Abstract
Sinorhizobium meliloti NRG247 has a Fix+ phenotype on Medicago truncatula A20 and is Fix− on M. truncatula A17, and the phenotype is reversed with S. meliloti NRG185. As the succinoglycan was shown to impact host specificity, an analysis of the succinoglycan oligosaccharides produced by each strain was conducted. The symbiotically active succinoglycan trimeric oligosaccharides (STOs) from the two S. meliloti strains were compared by chromatography and mass spectrometry, and the analysis of the S. meliloti NRG247 oligosaccharides showed that this strain produces an abundance of STO trimer 1 (T1), containing no succinate (i.e., three nonsuccinylated repeats), yet the low-molecular-weight pool contained no nonsuccinylated monomers (potential repeats). This showed that STO T1 is likely to be the active signal on M. truncatula A20 and that the biosynthesis of the STOs is not a random polymerization of the monomer population. The results also suggest that the fully succinylated STO T7 is required for the infection of M. truncatula A17.
INTRODUCTION
Gram-negative, nitrogen-fixing soil bacteria form a symbiotic association with many legumes. The establishment of the symbiosis involves a complex signal exchange between host and symbiont, resulting in the formation of the root nodule in which the bacteria reduce atmospheric N2 to ammonia (1). This symbiotic relationship is of agronomic significance because of the decreased requirement of nitrogen fertilizer for agriculturally important plants (e.g., soybean and alfalfa) (2). The plant and bacteria go through extensive physiological and physical alterations to reach this state, and this complex relationship is regulated by molecular signals and involves many plant genes in both the host and microsymbiont (1). Molecular signals have essential roles in the establishment of nitrogen-fixing nodules, and many studies have shown that bacterial exopolysaccharides (EPS) and K antigens may play a role in symbiotic infection (3–8). Sinorhizobium meliloti produces two forms of exopolysaccharides, succinoglycan and galactoglucan, and both have been shown to promote nodule infection in alfalfa (9–13), even at very low concentrations (14).
Succinoglycan is composed of octasaccharide repeating units with acetyl, pyruvyl, and succinyl substitutions (15, 16), and succinoglycan oligosaccharides consisting of three repeats (succinoglycan trimeric oligosaccharides [STOs]) act as signal molecules during the infection of alfalfa (17). The degree of succinylation varies among the different oligosaccharides produced by a particular strain (17), and the pattern of substitution differs among the various S. meliloti strains (18). The production of succinoglycan oligosaccharides is essential for Fix+ phenotype nodule formation in plants. In fact, researchers demonstrated that enhanced succinoglycan production improves the symbiotic productivity of S. meliloti 1021 with the host plant Medicago truncatula cv. Jemalong A17 (19). Overexpression of the exoY gene (encoding the enzyme that controls the first step in succinoglycan biosynthesis) was responsible for the increased succinoglycan production in S. meliloti Rm1021. Thus, the amount of the symbiotically active exopolysaccharide (succinoglycan oligosaccharides) produced by a rhizobial species is one of the elements involved in regulating the interaction with plant hosts (19).
Host specificity in the M. truncatula-S. meliloti interaction for nitrogen fixation has been well studied (18, 20–23). In one of the studies, a single gene (Mt-Sym6) that controls the host-specific nitrogen fixation upon inoculation with the S. meliloti strain A145 has been identified by genetic analysis (21). In Rhizobium leguminosarum bv. trifolii, strains display different host specificities with white clover than with Caucasian clover. Strain ICC105 produces nonfixing nodules on white clover but can produce functional nodules on Caucasian clover (24). For M. truncatula-S. meliloti interactions, the difference in succinoglycan substitution patterns has been shown to determine host specificity (i.e., strain-ecotype compatibility). S. meliloti strains Rm41 and NRG247 are Fix+ (compatible) on M. truncatula A20 but are Fix− (incompatible) on M. truncatula A17, and the phenotypes are reversed with S. meliloti strains NRG185 and NRG34. The compatibility, or host specificity, was shown to be correlated to the structural nature of the succinoglycan oligosaccharides produced by each strain (18).
In spite of the latest improvements in our understanding of the signaling pathways leading to initial communication between the organisms and to nodule development (25, 26), the molecular mechanisms underlying strain-specific nitrogen fixation are still mostly unknown. In this study, the succinoglycan oligosaccharides from two of the S. meliloti strains that displayed different compatibility patterns on M. truncatula were analyzed, and it was determined that the distribution of oligosaccharides in each strain correlates to compatibility with M. truncatula A17 versus A20. It was also shown that the biologically active oligosaccharides were not the result of random polymerization of the available monomeric octasaccharide subunits but resulted from a specific biosynthesis of the trimeric oligosaccharides.
MATERIALS AND METHODS
Nomenclature of succinoglycan oligosaccharides.
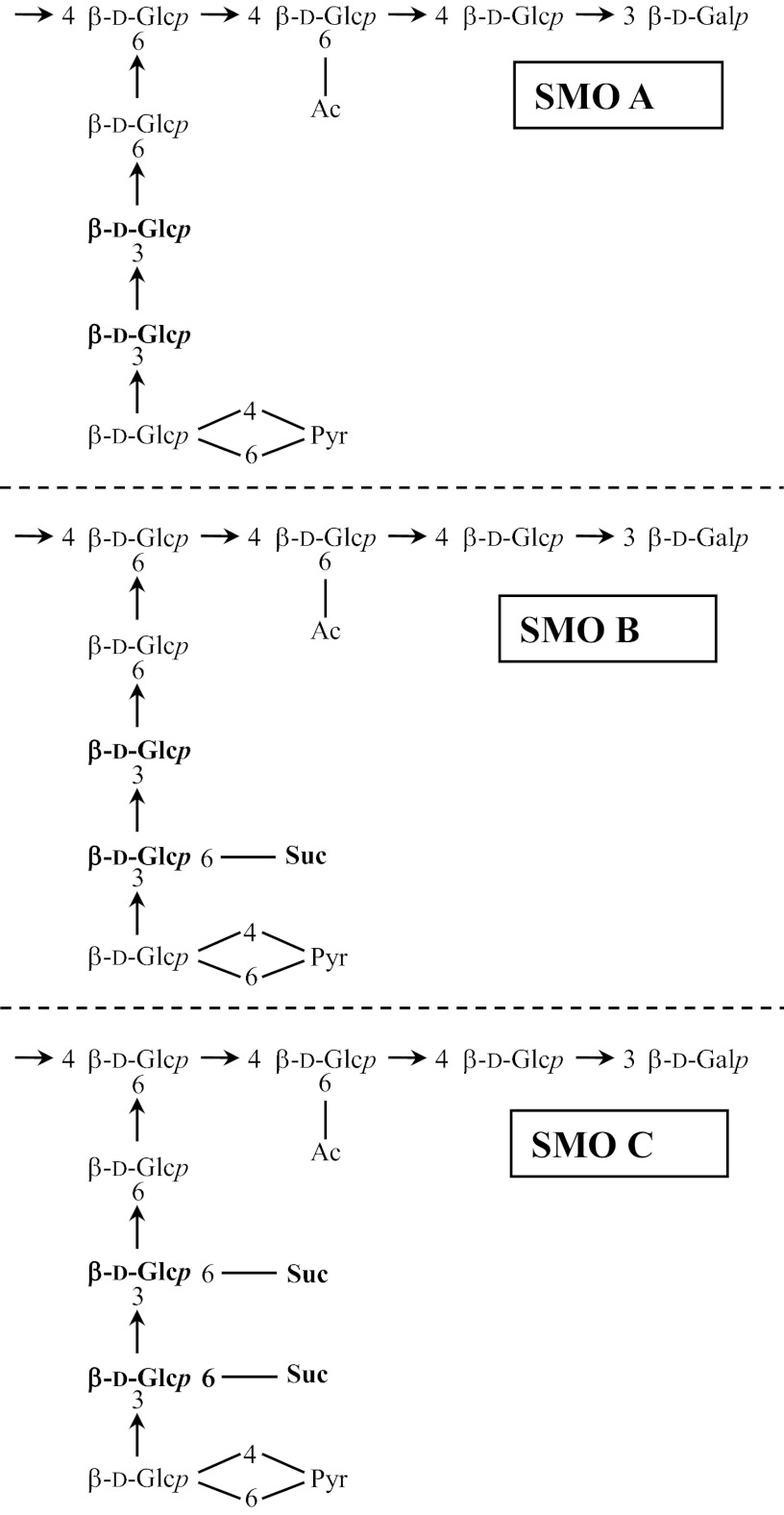
The succinoglycan oligosaccharides are termed STOs, succinoglycan dimeric oligosaccharides (SDOs), and succinoglycan monomeric oligosaccharides (SMOs), depending on the number of repeats. A monomer (potential repeat) with no succinate is designated “A,” a monomer with one succinate is “B,” and a monomer with two succinates is “C” (Fig. 1) (18), and thus, a fully succinylated STO would be C-C-C (Table 1).
Fig 1.
The succinoglycan repeating units. The potential succinylation sites are shown in boldface. Glc, glucose; Gal, galactose; Ac, acetyl; Pyr, pyruvyl; Suc, succinyl.
Table 1.
Distribution of succinoglycan oligosaccharides isolated from S. meliloti strains NRG247 and NRG185a
| Oligosaccharide | Possible STO(s)b | % of oligosaccharide estimated in strain: |
|
|---|---|---|---|
| NRG247 | NRG185 | ||
| STO | 55 | 50 | |
| SDO | 25 | 32 | |
| SMO | 20 | 18 | |
| STO trimer | |||
| T1 | A-A-A | 17 | NDc |
| T2 | A,A,B | Trace amt | ND |
| T3 | A,A,C; A,B,B | ND | ND |
| T4 | B-B-B; A,B,C | 21 | 5 |
| T5 | B,B,C; A,C,C | 34 | 30 |
| T6 | B,C,C | 22 | 38 |
| T7 | C-C-C | 6 | 27 |
The HMW/LMW succinoglycan ratios for strains NRG247 and NRG185 were 1.56 and 0.5, respectively.
Structures of SMOs A, B, and C are shown in Fig. 1. The possible STOs indicated with commas have multiple possible arrangements.
ND, none detected.
Bacterial strains, growth conditions, and production of succinoglycan.
S. meliloti NRG185 and S. meliloti NRG247 were grown in a salts-glutamate-mannitol (SGM) medium (pH 7.0) complemented with biotin, thiamine, and trace elements, as described by Simsek et al. (27) and Zevenhuizen and van Neerven (28). One liter of the SGM medium in a 2.8-liter Fernbach culture flask was inoculated with 10 ml of the strain from an overnight culture. The cells were cultured at 28°C for 5 days and then centrifuged at 20,000 × g for 30 min, and the supernatant, which contains the secreted exopolysaccharides, was concentrated by rotary evaporation and dialyzed against distilled H2O at 4°C for 2 days, with several changes of distilled H2O. It was again concentrated by rotary evaporation and then freeze-dried.
Some samples were dissolved into 200 ml of 0.1 M NaCl, and the high-molecular-weight (HMW) exopolysaccharide was precipitated with 3 volumes of ethanol. The supernatant was collected by centrifugation at 10,000 × g (20 min), and 7 volumes of ethanol were added to precipitate low-molecular-weight (LMW) oligosaccharides, which were collected by centrifugation (as described above), redissolved in distilled water, and freeze-dried (18).
SEC.
Size exclusion chromatography (SEC) analysis was performed with a Dionex BioLC chromatographic system (Dionex Corp., Sunnyvale, CA) equipped with a Hewlett-Packard 1047A (Hewlett-Packard Company, Palo Alto, CA) refractive index detector and a Superose 12 column (Amersham Bioscience, Piscataway, NJ) or a Sephadex G-75 column as previously described (17, 29). The sample (500 μg) was dissolved in 100 μl of ammonium formate eluent (50 mM; pH 5.5) and injected into the system. The sample was eluted with the same eluent at a flow rate of 0.4 ml/min. Fractions were assayed by the phenol-sulfuric acid method (30), pooled according to peak cutoff, and lyophilized.
DEAE anion-exchange chromatography.
Anion exchange chromatography was used for detailed analysis of the oligosaccharides separated by Sephadex G-75 SEC. A column (1.5 by 30 cm) of DEAE Sephadex A-25 (Sigma) was eluted with a gradient eluent system as described by Wang et al. (17). Fractions were collected (2 ml), and carbohydrate contents were analyzed as described above.
MALDI-TOF MS and FAB-MS.
The STO fractions were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using an Applied Biosystems (Framingham, MA) Voyager DE PRO mass spectrometer with a nitrogen laser (337 nm UV laser) as described previously (18). Fast atom bombardment (FAB)-MS was performed using a ZAB-SE instrument (VG, Manchester, England) in the negative mode, with an ionizing voltage of 70 eV and an accelerating voltage of 10 kV, operating at low resolution (ca. 1:800). The samples were dissolved in ultrapure H2O, and 1-μl amounts were added to a matrix of thioglycerol (2 μl). The scan range was 300 to 3,000 atomic mass units (amu).
RESULTS
Separation of HMW and LMW succinoglycans by SEC.
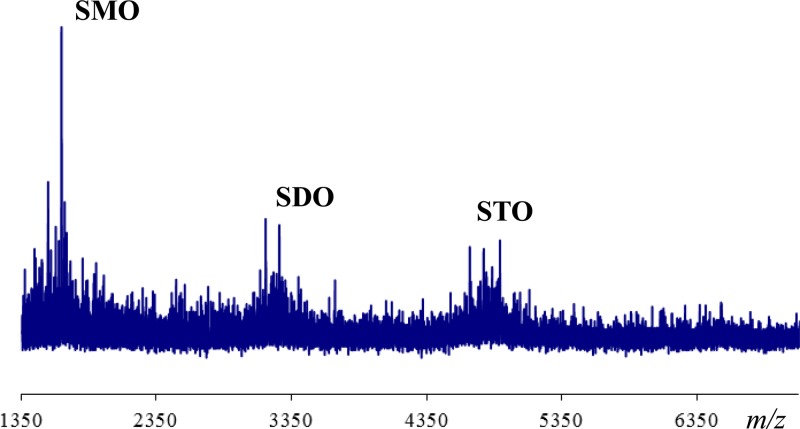
Our previous work has shown that M. truncatula-S. meliloti interactions may involve ecotype-strain specificity, as S. meliloti strains NRG247 and Rm41 are Fix+ (compatible) with M. truncatula A20 and Fix− (incompatible) with M. truncatula A17 and the Fix phenotypes are reversed with S. meliloti strains NRG185 and NRG34 (NRG185 and NRG34 are Fix+ [compatible] with M. truncatula A17 and Fix− [incompatible] with M. truncatula A20) (18). Based on the differences in host specificity between these S. meliloti strains, we undertook a detailed chemical analysis to investigate the fine structure of the succinoglycans produced by the two S. meliloti strains (NRG185 and NRG247) that display different host specificities. After these strains were grown in SGM cultures, crude succinoglycan preparations were prepared and analyzed by size exclusion chromatography (SEC), using a Superose 12 column (Fig. 2A and B). Both strains produce high-molecular-weight (HMW) and low-molecular-weight (LMW) succinoglycans, although the ratio of HMW to LMW succinoglycans was lower for strain NRG185 than for NRG247 (based on integration values), at ∼0.50 and ∼1.50 for HMW and LMW succinoglycans, respectively (Table 1). The LMW oligosaccharide pools were analyzed by MALDI-MS for both strains. The spectrum from NRG185, shown as an example in Figure 3 (NRG247 data are not shown), confirmed that the LMW fraction was composed of succinoglycan monomeric, dimeric, and trimeric oligosaccharides (SMOs, SDOs, and STOs, respectively; see Materials and Methods).
Fig 2.
Chromatographic analysis of succinoglycans from S. meliloti NRG247 and S. meliloti NRG185. (A) Analysis of overall succinoglycan production by S. meliloti NRG247 with size exclusion chromatography (SEC) using a Superose 12 column. (B) Analysis of overall succinoglycan production by S. meliloti NRG185 with SEC using a Superose 12 column. (C) Analysis of low-molecular-weight (LMW) succinoglycan from S. meliloti NRG247 by SEC using a Sephadex G-75 column. (D) Analysis of LMW succinoglycan from S. meliloti NRG185 with SEC using a Sephadex G-75 column. High-molecular-weight (HMW) succinoglycan and succinoglycan trimeric, dimeric, and monomeric oligosaccharides (STO, SDO and SMO, respectively) are labeled in the chromatograms. (E) Analysis of S. meliloti NRG247 STOs from Sephadex G-75 column using a DEAE Sephadex A-25 column. (F) Analysis of S. meliloti NRG185 STOs from Sephadex G-75 column using a DEAE Sephadex A-25 column. STOs are labeled as described in Table 1.
Fig 3.
Analysis of the low-molecular-weight fraction of S. meliloti NRG185 succinoglycan from Superose 12 size exclusion chromatography column by MALDI-TOF MS.
Analysis of SMO fractions by mass spectrometry.
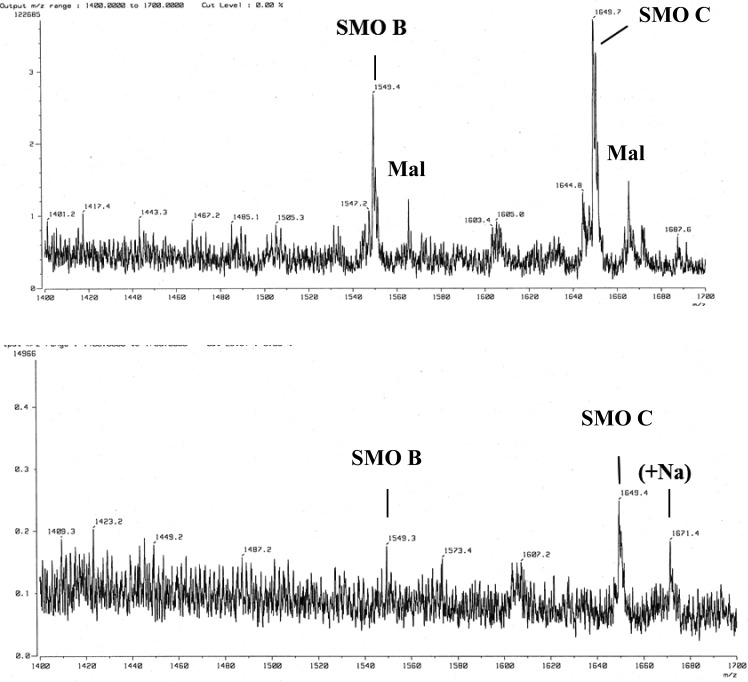
The SMO populations produced by S. meliloti NRG247 and NRG185 were analyzed by fast atom bombardment (FAB)-MS for high resolution (Fig. 4), since FAB-MS seemed to provide higher-resolution data for SMO populations. The analysis showed that NRG247 produces an abundance of both SMO C and SMO B, as well as the malylated species (Fig. 4, top), which are also produced by S. meliloti Rm41, another strain that is compatible (Fix+) with M. truncatula A17 (18). The mass spectrum of the SMO population from NRG185 showed that it produces primarily C (Fig. 4, bottom). There was no SMO A detected in any preparation from either strain, although SMO A was detected in the LMW EPS from Rm41 (18).
Fig 4.
FAB-MS analysis of succinoglycan monomeric oligosaccharides (SMO) from S. meliloti NRG247 (top) and S. meliloti NRG185 (bottom). All mass ions are mass + Na − 1. Additional salt adducts are also present (e.g., 1671 in the bottom panel). Ions were detected at m/z 1565 and m/z 1665, which represents the presence of an additional O in each oligosaccharide (i.e., 1549 + 16 and 1649 + 16) and has been shown to be due to the presence of malate (Mal) instead of succinate (18).
Separation of LMW oligosaccharides by SEC.
The LMW succinoglycan from the ethanol-precipitated sample was fractionated by SEC using Sephadex G-75, with a separation range of 1,000 to 50,000 Da. This resulted in the separation of the STO, SDO, and SMO oligosaccharides (Fig. 2C and D). Four primary components were detected, pooled, and analyzed by MALDI-TOF MS (data not shown). Peak 1, which eluted in the void volume of the column, yielded no detectable mass ions, indicating that it was residual HMW succinoglycan and beyond the mass range of the instrument. Peak 2 was identified as STO, peak 3 as SDO, and peak 4 as SMO for both preparations. The percentages for each fraction were calculated from the peak areas from the SEC (Table 1).
Separation of STOs by anion exchange chromatography.
The STO pools were further fractionated on a DEAE Sephadex A-25 to separate the distinct STOs (Fig. 2E and F). The elution times for each STO present were similar for both strains; however, the fractionation pattern was significantly different, as highly charged STOs were predominant in the column profile of NRG185, whereas oligosaccharides with a low charge density were more abundant in the column profile of NRG247. The nonrandom patterns differ significantly from the chromatography profile of the STOs from S. meliloti Rm1021, which show a nearly normal distribution, having both high and low charged STOs (17). The quantifications of the trimers were determined using the integration values of the peaks from DEAE Sephadex A-25 column chromatography (Table 1).
Analysis of STO fractions by mass spectrometry.
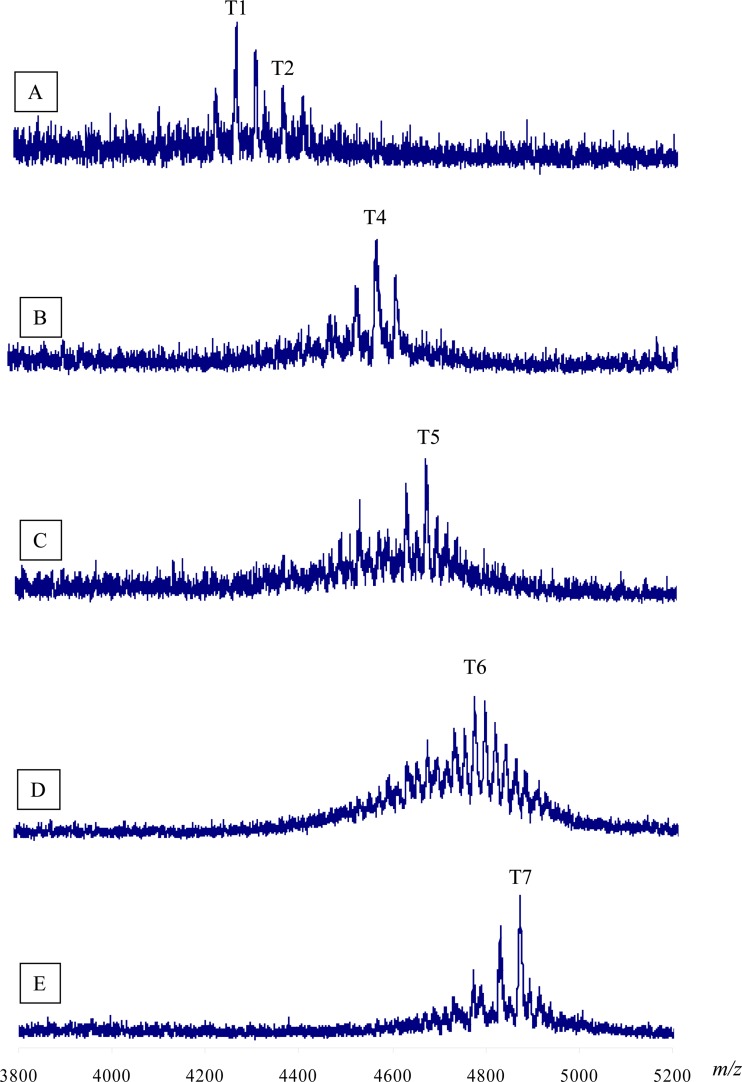
The distinct STOs were identified by the Na+ adducts of the mass ions in MALDI-TOF MS analyses of the oligosaccharides from S. meliloti NRG247 (Fig. 5 and Table 1); the other ions in each of the spectra are due to multiple salt adducts and lack of acetate or both, as well as some cross contamination of other STOs during the separation process. This complexity is to be expected in the mass range of these analyses. The peak 1 material from the DEAE Sephadex A-25 separation of the NRG247 oligosaccharides was shown to contain trimer 1 (T1) (A-A-A), which has no succinate in any repeat. There were also additional salt adducts, as well as ions corresponding to the presence of additional acetyl groups. Also present in peak 1 was a trace of T2, with one succinate group on one of the octasaccharide repeats (A,A,B [commas indicate multiple possible arrangements]).
Fig 5.
Analysis of succinoglycan trimeric oligosaccharides produced by S. meliloti NRG247 using MALDI-TOF mass spectroscopy. STOs were collected from a DEAE Sephadex A-25 column. Each of the fractions is labeled above the spectrum. Other ions in each spectrum are due to additional Na adducts, mass ions (not Na adducts), nonacetylated oligosaccharides, Na adducts of the nonacetylated oligosaccharides, and cross-contamination of other oligosaccharides during chromatography. The latter three classes of ions were most abundant in spectrum D.
Peak 2 contained T4, which has three succinyl groups (i.e., either B-B-B or A,B,C); there were also ions corresponding to an additional acetyl group, as well as the absence of acetyl groups. The mass spectrum of peak 3 contained the T5 oligosaccharides (B,B,C or A,C,C). Each subsequent peak contained the other STOs (Table 1), as well as the associated salt adducts and differences in acetylation. The estimated relative abundance of each oligosaccharide (based on the peak areas in the results shown in Figure 2) is given in Table 1. Note that approximately 50% of the T7 oligosaccharides appear to lack one or more acetyl groups. In contrast to NRG247, strain NRG185 produced no detectable T1, T2, or T3, and T5, T6, and T7 accounted for approximately 95% of the total STOs.
DISCUSSION
The results of this study showed that the production of succinoglycan oligosaccharides by S. meliloti is strain specific. The biosynthesis of succinoglycan in some of the S. meliloti strains, for example, S. meliloti Rm1021, has been well characterized, but the molecular mechanisms behind the succinoglycan-mediated nodule invasion in plant have not been completely understood (31). The presence of plant systems that sense structural features of symbiotically active EPS is suggested by the ability of a particular exopolysaccharide to promote nodule invasion by a rhizobial species on some of its plant hosts but not on other hosts (12, 32). In reporting their observations, Jones et al. (31) discussed the fact that LMW exopolysaccharides (defined in this study as STOs, SDOs, and SMOs) may act as signals to the plant that allow the initiation of critical reactions necessary for symbiosis (31). The requirement for LMW EPS I for the Fix+ symbiotic relationship might occur because LMW forms can reach the root-hair cell membrane to deliver a signal, while the plant cell wall stops the entrance of high-molecular-weight forms into the cell membrane.
Further understanding of the molecular basis of symbiotic interactions between rhizobia and leguminous plants that result in symbiotic nitrogen fixation is important, and ultimately it might be possible to transfer such symbiotic nitrogen fixation capacities to crops of agricultural importance. Here, we report that S. meliloti strain NRG247 displays a Fix+ phenotype on M. truncatula A20 and a Fix− phenotype on M. truncatula A17. On the other hand, these phenotypes are reversed for S. meliloti NRG185. Our study provides insight into the molecular basis of this difference in host compatibility, as S. meliloti species NRG247 and NRG185 display not only quantitative but also qualitative differences in their succinoglycan populations. Furthermore, the production of a significant amount of STO A-A-A by S. meliloti NRG247, without any detectable SMO A, indicates that all the nonsuccinylated repeats were used to produce T1 (and a trace of T2), demonstrating a specific biosynthetic pathway for the active oligosaccharides, not a random polymerization of the available subunits. After the first fraction, the next major fraction was T4, which also indicates that STO polymerization is not random.
It appears that an STO containing at least two doubly succinylated repeating units (T6 or T7) is the structure required for compatibility on M. truncatula A17, and given the fact that NRG247 produces a relative abundance of T5 and T6, it is likely to be the completely succinylated STO, T7, that is the active signal on that host plant. Of course, there may be other structural factors involved: if T6 were active in M. truncatula A17, it could be the order of the subunits that is essential, or the presence of a malylated repeat unit, and these analyses were beyond the scope of this work. In contrast to NRG247, strain NRG185 does not produce any STO without any succinate (T1, T2, or T3), so if T1 is the active signal in M. truncatula A20, there would be none secreted by NRG185.
ACKNOWLEDGMENT
This work was supported by grant MCB-0316785 from the National Science Foundation.
Footnotes
Published ahead of print 1 March 2013
REFERENCES
- 1. Fisher RF, Long SR. 1992. Rhizobium-plant signal exchange. Nature 357:655–660 [DOI] [PubMed] [Google Scholar]
- 2. Stacey G, Libault M, Brechenmacher L, Wan J, May GD. 2006. Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9:110–121 [DOI] [PubMed] [Google Scholar]
- 3. Carlson RW, Reuhs BL, Forsberg LS, Kannenberg EL. 1999. Rhizobial cell surface carbohydrates, p 53–90 In Goldberg JB. (ed), Genetics of bacterial polysaccharides. CRC Press, Boca Raton, FL [Google Scholar]
- 4. Kijne JW. 1992. The rhizobium infection process, p 349–398 In Stacey G, Burris RH, Evans HJ. (ed), Biological nitrogen fixation. Chapman & Hall, New York, NY [Google Scholar]
- 5. Leigh JA, Coplin DL. 1992. Exopolysaccharides in plant-bacterial interactions. Annu. Rev. Microbiol. 46:307–346 [DOI] [PubMed] [Google Scholar]
- 6. Noel KD. 1992. Rhizobial polysaccharides required in symbioses with legumes, p 341–357 In Verma DPS. (ed), Molecular signals in plant-microbe communications. CRC Press, Boca Raton, FL [Google Scholar]
- 7. Gibson KE, Kobayashi H, Walker GC. 2008. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42:413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leigh JA, Walker GC. 1994. Exopolysaccharides of rhizobium—synthesis, regulation and symbiotic function. Trends Genet. 10:63–67 [DOI] [PubMed] [Google Scholar]
- 10. Levery SB, Zahn H, Lee CC, Leigh JA, Hakomori S. 1991. Structural analyses of a second acidic exopolysaccharide of Rhizobium meliloti that can function in alfalfa root nodule invasion. Carbohydr. Res. 210:339–348 [DOI] [PubMed] [Google Scholar]
- 11. Zhan H, Levery SB, Lee CC, Leigh JA. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. U. S. A. 86:3055–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672 [DOI] [PubMed] [Google Scholar]
- 13. Reuber TL, Reed J, Glazebrook J, Glucksmann MA, Ahmann D, Marra A, Walker GC. 1991. Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance. Biochem. Soc. Trans. 19:636–641 [DOI] [PubMed] [Google Scholar]
- 14. Gonzalez JE, Reuhs BL, Walker GC. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. U. S. A. 93:8636–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Åman P, McNeil M, Franzén L-E, Darvill AG, Albersheim P. 1981. Structural elucidation, using h.p.l.c.-m.s. and g.l.c.-m.s., of the acidic polysaccharide secreted by Rhizobium meliloti strain 1021. Carbohydr. Res. 95:263–282 [Google Scholar]
- 16. Reinhold BB, Chan SY, Reuber TL, Marra A, Walker GC, Reinhold VN. 1994. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J. Bacteriol. 176:1997–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L-X, Wang Y, Pellock BJ, Walker GC. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 181:6788–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simsek S, Ojanen-Reuhs T, Stephens SB, Reuhs BL. 2007. Strain-ecotype specificity in Sinorhizobium meliloti-Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 189:7733–7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones KM. 2012. Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula. J. Bacteriol. 194:4322–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snyman P, Strijdom BW. 1980. Symbiotic characteristics of lines and cultivars of Medicago truncatula inoculated with strains of Rhizobium meliloti. Phytophylactica 12:173–176 [Google Scholar]
- 21. Tirichine L, de Billy F, Huguet T. 2000. Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol. 123:845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parra-Colmenares A, Kahn ML. 2005. Determination of nitrogen fixation effectiveness in selected Medicago truncatula isolates by measuring nitrogen isotope incorporation into pheophytin. Plant Soil 270:159–168 [Google Scholar]
- 23. Rangin C, Brunel B, Cleyet-Marel JC, Perrineau MM, Bena G. 2008. Effects of Medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural Sinorhizobium species community. Appl. Environ. Microbiol. 74:5653–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller SH, Elliot RM, Sullivan JT, Ronson CW. 2007. Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153:3184–3195 [DOI] [PubMed] [Google Scholar]
- 25. Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. 2010. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oldroyd GED, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45:119–144 [DOI] [PubMed] [Google Scholar]
- 27. Simsek S, Ojanen-Reuhs T, Marie C, Reuhs BL. 2009. An apigenin-induced decrease in K-antigen production by Sinorhizobium sp. NGR234 is y4gM- and nodD1-dependent. Carbohydr. Res. 344:1947–1950 [DOI] [PubMed] [Google Scholar]
- 28. Zevenhuizen LPTM, van Neerven ARW. 1983. Gel-forming capsular polysaccharide of Rhizobium leguminosarum and Rhizobium trifolii. Carbohydr. Res. 124:166–171 [Google Scholar]
- 29. Reuhs BL, Glenn J, Stephens SB, Kim JS, Christie DB, Glushka JG, Zablackis E, Albersheim P, Darvill AG, O'Neill MA. 2004. l-Galactose replaces l-fucose in the pectic polysaccharide rhamnogalacturonan II synthesized by the l-fucose-deficient mur1 Arabidopsis mutant. Planta 219:147–157 [DOI] [PubMed] [Google Scholar]
- 30. Dubois M, Gilles K, Hamilton J, Rebers P, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 20:350–356 [Google Scholar]
- 31. Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc. Natl. Acad. Sci. U. S. A. 105:704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Staehelin C, Forsberg LS, D'Haeze W, Gao MY, Carlson RW, Xie ZP, Pellock BJ, Jones KM, Walker GC, Streit WR. 2006. Exo-oligosaccharides of Rhizobium sp. strain NGR234 are required for symbiosis with various legumes. J. Bacteriol. 188:6168–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]