Abstract
The first step in bacterial cytokinesis is the assembly of a stable but dynamic cytokinetic ring made up of the essential tubulin homolog FtsZ at the future site of division. Although FtsZ and its role in cytokinesis have been studied extensively, the precise architecture of the in vivo medial FtsZ ring (Z ring) is not well understood. Recent advances in superresolution imaging suggest that the Z ring comprises short, discontinuous, and loosely bundled FtsZ polymers, some of which are tethered to the membrane. A diverse array of regulatory proteins modulate the assembly, stability, and disassembly of the Z ring via direct interactions with FtsZ. Negative regulators of FtsZ play a critical role in ensuring the accurate positioning of FtsZ at the future site of division and in maintaining Z ring dynamics by controlling FtsZ polymer assembly/disassembly processes. Positive regulators of FtsZ are essential for tethering FtsZ polymers to the membrane and promoting the formation of stabilizing lateral interactions, permitting assembly of a mature Z ring. The past decade has seen the identification of several factors that promote FtsZ assembly, presumably through a variety of distinct molecular mechanisms. While a few of these proteins are broadly conserved, many positive regulators of FtsZ assembly are limited to small groups of closely related organisms, suggesting that FtsZ assembly is differentially modulated across bacterial species. In this review, we focus on the roles of positive regulators in Z ring assembly and in maintaining the integrity of the cytokinetic ring during the early stages of division.
INTRODUCTION
Bacterial cytokinesis is mediated by a macromolecular protein machine that is accurately positioned in space and time during the cell cycle. The earliest defined event during cytokinesis is the assembly of a polymeric FtsZ structure at the site of future division known as the FtsZ ring, or Z ring (1). The Z ring serves as a scaffold for the recruitment of other division proteins (2–5). FtsZ, a tubulin homolog, forms homopolymeric linear protofilaments upon binding GTP, and these filaments subsequently disassemble upon hydrolysis of the bound nucleotide (6–9). Approximately 30% of cellular FtsZ is present in the ring, and fluorescence recovery after photobleaching (FRAP) studies reveal that there is dynamic exchange between FtsZ molecules in the ring polymers and FtsZ monomers or short oligomers in the cytoplasm (10, 11).
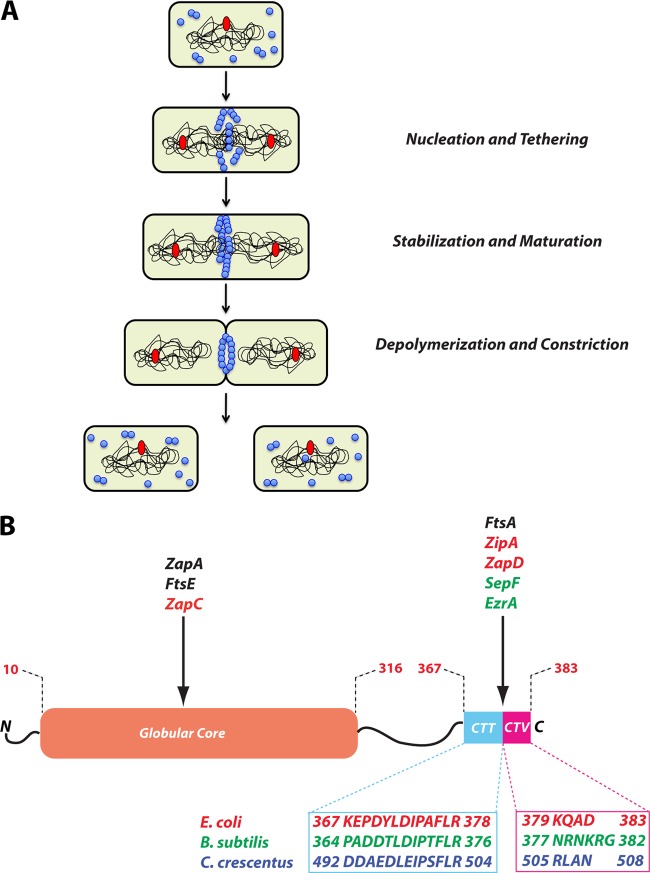
In Escherichia coli and Bacillus subtilis, cytokinesis can be separated chronologically into the following three steps: (i) assembly and stabilization of the Z ring at the future division site and, following a lag, (ii) recruitment of downstream division proteins, many of which are essential to form a constriction-competent division complex, and lastly, (iii) constriction of the Z ring coordinated with synthesis and splitting of septal peptidoglycan, leading to invagination of the cell membrane and division into two daughter cells (Fig. 1A) (2, 12, 13). The force required for constriction is likely generated, at least in part, by the remodeling of the nucleotide-bound FtsZ filaments tethered to the membrane (14–16).
Fig 1.
(A) Schematic of FtsZ assembly, maturation, and disassembly, depicted during different stages of the bacterial cell cycle. In a newborn cell, FtsZ is present as unassembled monomers or short oligomers. The chromosome (black curved lines) with a single origin of replication (red oval) is shown. Toward the end of chromosome replication and segregation, FtsZ (blue circles) polymerizes at the future site of division and is tethered to the membrane by specific FtsZ-interacting proteins. During this first step in cytokinesis, early divisomal proteins that directly bind FtsZ and promote lateral associations among FtsZ polymers stabilize the Z ring. During the second step in cytokinesis, maturation of the initial Z ring complex takes place by recruitment of downstream division proteins to the division site. The final step in cytokinesis constitutes the depolymerization and constriction of the Z ring, followed by separation into two daughter cells. (B) Domain organization of FtsZ with interaction sites for species-specific FtsZ regulators affecting the assembly and stability of the nascent Z ring during cytokinesis. The domain structure is not drawn to scale and shows amino acid residue numbers from the E. coli FtsZ sequence. The amino acid residues of the conserved CTT (C-terminal tail) and the CTV (C-terminal variable) regions of FtsZ in the three well-studied species are indicated below. FtsZ regulators conserved in all three species are shown in black, those found in E. coli are shown in red, and those in B. subtilis are shown in green. EzrA, a negative regulator of FtsZ in Bacillus, is included in this diagram because of its potential role in tethering FtsZ to the membrane. The FtsZ interaction sites of ZapB from E. coli and FzlA and FzlC from Caulobacter are currently undetermined.
In Caulobacter crescentus (a model species for studying asymmetric cell division, cell polarity, and cellular differentiation), the division machinery assembles in at least seven different stages, coordinated with cellular events such as chromosome segregation, cell elongation, septal invagination, and cell separation (17). For excellent recent reviews on FtsZ and the overall process of bacterial cytokinesis, the reader is referred to references 2–5 and 18.
IN VIVO ULTRASTRUCTURE OF THE Z RING
Conventional fluorescence microscopy data from E. coli and B. subtilis describe dynamic helical precursors of FtsZ which coalesce into a single tight-pitched transverse ring at the site of future division (19–22), suggesting that FtsZ may retain a conserved helical character (3). In vitro, FtsZ alone forms multiple rings when artificially tethered to tubular liposomes, indicating the capacity of membrane-bound FtsZ to assemble independent of other division proteins (14, 15). However, superresolution imaging of the Z ring suggests that FtsZ polymers do not exist in a continuous closed confirmation and also fails to describe extended helical FtsZ structures. Instead, the superresolution microscopy findings indicate that the precise configuration of the Z ring may vary across bacterial species. In E. coli, photo-activated light microscopy (PALM) images of FtsZ are consistent with a loose bundle of randomly overlapping FtsZ protofilaments at the midcell division site (23). In Bacillus, recent three-dimensional simulated illumination microscopy (3D-SIM) imaging reveals the Z ring to also be discontinuous, with FtsZ arranged in a “bead-like” pattern wherein densely populated regions of FtsZ polymers are interspersed with regions devoid of or with very few FtsZ polymers (24). In Caulobacter, cryotomography images of cells show sparse, short, nonoverlapping FtsZ protofilaments that do not form a continuous ring structure at midcell (25).
A pressing question in the field is how the Z ring is organized such that it forms a dynamic yet stable structure during cytokinesis. Two lines of evidence support the idea that Z ring polymers are likely held together by lateral associations in the cell. First, a myriad of regulatory proteins have been discovered in many bacteria that colocalize with FtsZ at midcell and stabilize the Z ring. These FtsZ regulatory proteins bind and promote lateral associations among FtsZ polymers in vitro. Second, a recent study indicates that FtsZ homologs from E. coli and B. subtilis have inherently varying assembly properties in vitro, and when these inherent bundling propensities are altered in mutant and chimeric proteins in vivo, cytokinesis is impaired (26). While FtsZ-FtsZ lateral bond energy is weak to nonexistent, the precise molecular nature of FtsZ lateral associations as mediated by FtsZ modulatory proteins is undetermined (5).
FtsZ REGULATORY PROTEINS AND Z RING FORMATION
The cellular levels of FtsZ remain essentially constant throughout the cell cycle in E. coli and B. subtilis, and therefore, spatial and temporal regulation of bacterial cytokinesis must largely be via control of FtsZ assembly and disassembly in these species (27). In Caulobacter, the abundances of FtsZ transcript and protein are tightly linked to cell cycle regulation, and this linkage constitutes an additional layer of control on FtsZ assembly/disassembly processes in this species (28, 29). Several regulatory proteins bind FtsZ directly and modulate the spatiotemporal integrity of the Z ring in vivo. FtsZ regulatory proteins can be broadly classified as either negative or positive. (i) Negative factors, also known as FtsZ inhibitors, prevent Z ring polymerization at the poles or over the nucleoid and maintain the dynamics of the Z ring at the future division site. (ii) Positive factors, also known as FtsZ stabilizers, aid in the assembly and maturation of a stable Z ring. Generally, positive factors would be required to perform any of three functions—to nucleate, tether, and/or stabilize FtsZ polymers at the site of future division. The existence of an FtsZ nucleator is considered likely given that negative regulation of FtsZ polymerization by FtsZ inhibitors alone is insufficient for precise localization of the Z ring at the future site of division in B. subtilis (30). Moreover, in certain bacteria that lack the canonical negative regulators of FtsZ, accurate Z ring placement at the future division site takes place by positive regulation. In sporulating cells of Streptomyces coelicolor, the actinomycete-specific membrane-associated divisome protein SsgB recruits and tethers FtsZ to the division site via direct interactions (31). The orthologous SsgA, which is also restricted to the Actinomycetes, mediates the localization of SsgB (31, 32). In vegetatively growing Myxococcus xanthus (deltaproteobacterium), a positive regulator of FtsZ, PomZ, is proposed to identify the future division site, recruit FtsZ to this site, and stabilize the Z ring (33).
In addition to regulation of FtsZ polymerization, interactions between FtsZ polymers and the cell membrane and of FtsZ polymers with each other are both critical to the formation of a functional Z ring. FtsZ has no intrinsic affinity for cell membrane, but tethering of FtsZ to the membrane is essential for formation of Z rings (34). Depletion of either essential division protein, FtsA or ZipA, which serve to link FtsZ to the membrane, is lethal in E. coli (34, 35). Maintaining lateral interactions among FtsZ polymers in the Z ring is also critical. Deletion of two or more Z ring stabilizers known to promote FtsZ polymer associations in vitro reduces cell viability (36–38).
EVOLUTION OF FtsZ AND POSITIVE REGULATORS OF FtsZ
FtsZ is a highly conserved ancient protein present in most bacteria, many archaea, all chloroplasts, and some primitive mitochondria (5). FtsZ consists of four discrete domains: a short, unstructured N-terminal region consisting of the first 10 amino acids, a globular core containing the nucleotide binding region and the T7 synergy loop required for nucleotide hydrolysis, an unstructured linker of variable length, and a C-terminal tail (CTT) consisting of a highly conserved set of residues and acting as a landing pad for both conserved and diverse FtsZ-interacting proteins (Fig. 1B) (5). Recently, the extreme C terminus of FtsZ was defined as the C-terminal variable (CTV) domain due to its diversity in length and amino acid sequence across various species (26). The net charge of the CTV region was shown to impact the assembly properties of FtsZ molecules in vitro. For instance, FtsZ of B. subtilis with its positively charged CTV bundled FtsZ protofilaments more efficiently than E. coli FtsZ with a net neutral CTV (26). The importance of CTV charge and perhaps FtsZ assembly properties in vivo was revealed when a B. subtilis FtsZ fused to an E. coli CTV displayed impaired Z ring formation and cytokinesis (26). Although FtsZ is highly conserved among various bacteria, positive regulators of FtsZ, many of which appear to have overlapping or redundant functions, display remarkable variability. Divergence in FtsZ CTV sequences and perhaps FtsZ assembly properties may offer a partial explanation for the diversity in FtsZ stabilizers.
Below, we discuss the role of positive regulators in FtsZ assembly and stability in three well-studied species, E. coli, B. subtilis, and C. crescentus. For the purpose of this review, we define positive regulators of FtsZ as proteins that localize to the future site of division and aid in tethering FtsZ to the membrane and/or promoting lateral bundling among FtsZ protofilaments or inducing specific conformational changes in FtsZ protofilaments.
FtsA
FtsA is a widely conserved membrane binding protein that belongs to the actin/Hsp70/sugar kinase superfamily of proteins and can bind ATP with low affinity (39). As yet, it is unclear whether the FtsA-nucleotide interactions are physiologically relevant (40). The 3D structure of Thermotoga FtsA resembles actin, although it displays a different subdomain architecture: domains 1a, 1c, 2a, and 2b remain, with the deletion of domain 1b and addition of domain 1c (41). Very recently, it was demonstrated that Thermotoga FtsA can indeed polymerize into a canonical actin-like protofilament in the presence of nonhydrolyzable ATP or lipid monolayers (42). Of note, Streptococcus pneumoniae FtsA assembles into large polymers that are unlike actin and differ from the FtsA bundles and sheets observed in the Szwedziak et al. study (43). Moreover, the molecular details of how S. pneumoniae FtsA intersubunit contacts within a protofilament differ and the relevance of such FtsA structures in vivo are not yet determined.
In E. coli, FtsA localizes to midcell in an FtsZ-dependent manner and tethers FtsZ to the membrane via interactions with the FtsZ CTT (Fig. 1B and 2A) (34, 44–46). Genetic screens in E. coli and biochemical data from Thermotoga support a role for FtsA domain 2b in mediating the FtsA-FtsZ interaction (42, 46). The FtsA membrane-targeting sequence (MTS) can be replaced by the MalF transmembrane domain or MinD MTS and remain functional for cell division, suggesting that the role of this sequence is limited to membrane binding (47, 48). FtsA domain 1c has a role in self-interaction and the recruitment of downstream division proteins in E. coli (41, 48–54).
Fig 2.

Diagrammatic representation of proteins that localize at the site of future division in E. coli (A), B. subtilis (B), and Caulobacter (C) and promote the Z ring assembly and stability during the first step in bacterial cytokinesis. Z, FtsZ; E, FtsE; X, FtsX; IM, inner membrane; PG, peptidoglycan; OM, outer membrane. ZapB is oligomeric in solution and has been reported to form a ring-like structure (not shown) with a smaller radius than that of the Z ring in E. coli. EzrA, a negative regulator of FtsZ in Bacillus, is included because of its putative role in tethering FtsZ to the membrane. In Caulobacter, FtsA is a late recruit to the divisome.
In E. coli, in addition to FtsA, ZipA (discussed below) can tether FtsZ to the membrane, stabilize FtsZ, and recruit downstream division proteins. The assertion that FtsA is the primary factor required for anchoring FtsZ to the membrane in E. coli is supported by the following lines of evidence: the broad conservation of FtsA, the ability of Z rings to assemble in the absence of ZipA, the ability of a gain-of-function ftsA mutant, FtsA* (R286W), and other functional ftsA mutants with reduced self-interaction to bypass the need for ZipA, and the direct interaction between FtsA and some downstream proteins (3, 52, 55–58). In addition to its role in FtsZ assembly, genetic studies infer that FtsA and the FtsA* mutant enhance Z ring integrity in vivo (48, 56, 59–61).
E. coli FtsA lacking the membrane-targeting sequence can form cytoplasmic rods or filaments in vivo (48). In a screen for FtsA mutants that do not form such cytoplasmic filaments, several mutants, including ftsA*, were identified (55). These mutants displayed decreased FtsA self-interaction, as determined by protein-protein interaction assays in yeast, were able to bypass the requirement of ZipA, and mapped to the oligomerization interface in the FtsA protofilament, indicative of their involvement in FtsA self-interaction (42, 55). However, the FtsA mutants with reduced self-interaction behaved differently when their functions were characterized in the context of the full-length protein in E. coli and B. subtilis. In E. coli, self-interaction ftsA mutants displayed a gain-of-function phenotype, potentially due to an increased efficiency of downstream division protein recruitment by the free 1c domain of FtsA (55). In contrast, in B. subtilis, ftsA self-interaction mutations display a loss-of-function phenotype, suggesting that at least some level of FtsA self-interaction promotes cytokinesis (42). Therefore, while it is clear that FtsA is required to tether FtsZ to the membrane and that it likely plays a role in Z ring stability, at present, there are differing views about the precise role of FtsA-FtsA self-interaction in cytokinesis.
The biochemical and biophysical characterization of FtsA has long been hampered by the tendency of the native protein to form nonfunctional aggregates, although a recent advance in purifying FtsA from inclusion bodies may have the potential to change this (62). Intriguingly, purified FtsA* depolymerized FtsZ filaments in an ATP-dependent manner, a function that appears discordant with its role in promoting Z ring integrity in vivo (63).
In B. subtilis, FtsA colocalizes in a helical pattern with FtsZ during vegetative growth (64). While FtsA is not essential for vegetative division, cells lacking FtsA are defective for cytokinesis, forming long filamentous cells with very few Z ring assemblies (64–66). A deletion of the Caulobacter FtsZ CTT leads to a loss in FtsZ-FtsA interaction, leading to speckled FtsZ localization in cells, consistent with FtsZ CTT being the interaction site for FtsA (67) (Fig. 1B). However, unlike E. coli and B. subtilis, FtsA in Caulobacter is not an early recruit to the future site of division and arrives only after Z ring assembly has occurred (17, 68). In Caulobacter, FtsA levels are closely linked to the cell cycle, and this is likely why FtsA arrives late to the divisome (17, 69). In toto, results from B. subtilis and Caulobacter suggest that other proteins may have functionally overlapping roles with FtsA in tethering FtsZ to the membrane in these species (Fig. 2B and C) (5, 17).
ZipA
ZipA homologs are found only in Gram-negative gammaproteobacteria, although functional orthologs likely exist in other bacteria (57, 58, 70). ZipA is an essential bitopic integral inner membrane protein comprised of a large cytoplasmic domain linked to a single N-terminal transmembrane domain via an extended linker (58). ZipA functionally overlaps with FtsA in tethering FtsZ to the membrane in E. coli (Fig. 2A). Like FtsA, ZipA binds conserved residues in the FtsZ CTT and recruits downstream division proteins (Fig. 1B) (44, 45, 71, 72). The C-terminal domain of ZipA can localize to the midcell septum in an FtsZ-dependent manner (73).
In addition to its role in tethering FtsZ to the membrane, cytological and genetic evidence support a role for ZipA in stabilizing the Z ring. Although FtsZ rings can still assemble in ZipA-depleted filaments, they do so with lower frequency and greater variability than wild-type cells (35). Furthermore, a 2-fold increase in ZipA abundance suppresses the thermosensitivity of the FtsZ84 (Ts) polymerization-defective mutant (57). Of note, moderate overexpression of the ZipA bypass mutant FtsA* is unable to rescue the thermosensitivity of FtsZ84 (61), indicating that FtsA* does not substitute for all ZipA functions and that FtsA and ZipA likely have discrete functional roles with respect to E. coli FtsZ. However, an FtsA-FtsA tandem fusion is able to suppress FtsZ84 thermosensitivity, providing evidence in support of a model that some level of FtsA self-interaction promotes division efficiency in E. coli.
In vitro, ZipA stabilizes polymers of both wild-type FtsZ and FtsZ84 in sedimentation assays mediating the formation of large bundled networks of FtsZ polymers (Fig. 3) (57, 73). The precise mechanism by which ZipA promotes FtsZ bundling is unclear, although it is hypothesized that the ZipA-FtsZ interaction is predominantly hydrophobic (74). The C-terminal domain of ZipA alone is necessary and sufficient to interact with and bundle FtsZ in vitro (73). The structure of the ZipA C-terminal domain was determined by solution nuclear magnetic resonance (NMR) and revealed a split beta-alpha-beta motif, common among RNA binding proteins, with a six-stranded beta sheet aligned against three alpha helices (75). A structure of the 17-amino-acid FtsZ CTT peptide in complex with ZipA revealed that the FtsZ peptide binds a solvent-exposed hydrophobic cleft in ZipA, with a majority of the binding affinity provided by a few key conserved residues (75). Interestingly, the structure of the FtsZ CTT peptide bound to ZipA is not identical to the structure of FtsZ CTT bound to FtsA (42, 75). The binding sites of FtsZ CTT on FtsA and ZipA are also dissimilar (42, 74, 75).
Fig 3.
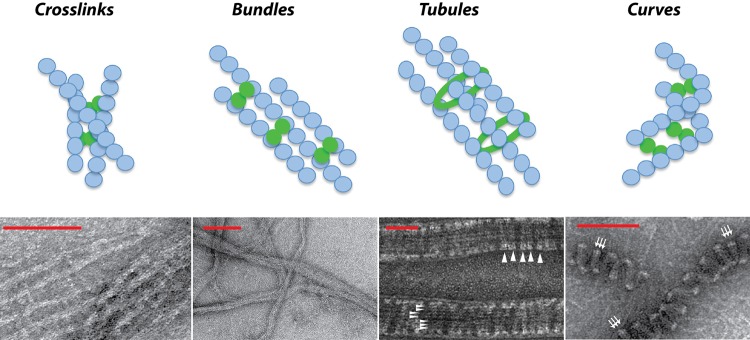
Diagram (top) and representative electron micrographs (bottom) of the effects of various positive regulators on the higher-order structures of FtsZ polymers in the Z ring. Blue circles represent FtsZ monomers, and solid green circles or rings represent various FtsZ stabilizers in different species. From left to right: FtsZ in the presence of E. coli ZapA shows cross-linked or bundled polymers, reprinted from reference 85 with permission from the American Chemical Society; FtsZ in the presence of E. coli ZapD shows bundled FtsZ polymers, reprinted from reference 37 with permission; FtsZ in the presence of Bacillus SepF shows FtsZ tubules (horizontal white arrowheads) and SepF rings (vertical white arrowheads), with images courtesy of Leendert Hamoen and reprinted from reference 76 with permission from Nature Publishing Group; and FtsZ in the presence of Caulobacter FzlA shows curved FtsZ polymers (white arrows), reprinted from reference 105 with permission from Elsevier. Note that ZapD-mediated FtsZ bundles are shown as a representative image of FtsZ polymer bundling by various FtsZ regulators. Scale bars from left to right: 100 nm, 200 nm, 50 nm, and 100 nm.
It is postulated that in Bacillus, which lacks ZipA, the FtsZ-interacting protein SepF (discussed below), and the FtsZ inhibitor EzrA, which is topologically very similar to ZipA, may perform a role analogous to that of ZipA by tethering FtsZ to the membrane (Fig. 2B) (5, 76). However, the narrow conservation of ZipA, in addition to the ability of ftsA* and other ftsA mutants to bypass its requirement in E. coli, leaves the basis of ZipA essentiality unclear. A recent study has addressed this question. Observing that ftsA mutants that bypass ZipA display lower self-interaction, it was reasoned that ZipA might disrupt FtsA self-interaction in wild-type E. coli and that such ZipA-mediated reduction of FtsA self-interaction is perhaps essential for cytokinesis (55).
FtsEX
FtsE (an ATPase) and FtsX (membrane binding domain) form an ATP binding transporter-like complex and are broadly conserved in bacteria (77). E. coli FtsEX is a divisomal component essential only under low-osmolarity conditions (77, 78), and recently, its precise molecular role in cytokinesis was elucidated (79). FtsX recruits EnvC, an activator of cell wall amidases AmiA and AmiB, to the midcell division site (79). FtsX regulates EnvC-mediated activation of the amidases through conformational changes induced by the ATPase activity of FtsE (79). A similar role for FtsEX in peptidoglycan separation in Gram-positive Streptococcus pneumoniae has also been described, indicating that regulation of cell wall remodeling may be a conserved feature of FtsEX (80).
In addition, various lines of evidence argue for the involvement of FtsEX in promoting FtsZ assembly and stability at the site of future division. FtsX bearing an N-terminal green fluorescent protein (GFP) tag localizes to midcell dependent on FtsZ, FtsA, or ZipA but independent of downstream division proteins (Fig. 2A) (77). Overexpression of FtsQAZ can rescue growth defects in ftsEX mutants in low-osmolarity medium (78). Deletion of ftsEX exacerbates growth defects in most cell division genes, including ftsA12 (Ts) (78). Finally, FtsE has been observed to coimmunoprecipitate with FtsZ (81). However, FtsE does not appear to interact with FtsZ CTT in E. coli, a docking site shared by many positive and negative FtsZ regulatory proteins (81) (Fig. 1B). This suggests a distinct mode for FtsE-mediated Z ring stability.
In B. subtilis, FtsEX contributes to the precise spatiotemporal activation of sporulation and promotes the switch from a medial to a polar septum formation by a molecular mechanism that is as yet undetermined (82). Nonetheless, consistent with its distinct role in B. subtilis versus E. coli, B. subtilis FtsEX is localized all around the cell membrane and not exclusively to the future site of division (82). Under vegetative growth conditions, an ftsEX mutant does not exhibit impaired cell division or growth phenotypes (82). Furthermore, high salt concentrations do not rescue the sporulation phenotype of an ftsEX mutant (82). Strikingly, in Caulobacter, FtsEX is recruited to the division site prior to FtsA (17). Therefore, in Caulobacter, FtsEX makes for an attractive candidate for tethering FtsZ to the membrane during the initial stages of Z ring assembly (Fig. 2C) (17). Taken together, these results suggest that while broadly conserved, FtsEX may serve specialized roles in cytokinesis in diverse bacterial species. An alternative possibility is that FtsEX performs the same function across different species, but mutations in ftsEX have distinct outcomes, as the divisome machinery is assembled differently in diverse bacteria.
ZapA
ZapA was first identified in B. subtilis, and orthologs are present in most bacteria, including in mitochondria of a primitive red alga (83, 84). ZapA localizes to the midcell dependent on a direct interaction with FtsZ and remains dynamically associated with the cytokinetic machinery throughout the cell cycle but is not essential for cell division or viability (Fig. 2) (83, 85–87).
Genetic data from E. coli support a role for ZapA in promoting Z ring stability. Cells overexpressing or lacking ZapA exhibit loose helical Z rings (85, 87, 88). In the absence of ZapA, cells show mild elongation phenotypes (85, 87). Furthermore, a zapA mutant, when combined with conditional mutations in ftsA or zipA or with mutations in other positive FtsZ regulators—zapB, zapC, or zapD (discussed below)—or in the presence of the polymerization-defective FtsZ84, reveals synthetic sick phenotypes in E. coli (36, 37, 87). These synergistic division phenotypes highlight the critical need for maintaining a precise balance among the levels of FtsZ regulators for optimal division efficiency. Additionally, the gain-of-function ftsA* mutant is able to suppress the zapA defect in cell division, suggesting that it may have a partially redundant role with ZapA. In vitro, however, the effects of ZapA and FtsA* on FtsZ polymers are different. ZapA promotes FtsZ stability (discussed below), while FtsA* stimulates FtsZ depolymerization in vitro (63, 87).
As predicted from its in vivo function, ZapA promotes Z ring stability by bundling FtsZ protofilaments in vitro with a concomitant decrease in FtsZ GTPase activity but with no conformational changes in the bound nucleotide (85, 89). Consistent with a role for ZapA in FtsZ lateral bundling, in vitro evidence suggests that ZapA binds to FtsZ in competition with the FtsZ inhibitor MinC, which, in part, prevents FtsZ lateral associations (Fig. 1B) (90). Recently, ZapA's role in cross-linking FtsZ protofilaments was described from quantitative rheometry and measurements of phase angles of FtsZ networks (87). Notably, electron microscopy images of ZapA-mediated FtsZ bundles reveal parallel FtsZ polymers, suggesting that ZapA-mediated cross-linking perhaps does not align FtsZ filaments at various acute angles (Fig. 3).
Depending on concentration, ZapA is either a dimer or tetramer in solution and does not form higher-order structures by itself (91). The crystal structure of ZapA reveals two antiparallel dimers interacting with each other through coiled-coil C-terminal domains (91). ZapA interacts with another FtsZ regulator, ZapB (discussed below), through its coiled-coil domain and with FtsZ through its globular domain (88, 91). Based on the endogenous concentrations of ZapA in E. coli, the association of nearly all cellular ZapA with the midcell Z ring, the molar concentrations which increase the stiffness of FtsZ networks in biophysical experiments, and the ability of a dimeric ZapA mutant to bind but not bundle FtsZ polymers, it is likely that the ZapA tetramer is the physiologically relevant oligomeric form in the cell (85, 87, 89).
In B. subtilis, cells lacking ZapA are sensitive to reduced levels of FtsZ or the absence of the FtsZ inhibitors MinC, EzrA, and DivIVA, suggesting that the role for ZapA in promoting Z ring stability may be conserved (83). Furthermore, in B. subtilis, overexpression of ZapA is able to suppress the loose helical Z ring phenotype of the FtsZ (Ts1) mutant at the nonpermissive temperature (92). This suggests that ZapA mediates bundling of FtsZ polymers in vivo. In contrast, the division defect of the FtsZ (Ts1) mutant cannot be rescued by deletion of the FtsZ inhibitor minC, which has been shown to disrupt FtsZ lateral bundling (92, 93). These data suggest that perhaps ZapA-mediated FtsZ lateral bundling alone is not sufficient to account for the suppression of the FtsZ (Ts1) phenotype. Given the role of ZapA in cross-linking FtsZ protofilaments in E. coli, it was proposed that it is not the bundling activity of Bacillus ZapA per se but perhaps the cross-linking function that plays a more prominent role in coalescing a helical Z ring in the cell (87). Interestingly, the abundance of ZapA in B. subtilis is much lower than that in E. coli, suggesting that ZapA perhaps does not play as key a role in FtsZ polymer bundling in B. subtilis. In support of this suggestion, it was recently shown that a chimeric E. coli FtsZ fused to B. subtilis CTV residues is able to form lateral associations independent of ZapA in vitro (26).
ZapB
The gene encoding ZapB, which is restricted to the gammaproteobacterial class, was initially isolated in a plasmid destabilization screen to identify host factors that in multicopy would interfere with plasmid partitioning systems (86). ZapB was subsequently determined to be a Z ring stabilizer that colocalizes with FtsZ at midcell and is recruited to the divisome by ZapA (Fig. 2A) (38, 86). Overproduction of ZapA but not ZapC, another positive regulator of FtsZ (discussed below), delocalizes ZapB, consistent with a role for ZapA in recruiting ZapB to the midcell (38). Although cells are viable, a deletion of ZapB results in modest cell elongation and aberrant Z ring assemblies in a fraction of the population (86). In the presence of the inefficiently polymerizing FtsZ84, cells lacking ZapB display exacerbated defects consistent with a role for ZapB in stabilizing the Z ring during the early stages of division (86). ZapB can mediate Z ring stability even in the absence of ZapA, suggesting that ZapB may function by two separate mechanisms in cytokinesis (38).
ZapB, unlike ZapA, is a homodimeric antiparallel coiled-coil protein without globular domains (86). Purified ZapB spontaneously forms oligomeric structures in vitro (88). The ZapB N-terminal domain interacts directly with ZapA; ZapA-ZapB binding leads to increased ZapB sedimentation and bundling in vitro (88). ZapB copellets with FtsZ but only in the presence of ZapA, indicating that ZapA links the FtsZ-ZapB interaction (38). Sedimentation and light scattering assays with purified ZapA, ZapB, and FtsZ show that increased concentrations of ZapB, but not of a ZapB mutant (ZapB S4A L5A) that is unable to bind ZapA, reduced the quantity of polymeric FtsZ in the pellet, indicating that the ZapA-ZapB interaction competes with the ZapA-FtsZ interaction (88). While the endogenous concentrations of ZapA, ZapB, and FtsZ are nearly equal to each other in E. coli, the physiological implications of the predicted stronger association of ZapA with ZapB on ZapA-mediated FtsZ lateral associations are yet to be determined.
In addition to aberrant Z ring assemblies and division defects, overexpression of ZapB induces nucleoid condensation (86). ZapB, anchored to the Z ring by ZapA, was recently shown to interact with the chromosome-structuring factor MatP (94). MatP binds and compacts the Ter macrodomain (Ter MD) of the E. coli chromosome, ensuring proper mobility and segregation of this region of the DNA (94). The ZapB-MatP interaction may therefore link replicated chromosomes by their Ter MD to the midcell Z ring in E. coli (94). Furthermore, the double mutants matP zapA and matP zapB display exacerbated defects in nucleoid segregation, indicating a potential role for ZapB in coordinating chromosome segregation and division (94). Interestingly, ZapB-GFP fusions accumulate into rings of a smaller diameter than Z rings in E. coli (38). The precise physiological relevance of such a ZapB ring structure is unclear but may serve to bridge chromosome segregation with septum constriction in this species. However, the ZapB-MatP interaction is not essential for chromosome segregation, suggesting that it may be critical only under specific growth conditions (94). Gammaproteobacterial species, such as E. coli, lack the parABS-type mitotic spindle (as seen in Caulobacter), and the capture extrusion model of chromosome segregation (as seen in B. subtilis) is unlikely in these species (95, 96). Therefore, the putative additional role of ZapB in chromosome segregation can perhaps explain the limited conservation of ZapB in E. coli and related species.
ZapC AND ZapD
Recently, two additional nonessential division proteins, ZapC and ZapD, have been identified in E. coli (36, 37, 97). Although ZapABCD proteins have overlapping functions in stabilizing FtsZ protofilaments, they do not share any primary sequence identity and belong to the growing group of proteins classified as FtsZ ring-associated proteins (Zaps). ZapC is restricted to gammaproteobacteria, while ZapD is found in gammaproteobacteria and some betaproteobacteria (36, 37, 97). ZapC and ZapD localize to the midcell division site dependent on FtsZ but not on downstream division proteins, indicating that they are part of the early divisomal complex (Fig. 2A) (36, 97).
ZapC is not essential for viability, but cells lacking ZapC exhibit mild increases in cell lengths (36, 97). Absence of ZapC in cells lacking ZapA, ZapB, or both leads to mild elongation in cell lengths, suggesting a role for ZapC in Z ring stability (36, 37). Consistent with this contention, cells lacking ZapC display enhanced sensitivity to overexpression of the FtsZ inhibitor MinC compared to the wild type (36). Overexpression of ZapC causes aberrant hyperstable FtsZ structures but does not affect localization of ZapB, suggesting that ZapB and, therefore, ZapA localize independently of ZapC to the midcell divisome (36, 88, 97).
In vitro, ZapC binds FtsZ directly, enhances FtsZ assembly into protofilament bundles (Fig. 3), and reduces FtsZ GTPase activity (36, 37). ZapC appears monomeric, but given its role in bridging FtsZ protofilaments, it is likely to form higher-order structures in vivo (36, 97). A ZapC mutant (ZapC L22P) that oligomerizes in solution and does not promote FtsZ bundling in vitro is unable to interact with FtsZ in vivo (36). Interestingly, a genome-wide proteomic analysis of ClpXP substrates in E. coli revealed that ZapC might be a target of this protease (98). Given that FtsZ itself is a ClpXP substrate in E. coli (99), proteolysis of FtsZ-interacting proteins, such as ZapC, may add another layer of regulatory control to FtsZ assembly/disassembly processes in E. coli.
Similar to cells lacking individual Zap proteins, cells with a zapD deletion display no significant changes in cell viability and division (37). However, a zapD deletion in cells with the polymerization-defective FtsZ84 (Ts) shows decreased viability and filamentous cells at the permissive temperature, supporting the role of ZapD as a positive regulator of FtsZ in vivo (37). In accordance, overexpression of ZapD leads to filamentation with aberrant hyperstable Z ring morphologies (37). Furthermore, consistent with ZapD's role in Z ring stability, synergistic division defects are seen upon removal of ZapD and ZapA (37). In vitro, purified ZapD binds FtsZ directly and promotes FtsZ polymer bundling with a concomitant decrease in FtsZ GTPase activity (Fig. 3) (37). In agreement with the predicted role of ZapD in linking two FtsZ protofilaments, ZapD is oligomeric in solution and the crystal structure of a close homologue in Vibrio parahaemolyticus also reveals a dimer (100).
While ZapC and ZapD each bind FtsZ in a protein-protein interaction assay in yeast, only ZapD binds the FtsZ CTT, suggesting that each protein binds discrete FtsZ sites and may promote Z ring stability by distinct molecular mechanisms (Fig. 1B) (37). Whether ZapC and ZapD are differentially regulated and whether they perform additional roles in other cellular functions are as yet undetermined.
SepF
Initial studies in Streptococcus and Synechococcus showed that sepF mutants had marked cell division defects, suggesting a role for SepF in cell division (65, 101, 102). The gene for SepF (ylmF) is located in a broadly conserved gene cluster in most Gram-positive bacteria (103). SepF interacts with itself and FtsZ in a yeast two-hybrid assay, suggestive of a role in the Bacillus divisome (103). Bacillus sepF mutants display modest cell division defects with aberrant septa, suggesting a unique role for SepF compared to other known FtsZ regulators (103). The precise stage of division in which SepF acts is unclear. A SepF-GFP fusion localizes to the division plane in either transverse bands or foci dependent on FtsZ but not the late-assembling division components, suggesting that SepF is part of the early divisomal complex (Fig. 2B) (103). Moreover, sepF ftsA double mutants in B. subtilis are synthetic lethal and do not form Z rings, implying a role for SepF in Z ring assembly (65, 103). Furthermore, overproduction of SepF suppresses the division defects of ftsA null cells, consistent with partially overlapping roles for SepF and FtsA (65). In contrast, genetic evidence demonstrates that depletion of SepF in the absence of EzrA does not prevent Z ring assembly or recruitment of downstream division proteins, arguing for a role for SepF in the later stages of division (103). In addition, unlike other positive FtsZ regulators that act early during cytokinesis, removal of SepF in cells with reduced FtsZ levels does not lead to synergistic division phenotypes (103).
Purified SepF promotes polymeric FtsZ structures with no marked changes in FtsZ GTPase activity, which is unusual given that increases in FtsZ bundling via regulatory proteins, molecular crowding agents, or higher concentrations of FtsZ all lead to reduced GTPase activity (76). In vitro, SepF can bind the CTT of either B. subtilis or E. coli FtsZ, indicating that the SepF-FtsZ interaction is dependent on the secondary and tertiary structure of the FtsZ CTT rather than specific amino acid residues (Fig. 1B) (104). Electron micrographs of purified SepF show large regular ring-like structures (76). Remarkably, at a physiologically relevant ratio of SepF and FtsZ, SepF rings bind FtsZ protofilaments into regular tubule-like structures (Fig. 3) (76). SepF-promoted FtsZ tubules form curved ends that are similar to frayed microtubules (76). However, microtubule-like dynamic instability is not seen with SepF-mediated FtsZ tubules, as they disintegrate over their entire lengths after prolonged reaction times (76). FtsZ lacking CTT sequences cannot form tubules in the presence of SepF, consistent with FtsZ CTT being the SepF interaction site (76).
The structural organization of SepF-mediated FtsZ tubules and the precise function they orchestrate in vivo are not known. A C-terminal mutant SepF (G135N), which interacts with FtsZ but forms only linear, not toroidal, SepF structures in vitro, cannot support growth in the absence of ftsA, suggesting that SepF rings may organize FtsZ protofilaments in the Z ring (76). Perplexingly, overexpression of either wild-type SepF or SepFG135N compensates for an ftsA deletion, suggesting that FtsZ tubule formation function is likely not shared by FtsA (76). Perhaps under these conditions, SepF-FtsZ or SepFG135N-FtsZ interaction competes with FtsZ negative regulators for binding FtsZ and thereby suppresses the loss of FtsA. Nonetheless, the limited conservation of SepF in species with thick peptidoglycan cell walls suggests that SepF-mediated FtsZ filament arrangements are likely to be beneficial to the developmental life cycle in these bacteria (76).
FzlA AND FzlC
FzlA (FtsZ-localized protein), which belongs to the glutathione transferase (GST) family of proteins, and FzlC were discovered and characterized in Caulobacter and are conserved in alphaproteobacterial species (105). Purified FzlC binds FtsZ in vitro but is not essential for viability or growth of Caulobacter (105). On the other hand, depletion of FzlA leads to filamentous Caulobacter cells without constrictions, indicating that FzlA is critical for division (Fig. 2C) (105). Similar to other division proteins in Caulobacter, FzlA transcript and protein levels are cell cycle regulated; highest levels of FzlA relative to FtsZ are found prior to and during Z ring assembly (105). However, in cells lacking FzlA, FtsZ assembly and downstream division protein recruitment are not impacted, indicating that FzlA is not required for these steps (105). Overproduction of FzlA leads to hyperstable compact foci of FtsZ and inhibition of division (105). Consistent with its role in promoting FtsZ stability, FzlA antagonizes the role of MipZ, an FtsZ inhibitor in Caulobacter that spatially constrains FtsZ to the division site (105, 106).
In vitro, FzlA promotes FtsZ polymerization with a corresponding inhibition of FtsZ GTPase activity (105). Strikingly, visualization of FzlA-induced FtsZ polymers at a 2:1 ratio of FzlA/FtsZ reveals helical arcs and bundles with a defined geometry that suggests two FtsZ filaments sandwiching a FzlA filament (Fig. 3) (105). Whether FzlA-induced filament curvature occurs in the cell and whether such filament curvature generates a constriction force or has other physiological relevance remain to be elucidated. In addition to previously known bundlers and cross-linkers, FzlA represents a third category of FtsZ stabilizers: shapers of FtsZ filament curvature.
CONCLUDING REMARKS
The organization of a stable yet dynamic Z ring during the early stages of bacterial cytokinesis appears to be a conserved feature in bacteria. Yet the FtsZ-interacting proteins that enable FtsZ to coalesce into a tight-pitched cytokinetic ring at the site of future division vary considerably between bacterial species. The question remains why different species exhibit such diversity in regulating the assembly and stability of the highly conserved FtsZ. Plausible, but not mutually exclusive, possibilities include the following. (i) The role of different positive regulators on FtsZ may be in accordance with the specific needs of the organism. SepF, for instance, is restricted to bacteria with thick cell walls, and SepF-mediated Z ring organization may be of particular benefit in cell constriction in such species. (ii) Multiple stabilizers with the same apparent function in Z ring stability in one species, such as the Zap proteins in E. coli, may display significant differences in the molecular mechanisms of how they achieve such stability. Perhaps such distinctions serve to increase division efficiency and serve as fail-safe systems for a critical cellular process. (iii) FtsZ stabilizers may be regulated differentially, thereby imparting flexibility to the assembly of a stable division machinery under various environmental conditions. While ZapC and ZapD exhibit a similar function—that of promoting Z ring stability in E. coli—the gene encoding ZapD is part of a conserved operon with coaE, a dephospho-coenzyme A (CoA) kinase, and perhaps yacG, an endogenous DNA gyrase inhibitor, raising the possibility that zapD is regulated. (iv) Positive regulators of FtsZ may participate in additional cellular functions by mediating interactions of FtsZ with other proteins. It was recently determined that ZipA is required for FtsZ-dependent cell wall synthesis at midcell during the transition from cell wall elongation to septum invagination in wild-type E. coli (107). It is unlikely that a specific interaction between preseptal synthesis components and ZipA is needed since preseptal peptidoglycan synthesis takes place in the ftsA* bypass allele. Nonetheless, ZipA serves as an example of a positive FtsZ regulator that, in addition to its function in division, orchestrates a role in cell wall synthesis.
A common theme emerging from studies characterizing FtsZ positive regulators in bacteria is that associations of FtsZ polymers, through mechanisms of cross-linking, bundling, and curvature modulation, likely have critical implications in the organization of a stable Z ring during the initial stages of cytokinesis. Consequently, furthering the understanding of the biology of such positive FtsZ regulators and their interactions with FtsZ at molecular and atomic details will aid us in parsing universal versus species-specific aspects of Z ring assembly and stability during bacterial division.
ACKNOWLEDGMENTS
We thank Seth Goldman, Petra Levin, Debu RayChaudhuri, and Manjula Reddy for their critical reading of the manuscript.
This work was supported by NSF MCB 1152059 (to A.J.); core facility support at CCNY comes from NIH/NIMHD/RCMI 8G12MD007603-27.
Footnotes
Published ahead of print 1 March 2013
REFERENCES
- 1. Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- 2. de Boer PAJ. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 69:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 5. Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74:504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Boer PAJ, Crossley R, Rothfield L. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–256 [DOI] [PubMed] [Google Scholar]
- 7. RayChaudhuri D, Park JT. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–254 [DOI] [PubMed] [Google Scholar]
- 8. Mukherjee A, Lutkenhaus J. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romberg L, Levin PA. 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U. S. A. 99:3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, Den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631–1645 [DOI] [PubMed] [Google Scholar]
- 13. Gamba P, Veening J, Saunders N, Hamoen L, Daniel R. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191:4186–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osawa M, Erickson HP. 2011. Inside-out Z rings—constriction with and without GTP hydrolysis. Mol. Microbiol. 81:571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mingorance J, Rivas G, Velez M, Gomez-Puertas P, Vicente M. 2010. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18:348–356 [DOI] [PubMed] [Google Scholar]
- 17. Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80:1680–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirkpatrick CL, Viollier PH. 2011. New(s) to the (Z-)ring. Curr. Opin. Microbiol. 14:691–697 [DOI] [PubMed] [Google Scholar]
- 19. Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 93:12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peters PC, Migocki MD, Thoni C, Harry EJ. 2007. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 64:487–499 [DOI] [PubMed] [Google Scholar]
- 21. Thanedar S, Margolin W. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14:1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ben-Yehuda S, Losick R. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257–266 [DOI] [PubMed] [Google Scholar]
- 23. Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One 5:e12682 doi:10.1371/journal.pone.0012682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10:e1001389 doi:10.1371/journal.pbio.1001389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buske PJ, Levin PA. 2012. The extreme C-terminus of the bacterial cytoskeletal protein FtsZ plays a fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287:10945–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weart RB, Levin PA. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185:2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. 1998. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 12:880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sackett MJ, Kelly AJ, Brun YV. 1998. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol. Microbiol. 28:421–434 [DOI] [PubMed] [Google Scholar]
- 30. Rodrigues CD, Harry EJ. 2012. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 8:e1002561 doi:10.1371/journal.pgen.1002561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25:89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Traag BA, van Wezel GP. 2008. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Treuner-Lange A, Aguiluz K, van der Does C, Gomez-Santos N, Harms A, Schumacher D, Lenz P, Hoppert M, Kahnt J, Munoz-Dorado J, Sogaard-Andersen L. 2013. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol. Microbiol. 87:235–253 [DOI] [PubMed] [Google Scholar]
- 34. Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hale CA, de Boer PAJ. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PAJ. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193:1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. 2012. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194:3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galli E, Gerdes K. 2010. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 76:1514–1526 [DOI] [PubMed] [Google Scholar]
- 39. Bork P, Sander C, Valencia A. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and Hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feucht A, Lucet I, Yudkin MD, Errington J. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115–125 [DOI] [PubMed] [Google Scholar]
- 41. van den Ent F, Löwe J. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szwedziak P, Wang Q, Freund SMV, Löwe J. 2012. FtsA forms actin-like protofilaments. EMBO J. 31:2249–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krupka M, Rivas G, Rico AI, Vicente M. 2012. Key role of two terminal domains in the bidirectional polymerization of FtsA protein. J. Biol. Chem. 287:7756–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma X, Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haney SA, Glasfeld E, Hale CA, Keeney D, He Z, de Boer PAJ. 2001. Genetic analysis of the Escherichia coli FtsZ-ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980–11987 [DOI] [PubMed] [Google Scholar]
- 46. Pichoff S, Lutkenhaus J. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 64:1129–1138 [DOI] [PubMed] [Google Scholar]
- 47. Shiomi D, Margolin W. 2008. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol. Microbiol. 67:558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722–1734 [DOI] [PubMed] [Google Scholar]
- 49. Carettoni D, Gomez-Puertas P, Yim L, Mingorance J, Massidda O, Vicente M, Valencia A, Domenici E, Anderluzzi D. 2003. Phage-display and correlated mutations identify an essential region of subdomain 1C involved in homodimerization of Escherichia coli FtsA. Proteins 50:192–206 [DOI] [PubMed] [Google Scholar]
- 50. Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. 2004. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 53:1359–1371 [DOI] [PubMed] [Google Scholar]
- 51. Busiek KK, Eraso JM, Wang Y, Margolin W. 2012. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J. Bacteriol. 194:1989–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corbin BD, Geissler B, Sadasivam M, Margolin W. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, Ruysschaert JM, Vicente M. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182:6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pichoff S, Shen B, Sullivan B, Lutkenhaus J. 2012. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 83:151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Geissler B, Elraheb D, Margolin W. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 100:4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. RayChaudhuri D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hale CA, de Boer PAJ. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185 [DOI] [PubMed] [Google Scholar]
- 59. Bernard CS, Sadasivam M, Shiomi D, Margolin W. 2007. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64:1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shiomi D, Margolin W. 2007. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 66:1396–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Geissler B, Shiomi D, Margolin W. 2007. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martos A, Monterroso B, Zorrilla S, Reija B, Alfonso C, Mingorance J, Rivas G, Jimenez M. 2012. Isolation, characterization and lipid-binding properties of the recalcitrant FtsA division protein from Escherichia coli. PLoS One 7:e39829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Beuria TK, Mullapudi S, Mileykovskaya E, Sadasivam M, Dowhan W, Margolin W. 2009. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J. Biol. Chem. 284:14079–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jensen SO, Thompson LS, Harry EJ. 2005. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-Ring assembly. J. Bacteriol. 187:6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. 2006. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60:1364–1380 [DOI] [PubMed] [Google Scholar]
- 66. Beall B, Lutkenhaus J. 1992. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J. Bacteriol. 174:2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Din N, Quardokus EM, Sackett MJ, Brun YV. 1998. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27:1051–1063 [DOI] [PubMed] [Google Scholar]
- 68. Moll A, Thanbichler M. 2009. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol. Microbiol. 72:1037–1053 [DOI] [PubMed] [Google Scholar]
- 69. Martin ME, Trimble MJ, Brun YV. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 54:60–74 [DOI] [PubMed] [Google Scholar]
- 70. Du Y, Arvidson CG. 2003. Identification of ZipA, a signal recognition particle-dependent protein from Neisseria gonorrhoeae. J. Bacteriol. 185:2122–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu Z, Mukherjee A, Lutkenhaus J. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853–1861 [DOI] [PubMed] [Google Scholar]
- 72. Hale CA, de Boer PAJ. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hale CA, Rhee AC, de Boer PAJ. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kuchibhatla A, Bhattacharya A, Panda D. 2011. ZipA binds to FtsZ with high affinity and enhances the stability of FtsZ protofilaments. PLoS One 6:e28262 doi:10.1371/journal.pone.0028262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moy FJ, Glasfeld E, Mosyak L, Powers R. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39:9146–9156 [DOI] [PubMed] [Google Scholar]
- 76. Gündoğdu ME, Kawai Y, Pavlendova N, Ogasawara N, Errington J, Scheffers D-J, Hamoen LW. 2011. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 30:617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reddy M. 2007. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189:98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. 2011. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U. S. A. 108:E1052–E1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sham L-T, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. U. S. A. 108:E1061–E1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Corbin BD, Wang Y, Beuria TK, Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189:3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. 2008. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol. Microbiol. 69:1018–1028 [DOI] [PubMed] [Google Scholar]
- 83. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yoshida Y, Kuroiwa H, Hirooka S, Fujiwara T, Ohnuma M, Yoshida M, Misumi O, Kawano S, Kuroiwa T. 2009. The bacterial ZapA-like protein ZED is required for mitochondrial division. Curr. Biol. 19:1491–1497 [DOI] [PubMed] [Google Scholar]
- 85. Mohammadi T, Ploeger GEJ, Verheul J, Comvalius AD, Martos A, Alfonso C, Van Marle J, Rivas G, Den Blaauwen T. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68:720–735 [DOI] [PubMed] [Google Scholar]
- 87. Dajkovic A, Pichoff S, Lutkenhaus J, Wirtz D. 2010. Cross-linking FtsZ polymers into coherent Z rings. Mol. Microbiol. 78:651–668 [DOI] [PubMed] [Google Scholar]
- 88. Galli E, Gerdes K. 2012. FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pacheco-Gómez R, Cheng X, Hicks MR, Smith CJI, Roper DI, Addinall S, Rodger A, Dafforn TR. 2013. Tetramerisation of ZapA is required for FtsZ bundling. Biochem. J. 449:795–802 [DOI] [PubMed] [Google Scholar]
- 90. Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18:235–244 [DOI] [PubMed] [Google Scholar]
- 91. Low HH, Moncrieffe MC, Löwe J. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839–852 [DOI] [PubMed] [Google Scholar]
- 92. Monahan LG, Robinson A, Harry EJ. 2009. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 74:1004–1017 [DOI] [PubMed] [Google Scholar]
- 93. Scheffers D-J. 2008. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 582:2601–2608 [DOI] [PubMed] [Google Scholar]
- 94. Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. 2012. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31:3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lemon KP, Grossman AD. 2001. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 15:2031–2041 [DOI] [PubMed] [Google Scholar]
- 97. Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683 [DOI] [PubMed] [Google Scholar]
- 99. Camberg JL, Hoskins J, Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJR. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60:787–796 [DOI] [PubMed] [Google Scholar]
- 101. Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 185:6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Miyagishima SY, Wolk CP, Osteryoung KW. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126–143 [DOI] [PubMed] [Google Scholar]
- 103. Hamoen LW, Meile J-C, De Jong W, Noirot P, Errington J. 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59:989–999 [DOI] [PubMed] [Google Scholar]
- 104. Krol E, van Kessel SP, van Bezouwen LS, Kumar N, Boekema EJ, Scheffers DJ. 2012. Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS One 7:e43293 doi:10.1371/journal.pone.0043293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. 2010. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39:975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162 [DOI] [PubMed] [Google Scholar]
- 107. Potluri L-P, Kannan S, Young KD. 2012. ZipA is required for FtsZ-dependent pre-septal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 194:5334–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]