Abstract
Bacterial endospores are the most resistant cell type known to humans, as they are able to withstand extremes of temperature, pressure, chemical injury, and time. They are also of interest because the endospore is the infective particle in a variety of human and livestock diseases. Endosporulation is characterized by the morphogenesis of an endospore within a mother cell. Based on the genes known to be involved in endosporulation in the model organism Bacillus subtilis, a conserved core of about 100 genes was derived, representing the minimal machinery for endosporulation. The core was used to define a genomic signature of about 50 genes that are able to distinguish endospore-forming organisms, based on complete genome sequences, and we show this 50-gene signature is robust against phylogenetic proximity and other artifacts. This signature includes previously uncharacterized genes that we can now show are important for sporulation in B. subtilis and/or are under developmental control, thus further validating this genomic signature. We also predict that a series of polyextremophylic organisms, as well as several gut bacteria, are able to form endospores, and we identified 3 new loci essential for sporulation in B. subtilis: ytaF, ylmC, and ylzA. In all, the results support the view that endosporulation likely evolved once, at the base of the Firmicutes phylum, and is unrelated to other bacterial cell differentiation programs and that this involved the evolution of new genes and functions, as well as the cooption of ancestral, housekeeping functions.
INTRODUCTION
Bacterial endospores, such as those formed by species of the Bacillus and Clostridium genera, are arguably the most resistant cellular structures known to scientists. Endospores resist physical and chemical changes, such as exposure to solvents, oxidizing agents, and lytic enzymes, high temperatures, vacuum, acceleration, and irradiation, that would rapidly destroy the vegetative form of the bacterium (1–3). The extreme conditions endured by bacterial endospores include simulated and actual extraterrestrial environments (2). The resilience of the endospore allows it to remain viable in the environment for long periods of time, contributing to the wide geographic distributions of spores in Earth's ecosystems (2). It also allows endospore formers to occupy niches in the gastrointestinal (GI) tract of metazoans, establishing either symbiotic or commensal relationships or pathogenic interactions, in which case the spore often serves as the infectious vehicle (4–7). The robustness of endospores is also the basis for several applications of endospores in biomedicine and biotechnology, including their use in probiotic formulations or as platforms for the display of enzymes or antigens (8–10).
Most of the previously described endosporulating bacteria belong to the Clostridia (anaerobic) and Bacilli (aerobic) classes of the Firmicutes phylum, one of the two eubacterial phyla that groups Gram-positive organisms (11). However, endospore formers are found in other classes within the Firmicutes and encompass aerobic or anaerobic organisms with a wide range of morphologies, lifestyles, and metabolic traits, including rods, cocci, branching species, plant or animal pathogens or symbionts, syntrophs, sulfate reducers, phototrophs (11–17), and remarkably, didermic species (18). Despite the extreme diversity of endospore-forming bacteria, the basic architecture of an endospore is conserved across species (19). The endospore consists of a core compartment, delimited by a lipid bilayer, that contains one copy of the genome. The core is surrounded by a series of concentric structures that function together for the maintenance of dormancy and protection. Two conspicuous endospore structures are the cortex peptidoglycan (PG), which is essential for the acquisition and maintenance of heat resistance, and a protein coat (in some species further encircled by an exosporium), which surrounds the cortex and protects it from the action of PG-breaking enzymes. The spore surface layers (the coat and the exosporium, when present) also mediate environmental interactions of the endospore, including adhesion and recognition of compounds that trigger germination (19).
Endospore differentiation is best known with the model organism B. subtilis (20–22). In this organism, a rod-shaped bacterium that grows vegetatively by binary division, entry into sporulation is a response to severe nutritional stress. Entry into sporulation requires the phosphorylation of the response regulator Spo0A, which also controls a number of other adaptations to stationary-phase conditions (23, 24). Above a certain threshold level of phosphorylation, Spo0A triggers sporulation (25). A key event controlled by Spo0A is the switch to an asymmetric mode of cell division, during which a larger mother cell and a smaller forespore (the future spore) are formed. Once the two chambers of the sporangial cell have formed, the differentiation program relies on the cell-type-specific activation of 4 alternative σ subunits of RNA polymerase (22, 26, 27). These 4 σ factors are activated sequentially, alternating between the two cells. Their activation is linked to the completion of key morphological intermediates in the process and also relies on signaling pathways between the two cells. The result is that the forespore and mother cell-specific programs are coordinately deployed and in sequence with the course of morphogenesis. σF and σE control the early stages of development in the forespore and the mother cell, respectively. Engulfment of the forespore by the mother cell, the process by which the forespore is isolated from the external milieu and is released as a free protoplast with the mother cell cytoplasm, is a hallmark of endosporulation (20, 22). Coincident with engulfment completion, σF and σE are replaced by σG and σK in the forespore and in the mother cell, respectively (22, 26). The genes that compose each of the σ regulons drive morphogenesis through its successive morphological stages (22, 26). Contrary to genes not known to be involved in endosporulation, the key regulators of sporulation in B. subtilis are more conserved than the genes that define the endosporulation machinery (28). Within each σ regulon, additional regulatory proteins help control gene expression, creating feed-forward loops that fine-tune gene expression and contribute to the fidelity of the morphogenetic process (28). These additional regulatory proteins show an intermediate degree of conservancy (28).
The recognition of an organism's ability to form endospores from genomic information is an important goal, given the prominent position of endospore-forming bacteria in human health and disease, as are the insights into prokaryotic biology that have and will arise from studying organisms other than model bacteria. The importance of establishing a robust genomic signature for endosporulation is also well exemplified by the claims, dismissed by some authors (29) but which are recurrent, that endospores can be formed by certain pathogenic Mycobacterium spp., explaining, for example, the long-term persistence of these organisms in infected hosts (30, 31), or by Gram-negative organisms, such as Rhodobacter johrii or Serratia marcescens, the latter a close relative of Escherichia coli (32, 33).
Several studies have led to the identification of a core of endosporulation genes (34–38). However, it is unclear whether the key endosporulation regulatory proteins identify an endospore-forming organism(s) and whether the core set can predict new genes important for the process. Here, we traced the phylogenetic profile of Spo0A and the 4 cell-type-specific endosporulation σ factors. In addition, we defined a core of endosporulation genes, based on those genes known to participate in the process in B. subtilis. We used the core to define a robust genomic signature for this process. We employed the signature to predict new endospore-forming organisms, as well as new functionally important genes, which we show have a role or are expressed during endosporulation in the model organism B. subtilis. Overall, our results support the view that endosporulation evolved only once, at the base of the Firmicutes phylum, and that during its evolutionary history it has been involved in the evolution of new genes and the engagement of ancestral housekeeping genes.
MATERIALS AND METHODS
Data sets of species.
In our analysis, we divided the species data set into four groups: endosporulating, exosporulating, mycobacteria, and nonendosporulating. The data set included 31 endosporulating species, 5 exosporulating species, 7 Mycobacterium species, and 29 nonendosporulating species (see Table S1 in the supplemental material). For the nonendosporulating species, we constructed our data set with the objective of maximizing the genetic divergence between species, as measured by 16S RNA sequences. Therefore, species belonging to phyla or classes as divergent as Gammaproteobacteria, Alphaproteobacteria, Firmicutes, Cyanobacteria, Actinobacteria, etc., were included in the analysis.
Phylogenetic tree of bacteria 16S rRNA.
We constructed the phylogenetic tree for the 16S RNA by using nucleotide sequences collected from the Ribosomal Database Project website (39) and, as an outgroup species, Methanocaldococcus jannaschii (39). Sequences were aligned using ClustalW (40). A maximum likelihood phylogenetic tree was reconstructed with the PhyML software (41), using the best of nearest-neighbor interchange (NNI) and subtree pruning and regrafting (SBR) heuristic searches, based on an HKY85 nucleotide substitution model with a gamma distributed rate variation across sites. The reliability of the clustering was tested using the likelihood ratio test (LRT) as implemented in PhyML. Clades with an LRT of ≥0.95 were considered significant.
Phylogenetic analysis of spo0A and the sporulation-specific σ factors.
Putative homologues of spo0A were identified in the Superfamily database 1.73 of structural domain assignments (42) by querying for proteins that contained the same domains as the protein in B. subtilis (52172 and 46894). We generated multiple sequence alignments with ClustalW 2.0.10 (40) and edited the alignment to remove gaps and regions of low alignment score by using the editor Jalview 2.3 (43). A neighbor-joining tree was built with Phylip 3.68 using the JTT substitution matrix (44). A maximum likelihood tree was constructed using RaxML v7.2.6 and the JTT substitution matrix with gamma distributed rate categories across sites (44). The reliability of the clades was evaluated using 100 bootstrap replicates (45). We considered true spo0A orthologues to be all the sequences that were monophyletic with the B. subtilis spo0A sequence in both trees.
We used a similar approach for σ factors. We defined putative σ factors in complete proteomes as all those proteins that had the two structural domains that characterize the σ70 family of σ factors (Superfamilies 88946 and 88659). This definition misses σK of B. subtilis, in which the final protein results from the fusion of two halves encoded by different genes (22, 26). To overcome this problem, we manually included the B. subtilis σK sequence for the alignment. We then aligned the sequences and estimated monophyly as described above. We considered true σF, σE, σG, and σK orthologues as all the sequences that were monophyletic with the corresponding B. subtilis sequence in both trees.
Large-scale orthology mapping and definition of a minimal endosporulation core.
We manually compiled a list of 654 genes that are required for the initiation of sporulation, either leading to the production or activation of Spo0A, that are under the control of Spo0A but with established functions in sporulation, or that belong to each of the σ regulons (σF, σE, σG, and σK) in B. subtilis (26, 46–63). We then mapped orthologues of these genes by searching for bidirectional best hits (BDBH) in other bacterial genomes (64). BDBH is a heuristic method to identify orthologues that has been shown to produce few false positives (64). A bidirectional best hit occurs when, in a BLAST search, given two genomes A and B, containing two genes a and b, gene a has gene b as a best match in genome B and gene b has gene a as a best match in genome A. To define a “hit,” we used a cutoff of E <10−2 in BLASTP. We assumed that genes present in more than 90% of the endosporulating species but absent from nonendosporulating species are essential and exclusive of sporulation. All comparisons of the number of orthologous genes were made with the nonparametric Kruskal-Wallis test with a Bonferroni correction for multiple comparisons.
Strains and general methods.
The B. subtilis strains used in this work (see Table S2 in the supplemental material) are congenic derivatives of the Spo+ strain MB24 (trpC2 metC3). LB medium was used for growth or maintenance of E. coli and B. subtilis, and sporulation was induced in Difco sporulation medium (DSM) (57). The efficiency of sporulation was determined 18 h after the onset of sporulation, as described previously (57).
Mutant construction.
DNA fragments internal to the coding regions of the ytaF and ytaG genes (343 bp and 345 bp, respectively) were PCR amplified from MB24 genomic DNA with primer pairs ytaF1030D/ytaF1373R and ytaG312D/ytaG657R (all primers used herein are listed in Table S3 in the supplemental material). The PCR products were digested with BamHI and inserted into BamHI-cut pAH256 (57) to yield plasmid pMS399 (ytaF) or with BglII and inserted into BamHI-cleaved pUS19 (57) to yield pMS403 (ytaG). Next, competent cells of MB24 were transformed with pMS399 or pMS403, with selection to spectinomycin-resistant (Spr) cells. These crosses produced the ytaF insertional mutant AH6676, which was shown by PCR analysis to result from the integration of pMS399 into the corresponding region of homology with the host chromosome by a single reciprocal crossover event (Campbell-type recombination). Attempts to inactivate ytaG by a single recombinant event using pMS403 were unsuccessful.
To construct a ylmC deletion mutant, a 992-bp PCR fragment upstream of the ylmC coding region was amplified with primers ylmC93D and ylmC1085R. The PCR product was digested with BglII, and a 285-bp fragment was inserted between the BamHI and EcoRV sites of pAH256 (57), yielding plasmid pCC1. Second, a 525-bp DNA fragment, downstream to the ylmC coding region, was PCR amplified with primers ylmC1200D and ylmC1725R, digested with EcoRI and XhoI, and inserted between the same sites of pCC1, yielding pMS397. Competent cells of MB24 were transformed with pMS397, with selection to Spr. A double-crossover event (marker replacement), verified by PCR, produced the ylmC deletion mutant AH6675.
To construct a ylzA deletion mutant, a 604-bp DNA fragment, upstream to the ylzA coding region, was PCR amplified with primers ylzA446D and ylzA1050R. The PCR product was digested with HindlIII, and a 192-bp fragment was inserted between the HindIII and EcoRV sites of pAH256, to give pMS401. Next, a 517-bp DNA fragment, downstream of the ylzA coding region, was PCR amplified with primers ylzA1240D and ylzA1757R, digested with BglII and XhoI, and inserted between the same sites of pMS401, producing pMS402. Competent cells of MB24 were transformed with pMS402, with selection to Spr. This produced the ylzA deletion mutant AH6700, the result of a double-crossover event, which was verified by PCR.
To construct a ymxH deletion mutant, a 341-bp DNA fragment upstream to the ymxH coding region was PCR amplified with primers ymxH601D and ymxH942R. The PCR product was digested with HindIII and PstI and inserted between the same sites of pMS38 (65), yielding plasmid pMS448. Second, a 534-bp DNA fragment, downstream to the ymxH coding region, was PCR amplified with primers ymxH1117D and ymxH1651R, digested with BglII, and inserted between the SnaBI and BglII sites of pMS448, yielding pMS449. Competent cells of MB24 and AH6675 were transformed with pMS449, with selection to Cmr, producing as the result of a double-crossover event, which was verified by PCR, the ymxH deletion mutant AH6870. Transformation of AH6870 with chromosomal DNA from strain AH6675 produced the double ymxH ylmC mutant AH6871.
Transcriptional gfp fusions.
To create transcriptional fusions of the ylmC, ylzA, and ytaF promoter regions to gfp, the following PCR products were generated: a 306-bp fragment using primers ytaF699D and ytaF1005R; a 276-bp fragment using primers ylmC732D and ylmC1008R; a 163-bp fragment using primers ylzA842D and ylzA1005R. The PCR fragments carrying the ytaF, ylmC, and ylzA promoter regions were digested with HindIII and BamHI and ligated to pMS157 (49) digested with the same enzymes to produce pMS404, pMS405, and pMS406, respectively. Mutations in the promoter region of ylzA were constructed using primers PsigFylzAD and PsigFylzAR for the −10 region of the σF promoter and plasmid pMS406 (see above), yielding plasmid pMS460 (mutations in the σF promoter). ScaI-digested pMS404, pMS405, pMS406, and pMS460 were used to transform the parental strain MB24, as well as a panel of congenic mutants with deletions in the genes for σF, σE, σG, σK, and spoIIID. Neomycin-resistant and AmyE− transformants, the result of a double crossover at the amyE locus, were kept for further studies (see Table S2 in the supplemental material).
Fluorescence microscopy.
Samples (0.6 ml) of DSM cultures were collected during sporulation and resuspended in 0.2 ml of phosphate-buffered saline, and the membrane dye FM4-64 (Molecular Probes) was added at a final concentration of 10 μg ml−1 for visualization of membranes. Microscopy was carried out as described previously (66).
RESULTS
Data set and strategy used for analysis.
We started this investigation by calculating the phylogenetic extent of the endosporulation machinery, including its main regulatory proteins, across the tree of life, and contrasted it with the existence of endosporulation, exosporulation (as an example of a reportedly unrelated cell differentiation), or no-spore differentiation programs. This required a manual compilation of lifestyles of bacteria regarding spore formation (see the list in Table S1 in the supplemental material). It also required the definition of the endosporulation machinery based on published data for the model organism Bacillus subtilis (reviewed in references 19, 21, 22, 26, and 28). Detailed descriptions of the data sets and sources of information are provided above in Materials and Methods and in the supplemental material.
The identification of orthologues of the endosporulation program is central to our analysis. For the critical regulatory proteins governing entry and cell-type-specific gene expression during endosporulation (see also below), which belong to large paralogous families, we used a sensitive method to identify putative homologues and then used phylogenetic analysis to identify orthologues. For the hundreds of genes that comprise the endosporulation regulons, this approach was not practical, and so we defined orthologues as the bidirectional or reciprocal best BLAST hits to the B. subtillis regulon proteins. This is a widely used heuristic method that is well suited for mapping orthologues for many genes and has been shown recently to produce a low rate of false positives in the identification of true orthologues, although at the expense of sensitivity (67) (details for both approaches are provided in Materials and Methods).
Evolutionary specificity of the endosporulation machinery: transcriptional regulators.
Spo0A is the master regulator governing entry into endosporulation, while 4 RNA polymerase σ factors govern gene expression in the two cells involved in endospore differentiation. σE and σK (active in the larger mother cell) and σF and σG (active in the forespore) define four regulons that act sequentially to govern progress through the morphological stages of sporulation (21, 22, 26, 28). We investigated their phylogenetic extent and whether these proteins are specific to endosporulating organisms. We noted that σH is also active in predivisional cells, is required for sporulation, and is conserved in spore-forming Bacillus and Clostridium organisms (36) (see Fig. 2, below). However, orthologues of this σ factor are also present in non-spore-forming organisms (36).
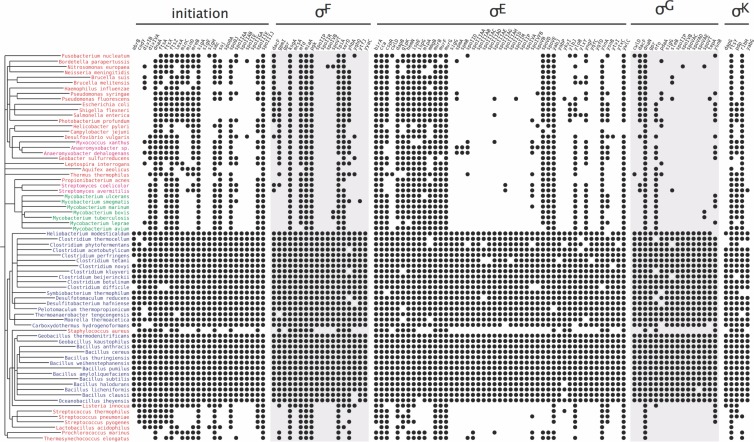
Fig 2.
The phylogenetic extent of the core endosporulation machinery reveals that a substantial part of the machinery is restricted to endosporulating species. Proteins are grouped according to the regulatory protein that controls their production (top), species are shown on the NCBI taxonomic tree (left), and lifestyle is denoted by color, with endosporulators, exosporulators, mycobacteria, and nonendosporulators shown in blue, pink, green, and red, respectively. The presence of an orthologue for a given protein in a given species is indicated by a black dot.
We first calculated a phylogenetic tree of the putative homologues of Spo0A, defined as all proteins with the same domain architecture as spo0A, which has a distinctive DNA-binding domain (68). These included, besides known Spo0A proteins, other transcription factors, such as CreB, CssR, DevR, and VraR, to name but a few, ranging from 0 to 94 sequences per species. In Fig. 1A we show the maximum likelihood tree of this family, which revealed a strongly supported monophyletic cluster of 44 sequences encompassing all Spo0A sequences from actual or putative endosporulating organisms but no other transcription factor. This tree topology unambiguously identified Spo0A and revealed that it is present in all known endospore formers, consistent with the notion that the protein is a synapomorphic trait (69; but see reference 36 and further discussion below). Moreover, the tree topology suggested a single origin from an early duplication, likely at the base of the Firmicutes. The observations that there is a single known or predicted Spo0A protein per endosporulating species in the Spo0A cluster and that this cluster is congruent with a 16S RNA tree (see Fig. S1 in the supplemental material) support a single origin with neither additional duplication nor horizontal gene transfer events. Some of the genomes in the cluster correspond to a class of 30 presumed non-endospore-forming organisms that do code for Spo0A, as previously noted (36) (see below).
Fig 1.
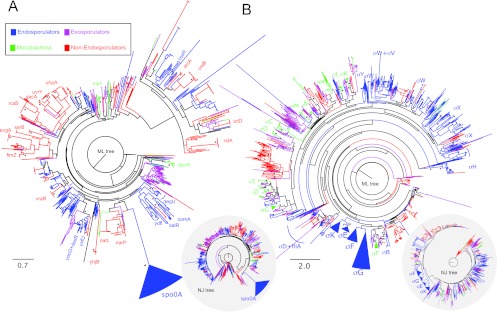
Phylogenetic analysis of the response regulator Spo0A (A) and the cell-type-specific σ70 factors that control endosporulation in B. subtilis (B) revealed that these regulatory factors are specific to endosporulating species. The trees were constructed from amino acid sequences in the species data set that contained the same structural domain architecture as each of the types of regulators and were inferred by maximum likelihood (ML) and neighbor joining (NJ) (see Materials and Methods for details). Tree scales are in evolutionary distances according to the Jones-Taylor-Thornton (JTT) amino acid substitution model. Asterisks indicate nodes supported by a bootstrap value higher than 70%.
We next focused on the RNA polymerase σ subunits that control cell-type-specific gene expression during sporulation. As described for Spo0A, we used structural domain assignments to identify all σ factors in each genome, finding between 1 and 72 per species. Each of the four σ factors involved in endosporulation formed one monophyletic cluster within the Firmicutes, exclusively composed of known endospore-forming species (Fig. 1B). Thus, they are specific for endosporulators, and as for Spo0A, they show synapomorphy. Of note, the phylogenetic analysis performed here did not support the idea that mycobacteria form endospores (29, 30, 31), as sequences annotated as σ factors for representatives of these organisms fall in clusters distantly related to any of the σ factors present in endosporulators. The only exception was the cluster formed by σF of Mycobacterium spp. and σB of endosporulators, which are both involved in the stress response (70). Importantly, σB of B. subtilis is not required for sporulation, although σB-inducing signals can influence activation of Spo0A (71).
Thus, our results showed that the main cell-type-specific transcriptional regulators of endosporulation in B. subtilis are only found in known endosporulating species, all of which are within the Firmicutes phylum. Moreover, they are absent from the other species investigated here, including the exospore-forming Streptomyces spp. and mycobacteria. In all, the confinement of Spo0A and the compartment-specific σ factors to known or putative endosporulators within the Firmicutes is in line with the idea that the regulatory proteins of endosporulation are highly conserved (28) and is against the view that the mycobacteria form endospores.
Evolutionary specificity of the endosporulation machinery: regulons.
In contrast, and contrary to non-endosporulation-related genes, the genes coding for the machinery that governs morphogenesis of the endospore are less conserved, with the proteins forming the feed-forward loops showing an intermediate degree of conservancy (28). Conceivably, this machine could be quite old but under the control of different sets of regulators in the different branches of the bacterial tree. To test this hypothesis, we computed the phylogenetic extent of the minimal core of the sporulation machinery. This machinery was defined based on literature searches, through which a compilation of 654 sporulation genes was produced (26, 46–63), in good agreement with the recent compilation reported by Galperin and colleagues (654 genes also [36]). We then reached a core of 111 genes, defined as the subset of the genes that were present in at least 90% of the endosporulating bacteria. The phylogenetic profile of these 111 core genes is shown in Fig. 2. It is immediately obvious that the minimal core of the endosporulation machinery is poorly conserved in all other organisms: nonsporulating organisms have a median number of orthologues of 45, compared to known endosporulators (P = 1.4 × 10−10; Kruskal-Wallis test). Exosporulating organisms do not share any more of the endosporulation machinery (median conservation of 50; P = 1.8 × 10−3). Finally, mycobacteria have a median number of orthologues that is even smaller (44; P = 2.0 × 10−04) (see Fig. S2 in the supplemental material).
There are 45 genes present in more than 90% of all bacteria analyzed, and these are found in many nonsporulating bacteria. These 45 genes, together with all analyzed core genes, are delineated in Fig. S3 in the supplemental material for each of the regulons considered. An analysis of the annotated function of these 45 genes showed that they play roles essential for bacterial life that are also important for spore differentiation. In the Spo0A regulon, examples include ftsZ, which is essential both for division at midcell during vegetative growth and for asymmetric division at the onset of sporulation, the ParA/B-type chromosome-partitioning proteins Spo0J/Soj and the DNA translocase SpoIIIE (which also participate in forespore chromosome segregation) (21, 26), and spoIIIJ, which encodes a membrane protein translocase essential for growth in most organisms (a redundant paralogue exists in B. subtilis) that has an irreplaceable role in the assembly of a cell-cell signaling complex during sporulation (66) (see below). In the σE regulon, several of the non-sporulation-specific genes code for metabolic functions, including peptidoglycan synthesis; spoVE, for example, codes for a cortex-specific SEDS family member whose vegetative counterparts control peptidoglycan biosynthesis during cell elongation or cell division, most likely acting as precursor flipases (72). A few genes in the σF and σG regulons also code for enzymes involved in peptidoglycan remodeling with vegetative counterparts (dacF and pdaA) or general functions of the cell (tpiA, involved in protein secretion; pgk, coding for phosphoglycerate kinase, an enzyme of the glycolytic pathway; or mfd, coding for a transcription-repair coupling factor). In the σK regulon, ftsY codes for the signal recognition particle receptor, ribH codes for a riboflavin synthase, and yabG codes for a serine-type protease involved in maturation of the spore coat (see Fig. S3). Very few σK target genes are sporulation specific, presumably because many code for species-specific spore surface proteins (19, 28, 36). In general, the non-sporulation-specific genes are equally distributed among non-endospore formers (exosporulaters and mycobacteria alike) (see Fig. S3). In consonance with the results of a previous study (36), about 60 genes code for proteins that are specific for sporulation.
A genomic signature of endosporulation.
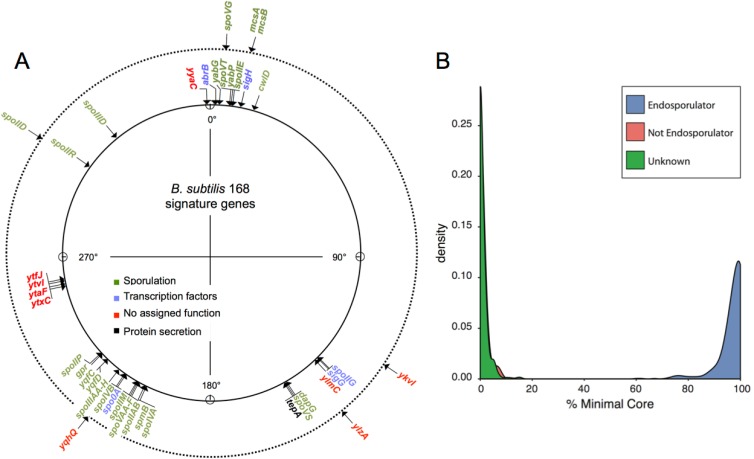
Equipped with the knowledge that many components of the endosporulation machinery are specific to endosporulating bacteria, we next defined a genomic signature for endosporulation as those genes that are found in 90% of endospore-forming organisms and present in no more than 5% (or in no more than 10%) of the nonendosporulators. The choice of these values, as opposed to an all-or-nothing 100% versus 0%, allows for potential errors that may arise out of false-negative or -positive detection of orthologues, lower-quality genome sequences, etc. This signature takes the form of a binary vector, where each coordinate, 0 or 1, defines the presence or absence of an orthologue for a specific gene in the signature. It includes 40 genes when a cutoff of 5% is used and an additional 8 genes when a cutoff of 10% is used (Fig. 3A). Genes with a known function in endosporulation prevail in both lists.
Fig 3.
Genomic signature for endosporulation. (A) The signature is defined as those genes present in 90% of endosporulating bacteria and in no more than 5% (inner circle) or 10% (outer circle) of the remaining bacterial species. Note that the first gene of the spoIIIA operon is only found at the 10% cutoff. Genes with an established function in sporulation, genes coding for the RNA polymerase σ factors that control gene expression during sporulation and genes encoding global transcriptional regulators are shown, as is one gene, tepA, with a predicted function in protein secretion but which has not yet been implicated in sporulation. Also shown are genes with no assigned function. The positions of the genes are shown in degrees in the B. subtilis 168 chromosome. (B) Percentage of the minimal core in genomes, showing that endosporulating organisms all have a high proportion of this signature, and that all mycobacteria and exosporulating organisms have less than 20% of this signature. Those organisms not known to sporulate but that show a proportion of the signature comparable to the known endosporulators are predicted to be able to form endospores.
We then used this signature to query a larger data set of bacterial genomes and computed the fraction of the signature that was detected in each genome. The results of this analysis (Fig. 3B) clearly showed that known endospore-forming bacteria, with higher-than-75% conservation of the signature, are clearly distinguishable from non-endospore formers, which have less than 10% of the signature. Our signature is sufficiently robust to identify Carboxydothermus hydrogenoformans as a spore former (95% of the signature detected), one of the endospore-forming organisms with the smallest genome (73). It is also robust to phylogenetic proximity, as it predicts that Clostridium sticklandii and Bacillus selenitireducens (12.5% and 15% of the signature detected) are not spore formers, even though their phylogenetic proximity would suggest otherwise. C. stiklandii was originally isolated from anoxic black muds in the San Francisco Bay area (74); B. selenitrireducens is a haloalkaliphilc organism that requires a hypersaline environment with a pH in the range of 8.5 to 10 and was found in anoxic muds of lake sediments (75). Life in a stable, predictable environment may explain the loss of the ability to sporulate (76).
Predicting new endosporulating organisms.
With the endosporulation spore signature, we can now predict how likely it is that a given bacterium will be able to form endospores. The set of predictions included several polyextremophiles and gut-associated bacteria. A selection of these predictions is shown in Table 1 for those cases where the sporulation capability was not easily predictable by phylogenetic proximity to other spore-forming species or was considered absent. While several anaerobic halophilic, alkalithermophilic bacteria are known to form spores (102), other thermophiles, like the saccharolytic Thermoanaerobacter pseudethanolicus (77) or the plant biomass-degrading organisms of the Caldicellulosiruptor genus, reportedly do not. Our results suggest that the first group does form endospores, as it matches 100% of the endosporulation signature, and that species of Caldicellulosiruptor will also form spores, as between 77.5 and 82.5% of the signature is present (Table 1). This in turn suggests that endosporulation may be a general feature of this genus, even though the conditions for its occurrence have not been identified yet. In fact, this statement may be true for several other bacteria that have been considered non-spore-forming bacteria (Table 1).
Table 1.
Putative new endosporulating species
| Species | % of signature present | Reference(s) |
|---|---|---|
| Thermoanaerobacter sp. strain X513 | 100 | 77 |
| Thermoanaerobacter pseudethanolicus ATCC 33223 | 100 | 77 |
| Thermoanaerobacter sp. strain X514 | 100 | 77 |
| Thermincola potens JR | 97.5 | 78 |
| Thermosediminibacter oceani DSM 16646 | 97.5 | 79, 80 |
| Halothermothrix orenii H 168 | 97.5 | 81, 82* |
| Natranaerobius thermophilus JWNM-WN-LF | 95 | 83, 84 |
| Syntrophothermus lipocalidus DSM 12680 | 95 | 85, 86 |
| “Candidatus Desulforudis audaxviator” MP104C | 95 | 13** |
| Ammonifex degensii KC4 | 92.5 | 87 |
| Thermaerobacter marianensis DSM 12885 | 90 | 88, 89 |
| Anaerocellum thermophilum DSM 6725 | 85 | 90, 91 |
| Ethanoligenens harbinense YUAN-3 | 85 | 92 |
| Caldicellulosiruptor hydrothermalis 108 | 82.5 | 93, 94 |
| Caldicellulosiruptor kristjanssonii 177R1B | 80 | 93, 95 |
| Caldicellulosiruptor kronotskyensis 2002 | 80 | 93–95 |
| Caldicellulosiruptor owensensis OL | 77.5 | 93, 96 |
| Caldicellulosiruptor obsidiansis OB47 | 77.5 | 97, 98 |
| Caldicellulosiruptor saccharolyticus DSM 8903 | 77.5 | 145 |
| Eubacterium eligens ATCC 27750 | 72.5 | 99, 100 |
| Eubacterium rectale ATCC 33656 | 62.5 | 99, 100 |
| Ruminococcus albus 7 | 60 | 101 |
a*, genomic study revealed many sporulation genes (82);
, spore-like structures were detected.
Other thermophiles or thermohalophiles, such as Thermincola potens (78), Thermosediminibacter oceanii (79), Therrnaerobacter rnarianensis (88), Synthrophothermus lipocalidus (85), or Ammonifex degensii (87), share at least 92.5% of the signature and most likely are able to form endospores (Table 1). The halophilic alkalithermophile Natranaerobius thermophiles (83) shares 95% of the signature and is also a possible endospore former (Table 1). Of note, T. oceanii has a Gram-negative-type envelope, like the recently characterized Acetonema longum, which during germination converts the inner spore membrane into an outer membrane (18).
Interestingly, the group of predicted endospore formers also includes several organisms that are found in association with the gastrointestinal tract of various animals. Eubacterium rectale (62% of the signature) and Eubacterium eligens (72%), for example, are predicted to form endospores (Table 1). Importantly, these organisms, which belong to Clostridium cluster XIVa, one of the most common gut Firmicute clades, are prevalent in the human gut (99, 100). Also, the finding that Ruminococcus albus 7, a prevalent rumen cellulolytic species, carries 60% of the signature suggests that it forms endospores, despite the fact that the organism is labeled as a non-spore-forming bacterium (101). Several of the organisms herein predicted to form endospores were also found in a recent study to be apparent non-spore-forming organisms (but that code for Spo0A) (36). However, and as shown here, the presence of a significant part of the genomic signature clearly suggests that some of these organisms are capable of endospore differentiation.
Signature genes.
Our sporulation signature includes, as expected, many known components of the sporulation machinery as defined by studies with B. subtilis (Table 1). The signature includes the genes for regulatory proteins Spo0A, σE, σG, and the ancillary transcription factors SpoIIID and SpoVT, the engulfment genes spoIID, spoIIM, and spoIIP, the gene coding for the SpoIVA ATPase involved in assembly of the spore surface layers, and the spoVA operon, which is required for transport of dipicolonic acid, a compound characteristic of bacterial endospores, into the forespore (reviewed in references 19, 26, 22, 28, and 103). It also includes genes for proteins that function in cell-type-specific activation of the sporulation σ factors, including the anti-σF/σG factor SpoIIAB, the SpoIIE phosphatase involved in antagonizing SpoIIAB in the forespore, the σF-controlled SpoIIR protein, which is secreted to the intermembrane space and required for the activation of pro-σE in the mother cell (reviewed in reference 26), and the σE-controlled spoIIIA operon, which codes for a transport system that connects the mother cell to the forespore (66, 104–107).
Uncharacterized signature genes.
For a group of 3 genes, ytaF, ylmC, and ylzA, however, a function in sporulation had not been tested when we started our study. In recent work, however, the ylmC and ylzA genes were also identified through phylogenetic analysis (38). Secondary structure predictions suggest that YtaF is a membrane protein, with at least 4 transmembrane (TM) domains (see Fig. S4A in the supplemental material). YtaF also shows structural similarity to several calmodulin-like Ca2+-binding proteins, in particular to EhCaBP1 from Entamoeba histolytica (PDB 3LI6). EhCaBP1 is a Ca2+-binding protein with four canonical EF-hand Ca2+-binding motifs (108, 109). A stretch of 12 amino acid residues in YtaF aligns with the Ca2+-binding motifs of EhCaBP1 as well as with the 12-residue-long consensual Ca2+-binding motif of EF-hand proteins (see Fig. S4A). Moreover, the Ca2+-binding motif in YtaF is located in a loop that connects two predicted α-helices, presumably corresponding to helices E and F of the motif, with helix F, corresponding to the fourth TM domain of the protein (see Fig. S4A). Thus, it appears likely that YtaF is a Ca2+-binding protein.
The ylzA (or remA) gene has been shown to be required for activating expression of the extracellular matrix biosynthetic operons during biofilm development by undomesticated strains of B. subtilis (110). A search for structural homologues revealed the similarity of YlzA to ribose-5-phosphate isomerases (RPI), with the highest similarity shared with the protein from Bartonella henselae (PDB 3HHE) (see Fig. S4B in the supplemental material).
As also noted by Traag and coauthors (38), YlmC shows structural similarity to several proteins containing a PRC barrel domain (see Fig. S4C). The PRC barrel is an all-β-sheet fold domain originally found in the H subunit of the purple photosynthetic reaction center of Rhodopseudomnas viridis and Rhodopseudomonas sphaeroides (111). The H subunit has a founder role in the assembly of the complex, interacting with other subunits via different surfaces (111). The H subunit also participates directly in the electron transfer reactions during the photosynthetic reaction, with an acidic residue (E173 in the PRC H protein of R. viridis) directly participating in the reaction (111). A homologous residue may be involved in nonphotosynthetic electron transfer reactions in nonphotosynthetic organisms (111). However, a homologous acidic residue is absent from many PRC barrel proteins, including those of the RimM family, which are involved in maturation of the ribosome and 16S rRNA processing (111). Together with the founder role of PRC H, this has led to the suggestion that the PRC barrel domain also has a role in mediating protein-protein interactions (111). The functionally important acidic residue is not conserved in the YlmC (see Fig. S4C) or YmxH (see below) proteins of B. subtilis. YlmC showed the highest structural similarity to a protein from Methanobacterium thermoautotrophicum (PDB 1PM3). Strikingly, this protein, which lacks the conserved acidic residue, oligomerizes to form a large honeycomb-like reticular structure that is thought to serve as an adaptor for the assembly of protein complexes (112).
The genomic signature reveals new genes required for endosporulation.
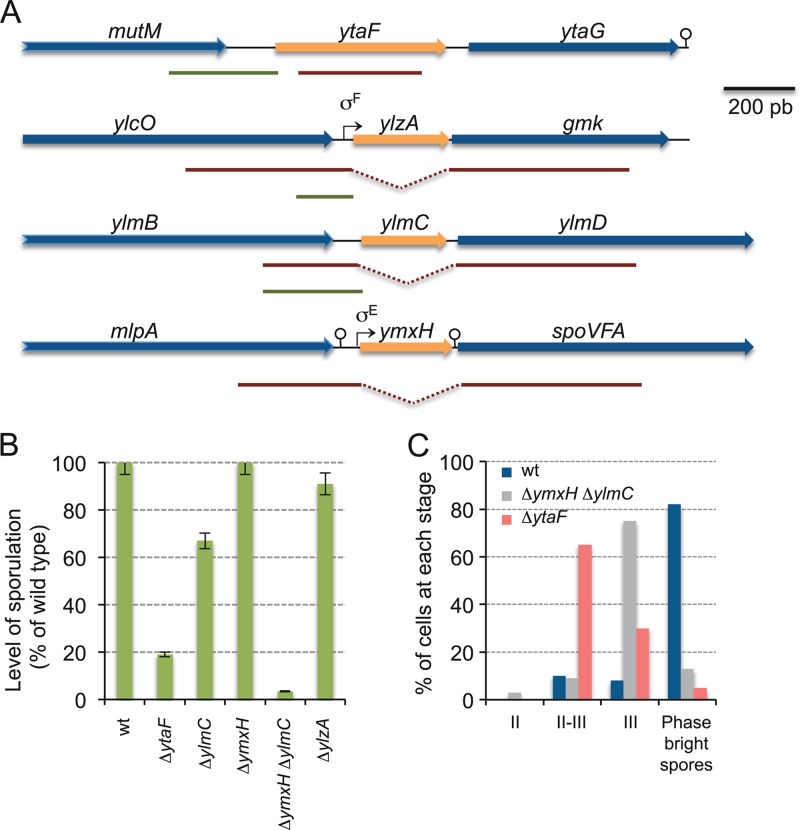
To test for a function(s) of ylzA, ylmC, and ytaF in sporulation, these 3 genes were individually disrupted in B. subtilis (Fig. 4A). None of the 3 mutations interfered with the ability of B. subtilis to form colonies or to grow on rich media (data not shown). However, when tested in sporulation medium in parallel with a congenic reference strain, disruption of the ytaF gene caused a 10-fold reduction in the efficiency of sporulation, as assessed by the titer of heat-resistant spores produced in DSM (Fig. 4B). To determine the stage blocked in the mutant, we used fluorescence and phase-contrast microscopy. Progress through the early stages of sporulation was followed using the membrane stain FM4-64, which enables the forespore to be differentiated from the mother cell until the end of the engulfment process. Following engulfment completion, the stage of forespore maturation was determined by phase-contrast microscopy. Cells of the ytaF mutant divided asymmetrically (morphological stage II) but failed to complete engulfment of the forespore by the mother cell (stage III) (Fig. 4C). Thus, the mutant was blocked at stage II-III of sporulation. The ytaF gene is followed by a gene of unknown function, ytaG, which in turn is separated by a putative transcriptional terminator from the convergent gene for a putative dehydrogenase (Fig. 4A). To test for a polar effect caused by insertion into the ytaF gene, we repeatedly but unsuccessfully tried to disrupt the ytaG gene by the single-reciprocal crossover integration of a plasmid (pMS403) (Fig. 4A). Our failure to obtain transformants is in line with the results of a previous study in which ytaG was found essential for growth and viability of B. subtilis (113). It also suggests that the sporulation phenotype imposed by the ytaF mutation is not due to a polar effect on the expression of ytaG.
Fig 4.
Identification of new endosporulation genes and their functional analysis in B. subtilis. (A) The region of the ytaF, ylzA, ylmC, and ymxH genes in the B. subtilis chromosome, with possible promoters and transcriptional terminators represented by broken arrows and stem-loop structures, respectively. The red lines below the physical map represent the inserts in the plasmids used to disrupt the indicated genes by means of a single-reciprocal (ytaF) or a double-crossover (ylmC, ymxH, and ylzA) event. The green lines represent the sequences transcriptionally fused to the gfp gene. (B) Impacts of insertional mutations in the indicated genes on the ability of the resulting strains to form spores, as assessed by comparing the titers of heat-resistant spores to the total (viable) cell count. The results presented are the averages of three independent experiments, and are shown, for each mutant, as the percentage of the sporulation level obtained for the congenic reference (Spo+) strain MB24 in DSM. (C) Quantitative analysis of the stage at which the indicated mutations affect sporulation, as determined by fluorescence (following staining with FM 4-64) and phase-contrast microscopy of cells collected at hour 8 of sporulation in DSM. The wild type is shown as a reference. Stage II, septation completed; stage III, engulfment completed.
In contrast, and as also found by Traag and coauthors (38), inactivation of ylzA or ylmC did not affect sporulation. The lack of a sporulation phenotype for the ylmC mutant could be due to the presence in the genome of B. subtilis a paralogous gene, ymxH (53, 114), which also codes for a PRC barrel protein (highest similarity detected with an uncharacterized protein from Rhodopseudomonas palustris [PDB 3HTR]) (data not shown). To test the idea that these two genes have a redundant function, we disrupted the ymxH gene (Fig. 4A) and we also constructed a double ylmC ymxH mutant. While the ymxH single mutant sporulated to wild-type levels, the double mutant showed a reduction of 100-fold in the efficiency of sporulation (Fig. 4B). By using fluorescence and phase-contrast microscopy, we were able to determine that most sporangia of the ylmC ymxH mutant completed engulfment of the forespore by the mother cell but that the engulfed forespores did develop refractility (Fig. 4C). Within the limits of light microscopy, we inferred that the mutant is blocked at morphological stage III of sporulation. In the case of the ylzA gene, we also transferred the mutation to an undomesticated strain of B. subtilis. As expected, the resulting ylzA mutant was defective in biofilm formation and also showed a delay in spore formation during biofilm formation, which is typical of mutants with blocks in matrix production (110) (data not shown). Both phenotypes, defective biofilm and spore formation, were corrected when a copy of the ylzA gene was introduced at the nonessential amyE locus (data not shown). However, the ylzA mutation did not detectably interfere with sporulation of the undomesticated strain in DSM (Fig. 4B).
In all, and in consonance with earlier work (38), the results of this analysis identified 3 new sporulation loci in B. subtilis, ytaF, ylmC, and ylzA, based on the prediction that gene conservancy among spore formers is an indication for an essential function in spore development.
The novel sporulation genes show cell-type-specific expression.
To localize the expression of the newly identified sporulation genes, as well as ylzA, we constructed transcriptional fusions of the putative promoters of the ylmC, ylzA, and ytaF genes to gfp. The fusions were inserted in trans at the nonessential amyE locus, and the time and compartment of expression during sporulation were examined by fluorescence microscopy. Fluorescence from the PylzA-gfp fusion was detected during vegetative growth and in the mother cell following asymmetric division at the onset of sporulation (Fig. 5A and B, top row). In addition, green fluorescent protein (GFP) accumulated strongly in the forespore soon after asymmetric division. Inspection of the region upstream of the ylzA gene revealed possible −35 (GTTTA) and −10 (GGCTAAACTA) elements (located between −162 and −132 relative to the start codon) recognized by σF, the early prespore-specific σ factor (−35, GYATA; −10, GG--A-AHTR, where Y is C or T, H is A, C, or T, and R is A or G; hyphens in the −10 sequence indicate spaces) (50). This suggested that expression of ylzA in the forespore could be governed by σF. Three observations are in line with this idea. First, disruption of the sigF gene abolished expression of the PylzA-gfp fusion (Fig. 5A, top). Second, point mutations designed to eliminate the −10 element of the putative σF promoter (changing it to GGCTCACCA) also abolished expression of the PylzA-gfp fusion (Fig. 5A, top). Third, expression of ylzA increased and became confined to the forespore at late times in development (Fig. 5A and B). In contrast, disruption of the gene for the first mother cell-specific σ factor, σE, did not eliminate fluorescence in this compartment (Fig. 5A, top), suggesting that accumulation of GFP in the mother cell results from expression of ylzA in predivisional cells. Using lacZ fusions to analyze expression of the ylzA gene, Traag and coauthors came to the conclusion that expression was not specifically induced during sporulation (38). However, because the expression level of ylzA during growth is high (Fig. 5A), the specific increase we noted here in the forespore compartment may have been missed in that previous study.
Fig 5.
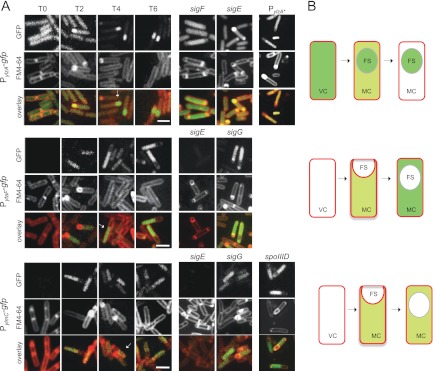
ylmC, ytaF, and ylzA are expressed during spore development. (A) Expression of transcriptional fusions of ylmC, ytaF, and ylzA to gfp, inserted at the nonessential amyE locus of a wild-type strain or the indicated mutant, during sporulation in DSM. PylzA* refers to a fusion bearing two substitutions in the −10 promoter element of a putative σF-dependent promoter (see the text for details). Samples were withdrawn from cultures at the represented times (in hours) after the onset of sporulation (or T0). The cells were stained with the membrane dye FM4-64 (middle column for each strain), prior to observation by fluorescence microscopy. The arrowheads point to the position of the mother cell (white) and forespore (yellow) compartments in the selected cells. Bar, 2 μm. (B) Schematic representation of the expression patterns found for the ylmC, ytaF, and ylzA genes; pale green denotes weak expression, and the darker green indicates stronger accumulation of GFP.
ytaF and ylmC showed weak expression in the mother cell soon after asymmetric division (Fig. 5A and B, middle and bottom rows, respectively). Expression of both genes continued in the mother cell following engulfment completion but became much stronger for ytaF (Fig. 5A). The expression from both fusions was dependent on σE, which is in agreement with the time and localization of the GFP signal (Fig. 5). Using lacZ fusions, Traag and coauthors also found the expression of ylmC to be under σE control (108). However, a contribution from σK, the late mother cell-specific σ factor, to the expression of the ylmC and ytaF genes cannot be ruled out. Surprisingly, we were not able to find, in the regulatory regions of ylmC or ytaF, consensus sequences for either of the mother cell-specific σ factors (data not shown). However, the expression of ylmC appeared to be repressed by SpoIIID (Fig. 5A). We also note that the ylmC paraloguue ymxH has been found to be under the control of σE (52).
DISCUSSION
The divergence of Firmicutes from other prokaryotic phyla has been estimated to have occurred somewhere around 2.5 to 3.0 billion years ago (115). As the Great Oxidation Event occurred ∼2.3 billion years ago (116), endosporulation could have developed as a way to cope with rising O2 levels or other changing environmental conditions associated with this period of Earth's history. It is interesting that the group of known or predicted endosporulating bacteria includes many thermophilic organisms (Table 1). Several of these extremophile organisms cluster in a group that seems more ancient than Bacillus or Clostridium (see Fig. S1 in the supplemental material). Endosporulation may have evolved through strong selective pressure for survival under a set of extreme conditions, and perhaps several lines used the faculty to form highly resistant endospores to survive new environmental challenges and to explore with success new niches. For instance, protozoan grazing is a major force that shapes the structure of bacterial populations, but endospores are resistant to digestion (117, 118), and several endospore formers live in close association with the gastrointestinal tract of their hosts. This is well documented for the guinea pig symbiont Metabacterium polyspora and certain morphotypes of the surgeonfish symbionts Epulopiscium spp. (37, 119, 120) for the “segmented filamentous bacteria,” which are mammalian intestinal symbionts related to Clostridium (121–126), and for many species that colonize the insect gut, including the termite symbiont, didermic organism Acetonema longum (18). Species like B. subtilis may cycle continuously between the soil and the GI tract but can nevertheless complete its entire life cycle in the gut (127). Our suggestion that E. rectale and E. eligens, prevalent in the human gut (99, 100), are able to endosporulate emphasizes the connection between endospore formers and the gut (Table 1). The association of spore-forming bacteria with the GI tract also extends to pathogenic species, as well typified by the strictly anaerobic human and animal pathogen Clostridium difficile, which uses the anaerobic colon to propagate (producing two potent cytotoxins) and to form spores through which the organism disseminates and can reinfect its hosts (7).
The clustering of nonendosporulating species interspersed among the endosporulating species cluster (see Fig. S1 in the supplemental material) has been attributed to the loss of the ability to endosporulate (28). One explanation is that loss of endosporulation is advantageous for organisms living in a fairly constant environment (76). The genes present in the minimal endosporulation core are specific to the endosporulating species of the Firmicutes phylum (references 35 to 37 and this work), indicating that no significant horizontal gene transfer events occurred for this developmental program. Moreover, the lack of orthologues of ∼60 genes known to be involved in endosporulation in other nonendosporulating species indicates that many protein families exist exclusively in endosporulators (reference 36 and this work). Together, these facts indicate that endosporulation is specific to endosporulating species of the Firmicutes phylum, provides evidence in favor of an independent, single origin of the endosporulating program at the basis of the Firmicutes phylum, and in line with other analyses (29), evidence against the reported capacity of Mycobacterium spp. to endosporulate (30, 31). If Mycobacterium can indeed endosporulate, then the origin of the endosporulating program could lie further back in the tree of life (see Fig. S1), with the implication that many other organisms are perhaps able to endosporulate. We also noted that mycobacteria belong to the Actinobacteria phylum, which also includes the Streptomyces genus, members of which differentiate exospores in a process unrelated to endosporulation (128, 129).
The signature genes herein identified point to important features of endosporulation. For instance, the inclusion of the genes for a mother cell (σE) and a forespore-specific σ factor (σG; but σF/σG and σE/σK are very similar proteins) as well as genes (spoIIID, mother cell specific, and the forespore-specific spoVT) involved in establishing the feed-forward loops that characterize the regulatory network of sporulation in B. subtiis (28). It is the specific activation of σF in the forespore that sets in motion the program of cell-type-specific gene expression. The pathway leading to the activation of σF involves spoIIAA, spoIIAB (the first and second cistrons of an operon that also codes for σF), and spoIIE (reviewed in reference 130). In the presence of ATP, SpoIIAB can bind to and inhibit σF, preventing its interaction with core RNA polymerase. This inhibition is counteracted by SpoIIAA, which acts as an anti-anti-σ factor. SpoIIAB can form an ADP-dependent complex with SpoIIAA and is also a serine protein kinase, which phosphorylates and thereby inactivates SpoIIAA. SpoIIAB inhibits σF in predivisional cells and in the mother cell compartment of the sporulating cell. Dephosphorylation of SpoIIAA by SpoIIE, which localizes to the asymmetric septum, allows it to bind to SpoIIAB, releasing σF and triggering forespore-specific gene expression. Although SpoIIAA is found in more non-spore-forming organisms than SpoIIE, both proteins appear to be conserved to the same extent among spore formers (Fig. 2; see also Fig. S3 in the supplemental material), suggesting conservation of the σF-activating pathway. It is possible that other important elements required for the proper activation of σF, including the instability of the protein and transient genetic asymmetry (the exclusion of spoIIAB from the forespore at an early stage in chromosome segregation) (132, 133) have also been conserved. SpoIIE also promotes polar septum formation, a function that requires its phosphatase domain (although not its catalytic activity) (131), and that may also have been conserved.
Another important signature gene is spoIIR, which is transcribed in the forespore as soon as σF is activated (134, 135). The SpoIIR protein is secreted to the intermembrane space and triggers the proteolytic activation of σE by SpoIIGA protease (both are encoded by the signature operon spoIIG). Therefore, the pathways leading to the activation of the forespore and mother cell lines of gene expression are part of the endosporulation signature. The presence of spoIIR in the signature is also of interest, because changes in its level or timing of expression can lead to alternative cell fates, including the formation of viable “twin” spores, in what has been recognized as a potential pathway for the evolution of endosporulation (136). The isolation of the forespore from the external milieu upon engulfment completion is a hallmark of endosporulation. A novel type of transport system coded for by the σE-controlled spoIIIA operon allows the mother cell to feed the forespore, maintaining forespore-specific gene expression, when its engulfment is finalized (66, 104–107). A central part of the complex is formed by the SpoIIIAH protein and by SpoIIQ, a σF-controlled, forespore-specific protein, which interact directly across the intermembrane space, forming a channel that links the cytoplasm of the two cells (104–106, 137). The absence of spoIIQ from the signature most likely is explained by nonorthologous gene replacement events (36), and thus the channel appears to be a universal requirement of endosporulation. Cell-cell signaling is therefore a characteristic of endosporulation that is well represented in the genomic signature.
The endosporulation signature also allowed the identification of new genes involved in sporulation in the model organism B. subtilis. The ylzA gene was previously shown to be required for expression of two key operons envolved in the synthesis of the extracellular matrix during biofilm formation (110). YlzA shows structural similarity to ribose-5-phosphate-isomerase. However, this similarity does not include the region encompassing the active site residues (138, 139). It is not known whether in B. subtilis YlzA functions as a ribose-5-phosphate isomerase. If so, that disruption of ylzA prevents transcription of the eps and yqxM operons, suggesting that a metabolite in the pentose-phosphate pathway controls the transcription of the biofilm matrix operons (110). In any event, our results suggest that ylzA is expressed during sporulation. Spore formation takes place within biofilms (140), and the identification of ylzA as a signature gene suggests an important evolutionary link between biofilm formation and sporulation. During sporulation, expression of ylzA seems to be confined to the forespore. In contrast to the mother cell, very few metabolic genes are part of the σF or σG regulons (49, 50). Clearly, the mother cell provides most of the metabolic activity required to drive spore differentiation. Therefore, the expression of ylzA in the forespore, even though we have not found a sporulation phenotype under laboratory conditions, suggests it has an important role. The group of the few metabolic genes that are expressed in the forespore include the glcU-gdh operon for glucose uptake and the pgk-tpiA-pgm operon, coding for enzymes of the glycolytic pathway (49, 50, 52). If ylzA functions as a ribose-5-phosphate isomerase, it seems plausible that coordinated expression of the glycolytic and pentose-phosphate pathways in the forespore is important for spore morphogenesis or germination under conditions other than those commonly used in the laboratory.
YtaF is most likely a membrane-associated EF-hand Ca2+-binding protein. Disruption of the ytaF gene in our strain background causes a reduction in the titer of heat-resistant spores. By analogy with calmodulin-like proteins, YtaF could be involved in the activation of as-yet-unknown Ca2+-dependent proteins required for sporulation. Another possibility is that YtaF is part of the pathway by which the Ca2+-chelate of dipicolinic acid (DPA) accumulates in spores. DPA is characteristic of endospores and accumulates to about 10% of the total spores dry weight (3, 103). The enzymes involved in its biosynthesis, SpoVFA and SpoVFB, are expressed in the mother cell, under the control of σK (141), and DPA is then imported into the forespore through the action of the products of the σG-controlled spoVA operon (142, 143). Perhaps significantly, the spoVA operon is also part of the endosporulation signature. DPA is essential for spore heat resistance, and disruption of the spoVF or spoVA operons, as found for ytaF, impairs sporulation (141, 144). Secondary structure predictions place the Ca2+-binding motif of YtaF in an extracytoplasmic compartment (see Fig. S4 in the supplemental material). It will be interesting to determine whether YtaF localizes to the spore membranes or to the cell membrane.
Finally, in the background of our reference strain, disruption of ylmC only impaired sporulation in combination with a mutation in ymxH. Like YlmC, the PRC barrel protein YmxH also lacks the acidic reside thought to be important in electron transfer reactions (111). Thus, YlmC and YmxH may serve a redundant role in the mother cell (references 52 and 108 and this work), possibly as scaffolds for the assembly of as-yet-unidentified protein complexes required for spore morphogenesis. Interestingly, Traag and coauthors found that deletion of ylmC caused a slight competitive advantage over a wild-type strain under sporulation-inducing conditions. Those authors suggested that YlmC, although causing a cost on spore formation, must confer some fitness advantage under unknown environmental conditions (38).
In all, our analysis shows that the identification of a genomic signature for endosporulation has uses both in identifying new organisms as capable of endospore differentiation as well as identifying new genes important for the process. Further studies are needed to establish the function of the genes identified here, based on the genomic signature for endosporulation, their phenotype, and expression patterns in a laboratory strain of B. subtilis. It will also be of interest to extend gene inactivation and expression studies such as those reported here to other signature genes in species of spore-forming organisms for which genetic tools are available.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Losick for helpful discussions and sharing information prior to publication.
This work was funded by grants ERA-PTG/SAU/0002/2008 and PEst-OE/EQB/LA0004/2011 from the Fundação para a Ciência e a Tecnologia (FCT) to A.O.H. and PTDC/EBB-BIO/119006/2010 to J.B.P.-L. M.S. was the recipient of a Ph.D. fellowship (SFRH/BPD/36328/2007) from FCT. A.B.A. was funded by an Instituto Gulbenkian de Ciência research fellowship and by a postdoc fellowship (SFRH/BPD/65605/2009) from the FCT.
Footnotes
Published ahead of print 8 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02110-12.
REFERENCES
- 1. Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nicholson WL. 2004. Ubiquity, longevity, and ecological roles of Bacillus spores, p 1–15 In Ricca E, Henriques AO, Cutting SM. (ed), Bacterial spore formers: probiotics and emerging applications. Horizon Scientific Press, London, England [Google Scholar]
- 3. Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525 [DOI] [PubMed] [Google Scholar]
- 4. Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579–606 [DOI] [PubMed] [Google Scholar]
- 5. Fouet A, Mock M. 2006. Regulatory networks for virulence and persistence of Bacillus anthracis. Curr. Opin. Microbiol. 9:160–166 [DOI] [PubMed] [Google Scholar]
- 6. Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 7. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 8. Knecht LD, Pasini P, Daunert S. 2011. Bacterial spores as platforms for bioanalytical and biomedical applications. Anal. Bioanal. Chem. 400:977–989 [DOI] [PubMed] [Google Scholar]
- 9. Hong HA, Duc le H, Cutting HSM. 2005. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 29:813–835 [DOI] [PubMed] [Google Scholar]
- 10. Cutting SM, Hong HA, Baccigalupi L, Ricca E. 2009. Oral vaccine delivery by recombinant spore probiotics. Int. Rev. Immunol. 28:487–505 [DOI] [PubMed] [Google Scholar]
- 11. Ludwig W, Schleifer K-H, Whitman WB. 2009. Revised road map for the phylum Firmicutes, 2nd ed Springer-Verlag, New York, NY [Google Scholar]
- 12. Pester M, Brambilla E, Alazard D, Rattei T, Weinmaier T, Han J, Lucas S, Lapidus A, Cheng JF, Goodwin L, Pitluck S, Peters L, Ovchinnikova G, Teshima H, Detter JC, Han CS, Tapia R, Land ML, Hauser L, Kyrpides NC, Ivanova NN, Pagani I, Huntmann M, Wei CL, Davenport KW, Daligault H, Chain PS, Chen A, Mavromatis K, Markowitz V, Szeto E, Mikhailova N, Pati A, Wagner M, Woyke T, Ollivier B, Klenk HP, Spring S, Loy A. 2012. Complete genome sequences of Desulfosporosinus orientis DSM765T, Desulfosporosinus youngiae DSM17734T, Desulfosporosinus meridiei DSM13257T, and Desulfosporosinus acidiphilus DSM22704T. J. Bacteriol. 194:6300–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin LH, Lowry SR, Moser DP, Richardson PM, Southam G, Wanger G, Pratt LM, Andersen GL, Hazen TC, Brockman FJ, Arkin AP, Onstott TC. 2008. Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322:275–278 [DOI] [PubMed] [Google Scholar]
- 14. Sattley WM, Madigan MT, Swingley WD, Cheung PC, Clocksin KM, Conrad AL, Dejesa LC, Honchak BM, Jung DO, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Page LE, Taylor HL, Wang ZT, Raymond J, Chen M, Blankenship RE, Touchman JW. 2008. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J. Bacteriol. 190:4687–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72 [DOI] [PubMed] [Google Scholar]
- 16. Kimble-Long LK, Madigan MT. 2001. Molecular evidence that the capacity for endosporulation is universal among phototrophic heliobacteria. FEMS Microbiol. Lett. 199:191–195 [DOI] [PubMed] [Google Scholar]
- 17. Ueda K, Yamashita A, Ishikawa J, Shimada M, Watsuji TO, Morimura K, Ikeda H, Hattori M, Beppu T. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen GJ. 2011. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146:799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henriques AO, Moran CP., Jr 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588 [DOI] [PubMed] [Google Scholar]
- 20. Piggot PJ, Coote JG. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579–586 [DOI] [PubMed] [Google Scholar]
- 22. Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341 [DOI] [PubMed] [Google Scholar]
- 23. Fawcett P, Eichenberger P, Losick R, Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 25. Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hilbert DW, Piggot PJ. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Hoon MJ, Eichenberger P, Vitkup D. 2010. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 20:R735–R745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Traag BA, Driks A, Stragier P, Bitter W, Broussard G, Hatfull G, Chu F, Adams KN, Ramakrishnan L, Losick R. 2010. Do mycobacteria produce endospores? Proc. Natl. Acad. Sci. U. S. A. 107:878–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghosh J, Larsson P, Singh B, Pettersson BM, Islam NM, Sarkar SN, Dasgupta S, Kirsebom LA. 2009. Sporulation in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 106:10781–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamont EA, Bannantine JP, Armien A, Ariyakumar DS, Sreevatsan S. 2012. Identification and characterization of a spore-like morphotype in chronically starved Mycobacterium avium subsp. paratuberculosis cultures. PLoS One 7:e30648 doi:10.1371/journal.pone.0030648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Girija KR, Sasikala C, Ramana Ch V, Sproer C, Takaichi S, Thiel V, Imhoff JF. 2010. Rhodobacter johrii sp. nov., an endospore-producing cryptic species isolated from semi-arid tropical soils. Int. J. Syst. Evol. Microbiol. 60:2099–2107 [DOI] [PubMed] [Google Scholar]
- 33. Ajithkumar B, Ajithkumar VP, Iriye R, Doi Y, Sakai T. 2003. Spore-forming Serratia marcescens subsp. sakuensis subsp. nov., isolated from a domestic wastewater treatment tank. Int. J. Syst. Evol. Microbiol. 53:253–258 [DOI] [PubMed] [Google Scholar]
- 34. Stragier P. 2002. A gene odyssey: exploring the genomes of endospore-forming bacteria, p 519–525 In Sonenshein AL. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 35. Onyenwoke RU, Brill JA, Farahi K, Wiegel J. 2004. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes). Arch. Microbiol. 182:182–192 [DOI] [PubMed] [Google Scholar]
- 36. Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. 2012. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 14:2870–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller DA, Suen G, Clements KD, Angert ER. 2012. The genomic basis for the evolution of a novel form of cellular reproduction in the bacterium Epulopiscium. BMC Genomics 13:265 doi:10.1186/1471-2164-13-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Traag BA, Pugliese A, Eisen JA, Losick R. 2 November 2012, posting date Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J. Bacteriol. 195:253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 42. Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. 2009. SUPERFAMILY: sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37:D380–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 45. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 46. Sonenshein AL. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566 [DOI] [PubMed] [Google Scholar]
- 47. Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:21027–21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39 [DOI] [PubMed] [Google Scholar]
- 49. Steil L, Serrano M, Henriques AO, Volker U. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399–420 [DOI] [PubMed] [Google Scholar]
- 50. Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16–37 [DOI] [PubMed] [Google Scholar]
- 51. Kuwana R, Kasahara Y, Fujibayashi M, Takamatsu H, Ogasawara N, Watabe K. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971–3982 [DOI] [PubMed] [Google Scholar]
- 52. Feucht A, Evans L, Errington J. 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149:3023–3034 [DOI] [PubMed] [Google Scholar]
- 53. Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945–972 [DOI] [PubMed] [Google Scholar]
- 54. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328 doi:10.1371/journal.pbio.0020328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wei Y, McPherson DC, Popham DL. 2004. A mother cell-specific class B penicillin-binding protein, PBP4b, in Bacillus subtilis. J. Bacteriol. 186:258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gomez M, Cutting SM. 1997. BofC encodes a putative forespore regulator of the Bacillus subtilis sigma K checkpoint. Microbiology 143:157–170 [DOI] [PubMed] [Google Scholar]
- 57. Henriques AO, Beall BW, Moran CP., Jr 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zheng LB, Losick R. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645–660 [DOI] [PubMed] [Google Scholar]
- 59. Horsburgh GJ, Atrih A, Foster SJ. 2003. Characterization of LytH, a differentiation-associated peptidoglycan hydrolase of Bacillus subtilis involved in endospore cortex maturation. J. Bacteriol. 185:3813–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kodama T, Takamatsu H, Asai K, Ogasawara N, Sadaie Y, Watabe K. 2000. Synthesis and characterization of the spore proteins of Bacillus subtilis YdhD, YkuD, and YkvP, which carry a motif conserved among cell wall binding proteins. J. Biochem. 128:655–663 [DOI] [PubMed] [Google Scholar]
- 61. Fujita M. 1999. Identification of new sigma K-dependent promoters using an in vitro transcription system derived from Bacillus subtilis. Gene 237:45–52 [DOI] [PubMed] [Google Scholar]
- 62. Kakeshita H, Oguro A, Amikura R, Nakamura K, Yamane K. 2000. Expression of the ftsY gene, encoding a homologue of the alpha subunit of mammalian signal recognition particle receptor, is controlled by different promoters in vegetative and sporulating cells of Bacillus subtilis. Microbiology 146:2595–2603 [DOI] [PubMed] [Google Scholar]
- 63. Reischl S, Thake S, Homuth G, Schumann W. 2001. Transcriptional analysis of three Bacillus subtilis genes coding for proteins with the alpha-crystallin domain characteristic of small heat shock proteins. FEMS Microbiol. Lett. 194:99–103 [DOI] [PubMed] [Google Scholar]
- 64. Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U. S. A. 96:2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zilhao R, Serrano M, Isticato R, Ricca E, Moran CP, Jr, Henriques AO. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Serrano M, Vieira F, Moran CP, Jr, Henriques AO. 2008. Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J. Bacteriol. 190:7786–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Altenhoff AM, Dessimoz C. 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 5:e1000262 doi:10.1371/joural.pcbi.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lewis RJ, Krzywda S, Brannigan JA, Turkenburg JP, Muchova K, Dodson EJ, Barak I, Wilkinson AJ. 2000. The trans-activation domain of the sporulation response regulator Spo0A revealed by X-ray crystallography. Mol. Microbiol. 38:198–212 [DOI] [PubMed] [Google Scholar]
- 69. Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Humpel A, Gebhard S, Cook GM, Berney M. 2010. The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192:2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reder A, Gerth U, Hecker M. 2012. Integration of σB activity into the decision-making process of sporulation initiation in Bacillus subtilis. J. Bacteriol. 194:1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235–247 [DOI] [PubMed] [Google Scholar]
- 73. Wu M, Ren Q, Durkin AS, Daugherty SC, Brinkac LM, Dodson RJ, Madupu R, Sullivan SA, Kolonay JF, Haft DH, Nelson WC, Tallon LJ, Jones KM, Ulrich LE, Gonzalez JM, Zhulin IB, Robb FT, Eisen JA. 2005. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1:e65 doi:10.1371/journal.pgen.0010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, Le Paslier D, Weissenbach J, Cohen GN, Kreimeyer A. 2010. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics 11:555 doi:10.1186/1471-2164-11-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Switzer Blum J, Burns Bindi A, Buzzelli J, Stolz JF, Oremland RS. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19–30 [DOI] [PubMed] [Google Scholar]
- 76. Maughan H, Birky CW, Jr, Nicholson WL. 2009. Transcriptome divergence and the loss of plasticity in Bacillus subtilis after 6,000 generations of evolution under relaxed selection for sporulation. J. Bacteriol. 191:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hemme CL, Mouttaki H, Lee YJ, Zhang G, Goodwin L, Lucas S, Copeland A, Lapidus A, Glavina del Rio T, Tice H, Saunders E, Brettin T, Detter JC, Han CS, Pitluck S, Land ML, Hauser LJ, Kyrpides N, Mikhailova N, He Z, Wu L, Van Nostrand JD, Henrissat B, He Q, Lawson PA, Tanner RS, Lynd LR, Wiegel J, Fields MW, Arkin AP, Schadt CW, Stevenson BS, McInerney MJ, Yang Y, Dong H, Xing D, Ren N, Wang A, Huhnke RL, Mielenz JR, Ding SY, Himmel ME, Taghavi S, van der Lelie D, Rubin EM, Zhou J. 2010. Sequencing of multiple clostridial genomes related to biomass conversion and biofuel production. J. Bacteriol. 192:6494–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Byrne-Bailey KG, Wrighton KC, Melnyk RA, Agbo P, Hazen TC, Coates JD. 2010. Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J. Bacteriol. 192:4078–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pitluck S, Yasawong M, Munk C, Nolan M, Lapidus A, Lucas S, Glavina Del Rio T, Tice H, Cheng JF, Bruce D, Detter C, Tapia R, Han C, Goodwin L, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Rohde M, Spring S, Sikorski J, Goker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. Complete genome sequence of Thermosediminibacter oceani type strain (JW/IW-1228P). Stand. Genomic Sci. 3:108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee YJ, Wagner ID, Brice ME, Kevbrin VV, Mills GL, Romanek CS, Wiegel J. 2005. Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter litoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru Margin. Extremophiles 9:375–383 [DOI] [PubMed] [Google Scholar]
- 81. Cayol JL, Ollivier B, Patel BK, Prensier G, Guezennec J, Garcia JL. 1994. Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. Int. J. Syst. Bacteriol. 44:534–540 [DOI] [PubMed] [Google Scholar]
- 82. Mavromatis K, Ivanova N, Anderson I, Lykidis A, Hooper SD, Sun H, Kunin V, Lapidus A, Hugenholtz P, Patel B, Kyrpides NC. 2009. Genome analysis of the anaerobic thermohalophilic bacterium Halothermothrix orenii. PLoS One 4:e4192 doi:10.1371/journal.pone.0004192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao B, Mesbah NM, Dalin E, Goodwin L, Nolan M, Pitluck S, Chertkov O, Brettin TS, Han J, Larimer FW, Land ML, Hauser L, Kyrpides N, Wiegel J. 2011. Complete genome sequence of the anaerobic, halophilic alkalithermophile Natranaerobius thermophilus JW/NM-WN-LF. J. Bacteriol. 193:4023–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mesbah NM, Hedrick DB, Peacock AD, Rohde M, Wiegel J. 2007. Natranaerobius thermophilus gen. nov., sp. nov., a halophilic, alkalithermophilic bacterium from soda lakes of the Wadi An Natrun, Egypt, and proposal of Natranaerobiaceae fam. nov. and Natranaerobiales ord. nov. Int. J. Syst. Evol. Microbiol. 57:2507–2512 [DOI] [PubMed] [Google Scholar]
- 85. Djao OD, Zhang X, Lucas S, Lapidus A, Del Rio TG, Nolan M, Tice H, Cheng JF, Han C, Tapia R, Goodwin L, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Ovchinnikova G, Pati A, Brambilla E, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Rohde M, Sikorski J, Spring S, Goker M, Detter JC, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. Complete genome sequence of Syntrophothermus lipocalidus type strain (TGB-C1). Stand. Genomic Sci. 3:268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. 2000. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int. J. Syst. Evol. Microbiol. 50:771–779 [DOI] [PubMed] [Google Scholar]
- 87. Huber R, Rossnagel P, Woese CR, Rachel R, Langworthy TA, Stetter KO. 1996. Formation of ammonium from nitrate during chemolithoautotrophic growth of the extremely thermophilic bacterium Ammonifex degensii gen. nov. sp. nov. Syst. Appl. Microbiol. 19:40–49 [DOI] [PubMed] [Google Scholar]
- 88. Han C, Gu W, Zhang X, Lapidus A, Nolan M, Copeland A, Lucas S, Del Rio TG, Tice H, Cheng JF, Tapia R, Goodwin L, Pitluck S, Pagani I, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Schneider S, Rohde M, Goker M, Pukall R, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Detter JC. 2010. Complete genome sequence of Thermaerobacter marianensis type strain (7p75a). Stand. Genomic Sci. 3:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Takai K, Inoue A, Horikoshi K. 1999. Thermaerobacter marianensis gen. nov., sp. nov., an aerobic extremely thermophilic marine bacterium from the 11,000 m deep Mariana Trench. Int. J. Syst. Bacteriol. 49:619–628 [DOI] [PubMed] [Google Scholar]
- 90. Kataeva IA, Yang SJ, Dam P, Poole FL, II, Yin Y, Zhou F, Chou WC, Xu Y, Goodwin L, Sims DR, Detter JC, Hauser LJ, Westpheling J, Adams MW. 2009. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J. Bacteriol. 191:3760–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang SJ, Kataeva I, Wiegel J, Yin Y, Dam P, Xu Y, Westpheling J, Adams MW. 2010. Classification of “Anaerocellum thermophilum” strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int. J. Syst. Evol. Microbiol. 60:2011–2015 [DOI] [PubMed] [Google Scholar]
- 92. Xing D, Ren N, Li Q, Lin M, Wang A, Zhao L. 2006. Ethanoligenens harbinense gen. nov., sp. nov., isolated from molasses wastewater. Int. J. Syst. Evol. Microbiol. 56:755–760 [DOI] [PubMed] [Google Scholar]
- 93. Blumer-Schuette SE, Ozdemir I, Mistry D, Lucas S, Lapidus A, Cheng JF, Goodwin LA, Pitluck S, Land ML, Hauser LJ, Woyke T, Mikhailova N, Pati A, Kyrpides NC, Ivanova N, Detter JC, Walston-Davenport K, Han S, Adams MW, Kelly RM. 2011. Complete genome sequences for the anaerobic, extremely thermophilic plant biomass-degrading bacteria Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor owensensis, and Caldicellulosiruptor lactoaceticus. J. Bacteriol. 193:1483–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Miroshnichenko ML, Kublanov IV, Kostrikina NA, Tourova TP, Kolganova TV, Birkeland NK, Bonch-Osmolovskaya EA. 2008. Caldicellulosiruptor kronotskyensis sp. nov. and Caldicellulosiruptor hydrothermalis sp. nov., two extremely thermophilic, cellulolytic, anaerobic bacteria from Kamchatka thermal springs. Int. J. Syst. Evol. Microbiol. 58:1492–1496 [DOI] [PubMed] [Google Scholar]
- 95. Bredholt S, Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK. 1999. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int. J. Syst. Bacteriol. 49:991–996 [DOI] [PubMed] [Google Scholar]
- 96. Huang CY, Patel BK, Mah RA, Baresi L. 1998. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int. J. Syst. Bacteriol. 48:91–97 [DOI] [PubMed] [Google Scholar]
- 97. Elkins JG, Lochner A, Hamilton-Brehm SD, Davenport KW, Podar M, Brown SD, Land ML, Hauser LJ, Klingeman DM, Raman B, Goodwin LA, Tapia R, Meincke LJ, Detter JC, Bruce DC, Han CS, Palumbo AV, Cottingham RW, Keller M, Graham DE. 2010. Complete genome sequence of the cellulolytic thermophile Caldicellulosiruptor obsidiansis OB47T. J. Bacteriol. 192:6099–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M, Carroll S, Allman S, Phelps TJ, Keller M, Elkins JG. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76:1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. 2012. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 78:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Suen G, Stevenson DM, Bruce DC, Chertkov O, Copeland A, Cheng JF, Detter C, Detter JC, Goodwin LA, Han CS, Hauser LJ, Ivanova NN, Kyrpides NC, Land ML, Lapidus A, Lucas S, Ovchinnikova G, Pitluck S, Tapia R, Woyke T, Boyum J, Mead D, Weimer PJ. 2011. Complete genome of the cellulolytic ruminal bacterium Ruminococcus albus 7. J. Bacteriol. 193:5574–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mesbah NM, Wiegel J. 2012. Life under multiple extreme conditions: diversity and physiology of the halophilic alkalithermophiles. Appl. Environ. Microbiol. 78:4074–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 104. Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U. S. A. 105:15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5:e1000566 doi:10.1371/journal.pgen.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kumar S, Ahmad E, Mansuri MS, Jain R, Khan RH, Gourinath S. 2010. Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica. Biophys. J. 98:2933–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jain R, Kumar S, Gourinath S, Bhattacharya S, Bhattacharya A. 2009. N- and C-terminal domains of the calcium binding protein EhCaBP1 of the parasite Entamoeba histolytica display distinct functions. PLoS One 4:e5269 doi:10.1371/journal.pone.0005269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Winkelman JT, Blair KM, Kearns DB. 2009. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J. Bacteriol. 191:3981–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Anantharaman V, Aravind L. 2002. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 3:RESEARCH0061. doi:10.1186/gb-2002-3-11-research0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ye H, Chen TC, Xu X, Pennycooke M, Wu H, Steegborn C. 2004. Crystal structure of the putative adapter protein MTH1859. J. Struct. Biol. 148:251–256 [DOI] [PubMed] [Google Scholar]
- 113. Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sogaard-Andersen L, Overgaard M, Lobedanz S, Ellehauge E, Jelsbak L, Rasmussen AA. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1–8 [DOI] [PubMed] [Google Scholar]
- 115. Battistuzzi FU, Feijao A, Hedges SB. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4:44 doi:10.1186/1471-2148-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bekker A, Holland HD, Wang PL, Rumble D, III, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ. 2004. Dating the rise of atmospheric oxygen. Nature 427:117–120 [DOI] [PubMed] [Google Scholar]
- 117. Klobutcher LA, Ragkousi K, Setlow P. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 103:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Laaberki MH, Dworkin J. 2008. Role of spore coat proteins in the resistance of Bacillus subtilis spores to Caenorhabditis elegans predation. J. Bacteriol. 190:6197–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Angert ER, Losick RM. 1998. Propagation by sporulation in the guinea pig symbiont Metabacterium polyspora. Proc. Natl. Acad. Sci. U. S. A. 95:10218–10223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Angert ER, Clements KD. 2004. Initiation of intracellular offspring in Epulopiscium. Mol. Microbiol. 51:827–835 [DOI] [PubMed] [Google Scholar]
- 121. Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. 2011. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10:260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, Ishifune C, Maekawa Y, Yasutomo K, Hattori M, Hayashi T. 2011. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 18:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, Taylor TD, Ohno H, Umesaki Y, Hattori M. 2011. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe 10:273–284 [DOI] [PubMed] [Google Scholar]
- 124. Chase DG, Erlandsen SL. 1976. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J. Bacteriol. 127:572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ferguson DJ, Birch-Andersen A. 1979. Electron microscopy of a filamentous, segmented bacterium attached to the small intestine of mice from a laboratory animal colony in Denmark. Acta Pathol. Microbiol. Scand. B 87:247–252 [DOI] [PubMed] [Google Scholar]
- 126. Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int. J. Syst. Bacteriol. 45:780–782 [DOI] [PubMed] [Google Scholar]
- 127. Tam NK, Uyen NQ, Hong HA, Duc le H, Hoa TT, Serra CR, Henriques AO, Cutting SM. 2006. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 188:2692–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chater KF, Chandra G. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651–672 [DOI] [PubMed] [Google Scholar]
- 129. Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 130. Yudkin MD, Clarkson J. 2005. Differential gene expression in genetically identical sister cells: the initiation of sporulation in Bacillus subtilis. Mol. Microbiol. 56:578–589 [DOI] [PubMed] [Google Scholar]
- 131. Carniol K, Ben-Yehuda S, King N, Losick R. 2005. Genetic dissection of the sporulation protein SpoIIE and its role in asymmetric division in Bacillus subtilis. J. Bacteriol. 187:3511–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Dworkin J, Losick R. 2001. Differential gene expression governed by chromosomal spatial asymmetry. Cell 107:339–346 [DOI] [PubMed] [Google Scholar]
- 133. Pan Q, Garsin DA, Losick R. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell 8:873–883 [DOI] [PubMed] [Google Scholar]
- 134. Karow ML, Glaser P, Piggot PJ. 1995. Identification of a gene, spoIIR, that links the activation of sigma E to the transcriptional activity of sigma F during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 92:2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Londono-Vallejo JA, Stragier P. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503–508 [DOI] [PubMed] [Google Scholar]
- 136. Eldar A, Chary VK, Xenopoulos P, Fontes ME, Loson OC, Dworkin J, Piggot PJ, Elowitz MB. 2009. Partial penetrance facilitates developmental evolution in bacteria. Nature 460:510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18:2916–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rangarajan ES, Sivaraman J, Matte A, Cygler M. 2002. Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli. Proteins 48:737–740 [DOI] [PubMed] [Google Scholar]
- 139. Zhang R, Andersson CE, Savchenko A, Skarina T, Evdokimova E, Beasley S, Arrowsmith CH, Edwards AM, Joachimiak A, Mowbray SL. 2003. Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle. Structure 11:31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152–163 [DOI] [PubMed] [Google Scholar]
- 141. Daniel RA, Errington J. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J. Mol. Biol. 232:468–483 [DOI] [PubMed] [Google Scholar]
- 142. Tovar-Rojo F, Chander M, Setlow B, Setlow P. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184:584–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li Y, Davis A, Korza G, Zhang P, Li YQ, Setlow B, Setlow P, Hao B. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194:1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Errington J, Mandelstam J. 1984. Genetic and phenotypic characterization of a cluster of mutations in the spoVA locus of Bacillus subtilis. J. Gen. Microbiol. 130:2115–2121 [DOI] [PubMed] [Google Scholar]
- 145. van de Werken HJG, Verhaart MRA, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EWJ, Stams AJM, Ward DE, de Vos WM, van der Oost J, Kelly RM, Kengen SWM. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74:6720–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.