Abstract
The G-protein coupled receptor CXCR4 is a co-receptor for HIV-1 infection and is involved in signaling cell migration and proliferation. In a previous study of non-peptide, guanide-based CXCR4-binding compounds, spermine and spermidine phenylguanides inhibited HIV-1 entry at low micromolar concentrations. Subsequently, crystal structures of CXCR4 were used to dock a series of naphthylguanide derivatives of the polyamines spermidine and spermine. Synthesis and evaluation of the naphthylguanide compounds identified our best compound, spermine tris-1-naphthylguanide, which bound CXCR4 with an IC50 of 40nM and inhibited the infection of TZM-bl cells with X4, but not R5, strains of HIV-1 with an IC50 of 50–100nM.
Keywords: Human immunodeficiency virus, co-receptor, CXCR4, guanide, inhibitor
Chemokine receptor type 4 (CXCR4) is an integral membrane protein of the G-protein coupled receptor family. CXCR4 signaling is induced by the binding of its natural ligand, stromal-derived-factor-1 (SDF-1), and can lead to physiological effects such as stem cell homing to the bone marrow and lymphocyte chemotaxis. The expression of CXCR4 by certain tumor cells has also been shown to be an important factor in metastasis of those cells to tissues with high SDF-1 expression such as bone marrow, lungs, lymph nodes and liver.1 CXCR4 is also one of the co-receptors used by human immunodeficiency virus type 1 (HIV-1) for infection of human lymphocytes.
The structure of CXCR4 has been solved bound to two different inhibitors: CVX15, an analog of the peptide T140, and IT1t, a small-molecule isothiourea derivative.2 CXCR4 was found to have a large, open, extracellular binding pocket capable of holding the entire cyclic CVX15 peptide, and this large pocket was similarly found when IT1t was bound to CXCR4, even though the molecule only inhabits a small part of the bottom of the binding pocket.
Maraviroc, a small molecule antagonist of the CCR5 receptor was the first drug of its type to be approved, and it has proven to be effective against “R5” HIV-1 that uses the CCR5 co-receptor.3 However, most patients that have failed conventional therapies also harbor “X4” HIV-1 that uses CXCR4 or dual-tropic viruses that can use either CCR5 or CXCR4. The appearance of the X4 viruses is associated with progression to AIDS,4 and drug resistance is more often linked to X4 than to R5 HIV-1.5 Despite the need for an effective drug that blocks CXCR4, no CXCR4 antagonists have shown promise in HIV-1 clinical trials to date.
T-140 is a 14-residue peptide antagonist of CXCR4 that displaces the natural agonist ligand SDF-1α with nanomolar affinity and blocks binding of X4, but not R5, strains of HIV-1 in vitro 6–8, but is rapidly degraded in serum8 and has not been pursued as an HIV-1 therapeutic. The structure of T-140 contains 5 Arg and 2 Lys residues; the latter can be substituted with uncharged sidechains without loss of activity.7, 9 In a previous study,10 we developed a series of small molecule inhibitors containing multiple guanide or biguanide groups to test their ability to inhibit HIV-1 binding to CXCR4. Among these non-peptide compounds with 2–8 guanide groups, spermine and spermidine phenylguanides (PheG) were identified as CXCR4 ligands that inhibit T-140 binding with IC50s around 200 nM and inhibit HIV-1 entry at low micromolar concentrations. In the present study, we have improved on these results by synthesizing naphthylguanide (NapG) compounds based on predicted affinity increases in docking simulations with the published CXCR4 structures and then examining their ability to inhibit the binding of T140 to CXCR4, block binding of anti-CXCR4 antibodies to CXCR4+ cells, and inhibit HIV-1 infection. These new compounds inhibited infection by X4, but not R5, strains of HIV-1 with IC50s 5–10 fold lower than the previously reported compounds. The results demonstrate the potential of these compounds to be further improved and lead to the development of an effective X4 HIV-1 entry inhibitor.
Making naphthylguanide derivatives of spermine and spermidine seemed an obvious extension of our initial study in which the phenylguanide derivatives showed increased activity compared to the related guanide and biguanide derivatives, and the linear oligoamine scaffolds (spermine and spermidine) showed the best CXCR4 binding and HIV-1 inhibition. Additionally, the CXCR4 inhibitor T140 includes a naphthyl-alanine residue with a 2-naphthyl substituent, which has been shown to be critical for CXCR4 binding9 as well as being critical in cyclic pentapeptides based on T140.11
Initially, we examined the potential CXCR4 binding of the naphthylguanide derivatives of spermidine and spermine by docking model compounds to published crystal structures of CXCR4 as ‘rigid’ receptors with no flexible side chains using Autdock Vina.12 In order to minimize the effect of any small changes in the crystal structures caused by the binding of the two structurally diverse ligands, two of the published crystal structures were used for the docking simulations: 3OE0 which has the T140 peptide analog, CVX15, bound and 3ODU which includes the bound small molecule IT1t.2 The ligand guanidinium bonds were set to non-rotatable, but care was taken to allow the C-N bond between the guanidine group and the naphthyl group to rotate freely. The affinity of the best docking models (kcal/mole) for each compound are listed in Table 1. The docking studies suggested that the naphthylguanide derivatives should bind more tightly to CXCR4 compared to the previously tested phenylguanide compounds. There was not a clear preference for 1-naphthyl vs. 2-naphthyl substitution on the aromatic rings among all of the models examined, but it appeared that the affinity generally increased with the addition of more naphthyl groups.
Table 1.
Compound affinities determined by Autdock Vina compared to experimental IC50 values for fluorescent T-140/CXCR4 cross-link inhibition and in vitro cytotoxicities against a CXCR4 expressing human cell line (MDA-MB-231).
| Compound | Docking Affinity | CXCR4-T-140 Inhibition (nM ± SEM) | Cytotoxicity CC50 (nM) | ||
|---|---|---|---|---|---|
| 3ODU (IT1t) rigid (kcal/mole) | 3OE0 (T-140) rigid (kcal/mole) | 3ODU flexible (kcal/mole) | |||
| spermidine bis-PheG | −7.8 | −7.2 | −9.8 | 260 ± 60 | > 1,000,000 |
| spermidine tris-PheG | −9.4 | −7.9 | −11.7 | 280 ± 160 | 510,000 |
| spermine tetra-PheG | −10.0 | −8.9 | −11.6 | 510 ± 300 | > 100,000 |
| spermidine bis-1-NapG | −9.5 | −7.2 | −11.7 | 220 ± 40 | 120,000 |
| spermidine bis-2-NapG | −9.0 | −7.1 | −12.1 | 62 ± 23 | 76,000 |
| spermidine tris-1-NapG | −11.0 | −9.5 | ND | 110 ± 30 | ND |
| spermidine tris-2-NapG | −10.7 | −7.6 | ND | 190 ± 120 | ND |
| spermine bis-1-NapG | −8.7 | −6.7 | −12.3 | 180 ± 30 | 300,000 |
| spermine bis-2-NapG | −9.6 | −7.3 | −11.7 | 110 ± 20 | 17,000 |
| spermine tris-1-NapG | −10.6 | −8.0 | −13.1 | 40 ± 8 | 18,000 |
| spermine tris-2-NapG | −10.2 | −7.5 | −14.5 | 93 ± 21 | 15,000 |
| spermine tetra-1-NapG | −11.9 | −8.1 | −15.3 | 380 ± 40 | 7,300 |
| spermine tetra-2-NapG | −12.7 | −8.0 | −14.3 | 300 ± 30 | 20,000 |
Subsequently, the compounds were docked to one of the CXCR4 crystal structures (3ODU) in which the side chains of the amino acids lining the binding cavity of the protein were allowed to be ‘flexible’ and sample the amino acid rotamers during the docking process. Not surprisingly, this led to models with higher affinity binding values, but the overall trends between the phenylguanide and various naphthylguanide compounds remained the same when using a ‘flexible’ receptor or a ‘rigid’ receptor (Table 1).
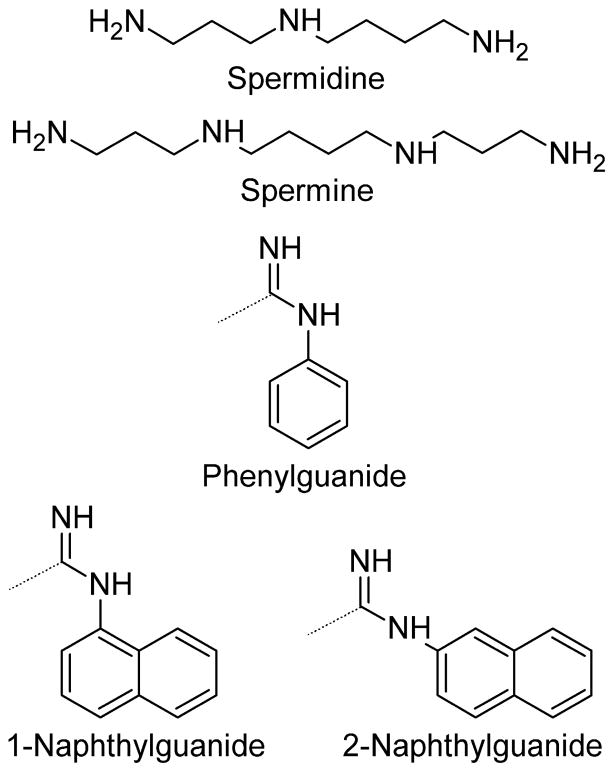
Naphthylguanide derivatives of the oligoamine compounds spermidine and spermine (Figure 1) were synthesized similarly to the previously published phenylguanide derivatives.10 1-(2-naphthyl)-2-thiourea was prepared from 2-naphthylamine as previously published.13,14 S-methyl-N-(1- or 2-)-naphthylisothiouronium iodide was prepared as previously published10 by the addition of methyl iodide to the thioureas. Addition of the naphthylguanide reagent (S-methyl-naphthylisothiouronium iodide, in excess) to the amine compounds, spermine and spermidine, was performed in 1:1 acetonitrile:water at reflux for approximately 24 h. The pH of the reactions were monitored and adjusted with 1 M NaOH when it fell below pH~8. Reaction progress was monitored by MALDI analysis. Due to loss of naphthylguanide reagent through hydrolysis, additional reagent was added when necessary as observed by MALDI. Completed reactions were acidified by the addition of 100% trifluoroacetic acid and lyophilized. The reactions were resuspended in water/acetonitrile with the minimal amount of acetonitrile required for solubility and purified by reverse phase HPLC.
Figure 1.
Guanide derivative structures. Spermidine and spermine were derivatized to make the phenylguanide, 1-naphthylguanide, or 2-naphthylguanide derivatives. The dotted lines on the guanide functional groups show the attachment point to the nitrogens of the spermidine and spermine starting amines.
Naphthylguanide derivatization reactions were not pushed to completion in order to examine the structure-activity relationship relative to the number of groups added. This allowed us to isolate and screen the bis- and trisnaphthyl derivatives of spermidine and the bis-, tris-, and tetranaphthyl derivatives of spermine. The bisnaphthyl derivatives of both spermidine and spermine appear to be mainly derivatized on the terminal primary amines, due to the difference in reactivity between the primary and secondary amines. The 1H NMR data suggest that in each case at least 80–90% of the isolated products are derivatized on the two primary amines, but there is a small amount of each bisnaphthyl derivative in which one of the naphthylguanide groups is attached to an internal, secondary amine. The spermine trisnaphthyl derivatives appear to be only formed as the products with naphthylguanides on the two terminal amines and a single naphthylguanide on one of the internal nitrogens. The symmetric nature of spermine thus yields only a single trisnaphthyl derivative. The series of derivatives for spermidine and spermine were synthesized for both the 1-naphthyl and 2-naphthyl structural isomers (Figure 1).
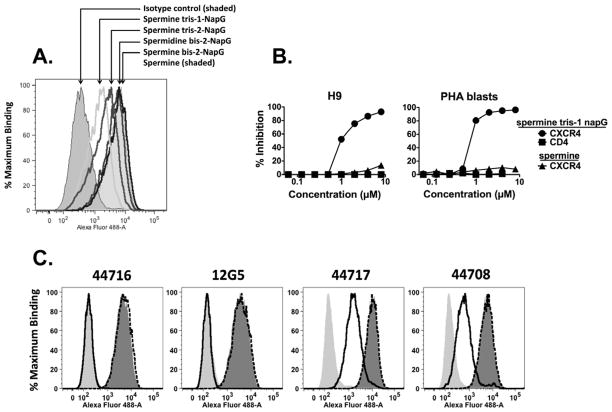
The compounds were tested for CXCR4 binding by inhibiting the cross-linking of a photoactive, fluorescent derivative of the known CXCR4 binding peptide T140 as previously described.10 The results of the CXCR4-T140 cross-link inhibition assay are shown in Table 1. As a whole, the naphthylguanide derivatives of spermidine and spermine have lower IC50 values in the CXCR4-T140 cross-link inhibition screening assay than the previous phenylguanides. Five of the compounds tested were notably more active than the phenylguanides and the other naphthylguanides: spermidine bis-2-naphthylguanide, spermidine tris-1-naphthylguanide, spermine bis-2-naphthylguanide, spermine tris-1-naphthylguanide, and spermine tris-2-naphthylguanide.
The toxicity of the compounds was evaluated after 48 to 72 hours of exposure to a CXCR4 expressing human breast cancer cell line (MDA-MB-231) for all of the compounds using Promega’s CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay (MTS) following the manufacturer’s instructions (Table 1). The naphthylguanide derivatives appear to be more cytotoxic to mammalian cells compared to the phenylguanide derivatives. The cytotoxicity appears to roughly correlate with the number of naphthyl rings on the compounds with the bis-naphthylguanide derivatives being less toxic than the more heavily derivatized tris- and tetra- derivatives. Toxicity of spermine tris-1-NapG was also tested on TZM-bl, H9, and PHA blast cultures. There was no significant inhibition in MTS dye reduction at 72 h in concentrations up to 4 μM, greater than the highest concentration used in the HIV-1 inhibition assays (data not shown).
The isolation of the bisnaphthylguanide derivatives of spermidine and spermine as mixtures raises the possibility that one of the minor isomers (with one of the groups added to an internal, secondary amine) may be significantly more active and/or cytotoxic than the major isomer (with both groups on the terminal primary amines). We examined the docking of all possible isomers using Autodock Vina. Only for the spermine bis-2-NapG derivative was there a significantly higher (> 0.5 kcal/mole) predicted affinity for a minor isomer compared to that of the major isomers reported in Table 1 consistent between the two CXCR4 structures. However, the spermine bis-2-NapG was one of the most pure of the bis derivative mixtures (≥90% by 1H NMR); thus, the minor isomer would have to be many times more active and/or more cytotoxic than the major isomer to significantly impact the observed results. Since spermidine is asymmetric, there are 3 isomers of each bisnaphthylguanide, and separation of these by HPLC could not be achieved. The analyzed compound was ~80% derivatized on the terminal amines, but the modeling did not predict significant differences in binding for the minor isomers.
Four of the best naphthylguanide derivatives were tested for their ability to interfere with the binding of anti-CXCR4 mAbs to the CD4+, CXCR4+ lymphoma cell line H9 and primary PHA-induced T-cell blast cultured cells15 (Figure 2). Four different anti-CXCR4 44716, 44717, 44708, and 12G516 were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP), as was the specificity control anti-CD4 mAb Sim.2.17 The isotype control Ab was anti-ricin mAb RAC18.18 Cells were incubated with naphthylguanide derivatives in PBS, 1% bovine serum albumin, 0.01% sodium azide (PBS/BSA/Azide) for 20–60 min at 4°C, and then mAb was added to a final concentration of 1 μg/ml. Cells were incubated for an additional 1 h, washed, and resuspended in Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L chain, Invitrogen) at 2 μg/ml. After 1 h the cells were washed and fixed in 2% paraformaldehyde. Cells (10,000 events) were analyzed in an LSR-II flow cytometer (BD Biosciences, San Jose CA), and percent inhibition was determined using median fluorescent intensity (MFI) according to the formula:
Figure 2. Inhibition of binding of anti-CXCR4 mAbs to cells by naphthylguanide derivitives.
a. H9 lymphoma cells were incubated at 4°C with 1 μM naphthylguanide derivatives in PBS/BSA/azide. After 20 min, anti-CXCR4 mAb 44716 or an isotype control was added to 1 μg/ml. After 1 h at 4°C, the cells were washed and Alexa-fluor 488-conjugated anti-mouse IgG was added. Cells were incubated at 4°C for one hour, washed, and fixed in 2% paraformaldehyde. Fluorescence of the cells (10,000 events) was then analyzed by flow cytometry. b. H9 cells or primary T cell blasts were incubated with varying concentrations of spermine tris-1-NapG or spermine alone, and then stained with anti-CXCR4 mAb 12G5 or, as a control, anti-CD4 mAb Sim.2 as described in panel A. c. Inhibition of four different anti-CXCR4 mAbs by 3 μM spermine tris-1-NapG or spermine. Cells were stained as in panel A.
Spermine tris-1-NapG (Fig. 2A) was the most effective at blocking mAb binding, spermine tris-2-NapG was next best, whereas the bis- derivatives had marginal effects. Panel B shows a titration of spermine tris-1-NapG and spermine on the inhibition of binding of Abs to CXCR4 and CD4 expressed on two different cell types. The naphthylguanide derivative inhibits anti-CXCR4 binding, but not anti-CD4; spermine does not inhibit anti-CXCR4. In panel C, we tested the inhibition of binding of four different anti-CXCR4 mAbs by spermine tris-1-NapG or spermine. No inhibition of any mAb was observed with spermine. The binding of mAbs 44716 and 12G5 was completely inhibited by 3μM spermine tris-1-NapG, while that of 44717 and 44708 was only partly inhibited, suggesting that these two sets of mAbs bind to somewhat different epitopes on CXCR4. This information could give insight into the binding mode of spermine tris-1-NapG. It is known that these antibodies show differential reactivity to various CXCR4 point mutations and interspecies chimeric constructs.19–20 The limited mutational data in those studies suggest that the epitopes of 44708 and 44716 are more similar to each other than they are to 44717 and 12G5, but are not inconsistent with the observation that spermine tris-1-NapG reduced the binding of 12G5 and 44716 more effectively than 44708 and 44717 (figure 2C). This may be due to the latter having more interactions with CXCR4 in the parts of their epitope that are not blocked by the binding of spermine tris-1-NapG. Although the binding studies presented here were performed with the naphthylguanide compounds present at the same time as the anti-CXCR4 mAb, we obtained similar results when the cells were washed prior to the addition of the Ab (not shown).
The four most active compounds were selected for testing as HIV-1 infection inhibitors using a standard TZM-bl HIV-1 neutralization assay (Fig. 3A). HIV-1 isolates 92-HT-599, 92-UG-029, and BZ-167 were obtained from ARRRP and the UNAIDS Network for HIV-1 Isolation and Characterization.21 Cell-free virus supernatants were prepared in PHA blast cultures as described elsewhere.15 TZM-bl cells22 were obtained from the ARRRP. CXCR4 inhibitors were added to the cells (4 × 104 cells per well) for 1 hour at 37°C. Predetermined dilutions of the virus supernatants, representing 10–20 Tissue Culture Infective Doses (TCID50) in 15 μg/ml DEAE-dextran, were then added to the wells containing the cells plus inhibitor. Three days later, the cells were lysed and luciferase activity measured (Bright-Glo Luciferase assay, Promega) in relative luminescence units (RLU) measured at a sensitivity of 125 on a Bio-Tek Synergy plate reader. The percent inhibition was determined according to the formula:
Figure 3. Neutralization of HIV infectivity by naphthylguanide derivatives.
Neutralization of X4 isolates of HIV-1 was measured using TZM-bl cells. Compounds were added to cells, followed by a virus dilution representing 10–20 TCID50. Cells, drug, and virus were incubated for 72 h. The cells were lysed and chemiluminescence measured in relative luminescence units. Samples were run in duplicate. Percent inhibition was calculated and is displayed as mean and SEM. a. Isolate 92-HT-599 was tested against four naphthylguanide derivitives, spermine, and spermidine. b. Four different X4 isolates were tested against spermine tris-1-NapG.
Consistent with the results observed in live cell binding, spermine tris-1-NapG was the most effective, followed by spermine tris-2-NapG. We then tested spermine tris-1-NapG against a panel of four X4-tropic viruses (figure 3B), and showed that all were inhibited at submicromolar concentrations. The compounds were also tested against R5-tropic HIV-1 isolates and had no effect (data not shown).
One of the new compounds reported in this study, spermine tris-1-naphthylguanide, bound CXCR4 with an IC50 of 40 nM, a 5-fold higher affinity than the previously published phenylguanides. This is better than some other non-peptide analogs of T-140 that have been reported,23 though it does not approach the nanomolar affinity of T-140 itself. Spermine tris-1-naphthylguanide inhibited X4 HIV-1 infection by multiple X4 strains at 50–100 nM, at least a 30-fold lower concentration than observed previously with the phenylguanides.
The optimal structure for a non-peptide CXCR4 antagonist cannot be readily predicted from the crystal structures of CXCR4 in complex with a T-140 analog and a small molecule inhibitor2 because the binding cavity is very large and allows for many interactions with ligands. Attempts to develop small cyclic peptide and non-peptide analogs from T-14011, 23–24 have pointed to the importance of both a guanide functional group (in the form of an arginine sidechain) and a naphthyl group. This led us to model the docking of a series of guanide derivatives of polyamines to two of the published CXCR4 structures. Although the simulations allowed for many different binding conformations of these compounds, most of which are much smaller than T-140 and have plenty of room in the binding cavity, the results were in general agreement with the relative binding affinities observed in our T-140 competition assays. There were clear trends indicating that the spermine and spermidine naphthylguanide derivatives would have higher affinity than their phenylguanide counterparts, which had been the most effective inhibitors of HIV-1 infection in our previous study. When the naphthylguanides were synthesized and tested, their observed affinities were indeed generally higher than the phenylguanides, but the predicted binding energies among the various naphthylguanides did not correlate completely with their observed IC50s, pointing out the limitations of the docking models. The tetranaphthylguanide derivatives in particular had lower affinities than predicted; they tended to fold into conformations that stacked the aromatic rings in the simulations, which may have biased those results. The balance between charge and hydrophobic character appeared to be important in our previous study, and the tetranaphthyl derivatives may be too hydrophobic and interact nonspecifically with other sites, limiting their effectiveness. Alternatively, the bulky naphthyl groups may be preventing some important polar interactions between the receptor and the guanide or amine groups of the ligands which are not being adequately accounted for in the models.
We have shown that we can selectively derivatize the terminal primary amines of spermine and spermidine with naphthylguanide groups. These derivatives can be used to synthesize additional compounds with the secondary amines changed to guanide or phenylguanide groups. Additional structures can also be synthesized to create molecules with a mixture of naphthyl, phenyl, and guanide groups at different spacings that more closely mimic those in T-140 and/or are predicted to have higher binding energies in more sophisticated docking simulations than those reported here. Previous studies have also shown that spacing between the functional groups is important, and this can be explored systematically by using synthetic polyamine precursors other than spermine and spermidine to make these guanide compounds. Docking simulations with such constructs suggest that further improvements in affinity may be possible (data not shown).
The naphthylguanide compounds have significantly improved effectiveness compared to the phenylguanides in inhibiting infection of CXCR4 positive cells by X4 strains of HIV-1. Therefore, they represent more promising leads for developing non-peptide inhibitors of X4 HIV-1 infection. Their cytotoxicity is somewhat higher than the phenylguanides; however, reducing the overall hydrophobicity by adding a guanide or phenylguanide group to a spermine or spermidine mono- or bis-naphthylguanide may reverse this trend and make them promising candidates for future animal and clinical studies; their non-peptide structures may also give them greater serum stability than T140.
Acknowledgments
Funding Sources
This work was supported by Public Health Service grant AI076965 (ML) from the National Institute of Allergy and Infectious Diseases (NIAID), by the Louisiana Vaccine Center, and by the Research Institute for Children.
We thank the Research Computing Group in the Center for Computational Biology at MSU for computing resources to run the ‘flexible’ receptor Autodock runs.
Abbreviations
- CXCR4
CXC chemokine receptor 4
- SDF-1
stromal-derived-factor-1
- HIV-1
human immunodeficiency virus type 1
- NapG
naphthylguanide
- PheG
phenylguanide
- ARRRP
AIDS Research and Reference Reagent Program
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Royce A. Wilkinson, Email: roycew@montana.edu.
Seth H. Pincus, Email: spincus@chnola-research.org.
Kejing Song, Email: ksong@chnola-research.org.
Joyce B. Shepard, Email: brewer.joyce01@gmail.com.
Alan J. Weaver, Jr., Email: alan.weaver@msu.montan.edu.
Mohamed E. Labib, Email: labib@novaflux.com.
Martin Teintze, Email: mteintze@montana.edu.
References
- 1.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Nature. 2001;410:50. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 2.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Science. 2010;330:1066. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Antimicrob Agents Chemother. 2005;49:4721. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. J Exp Med. 1997;185:621. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner TA, Frenkel LM. Aids. 2008;22:2393. doi: 10.1097/QAD.Ob013e328312c72c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamamura H, Xu Y, Hattori T, Zhang X, Arakaki R, Kanbara K, Omagari A, Otaka A, Ibuka T, Yamamoto N, Nakashima H, Fujii N. Biochem Biophys Res Commun. 1998;253:877. doi: 10.1006/bbrc.1998.9871. [DOI] [PubMed] [Google Scholar]
- 7.Tamamura H, Sugioka M, Odagaki Y, Omagari A, Kan Y, Oishi S, Nakashima H, Yamamoto N, Peiper SC, Hamanaka N, Otaka A, Fujii N. Bioorg Med Chem Lett. 2001;11:359. doi: 10.1016/s0960-894x(00)00664-8. [DOI] [PubMed] [Google Scholar]
- 8.Tamamura H, Hiramatsu K, Kusano S, Terakubo S, Yamamoto N, Trent JO, Wang Z, Peiper SC, Nakashima H, Otaka A, Fujii N. Org Biomol Chem. 2003;1:3656. doi: 10.1039/b306473p. [DOI] [PubMed] [Google Scholar]
- 9.Tamamura H, Omagari A, Oishi S, Kanamoto T, Yamamoto N, Peiper SC, Nakashima H, Otaka A, Fujii N. Bioorg Med Chem Lett. 2000;10:2633. doi: 10.1016/s0960-894x(00)00535-7. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson RA, Pincus SH, Shepard JB, Walton SK, Bergin EP, Labib M, Teintze M. Antimicrob Agents Chemother. 2011;55:255. doi: 10.1128/AAC.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda S, Oishi S, Wang ZX, Araki T, Tamamura H, Cluzeau J, Ohno H, Kusano S, Nakashima H, Trent JO, Peiper SC, Fujii N. J Med Chem. 2007;50:192. doi: 10.1021/jm0607350. [DOI] [PubMed] [Google Scholar]
- 12.Trott O, Olson AJ. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair G, Vijayakumaran Journal of the Indian Chemical Society. 1963;40:953. [Google Scholar]
- 14.Frank RL, Smith PV. Organic Syntheses. 1955;3:753. [Google Scholar]
- 15.Krowicka H, Robinson J, Clark R, Hager S, Broyles S, Pincus S. AIDS Res Hum Retroviruses. 2008;24:957. doi: 10.1089/aid.2007.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, McKnight A, Thomas JF, Stoebenau-Haggarty B, Choe S, Vance PJ, Wells TN, Power CA, Sutterwala SS, Doms RW, Landau NR, Hoxie JA. Cell. 1996;87:745. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 17.McCallus DE, Ugen KE, Sato AI, Williams WV, Weiner DB. Viral Immunol. 1992;5:163. doi: 10.1089/vim.1992.5.163. [DOI] [PubMed] [Google Scholar]
- 18.Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, Pincus SH. J Immunol. 2004;172:6221. doi: 10.4049/jimmunol.172.10.6221. [DOI] [PubMed] [Google Scholar]
- 19.Carnec X, Quan L, Olson WC, Hazan U, Dragic T. J Virol. 2005;79:1930. doi: 10.1128/JVI.79.3.1930-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baribaud F, Edwards TG, Sharron M, Brelot A, Heveker N, Price K, Mortari F, Alizon M, Tsang M, Doms RW. J Virol. 2001;75:8957. doi: 10.1128/JVI.75.19.8957-8967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louwagie J, Delwart EL, Mullins JI, McCutchan FE, Eddy G, Burke DS. AIDS Res Hum Retroviruses. 1994;10:561. doi: 10.1089/aid.1994.10.561. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, Decker J, Liu H, Zhang Z, Arani R, Kilby J, Saag M, Wu X, Shaw G, Kappes J. Antimicrob Agents Chemother. 2002;46:1896. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda S, Kato M, Inuki S, Ohno H, Evans B, Wang ZX, Peiper SC, Izumi K, Kodama E, Matsuoka M, Nagasawa H, Oishi S, Fujii N. Bioorg Med Chem Lett. 2008;18:4124. doi: 10.1016/j.bmcl.2008.05.092. [DOI] [PubMed] [Google Scholar]
- 24.Fujii N, Oishi S, Hiramatsu K, Araki T, Ueda S, Tamamura H, Otaka A, Kusano S, Terakubo S, Nakashima H, Broach J, Trent J, Wang Z, Peiper S. Angew Chem Int Ed Engl. 2003;42:3251. doi: 10.1002/anie.200351024. [DOI] [PubMed] [Google Scholar]