI] [PubMed] [Google Scholar]
- 2. Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243–254 [DOI] [PubMed] [Google Scholar]
- 3. Inui M, Terasawa M, Yukawa H. 1999. Encyclopedia of bioprocess technology: fermentation, biocatalysis, and bioseparation. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 4. Kumagai H. 2000. Microbial production of amino acids in Japan. Adv. Biochem. Eng. Biotechnol. 69:71–85 [DOI] [PubMed] [Google Scholar]
- 5. Hermann T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155–172 [DOI] [PubMed] [Google Scholar]
- 6. Smith KM, Cho KM, Liao JC. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87:1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Pühler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25 [DOI] [PubMed] [Google Scholar]
- 8. Ikeda M, Nakagawa S. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99–109 [DOI] [PubMed] [Google Scholar]
- 9. Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058 [DOI] [PubMed] [Google Scholar]
- 10. Teramoto H, Inui M, Yukawa H. 2011. Transcriptional regulators of multiple genes involved in carbon metabolism in Corynebacterium glutamicum. J. Biotechnol. 154:114–125 [DOI] [PubMed] [Google Scholar]
- 11. Teramoto H, Inui M, Yukawa H. 2010. Regulation of genes involved in sugar uptake, glycolysis and lactate production in Corynebacterium glutamicum. Future Microbiol. 5:1475–1481 [DOI] [PubMed] [Google Scholar]
- 12. Krin E, Sismeiro O, Danchin A, Bertin PN. 2002. The regulation of enzyme IIAGlc expression controls adenylate cyclase activity in Escherichia coli. Microbiology 148:1553–1559 [DOI] [PubMed] [Google Scholar]
- 13. Reddy P, Kamireddi M. 1998. Modulation of Escherichia coli adenylyl cyclase activity by catalytic-site mutants of protein IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system. J. Bacteriol. 180:732–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim HJ, Kim TH, Kim Y, Lee HS. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 186:3453–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toyoda K, Teramoto H, Inui M, Yukawa H. 2011. Genome-wide identification of in vivo binding sites of GlxR, a cyclic AMP receptor protein-type regulator in Corynebacterium glutamicum. J. Bacteriol. 193:4123–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cha PH, Park SY, Moon MW, Subhadra B, Oh TK, Kim E, Kim JF, Lee JK. 2010. Characterization of an adenylate cyclase gene (cyaB) deletion mutant of Corynebacterium glutamicum ATCC 13032. Appl. Microbiol. Biotechnol. 85:1061–1068 [DOI] [PubMed] [Google Scholar]
- 17. Cocaign-Bousquet M, Monnet C, Lindley ND. 1993. Batch kinetics of Corynebacterium glutamicum during growth on various carbon substrates: use of substrate mixtures to localise metabolic bottlenecks. Appl. Microbiol. Biotechnol. 40:526–530 [Google Scholar]
- 18. Dominguez H, Cocaign-Bousquet M, Lindley ND. 1997. Simultaneous consumption of glucose and fructose from sugar mixtures during batch growth of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 47:600–603 [Google Scholar]
- 19. Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claes WA, Pühler A, Kalinowski J. 2002. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 184:2728–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merkens H, Beckers G, Wirtz A, Burkovski A. 2005. Vanillate metabolism in Corynebacterium glutamicum. Curr. Microbiol. 51:59–65 [DOI] [PubMed] [Google Scholar]
- 22. Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF. 2005. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71:5920–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67:305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teramoto H, Inui M, Yukawa H. 2009. Regulation of expression of genes involved in quinate and shikimate utilization in Corynebacterium glutamicum. Appl. Environ. Microbiol. 75:3461–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arndt A, Eikmanns BJ. 2007. The alcohol dehydrogenase gene adhA in Corynebacterium glutamicum is subject to carbon catabolite repression. J. Bacteriol. 189:7408–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kronemeyer W, Peekhaus N, Krämer R, Sahm H, Eggeling L. 1995. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J. Bacteriol. 177:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kotrbova-Kozak A, Kotrba P, Inui M, Sajdok J, Yukawa H. 2007. Transcriptionally regulated adhA gene encodes alcohol dehydrogenase required for ethanol and n-propanol utilization in Corynebacterium glutamicum R. Appl. Microbiol. Biotechnol. 76:1347–1356 [DOI] [PubMed] [Google Scholar]
- 28. Hayashi M, Mizoguchi H, Shiraishi N, Obayashi M, Nakagawa S, Imai J, Watanabe S, Ota T, Ikeda M. 2002. Transcriptome analysis of acetate metabolism in Corynebacterium glutamicum using a newly developed metabolic array. Biosci. Biotechnol. Biochem. 66:1337–1344 [DOI] [PubMed] [Google Scholar]
- 29. Muffler A, Bettermann S, Haushalter M, Horlein A, Neveling U, Schramm M, Sorgenfrei O. 2002. Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. J. Biotechnol. 98:255–268 [DOI] [PubMed] [Google Scholar]
- 30. Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99–122 [DOI] [PubMed] [Google Scholar]
- 31. Toyoda K, Teramoto H, Inui M, Yukawa H. 2009. Involvement of the LuxR-type transcriptional regulator, RamA, in regulation of expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum. J. Bacteriol. 191:968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toyoda K, Teramoto H, Inui M, Yukawa H. 2008. Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl. Microbiol. Biotechnol. 81:291–301 [DOI] [PubMed] [Google Scholar]
- 33. Toyoda K, Teramoto H, Inui M, Yukawa H. 2009. Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding l-lactate dehydrogenase in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 83:315–327 [DOI] [PubMed] [Google Scholar]
- 34. Tanaka Y, Teramoto H, Inui M, Yukawa H. 2008. Regulation of expression of general components of the phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) by the global regulator SugR in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 78:309–318 [DOI] [PubMed] [Google Scholar]
- 35. Engels V, Lindner SN, Wendisch VF. 2008. The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J. Bacteriol. 190:8033–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaigalat L, Schlueter JP, Hartmann M, Mormann S, Tauch A, Pühler A, Kalinowski J. 2007. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 8:104 doi:10.1186/1471-2199-8-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Engels V, Wendisch VF. 2007. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 189:2955–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cramer A, Gerstmeir R, Schaffer S, Bott M, Eikmanns BJ. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 188:2554–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohl TA, Tauch A. 2009. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J. Biotechnol. 143:239–246 [DOI] [PubMed] [Google Scholar]
- 40. Kohl TA, Baumbach J, Jungwirth B, Pühler A, Tauch A. 2008. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J. Biotechnol. 135:340–350 [DOI] [PubMed] [Google Scholar]
- 41. Auchter M, Cramer A, Huser A, Ruckert C, Emer D, Schwarz P, Arndt A, Lange C, Kalinowski J, Wendisch VF, Eikmanns BJ. 2011. RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J. Biotechnol. 154:126–139 [DOI] [PubMed] [Google Scholar]
- 42. Blombach B, Arndt A, Auchter M, Eikmanns BJ. 2009. l-Valine production during growth of pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum in the presence of ethanol or by inactivation of the transcriptional regulator SugR. Appl. Environ. Microbiol. 75:1197–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cramer A, Eikmanns BJ. 2007. RamA, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to negative autoregulation. J. Mol. Microbiol. Biotechnol. 12:51–59 [DOI] [PubMed] [Google Scholar]
- 44. Jungwirth B, Emer D, Brune I, Hansmeier N, Pühler A, Eikmanns BJ, Tauch A. 2008. Triple transcriptional control of the resuscitation promoting factor 2 (rpf2) gene of Corynebacterium glutamicum by the regulators of acetate metabolism RamA and RamB and the cAMP-dependent regulator GlxR. FEMS Microbiol. Lett. 281:190–197 [DOI] [PubMed] [Google Scholar]
- 45. Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 47. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 48. Inui M, Suda M, Okino S, Nonaka H, Puskás LG, Vertès AA, Yukawa H. 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504 [DOI] [PubMed] [Google Scholar]
- 49. Ozaki A, Katsumata R, Oka T, Furuya A. 1984. Functional expression of the genes of Escherichia coli in gram-positive Corynebacterium glutamicum. Mol. Gen. Genet. 196:175–178 [DOI] [PubMed] [Google Scholar]
- 50. Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182–196 [DOI] [PubMed] [Google Scholar]
- 51. Toyoda K, Teramoto H, Inui M, Yukawa H. 2009. The ldhA gene, encoding fermentative l-lactate dehydrogenase of Corynebacterium glutamicum, is under the control of positive feedback regulation mediated by LldR. J. Bacteriol. 191:4251–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H. 1993. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res. Microbiol. 144:181–185 [DOI] [PubMed] [Google Scholar]
- 53. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 54. Park SY, Moon MW, Subhadra B, Lee JK. 2010. Functional characterization of the glxR deletion mutant of Corynebacterium glutamicum ATCC 13032: involvement of GlxR in acetate metabolism and carbon catabolite repression. FEMS Microbiol. Lett. 304:107–115 [DOI] [PubMed] [Google Scholar]
- 55. Dietrich C, Nato A, Bost B, Le Marechal P, Guyonvarch A. 2009. Regulation of ldh expression during biotin-limited growth of Corynebacterium glutamicum. Microbiology 155:1360–1375 [DOI] [PubMed] [Google Scholar]
- 56. Schröder J, Tauch A. 2010. Transcriptional regulation of gene expression in Corynebacterium glutamicum: the role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol. Rev. 34:685–737 [DOI] [PubMed] [Google Scholar]
- 57. Shen-Orr SS, Milo R, Mangan S, Alon U. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64–68 [DOI] [PubMed] [Google Scholar]
- 58. Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298:824–827 [DOI] [PubMed] [Google Scholar]
- 59. Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U. S. A. 100:11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mangan S, Zaslaver A, Alon U. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197–204 [DOI] [PubMed] [Google Scholar]
- 61. Mangan S, Itzkovitz S, Zaslaver A, Alon U. 2006. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J. Mol. Biol. 356:1073–1081 [DOI] [PubMed] [Google Scholar]
- 62. van Ooyen J, Emer D, Bussmann M, Bott M, Eikmanns BJ, Eggeling L. 2011. Citrate synthase in Corynebacterium glutamicum is encoded by two gltA transcripts which are controlled by RamA, RamB, and GlxR. J. Biotechnol. 154:140–148 [DOI] [PubMed] [Google Scholar]
- 63. Emer D, Krug A, Eikmanns BJ, Bott M. 2009. Complex expression control of the Corynebacterium glutamicum aconitase gene: identification of RamA as a third transcriptional regulator besides AcnR and RipA. J. Biotechnol. 140:92–98 [DOI] [PubMed] [Google Scholar]
- 64. Bussmann M, Emer D, Hasenbein S, Degraf S, Eikmanns BJ, Bott M. 2009. Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J. Biotechnol. 143:173–182 [DOI] [PubMed] [Google Scholar]
- 65. Tanaka Y, Okai N, Teramoto H, Inui M, Yukawa H. 2008. Regulation of the expression of phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) genes in Corynebacterium glutamicum R. Microbiology 154:264–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.