Abstract
Human mesenchymal stem cell (hMSC) migration and recruitment play a critical role during bone fracture healing. Within the complex 3D in vivo microenvironment, hMSC migration is regulated through a myriad of extracellular cues. Here, we use a thiol-ene photopolymerized hydrogel to recapitulate structural and bioactive inputs in a tunable manner to understand their role in regulating 3D hMSC migration. Specifically, peptide-functionalized poly(ethylene glycol) hydrogels were used to encapsulate hMSC while varying the crosslinking density, 0.18 ± 0.02 - 1.60 ± 0.04 mM, and the adhesive ligand density, 0.001 to 1.0 mM. Using live cell videomicroscopy migratory cell paths were tracked and fit to a persistent random walk model. It was shown that hMSC migrating through the lowest crosslinking density and highest adhesivity had more sustained polarization, higher migrating speeds (17.6 ± 0.9 μm/hr), and higher cell spreading (Elliptical Form Factor = 3.9 ± 0.2). However, manipulation of these material properties did not significantly affect migration persistence. Further, there was a monotonic increase in cell speed and spreading with increasing adhesivity showing a lack of the biphasic trend seen in two dimensional cell migration. Immunohistochemistry showed well-formed actin fibers and β1 integrin staining at the ends of stress fibers. This thiol-ene platform provides a highly tunable substrate to characterize 3D hMSC migration with application as an implantable cell carrier platform or for the recruitment of endogenous hMSC in vivo.
1. Introduction
Bone marrow derived human mesenchymal stem cells (hMSC) are multipotent cells that are known to be recruited to bone fracture sites and play a significant role in bone regrowth [1, 2]. As a result, there is a growing interest within the orthopedic tissue engineering community to develop materials that could serve as carriers for hMSCs or to recruit endogenous hMSCs to promote the healing of bone fractures and defects [3, 4]. Minimally, the targeted materials must be engineered to permit or promote cell infiltration, so understanding the role of various matricellular signals in regulating migration of hMSC populations (e.g., their speed, persistence) is often essential [5]. In this regard, many synthetic materials have proven useful as tools to study cell motility in three-dimensions, as they can be fabricated to recapitulate desired aspects of extracellular matrix (ECM) properties in a highly tunable and reproducible manner [6, 7]. Here, we exploit one such system, peptide functionalized poly(ethylene glycol) (PEG) hydrogels formed via thiol-ene chemistry, as a versatile matrix to study hMSC migration ex vivo and in real time.
During development, wound healing, and other in vivo processes, cell migration is governed by a complex milieu of structural [8-10] and bioactive [11, 12] extracellular signals [5]. Pioneering work performed in two dimensions (2D) has provided the field with an understanding of how cell motility depends on these cell-matrix interactions such as substrate stiffness [13-15] and adhesivity [11, 15, 16]. However, more recent literature demonstrates that two-dimensional phenomena may not fully translate to three dimensions, for example Doyle et al. compared fibroblast migration on 2D fibronectin substrates with 3D cell derived matrix and saw significant differences in migration speed and leading edge protrusion rate when differing dimensionality [8]. 3D substrates can better recapitulate aspects of the in vivo microenvironment and allow for further understanding of cell migration as 3D does not un-naturally polarize cells and can be designed to require degradation of the matrix by cell-secreted enzymes, such as matrix metalloproteinases (MMP), for local spreading and migration. Natural materials, such as collagen [17] and matrigel [18], provide a heterogeneous, fibrillar platform to observe the role of extracellular cues in regulating cell migration and to discern valuable insight into 3D cell migration; however, there can be a large batch-to-batch variability, as well as complex relationships between adhesivity and mechanical properties that are difficult to deconvolute. Synthetic materials can reproduce many of these cues in a controlled manner in vitro and allow one to study systematically their effects on cell migration, but lack biological signals. Therefore, peptide-functionalized synthetic hydrogels can serve as an attractive option as extracellular matrix analogs, providing a platform with tailorable biophysical and biochemical cues to study 3D cell migration [7].
In work by Ehrbar et al., 8-arm poly (ethylene glycol) hydrogels were used to show that the speed of migration of pre-osteoblastic cells was reduced with increasing polymer density[19]. Further, mouse MSCs were observed to undergo collective migration when entrapped in a fibrous clot embedded in a hyaluronic acid gel; the cells also showed reduced migration speeds when surrounded by a matrix of increasing polymer density [20]. Beyond matrix density, another extracellular material property of importance is the mechanism by which the material degrades and its interplay with how a cell locally migrates in relation to local matrix remodeling. Early work by Raeber et al. used a 3D culture platform from 4-arm PEG-vinyl sulfone reacted with an MMP-degradable peptide to demonstrate that human foreskin fibroblasts relied on the secretion of proteases to spread and migrate in a density dependent manner [21]. These results may appear somewhat intuitive, as matrix density often necessitates degradation of the local microenvironment by cell secreted proteases [22], but the relationship can be complex as matrix density also influences the local adhesive ligand density and matrix mechanical properties. Each of these ECM properties can influence cellular functions, such as MMP activity [23, 24] and matrix deposition [25], which in turn can affect cell reorganization of its local matrix density.
Along with mechanical properties of a matrix, the adhesivity of the network plays a significant role in regulating cell spreading and migration. Many cell types simultaneously rely on adhesion to proteins within their extracellular space in addition to matrix degradation before they can migrate through a 3D material microenvironment. Using model surfaces and protein matrices, such as inclusion of the fibronectin peptide mimic RGDS, adhesive ligand density has been shown to play a crucial role in regulating migration of multiple cell types including, fibroblasts [26] and prostate cancer cells [18]. For example, in seminal work from Palecek et al, CHO cells cultured on 2D substrates coated with either fibronectin or fibrinogen had a biphasic response in their mean cell speed in relation to substrate adhesivity [11]. This trend was observed also when a fibroblast cell line, HT-1080s, was encapsulated in a three-dimensional MMP-degradable PEG gel [27]. While the speed of migration was dependent on the concentration of the RGDS epitope, the effect was much smaller in three-dimensions and potentially attributed to weaker matrix interactions of this cancer cell line. Early work, studied 3D MSC migration in a polarizing macroporous environment and implicated stiffness and ligand density as potent regulators of speed [9]. Finally, collectively migrating, mouse MSCs demonstrated a slight monotonic increase in distance traveled from a fibrous clot when the CRGDS concentration range was increased from 0.1 to 1.0 mM in a surrounding hyaluronic acid based gel [20].
Despite this foundational work in correlating cell migration to microenvironmental cues, there is a gap in knowledge as to how these chemical and physical cues regulate individual hMSC migration in 3D, especially over a broader range of material properties. To address this issue, we used photoinitiated thiol-ene reaction to create peptide functionalized PEG hydrogels of varying crosslinking density (0.18 ± 0.02 - 1.60 ± 0.04 mM) and ligand density (0.001 – 1.000 mM of CRGDS) [28]. Specifically, a 4-arm PEG macromolecule was end-capped with norbornene functionalities and crosslinked with cysteine-containing peptides (KCGPQG↓IWGQCK) through radical-mediated, step-growth photopolymerization to form highly swollen gels (97.3 – 98.4% water). The linker peptide sequence is susceptible to cleavage by matrix metalloproteinases (MMPs) 1,2,3,8, and 9 [29, 30], of which hMSC have been observed to secrete all of these MMPs [31-34], thus providing a substrate that hMSCs can locally degrade. Further, this thiol-ene reaction allows for covalently tethering pendant peptide sequences, such as CRGDS, to promote cell-matrix interactions. Further analysis of actin structure and β1 integrin spatial distribution is characterized using immunohistochemistry to better understand local cell-matrix interactions. Through manipulation of the macromer solution composition, microenvironments with tailored crosslinking density and adhesive ligand concentration were prepared for 3D hMSC culture, and morphology, speed, polarity, and persistence were monitored in real time as a function of systematic changes to the matrix properties. These results provide valuable insight into hMSC migration, including how individual hMSC interact and attach with their local microenvironment, and how these materials can be manipulated to control the migration phenotype and provide further insight into the role of matricellular signaling on hMSC migration. We believe that this quantitative characterization should prove useful for the development of scaffold design for hMSC, especially those developed as cell carriers and/or platforms for cell colonization and infiltration.
2. Materials and Methods
2.1 Macromer Synthesis
Norbornene acid was conjugated to 4-Arm PEG-hydroxyl (JenKem, USA) as previously described to create 4-arm PEG-Norbornene [28]. In brief, this occurs through the addition of norbornene acid to the PEG-OH by the symmetric anhydride N,N’-dicyclohexylcarbodiimide (DCC) coupling. Norbornene acid was dissolved in dichloromethane and reacted with DCC to couple the norbornenes. This was transferred anhydrously to a second reaction flask. The second reaction flask contained 20 kDa 4-arm PEG-OH dissolved in DCM, 4-(Dimethylamino)pyridine, and pyridine and was stirred overnight. The urea byproduct was filtered using a glass-fritted funnel. The filtrate was then washed with alternating glycine buffer and brine wash to remove un-reacted norbornenes. The final product was precipitated and washed with ice-cold diethyl ether. HNMR was used to characterize the purity and functionality (>95% for these studies) of the product (Supplemental Figure 1).
Synthesis of the peptide sequences, KCGPQG↓IWGQCK, CRGDS, and CRDGS, was completed on either a 433A solid phase peptide synthesizer (Applied Biosystems) or a Tribute peptide synthesizer (Protein Technologies) using standard solid-phase peptide reagents with Fluorenylmethoxycarbonyl (Fmoc) chemistry on a Rink-amide resin. Peptides were cleaved from the resin and purified using reverse-phase high-performance liquid chromatography (HPLC), and molecular weights were confirmed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry.
2.2 Cell Culture
Human mesenchymal stem cells (hMSCs) were isolated from bone marrow aspirates (Lonza). Bone marrow from a single donor was combined with the ammonium chloride solution (Stem Cell Technologies) vortexed and iced for ten minutes to lyse red blood cells. Cells were then washed twice and plated with growth media (low-glucose DMEM, 10% fetal bovine serum (FBS), 50 U mL−1 each penicillin/streptomycin, 1 μg mL−1 Fungizone antimycotic with 1 ng mL−1 recombinant human fibroblast growth factor-basic (FGF-2, Peprotech)). hMSCs were distinguished as the plastic adherent cells and were cultured at 37°C and 5% CO2. After 72 hours, non-adherent cells were washed away using growth media. Subsequently, media was changed every 2-3 days to remove non-adherent cells and the media was replenished. After cells reached ~70 to 80% confluency, they were trypsinized and frozen in a solution of 95% FBS and 5% dimethyl sulfoxide. For all experiments, hMSCs were thawed and passaged at ~70 to 80% confluence with media changes every 2-3 days. For all of these studies, hMSCs from passages two or three were used.
2.3 Gel Formulation and Characterization
The pre-polymerized solution consists of 6 wt% PEG-Norbornene (3 mM, f = 4), a specific off-stoichiometric thiol to -ene ratio 0.65, 0.725, and 0.85 (3.9 mM, 4.35 mM, and 5.1 mM respectively) amount of peptide crosslinker (KCGPQG↓IWGQCK, f = 2), a 1 mM ratio of CRGDS and CRDGS (f = 1), and 0.05% lithium phenyl-2,4,6-trimethylbenzoylphosphinate [35]. CRDGS is a non-bioactive scramble of CRGDS that allows for similar material properties between gels of varying concentration of bioactive CRGDS by keeping the pendant peptide concentration equivalent between all samples. For 3D cell studies, cells were suspended in phosphate buffered saline and added to the pre-polymer solution at a density of 2×105 cells/mL. The solutions were placed under 365 nm light at 10 mW/cm2 for 3 minutes. In situ rheometry demonstrated full gel conversion after 3 minutes (data not shown). These gels were then placed in experiment media (growth media without 1 ng mL−1 recombinant human fibroblast growth factor-basic (FGF-2, Peprotech)) for swelling overnight.
The swollen mass of the gels was measured and divided by the theoretical dry mass to calculate the swollen mass ratio (q). Parallel plate rheometry was also performed on an Ares 4400 rheometer (TA instruments) with 10% strain frequency sweep and a 10 rad s−1 strain sweep. Elastic modulus was determined when the gel was in the linear viscoelastic regime for both the frequency and strain sweep.
2.4 Real-time Cell Motility and Spreading Studies
Cell motility in cell-laden hydrogels was characterized using a Nikon TE2000-E microscope with a Nikon environmental chamber and an external heater (In vivo Scientific) and CO2 regulator (In vivo Scientific). Hydrogels were polymerized and swollen as described previously and then placed in a 24-well culture insert plate (BD Falcon, Fisher) and held in place by a transwell insert (Becton Dickinson) with the bottom removed by a 5 mm biopsy punch. Fresh experimental media was placed in the well at the beginning of each experiment. Real-time tracking was performed using Metamorph software for automated stage control, image collecting and positional cell tracking. After 30 hours in culture, cell centers tracking was commenced and followed for 7 hours. A Matlab (Mathworks) program was used to analyze the positional outputs and fit the data to a Persistent Random Walk model [36]:
| Eq.1 |
where < d2(t) > is the mean-square displacement (MSD) of the cell calculated using the overlap method, S is the speed of the cell which is calculated by the total distance covered over the tracking window divided by 7 hours, t is time, and P is persistence which is the characteristic time scale a cell migrates in a certain direction. All cell migration tracks were assumed to be isotropic and appropriate scaling of the MSD was applied to account for the z-directional migration. The individual cell’s mean free path, the characteristic length scale a cell migrates in a certain direction, can then be calculated by multiplying the speed by the persistence. Only cells that had a mean free path greater than 3 μm were considered migrating and used to calculate population averages. Metamorph was also used to measure cell spreading within the different gel systems. The Elliptical Form Factor (EFF) was calculated by taking the ratio of the major axis length to the perpendicular minor axis width. This ratio allows for a quantified comparison of cell spreading, where a cell with an EFF of 1 would be completely rounded and a cell with an EFF > 1 would be more spread.
MSD for each condition were calculated by averaging each cell’s individual MSD for a given condition. These were compared to ballistic migration (straight-line migration) and diffusion migration (random migration). These scale with time to the second power and first power, respectively. Turning probabilities were compiled by calculating the change in angle of each cell’s movement at each time point. These were then placed in a histogram with bin size of 10 degrees. The percent of forward movements, defined as a turning angle less than 40°, was calculated to more fully elucidate differences between the systems.
2.5 Immunostaining
Hydrogels with encapsulated cells were fabricated as previously described. Cells were cultured in these hydrogels for 48 hours, fixed using 4% PFA in PBS for 15 min, rinsed twice with PBS for 5 minutes, permeabilized with 1% Triton X-100 for 1 hour, rinsed twice with PBS for 10 minutes, and blocked using 5% BSA in PBS for 1 hour at room temperature and overnight at 4°C. All steps, unless noted otherwise, were performed at room temperature. Cells were then incubated with β1 primary antibody solution (Abcam, 1:20 dilution) with 3% BSA and 0.1% Tween-20 overnight at 4°C. The gels were washed three times with wash buffer, then incubated with goat anti-mouse 488 secondary antibody (Life Technologies, 1:200 dilution in 3% BSA and 0.1% Tween-20 in PBS overnight at 4°C. Gels were washed twice using 3% BSA and 0.1% Tween 20 for an hour at room temperature and then overnight at 4°C. Gels were then incubated with TRITC-Phalloidin (1:200 dilution) and DAPI (1:1000 dilution) for 1 hour at room temperature. Confocal imaging was performed on an LSM 710 (Zeiss).
2.6 Statistical Analysis
A one-way ANOVA with a Tukey’s test (alpha = 0.05) was performed on all data sets to determine statistical significance. For persistence their was extremely high variability for a few points, therefore all points outside of 3 standard deviations from the mean were considered outliers and not used for statistical measurements. For all conditions tested this removed only three cells from the calculations. All experiments were performed three times, and cell metrics were pooled from these different biological replicates for statistical calculations.
3. Results and Discussion
3.1 Peptide-Functionalized PEG Hydrogels: Controlling physical and chemical properties
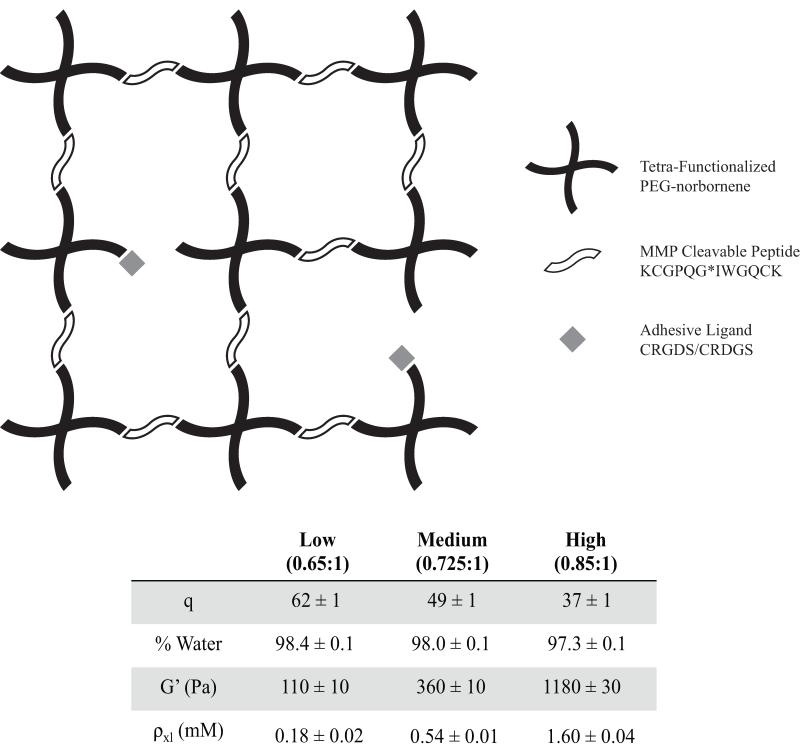
hMSCs were encapsulated within 4-arm PEG-norbornene hydrogels through photopolymerization with a di-thiol, MMP-degradable peptide (KCGPQG↓IWGQCK) linker and the introduction of an adhesive ligand mimic, CRGDS, and its non-bioactive scramble, CRDGS. To control the initial network crosslinking density, the di-thiol linker was added at three different stoichiometric ratios of 0.65, 0.725 and 0.85 thiol to -ene to render low, medium, and high crosslinking density gels, respectively. For organization and simplicity, the gel crosslinking density will be referred to as Low, Medium, or High for the duration of this discussion. The different crosslinking densities showed a decreasing trend in swelling ratio with increasing di-thiol linker concentration from Low to High of 62±1 (98.4% water), 49±1 (98.0% water), and 37±1 (97.3% water), (Figure 1). In general, the higher the gel crosslinking density, which is achieved by increasing the peptide crosslinker content, the denser the polymer network that cells must locally degrade in order to spread and migrate. Note that the network mesh size is orders of magnitude smaller (nanometer size-scale) than the size of a cell (micrometer size-scale). The elastic modulus of gels is directly proportional to the network crosslinking density, and the Low, Medium and High gels demonstrated an increase in elastic modulus with increasing di-thiol concentration of 110 ±10 Pa, 360 ±10 Pa, and 1180 ± 30 Pa, respectively (Figure 1). Over large ranges of elasticity, material stiffness has been shown to affect hMSC attachment, spreading, and differentiation on 2D substrates [37]; however, less is known about how substrate elasticity influences hMSC morphology, phenotype, and migration in 3D.
Figure 1.
Thiol-ene photopolymerization of a tetra-functionalized PEG-norbornene, MMP cleavable peptide (KCGPQG↓IWGQCK), and adhesive ligands (CRGDS, CRDGS) occurs through a radically mediated step growth reaction to encapsulate hMSC within a 3D tunable microenvironment. The table presents mass swelling ratio (q), % water, shear modulus (G’), and crosslinking density (ρxl, calculated from Rubber Elasticity Theory) while the thiol:ene ratio was varied to produce Low, Medium, and High gels. As thiol:ene ratio increases, mass swelling decreases, % water slightly decreases, and shear modulus and crosslinking density increase.
By applying rubber elasticity theory and the experimentally measured equilibrium swelling ratio and elastic modulus, a crosslinking density was calculated for each of the three systems and found to be 0.18 ± 0.02 mM, 0.54 ± 0.01 mM, and 1.60 ± 0.04 mM with increasing di-thiol concentration [38]. These calculated values varied from what is expected theoretically for ideal networks that are free of loops and entanglements and completely reacted at these stoichiometric levels. The predicted crosslinking density for ideal networks is 0.43 mM for the Low, 0.74 mM for the Medium, and 1.44 mM for the High. This implies that there are non-idealities present in the system for the lower two gel systems, which results in a larger mesh size than expected and potential gel defects, such as dangling ends or loops. Interestingly, at the highest stoichiometric ratio/crosslinking density, the value calculated based on experimental measurements of the gel properties was higher than that predicted, possibly indicating the role of entanglements acting as effective crosslinks. Regardless, these defects are still orders of magnitude smaller than a cell body and do not interfere with the degradability of the gel. On a relative basis, these networks are much more ideal than chain polymerized PEG gels that are widely used for 3D cell culture [39-41]. Despite the non-idealities and deviation from theoretical predictions, the characterization of these gels indicates that one can systematically manipulate the polymer density and elasticity of the networks by simple variations in the amount of di-thiol peptide in the pre-polymer solution.
3.2 Effect of Crosslinking Density on Cell Spreading and Morphology
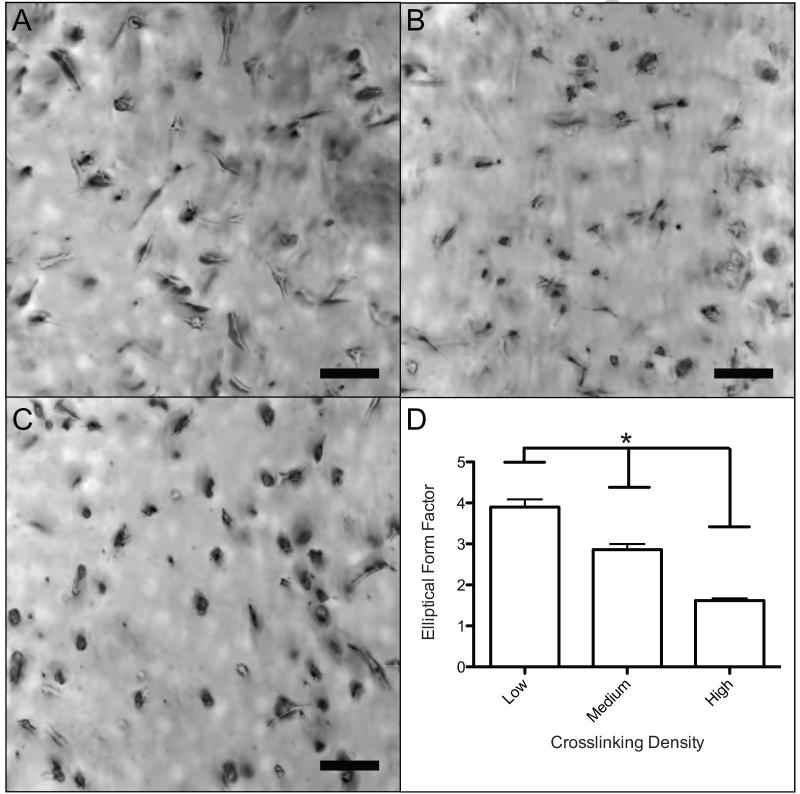
To understand the effect of crosslinking density on hMSC morphology and spreading in three-dimensions, hMSC were photoencapsulated in PEG-norbornene hydrogels at Low, Medium, and High crosslinking densities with 1 mM CRGDS to promote cell-matrix interactions. At early time points (30 hours), the high crosslinking density in the gels limits cell spreading, as observed by a decrease in the elliptical form factor (EFF) with increasing crosslinking density (Figure 2). Specifically, the EFF decreased with increasing crosslinking density from 3.9 ± 0.2 (Low) to 2.9 ± 0.1 (Medium) to 1.6 ± 0.1 (High) (Figure 2D). For comparison, fibroblasts within collagen, fibrin and cell derived matrix were reported to have an EFF of ~4, which correlates well with the Low crosslinked gel system.27 In loosely crosslinked gels, hMSCs appeared more uniaxial and highly elongated (Figure 2A); however, in Medium crosslinked gels, cells were less spread and only smaller protrusions were observed (Figure 2B). Cells encapsulated in the High crosslinked gels were mostly rounded with few if any protrusions (Fig 2C). These data indicate that although the polymer density is relatively low in all of the systems (1.6 to 2.7 weight % polymer, Figure 1), an increase in the number of crosslinks that a cell must degrade impedes cell spreading.
Figure 2.
Morphology of encapsulated hMSCs cultured for 30 hours in gel systems with varying crosslinking density. A) Brightfield image (10× magnification) of hMSCs in a Low (65%) gel. Cells were found to be well spread and linear. B) Brightfield image (10× magnification) of hMSCs in a Medium (72.5%) gel. Cells were found spread with many protrusions. C) Brightfield image (10× magnification) of hMSCs in a High (85%) gel. Cells were rounded with little or no protrusions. D) An elliptical form factor was calculated by dividing the length and perpendicular width of each cell. There is a significant (*p < 0.05) decreasing trend found from a one-way ANOVA and Tukey’s Test. Scale bar represents 100 μm.
3.3 Effect of Crosslinking Density on hMSC Motility
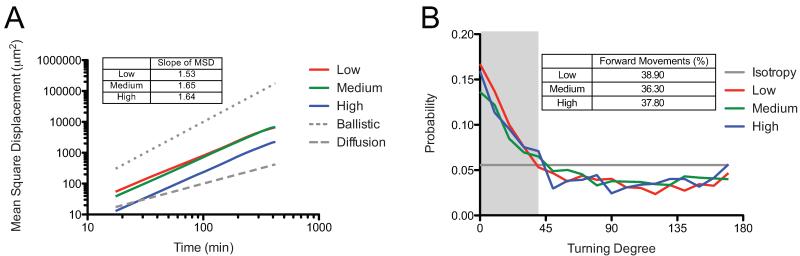
Along with cell morphology, single cell migration was analyzed using live-cell videomicroscopy in each of the hydrogels. Cell tracks were followed for a continuous 7 hours. Tracking was commenced after the cells had been cultured for 30 hours post-encapsulation in the gels; this time point was selected to allow cells to recover from trypsinization and encapsulation, as well as to allow for some local matrix remodeling before monitoring migration. Isotropic motility was observed over the range of crosslinking densities studied from 0.18 ± 0.02 mM - 1.60 ± 0.04 mM, as hypothesized based on the lack of polarizing cues within the material or media. Mean square displacements (MSD) were calculated from the cell tracks for each cell and were averaged over all migrating cells for an aggregate MSD for each gel composition. Comparisons are shown for ballistic migration, which scales to the second power with time, and diffusive migration, which scales to the first power with time. Ballistic migration is a cell that migrates persistently in a straight line, whereas diffusive migration describes a cell moving in a completely random path. All three slopes fall in-between a completely persistent and completely random migration, prompting further analyses of the persistence of the cells.
During cell migration, motile cells polarize to direct their migration spatially, and this process plays a key role in how persistent a cell migrates. The degree of cell polarization can be quantified indirectly within our system by calculating the probability of a cell to migrate in a specific direction relative to the previous direction it migrated. Using this type of analysis, polarized cells sustain their polarity in a fixed direction while randomly migrating cells have unbiased isotropic turning probabilities. Therefore, a cell that has a sustained polarization in a certain direction during migration will have a high probability of very low turning angles. Conversely, a cell that has a highly transient polarization will have an equal probability to turn in any direction. All three systems demonstrated a similar bias toward polarized cells with migration continuing in a certain direction (Figure 3B). To quantify this further, a critical angle of 40° was defined to calculate the percent of turns where a cell continued in a similar direction to their previous movement and therefore stayed polarized (Grey area on Figure 3B). The percent of turning angles that were 40° or less were 38.9%, 36.3%, and 37.8% with increasing crosslinking density. These values are quite similar and argue that varying the crosslinking density does not manipulate a cell’s ability to sustain polarization while migrating in a 3D network. This result is a bit counterintuitive in that it would seem that in increasing the network density, a cell would need to degrade more of the local network to continue in a sustained direction and therefore would be less likely to stay polarized. However, the observed independence of polarization on crosslinking density may instead be correlated to the fact that speed is affected by network density, but hMSC remain polarized for equivalent time frames. Given that there are no external chemotactic factors, such as growth factors or cytokines, to induce polarization, it is possible that the sustained polarization is a result of local upregulation of secreted proteolytic factors at a particular location on the cell, but this is difficult to measure [42]. Greater insight into local quantification of the network’s stiffness and crosslinking density (i.e., how hMSCs have remodeled and altered the local material properties) would help better elucidate these effects on cell migration.
Figure 3.
hMSC migration was followed over 7 hours using live cell videomicroscopy. Effect of varying crosslinking density on hMSC mean square displacement and sustained cell polarity was calculated from the cell tracks measured using Metamorph. (A) Mean-square displacement was similar for Low and Medium gels, but differed for cells in High crosslinked gels. All three gels had slopes that fell between 1 (random migration) and 2 (ballistic migration) (B) Sustained cell polarity showed a similar bias for all three systems, and a similar percentage of sustained steps.
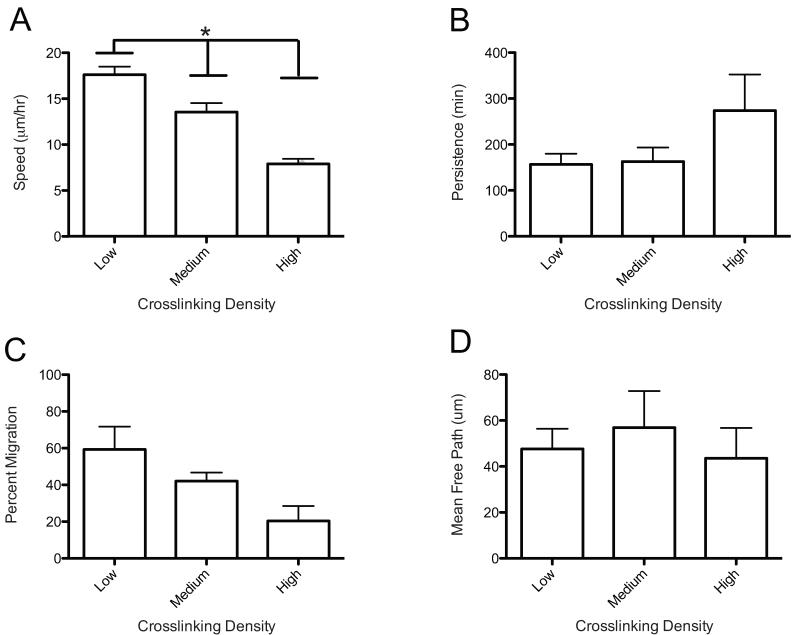
Complementary to the above analyses, key metrics from this data were obtained by fitting the MSD data to a Persistent Random Walk model. This approach allows for the calculation of the speed, persistence, and mean free path of migrating hMSCs as a function of gel properties [36]. hMSC speed correlated similarly with morphology, as average speed decreased from 17.6 ± 0.9 μm/hr to 13.6 ± 1.0 μm/hr to 7.9 ± 0.5 μm/hr as the network density decreased (Figure 4A). The percentage of cells migrating followed the same trend, as migrating cells decreased from 59%, 42%, and 20% with increasing crosslinking density (Figure 4C). Cell survival has been previously reported to be very high (>95%) in similar gel systems for up to two weeks [28, 43], so the non-migrating population was likely not due to loss of cell viability. Collectively, these data corroborate the hypothesis that denser networks inhibit hMSC migration, as cells must locally degrade more crosslinks in their pericellular space in order to spread and migrate. This results in slowing both the speed at which hMSCs can migrate and the overall number of cells that can overcome the physical barrier to migrate. Interestingly, cell persistence over the range of crosslinking densities studied did not show any statistically significant differences (Figure 4B). This correlates well with the cell polarity data (Figure 3B). The mean free path of the cell’s motility was also equivalent for the three crosslinking densities (Figure 4D). This lack of statistical significance found in cell motility might also relate to the higher amount of heterogeneity of persistences observed for each cell population; hMSCs are primary cells and there is typically heterogeneity in the differentiation potential of the populations as well [1]. Interestingly, despite slower speeds in more highly crosslinked networks, it was still observed that cells moved on a similar characteristic time and length scales over this range of crosslinking density.
Figure 4.
Cell tracks from hMSC migration in 3D are modeled using a Persistent Random Walk model (PRW). Effect of varying crosslinking density on hMSC migration. (A) Cell speed shows a decreasing trend with increasing crosslink density. (*Significance P< 0.05) (B) Persistence shows no statistical difference over the varying crosslink densities. (C) Percent migration shows a decreasing although not statistically significant difference. (D) Mean free path was not affected by varying the crosslinking density.
3.4 Actin Cytoskeleton and β1 Integrin Organization and Structure
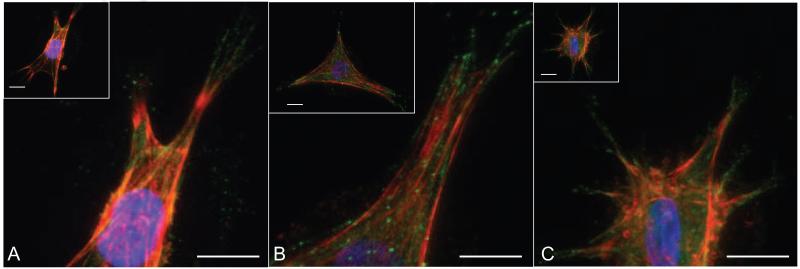
Cells spread and migrate within 3D networks through adhesion to the matrix via integrin binding. To further understand how hMSCs interact with pendant adhesion functionalities (i.e., R3GDS) when encapsulated in thiol-ene hydrogels, immunohistochemistry was used to visualize actin cytoskeleton organization and β1 integrin distribution for cells entrapped in gels of varying crosslink densities. As mentioned previously (Figure 2), there was a distribution of cell spreading and morphologies within the three different systems; and Figure 5 provides more detailed characterization of the cell cytoskeleton. In brief, hMSCs had numerous actin stress fibers in all gels, regardless of crosslinking density.
Figure 5.
Encapsulated hMSCs were cultured in gels at the three different crosslinking densities for 48 hours and immunostained for actin (red), β1 integrin (green) and DAPI (blue). A) Low Crosslinking B) Medium Crosslinking C) High Crosslinking. Spread hMSC in all three gels systems show actin fiber formation and punctate β1 integrin at the ends of these fibers over this range of crosslinking densities. Scale bars represent 50 μm.
Immunostaining for β1 integrin was performed to better understand the effect of crosslinking density on the ability of hMSCs to adhere and interact with matricellular cues in the gels. β1 integrin plays a prominent role in cell motility and is one of the primary integrins that binds to the adhesive RGD motif [44]. β1 integrin was localized to punctate foci at the end of actin stress fibers for all three crosslinking densities, illustrating strong cell adhesion to the network (Figure 5). The actin stress fibers and β1 integrins appear similar to what has been reported for fibroblasts in 3D environments composed of cell derived matrix, collagen and fibrin [45]. Therefore, although the thiol-ene gels are synthetic matrices with a smaller mesh size, they appear to appropriately recapitulate some of the critical factors present in natural materials to promote cell attachment and spreading in these 3D environments. Furthermore, the observance of similar integrin staining in 3D for all of the three gel systems prompted us to pursue a more in-depth study into varying the adhesivity of the network and its effect on cell migration.
3.5 Effect of RGD Density on Cell Morphology
Along with substrate stiffness and crosslinking density, substrate adhesivity has been shown to affect MSC migration in 2D and collective MSC migration in 3D. Wu et al. showed that in 2D the concentration of fibronectin, vitronectin and collagen coated on glass slides affected MSC speed [46]. Also, Lei et al. reported that collective migration of mouse MSCs in 3D hyaluronan gels showed slight differences in overall distance migrated from a fibrin clot while varying the adhesivity of the network [20]. Here, hMSCs were encapsulated in gels containing varying CRGDS concentrations to study how ligand concentration alters single cell morphology and migration in an isotropic network. While this work provides one of the first characterizations of MSC migration in 3D, experiments with natural materials are confounded by the fact that material density and adhesivity are fully coupled in natural gels. In the thiol-ene gel system, these parameters can be decoupled, enabling the systematic investigation of ligand density on cell migration for each gel composition. Here, a non-bioactive scramble of CRGDS was introduced to keep the total pendant peptide concentration equivalent while varying the concentration of the bioactive CRGDS so that the crosslinking density and resulting gel elasticity remained similar for the various systems. Swelling studies were also performed on all gels to ensure similar water contents.
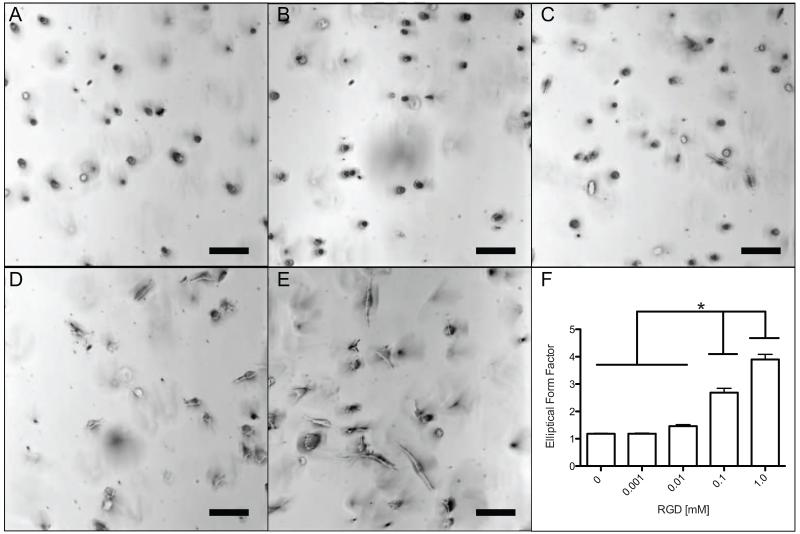
The effect of changing CRGDS ligand density on hMSC morphology, migration, and cellular interaction was observed in the Low crosslinked gels, as these gels facilitated the highest degree of spreading and migration. This selection allowed for easier elucidation of differences in outputs when ligand density was varied. Brightfield images and elliptical form factor calculations of the cells show significant increases in cell spreading with increasing CRGDS after 30 hours (Figure 6). At 0 mM and 0.001 mM CRGDS concentrations, hMSCs were rounded with no visible protrusions (Figure 6A, 6B). An increase to 0.01 mM CRGDS showed cells that still were quite rounded but had protrusions emerging from the rounded cells (Figure 6C). At 0.1 mM and 1.0 mM, the large majority of cells were highly spread, elongated, and uniaxial (Figure 6D, 6E). These changes in cell shape are also reflected through quantification of EFF values with significant increases between 0.01 mM to 0.1 mM and 0.1 mM to 1.0 mM (Figure 6F). In general, below a certain threshold at low CRGDS concentration, cells are rounded, presumably because of limited adhesions to the matrix, but as the CRGDS concentration is increased to 0.1 mM and beyond, cell attachment appears to increase as observed through dramatic changes in morphology and spreading.
Figure 6.
Morphology of encapsulated hMSCs cultured for 30 hours in gel systems with varying CRGDS. A) Brightfield image (10× magnification) of hMSCs in 0 mM CRGDS gel. Cells were rounded. B) Brightfield image (10× magnification) of hMSCs in 0.001 mM CRGDS gel. Cells were rounded. C) Brightfield image (10× magnification) of hMSCs in 0.01 mM gels. Cells were rounded with little or no protrusions. D) Brightfield image (10× magnification) of hMSCs in 0.1 mM CRGDS gel. Cells were spread and linear. E) Brightfield image (10× magnification) of hMSCs in 1.0 mM CRGDS gel. Cells were more spread and linear. F) An elliptical form factor was calculated by dividing the length and perpendicular width of each cell. There is a significant (*p < 0.05) increasing trend found from a one-way ANOVA and Tukey’s Test between the first three systems with the 0.1 mM and 1.0 mM systems. Scale bar represents 100 μm.
3.6 Actin Cytoskeleton and β1 Integrin Organization and Structure
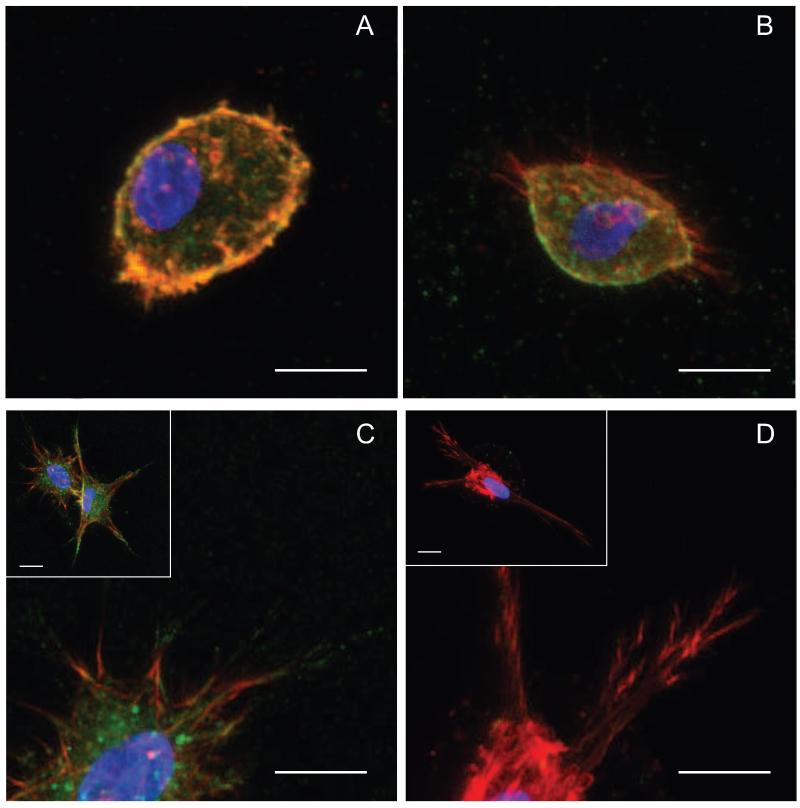
To more fully characterize and understand the implications of these changes in hMSC morphology, immunohistochemistry was performed to study actin cytoskeletal formation and β1 integrin distribution as indicators of cell-material interactions. Decreasing RGD concentration resulted in a notable decrease in actin stress fiber formation and β1 integrin staining at the end of actin stress fibers (Figure 7). For cells within 0 mM and 0.001 mM, no spread cells were observed in the representative images, and cells were primarily rounded with only diffuse actin staining and little to no β1 integrin staining (Figure 7A). Within 0.01 mM gels, hMSCs showed small protrusions, but little if any β1 staining at the end of the small actin fibers with much of it found on the rounded cell edges (Figure 7B). Increasing to 0.1 and 1.0 mM, spread cells have well-formed actin fibers with punctate β1 staining at the end of fibers significantly present for the 1.0 mM gels (Figure 7C, 7D). This correlates well with the EFF data, and further argues that over this large range of CRGDS that lower concentrations of CRGDS lead to low levels of cell interactions with the matrix resulting in a less-developed actin cytoskeleton, whereas higher concentrations promoted high cellular interactions and development of numerous actin stress fibers. With these significant changes in cell morphology and interactions as a function of matrix composition, we hypothesized that significant variations may exist in hMSC migration over this range of adhesive ligand density.
Figure 7.
Encapsulated hMSCs were cultured in gels with varying CRGDS concentration for 48 hours and immunostained for actin (red), β1 integrin (green) and DAPI (blue). A) For 0.001 mM gels, cells remain rounded and show little if any β1 integrin even on the rounded edges of the cell B) For 0.01 mM gels, actin protrusions are present although limited with few β1 integrin staining on actin fibers C) For 0.1 mM gels, cells are spread with large, protrusive actin fiber formation with β1 integrin staining on protrusions. D) For 1.0 mM gels, cells are similar to the 0.1 mM cells with protrusive actin fibers however, the β1 integrin staining seems more localized to the ends of actin fibers. Scale bar represents 50 μm.
3.7 Effect of RGD on hMSC Motility
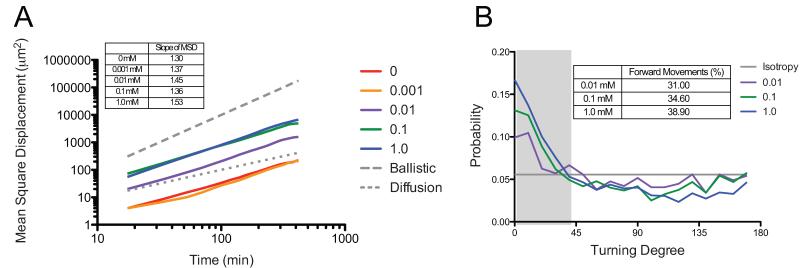
hMSC migration was followed through live-cell videomicroscopy over the same range of CRGDS densities for 7 hours. Again, cell tracks were monitored and MSD was calculated using the overlapping method [36]. In contrast to the results with crosslinking density, we observed an increasing trend in the MSD slopes with increasing CRGDS concentration, except for the interesting decrease in slope for cells encapsulated within the 0.1 mM CRGDS functionalized gels (Figure 8A). All slopes similarly fall in-between the ballistic and diffusive slopes prompting further quantification and study of the persistence of individual cell migration.
Figure 8.
hMSC migration was followed over 7 hours using live cell videomicroscopy. The effect of varying CRGDS concentration on hMSC mean square displacement and sustained cell polarity was calculated from the cell tracks measured using Metamorph. A) Mean-square displacement was similar for 1.0 mM and 0.1 mM gels, and 0 mM and 0.001 mM gels were also similar. All five gels had slopes that fell between 1 (random migration) and 2 (ballistic migration) B) Sustained cell polarity showed a similar bias for all three systems, and a similar percentage of sustained steps.
The directional turning probabilities for the three highest CRGDS concentrations are presented in Figure 8B. The hMSCs encapsulated in gel formulations with CRGDS concentrations between 0 mM and 0.001 mM were not plotted in this figure due to too few migrating cells (n=6, n=5, respectively), as well as the fact that hMSCs in these gels showed very low cell speeds (~5 μm/hr). Speeds of ~5 μm/hr correlate with cells that are essentially not moving but have non-zero speeds due to error from inadequately tracking the exact center of a cell. The plot shows an increasing trend in sustained cell polarity with increasing CRGDS concentration. Again, looking at the percent of forward movements (turning angle < 40°), results illustrate a decrease in polarization with decreasing CRGDS content from 38.9% (1 mM) to 34.6% (0.1 mM) to 31.0% (0.01 mM). These results suggest that hMSCs are more polarized with increasing CRGDS concentration, presumably related to increased cell-matrix interactions, which should in turn promote higher cell persistence.
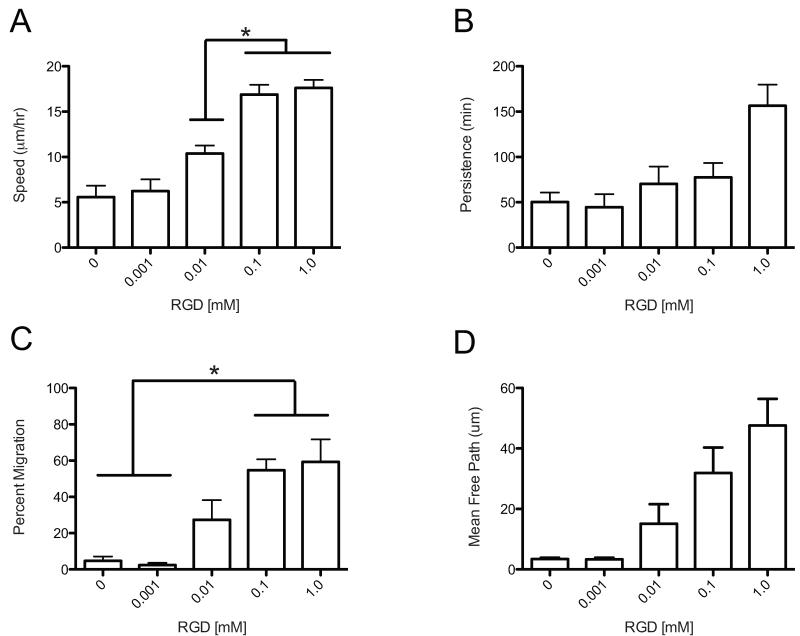
The MSD for each cell was then fit using a non-linear least-squares regression to the Persistent Random Walk model to gain insight into cell speed, persistence and mean free path while varying CRGDS concentration. Trends in cell speed indicated increasing speed with increasing CRGDS concentration at lower concentrations with a plateau at 16.9 ± 1.1 μm/hr and 17.6 ± 0.9 μm/hr (0.1 mM and 1.0, respectively) (Figure 9a). It is speculated that this occurs through the stochastic ability of a cell to garner significant attachment to the network, allowing actin cytoskeletal tension to produce migration. Put differently, when the network is more adhesive, a cell can likely attach and spread more easily in order to provide the necessary tension to perform persistent migration. The classic biphasic trend was not observed over this range of ligand densities and is more similar to the monotonic increase observed in the distance migrated for collective mouse MSC migration in 3D [20]. Not surprisingly, these speeds are less than half the speed of hMSCs migrating in 2D found by Wu et al., as cells in 2D do not need to degrade a matrix as part of their motility mechanism [47]. Requiring cells to locally degrade their microenvironment, which is typical of many in vivo tissues, provides a significant barrier to their migration speed in comparison to cell migration on 2D surfaces.
Figure 9.
Cell tracks from hMSC migration in 3D are modeled using a Persistent Random Walk model (PRW). Effect of varying CRGDS concentration on hMSC migration was studied. A) Cell speed shows an increasing trend with increasing CRGDS concentration.(*Significance P< 0.05) B) Persistence shows no statistical difference over the varying CRGDS C) Percent migration shows an increasing trend with increasing CRGDS concentration that plateaus for 0.1 mM and 1.0 mM gels. (*Significance P< 0.05) D) Mean free path was not affected by varying the CRGDS concentration.
The percentage of cells migrating also increased over this range with a plateau observed in the number of migration cells, slightly less than 60% for cells in gels with 0.1 and 1.0 mM CRGDS (Figure 9C). Very few cells migrated within the 0 mM and 0.001 mM, which correlates well with the spreading data in Figure 6 (n = 6, n=5, respectively). Therefore, it is believed that the concentration of 0.001 mM CRGDS is below a threshold where cells interact with and form significant attachments to the network to allow spreading, and therefore, lose their ability to migrate as well. Persistence and mean free path were not found to be statistically different over the range of CRGDS concentrations studied (Figure 9B, 9D). The large variability in persistence is partially explained by the heterogeneity in the cell population. The 3D values of persistence found for cells in this study are roughly twice as large as values observed in 2D for hMSCs, and a similarly high variability was observed in both platforms [46]. Finally, it should be noted that the 2D studies used full proteins instead of peptide mimics, which certainly affects the strength of integrin interactions, so more in-depth comparisons of these numbers are potentially confounded.
Thiol-ene networks allow for facile synthesis of peptide functionalized PEG hydrogels with readily manipulated material properties. Since the chemistry is cytocompatible, it provides for a versatile system to study and observe cell migration, and here we report on hMSC morphology and migration speed as a function of matricellular properties [28]. We believe that such fundamental knowledge will prove helpful in the engineering of materials to serve as cell carriers or for the colonization of endogenous hMSCs into a bone defect site. Interestingly, the structural and bioactive inputs tested here do not manipulate the time and length scales over which hMSCs migrate in a persistent manner. However, these results are for hMSCs isolated from a single donor, and it would be interesting to examine potential differences as a function of age and/or gender [48]. This motivates future work to design more complex systems, such as gradient presentation of soluble growth factors or specific tethered cues to promote recruitment and persistent anisotropic hMSC migration. Further, thiol-ene hydrogels provide an interesting platform as they are photochemically controlled, which can enable spatial and temporal control of mechanical properties and bioactive presentation through photopatterning [49]. This might allow for further study of physical and biological complexity on different time scales to be introduced within the network, and it would be interesting to use such an approach to manipulate cellular microenvironment to elicit desired migration responses.
4. Conclusion
Thiol-ene photopolymerized hydrogels allow for the facile introduction of important structural and bioactive inputs that govern hMSC migration within the in vivo microenvironment. In this work, the effects of matrix crosslinking density and ligand density on hMSC migration were studied to provide insight for strategies to better engineer materials for hMSC carrier platforms and materials to recruit endogenous hMSC to fracture sites. It was shown that hydrogels with low crosslinking density (0.18 ± 0.02 mM) and high adhesivity (1 mM CRGDS) permitted higher cell speeds (17.6 ± 0.9 μm/hr), higher cell spreading (EFF = 3.9 ± 0.2), and more sustained polarization. Also, there was a monotonic increase in cell speed relative to an increase in ligand density, lacking the biphasic trend seen in 2D. Further, it was shown that manipulating crosslinking density and ligand density did not significantly affect persistence. Thiol-ene photopolymerized hydrogels provide a valuable tool to study hMSC migration in 3D environments, and this knowledge should provide valuable insight into future engineered materials.
Supplementary Material
Supplemental Figure 1. hNMR spectra of PEG-Norbornene. Functionalization was calculated by dividing the norbornene alkene peaks by the PEG alkyl peaks.
Acknowledgments
The authors would like to thank Dr. Mark W. Tibbitt and Jared Young for valuable discussion and input on manuscript preparation. Funding for these studies was provided in part by grants from the National Institutes of Health (R01DE016523) and National Science Foundation (CBET 1236662).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nature Reviews Immunology. 2008;8:726–36. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- [2].Salem HK, Thiemermann C. Mesenchymal Stromal Cells: Current Understanding and Clinical Status. Stem Cells. 2010;28:585–96. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Discher DE, Mooney DJ, Zandstra PW. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science. 2009;324:1673–7. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen F-M, Wu L-A, Zhang M, Zhang R, Sun H-H. Homing of endogenous stem/progenitor cells for in situ tissue regeneration: Promises, strategies, and translational perspectives. Biomaterials. 2011;32:3189–209. doi: 10.1016/j.biomaterials.2010.12.032. [DOI] [PubMed] [Google Scholar]
- [5].Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–9. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–41. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kim H-D, Peyton SR. Bio-inspired materials for parsing matrix physicochemical control of cell migration: A Review. Integrative Biology. 2012;4:37–52. doi: 10.1039/c1ib00069a. [DOI] [PubMed] [Google Scholar]
- [8].Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-imensional fibrillar cell migration. J Cell Biol. 2009;184:481–90. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA, et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnology and Bioengineering. 2011;108:1181–93. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- [11].Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–40. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- [12].Purcell BP, Elser JA, Mu A, Margulies KB, Burdick JA. Synergistic effects of SDF-1α chemokine and hyaluronic acid release from degradable hydrogels on directing bone marrow derived cell homing to the myocardium. Biomaterials. 2012;33:7849–57. doi: 10.1016/j.biomaterials.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pelham RJ, Jr., Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lo C-M, Wang H-B, Dembo M, Wang Y-l. Cell Movement Is Guided by the Rigidity of the Substrate. Biophysical Journal. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophysical Journal. 2004;86:617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- [17].Grinnell F, Petroll WM. Cell Motility and Mechanics in Three-Dimensional Collagen Matrices. Annual Review of Cell and Developmental Biology. 2010;26:335–61. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- [18].Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. PNAS. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, et al. Elucidating the Role of Matrix Stiffness in 3D Cell Migration and Remodeling. Biophysical Journal. 2011;100:284–93. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lei Y, Gojgini S, Lam J, Segura T. The spreading, migration and proliferation of mouse mesenchymal stem cells cultured inside hyaluronic acid hydrogels. Biomaterials. 2011;32:39–47. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Raeber GP, Lutolf MP, Hubbell JA. Molecularly Engineered PEG Hydrogels: A Novel Model System for Proteolytically Mediated Cell Migration. Biophysical Journal. 2005;89:1374–88. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Raeber GP, Lutolf MP, Hubbell JA. Mechanisms of 3-D migration and matrix remodeling of fibroblasts within artificial ECMs. Acta Biomaterialia. 2007;3:615–29. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- [23].Zhang YH, Zhao CQ, Jiang LS, Dai LY. Substrate stiffness regulates apoptosis and the mRNA expression of extracellular matrix regulatory genes in the rat annular cells. Matrix Biology. 2011;30:135–44. doi: 10.1016/j.matbio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- [24].Hanjaya-Putra D, Yee J, Ceci D, Truitt R, Yee D, Gerecht S. Vascular endothelial growth factor and substrate mechanics regulate in vitro tubulogenesis of endothelial progenitor cells. Journal of Cellular and Molecular Medicine. 2010;14:2436–47. doi: 10.1111/j.1582-4934.2009.00981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gould ST, Darling NJ, Anseth KS. Small peptide functionalized thiol-ene hydrogels as culture substrates for understanding valvular interstitial cell activation and de novo tissue deposition. Acta Biomaterialia. 2012;8:3201–9. doi: 10.1016/j.actbio.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gaudet C, Marganski WA, Kim S, Brown CT, Gunderia V, Dembo M, et al. Influence of type I collagen surface density on fibroblast spreading, motility, and contractility. Biophysical Journal. 2003;85:3329–35. doi: 10.1016/S0006-3495(03)74752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schwartz MP, Fairbanks BD, Rogers RE, Rangarajan R, Zaman MH, Anseth KS. A synthetic strategy for mimicking the extracellular matrix provides new insight about tumor cell migration. Integrative Biology. 2010;2:32–40. doi: 10.1039/b912438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Advanced Materials. 2009;21:5005–10. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- [30].Patterson J, Hubbell JA. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 2010;31:7836–45. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- [31].Ho IAW, Chan KYW, Ng W-H, Guo CM, Hui KM, Cheang P, et al. Matrix metalloproteinase 1 is necessary for the migration of human bone marrow-derived mesenchymal stem cells toward human glioma. Stem Cells. 2009;27:1366–75. doi: 10.1002/stem.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Halfon S, Abramov N, Grinblat B, Ginis I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011;20:53–66. doi: 10.1089/scd.2010.0040. [DOI] [PubMed] [Google Scholar]
- [33].Parikka V, Väänänen A, Risteli J, Salo T, Sorsa T, Väänänen HK, et al. Human mesenchymal stem cell derived osteoblasts degrade organic bone matrix in vitro by matrix metalloproteinases. Matrix Biol. 2005;24:438–47. doi: 10.1016/j.matbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- [34].Djouad F, Delorme B, Maurice M, Bony C, Apparailly F, Louis-Plence P, et al. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res Ther. 2007:9. doi: 10.1186/ar2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–7. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dickinson RB, Tranquillo RT. Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE Journal. 1993;39:1995–2010. [Google Scholar]
- [37].Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- [38].Rubinstein M, Colby RH. Polymer Physics. 9ed. Oxford University Press Inc.; New York: 2003. [Google Scholar]
- [39].Sakai T, Matsunaga T, Yamamoto Y, Ito C, Yoshida R, Suzuki S, et al. Design and fabrication of a high-strength hydrogel with ideally homogeneous network structure from tetrahedron-like macromonomers. Macromolecules. 2008;41:5379–84. [Google Scholar]
- [40].Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, et al. Synthesis of well-defined hydrogel networks using Click chemistry. Chemical Communications. 2006:2774–6. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- [41].Yang T, Long H, Malkoch M, Gamstedt EK, Berglund L, Hult A. Characterization of Well-Defined Poly(ethylene glycol) Hydrogels Prepared by Thiol-ene Chemistry. Journal of Polymer Science Part a-Polymer Chemistry. 2011;49:4044–54. [Google Scholar]
- [42].Packard BZ, Artym VV, Komoriya A, Yamada KM. Direct visualization of protease activity on cells migrating in three-dimensions. Matrix Biology. 2009;28:3–10. doi: 10.1016/j.matbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Anderson SB, Lin CC, Kuntzler DV, Anseth KS. The performance of human mesenchymal stem cells encapsulated in cell-degradable polymer-peptide hydrogels. Biomaterials. 2011;32:3564–74. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–24. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu S, Wells A, Griffith LG, Lauffenburger DA. Controlling multipotent stromal cell migration by integrating “course-graining” materials and “fine-tuning” small molecules via decision tree signal-response modeling. Biomaterials. 2011;32:7524–31. doi: 10.1016/j.biomaterials.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu S, Wells A, Griffith LG, Lauffenburger DA. Controlling multipotent stromal cell migration by integrating “course-graining” materials and “fine-tuning” small molecules via decision tree signal-response modeling. Biomaterials. 2011;32:7524–31. doi: 10.1016/j.biomaterials.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Phinney DG, Kopen G, Righter W, Webster S, Tremain N, Prockop DJ. Donor variation in the growth properties and osteogenic potential of human marrow stromal cells. Journal of Cellular Biochemistry. 1999;75:424–36. [PubMed] [Google Scholar]
- [49].DeForest CA, Anseth KS. Photoreversible Patterning of Biomolecules within Click-Based Hydrogels. Angewandte Chemie International Edition. 2012;51:1816–9. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. hNMR spectra of PEG-Norbornene. Functionalization was calculated by dividing the norbornene alkene peaks by the PEG alkyl peaks.