Abstract
Amino acid catabolism in Thermococcales is presumed to proceed via three steps: oxidative deamination of amino acids by glutamate dehydrogenase (GDH) or aminotransferases, oxidative decarboxylation by 2-oxoacid:ferredoxin oxidoreductases (KOR), and hydrolysis of acyl-coenzyme A (CoA) by ADP-forming acyl-CoA synthetases (ACS). Here, we performed a genetic examination of enzymes involved in Glu catabolism in Thermococcus kodakarensis. Examination of amino acid dehydrogenase activities in cell extracts of T. kodakarensis KUW1 (ΔpyrF ΔtrpE) revealed high NADP-dependent GDH activity, along with lower levels of NAD-dependent activity. NADP-dependent activities toward Gln/Ala/Val/Cys and an NAD-dependent threonine dehydrogenase activity were also detected. In KGDH1, a gene disruption strain of T. kodakarensis GDH (Tk-GDH), only threonine dehydrogenase activity was detected, indicating that all other activities were dependent on Tk-GDH. KGDH1 could not grow in a medium in which growth was dependent on amino acid catabolism, implying that Tk-GDH is the only enzyme that can discharge the electrons (to NADP+/NAD+) released from amino acids in their oxidation to 2-oxoacids. In a medium containing excess pyruvate, KGDH1 displayed normal growth, but higher degrees of amino acid catabolism were observed compared to those for KUW1, suggesting that Tk-GDH functions to suppress amino acid oxidation and plays an anabolic role under this condition. We further constructed disruption strains of 2-oxoglutarate:ferredoxin oxidoreductase and succinyl-CoA synthetase. The two strains displayed growth defects in both media compared to KUW1. Succinate generation was not observed in these strains, indicating that the two enzymes are solely responsible for Glu catabolism among the multiple KOR and ACS enzymes in T. kodakarensis.
INTRODUCTION
Among the archaea, members of the Thermococcales are anaerobic heterotrophs that utilize a wide range of organic compounds, including amino acids, a variety of sugars, and organic acids such as pyruvate. When available, they use elemental sulfur as the terminal electron acceptor and are also capable of hydrogen fermentation (1, 2). Extensive research has been carried out on the metabolism of these organisms, and unique enzymes and pathways have been discovered (3–5).
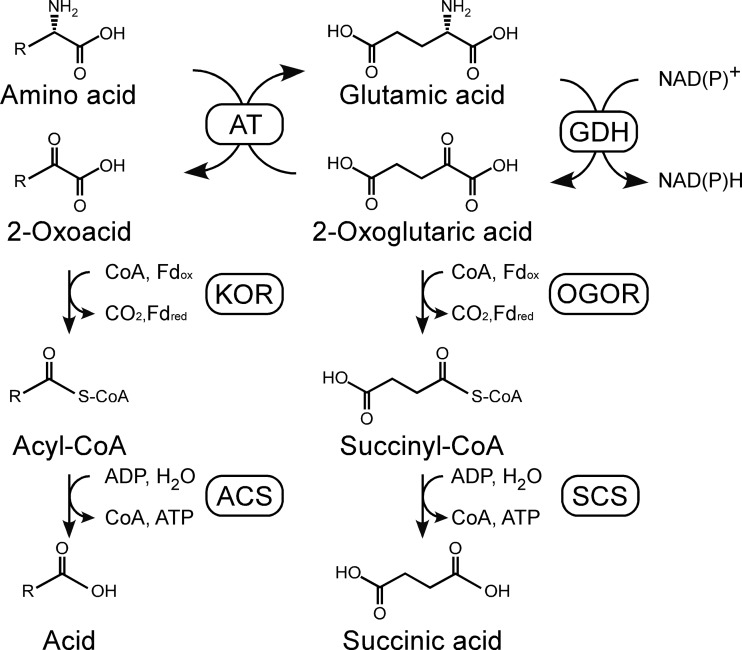
In terms of amino acid catabolism in Thermococcales, biochemical examinations have been carried out on a number of enzymes, including glutamate dehydrogenase (GDH), amino acid:2-oxoacid aminotransferase (AT), 2-oxoacid (2-ketoacid):ferredoxin oxidoreductase (KOR), and NDP-forming acyl-coenzyme A (CoA) synthetase (ACS). Based on these studies, amino acid catabolism in Thermococcales is presumed to proceed via three steps (6): (i) the oxidative deamination of amino acids, resulting in the formation of 2-oxoacids (catalyzed by GDH or AT), (ii) oxidative decarboxylation of 2-oxoacids to their corresponding acyl-CoAs (catalyzed by KOR), and (iii) hydrolysis of acyl-CoA coupled to substrate-level phosphorylation (catalyzed by ACS) (Fig. 1).
Fig 1.
Illustration of the enzymes presumed to be involved in amino acid catabolism in T. kodakarensis. ACS, ADP-forming acyl-CoA synthetase; AT, amino acid aminotransferase; GDH, glutamate dehydrogenase; KOR, 2-oxoacid:ferredoxin oxidoreductase; OGOR, 2-oxoglutarate:ferredoxin oxidoreductase; SCS, ADP-forming succinyl-CoA synthetase.
GDH is considered to play a central role in metabolism, as it is one of the most abundant proteins in Thermococcales cells, exceeding 10% of total cytoplasmic protein in the case of P. furiosus and T. kodakarensis (7, 8). In general, the Thermococcales GDHs are hexamers and accept both NAD and NADP as electron carriers, mostly with a preference for NADP (7–17). Concerning the ATs, a number of enzymes with different substrate specificities have been examined, including alanine aminotransferases (AlaAT) (18), aspartate aminotransferases (AspAT) (19), aromatic aminotransferases (AroAT) (19–22), and alanine:glyoxylate aminotransferases (23).
After the initial oxidative deamination of amino acids, the resulting 2-oxoacids are converted to acyl-CoA via the function of CoA-dependent KORs (6). KORs consist of four subunits (α, β, γ, and δ) or their fusion proteins, and multiple KOR gene clusters are present on the genomes of Thermococcales. Extensive examinations of the individual KORs from P. furiosus and/or T. litoralis have been performed by the group of Michael Adams, revealing the substrate preferences of four KORs. Studies have also been performed on enzymes from T. kodakarensis and T. profundus. Pyruvate:ferredoxin oxidoreductase (POR) is active toward the 2-oxoacids generated from Ala (pyruvate) and, to a lesser extent, Leu (24–28). 2-Oxoisovalerate:ferredoxin oxidoreductase (VOR) prefers 2-oxoacids generated from a relatively wide range of hydrophobic amino acids (29, 30). Indolepyruvate:ferredoxin oxidoreductase (IOR) recognizes 2-oxoacids deriving from aromatic amino acids (31–33), and 2-oxoglutarate:ferredoxin oxidoreductase (OGOR or KGOR) is relatively specific for 2-oxoglutarate generated from Glu (34). In addition, the genome sequences suggest the presence of a fifth KOR (XOR) in Thermococcales (6), which is, at present, still uncharacterized and whose substrate specificity is unknown.
The final reaction, which is the hydrolysis of acyl-CoA coupled to substrate-level phosphorylation, is catalyzed by ADP-forming acyl-CoA synthetases (35–39). These enzymes are tetrameric enzymes consisting of two distinct α and β subunits (α2β2). Five α subunit homologs and two β subunit homologs are found on all of the sequenced genomes of Pyrococcus and Thermococcus species, with the α subunit presumed to be responsible for specificity toward the acyl moiety (40). Among the five α subunits, two have been designated to be components of acetyl-CoA synthetases I and II (ACS I and II), based on studies of the enzymes from P. furiosus (35, 37, 41, 42). ACS I and II exhibit activity not only toward acetyl-CoA but also toward branched-chain acyl-CoAs. ACS II displays further activity toward aryl-CoAs (37). A third α subunit (TK1880) from T. kodakarensis, along with a β subunit, forms an acyl-CoA synthetase with a strict preference for succinyl-CoA, and it has been designated succinyl-CoA synthetase (SCS) (40). The substrate specificities of the remaining two members of the ACSs have not been published.
In this study, we have carried out a genetic examination of the three major enzymes considered to be involved in Glu catabolism, GDH, OGOR, and SCS. The GDH from T. kodakarensis (Tk-GDH) is shown to be the only relevant amino acid dehydrogenase in this organism involved in amino acid catabolism. Our results also indicate that OGOR and SCS are the only relevant KOR and ACS, respectively, involved in the conversion of Glu to succinate.
MATERIALS AND METHODS
Strains and culture conditions.
Routine cultivation of T. kodakarensis KOD1 (43–45) and mutant strains (Table 1) was performed under anaerobic conditions at 85°C in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA). ASW-YT medium was composed of 0.8× artificial seawater (ASW), 5.0 g liter−1 yeast extract, and 5.0 g liter−1 tryptone. Sodium pyruvate (5 g liter−1) and elemental sulfur (2 g liter−1) were supplemented prior to inoculation to prepare ASW-YT-Pyr and ASW-YT-S0 medium, respectively. The composition of ASW-AA medium is described elsewhere (46, 47). In all liquid media, resazurin sodium salt (0.5 mg liter−1) was added as an oxygen indicator, and 5.0% Na2S · 9H2O was added until the medium became colorless. In the case of plate culture, Gelrite (10 g liter−1) was added to solidify ASW-YT medium. Specific modifications of the medium to select individual mutant strains are described below. Escherichia coli strain DH5α, used for plasmid construction and amplification, was cultivated at 37°C in LB medium (5 g liter−1 yeast extract, 10 g liter−1 tryptone, 10 g liter−1 NaCl) containing ampicillin (100 mg liter−1). Unless mentioned otherwise, all components were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Table 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| T. kodakaraensis | ||
| KOD1 | Wild type | 43, 45 |
| KU216 | ΔpyrF | 49 |
| KUW1 | KU216 ΔtrpE | 49 |
| KGDH1 | KUW1 Δgdh | This study |
| KGDH1C | KGDH1 pLC64-gdh | This study |
| KOGOR1 | KU216 Δogor | This study |
| KSCS1 | KU216 Δscs | This study |
| Plasmids | ||
| pUD3 | pUC118 (TaKaRa) derivative; pyrF marker cassette (PpyrF::pyrF) | 48 |
| pUD3-Δgdh | pUD3 derivative; Δgdh | This study |
| pUD3-Δogor | pUD3 derivative; Δogor | This study |
| pUD3-Δscs | pUD3 derivative; Δscs | This study |
| pLC64 | trpE | 50 |
| pLC64-gdh | pLC64 derivative; gdh expression cassette (Pgdh::gdh::Tgdh) | This study |
Disruption of the GDH, OGOR, and SCS genes.
In disrupting the Tk-GDH (TK1431) gene, the entire coding region, along with 87 bp of the 5′-flanking region and 85 bp of the 3′-flanking region, was deleted. In the case of OGOR (α:TK1125, β:TK1124, γ:TK1123, δ:TK1131), as the genes were clustered with other genes, a region starting from base number 109 of TK1125 and ending in base number 435 of TK1123 was deleted. In the case of SCS (α:TK1880, β:TK0943), the entire coding region of TK1880 was deleted. Each region we intended to disrupt was amplified along with their 5′- and 3′-flanking regions (984 to 1,100 bp each) from T. kodakarensis KOD1 genomic DNA using the primer sets Dgdh-F1/Dgdh-R1, Dogor-F1/Dogor-R1, and Dscs-F1/Dscs-R1, (see Table S1 in the supplemental material). The fragments were inserted into the multiple cloning site of plasmid pUD3, which derives from pUC118 and harbors the pyrF gene of T. kodakarensis inserted in the ApaI site (48). In order to remove the regions to be deleted, inverse PCR was performed on the three plasmids with the corresponding primer sets Dgdh-F2/Dgdh-R2, Dogor-F2/Dogor-R2, and Dscs-F2/Dscs-R2, followed by self-ligation. The sequences of the inserted regions were confirmed for the absence of unintended mutations, and the plasmids were introduced into T. kodakarensis KUW1 (ΔpyrF ΔtrpE) for Tk-GDH disruption and KU216 (ΔpyrF) for OGOR and SCS disruption. Restriction and modification enzymes were purchased from Toyobo (Osaka, Japan) and TaKaRa (Otsu, Japan). Plasmid DNA was isolated with Quantum Prep from Bio-Rad (Hercules, CA). KOD plus (Toyobo) was used as a polymerase for PCR, and a Wizard SV gel and PCR clean-up system (Promega, Tokyo, Japan) was used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed using a BigDye Terminator cycle sequencing kit, version 3.1, and a model 3130 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Transformation of T. kodakarensis KUW1 and KU216 (49) was performed as follows. Cells grown in ASW-YT-S0 until the late log phase were harvested, washed, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After treatment with 3.0 μg of plasmid DNA and further incubation on ice for 1 h, cells were cultivated in ASW-AA medium (without uracil) with elemental sulfur for 2 to 5 days at 85°C. Cells were consecutively cultivated under the same conditions to enrich the desired transformants that display uracil prototrophy. Cells were then spread onto ASW-YT solid medium supplemented with 5-fluoroorotic acid (5-FOA; 10 g liter−1) and 60 mM NaOH. For Tk-GDH disruption, amycol (5 g liter−1), a mixture of maltooligosaccharides (Nippon Starch Chemical, Osaka, Japan), was further added to the medium. For OGOR and SCS disruption, sodium pyruvate (5 g liter−1), 0.2% (vol/vol) polysulfide solution, and sodium glutamate (1 g liter−1) were further added. Only cells that have undergone a second recombination that removes the pyrF gene can grow in the presence of 5-FOA. Cells were grown for 2 days at 85°C for colony formation, and transformants were isolated and cultivated in ASW-YT medium supplemented with amycol (Tk-GDH disruption) or in ASW-YT-S0 (OGOR and SCS disruption). The genotypes of the transformants were analyzed by PCR using primer set Dgdh-F3/Dgdh-R3, Dogor-F3/Dogor-R3, or Dscs-F3/Dscs-R3. Transformants whose amplified DNA products displayed the expected size were chosen, and relevant sequences were confirmed. The strains were designated T. kodakarensis KGDH1, KOGOR1, and KSCS1, respectively.
Introduction of the GDH gene.
In order to reintroduce the Tk-GDH gene into strain KGDH1, a Tk-GDH expression cassette was inserted into the autonomously replicating plasmid pLC64, which harbors a trpE gene for selection (50). The plasmid was a kind gift from Thomas Santangelo and John Reeve at Ohio State University. The entire coding region of the Tk-GDH gene, along with 400 bp of 5′-flanking region and 110 bp of 3′-flanking region, was amplified from T. kodakarensis genomic DNA with the primer set Egdh-F1/Egdh-R1. This fragment was inserted in the EcoRV site of pLC64, resulting in the plasmid pLC64-gdh, and sequenced. Transformation of KGDH1 with pLC64-gdh was performed as follows. Cells at the late log phase were harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After addition of 3.0 μg plasmid and further incubation on ice for 1 h, the samples were incubated for 1 min at 85°C. After incubating on ice for 15 min, cells were cultivated in ASW-AA medium with 2 g liter−1 elemental sulfur and 10 μg ml−1 uracil but without Trp for 5 days at 85°C in order to enrich cells displaying Trp prototrophy. Cells were subsequently spread onto ASW-YT plate medium supplemented with 0.2% (vol/vol) polysulfide solution. After cultivation for 2 days at 85°C, transformants were isolated and cultivated in ASW-YT-S0 medium. The presence of the plasmid in the transformants was confirmed by PCR using the primer set Egdh-F2/Egdh-R2, and one transformant, designated T. kodakarensis KGDH1C, was chosen for further analysis.
Activity measurements of amino acid dehydrogenase in cell extracts of T. kodakarensis.
Amino acid dehydrogenase activity was examined in the direction of amino acid oxidation [amino acid + NAD(P)+ → 2-oxoacid + NAD(P)H + NH3] (8). The generation rate of NAD(P)H was measured by monitoring the increase in absorbance at 340 nm. Measurements were performed at 80°C with a UV-1650PC spectrophotometer (Shimadzu, Kyoto, Japan) equipped with a thermal control unit. Reaction mixtures contained 10 mM each amino acid, 1 mM NAD+ or NADP+ (Oriental Yeast, Tokyo, Japan), and cell extracts of T. kodakarensis (10 or 100 μg ml−1) in 100 mM sodium phosphate (pH 8.0). Due to low solubility, Tyr was examined at a concentration of 1 mM. Reactions were initiated by the addition of the cell extracts. In order to obtain cell extracts, T. kodakarensis KUW1, KGDH1, and KGDH1C cells were cultivated in ASW-YT-S0 or ASW-YT-Pyr media. After harvesting, cells were suspended in 50 mM Tris-HCl buffer (pH 7.5), sonicated, and centrifuged (20,000 × g, 15 min). The supernatant was ultrafiltrated with 50 mM Tris-HCl buffer (pH 7.5) using an Amicon Ultra-4 Centrifugal Filter Unit 10K (Millipore, Bedford, MA) in order to remove low-molecular-weight compounds, such as amino acids. Protein concentrations were determined using a protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
Analyses of metabolites in the growth medium.
T. kodakarensis KUW1, KGDH1, KOGOR1, and KSCS1 cells were cultivated in ASW-YT-Pyr media at 85°C. After 4 and 8 h, aliquots of the cultures were taken, cooled on ice, and centrifuged (17,000 × g, 5 min, 4°C). The supernatants were filtered with USY-1 disposable ultrafilter units (Advantec, Tokyo, Japan). In analyses of amino acids, 10-μl aliquots were applied to a Shim-pack Amino-Li column (100 by 6.0 mm; Shimadzu) at 39°C. The mobile phase was a gradient of 7% 2-methoxyethanol in 50 mM lithium citrate buffer (pH 2.6) and 100 mM lithium citrate buffer (pH 10) at a flow rate of 0.6 ml min−1. Amino acids were subjected to postcolumn derivatization using o-phthalaldehyde and detected with a fluorescence detector (RF-10AXL; Shimadzu) with excitation and emission wavelengths of 350 and 450 nm, respectively. For analyses of organic acids, 50-μl aliquots were applied to two consecutive Shim-pack SCR-102H (300 by 8 mm) columns at 45°C. The mobile phase was 5 mM p-toluenesulfonic acid at a flow rate of 0.8 ml min−1. An electric conductivity detector (CDD-6A; Shimadzu) was used for detection.
Thermostabilities of metabolites.
For Glu, Ala, Val, Leu, pyruvate, succinate, and acetate, whose concentrations in the medium could clearly be quantified by high-performance liquid chromatography (HPLC) with the methods described above, thermal degradation was measured after 4 and 8 h of incubation of the medium at 85°C. For 2-oxoglutarate, a reductive GDH reaction was applied. 2-Oxoglutaric acid (2 mM) was incubated at 85°C in 200 mM sodium phosphate buffer (pH 7.0). After cooling on ice, a 50-μl aliquot was added into 950 μl of a GDH reaction mixture consisting of 0.2 mM NADPH (Oriental Yeast), 5 mM NH4Cl, and 12 U l-glutamic dehydrogenase from bovine liver (Sigma-Aldrich, St. Louis, MO) in 200 mM sodium phosphate buffer (pH 7.0). The mixture was incubated at 25°C, and the amount of 2-oxoglutarate was determined spectrophotometrically at 340 nm. For succinyl-CoA, the hydroxamate method was applied (40, 51). After incubation of 10 mM succinyl-CoA at 85°C in 200 mM sodium phosphate buffer (pH 7.0), a 100-μl aliquot was mixed with 250 μl of 50 mM hydroxylamine and incubated for 30 min at room temperature. After the reaction, 200 μl of 20% trichloroacetic acid and 150 μl of 1 M FeCl3 were added to the mixture, and formation of the iron succinohydroxymate complex derived from succinyl-CoA was determined spectrophotometrically at 520 nm.
RESULTS
Examination of amino acid dehydrogenase activities in T. kodakarensis.
We first examined whether there were any other amino acid dehydrogenase activities in the cell extracts of T. kodakarensis in addition to GDH. Cells were grown in ASW-YT-S0 and ASW-YT-Pyr. Cell growth in the former medium can be considered to be dependent on amino acid catabolism, whereas the metabolism of pyruvate, a 2-oxoacid, is dominant in the latter medium. NAD+ and NADP+ were used as electron acceptors. As expected, high levels of dehydrogenase activity toward Glu were observed in the extracts of cells grown in ASW-YT-S0 (see Table S2 in the supplemental material). NADP+-dependent activity was three orders of magnitude higher than that with NAD+. Similar levels of activity were also observed in ASW-YT-Pyr. Concerning other amino acids, although much lower than the GDH activity, a significant level of NAD+-dependent threonine dehydrogenase (ThrDH) activity was observed. We also observed low levels of NADP+-dependent dehydrogenase activities toward Gln and Cys in both media. Interestingly, NADP+-dependent activity toward Ala and Val was detected only in ASW-YT-Pyr medium. Activities on other amino acids were not detected.
Gene disruption of the GDH gene.
As activity measurements confirmed that GDH was the major amino acid dehydrogenase in T. kodakarensis cells, the gdh gene (TK1431) was disrupted, and its effect on the amino acid metabolism of T. kodakarensis was examined. T. kodakarensis KUW1 (ΔpyrF ΔtrpE) was used as the host strain, and as the gdh gene was not clustered with other genes, the entire coding region was deleted. By using pyrF as a selection/counterselection marker, a Δgdh strain was selected based on uracil prototrophy and resistance to 5-fluoroorotic acid (see Fig. S1A in the supplemental material). Gene disruption was confirmed by PCR (see Fig. S1B) and sequencing, and the strain was designated T. kodakarensis KGDH1.
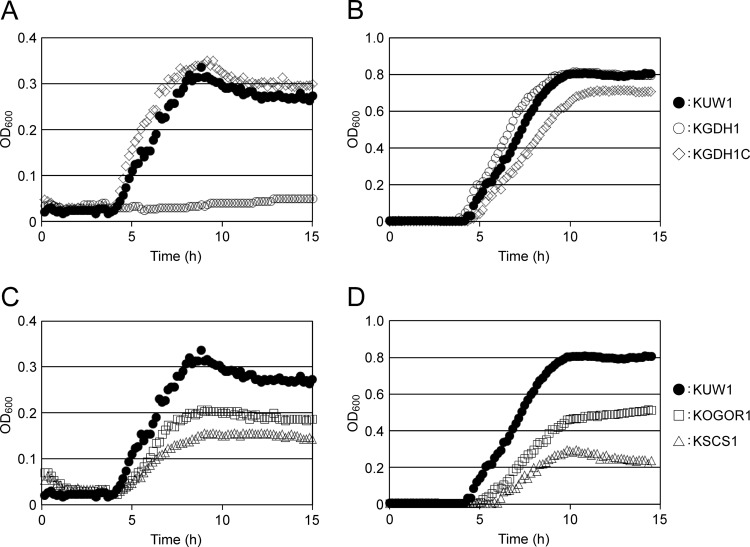
KGDH1 cells were grown in ASW-YT-S0 and ASW-YT-Pyr media. Although high levels of GDH activity were observed in cell extracts of the host strain KUW1 grown in ASW-YT-Pyr medium (see Table S2 in the supplemental material), disruption of the gdh gene did not have effects on cell growth in this medium (Fig. 2). In contrast, KGDH1 cells could not grow at all in ASW-YT-S0 medium. In order to exclude the possibility that the phenotypic changes observed in KGDH1 cells were due to polar effects brought about by gdh disruption, the gdh gene, with its flanking regions, was inserted into the autonomously replicating plasmid pLC64, and the plasmid (pLC64-gdh) was introduced into KGDH1 (see Fig. S1). Cells harboring pLC64-gdh were selected by Trp prototrophy, and the strain was designated KGDH1C. As expected, growth in ASW-YT-S0 medium was restored by reintroduction of the gdh gene (Fig. 2), confirming that Tk-GDH is essential for T. kodakarensis to grow on amino acids coupled with the reduction of elemental sulfur. The absence of Tk-GDH protein from KGDH1 cells and its presence in KGDH1C cells were confirmed by SDS-PAGE followed by Coomassie brilliant blue staining and Western blot analysis using antisera raised against Tk-GDH (Fig. 3).
Fig 2.
Growth of T. kodakarensis KUW1, KGDH1, and KGDH1C (A and B), as well as KOGOR1 and KSCS1 (C and D), in ASW-YT-S0 (A and C) and ASW-YT-Pyr (B and D). KUW1, closed circles; KGDH1, open circles; KGDH1C, open diamonds; KOGOR1, open squares; KSCS1, open triangles.
Fig 3.
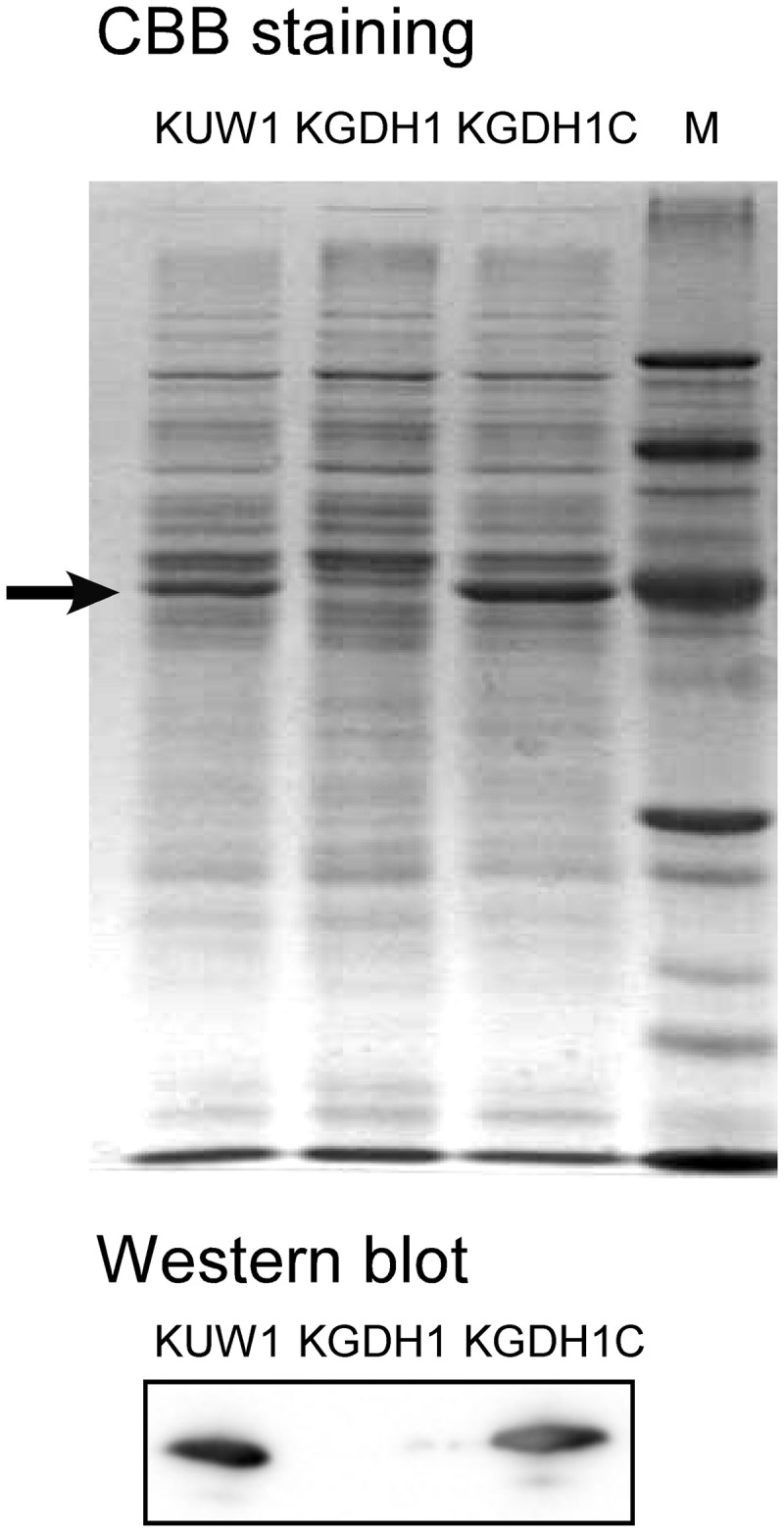
SDS-PAGE gel stained with Coomassie brilliant blue (CBB) (upper) and Western blot analysis using antisera against GDH (lower) to confirm GDH protein expression in the host strain, the gdh disruption strain KGDH1, and the gdh disruption strain complemented with pLC64-gdh, strain KGDH1C. The arrow indicates the position of the GDH protein in T. kodakarensis.
As KGDH1 could not grow in ASW-YT-S0, cells were grown in ASW-YT-Pyr medium, and their cell extracts were examined for amino acid dehydrogenase activity. Neither NAD+- nor NADP+-dependent GDH activity could be observed, confirming that the TK1431 protein was responsible for these activities (see Table S2 in the supplemental material). In addition, the low levels of activity toward Gln, Ala, Val, and Cys observed in KUW1 cells were not detected in KGDH1 cells, indicating that these activities were dependent on Tk-GDH. This could be a result of Tk-GDH directly recognizing these amino acids and/or the coupling of other enzymes, such as aminotransferases, with Tk-GDH. When the Tk-GDH gene was reintroduced into KGDH1, activities toward Glu, Gln, Ala, Val, and Cys all were restored, further supporting the involvement of Tk-GDH in these activities. On the contrary, the NAD+-dependent ThrDH activity was still observed in KGDH1 cells, and we could even observe a slight increase in KGDH1 cells compared to levels observed in KUW1 cells. This indicates that a ThrDH activity, which is independent of Tk-GDH, is present in T. kodakarensis at significant levels. Although low, activity toward Ser was detected specifically in KGDH1 cells. Interestingly, ThrDH activity seems to respond to the presence/absence of Tk-GDH, as the increase observed in KGDH1 cells is no longer observed in KGDH1C cells.
Disruption of genes encoding OGOR and SCS.
After the initial oxidative deamination of Glu by GDH or aminotransferases, the resulting 2-oxoglutarate is presumed to be converted to succinyl-CoA via the function of OGOR. The OGOR of P. furiosus is encoded by PF1771 (α), PF1772 (β), PF1773 (γ), and PF1767 (δ). Genes in T. kodakarensis that most resemble the P. furiosus OGOR genes are α:TK1125, β:TK1124, γ:TK1123, and δ:TK1131. Succinyl-CoA is further hydrolyzed to succinate and CoA coupled to substrate-level phosphorylation by the function of SCS. An ACS that is specific toward succinyl-CoA and encoded by α:TK1880 and β:TK0943 has previously been identified in T. kodakarensis (40).
In the case of OGOR, a region, starting from base number 109 of TK1125 and ending in base number 435 of TK1123, was deleted (see Fig. S1 in the supplemental material). The reason the genes were not deleted in their entirety was that TK1125 and TK1123 overlapped with their adjacent genes, TK1126 and TK1122, respectively, and care was taken not to disturb their transcriptional termination. In the case of SCS, the gene did not cluster with other genes, and the entire coding region of TK1880 was deleted from the genome (see Fig. S1). In both cases, T. kodakarensis KU216 was used as the host strain, and selection of the gene disruption strains was performed based on the methods applied for gdh disruption. The Δogor and Δscs strains were designated T. kodakarensis KOGOR1 and KSCS1, respectively.
The growth characteristics of the strains KOGOR1 and KSCS1 were examined in ASW-YT-S0 and ASW-YT-Pyr media. In ASW-YT-S0 medium, whereas KGDH1 could not grow at all, KOGOR1 and KSCS1 displayed growth (Fig. 2). The specific growth rates and final cell yield, however, were lower than those of T. kodakarensis KUW1. This tendency was also observed in ASW-YT-Pyr medium, with SCS gene disruption leading to slightly greater defects than that of OGOR.
Analyses of consumption and production of metabolites in the growth medium.
In order to gain insight on the involvement of GDH, OGOR, and SCS in amino acid catabolism, we examined the consumption of amino acids in the medium and the production of the organic acids expected to be formed by amino acid degradation. The amino acids examined were Glu, Ala, Val, and Leu, and the corresponding acids were succinate, acetate, isobutyrate, and isovalerate. Pyruvate was also examined, as ASW-YT-Pyr was used as the growth medium. T. kodakarensis KGDH1, KOGOR1, KSCS1, and the host cell line KUW1 were grown in ASW-YT-Pyr, and aliquots of the cultures were taken after 4 and 8 h of growth. All experiments were performed at least in triplicate. In ASW-YT-Pyr, it can be presumed that pyruvate is directly catabolized via pyruvate:ferredoxin oxidoreductase and ACS I and II, generating reduced ferredoxin and ATP. The high concentrations of pyruvate in the medium should also promote the conversion of amino acids to their corresponding 2-oxoacids (coupled to the formation of Ala from pyruvate) via aminotransferases.
When medium was incubated without cells, the amino acids and acetate were stable after 4 to 8 h at 85°C with no apparent decrease in concentration. In the case of pyruvate, however, thermal degradation corresponding to a half-life of 31 ± 3 h at 85°C was observed (Fig. 4). In the case of succinate, its half-life at 85°C was estimated to be 94 ± 7 h. In addition, the thermostability of 2-oxoglutarate and succinyl-CoA, the two intermediates involved in the oxidative catabolism of Glu to succinate, was examined. In sodium phosphate buffer (pH 7.0) at 85°C, half-lives of 2-oxoglutarate and succinyl-CoA were 300 ± 20 h and 89 ± 3 s, respectively, revealing a surprisingly high thermostability of 2-oxoglutarate and thermolabile nature of succinyl-CoA.
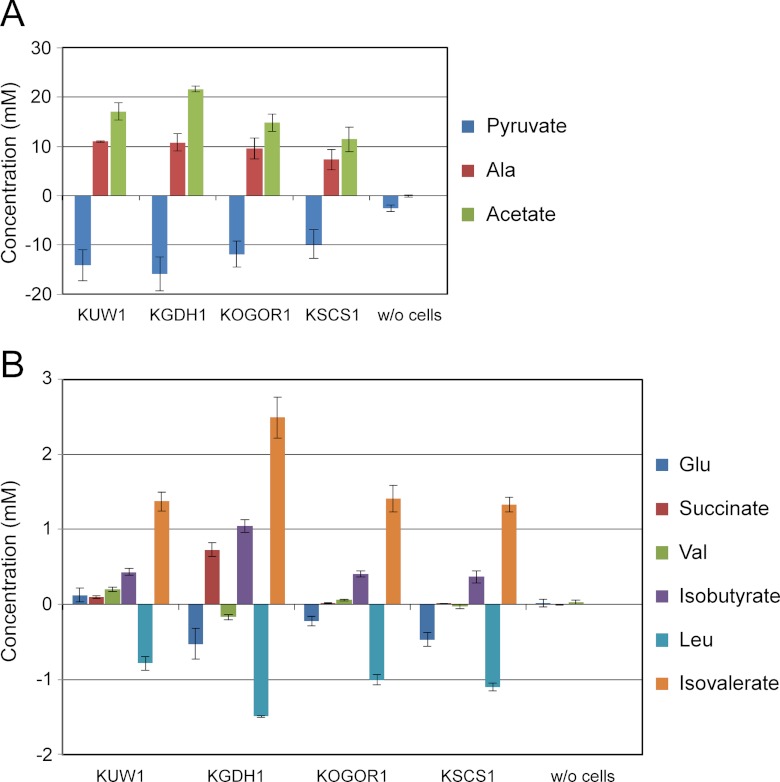
Fig 4.
Consumption or generation of metabolites by T. kodakarensis KUW1, KGDH1, KOGOR1, and KSCS1 grown in ASW-YT-Pyr. Changes in concentration observed between 4 and 8 h of culture are shown. Positive values indicate generation of the metabolite, and negative values represent consumption. Medium without cell inoculation (w/o cells) was also examined. All experiments were performed at least in triplicate. The metabolites analyzed in this study are pyruvate, Ala, and acetate (A) and Glu, succinate, Val, isobutyrate, Leu, and isovalerate (B).
The changes in concentrations of amino acids/organic acids produced or consumed in the period between 4 and 8 h of cultivation of T. kodakarensis KUW1 and the three gene disruption strains were measured. In the case of host strain KUW1, approximately 14 ± 3 mM pyruvate in the medium was consumed between 4 and 8 h. During this period, an increase in the concentration of acetate (17 ± 2 mM) was observed, as well as an 11 ± 0 mM increase in Ala concentration. If acetate and Ala were generated solely from pyruvate, the decrease in pyruvate concentration can be expected to be similar to the sum of acetate and Ala formation. We cannot identify the specific reason for this discrepancy, but generation of Ala from peptide components in the tryptone and pyruvate generation from sugar components in the medium are likely to have effects on the stoichiometry of pyruvate, acetate, and Ala in batch cultures. As for the disruption strains, in KGDH1, the levels of pyruvate consumption and, in particular, acetate generation were higher than those in KUW1, but the levels of Ala generation were comparable. This indicates that in the absence of Tk-GDH, the oxidative degradation of pyruvate is enhanced. In KOGOR1 and KSCS1, the strains displayed lower extents of Ala/acetate production and pyruvate consumption than KUW1. This is due to the lower cell densities of these strains in ASW-YT-Pyr medium, but the ratios were similar to those observed for KUW1. On the other hand, changes in Glu metabolism were also observed. The disruption of gdh resulted in an increase in Glu consumption, 0.53 mM consumption in KGDH1 cells compared to 0.12 mM generation in KUW1 cells. An increase in succinate production (KGDH1, 0.73 mM generation; KUW1, 0.10 mM generation) was also observed, indicating that the levels of oxidative degradation of Glu to succinate increase in the absence of Tk-GDH. In KOGOR1 and KSCS1, an increase in Glu consumption was observed, but importantly, the production of succinate was no longer detectable (Fig. 4). This indicates that OGOR and SCS are the major 2-oxoacid:ferredoxin oxidoreductase and acyl-CoA synthetase, respectively, responsible for the breakdown of Glu to succinate in T. kodakarensis. Interestingly, KGDH1 displayed higher levels of Val/Leu consumption and isobutyrate/isovalerate production than KUW1. This indicates that the absence of Tk-GDH also leads to an increase in the oxidative degradation of amino acids other than Glu. In KOGOR1 and KSCS1, the oxidative degradation of Val/Leu also increased. Val/Leu consumption and isobutyrate/isovalerate production levels were equivalent to or higher than those observed for KUW1 cells, even though the cell densities of the two mutant strains were lower than that of KUW1.
DISCUSSION
In this study, the presence/absence of amino acid dehydrogenase activity in T. kodakarensis toward all 20 amino acids has been examined. An extremely high level of NADP+-dependent GDH activity, along with a relatively lower level of NAD+-dependent GDH activity, was observed. As the TK1431 (Tk-GDH) gene disruption strain KGDH1 did not display any GDH activity, it is clear that Tk-GDH is responsible for both activities. Besides activity toward Glu, we also detected activities with Gln, Ala, Val, and Cys, but the absence of these activities in KGDH1 strongly suggests that these are also dependent on Tk-GDH. The activities may be due to Tk-GDH directly recognizing these amino acids, but they are most likely the result of coupling of Tk-GDH with an amino acid aminotransferase(s). We have observed that transcript levels of TK1094, encoding alanine aminotransferase, and TK0186, encoding multiple-substrate aminotransferase, increase more than 4-fold in cells grown in ASW-YT-Pyr compared to those grown in ASW-YT-S0 (unpublished data), which may explain why the dehydrogenase activity toward Gln, Ala, Val, and Cys was observed only in cells grown in ASW-YT-Pyr. The only Tk-GDH-independent amino acid dehydrogenase activities present in T. kodakarensis cell extracts were ThrDH activity and serine dehydrogenase activity. The latter was observed only in KGDH1. It is most likely that the activities toward Thr/Ser derive from the threonine 3-dehydrogenase of T. kodakarensis, encoded by TK0916 (52, 53). The enzyme has been characterized, has been shown to be dependent on NAD+, and displays a low level of activity toward Ser. The fact that the serine dehydrogenase activity was observed only in KGDH1 cells most likely is due to the increase in ThrDH activity observed in KGDH1 cells compared to that in KUW1 and KGDH1C cells. The increase in ThrDH activity in response to Tk-GDH disruption is intriguing. One possibility is that, although low compared to NADP+-dependent activity, the NAD+-dependent activity of Tk-GDH is physiologically relevant. Disruption of Tk-GDH may result in a decrease in NADH generation, and increases in ThrDH activity may reflect a response to compensate for this decrease. This is of interest, as the enzyme(s) responsible for NADH generation in T. kodakarensis is still not well understood. As clearly shown in this study, there are no relevant amino acid dehydrogenase activities, other than the GDH and ThrDH activities, in T. kodakarensis that are dependent on NAD+.
That KGDH1 cells could not grow in ASW-YT-S0 strongly suggests that Tk-GDH is the only enzyme that can discharge the electrons (to NADP+/NAD+) released from amino acids in their oxidation to 2-oxoacids. On the other hand, disruption of Tk-GDH had no apparent effect on growth in ASW-YT-Pyr medium. However, when the metabolites in the medium were examined, a significant difference in the extent of amino acid catabolism was observed between KUW1 and KGDH1 cells. Higher concentrations of carboxy acids, such as succinate, isobutyrate, and isovalerate, were generated during growth. This suggests that when cells are grown in ASW-YT-Pyr medium, Tk-GDH plays an anabolic role, converting 2-oxoacids to amino acids. In this medium, one can expect that a considerable amount of amino acids are converted to their corresponding 2-oxoacids by the excess levels of pyruvate via aminotransferases. Under these conditions, Tk-GDH acts to suppress the oxidation of amino acids. The electrons, or NADPH, needed to reduce 2-oxoacids may be supplied by the NADP-dependent cytosolic hydrogenase (Hyh), which can utilize the molecular hydrogen generated by T. kodakarensis in ASW-YT-Pyr medium (54, 55). This mechanism does provide an advantage, as cells would be able to prevent excess consumption of the amino acid pool and rely more on the oxidation of the excess pyruvate for reducing equivalents and ATP synthesis. In ASW-YT-Pyr medium, the amount of amino acids in the medium is probably high enough that KGDH1 cells do not exhibit growth defects in our experiments.
In terms of OGOR and SCS, disruption of either of these genes resulted in the abolishment of succinate generation. The results clearly indicate that OGOR and SCS are the only KOR and ACS involved in the oxidation of 2-oxoglutarate, respectively. This provides genetic evidence suggesting that the other KORs and ACSs of T. kodakarensis are not involved in oxidation of 2-oxoglutarate. In particular, there are still two ACSs in T. kodakarensis that have not been characterized, but the results shown here suggest that they are not involved in oxidation of 2-oxoglutarate. The results also suggest that the XOR in T. kodakarensis does not function in Glu catabolism. It is not obvious why KOGOR1 and KSCS1 cells displayed growth defects in ASW-YT-S0 and ASW-YT-Pyr media. As we have demonstrated that succinyl-CoA is extremely thermolabile (half-life of 89 ± 3 s at 85°C), accumulation of succinyl-CoA in KSCS1 cells can be expected to lead to a larger portion of this metabolite that is thermally degraded, thus resulting in a decrease in the efficiency of energy conservation during amino acid catabolism. In KOGOR1 cells, we can presume that an accumulation of 2-oxoglutarate occurs, as this metabolite is thermostable. Accumulation of 2-oxoglutarate is consistent with the observation of increases in oxidative degradation of Val and Leu, as an increase in 2-oxoglutarate levels would promote the oxidation of these amino acids via aminotransferases. However, this does not explain why growth is disturbed in KOGOR1 cells. Concerning the growth defects in KSCS1 and KOGOR1 cells, as they were observed in both ASW-YT-S0 and ASW-YT-Pyr, there may be a metabolic importance in generating succinyl-CoA and succinate to exhibit optimal growth.
Taken together with previously reported biochemical studies, the genetic analyses performed here have revealed the physiological importance of GDH, OGOR, and SCS in T. kodakarensis. As homologs of the genes examined in this study are present in all members of the Thermococcales and whose genome sequences have been published, our results most likely apply for many, if not all, members of the Thermococcales. Based on the genome sequence and previous biochemical studies, there are several other enzymes that may be involved in Glu metabolism in T. kodakarensis. Glutamine synthetase, which has been biochemically characterized (56), is most likely involved in the biosynthesis of Gln from Glu. Glutamate synthase from T. kodakarensis has also been characterized (57). The function of this protein, however, will need further examination, as closely related homologs from Pyrococcus species have been demonstrated to exhibit different roles, such as the electron transfer component of sulfide dehydrogenase (58, 59). One more gene that should be studied is a putative glutamate decarboxylase gene. Future genetic analysis of these genes should reveal further aspects of Glu metabolism, particularly the de novo biosynthetic pathways of Glu/Gln, which is currently unknown in T. kodakarensis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the Japan Science and Technology Agency to H.A. and T.I. and the Japan Society for the Promotion of Science (Grant-in-Aid for JSPS Fellows 23.3910 to Y.Y.).
Footnotes
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01979-12.
REFERENCES
- 1. Itoh T. 2003. Taxonomy of nonmethanogenic hyperthermophilic and related thermophilic archaea. J. Biosci. Bioeng. 96:203–212 [PubMed] [Google Scholar]
- 2. Schut GJ, Boyd ES, Peters JW, Adams MW. 2013. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37:182–203 [DOI] [PubMed] [Google Scholar]
- 3. Sato T, Atomi H. 2011. Novel metabolic pathways in Archaea. Curr. Opin. Microbiol. 14:307–314 [DOI] [PubMed] [Google Scholar]
- 4. Siebers B, Schönheit P. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8:695–705 [DOI] [PubMed] [Google Scholar]
- 5. Verhees CH, Kengen SW, Tuininga JE, Schut GJ, Adams MW, De Vos WM, van der Oost J. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schut GJ, Menon AL, Adams MW. 2001. 2-Keto acid oxidoreductases from Pyrococcus furiosus and Thermococcus litoralis. Methods Enzymol. 331:144–158 [DOI] [PubMed] [Google Scholar]
- 7. Consalvi V, Chiaraluce R, Politi L, Vaccaro R, De Rosa M, Scandurra R. 1991. Extremely thermostable glutamate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Eur. J. Biochem. 202:1189–1196 [DOI] [PubMed] [Google Scholar]
- 8. Rahman R, Fujiwara S, Takagi M, Imanaka T. 1998. Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and comparison of the enzymatic characteristics of native and recombinant GDHs. Mol. Gen. Genet. 257:338–347 [DOI] [PubMed] [Google Scholar]
- 9. DiRuggiero J, Robb FT, Jagus R, Klump HH, Borges KM, Kessel M, Mai X, Adams MW. 1993. Characterization, cloning, and in vitro expression of the extremely thermostable glutamate dehydrogenase from the hyperthermophilic archaeon, ES4. J. Biol. Chem. 268:17767–17774 [PubMed] [Google Scholar]
- 10. Klump H, Di Ruggiero J, Kessel M, Park JB, Adams MW, Robb FT. 1992. Glutamate dehydrogenase from the hyperthermophile Pyrococcus furiosus. Thermal denaturation and activation. J. Biol. Chem. 267:22681–22685 [PubMed] [Google Scholar]
- 11. Kobayashi T, Higuchi S, Kimura K, Kudo T, Horikoshi K. 1995. Properties of glutamate dehydrogenase and its involvement in alanine production in a hyperthermophilic archaeon, Thermococcus profundus. J. Biochem. 118:587–592 [DOI] [PubMed] [Google Scholar]
- 12. Lee MK, Gonzalez JM, Robb FT. 2002. Extremely thermostable glutamate dehydrogenase (GDH) from the freshwater archaeon Thermococcus waiotapuensis: cloning and comparison with two marine hyperthermophilic GDHs. Extremophiles 6:151–159 [DOI] [PubMed] [Google Scholar]
- 13. Ma K, Loessner H, Heider J, Johnson MK, Adams MW. 1995. Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J. Bacteriol. 177:4748–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma K, Robb FT, Adams MW. 1994. Purification and characterization of NADP-specific alcohol dehydrogenase and glutamate dehydrogenase from the hyperthermophilic archaeon Thermococcus litoralis. Appl. Environ. Microbiol. 60:562–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohshima T, Nishida N. 1993. Purification and properties of extremely thermostable glutamate dehydrogenases from two hyperthermophilic archaebacteria, Pyrococcus woesei and Pyrococcus furiosus. Biosci. Biotechnol. Biochem. 57:945–951 [DOI] [PubMed] [Google Scholar]
- 16. Rahman R, Fujiwara S, Nakamura H, Takagi M, Imanaka T. 1998. Ion pairs involved in maintaining a thermostable structure of glutamate dehydrogenase from a hyperthermophilic archaeon. Biochem. Biophys. Res. Commun. 248:920–926 [DOI] [PubMed] [Google Scholar]
- 17. Yip KS, Stillman TJ, Britton KL, Artymiuk PJ, Baker PJ, Sedelnikova SE, Engel PC, Pasquo A, Chiaraluce R, Consalvi V. 1995. The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures. Structure 3:1147–1158 [DOI] [PubMed] [Google Scholar]
- 18. Ward DE, Kengen SW, van der Oost J, de Vos WM. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182:2559–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward DE, de Vos WM, van der Oost J. 2002. Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andreotti G, Cubellis MV, Nitti G, Sannia G, Mai X, Adams MW, Marino G. 1995. An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1247:90–96 [DOI] [PubMed] [Google Scholar]
- 21. Andreotti G, Cubellis MV, Nitti G, Sannia G, Mai X, Marino G, Adams MW. 1994. Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 220:543–549 [DOI] [PubMed] [Google Scholar]
- 22. Matsui I, Matsui E, Sakai Y, Kikuchi H, Kawarabayasi Y, Ura H, Kawaguchi S, Kuramitsu S, Harata K. 2000. The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii. J. Biol. Chem. 275:4871–4879 [DOI] [PubMed] [Google Scholar]
- 23. Sakuraba H, Kawakami R, Takahashi H, Ohshima T. 2004. Novel archaeal alanine:glyoxylate aminotransferase from Thermococcus litoralis. J. Bacteriol. 186:5513–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blamey JM, Adams MW. 1993. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161:19–27 [DOI] [PubMed] [Google Scholar]
- 25. Kletzin A, Adams MW. 1996. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma K, Hutchins A, Sung SJ, Adams MW. 1997. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. U. S. A. 94:9608–9613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menon AL, Hendrix H, Hutchins A, Verhagen MF, Adams MW. 1998. The δ-subunit of pyruvate ferredoxin oxidoreductase from Pyrococcus furiosus is a redox-active, iron-sulfur protein: evidence for an ancestral relationship with 8Fe-type ferredoxins. Biochemistry 37:12838–12846 [DOI] [PubMed] [Google Scholar]
- 28. Smith ET, Blamey JM, Adams MW. 1994. Pyruvate ferredoxin oxidoreductases of the hyperthermophilic archaeon, Pyrococcus furiosus, and the hyperthermophilic bacterium, Thermotoga maritima, have different catalytic mechanisms. Biochemistry 33:1008–1016 [DOI] [PubMed] [Google Scholar]
- 29. Heider J, Mai X, Adams MW. 1996. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178:780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozawa Y, Nakamura T, Kamata N, Yasujima D, Urushiyama A, Yamakura F, Ohmori D, Imai T. 2005. Thermococcus profundus 2-ketoisovalerate ferredoxin oxidoreductase, a key enzyme in the archaeal energy-producing amino acid metabolic pathway. J. Biochem. 137:101–107 [DOI] [PubMed] [Google Scholar]
- 31. Mai X, Adams MW. 1994. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 269:16726–16732 [PubMed] [Google Scholar]
- 32. Ozawa Y, Siddiqui MA, Takahashi Y, Urushiyama A, Ohmori D, Yamakura F, Arisaka F, Imai T. 2012. Indolepyruvate ferredoxin oxidoreductase: an oxygen-sensitive iron-sulfur enzyme from the hyperthermophilic archaeon Thermococcus profundus. J. Biosci. Bioeng. 114:23–27 [DOI] [PubMed] [Google Scholar]
- 33. Siddiqui MA, Fujiwara S, Imanaka T. 1997. Indolepyruvate ferredoxin oxidoreductase from Pyrococcus sp. KOD1 possesses a mosaic structure showing features of various oxidoreductases. Mol. Gen. Genet. 254:433–439 [DOI] [PubMed] [Google Scholar]
- 34. Mai X, Adams MW. 1996. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178:5890–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glasemacher J, Bock AK, Schmid R, Schönheit P. 1997. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 244:561–567 [DOI] [PubMed] [Google Scholar]
- 36. Hutchins AM, Mai X, Adams MW. 2001. Acetyl-CoA synthetases I and II from Pyrococcus furiosus. Methods Enzymol. 331:158–167 [DOI] [PubMed] [Google Scholar]
- 37. Mai X, Adams MW. 1996. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:5897–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schäfer T, Schönheit P. 1991. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Arch. Microbiol. 155:366–377 [Google Scholar]
- 39. Schäfer T, Selig M, Schönheit P. 1993. Acetyl-CoA synthetase (ADP forming) in archaea, a novel enzyme involved in acetate formation and ATP synthesis. Arch. Microbiol. 159:72–83 [Google Scholar]
- 40. Shikata K, Fukui T, Atomi H, Imanaka T. 2007. A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal nucleoside diphosphate-forming acetyl-CoA synthetases. J. Biol. Chem. 282:26963–26970 [DOI] [PubMed] [Google Scholar]
- 41. Bräsen C, Schmidt M, Grötzinger J, Schönheit P. 2008. Reaction mechanism and structural model of ADP-forming acetyl-CoA synthetase from the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a second active site histidine residue. J. Biol. Chem. 283:15409–15418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Musfeldt M, Selig M, Schönheit P. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robb FT, Place AR. 1995. Media for thermophiles, p 167–168 In Robb FT, Place AR. (ed), Archaea: a laboratory manual-thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47. Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yokooji Y, Tomita H, Atomi H, Imanaka T. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284:28137–28145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santangelo TJ, Čuboňová L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeda Y, Suzuki F, Inoue H. 1969. ATP citrate lyase (citrate-cleavage enzyme). Methods Enzymol. 13:153–160 [Google Scholar]
- 52. Bashir Q, Rashid N, Jamil F, Imanaka T, Akhtar M. 2009. Highly thermostable L-threonine dehydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Biochem. 146:95–102 [DOI] [PubMed] [Google Scholar]
- 53. Bowyer A, Mikolajek H, Stuart JW, Wood SP, Jamil F, Rashid N, Akhtar M, Cooper JB. 2009. Structure and function of the L-threonine dehydrogenase (TkTDH) from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Struct. Biol. 168:294–304 [DOI] [PubMed] [Google Scholar]
- 54. Kanai T, Ito S, Imanaka T. 2003. Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:1705–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanai T, Matsuoka R, Beppu H, Nakajima A, Okada Y, Atomi H, Imanaka T. 2011. Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193:3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adul Rahman RN, Jongsareejit B, Fujiwara S, Imanaka T. 1997. Characterization of recombinant glutamine synthetase from the hyperthermophilic archaeon Pyrococcus sp. strain KOD1. Appl. Environ. Microbiol. 63:2472–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jongsareejit B, Rahman R, Fujiwara S, Imanaka T. 1997. Gene cloning, sequencing and enzymatic properties of glutamate synthase from the hyperthermophilic archaeon Pyrococcus sp. KOD1. Mol. Gen. Genet. 254:635–642 [DOI] [PubMed] [Google Scholar]
- 58. Hagen WR, Silva PJ, Amorim MA, Hagedoorn PL, Wassink H, Haaker H, Robb FT. 2000. Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence for a [2Fe-2S] cluster with Asp(Cys)3 ligands. J. Biol. Inorg. Chem. 5:527–534 [DOI] [PubMed] [Google Scholar]
- 59. Dincturk HB, Cunin R, Akce H. 2011. Expression and functional analysis of glutamate synthase small subunit-like proteins from archaeon Pyrococcus horikoshii. Microbiol. Res. 166:294–303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.