Abstract
A striking characteristic of mycobacteria is the presence of an unusual outer membrane which forms a thick permeability barrier and provides resistance to many antibiotics. Although specialized proteins must reside in this layer, only few mycolate outer membrane (MOM) proteins have been identified to date. Their discovery is complicated by difficulties in obtaining good separation of mycobacterial inner and outer membranes. During our efforts to identify novel mycobacterial outer membrane proteins (MOMPs), we discovered that we can enrich for MOMPs using differential solubilization of mycobacterial cell envelopes. Subsequently, these different fractions were analyzed by nano liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS). This proteomic analysis confirmed that our marker proteins for inner membrane and MOM were found in their expected fractions and revealed a few interesting candidate MOMPs. A number of these putative MOMPs were further analyzed for their expression and localization in the cell envelope. One identified MOMP, MMAR_0617 of Mycobacterium marinum, was purified and demonstrated to form a large oligomeric complex. Importantly, this protein showed a clear single-channel conductance of 0.8 ± 0.1 ns upon reconstitution into artificial planar lipid bilayers. The most surprising feature of MMAR_0617 is a long C-terminal threonine-rich domain with extensive modifications. In summary, we have identified a novel mycobacterial outer membrane porin with unusual properties.
INTRODUCTION
The architecture of the cell envelope of Mycobacterium tuberculosis has been an enigma for many years. This extraordinary hydrophobic cell envelope contains, in addition to a relatively normal cytoplasmic or inner membrane (IM), a polymer of peptidoglycan and arabinogalactan, which is covalently linked to beta-hydroxy branched-chain fatty acids of considerable size, the mycolic acids. These mycolic acids are the key components of the atypical outer membrane, which has been called the mycolate outer membrane (MOM) (1). Although the existence of a second membrane was already suggested by Minnikin (2), it was only recently unequivocally shown as a lipid bilayer structure by cryoelectron tomography (3–5). As the observed thickness of the MOM is 7 to 8 nm, mycolic acids are proposed to be present in a folded conformation (4, 6), which is in agreement with recent Langmuir assays (7). The outer leaflet of this membrane is probably formed by an array of unusual mycolate- or mycobacterium-specific (glyco)lipids, such as trehalose dimycolate, phtiocerol dimycoserosate, and sulfolipids.
The MOM forms an exceptionally dense barrier, with even lower permeability than most Gram-negative bacteria (8). In analogy with the Gram-negative outer membrane, specific outer membrane proteins should be present to accommodate processes such as nutrient uptake, protein secretion, and membrane biogenesis. Although more than 60 integral outer membrane proteins have been described for the Gram-negative Escherichia coli (9), not even a few mycobacterial outer membrane proteins (MOMPs) are known. Channel activity, indicative of specific porin proteins, has been identified in cell extracts of Mycobacterium bovis BCG. These experiments showed large cation-selective channels with a single-channel conductance of 4.0 ns and small anion-selective channels with single-channel conductance of approximately 0.8 ns (10). However, the nature of these proteins was never established. The best-studied MOMP is the major Mycobacterium smegmatis porin MspA. This protein forms an extremely stable octameric goblet-like β-barrel with a central 10-nm channel (11). Although these features significantly differ from the general monomeric or trimeric porins found in Gram-negative bacteria (12), all these proteins have in common is that they cross the membrane using a β-barrel structure. Unfortunately, no homologues of MspA are found in M. tuberculosis. For this species, the only proteins described to reside in the MOM are MctB (mycobacterial copper transport protein B; Rv1698) (13) and OmpATb (Rv0899). MctB is a putative porin (14) implicated to be involved in copper efflux (13), whereas OmpATb shows homology with the periplasmic domain of E. coli OmpA and has been the subject of a continuous debate about its function, localization, and structure. Originally, OmpATb has been described as a surface-accessible porin with channel activity (15), but recently these results have been questioned (16).
Other mycolic acid-producing bacteria, such as Corynebacterium glutamicum and Nocardia sp., provide further knowledge of the proteins residing in the MOM. Channel-forming porins resembling MspA have been identified in both these species (17). In addition, porins have been identified that are proposed to have an α-helical structure (18) or modified by O-mycoloylation (19). These results indicate that the classical model of outer membrane proteins as (predominantly) being β-barrels might not apply to all mycolate outer membrane proteins. Recently, various attempts were made to define proteins in the cell envelope or cell wall fractions of different strains of M. tuberculosis, M. bovis BCG, and Mycobacterium avium (20–22), but none were dedicated to the specific identification of MOMPs. In this study, we developed a method to specifically enrich for MOM proteins by differential detergent extraction of mycobacterial cell envelopes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Mycobacterium marinum strain E11 (23) and M. bovis BCG were routinely grown at 30°C in Middlebrook 7H9 (Difco) liquid medium and on Middlebrook 7H10 plates (Difco) supplemented with 10% Middlebrook ADC and 0.05% Tween 80 (BD Biosciences) or only oleic acid-albumin-dextrose-catalase (OADC), respectively. Escherichia coli strain DH5α was used for amplification and manipulation of plasmid DNA. Antibiotic concentration used was 50 μg/ml hygromycin.
Subcellular fractionation procedure.
A culture of 200 to 500 ml of M. marinum or M. bovis BCG was grown to mid-log phase. Prior to lysis, bacteria were killed by the addition of 100 μg/ml ciprofloxacin (Sigma) to the culture medium and incubation for an hour at 30°C. Out of several killing procedures tested, this was the one with the smallest effect on cell envelope isolation. Cells were harvested at 4°C by centrifugation at 4,000 × g, washed three times with phosphate-buffered saline (PBS), and resuspended in PBS supplemented with proteinase inhibitor (Complete, Roche), 1 mM ethylenediamine tetra acetic acid (EDTA), and 0.1 mg/ml DNase. Cells were broken either by a three-time passage through a French press (12,000 lb/inch) or two passages through a One-Shot cell disrupter (Constant Systems). After the first passage, 1 mM dithiothreitol (DTT) was added to the lysate. Lysates were subjected to low-speed centrifugation for 5 min at 3,000 × g to remove unbroken cells, which was repeated twice. Subsequently, cytosolic and cell envelope fractions were separated by centrifugation at 27,000 × g. The obtained pellet was resuspended in PBS and centrifuged again at 27,000 × g to obtain the final cell envelope fraction. In addition, the soluble fraction of the first centrifugation step, representing the cytosol fraction, was subsequently centrifuged at 100,000 × g to further purify this fraction.
Detergent extraction of cell envelope proteins.
Cell envelope fractions were resuspended in PBS or PBS containing either 1% (wt/vol) n-octyl-β-d-glucopyranoside (OBG; Calbiochem), 0.5% (vol/vol) Genapol X-80 (Fluka), 1% (wt/vol) sodium-lauryl sarcosinate (Sarkosyl, Sigma), or 1% (vol/vol) Triton X-100 (Sigma), all above critical micelle concentration (CMC) values, and incubated for 30 min at room temperature. Subsequently, samples were centrifuged twice at 27,000 × g to separate the soluble from the insoluble fraction. The final soluble fraction was subjected to TCA precipitation, whereas the pellet fraction was washed by PBS. Both fractions were dissolved in SDS solubilization buffer (50 mM Tris-HCl [pH 6.8], 5 mM EDTA, 10% [vol/vol] glycerol, 2% [wt/vol] SDS, 100 mM DTT, and bromophenol blue) and boiled at 95°C prior to analysis by SDS-PAGE and immunoblotting.
SDS-PAGE, blue native PAGE, and immunostaining.
Samples were loaded on SDS-PAGE gels (Bio-Rad/Hoefer) or NativePAGE gels (Invitrogen) and subsequently visualized by staining with Coomassie GT-250 (Bio-Rad) or PageSilver silver staining kit (Fermentas), by immunoblotting, or by incubation with horseradish peroxidase (HRP)-conjugated ConA (Sigma). Antisera used for immunostaining are rabbit antisera reactive to FtsH (24) or EccC5 (25) and mouse monoclonal antibodies against the influenza hemagglutinin (HA) epitope (HA.11; Covance), the His6 epitope (Roche), or GroEL2 (CS44; Colorado State University). Rabbit antiserum recognizing MctB was raised against a peptide corresponding to amino acids 123 to 135 of MctB (Rv1698), prepared by Innovagen (Sweden). The presence of bound HRP-conjugated secondary antibodies was detected via chemiluminescence (Pierce) using a charge-coupled-device (CCD) camera (Bio-Rad). Provided QuantityOne software (Bio-Rad) was used to (semi)quantify signals obtained via immunostaining.
Mass spectrometric analysis.
Protein lanes from Coomassie brilliant blue (CBB)-stained SDS-PAGE gels were excised and prepared for nano liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) analysis as previously described (5, 26), using Scaffold 3 for protein identification.
Bioinformatic analysis.
Proteins were quantified by spectral counting and exported to Microsoft Excel (see Table S2 in the supplemental material). All identified proteins were annotated and functionally classified using MarinoList (27). Prediction of possible signal sequences and transmembrane domains (TMDs) was performed using the SignalP 3.0 and TMHMM version 2.0, respectively. Subsequently, identified proteins were ordered according to the number of spectral counts detected in the detergent-solubilized fraction compared to the total number of counts. Putative MOMPs (i.e., >90% extracted in three biological replicates) were subjected to BLASTp analysis. Additionally, spectral counts were normalized by dividing the spectral counts per protein by the sum of all counts per sample, multiplied by the average sum across all samples. Subsequently, the fold change (Fc) and P values between counts for detergent pellet and detergent supernatant were calculated using the beta binominal test for spectral count data (28).
Molecular cloning.
Selected M. marinum genes mmar_3191 (ftsQ), mmar_4637 (ompA), mmar_2503 (mctB), dppA, mmar_0617, mmar_4057, mmar_4366, and mmar_5387 were amplified by PCR using Pfu polymerase (Fermentas) and specific anchored primers containing restriction sites and a C-terminal HA tag (YPYDVPDYA) or His6 tag (see Table S1 in the supplemental material). After restriction with NheI/XbaI/SpeI and BamHI/BglII (for specific details, see Table S1), the resulting PCR fragments were ligated into NheI/BamHI-digested pSMT3-LipYtb (29), directly behind the hsp60 promoter. Resulting vectors were checked by restriction digestion and nucleotide sequence analysis of the relevant part.
Analysis of putative MOMPs.
Putative MOMPs were tested for heat modifiability by resuspending mycobacterial cell envelopes in solubilization buffer containing either 2% SDS and 100 mM DTT or 0.2% SDS and no DTT, incubating these samples at 95°C or 37°C, respectively, and then running them on a polyacrylamide gel lacking SDS. To examine protease sensitivity of MOMPs, isolated cell envelopes were either directly incubated with different concentrations of proteinase K (Sigma) or trypsin (Sigma) or solubilized by 1% OBG prior to protease treatment. Protease activity was inhibited after incubation for 30 min at room temperature using phenylmethylsulfonyl fluoride or trypsin inhibitor (Sigma).
Immunogold electron microscopy (EM).
Fixation of bacterial cells, cryosectioning, and immunogold labeling of fixed cells were essentially done as described in reference 5. The HA.11 mouse monoclonal antibody (Covance), bridging rabbit anti-mouse antibody (Dako) and protein A attached to 10 nm gold (University Utrecht), was used to visualize the HA-tagged MOMPs. Localization of gold particles was quantified double-blind. For each strain, a representative 50 to 200 cells were counted for gold particles, which were assigned to three different compartments: intracellular (cytosol), associating with the observed membrane layers (membrane), or localized external to the membrane layers, including the electron transparent zone (cell wall).
Purification of MMAR_0617-His.
Cell envelope preparations of M. marinum E11 with or without pSMT3::MMAR_0617-His or pSMT3::MMAR_0617ΔC-His were incubated for 30 min at room temperature in phosphate buffer (20 mM Na phosphate [pH 7.8], 500 mM NaCl) containing 1% (vol/vol) Triton X-100 (Sigma) and 10 mM imidazole and centrifuged at 27,000 × g to obtain the solubilized proteins. Subsequently, these protein samples were loaded on HisTrap HP columns (GE Healthcare) equilibrated with wash buffer (phosphate buffer containing 10 mM imidazole and 1% [vol/vol] Triton X-100) coupled to an ÄKTA FPLC system (Amersham Biosciences) at 0.5 ml/min. Proteins were eluted by applying a linear gradient of 40 to 160 mM imidazole (buffer containing either 1% or 0.1% [vol/vol] Triton X-100). Fractions containing the Ni2+-purified protein were optionally precipitated using ammonium sulfate and further purified by size exclusion chromatography using a SuperdexTM 200 HR 10/30 column (Amersham Pharmacia no. 17-1088-01). In a separate approach, HA-labeled MMAR_0617 was isolated from solubilized cell envelope proteins using the HA tag immunoprecipitation (IP)/coimmunoprecipitation (Co-IP) kit (Pierce). Supplied buffers were supplemented with 1% (vol/vol) Triton X-100 to avoid protein aggregation.
Single-channel reconstitution and ion conductance measurements.
Virtually solvent-free lipid bilayers were formed as previously described (30, 31). 1,2-Diphytanoyl-sn-glycero-3-phosphatidylcholine (DPhPC), the most widely used lipid for the characterization of reconstituted channels, was used for membrane formation in a Teflon cell with an approximately 100-μm-diameter aperture in the 25-μm-thick Teflon partition. A buffer containing 1 M KCl and 10 mM HEPES, pH 7.4, served as the electrolyte. Standard silver-silver chloride electrodes from World Precision Instruments (WPI) were placed in each chamber to detect the ion current. Small amounts of purified MMAR_0617 stock solution were added to the cis side of the chamber (ground electrode). The addition of proteins into the aqueous phase resulted in a rapid dilution of the detergent below the CMC, causing channels to spontaneously insert into the lipid bilayers. An ion current flowing through the channel was induced by application of a transmembrane voltage. Conductance measurements were recorded by using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). The signal was filtered using a four-pole low-pass Bessel filter at a frequency of 10 kHz and sampled at 50 kHz, acquired using a Digidata 1440A digitizer, and analyzed using the pClamp 10.0 software (Axon Instruments, Foster City, CA). All measurements were repeated 10 times with different purification batches to establish confidence about the reproducibility.
RESULTS
Development of a differential detergent extraction method to enrich for MOMPs.
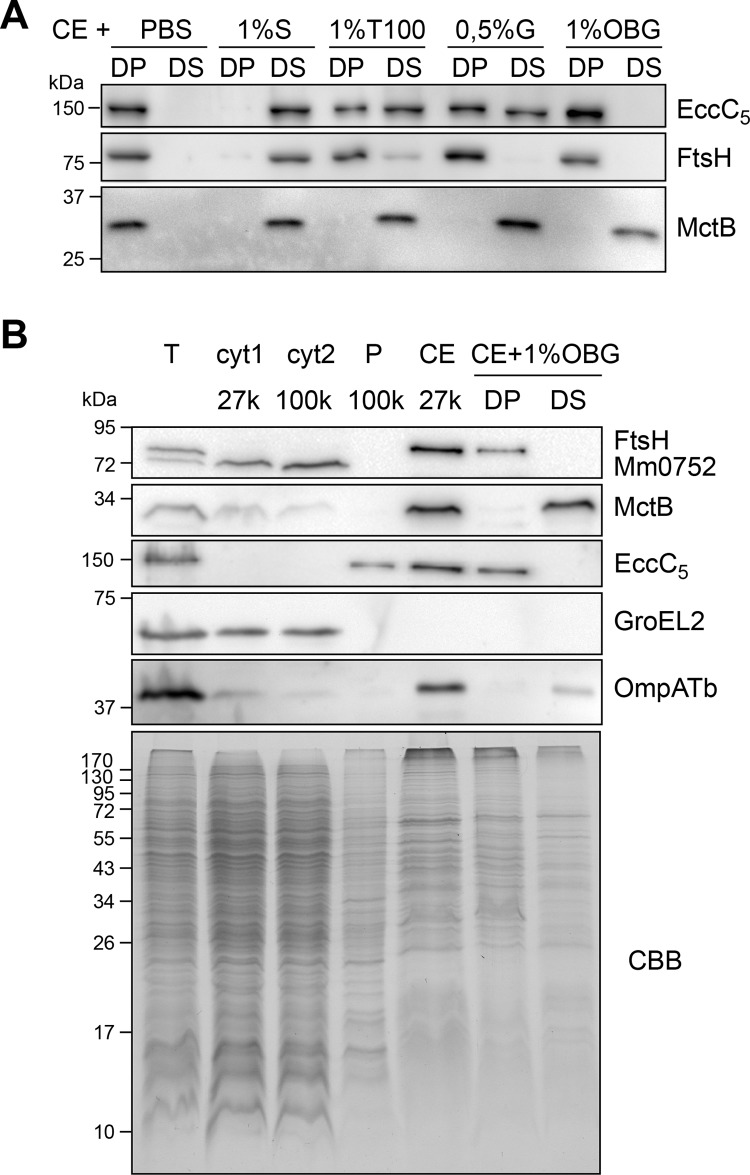
Several techniques have been developed for the subcellular fractionation of mycobacteria, but none of them have been successfully used to isolate and identify mycobacterial outer membrane proteins (32). In this work, we used a variety of different methods to identify inner and outer membrane proteins of M. marinum. As markers for successful fractionation, antibodies were used directed against the MOM protein MctB and against the inner membrane (IM) proteins EccC5 and FtsH (24); the latter antiserum additionally recognizes the cytosolic ATPase MMAR_0752 (33). The most commonly used method to isolate different cell envelope fractions is by differential centrifugation (34–36). Using our markers, we could show, in contrast to a previous study (36), that both IM and MOM proteins were pelleted by a centrifugation step at 27,000 × g (Fig. 1B). We also tried to separate IM and MOM proteins using sucrose gradient centrifugation (37), but this approach was not successful (results not shown). Next, we performed “shaving” of M. marinum cells using trypsin-coated beads that specifically cleave exposed protein domains (38). This procedure identified only abundant cytosolic and secreted proteins, such as GroEL and EsxA, and not any known or putative membrane proteins (results not shown). Specific labeling of surface proteins by whole-cell biotinylation using hydrophilic Sulfo-NHS-SS-biotin and subsequent purification of biotinylated proteins by streptavidin beads did not identify any known MOMP either. Finally, we succeeded to specifically isolate MOMPs using a differential detergent extraction protocol. Treatment of the cell envelope fraction with different detergents showed that most detergents, such as Triton X-100, Sarkosyl, and Genapol X-80, solubilized both IM and MOM marker proteins (Fig. 1A). However, by using n-octyl-β-d-glucopyranoside (OBG), we could differentially extract MctB, while leaving both IM proteins FtsH and EccC5 in the pellet fraction (Fig. 1A). Also, the putative MOMP OmpATb was extracted by this detergent (Fig. 1B). Interestingly, this specific detergent, OBG, has previously been used to specifically disrupt the MOM after treatment of whole mycobacterial cells (3). CBB staining of the detergent-treated fractions reproducibly showed that a large group of putative cell envelope proteins was specifically extracted with this detergent (Fig. 1B). To test the applicability of this method on other mycobacterial species, we decided to perform the same fractionation procedure, followed by differential detergent extraction with OBG, for M. bovis BCG. Also, here, we found that we could differentially solubilize MctB and OmpATb, while leaving the major fraction of FtsH in the detergent pellet (see Fig. S1A in the supplemental material).
Fig 1.
Mycobacterial outer membrane proteins can be specifically enriched for by extraction with OBG. Immunoblots probed with antisera recognizing the inner membrane proteins FtsH and EccC5, MOM proteins MctB and OmpATb, and cytosolic proteins MMAR_0752 and GroEL2. (A) Equal amounts of pellet (DP) and supernatant (DS) fractions after solubilization of M. marinum cell envelope (CE) with Sarkosyl (S), Triton X-100 (T100), Genapol X-80 (G), and n-octyl-β-d-glucopyranoside (OBG). (B) Immunoblot of fractions corresponding to equal OD units of total lysate (T), soluble fraction (cyt1 27k), and pellet fraction (cell envelopes [CE], 27k) after centrifugation at 27,000 × g, after an additional centrifugation step at 100,000 × g (cyt2 100k and P 100k), and detergent pellet (DP) and supernatant (DS) were obtained after solubilization of CE with 1% OBG. For the CBB-stained SDS-PAGE gel, all membrane fractions (last 4 lanes) were loaded in 3-fold excess compared to total and cytosolic fractions.
Identification of MOMP candidates.
To identify which proteins of M. marinum are specifically extracted by OBG, both the detergent pellet and supernatant fractions of three independent biological replicates were analyzed by nano liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) (see Fig. S1B and Table S2 in the supplemental material). Spectral counts of identified proteins were normalized, and supervised clustering of the fold change between detergent pellet and supernatant confirmed major differences in protein content (see Fig. S1C). Next, we determined for every detected protein the relative amount of peptides extracted by the detergent (i.e., found in the detergent supernatant fraction compared to the pellet fraction). Both known MOMPs, MctB and OmpATb, were found to be >90% extracted, whereas the IM markers FtsH and EccC5 were on average 29% and 32% extracted (see Table S2). Based on these findings, we divided the identified proteins into three groups: (i) a putative IM protein group, containing the proteins that were less than 30% extracted; (ii) a putative MOMP group with proteins, which were over 90% extracted; and (iii) a middle group with all the other proteins (Fig. 2A). In silico analysis for signal peptide, lipobox, PE/PPE, and transmembrane domain (TMD) predictions showed that the IM protein group had an enrichment of proteins with multiple predicted TMDs. The detergent-extracted group enriched for MOMPs consisted of 219 proteins and was enriched for lipoproteins, putative secreted proteins (i.e., containing predicted signal peptides), and proteins with one putative TMD (Fig. 2A). Functional classification analysis (27) indicated that the latter group contains more conserved hypothetical proteins and proteins involved in cell wall and cell processes compared to the other groups (Fig. 2B). Subsequently, we purged this list based on several criteria. Lipoproteins (which could be either in IM or MOM) and proteins with a known cytosolic or inner membrane function were excluded. An additional selection criterion is the absence of close homologs in closely related species that lack a MOM, such as Streptomyces. Notably, because MOMPs reside in a unique membrane and therefore might have unexpected features, we did not exclude proteins without a signal sequence or an overall β-sheet prediction. Based on these criteria, we obtained a shortlist of 64 candidate MOMPs (see Table S3 in the supplemental material), of which a number of proteins were selected for further characterization.
Fig 2.
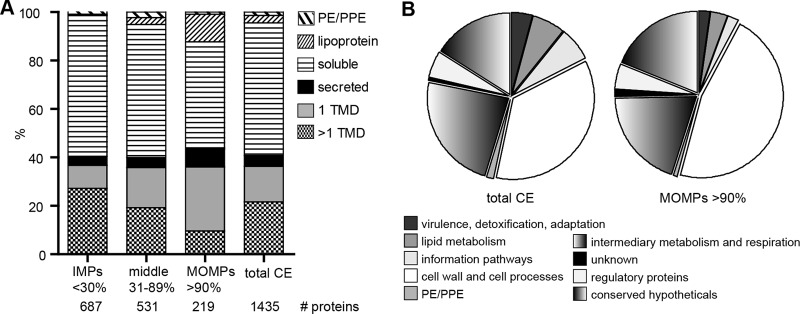
Mass spectrometry analysis of differentially detergent-extracted cell envelopes. (A) Relative detergent extractability of detected proteins, grouped according to the number of predicted TMDs, presence of predicted signal peptides, lack of TMDs (secreted proteins) and lipoboxes, and lack of TMDs (lipoproteins) and PE/PPE motifs. The remaining proteins are considered soluble. Proteins that are <30% extracted are considered putative inner membrane proteins (IMPs), 31 to 89% extracted are undetermined, and >90% extracted are putative mycobacterial outer membrane proteins (MOMPs). The relative abundance of protein groups is indicated. (B) Functional classification of detected proteins in total CE and MOMP groups, as described by the Mycobrowser (27).
Expression and characterization of putative MOMPs.
Four candidate MOMPs, all hypothetical proteins that were reproducibly detergent extractable in all biological replicates, were selected for further characterization, namely, MMAR_0617, MMAR_4057, MMAR_4366, and MMAR_5387 (Table 1; see also Fig. S2A in the supplemental material). The latter two candidates are described as core mycobacterial genes (39) that are highly conserved among mycolata but absent in Streptomyces species. MMAR_0617 was chosen for its exceptionally threonine-rich C terminus, indicating possible modification (see below), while MMAR_4057 represents one of the two Mce-associated proteins present on the MOMP candidate shortlist. Mce-associated proteins have been implicated in lipid import (40).
Table 1.
Characterization of marker proteins and candidate MOMPsa
| Protein | Orthologue H37Rv | No. of TMD | SP | Predicted molecular mass (kDa) | Observed molecular mass (kDa) | % extraction, endogenousb | % extraction, +HA tagc |
|---|---|---|---|---|---|---|---|
| FtsQ | Rv2151c | 1 | N | 34 | 40 | 24 | 36 |
| OmpATb | Rv0899 | 1 | N | 34 | 40 | 100 | 97 |
| MctB | Rv1698 | 1 | Y | 32 | 35 | 90 | 82 |
| MMAR_0617 | Rv0312d | 1 | N | 62 | 120, 80 | 100 | 75 |
| MMAR_4057 | Rv1972 | 1 | N | 26 | 34, 25 | 98 | 53 |
| MMAR_4366 | Rv1100 | 1 | N | 22 | 30, 24 | 100 | 93 |
| MMAR_5387 | Rv3835 | 1 | N | 48 | 75, 60, more | 100 | 87 |
TMD, predicted transmembrane domains (TMHMM); SP, predicted signal peptide (SignalP).
Extractability by OBG of endogenous protein quantified by nanoLC-MS/MS.
Extractability of HA-tagged protein by OBG as quantified from immunoblot signals (Fig. 3A) using Quantity One.
Closest homologue of MMAR_0617, orthologue of Rv0312c, is MMAR_0562.
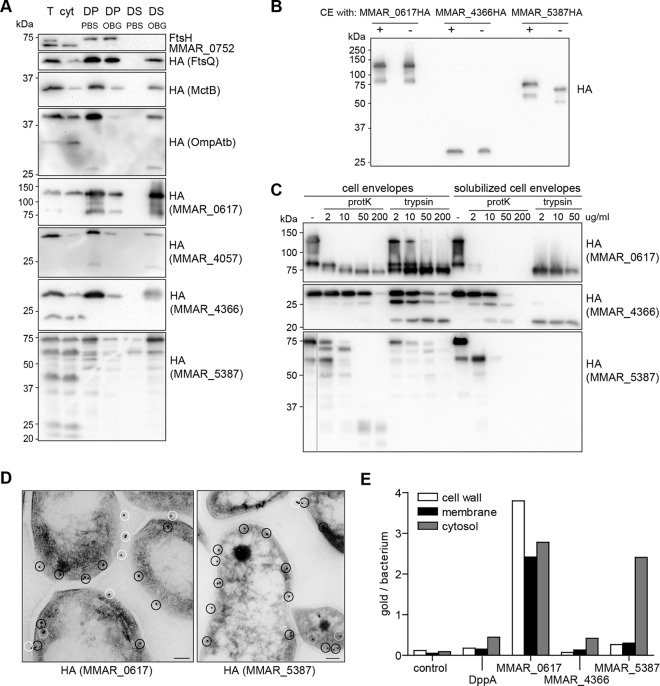
The corresponding genes were cloned into a shuttle vector behind an hsp60 promoter with a linker coding for a C-terminal hemagglutinin (HA) epitope. As controls, IM marker protein FtsQ and MOM marker proteins OmpATb and MctB were included. After introduction of the constructs into M. marinum E11, immunoblot analysis using anti-HA serum showed that the protein levels of the different HA-tagged proteins were highly variable (see Fig. S2B in the supplemental material). Expression ranged from hardly detectable (MMAR_4057-HA) to high (MMAR_0617-HA) (see below). Since all constructs contain the same promoter and ribosome binding site, this variation is probably caused by differences in mRNA or protein stability. Interestingly, the observed products for MMAR_0617-HA and MMAR_5387-HA were significantly larger than their expected molecular weights.
First, we set out to verify the results obtained by nanoLC-MS/MS. Subcellular fractionation and subsequent detergent extraction confirmed that three of the four HA-tagged MOMPs and also the two MOMP markers MctB and OmpATb are primarily localized to the cell envelope and >75% detergent extractable with OBG, whereas approximately 30% of IM marker FtsQ-HA is extracted (Fig. 3A and Table 1). The only conflicting result was obtained with MMAR_4057-HA, which was only 50% extractable. Because this protein was also very inefficiently expressed, MMAR_4057-HA was excluded from the follow-up experiments. As might be expected, the highly expressed MMAR_0617-HA and MMAR_5387-HA, while mainly cell envelope associated, also show considerable cytosolic localization (Fig. 3A).
Fig 3.
Candidate MOMPs localize to the detergent extractable cell envelope fraction. (A to C) Immunoblots were probed with antisera recognizing FtsH, MMAR_0752, or the HA epitope. (A) Subcellular fractions of M. marinum expressing HA-tagged versions of the control proteins and the candidate MOMPs. Total lysate (T), soluble lysate (cyt), detergent pellet (DP), and supernatant (DS) obtained after treatment of cell envelopes with PBS or 1% OBG were loaded in a 1:1:5:5:5:5 ratio. (B to C) Heat modifiability and protease accessibility of putative MOMPs. (B) Cell envelope fractions were denatured in 2% SDS at 95°C (+) or incubated in 0.2% SDS at 37°C (−) and loaded on an SDS-free PAGE gel. (C) MOMPs were treated with different concentrations of proteinase K (protK) or trypsin, without or after detergent extraction with OBG. (D) Representative Tokuyasu cryosections of M. marinum expressing MMAR_0617-HA or MMAR_5387-HA probed with HA antiserum followed by goat anti-mouse conjugate labeled with 10-nm gold particles. Scale bar is 100 nm. Particles are marked with color coding similar to that described in panel E. (E) Quantification of gold particles assigned to different subcellular compartments (see Material & Methods for definitions).
Heat modifiability and cell envelope association of MOMP candidates.
One of the common characteristics of β-barrel OM proteins of Gram-negative bacteria is their heat modifiability (41). This heat-dependent unfolding can be visualized by a shift in molecular mass on SDS-PAGE gels (42). Interestingly, while two of our proteins did not reveal a molecular shift upon heat treatment, the unboiled samples of MMAR_5387-HA migrated roughly 5 kDa lower than the denatured form (Fig. 3B). Bioinformatic analysis of the MMAR_5387 amino acid sequence does not confirm the presence of a β-barrel structure, which means that the nature of the heat modifiability of MMAR_5387 is still unclear. Notably, the marker MOMPs MctB and OmpATb did not exhibit heat modifiability (not shown).
To further investigate the membrane association of the putative MOMPs, we tested their protease accessibility. Unfortunately, protease treatment of whole cells did not affect any of the MOMPs, including OmpATb and MctB (not shown), possibly due to the thick capsule layer (5). We additionally performed limited proteolysis on isolated cell envelopes and on MOMPs solubilized from the cell envelope fractions using OBG. All tested proteins are more protease sensitive when solubilized from the cell envelope (Fig. 3C), indicating they are indeed embedded in the membrane. This is most apparent for MMAR_0617 and MMAR_4366. Interestingly, protease treatment of MMAR_0617 resulted in a C-terminal HA-tag-containing fragment of ∼70 kDa that is highly resistant to trypsin and, to a lesser extent, proteinase K. This proteolytic fragment, which is still larger than the predicted molecular mass of 62 kDa, was also identified after trypsinization of solubilized material (also see below). In contrast, MMAR_5387, the heat modifiable protein, is relatively sensitive to trypsin and proteinase K (Fig. 3C).
We also investigated the membrane localization of the candidate MOMPs by immunogold electron microscopy. None of the C-terminal HA-tagged constructs could be detected upon labeling of whole cells (not shown). This detection problem might again be caused by the thick capsular layer or by the possibility that the C-terminal epitope is exposed toward the periplasm instead of the cell surface. To circumvent these problems, we performed immunogold labeling on Tokuyasu cryosections. Since in these sections only a limited amount of membrane is accessible for antibody labeling, especially compared to the cytosol, the detection of a membrane protein is strongly linked to its expression levels, and the percentage of cell envelope association is also not absolute. While MMAR_4366 showed little labeling, MMAR_0617 and MMAR_5387 were very efficiently labeled (Fig. 3D). This is also confirmed in quantitative analysis of gold particles per cell (Fig. 3E). Wild-type cells and cells expressing DppA-HA were used as controls in this analysis. DppA is described as a periplasmic protein in E. coli (43) and was also found to be 100% extracted in our LC-MS analysis (see Table S2 in the supplemental material). The immunogold labeling showed clear association of the proteins with the cell envelope, although also some cytosolic localization was observed, which is in line with the biochemical fractionation results (Fig. 3A). Unfortunately, in these experiments, we were unable to distinguish between the inner and outer membrane fraction due to the size of the antiserum-gold complex (∼20 nm). Quantification of gold particles for their localization did show that especially MMAR_0617 was detected primarily at the cell wall and membrane fractions (Fig. 3E).
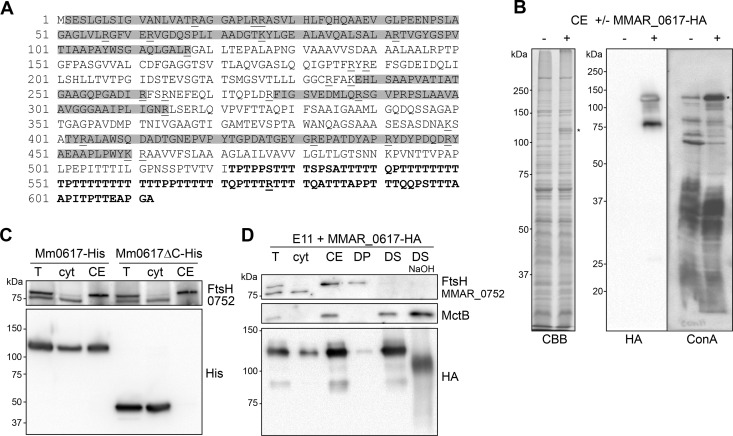
MMAR_0617 is extensively modified at its C terminus.
MMAR_0617 has a C-terminal domain that contains exceptionally large stretches of threonine residues (up to 11 consecutive residues; Fig. 4A). Curiously, this protein has a predicted molecular mass of 62 kDa, but the apparent molecular mass observed on SDS-PAGE is ∼120 kDa, with an additional minor form of ∼80 kDa (Fig. 3A). This large ∼120-kDa product of MMAR_0617-HA is abundantly present in the cell envelope fraction and is even visible by CBB staining (Fig. 4B). The appearance of this SDS-stable product was irrespective of the C-terminal label, as also the His-tagged protein migrated similarly (Fig. 4C). Deletion of the C-terminal 91 amino acids, containing 56 threonines (Fig. 4A), resulted in a truncated protein that migrated at the predicted molecular mass of ∼50 kDa (Fig. 4C). This indicates that (posttranslational modification of) the C-terminal Thr stretch of MMAR_0617 is responsible for the observed increase in molecular mass. Remarkably, the deletion of this domain also greatly affected the cell envelope localization of MMAR_0617, as nearly all of the truncated version was found in the cytosolic fraction (Fig. 4C). This result is surprising, because MMAR_0617 contains a predicted TMD that was preserved in the truncated version, suggesting that this domain does not function as a membrane anchor.
Fig 4.
Extensive modification of MMAR_0617. (A) Amino acid sequence of MMAR_0617. Highlighted in gray are peptide sequences that were detected by MS/MS analysis and therefore do not contain modifications. Underlined are trypsin digestion sites, and the Thr-rich C-terminal domain that is deleted in the truncated MMAR_0617ΔC is indicated in bold. (B) CBB staining and immunoblots using the HA antibody and horseradish peroxidase (HRPO)-conjugated ConA of cell envelope fractions (CE) without (−) and with (+) MMAR_0617-HA (indicated with *). (C) Immunoblot of total lysate (T), cytosol (cyt), and CE of M. marinum expressing MMAR_0617-His or MMAR_0617ΔC-His loaded in a 1:1:1 ratio. (D) Immunoblot of subcellular fractions of M. marinum expressing MMAR_0617-HA probed with anti-FtsH, anti-MctB, and anti-HA antibodies; T, cyt, CE, detergent pellet (DP), and supernatant (DS) obtained after solubilization of CE with 1% OBG and DS treated with 0.15 M NaOH (DS NaOH) loaded in a 1:1:3:5:5:5 ratio.
The excessive number of Thr residues in the C-terminal domain MMAR_0617 could be posttranslationally modified by O-glycosylation, as has been described for, e.g., the M. tuberculosis antigens Apa (44) and Mpb83 (45). Glycosylation was analyzed by incubating blots with horseradish peroxidase (HRP)-conjugated concanavalin A (ConA). The clear binding of this lectin to the 120-kDa band indicates the presence of α-linked glucose or, more likely, mannose residues on MMAR_0617 (Fig. 4B). A faint 120-kDa reacting band was observed in the negative control, which might represent endogenous MMAR_0617. Cell envelope-solubilized MMAR_0617-HA was also tested for its sensitivity to mild alkaline hydrolysis, which releases ester-linked modifications and O-linked carbohydrates. This treatment resulted in a distinct shift in molecular mass of ∼20 kDa of the 120-kDa product of MMAR_0617 (Fig. 4D), further indicating that MMAR_0617 is indeed extensively modified. The latter treatment also affected ConA reactivity of the protein (not shown), whereas the control protein MctB remained unaffected (Fig. 4D). Finally, MS/MS analysis of purified MMAR_0617 resulted in 38% peptide coverage (Fig. 4A), revealing the peptides without posttranslational modifications. The Thr-rich tail was not identified in this analysis, but it should be noted that a large part of the C-terminal domain lacks trypsin digestion sites and is therefore probably too large to be detected by mass spectrometry, even without modifications. In conclusion, the putative MOMP MMAR_0617 is extensively modified, likely at the Thr-rich C terminus, by O-linked glycosyl groups.
MMAR_0617 forms an oligomer with channel-forming activity in lipid bilayer experiments.
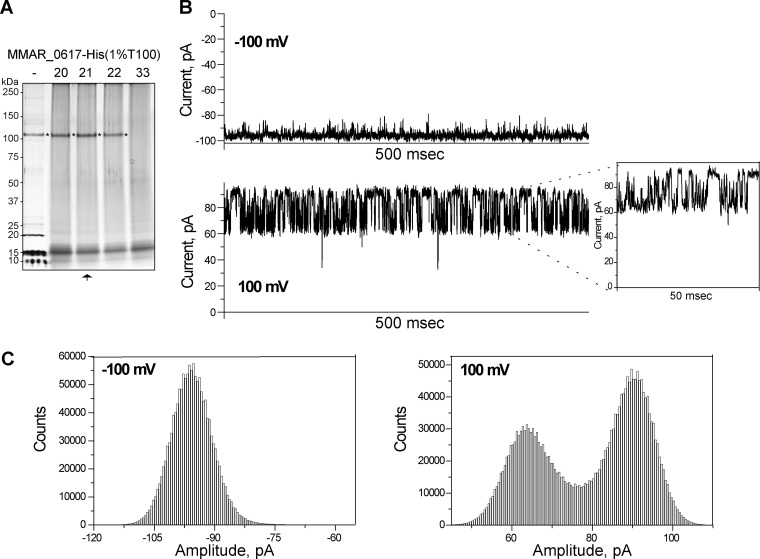
To examine whether MMAR_0617 could function as a porin in the MOM, the His-tagged version of this protein was purified from M. marinum cell envelopes using Ni-nitrilotriacetic acid (NTA) affinity chromatography, and channel activity was analyzed by reconstitution of purified protein into planar lipid membranes. As negative controls, the same purifications were performed for wild-type (WT) strains or strains expressing MMAR_0617-His with the C-terminal deletion (see Fig. S3A in the supplemental material). Analysis of purified MMAR_0617-His by blue native PAGE indicated that this protein forms two major oligomeric complexes with apparent molecular masses of ∼200 and ∼400 kDa (see Fig. S3B). Lowering the Triton X-100 concentration to 0.1% during the purification (see Materials and Methods) slightly decreased the apparent molecular mass of both complexes (see Fig. S3B). Further purification of these eluates by gel filtration chromatography also confirmed that MMAR_0617 forms an oligomeric complex of ∼400 kDa (see Fig. S3C).
Next, we used the planar lipid bilayer technique to characterize the single-channel properties of Ni-NTA-purified MMAR_0617-His, both before (not shown) and after gel filtration (Fig. 5A). Channel properties, such as conductance, selectivity, and gating, were investigated. When MMAR_0617 was reconstituted into stable 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (DPhPc) lipid bilayers, it formed a stable channel that allowed a specific ion current under applied transmembrane voltage. Notably, no activity was recorded in control experiments using the Ni-NTA purifications from the empty strain or expressing MMAR_0617ΔC-His (results not shown). Single MMAR_0617 channels in the bilayer membrane showed a characteristic conductance of 0.8 ± 0.1 ns in 1 M KCl (Fig. 5B), which was consistently observed using independent biological replicates. Moreover, the channel showed a slight asymmetry in the channel conductance with respect to the polarity of the applied voltage, i.e., at negative voltage, the channel conductance is slightly higher. However, the exact physical orientation of the channel could not be detected in this type of measurements. Further analysis of the data showed that MMAR_0617 forms a highly voltage-sensitive channel. In addition, single MMAR_0617 channels showed asymmetry in the channel closure with respect to the polarity of the applied voltage. For example, at positive voltage above 100 mV, the channel fluctuated between several different subconductance states, whereas at negative voltage, the channel remained in one open steady state (Fig. 5B). At present, this mechanism of voltage sensitivity is not understood. The corresponding current amplitude histograms showed that the MMAR_0617 channel fluctuates between two states at positive voltage, whereas it remains mostly in one open state at negative voltage (Fig. 5C). The ion selectivity was determined with reversal potential measurements after applying a 10-fold salt gradient (0.1 M KCl cis side/1 M KCl trans side). We used the Goldman-Hodgkin-Katz (GHK) equation to determine the ratio of cation and anion permeability. The calculated PK/Cl ratio around ∼2 indicates that the channel is slightly cation selective.
Fig 5.
Purified MMAR_0617-His shows channel activity in lipid bilayer experiments. (A) Silver-stained SDS-PAGE gel containing Ni-NTA-purified MMAR_0617-His in 1% Triton X-100 (T100) before (−) and after gel filtration chromatography. Obtained fractions 20, 21, 22, and 33 (negative control) were TCA precipitated before loading. A background signal of ∼15 kDa was observed in all fractions, which likely represents coprecipitated T100. MMAR_0617 is indicated by an asterisk, and the fraction used for single-channel activity recordings is marked by an arrow. (B and C) Single-channel activity recordings representative for three biological replicates. Experimental conditions were 1 M KCl, 10 mM HEPES, pH 7.4, at room temperature. (B) Ionic currents through a single channel at −100 mV and +100 mV. At −100 mV, the channel exists in one open conductance state, whereas at +100 mV, the channel fluctuates between different conductance states. (C) Corresponding amplitude histograms.
To further exclude that other proteins that copurify with MMAR_0617-His are responsible for the observed channel activity, we also purified C-terminally HA-labeled MMAR_0617 using a different technique, i.e., using beads coated with HA antibodies (see Fig. S4A in the supplemental material). Also, this purified MMAR_0617-HA preparation clearly showed channel activity in lipid bilayers (see Fig. S4B), although the channels fluctuated between different conductance states with very high noise. This relative instability might be caused by the HA tag or the purification procedure that involved elution at low pH. In conclusion, in three different MMAR_0617 purifications, we have observed that MMAR_0617 exhibits channel activity in artificial lipid bilayers, which indicates it forms a pore in the MOM.
DISCUSSION
Recent studies have revealed that mycobacteria contain a second lipid bilayer, the mycolate outer membrane (MOM) (3–5). The presence of this impermeable lipid bilayer indicates that channel proteins are necessary for transport of proteins and nutrients across the MOM. In this study, we have tried various methods to discover novel MOMPs, using marker proteins to ascertain successful separation. We failed to separate inner and outer membranes by differential centrifugation (36) or sucrose-gradient centrifugation of isolated cell envelope fractions. The latter method has been successfully used for C. glutamicum, for which two fractions of different density were observed in a gradient of 35 to 56% sucrose (46). However, multiple attempts for M. marinum always showed one fraction at a density of 31 to 34%, which contained both inner and outer membrane proteins (not shown). This disparity might reflect a difference in composition of the corynebacterial and mycobacterial MOM, perhaps caused by the length of their mycolic acids (i.e., mycobacterial mycolic acids are considerably longer). In the end, fractionation of IM and MOM proteins by differential detergent extraction with OBG was successful. Analysis of the obtained fractions by nanoLC-MS/MS identified a number of putative MOMP candidates. One of these candidates, MMAR_0617, indeed exhibited channel activity in lipid bilayer experiments, indicating it is a novel porin protein.
Protease accessibility experiments showed that MMAR_0617 is embedded in the membrane, probably via its C-terminal domain. Thus far, it is unknown how this protein is transported across the IM; it does not contain an N-terminal signal sequence nor the recently identified general type VII secretion motif (47). The MS/MS analysis of MMAR_0617 purified from cell envelopes also indicates that no N-terminal processing takes place. Reconstitution of this protein into planar lipid membranes clearly showed it has a single-channel conductance of 0.8 ± 0.1 ns. Furthermore, MMAR_0617 was shown to be highly voltage sensitive, exhibiting different conductance states when positive voltages were applied. Importantly, gel filtration chromatography and blue native PAGE analysis indicated that the channel is formed by an oligomer of MMAR_0617. Bioinformatic analysis with JPred3 (48) revealed that the protein contains some β-strands but also several α-helices (not shown). It thus seems unlikely that MMAR_0617 has a β-barrel structure comparable to MspA (11). The channel conductance is also considerably lower than that of MspA. Accordingly, the high expression level of MMAR_0617 does not significantly affect bacterial viability or antibiotic sensitivity (not shown). Interestingly, under a different purification condition (0.1% instead of 1% Triton X-100 during the Ni purification and before the gel filtration step), we observed two channel activities. We additionally measured channels with higher conductance of 4 to 10 ns (see Fig. S5 in the supplemental material), although less frequent than the ∼0.8-ns channel activity. This larger channel was voltage dependent, a behavior that is similar to MspA (49). Using the same sample, we also observed that few channels reconstituted into lipid bilayers fluctuated between different subconductance states with very high noise (data not shown). Importantly, in preparations using 1% Triton X-100 or including further purification by gel filtration in the presence of 0.1% Triton X-100, such channels were never observed, indicating that these channel activities are due to contaminating proteins. Future research will focus on identifying the proteins that are responsible for these activities.
MMAR_0617 is an unusual channel-forming protein. Its Thr-rich C terminus seems to be highly modified, resulting in an apparent molecular mass of 120 kDa on SDS-PAGE, while the predicted molecular mass is 62 kDa. Although we have not yet been able to specifically determine which modifications are present, the available evidence points toward O-linked glycosylation. Bacterial O-linked glycosylation is usually restricted to short stretches of glycans. Therefore, it seems unlikely that the increase in molecular mass of 60 kDa of MMAR_0617 is due solely to glycosylation. Interestingly, two outer membrane proteins of C. glutamicum, PorA and PorH, have recently been described to be modified by O-mycoloylation (19). Similarly, MMAR_0617 could contain O-linked mycolic acids, which would also explain that the C terminus of MMAR_0617 is responsible for the cell envelope interaction of this protein. Future studies will be dedicated to map the modifications of MMAR_0617.
Interestingly, although M. tuberculosis does not contain an orthologue of MMAR_0617, several conserved proteins have a similar organization, i.e., these proteins (Rv0312, Rv0538, and Rv2198c) have a single predicted TMD in the middle of the protein followed by a Thr-rich C-terminal domain. The gene encoding Rv0312 has also been described to be essential for growth (50). Furthermore, proteomic analysis showed that these proteins are located in the cell envelope (51). Their orthologues MMAR_0884 and MMAR_3358 are also highly detergent extractable (see Table S2 in the supplemental material). These data could indicate that all these Thr-rich proteins are in fact MOMPs.
By differential detergent solubilization, we could separate inner membrane proteins from a specific group of proteins that contains MOMPs. Curiously, the OMPs of Gram-negative bacteria generally resist detergent solubilization, and detergent treatment in these bacteria is used to specifically solubilize inner membrane proteins (52, 53). This discrepancy could indicate major structural differences between mycobacterial and Gram-negative outer membrane proteins, which reflects the difference in composition of their respective outer membranes. Alternatively, we have identified a specific relatively detergent-sensitive subpopulation of MOMPs, whereas others do resist detergent solubilization. This might be related to specific lipid patches, in which this MOMP subpopulation resides. Interestingly, most of the candidate MOMPs that we tested showed an increased molecular mass on SDS-PAGE, indicative of posttranslational modification (Table 1). We also confirmed a modification of MMAR_5387, as this protein showed a minor shift in molecular mass upon treatment with NaOH, indicating it contains O-linked modifications (not shown).
In conclusion, we have developed a method to specifically solubilize specific MOMPs, which led to the discovery of a novel mycobacterial channel-forming protein with extensive modifications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from ECHO grant (to A.D.V.D.W.) and VENI grant (to E.N.G.H.), both from the Netherlands Organization for Scientific Research.
We thank Ben Appelmelk and Yann Guerardel for helpful discussions and Annette Dreisbach, Brigitte Wevers, and Hilde Brouwers for technical support.
Footnotes
Published ahead of print 1 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02236-12.
REFERENCES
- 1. Bou Raad R, Meniche X, de Sousa-d'Auria C, Chami M, Salmeron C, Tropis M, Labarre C, Daffe M, Houssin C, Bayan N. 2010. A deficiency in arabinogalactan biosynthesis affects Corynebacterium glutamicum mycolate outer membrane stability. J. Bacteriol. 192:2691–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Minnikin DE. 1991. Chemical principles in the organization of lipid components in the mycobacterial cell envelope. Res. Microbiol. 142:423–427 [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190:5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 6:e1000794 doi:10.1371/journal.ppat.1000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhamidi S, Scherman MS, Jones V, Crick DC, Belisle JT, Brennan PJ, McNeil MR. 2011. Detailed structural and quantitative analysis reveals the spatial organization of the cell walls of in vivo grown mycobacterium leprae and in vitro grown M. tuberculosis. J. Biol. Chem. 286:23168–23177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Villeneuve M, Kawai M, Kanashima H, Watanabe M, Minnikin DE, Nakahara H. 2005. Temperature dependence of the Langmuir monolayer packing of mycolic acids from Mycobacterium tuberculosis. Biochim. Biophys. Acta 1715:71–80 [DOI] [PubMed] [Google Scholar]
- 8. Jarlier V, Nikaido H. 1990. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J. Bacteriol. 172:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871–2881 [DOI] [PubMed] [Google Scholar]
- 10. Lichtinger T, Heym B, Maier E, Eichner H, Cole ST, Benz R. 1999. Evidence for a small anion-selective channel in the cell wall of Mycobacterium bovis BCG besides a wide cation-selective pore. FEBS Lett. 454:349–355 [DOI] [PubMed] [Google Scholar]
- 11. Faller M, Niederweis M, Schulz GE. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189–1192 [DOI] [PubMed] [Google Scholar]
- 12. Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239–253 [DOI] [PubMed] [Google Scholar]
- 13. Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 108:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siroy A, Mailaender C, Harder D, Koerber S, Wolschendorf F, Danilchanka O, Wang Y, Heinz C, Niederweis M. 2008. Rv1698 of Mycobacterium tuberculosis represents a new class of channel-forming outer membrane proteins. J. Biol. Chem. 283:17827–17837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Senaratne RH, Mobasheri H, Papavinasasundaram KG, Jenner P, Lea EJ, Draper P. 1998. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J. Bacteriol. 180:3541–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song H, Huff J, Janik K, Walter K, Keller C, Ehlers S, Bossmann SH, Niederweis M. Expression of the ompATb operon accelerates ammonia secretion and adaptation of Mycobacterium tuberculosis to acidic environments. Mol. Microbiol. 80:900–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klackta C, Knorzer P, Riess F, Benz R. 2010. Hetero-oligomeric cell wall channels (porins) of Nocardia farcinica. Biochim. Biophys. Acta 1808:1601–1610 [DOI] [PubMed] [Google Scholar]
- 18. Ziegler K, Benz R, Schulz GE. 2008. A putative alpha-helical porin from Corynebacterium glutamicum. J. Mol. Biol. 379:482–491 [DOI] [PubMed] [Google Scholar]
- 19. Huc E, Meniche X, Benz R, Bayan N, Ghazi A, Tropis M, Daffe M. 2010. O-mycoloylated proteins from Corynebacterium: an unprecedented post-translational modification in bacteria. J. Biol. Chem. 285:21908–21912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malen H, Berven FS, Softeland T, Arntzen MO, D'Santos CS, De Souza GA, Wiker HG. 2008. Membrane and membrane-associated proteins in Triton X-114 extracts of Mycobacterium bovis BCG identified using a combination of gel-based and gel-free fractionation strategies. Proteomics 8:1859–1870 [DOI] [PubMed] [Google Scholar]
- 21. Malen H, De Souza GA, Pathak S, Softeland T, Wiker HG. 2011. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Z, De Buck J. 2010. Localization of proteins in the cell wall of Mycobacterium avium subsp. paratuberculosis K10 by proteomic analysis. Proteome Sci. 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puttinaowarat S, Thompson KD, Adams A. 2000. Mycobacteriosis: detection and identification of aquatic Mycobacterium species. Fish Vet. J. 5:6–21 [Google Scholar]
- 24. van Bloois E, Dekker HL, Froderberg L, Houben EN, Urbanus ML, de Koster CG, de Gier JW, Luirink J. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419–1424 [DOI] [PubMed] [Google Scholar]
- 25. Houben ENG, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 86:472–484 [DOI] [PubMed] [Google Scholar]
- 26. Piersma SR, Fiedler U, Span S, Lingnau A, Pham TV, Hoffmann S, Kubbutat MH, Jimenez CR. 2010. Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: method evaluation, differential analysis, and verification in serum. J. Proteome Res. 9:1913–1922 [DOI] [PubMed] [Google Scholar]
- 27. Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb.) 91:8–13 [DOI] [PubMed] [Google Scholar]
- 28. Pham TV, Piersma SR, Warmoes M, Jimenez CR. 2010. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26:363–369 [DOI] [PubMed] [Google Scholar]
- 29. Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, Peters PJ, Luirink J, Manganelli R, Bitter W. 2011. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J. Biol. Chem. 286:19024–19034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahendran KR, Chimerel C, Mach T, Winterhalter M. 2009. Antibiotic translocation through membrane channels: temperature-dependent ion current fluctuation for catching the fast events. Eur. Biophys. J. 38:1141–1145 [DOI] [PubMed] [Google Scholar]
- 31. Montal M, Mueller P. 1972. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. U. S. A. 69:3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. 2010. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 18:109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, Piersma SR, Jimenez CR, Luirink J, Bitter W, Houben EN. 2012. Specific chaperones for the type VII protein secretion pathway. J. Biol. Chem. 287:31939–31947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee BY, Hefta SA, Brennan PJ. 1992. Characterization of the major membrane protein of virulent Mycobacterium tuberculosis. Infect. Immun. 60:2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hirschfield GR, McNeil M, Brennan PJ. 1990. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J. Bacteriol. 172:1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rezwan M, Laneelle MA, Sander P, Daffe M. 2007. Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68:32–39 [DOI] [PubMed] [Google Scholar]
- 37. Niederweis M, Maier E, Lichtinger T, Benz R, Kramer R. 1995. Identification of channel-forming activity in the cell wall of Corynebacterium glutamicum. J. Bacteriol. 177:5716–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dreisbach A, van der Kooi-Pol MM, Otto A, Gronau K, Bonarius HP, Westra H, Groen H, Becher D, Hecker M, van Dijl JM. 2011. Surface shaving as a versatile tool to profile global interactions between human serum proteins and the Staphylococcus aureus cell surface. Proteomics 11:2921–2930 [DOI] [PubMed] [Google Scholar]
- 39. Marmiesse M, Brodin P, Buchrieser C, Gutierrez C, Simoes N, Vincent V, Glaser P, Cole ST, Brosch R. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483–496 [DOI] [PubMed] [Google Scholar]
- 40. Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burgess NK, Dao TP, Stanley AM, Fleming KG. 2008. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 283:26748–26758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olson ER, Dunyak DS, Jurss LM, Poorman RA. 1991. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J. Bacteriol. 173:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. 1996. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J. Bacteriol. 178:2498–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Michell SL, Whelan AO, Wheeler PR, Panico M, Easton RL, Etienne AT, Haslam SM, Dell A, Morris HR, Reason AJ, Herrmann JL, Young DB, Hewinson RG. 2003. The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1→3)-mannobiose moieties. J. Biol. Chem. 278:16423–16432 [DOI] [PubMed] [Google Scholar]
- 46. Marchand CH, Salmeron C, Bou Raad R, Meniche X, Chami M, Masi M, Blanot D, Daffe M, Tropis M, Huc E, Le Marechal P, Decottignies P, Bayan N. 2012. Biochemical disclosure of the mycolate outer membrane of Corynebacterium glutamicum. J. Bacteriol. 194:587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, Bitter W. 2012. General secretion signal for the mycobacterial type VII secretion pathway. Proc. Natl. Acad. Sci. U. S. A. 109:11342–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Engelhardt H, Heinz C, Niederweis M. 2002. A tetrameric porin limits the cell wall permeability of Mycobacterium smegmatis. J. Biol. Chem. 277:37567–37572 [DOI] [PubMed] [Google Scholar]
- 50. Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malen H, Pathak S, Softeland T, de Souza GA, Wiker HG. 2010. Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol. 10:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Filip C, Fletcher G, Wulff JL, Earhart CF. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schnaitman CA. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.