Abstract
Significance: Chronic obstructive pulmonary disease (COPD) is predominantly a tobacco smoke-triggered disease with features of chronic low-grade systemic inflammation and aging (inflammaging) of the lung associated with steroid resistance induced by cigarette smoke (CS)-mediated oxidative stress. Oxidative stress induces various kinase signaling pathways leading to chromatin modifications (histone acetylation/deacetylation and histone methylation/demethylation) in inflammation, senescence, and steroid resistance. Recent Advances: Histone mono-, di-, or tri-methylation at lysine residues result in either gene activation (H3K4, H3K36, and H3K79) or repression (H3K9, H3K27, and H3K20). Cross-talk occurs between various epigenetic marks on histones and DNA methylation. Both CS and oxidants alter histone acetylation/deacetylation and methylation/demethylation leading to enhanced proinflammatory gene expression. Chromatin modifications occur in lungs of patients with COPD. Histone deacetylase 2 (HDAC2) reduction (levels and activity) is associated with steroid resistance in response to oxidative stress. Critical Issues: Histone modifications are associated with DNA damage/repair and epigenomic instability as well as premature lung aging, which have implications in the pathogenesis of COPD. HDAC2/SIRTUIN1 (SIRT1)-dependent chromatin modifications are associated with DNA damage-induced inflammation and senescence in response to CS-mediated oxidative stress. Future Directions: Understanding CS/oxidative stress-mediated chromatin modifications and the cross-talk between histone acetylation and methylation will demonstrate the involvement of epigenetic regulation of chromatin remodeling in inflammaging. This will lead to identification of novel epigenetic-based therapies against COPD and other smoking-related lung diseases. Pharmacological activation of HDAC2/SIRT1 or reversal of their oxidative post-translational modifications may offer therapies for treatment of COPD and CS-related diseases based on epigenetic histone modifications. Antioxid. Redox Signal. 18, 1956–1971.
Introduction
Lung cells are constantly exposed to a variety of oxidants, either generated endogenously due to metabolic activity (mitochondrial electron transport during respiration, upon activation of phagocytes), or exogenously through cigarette smoke (CS), air pollutants, and noxious gases (119, 124). The lung is a direct target for oxidative injury from reactive oxygen species (ROS) and free radicals. Oxidative stress results in increased cellular signaling associated with post-translational modifications of histones and nonhistone proteins, and redox modifications of proteins implicated in chromatin remodeling (48, 152, 179, 181) (Fig. 1).
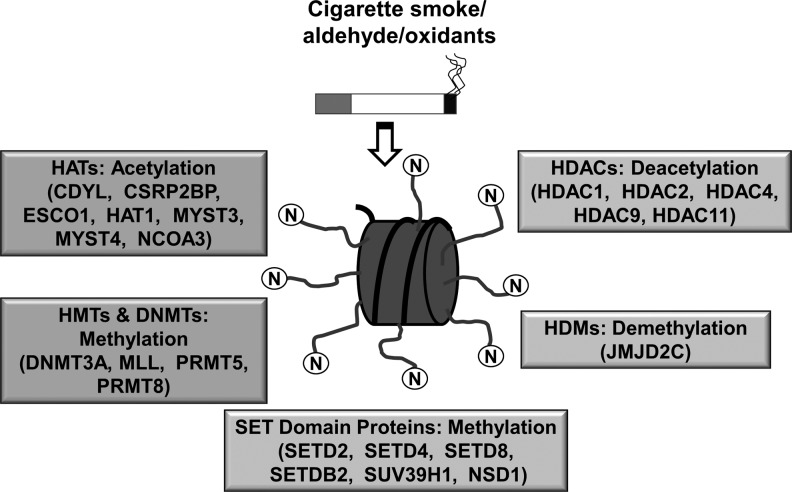
FIG. 1.
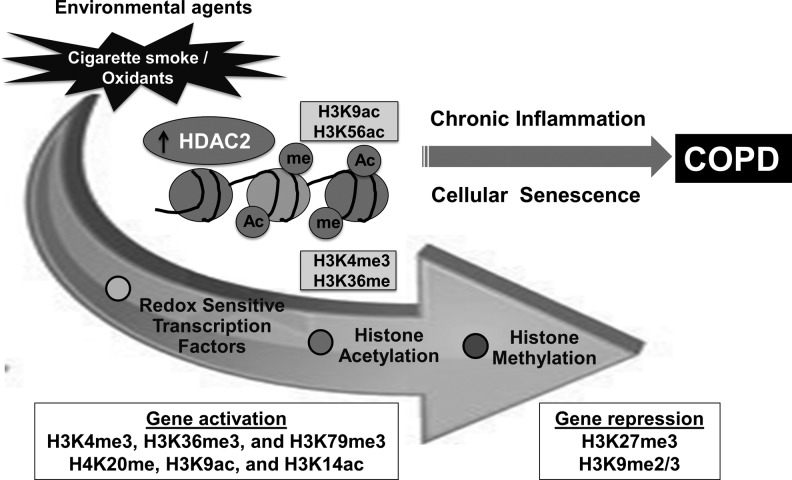
Differentially regulated chromatin modification genes and DNA methyltransferases are involved in cigarette smoke-mediated histone modifications. Unwinding and rewinding of DNA is regulated by epigenetic alterations, such as histone acetylation/deacetylation and histone methylation/demethylation. This includes histone acetylation by histone acetyltransferases (HATs), histone deacetylation by histone deacetylases (HDACs), histone methylation by SET domain proteins and histone methyltransferases (HMTs), histone demethylation by histone demethylases (HDMs), and DNA methylation by DNA methyltransferases (DNMTs), respectively. Cigarette smoke-mediated epigenetic changes and chromatin conformation changes can lead to alterations in DNA accessibility for transcription factors, coactivators, and polymerases, thereby, resulting in either transcriptional gene activation or gene repression of proinflammatory genes. Some examples of histone modification enzymes are given in parenthesis.
DNA is packed into chromatin by histones to form a tightly coiled structure in the nucleus. Chromatin remodeling by histone modifications plays a key role in the regulation of cellular processes, including gene transcription/repression, DNA repair, differentiation, and proliferation. Post-translational histone modifications predominantly occur via acetylation/deacetylation and methylation/demethylation reactions (41, 127) (Fig. 1).
Histone acetyltransferases (HATs, including p300- CREB-binding protein [CBP]-associated factor, p300/CBP, and GCN5) and histone deacetylases (HDACs e.g., HDAC2 and SIRTUIN1 [SIRT1]) maintain the balance between histone acetylation and deacetylation. HATs affect the binding of DNA sequence-specific transcription factors and, subsequently, recruit coactivators or corepressors on gene-specific regions to form either coactivator or corepressor complexes (166). It is well known that transcriptional coactivators possess intrinsic HAT (CBP/p300) and HDAC activities, suggesting that histone acetylation and deacetylation play a causal role in regulating gene transcription (72, 170, 171). It is generally accepted that increase in histone acetylation leads to increased gene transcription, while histone hypoacetylation is linked to decreased gene transcription (72, 170, 171). Histone acetylation by HATs and deacetylation by HDACs are also linked to cell-cycle progression, proliferation, senescence, DNA repair, and recombination events, as well as proinflammatory gene transcription, which can be affected by oxidative stress and redox signaling (15, 69, 146).
Methylation of histones occurs either on lysine (K) or arginine (R) residues in a reaction that is catalyzed by specific histone methyltransferases (HMTs) (11, 192). HMTs utilize S-adenosylmethionine as a cofactor during transmethylation and produce S-adenosylhomocysteine as a by-product. Methylation of specific lysine residues results in the formation of binding sites/interacting domains, which allow recruitment of other regulator proteins. HMTs are deregulated in several chronic lung diseases, thereby affecting the global methylation status. Histone demethylases (HDMs) catalyze the removal of methyl groups from lysine or arginine residue of histones. These include lysine-specific demethylase 1 and Jumonji C domain family proteins involved in the regulation of gene expression (149). These enzymes demethylate specific histone residues, thereby forming transcriptional repression complexes (133). HMTs and HDMs are subject to redox regulation such that post-translational modifications of histone proteins by oxidants and environmental stresses can trigger the transcription of genes that are involved in chronic inflammatory events. This review is focused on the role of ROS-mediated effects on chromatin modification enzymes and epigenetic chromatin modifications (histone acetylation/deacetylation and histone methylation/demethylation) in inflammatory response, steroid resistance, and cellular senescence that occurs in chronic inflammatory lung diseases associated with cigarette/tobacco smoking.
Cigarette Smoke and Oxidative Stress
ROS can affect the lung by directly oxidizing proteins, DNA, and lipids or induce changes indirectly through the generation of secondary metabolic reactive species. ROS can fragment the extracellular matrix, and cause mucus secretion, apoptosis, and alter cell proliferation (113). Relevant ROS generating enzyme systems in Chronic Obstructive Pulmonary Disease (COPD) include xanthine–xanthine oxidase, NADPH oxidase, and heme peroxidases (108, 117). CS, which contains 1015 free radicals and 4700 different chemical compounds per puff, is the major source of inhaled environmentally derived ROS. It has been estimated that 15%–20% of smokers develop COPD. The Global Initiative for Chronic Obstructive Lung Disease defines COPD as a syndrome of chronic, irreversible obstruction of lung airflow characterized by progressive decline in forced expiratory volume in one second. The inflammatory immune features of the COPD phenotype includes, activation of epithelial cells, and resident macrophages, as well as the recruitment and activation of neutrophils, monocytes and B and T lymphocytes into the lung. Inflammatory cells are activated in response to cytokines/chemokines/chemoattractants after recruitment into the interstitium, thereby generating various ROS. ROS cause lipid peroxidation (oxidation of membrane phospholipids) and the generation of malondialdehyde, 4-hydroxy-2-nonenal, acrolein, and F2-isoprostanes (114, 120).
Oxidative Stress and Kinases Signaling in Chromatin Remodeling
It is well known that ROS can activate signal transducing molecules through the effects on oxidation-prone cysteine-rich domains, thereby activating gene transcription (6, 118). These include the mitogen-activated protein kinase family, extracellular signal-regulated kinase, c-Jun N-terminal kinase (JNK), p38 kinase, and phosphoinositide 3-kinase (PI3K)/Akt. Based on the redox status of the cells, members of the MAPK family are activated, leading to a multifaceted transactivation of redox-sensitive transcription factors (ATF-2, CBP) (6, 147). Furthermore, activation of upstream kinases, such as nuclear factor-κB (NF-κB)-inducing kinase, mitogen- and stress-activated kinase 1, and IκB kinase-α, results in downstream chromatin remodeling events that alter the function of various gene promoters (27, 28, 138, 179, 181). This epigenetic mechanism modulates a series of specific proinflammatory gene transcription events that modulate apoptosis, autophagy, senescence, proliferation, transformation, and differentiation (53). CS induces activation of protein kinase C (PKC), mediates regulation of intracellular signaling pathways (174). PKCζ-knockout mice exposed to CS or lipopolysaccharide showed a reduction in lung inflammatory response due to chromatin modifications, suggesting that PKCζ is an important modifier of lung inflammatory response (Fig. 2) (186). Hence, activation of kinases by ROS and aldehyde/lipid peroxidation products can lead to a proinflammatory response by chromatin remodeling.
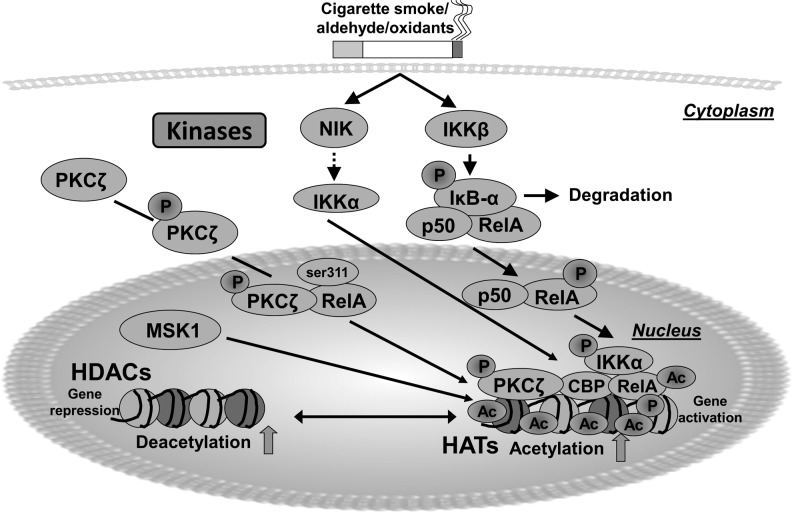
FIG. 2.
Role of redox kinase signaling in chromatin modifications. Cigarette smoke-derived oxidants/aldehydes activate several kinase signaling mechanisms, such as NIK, IKKα, MSK1, and PKCζ by redox modulation, and induce proinflammatory gene transcription via chromatin modifications. Upon activation of these kinases, NF-κB interacts with the master coactivator cAMP-response-element-binding protein (CBP/p300), and causes acetylation of specific lysine residues on core histones by displacing/decreasing histone deacetylases, leading to gene transcription. HATs, such as CBP/p300 and P/CAF, mediate histone acetylation (euchromatin) by loosening the nucleosomes, thereby, promoting access of cofactors and RNA polymerase II to mediate active gene expression. HDACs, such as SIRT1 and HDAC2, mediate deacetylation (heterochromatin) by tightly winding the nucleosomes, thereby, making the DNA inaccessible to transcription factors and other protein complexes, thus, resulting in gene repression.
The role of PI3K and p38 signaling pathways have been shown in pathogenesis of COPD and lung cancer (34, 37, 51). Inhibition of PI3K/p38 lowers tobacco/CS-induced lung inflammatory response (34). Interestingly, PI3K inhibition significantly increased the steroid efficacy to inhibit an inflammatory response under oxidative stress (14, 82, 151). Therefore, in addition to p38 MAPK and PKCζ, oxidants/CS-mediated activation of the PI3K signaling pathway may be a target for intervention in COPD. These oxidant- and redox-sensitive pathways (PI3K/p38) activate various kinases, such as NF-κB-inducing kinase, mitogen- and stress-activated kinase 1, and IκB kinase-α, resulting in histone acetylation and transcriptional activation of proinflammatory genes (e.g., IL-6, IL-8, and COX-2) (27, 138, 179) (Fig. 2).
Chromatin Remodeling: Histone Acetylation, Deacetylation, Methylation, and Demethylation
Histone acetyltransferases
HATs transfer acetyl groups from acetyl-CoA to the N-terminal lysine residues of histones (H3 and H4), thereby uncoiling DNA resulting in the transcription factor binding to promoters followed by transcriptional gene activation (56, 58, 119, 144). HATs and HDACs play opposing roles in regulating acetylation of core nucleosomal histones. HATs are classified into five distinct families. These include the CBP/p300 HATs, Gcn5-related acetyltransferases, the general transcription factor HATs (TFIID subunit TAF250), and the nuclear hormone-related HATs SRC1 and ACTR (SRC3) (144, 153). Each of these HAT families has diverse roles in regulating the chromatin assembly and structure (chromatin remodeling) although they share a common enzymatic activity (112).
CS/oxidant-mediated acetylation of histone H3 occurs in macrophages and epithelial cells, and in lungs of humans and rodents, suggesting that histone acetylation/deacetylation plays an important role in chromatin remodeling. Oxidative stress and/or glutathione depletion is associated with increased histone acetylation (119, 120). CS-mediated alterations in chromatin remodeling are shown to be involved in sustained lung inflammatory diseases, such as COPD (83, 140, 143, 179, 186). CBP and p300 are the known coactivators that possess intrinsic HAT activity. They are regulated by the JNK/p38 MAP kinase pathway, which activates redox-sensitive transcription factors, such as NF-κB and activator protein-1 (148). Therefore, histone acetylation by CBP/p300 has an impact on the activation of specific proinflammatory mediators linked with NF-κB/AP-1-mediated gene expression (22, 64, 115). Transcription factor NF-κB (RelA/p65) is acetylated at K310 residue by CBP/p300 HAT, but also causes histone acetylation by recruiting other cofactors/coactivators and chromatin remodeling complexes to activate proinflammatory gene transcription (e.g., IL-6 and IL-8) (46, 57, 75). It is shown that NF-κB-induced acetylation of histone H3, but not histone H2A, H2B, or H3, occurs in epithelial cells on specific lysine residues (Lys8 and Lys12) at NF-κB-responsive regulatory elements on proinflammatory genes (57). Oxidative stress and CS activate NF-κB, HAT, CBP/p300 leading to specific histone acetylation in macrophages and lung cells (60, 93, 178, 179, 187). Therefore, development of small molecules that target for inhibition of HATs (e.g., CBP/p300, PCAF, and GCN) may be useful for therapeutic intervention in COPD and other chronic inflammatory lung diseases associated with smoking mediated oxidative stress (8, 99).
Histone deacetylases
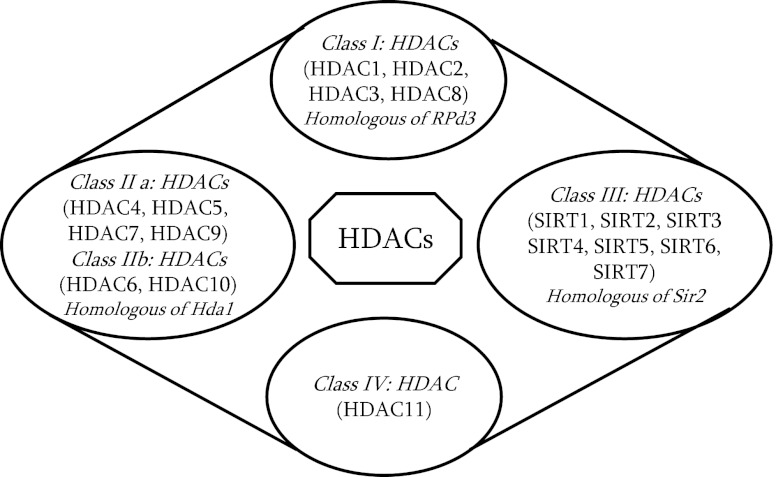
HDACs play a crucial role in removing the acetyl groups from histones, thus providing a regulatory role toward the epigenetic control of gene expression (17). Presence of HDACs within the gene regulatory regions leads to formation of closed chromatin complexes (heterochromatin) resulting in gene silencing. So far, eighteen mammalian HDAC enzymes are known, which are classified into four classes based on their homology to a prototypical HDAC found in yeast (32) (Fig. 3). Class I HDACs (HDACs 1, 2, 3, and 8) are ubiquitously expressed with the possible exception of HDAC3 and HDAC10, which are predominantly localized in the nucleus (145). Class II HDACs (HDACs 4, 5, 6, 7, 9, and 10) are expressed in a tissue-specific manner, and they have the ability to shuttle between the nucleus and cytoplasm (13, 50). The shuttling of class II HDACs from the nucleus is mainly regulated by nuclear export signaling 14-3-3 proteins (13). Based on the existence of tandem deacetylase domains, class II HDACs are further classified into class IIa (HDACs, 4, 5, 7, and 9) and class IIb (HDACs 6 and 10) (62). Class I and II HDACs share significant homology at the deacetylase domain, but differ in their N-terminal sequence (Fig. 3).
FIG. 3.
Classes of histone deacetylases (HDACs/SIRTs). HDACs are classified into four distinct classes I to IV. Class I, class II (subclasses: IIa and IIb), and class IV HDACs are zinc-dependent enzymes, whereas class III HDACs that includes sirtuins (SIRTs: SIRT1–7) require nicotinamide adenine dinucleotide (NAD+) for catalytic activity.
The class III HDACs or sirtuins are named based on their homology to the yeast Sir2 gene, which is a highly conserved gene family. In humans, sirtuins comprise seven members, SIRT 1–7 (86). Among these, SIRT1, SIRT2, SIRT3, and SIRT5 have an NAD+-dependent deacetylase domain (distinct from the zinc-dependent deacetylase domains of class I and II HDACs), which catalyze the deacetylation of histones and nonhistone proteins. Since sirtuins utilize NAD+ obtained from metabolic processes, it plays vital roles in linking molecular mechanisms between cellular metabolic status (NAD+/NADH levels) and several cellular processes (16). In contrast, SIRT4 and SIRT6 have an NAD+-dependent ADP ribosylation domain and catalyze protein ribosylation. The subcellular localizations of sirtuin family members are well defined. SIRT1, SIRT6, and SIRT7 are localized to the nucleus; SIRT2 to the cytoplasm and SIRT3, SIRT4, and SIRT5 are localized to the mitochondria. Class III HDACs share less homology compared to the class I and II HDACs, and are not inhibited by widely used HDAC inhibitors, such as butyrate, valproic acid, trichostatin A (TSA), or suberoylanilide hydroxamic acid (175) (Fig. 3). Instead, they are inhibited by nicotinamide, splitomicin, and sirtinol. SIRT1 is a redox-sensitive deacetylases, which requires intracellular NAD+ for its regulations (20). HDAC11 belongs to class IV HDACs, which share limited homology with class I and II HDACs (44).
HDACs are also reported to deacetylate nonhistone proteins, such as the redox-sensitive transcription factor NF-κB (RelA/p65) and forkhead box class O3 (FOXO3), thereby regulating transcription of NF-κB-dependent proinflammatory genes (132). The levels and activities of HDACs are significantly decreased in macrophages, and lung cells in vitro in response to oxidative/carbonyl stress, as well as in lungs of COPD patients via alterations in intracellular GSH/GSSG redox ratio (caused by H2O2 and CS exposures). Decreased HDAC2 activity is linked with COPD severity, asthma, and steroid resistance in asthmatics who smoke tobacco (5, 60, 93, 178, 187). Intracellular GSH maintains HDAC2 in a reduced environment, and increasing intracellular GSH inhibits HDAC2 oxidative post-translational modifications (5, 85, 178). A decrease in levels of HDAC2 in several cellular systems is due to post-translational modifications, such as oxidation/carbonylation, nitrosylation, acetylation, and phosphorylation in response to oxidants derived from CS (3, 5, 59, 93, 178). Hence, HDAC2 levels/activities and the status of oxidative post-translational modifications dictate the effects of coactivators on the chromatin status on various promoters to drive the gene activation or repression.
HDAC2 and steroid resistance in inflammation
Decreased HDAC2 occurring as a result of CS/oxidants/aldehydes is associated with activation of NF-κB RelA/p65 subunit (evident as increased levels of total and phospho-acetylated RelA/p65). RelA/p65 interacts with HDAC2, and nuclear accumulation of RelA/p65 result in transcription of proinflammatory genes when HDAC2 is post-translationally modified, ubiquitinated, and degraded (79, 178, 185, 187). HDAC2 modification is dependent on the cellular thiol (GSH/GSSG) redox status (85, 178). In vitro studies using trichostatin A (HDAC inhibitor) in different cell lines showed enhanced NF-κB-driven inflammatory gene transcription (24, 57). Hence, alteration of HDACs (particularly HDAC2) by CS/oxidative stress leads to increased acetylation of histones (histone H3 and H4) and activation of NF-κB, thus enhancing transcription of proinflammatory genes (4, 123, 178). In humans, a marked reduction of HDAC2 expression/activity in lung parenchyma, bronchial biopsies, and alveolar macrophages of patients with COPD is correlated with the severity of inflammation and the progression of the disease (60). The underlying mechanism for reduced HDAC2 levels/activities include post-translational modifications (i.e., nitrosylation, phosphorylation, and ubiquitination) leading to proteasome-dependent degradation, particularly, in response to CS (5, 43, 123), and/or oxidative/carbonyl modifications/degradation of HDAC2 (178). HDAC2 is important for the anti-inflammatory effects of glucocorticoid treatment. The levels/activity of HDAC2 is decreased in lungs of patients with COPD who are resistant to corticosteroid (60). Corticosteroid resistance is also associated with increased oxidative/carbonyl stress (aldehyde-mediated carbonylation of proteins on cysteine, histidine, and lysine residues). Corticosteroids do not block the inflammatory response when there is an increased oxidative stress, such as in severe asthmatics and patients with COPD. This inefficacy is not associated with alterations in nuclear HAT activity (60). Increasing HDAC activity, by dietary polyphenol curcumin, sulforaphane, or theophylline (HDAC2 activators), significantly enhances the steroid-induced suppression of IL-8 release in monocytes and alveolar macrophage from COPD patients; conversely, HDAC inhibitors have the opposing effects (30, 52, 85). This suggests a specific role of HDAC2 in steroid resistance. Furthermore, deacetylation of glucocorticoid receptor (GR) by HDAC2 enhanced the association of GR and RelA/p65, which resulted in the attenuation of proinflammatory gene transcription in monocytes and lung cells (61). Consequently, restoration or attenuation of decreased HDAC2 levels/activities will further enhance the glucocorticoid sensitivity by deacetylating RelA/p65 and GR. This can be achieved by reversing the post-translational modifications of HDAC2, such as decarbonylation or dephosphorylation, via inducing aldehyde dehydrogenases/reductases, thioredoxin reductase, and phosphatases, or inducing the antioxidant buffer systems by using Nrf2 activators and extracellular superoxide dismutase mimetics (2, 183). Thus, HDAC2 is a redox-sensitive protein, whose levels/activities are decreased under oxidative/carbonyl stress conditions, resulting in chromatin modifications associated with steroid resistance leading to lung inflammation.
HDAC2, DNA damage/repair, and cellular senescence
CS/oxidants can also induce cellular senescence (i.e., stress-induced premature senescence, SIPS) in alveolar epithelial cells and fibroblasts, which is independent of telomere shortening (95, 101, 102, 155–157). Cellular senescence (irreversible cell growth arrest) impairs the repair of damaged lung tissue, perhaps, explaining why COPD progresses even after cessation of smoking. The role of cellular senescence in COPD is supported by animal studies (74, 130, 137, 184), where CS is known to cause premature senescence. The senescent cells are metabolically active and prone to secrete proinflammatory mediators (i.e., IL-6 and IL-8) and matrix metalloproteinases (i.e., MMP-1, MMP-3, and MMP-9) (40, 125). These proinflammatory mediators may in turn initiate and maintain cellular senescence as the deficiency of IL-6, IL-6R, or CXCR2 prevents premature senescence (1, 71). Indeed, the percentage of proinflammatory senescent type II cells expressing both p16 and phosphorylated NF-κB (i.e., senescent-associated secretory phenotype) was increased in lungs of patients with COPD as compared to smokers and nonsmokers (157). However, the mechanism underlying premature senescence and associated secretory senescence phenotype (secretory senescent cells) in the development of COPD remains unknown.
Persistent DNA damage causes premature senescence and senescence-associated secretory phenotype via forming persistent foci, termed as DNA segments with chromatin alterations reinforcing senescence (29, 125, 126). Further study demonstrated the requirement of the upstream signals of DNA damage response, including ataxia telangiectasia mutated and its substrates Nijmegen breakage syndrome 1 and checkpoint kinase 2 for the NF-κB-dependent inflammatory phenotype in senescent cells (36, 125, 173). The double-strand break is the most dramatic form of DNA damage, which can be repaired either by homologous recombination, classical, or alternative nonhomologous end joining. CS-mediated oxidative stress has been shown to cause DNA damage, which is increased in patients with COPD (21, 23, 33, 106, 134). The nonhomologous end joining repair proteins, such as Ku70, Ku80/Ku86, are also impaired by CS, which may, in turn, accelerate the formation of persistent DNA segments with chromatin alterations reinforcing senescence leading to sustained DNA damage and an associated secretory senescence phenotype observed in patients with COPD (21, 126, 135). Ku70, Ku80/Ku86 proteins may be regulated by acetylation/deacetylation. However, it is unclear how chromatin remodeling influences premature senescence and associated secretory senescence phenotype during DNA damage/repair by CS/oxidants in lung cells.
HDAC2, not only functions in DNA damage response to promote repair, but also regulates cellular senescence and inflammation via deacetylating histones and transcription factors (e.g., NF-κB) (89, 150, 165, 169, 193). Indeed, histone acetylation and methylation as well as NF-κB activation play an important role in DNA damage repair and genomic instability as well as premature aging (159, 190). Additionally, other histone modifiers, such as CBP/p300, regulate DNA damage and repair (103, 129, 163, 167). Cellular senescence and its associated inflammatory phenotype occur in lungs of patients with COPD (156, 157). It is evident that HDAC2 regulates cellular senescence along with its complex, such as HDAC1, in response to stress (169). However, no information is available regarding the role of HDAC2 epigenetic modifications in DNA damage/repair, as well as premature senescence and associated secretory senescence phenotype, particularly, in response to CS exposure in lung cells.
Histone modifications, including acetylation and methylation, play an important role in DNA damage repair and genomic instability as well as premature aging (159, 163, 190). It is possible that HDAC2 regulates CS-mediated DNA damage, premature senescence, and associated secretory senescence phenotype through histone deacetylation (Fig. 4). HDAC2 can interact with protein methyltransferases to form a large and multiple-protein complex(es), thereby regulating transcriptional repression or activation (161). Our preliminary studies have recently showed that CS exposure alters histone methylation (e.g., H3K4me, H3K27me, H3K79me, H3K36ac/me) in cells and mouse lung (27, 177). CS/oxidant-mediated HDAC2 reduction may alter histone methylation by regulating enzyme methyltransferases. In addition to HDACs, CBP/p300 also has shown to acetylate histone H3 and H4 at the sites of DNA double-strand break, thereby regulating DNA damage and repair (103, 163). HDAC2 interacts with CBP in response to CS exposure (3). Therefore, other coactivators/repressors, such as CBP/p300, and NF-κB along with HDAC2 participate in the CS-mediated DNA damage and premature senescence and associated secretory senescence phenotype.
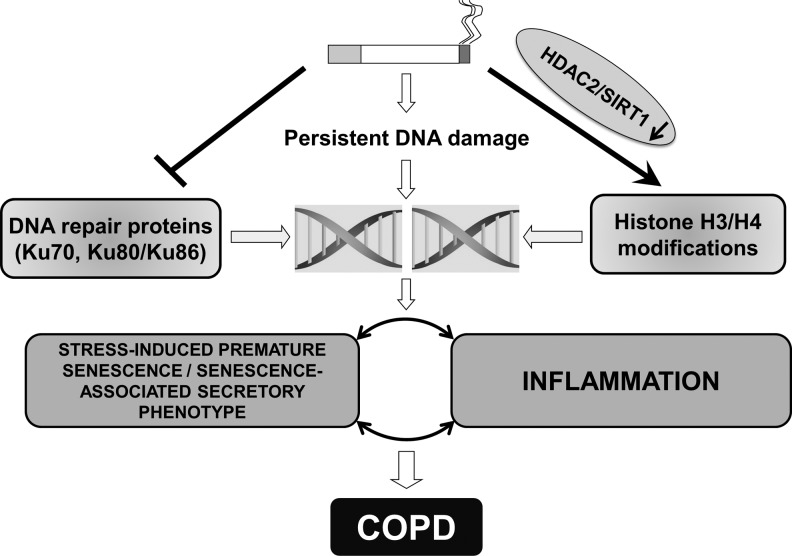
FIG. 4.
Epigenetic regulation in cigarette smoke-mediated persistent DNA damage leading to chronic obstructive lung disease. Cigarette smoke-induced reduction in HDAC2/SIRT1 and DNA damage response proteins, such as Ku70 and Ku80, resulting in persistent DNA damage response. Cigarette smoke also alters the expression levels of chromatin modification enzymes, thus, affects epigenetic regulation of gene expression due to site-specific histone modification in histone H3 and H4. Persistent DNA damage further leads to stress-induced premature senescence/senescence-associated secretory phenotype, which subsequently causes premature aging of the lung in pathogenesis of chronic obstructive pulmonary disease (COPD) and smoking related disorders.
SIRT1 deacetylase, FOXO3, NF-κB, and cellular senescence
Sirtuin 1 (SIRT1) belongs to class III HDACs, which possess anti-inflammatory, anti-aging/senescence, and anti-apoptotic/autophagy properties due to their ability to deacetylate both histones and nonhistone proteins, including transcription factors (e.g., NF-κB, FOXO3, p53, Ku70, and PGC-1α) (182) (Fig. 5). SIRT1 levels and activities were decreased in monocytes (MonoMac6 cells), bronchial epithelial cells (Beas-2B), mouse lungs exposed to CS, as well as in the lungs of COPD patients and smokers (54, 121, 122, 180), suggesting the involvement of SIRT1 in the pathogenesis of COPD. The mechanism for SIRT1 reduction involves post-translational modifications, such as phosphorylation, nitrosylation, and oxidation/carbonylation by ROS/carbonyls-aldehydes in response to CS/oxidative/carbonyl stress, ultimately leading to SIRT1 degradation (20, 121, 122). SIRT1 is also regulated by the intracellular redox thiol (GSH/GSSG) pool (20, 180). SIRT1 knockdown by siRNA leads to heightened NF-κB activation and inflammatory response. SIRT1 activators, such as SRT1720 and resveratrol diminished the release of proinflammatory mediators in response to oxidative stress/CS exposure (122). These findings suggest a role for SIRT1 activators (90, 91) in the therapeutic intervention of COPD. Administration of the SIRT1 activators, SRT2172 and SRT1720, attenuated CS-induced lung inflammation in mice (96). Furthermore, SIRT1-mediated protection in response to CS/oxidative stress against lung inflammatory response and injury are linked to the deacetylation of NF-κB RelA/p65 and FOXO3, and negative regulation of MMP-9 via the tissue inhibitor of metalloproteinases-1 (25, 96, 180, 184). Further studies are required to investigate the key molecular mechanisms to understand how SIRT1 can mediate protection against oxidative stress during COPD progression (184, 188).
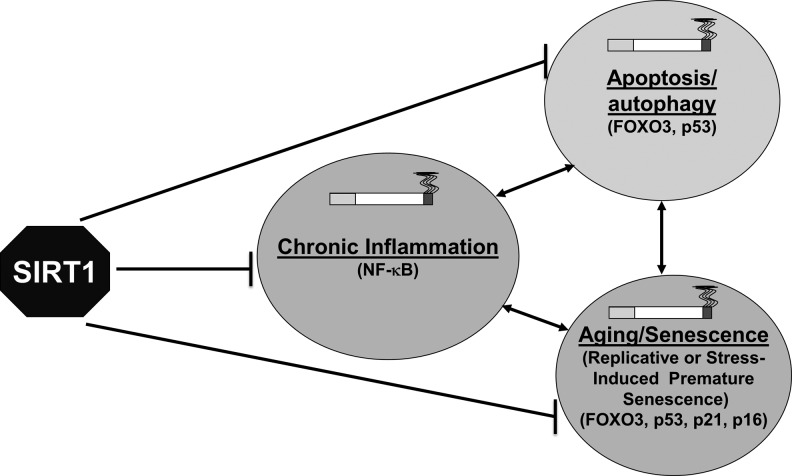
FIG. 5.
SIRT1 as master regulator of chronic inflammation, aging/senescence, and apoptosis/autopahgy. SIRT1 is a class III deacetylase that can deacetylate both histone and nonhistone proteins, including transcription factors, such as NF-κB, FOXO3, p53, p21, Ku70, and PGC1-α, thereby, regulating oxidative stress-induced chronic inflammation, cellular senescence, apoptosis, and autophagy, which play key roles in the pathogenesis of COPD. SIPS, stress-induced premature senescence.
SIRT1 also deacetylates other transcription factors, such as FOXO3, p53, and NF-κB, thereby regulating CS/oxidative stress-induced cell cycle arrest, apoptosis, and cellular senescence, which play important roles in the pathogenesis of COPD (Fig. 6) (188). Transcriptional activity of FOXO3 is regulated by its phosphorylation and acetylation status. SIRT1 deacetylates FOXO3, leading to its activation and to the induction of cell cycle arrest, thereby reducing premature senescence (94, 184, 189). FOXO3 is acetylated when SIRT1 levels/activity are reduced in response to CS in mouse lung (121). Furthermore, FOXO3 has a regulatory role in lung inflammatory response and regulation of antioxidant genes, such as manganese superoxide dismutase and catalase. Targeted disruption of FOXO3 results in downregulation of these antioxidant genes in mouse lungs and increases the susceptibility for the development of COPD/emphysema (55). Yao and colleagues have shown that the SIRT1-FOXO3-mediated pathway is important in pathogenesis of COPD independent of the NF-κB pathway (116, 184). Further studies on the SIRT1-FOXO3 pathway will elucidate the underlying mechanisms in response to oxidative/carbonyl stress, and will provide the possible therapeutic interventions in treatment of COPD based on SIRT1 activation.
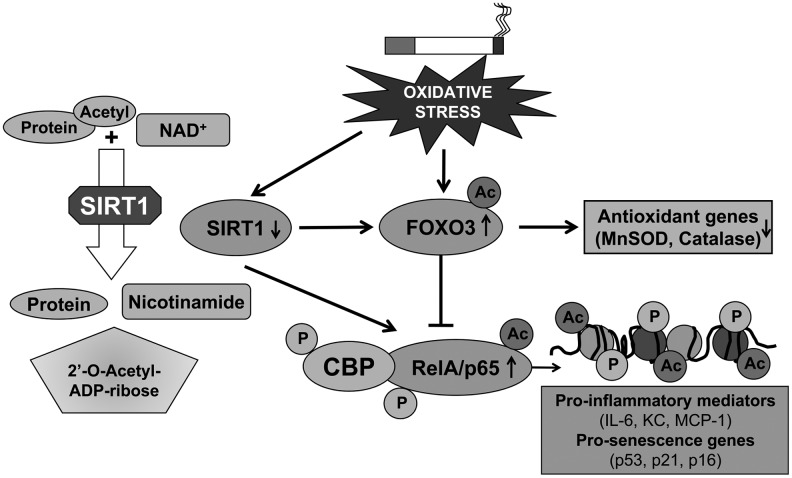
FIG. 6.
Role of SIRT1, FOXO3, and NF-κB in oxidants/cigarette smoke-mediated lung inflammation. Regulation of SIRT1 activity by its substrate acetylated protein in presence of NAD+ will result in deacetylated protein, nicotinamide and acetyl ADP-ribose molecules (Left panel). SIRT1 activity and level are reduced by oxidative/carbonyl stress imposed by cigarette smoke leading to RelA/p65 and FOXO3 acetylation (Right panel). This results in downregulation of antioxidant genes, such as MnSOD and catalase, and NF-κB activation via its interaction with CBP favoring expression of proinflammatory and prosenescence genes.
SIRT1 interacts with p53, and deacetylates lysine residue at the C-terminal regulatory domain (162). Decreasing SIRT1 levels/activity increases p53 acetylation, thereby promoting apoptosis and senescence (80, 162). Oxidative stress is shown to accelerate cellular senescence, due to increased p53 acetylation via decreasing the function of SIRT1 through NAD+ depletion (42) (104). Furthermore, blocking p53 by antisense oligonucleotides reversed the inhibitory effect of SIRT1 on cellular senescence (104). Earlier studies have shown that the nuclear levels of SIRT1 were decreased in response to CS both in vitro and in vivo in lungs (180), but still it remains unclear whether SIRT1-mediated regulation of p53 (acetylation/deacetylation) plays an important role in oxidants/CS-mediated apoptosis, autophagy, and cellular senescence/aging.
Other sirtuins have also been shown to modify cellular functions under oxidative stress. For example, a recent report demonstrated the role of SIRT6 in DNA repair under oxidative stress through the activation of poly [adenosine diphosphate-ribose] polymerase 1 (PARP1) (81). Oxidative stress in mammalian cells recruits SIRT6 to the sites of DNA double-strand breaks, where it interacts with PARP1 (redox sensitive), and mono-ADP-ribosylates PARP1 on Lys521, thus stimulating PARP1's poly-ADP-ribosylase activity, and repairing DNA double-strand break through both nonhomologous end joining and homologous recombination repair mechanisms (81). Therefore, SIRT6, in addition to SIRT1 is implicated to play crucial roles in oxidative stress-induced inflammatory response, senescence, genomic instability, DNA damage/repair, and aging (65, 81, 88, 158). Consequently, other sirtuin members possibly gain equal credence in understanding the pathogenesis of COPD and other chronic lung diseases associated with tobacco smoking.
SIRT1 is a key regulator of vascular endothelial homeostasis controlling angiogenesis, vascular tone, and endothelial dysfunction by regulating the endothelial nitric oxide synthase activity (109, 172). Endothelial dysfunction plays an important role in pathogenesis of emphysema, and CS-induced emphysematous alveolar destruction, which is almost avascular, associated with decreased expression of endothelial nitric oxide synthase (35, 38). SIRT1 deacetylates lysines at position K496 and K506 in the calmodulin-binding domain of endothelial nitric oxide synthase, leading to enhanced nitric oxide production, which is vital for endothelial-dependent vasorelaxation, cell survival, migration, and postnatal neovascularization (10, 84). NO activates the SIRT1 promoter, thereby increasing SIRT1 mRNA and protein levels (100, 105), suggesting that a positive feedback mechanism exists between SIRT1 and endothelial nitric oxide synthase (110). Therefore, the activation of SIRT1 by small molecules (such as SRT1720) may help in resetting the endothelial nitric oxide synthase activity during endothelial dysfunction in patients with COPD, where NO availability is limited (87). SIRT1 overexpression attenuated CS-induced apoptosis and inflammatory response in cultured coronary arterial endothelial cells (31). Therefore, SIRT1 acts as a possible epigenetic chromatin redox regulator in the treatment and prevention of chronic lung diseases, including COPD and comorbidities associated with endothelial dysfunction and cardiovascular problems by protecting endothelial cells from stress-induced premature senescence, autophagy, apoptosis, and inflammatory response.
Histone methyltransferase
Methylation of histones occurs either on lysine (K) or arginine (R) residues in a reaction that is catalyzed by specific HMTs (PRMT family, the SET-domain [Suppressor of variegation-Enhancer of zeste-Trithorax domain]-containing protein and the non-SET-domain proteins DOT1/DOT1L) (11,192). HMTs utilize S-adenosylmethionine as a cofactor during transmethylation and produce S-adenosylhomocysteine as a by-product. The substrate specificity and activity of HMT depends on SET domains and associated motifs (191). Methylation of specific lysine residues results in the formation of binding sites/interacting domains, which allow recruitment of other regulator proteins (coactivators/corepressors). HMTs are deregulated in several chronic lung diseases, thereby affecting the global methylation status. Histone H3K4, H3K9, and H3K27, and H4K20 are frequently and preferentially methylated as mono-, di-, or tri-methylated histone H3 and histone H4 (131). Methylation at H3K4, H3K36, and H3K79 is linked to gene activation, whereas H3K9, H3K20, and H3K27 methylation is associated with gene repression (49) (Fig. 7).
FIG. 7.
Redox-mediated chromatin modifications in chronic inflammation and cellular senescence. Cigarette smoke-derived oxidants/carbonyls can activate various redox signaling cascades, thereby, leading to histone modifications on proinflammatory gene promoters. Histone modifications can cross talk with CpG methylation to activate or repress gene transcription. Modulation of various genes due to specific histone acetylation/methylation can result in inflammation and cellular senescence observed in chronic lung inflammatory diseases. Some of the known methylation and acetylation histone marks that either cause gene repression (H3K27me3 and H3K9me2/3) or gene activation (H3K9ac, H3K14ac, H3K4me3, H3K36me3, H3K79me3, H4K20me) are summarized.
Histone modifications are regulated by specific chromatin modification enzymes, which have a residue-specific role in regulating gene expression with respect to histone modifications (9, 19, 68, 127). In vitro exposure of airway epithelial cells or immortalized bronchial epithelial cells to CS condensate/extract (oxidants) reduced H4K16ac and H4K20me3, but increased H3K27me3. This may have implications in tumor development in several cancer types associated with tobacco smoking (77). It is well known that COPD is associated with a significantly increased incidence of lung cancer and other comorbidities in susceptible chronic smokers. HDACs, including HDAC2 and SIRT1, play an important role in cancer cell epigenetic regulation and gene expression. Histone H4K16ac and H4K20me3 has been suggested as a hallmark in different kinds of human tumors and tumor-derived cell lines, thus characterized as known epigenetic marks in cancer (39). SIRT1 depletion by siRNA in mammalian cells resulted in hyperacetylation of histone H4K16 and decreased H3K9me3 and H4K20me3 (160). However, it remains to be seen whether the regulation of these enzymes and specific histone modifications via modulation by oxidative stress and/or redox status of the cells also occur in redox epigenetic chromatin regulation of proinflammatory genes in COPD and smoking-induced diseases.
The cross-talk between histone methylation sites also controls the transcriptional activation of target genes (26, 168). The positive and negative cross talk, ultimately, generates the complex pattern of gene- or locus-specific histone marks, which are linked with distinct chromatin states (euchromatin or heterochromatin), leading to transcriptional repression or activation. Cross-talk exists between various epigenetic events on histones and DNA methylation (Fig. 7). For example, the methylcytosine-binding protein recruits HDACs to methylated DNA as well as to methylated histones. These events are associated with histone deacetylation and chromatin condensation, leading to transcriptional silencing of genes (47, 98). On the contrary, inhibition of HDACs and histone acetylation/methylation leads to upregulation of proinflammatory mediators, such as the granulocyte–macrophage colony-stimulating factor by IL-1β in lung epithelial cells (63).
Although histone H3K4 tri-methylation has been reported to play a role in tumorigenesis and X-chromatin inactivation in human cells (73), little evidence is available for its role in inflammatory responses. Furthermore, hypermethylation of histone H3K4 was found to be unrelated to acute TNF-α-induced gene expression (128). In contrast, the involvement of H3K4 tri-methylation in IL-1β-induced gene expression has been reported (164). Furthermore, H3K4 tri-methylation plays a critical role in IL-1β stimulated secretory leukocyte protease inhibitor expression by modulating RNA polymerase II recruitment, and subsequent engagement of SET1 (164). Conversely, very few reports are available on cross-talk between histone acetylation and methylation in response to CS/oxidative stress-mediated lung inflammation and in pathogenesis of chronic inflammatory lung diseases. Studies on sequential events of histone acetylation/histone methylation will provide the understanding on the new epigenetics-based biomarkers and/or treatment for chronic inflammatory diseases, including COPD (Fig. 7).
Histone demethylases
HDMs catalyze the removal of methyl groups from lysine or arginine residue of histones. They are classified into two types, the lysine-specific demethylase 1 and JmjC domain family proteins involved in the regulation of gene expression (149). Lysine-specific demethylase 1 specifically demethylates histone H3K4me2, and is an important member of HDMs among other transcriptional repression complexes (133). Lysine-specific demethylase 1 utilizes oxygen as an electron acceptor to reduce methylated lysine to form lysine, formaldehyde, and hydrogen peroxide (133). JmjC demethylates mono-, di-, or trimethyl lysine residues by a different mechanism that requires cofactors, such as molecular oxygen, α-ketoglutarate, Fe2+, and ascorbate via a redox modulating process (66, 67). Aberrant expression of HDMs occurs during the course of tumor initiation and progression (76) possibly due to increased oxidative stress. However, the role of ROS/redox GSH status in modulation of HDMs in COPD and other chronic lung diseases remains unknown. Hypoxia is known to occur in tumor microenvironments, as well as in lungs of patients with COPD. The level of lysine-specific demethylase 4B is upregulated in response to hypoxia, which depends on hypoxia-inducible factor 1 alpha (176). Inhibition of H3K4 demethylase (JmjC domain-containing histone demethylation protein 1A) reduces tumor growth in vivo demonstrating its role in regulating histone methylation in hypoxia. Furthermore, induction of JmjC domain-containing histone demethylation protein 1A by hypoxia-inducible factor 1 alpha acts as an epigenetic signal amplifier to enhance hypoxic gene expression, thus facilitating tumor growth (70). In contrast, hypoxia increases global levels of H3K4me3 in alveolar A549 and bronchial Beas-2B cell lines, due to the inhibition of demethylation process, particularly, demethylase (Lysine-specific demethylase 5A). Therefore, hypoxia targets lysine-specific demethylase 5A, which induces H3K4me3 both globally and at gene-specific promoters (heme oxygenase-1 and decay-accelerating factor), thus promoting alterations in the pattern of gene expression and tumor progression (194). Understanding the molecular epigenetic mechanism that involves HDMs in COPD/lung cancer will influence treatment options and outcomes in COPD, lung cancer, and other chronic lung diseases associated with tobacco smoking (139).
DNA Methylation
DNA methylation is a nonhistone epigenetic event that plays an important role in transcriptional regulation, for example, gene silencing. Methylation occurs on cytosine residues, producing 5-meC (5-methylcytosine) (142). DNA methylation is associated with epigenetic silencing, and this effect is, in part, mediated by recruitment of HDACs through the methyl-DNA-binding motifs, including components of several HDAC-containing complexes (97). Environmental factors/agents that trigger oxidative stress, such as diet, genetic predisposition factors, and aging can gradually affect the promoter CpG methylation by recruiting methyl CpG-binding protein 2 and DNA methytransferases to various promoters along with HDACs. This leads to alterations in the expression of tumor suppressor, onocogenes, and pro- and anti-inflammatory genes. Gene-silencing occurs in several genes (e.g., GDNF, MTHFR, OPCML, TNFRSF25, TCF21, PAX8, PTPRN2, and PITX2) due to promoter-specific hypermethylation in lung cancer (7, 12). However, the relationship between gene-specific DNA methylation and smoking history/disease progression is not known (154). Georgiou et al. reported methylation of the p16 promoter in sputum of patients with COPD that significantly correlated with heavy cigarette smoking, suggesting DNA methylation is associated with CS/oxidant-mediated lung diseases (45). Array-based methylation screening from two family-based cohorts (n=1,085 and 396 subjects) revealed 349 CpG sites significantly associated with the presence and severity of COPD. Significant association was shown between SERPINA1 hypomethylation and pathogenesis of COPD with lower lung function. Similarly, Philibert et al. have recently shown that smoking altered DNA methylation in alveolar macrophages correlates with vascular endothelial growth factor 1 gene expression (107). This implicates that DNA methylation/hypomethylation that is caused by oxidative stress in specific genes may be an epigenetic event in pathogenesis of COPD (111).
CS affects DNA methylation on several genes on their CpG island directly or indirectly. CS downregulates DNA methyltransferase 3B in alveolar A549 epithelial cells resulting in demethylation of the CpG island in the oncogene synuclein-gamma and leading to high expression of this prometastatic oncogene in A549 cells (78). Sood et al. identified the promoter methylation from sputum samples of patients demonstrating wood smoke (biomass fuel burning) exposure associated with the COPD phenotype (decline in lung function, airflow obstruction, and chronic bronchitis), and smokers with aberrant p16 and GATA binding protein 4 methylation (136). It would be interesting to conduct studies on identifying epigenetic changes in DNA isolated from chronic CS or biomass/wood smoke exposed mouse lungs to identify the genes that are specifically modified by CS and/or biomass/wood smoke exposure that can be used as better biomarkers in smoke-mediated chronic lung diseases. Suzuki et al. demonstrated methylation of IL-12Rb2 and Wnt inhibitory factor-1 in COPD patients compared to the non-COPD group, suggesting the molecular epigenetic events that influence COPD-related nonsmall cell lung cancer 141). Another report described the association of prenatal exposure to tobacco smoke with significant changes in two types of DNA methylation (global methylation and promoter CpG island methylation) (18). Furthermore, children exposed to second-hand tobacco smoke had a lower methylation profile of their AluYb8 repetitive elements compared to unexposed children. Genetic susceptibility plays a vital role in smoking related effects based on differences in gene variants (methylation of LINE1 with the common GSTM1 null genotype and CpG-specific methylation with a common GSTP1 haplotype) among children involved in detoxification of tobacco smoke. This supports that epigenetic effects of in utero exposures may be due to alterations in DNA methylation patterns (18). Coordinated changes in DNA methylation, particularly, at the aryl hydrocarbon receptor repressor was observed in lymphoblasts and alveolar macrophages from smokers suggesting the role of epigenetic effects mediated by CS on carcinogenesis and other related comorbidities (92). The DNA methylation status on susceptibility genes may have implications in smokers, who are at high risk for COPD development. These studies suggest that DNA methylation may play an important role in the regulation of proinflammatory response. The role of various histone modifications versus promoter DNA methylation, particularly, in response to oxidative stress in inflammatory conditions, and in pathogenesis of COPD and other smoking related airway diseases is an emerging field of further research.
Translational Impact of Chromatin Remodeling in COPD and Smoking Related Diseases
COPD is predominantly a tobacco smoke-triggered disease characterized by premature lung aging and steroid resistance, which is, in part, due to CS-mediated chromatin modifications. The translational impact of chromatin remodeling in the pathogenesis of COPD and other smoking related diseases is as follows: (i) CS-mediated ROS production activates various kinases leading to histone acetylation and chromatin remodeling on proinflammatory genes. Inhibition of HATs by small molecule inhibitors would inhibit lung inflammation. (ii) HDAC2 reduction is associated with steroid resistance in the inflammatory response by CS-mediated oxidative stress. Activation of HDAC2 by theophylline and curcumin would reverse steroid resistance, and hence, provide protection against lung inflammatory response. (iii) SIRT1 undergoes oxidative/carbonyl post-translational modifications leading to degradation in response to CS. SIRT1 regulates several biological processes via FOXO3 and NF-κB-dependent mechanisms, which are involved in lung inflammaging. Activation of SIRT1 by pharmacological agents would protect lung cells against premature senescence and inflammaging. (iv) HDAC2-dependent epigenetic mechanism may be associated with DNA damage-induced premature senescence and associated secretory senescence phenotype in response to oxidative stress imposed by smoke. (v) CS/oxidative stress causes DNA damage, which is increased in patients with COPD/emphysema. Persistent DNA damage can lead to premature senescence with increased proinflammatory mediators release by secretory senescent cells via forming persistent DNA damage foci, termed as DNA segments with chromatin alterations reinforcing senescence (distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion). (vi) Chromatin remodeling, including histone modifications (e.g., histone acetylation/deacetylation and methylation/demethylation) are shown to regulate cellular senescence. Histone acetylation and NF-κB-dependent proinflammatory cytokines are increased in CS-exposed rodent lungs and in lungs of patients with COPD. Histone modifications are associated with DNA damage/repair and epigenomic instability as well as premature lung aging, which has implications in pathogenesis of COPD. The chromatin modification enzymes responsible for histone methylation/demethylation and acetylation/deacetylation (e.g., HDACs) also regulate cellular senescence and aging. Overall, reversing the CS-induced aberrant chromatin modifications on proinflammatory and prosenescent genes would have effects on attenuating the premature lung aging in the progression of COPD.
Conclusions and Future Directions
Oxidative stress by CS/oxidants is critical for the chronic lung inflammatory response via activation of stress kinases, redox-sensitive transcription factors, and modulation of epigenetic chromatin modifications, which result in gene transcription. Recent studies have focused on identifying genes that undergo epigenetic modifications; it is increasingly important to better understand the molecular mechanism that controls the overall epigenetic state of a cell, particularly, in response to environmental stresses. Another key area that will gain more importance is to understand whether these histone modifications enzymes, such as HATs/HDACs and HMTs/HDMs share any common targets, so as to design a common therapeutic agent. Recent studies have highlighted the pharmacological significance of HDACs and SIRT1 in regulation of lung inflammation. Identification of specific substrates/molecules targeted by HDACs and SIRT1 may have considerable therapeutic implications against chronic inflammatory lung diseases. Histone acetylation and deacetylation, as well as histone/DNA methylation play an important role in oxidative stress-induced chronic inflammatory diseases, such as asthma, COPD, and other smoking induced diseases. HDAC2 is required for glucocorticoids to function as anti-inflammatory agents. The level/activity of HDAC2 is reduced in inflammatory cells and lungs of patients with severe asthma and COPD (114, 187). Activation of HDAC2 or reversal of oxidative post-translational modifications of HDAC2 is another avenue to devise epigenetic-based therapy in treatment of severe asthma and COPD. Thus, understanding the molecular epigenetic redox regulation mechanisms will enable the development of newer epigenetic-based therapeutic strategies in the treatment of chronic inflammatory lung diseases.
Cross-talk exists between various epigenetic markers on histones and DNA methylation. Future studies will help to understand the extent to which these post-translational modifications, which are regulated by ROS in inflammation and steroid resistance, are linked to chronic inflammatory lung diseases associated with cigarette smoking. Studies on ROS and redox epigenetic regulation will further unravel the mechanisms and identify possible epigenetics-based therapies for chronic inflammatory diseases, particularly, COPD and smoking induced diseases.
Abbreviations Used
- CBP
CREB-binding protein
- COPD
chronic obstructive pulmonary disease
- CS
cigarette smoke
- FOXO3
forkhead box O3
- GR
glucocorticoid receptor
- GSH
glutathione
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
histone demethylase
- HMT
histone methytransferase
- JNK
c-Jun N-terminal kinase
- NF-κB
nuclear factor κB
- PI3K
phosphoinositide 3-kinase
- PKC
protein kinase C
- ROS
reactive oxygen species
- SIPS
stress-induced premature senescence
- SIRT1
sirtuin 1
- TSA
trichostatin A
Acknowledgments
This study was supported by NIH 1R01HL085613, 1R01HL097751, 1R01HL092842, and NIEHS Environmental Health Science Center grant P30-ES01247.
References
- 1.Acosta JC. O'Loghlen A. Banito A. Guijarro MV. Augert A. Raguz S. Fumagalli M. Da Costa M. Brown C. Popov N. Takatsu Y. Melamed J. d'Adda di Fagagna F. Bernard D. Hernando E. Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 2.Adenuga D. Caito S. Yao H. Sundar IK. Hwang JW. Chung S. Rahman I. Nrf2 deficiency influences susceptibility to steroid resistance via HDAC2 reduction. Biochem Biophys Res Commun. 2010;403:452–456. doi: 10.1016/j.bbrc.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adenuga D. Rahman I. Protein kinase CK2-mediated phosphorylation of HDAC2 regulates co-repressor formation, deacetylase activity and acetylation of HDAC2 by cigarette smoke and aldehydes. Arch Biochem Biophys. 2010;498:62–73. doi: 10.1016/j.abb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adenuga D. Yang SR. Rajendrasozhan S. Rahman I. HDAC2 degradation is associated with increased hyperphosphorylation and a proteasome-dependent mechanism in response to cigarette smoke in macrophages. Am J Respir Crit Care Med. 2008;177:A866. [Google Scholar]
- 5.Adenuga D. Yao H. March TH. Seagrave J. Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am J Respir Cell Mol Biol. 2009;40:464–473. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adler V. Yin Z. Tew KD. Ronai Z. Role of redox potential and reactive oxygen species in stress signaling. Oncogene. 1999;18:6104–6111. doi: 10.1038/sj.onc.1203128. [DOI] [PubMed] [Google Scholar]
- 7.Anglim PP. Galler JS. Koss MN. Hagen JA. Turla S. Campan M. Weisenberger DJ. Laird PW. Siegmund KD. Laird-Offringa IA. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer. 2008;7:62. doi: 10.1186/1476-4598-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arif M. Pradhan SK. Thanuja GR. Vedamurthy BM. Agrawal S. Dasgupta D. Kundu TK. Mechanism of p300 specific histone acetyltransferase inhibition by small molecules. J Med Chem. 2009;52:267–277. doi: 10.1021/jm800657z. [DOI] [PubMed] [Google Scholar]
- 9.Arrowsmith CH. Bountra C. Fish PV. Lee K. Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 10.Arunachalam G. Yao H. Sundar IK. Caito S. Rahman I. SIRT1 regulates oxidant- and cigarette smoke-induced eNOS acetylation in endothelial cells: role of resveratrol. Biochem Biophys Res Commun. 2010;393:66–72. doi: 10.1016/j.bbrc.2010.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannister AJ. Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 12.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 13.Bertos NR. Wang AH. Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 14.Bhavsar P. Khorasani N. Hew M. Johnson M. Chung KF. Effect of p38 MAPK inhibition on corticosteroid suppression of cytokine release in severe asthma. Eur Respir J. 2010;35:750–756. doi: 10.1183/09031936.00071309. [DOI] [PubMed] [Google Scholar]
- 15.Biswas SK. Rahman I. Environmental toxicity, redox signaling and lung inflammation: the role of glutathione. Mol Aspects Med. 2009;30:60–76. doi: 10.1016/j.mam.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bordone L. Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 17.Brandl A. Heinzel T. Krämer OH. Histone deacetylases: salesmen and customers in the post-translational modification market. Biology of the Cell. 2009;101:193–205. doi: 10.1042/BC20080158. [DOI] [PubMed] [Google Scholar]
- 18.Breton CV. Byun HM. Wenten M. Pan F. Yang A. Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler JS. Koutelou E. Schibler AC. Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caito S. Rajendrasozhan S. Cook S. Chung S. Yao H. Friedman AE. Brookes PS. Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caramori G. Adcock IM. Casolari P. Ito K. Jazrawi E. Tsaprouni L. Villetti G. Civelli M. Carnini C. Chung KF. Barnes PJ. Papi A. Unbalanced oxidant-induced DNA damage and repair in COPD: a link towards lung cancer. Thorax. 2011;66:521–527. doi: 10.1136/thx.2010.156448. [DOI] [PubMed] [Google Scholar]
- 22.Carrero P. Okamoto K. Coumailleau P. O'Brien S. Tanaka H. Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceylan E. Kocyigit A. Gencer M. Aksoy N. Selek S. Increased DNA damage in patients with chronic obstructive pulmonary disease who had once smoked or been exposed to biomass. Respir Med. 2006;100:1270–1276. doi: 10.1016/j.rmed.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Chen L. Fischle W. Verdin E. Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 25.Chen LF. Mu Y. Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung P. Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19:563–573. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 27.Chung S. Sundar IK. Hwang JW. Yull FE. Blackwell TS. Kinnula VL. Bulger M. Yao H. Rahman I. NF-kappaB Inducing Kinase, NIK Mediates Cigarette Smoke/TNFalpha-Induced Histone Acetylation and Inflammation through Differential Activation of IKKs. PLoS One. 2011;6:e23488. doi: 10.1371/journal.pone.0023488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung S. Sundar IK. Yao H. Ho YS. Rahman I. Glutaredoxin 1 regulates cigarette smoke-mediated lung inflammation through differential modulation of I{kappa}B kinases in mice: impact on histone acetylation. Am J Physiol Lung Cell Mol Physiol. 2010;299:L192–L203. doi: 10.1152/ajplung.00426.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coppe JP. Patil CK. Rodier F. Sun Y. Munoz DP. Goldstein J. Nelson PS. Desprez PY. Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cosio BG. Tsaprouni L. Ito K. Jazrawi E. Adcock IM. Barnes PJ. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J Exp Med. 2004;200:689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csiszar A. Labinskyy N. Jo H. Ballabh P. Ungvari Z. Differential proinflammatory and prooxidant effects of bone morphogenetic protein-4 in coronary and pulmonary arterial endothelial cells. Am J Physiol Heart Circ Physiol. 2008;295:H569–H577. doi: 10.1152/ajpheart.00180.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Ruijter AJ. van Gennip AH. Caron HN. Kemp S. van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deslee G. Woods JC. Moore C. Conradi SH. Gierada DS. Atkinson JJ. Battaile JT. Liu L. Patterson GA. Adair-Kirk TL. Holtzman MJ. Pierce RA. Oxidative damage to nucleic acids in severe emphysema. Chest. 2009;135:965–974. doi: 10.1378/chest.08-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doukas J. Eide L. Stebbins K. Racanelli-Layton A. Dellamary L. Martin M. Dneprovskaia E. Noronha G. Soll R. Wrasidlo W. Acevedo LM. Cheresh DA. Aerosolized phosphoinositide 3-kinase gamma/delta inhibitor TG100-115 [3-[2,4-diamino-6-(3-hydroxyphenyl)pteridin-7-yl]phenol] as a therapeutic candidate for asthma and chronic obstructive pulmonary disease. J Pharmacol Exp Ther. 2009;328:758–765. doi: 10.1124/jpet.108.144311. [DOI] [PubMed] [Google Scholar]
- 35.Edirisinghe I. Yang SR. Yao H. Rajendrasozhan S. Caito S. Adenuga D. Wong C. Rahman A. Phipps RP. Jin ZG. Rahman I. VEGFR-2 inhibition augments cigarette smoke-induced oxidative stress and inflammatory responses leading to endothelial dysfunction. FASEB J. 2008;22:2297–2310. doi: 10.1096/fj.07-099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elkon R. Rashi-Elkeles S. Lerenthal Y. Linhart C. Tenne T. Amariglio N. Rechavi G. Shamir R. Shiloh Y. Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol. 2005;6:R43. doi: 10.1186/gb-2005-6-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelman JA. Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 38.Ferrer E. Peinado VI. Diez M. Carrasco JL. Musri MM. Martinez A. Rodriguez-Roisin R. Barbera JA. Effects of cigarette smoke on endothelial function of pulmonary arteries in the guinea pig. Respir Res. 2009;10:76–87. doi: 10.1186/1465-9921-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraga MF. Ballestar E. Villar-Garea A. Boix-Chornet M. Espada J. Schotta G. Bonaldi T. Haydon C. Ropero S. Petrie K. Iyer NG. Perez-Rosado A. Calvo E. Lopez JA. Cano A. Calasanz MJ. Colomer D. Piris MA. Ahn N. Imhof A. Caldas C. Jenuwein T. Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 40.Freund A. Orjalo AV. Desprez PY. Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda H. Sano N. Muto S. Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- 42.Furukawa A. Tada-Oikawa S. Kawanishi S. Oikawa S. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD+ depletion. Cell Physiol Biochem. 2007;20:45–54. doi: 10.1159/000104152. [DOI] [PubMed] [Google Scholar]
- 43.Galasinski SC. Resing KA. Goodrich JA. Ahn NG. Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J Biol Chem. 2002;277:19618–19626. doi: 10.1074/jbc.M201174200. [DOI] [PubMed] [Google Scholar]
- 44.Gao L. Cueto MA. Asselbergs F. Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 45.Georgiou E. Valeri R. Tzimagiorgis G. Anzel J. Krikelis D. Tsilikas C. Sarikos G. Destouni C. Dimitriadou A. Kouidou S. Aberrant p16 promoter methylation among Greek lung cancer patients and smokers: correlation with smoking. Eur J Cancer Prev. 2007;16:396–402. doi: 10.1097/01.cej.0000236260.26265.d6. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh S. Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 47.Gilliland FD. Harms HJ. Crowell RE. Li YF. Willink R. Belinsky SA. Glutathione S-transferase P1 and NADPH quinone oxidoreductase polymorphisms are associated with aberrant promoter methylation of P16(INK4a) and O(6)-methylguanine-DNA methyltransferase in sputum. Cancer Res. 2002;62:2248–2252. [PubMed] [Google Scholar]
- 48.Gilmour PS. Rahman I. Donaldson K. MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am J Physiol Lung Cell Mol Physiol. 2003;284:L533–L540. doi: 10.1152/ajplung.00277.2002. [DOI] [PubMed] [Google Scholar]
- 49.Gosden RG. Feinberg AP. Genetics and epigenetics—nature's pen-and-pencil set. N Engl J Med. 2007;356:731–733. doi: 10.1056/NEJMe068284. [DOI] [PubMed] [Google Scholar]
- 50.Grozinger CM. Hassig CA. Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gustafson AM. Soldi R. Anderlind C. Scholand MB. Qian J. Zhang X. Cooper K. Walker D. McWilliams A. Liu G. Szabo E. Brody J. Massion PP. Lenburg ME. Lam S. Bild AH. Spira A. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hew M. Bhavsar P. Torrego A. Meah S. Khorasani N. Barnes PJ. Adcock I. Chung KF. Relative corticosteroid insensitivity of peripheral blood mononuclear cells in severe asthma. Am J Respir Crit Care Med. 2006;174:134–141. doi: 10.1164/rccm.200512-1930OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoshino S. Yoshida M. Inoue K. Yano Y. Yanagita M. Mawatari H. Yamane H. Kijima T. Kumagai T. Osaki T. Tachiba I. Kawase I. Cigarette smoke extract induces endothelial cell injury via JNK pathway. Biochem Biophys Res Commun. 2005;329:58–63. doi: 10.1016/j.bbrc.2005.01.095. [DOI] [PubMed] [Google Scholar]
- 54.Hwang JW. Chung S. Sundar IK. Yao H. Arunachalam G. McBurney MW. Rahman I. Cigarette smoke-induced autophagy is regulated by SIRT1-PARP-1-dependent mechanism: implication in pathogenesis of COPD. Arch Biochem Biophys. 2010;500:203–209. doi: 10.1016/j.abb.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang JW. Rajendrasozhan S. Yao H. Chung S. Sundar IK. Huyck HL. Pryhuber GS. Kinnula VL. Rahman I. FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. J Immunol. 2011;187:987–998. doi: 10.4049/jimmunol.1001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imhof A. Wolffe AP. Transcription: gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- 57.Ito K. Barnes PJ. Adcock IM. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol Cell Biol. 2000;20:6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito K. Charron CE. Adcock IM. Impact of protein acetylation in inflammatory lung diseases. Pharmacol Ther. 2007;116:249–265. doi: 10.1016/j.pharmthera.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Ito K. Hanazawa T. Tomita K. Barnes PJ. Adcock IM. Oxidative stress reduces histone deacetylase 2 activity and enhances IL-8 gene expression: role of tyrosine nitration. Biochem Biophys Res Commun. 2004;315:240–245. doi: 10.1016/j.bbrc.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 60.Ito K. Ito M. Elliott WM. Cosio B. Caramori G. Kon OM. Barczyk A. Hayashi S. Adcock IM. Hogg JC. Barnes PJ. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 61.Ito K. Yamamura S. Essilfie-Quaye S. Cosio B. Ito M. Barnes PJ. Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 63.Kagoshima M. Wilcke T. Ito K. Tsaprouni L. Barnes PJ. Punchard N. Adcock IM. Glucocorticoid-mediated transrepression is regulated by histone acetylation and DNA methylation. Eur J Pharmacol. 2001;429:327–334. doi: 10.1016/s0014-2999(01)01332-2. [DOI] [PubMed] [Google Scholar]
- 64.Kamei Y. Xu L. Heinzel T. Torchia J. Kurokawa R. Gloss B. Lin SC. Heyman RA. Rose DW. Glass CK. Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 65.Kawahara TL. Michishita E. Adler AS. Damian M. Berber E. Lin M. McCord RA. Ongaigui KC. Boxer LD. Chang HY. Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klose RJ. Kallin EM. Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 67.Klose RJ. Yamane K. Bae Y. Zhang D. Erdjument-Bromage H. Tempst P. Wong J. Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 68.Kooistra SM. Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 69.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 70.Krieg AJ. Rankin EB. Chan D. Razorenova O. Fernandez S. Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuilman T. Michaloglou C. Vredeveld LC. Douma S. van Doorn R. Desmet CJ. Aarden LA. Mooi WJ. Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 72.Kuo MH. Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 73.Lachner M. Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 74.Lee J. Reddy R. Barsky L. Scholes J. Chen H. Shi W. Driscoll B. Lung alveolar integrity is compromised by telomere shortening in telomerase-null mice. Am J Physiol Lung Cell Mol Physiol. 2009;296:L57–L70. doi: 10.1152/ajplung.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee KY. Ito K. Hayashi R. Jazrawi EP. Barnes PJ. Adcock IM. NF-kappaB and activator protein 1 response elements and the role of histone modifications in IL-1beta-induced TGF-beta1 gene transcription. J Immunol. 2006;176:603–615. doi: 10.4049/jimmunol.176.1.603. [DOI] [PubMed] [Google Scholar]
- 76.Lim S. Metzger E. Schule R. Kirfel J. Buettner R. Epigenetic regulation of cancer growth by histone demethylases. Int J Cancer. 2010;127:1991–1998. doi: 10.1002/ijc.25538. [DOI] [PubMed] [Google Scholar]
- 77.Liu F. Killian JK. Yang M. Walker RL. Hong JA. Zhang M. Davis S. Zhang Y. Hussain M. Xi S. Rao M. Meltzer PA. Schrump DS. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu H. Zhou Y. Boggs SE. Belinsky SA. Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26:5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 79.Londhe VA. Sundar IK. Lopez B. Maisonet TM. Yu Y. Aghai ZH. Rahman I. Hyperoxia impairs alveolar formation and induces senescence through decreased histone deacetylase activity and up-regulation of p21 in neonatal mouse lung. Pediatr Res. 2011;69:371–377. doi: 10.1203/PDR.0b013e318211c917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo J. Li M. Tang Y. Laszkowska M. Roeder RG. Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mao Z. Hine C. Tian X. Van Meter M. Au M. Vaidya A. Seluanov A. Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marwick JA. Caramori G. Stevenson CS. Casolari P. Jazrawi E. Barnes PJ. Ito K. Adcock IM. Kirkham PA. Papi A. Inhibition of PI3Kdelta restores glucocorticoid function in smoking-induced airway inflammation in mice. Am J Respir Crit Care Med. 2009;179:542–548. doi: 10.1164/rccm.200810-1570OC. [DOI] [PubMed] [Google Scholar]
- 83.Marwick JA. Kirkham PA. Stevenson CS. Danahay H. Giddings J. Butler K. Donaldson K. Macnee W. Rahman I. Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs. Am J Respir Cell Mol Biol. 2004;31:633–642. doi: 10.1165/rcmb.2004-0006OC. [DOI] [PubMed] [Google Scholar]
- 84.Mattagajasingh I. Kim CS. Naqvi A. Yamamori T. Hoffman TA. Jung SB. DeRicco J. Kasuno K. Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meja KK. Rajendrasozhan S. Adenuga D. Biswas SK. Sundar IK. Spooner G. Marwick JA. Chakravarty P. Fletcher D. Whittaker P. Megson IL. Kirkham PA. Rahman I. Curcumin restores corticosteroid function in monocytes exposed to oxidants by maintaining HDAC2. Am J Respir Cell Mol Biol. 2008;39:312–323. doi: 10.1165/rcmb.2008-0012OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michan S. Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michaud SE. Dussault S. Haddad P. Groleau J. Rivard A. Circulating endothelial progenitor cells from healthy smokers exhibit impaired functional activities. Atherosclerosis. 2006;187:423–432. doi: 10.1016/j.atherosclerosis.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 88.Michishita E. McCord RA. Berber E. Kioi M. Padilla-Nash H. Damian M. Cheung P. Kusumoto R. Kawahara TL. Barrett JC. Chang HY. Bohr VA. Ried T. Gozani O. Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller KM. Tjeertes JV. Coates J. Legube G. Polo SE. Britton S. Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Milne JC. Denu JM. The Sirtuin family: therapeutic targets to treat diseases of aging. Curr Opin Chem Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 91.Milne JC. Lambert PD. Schenk S. Carney DP. Smith JJ. Gagne DJ. Jin L. Boss O. Perni RB. Vu CB. Bemis JE. Xie R. Disch JS. Ng PY. Nunes JJ. Lynch AV. Yang H. Galonek H. Israelian K. Choy W. Iffland A. Lavu S. Medvedik O. Sinclair DA. Olefsky JM. Jirousek MR. Elliott PJ. Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monick MM. Beach SR. Plume J. Sears R. Gerrard M. Brody GH. Philibert RA. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moodie FM. Marwick JA. Anderson CS. Szulakowski P. Biswas SK. Bauter MR. Kilty I. Rahman I. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. FASEB J. 2004;18:1897–1899. doi: 10.1096/fj.04-1506fje. [DOI] [PubMed] [Google Scholar]
- 94.Motta MC. Divecha N. Lemieux M. Kamel C. Chen D. Gu W. Bultsma Y. McBurney M. Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 95.Muller KC. Welker L. Paasch K. Feindt B. Erpenbeck VJ. Hohlfeld JM. Krug N. Nakashima M. Branscheid D. Magnussen H. Jorres RA. Holz O. Lung fibroblasts from patients with emphysema show markers of senescence in vitro. Respir Res. 2006;7:32. doi: 10.1186/1465-9921-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakamaru Y. Vuppusetty C. Wada H. Milne JC. Ito M. Rossios C. Elliot M. Hogg J. Kharitonov S. Goto H. Bemis JE. Elliott P. Barnes PJ. Ito K. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. FASEB J. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- 97.Nan X. Ng HH. Johnson CA. Laherty CD. Turner BM. Eisenman RN. Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 98.Ng HH. Zhang Y. Hendrich B. Johnson CA. Turner BM. Erdjument-Bromage H. Tempst P. Reinberg D. Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 99.Nicodeme E. Jeffrey KL. Schaefer U. Beinke S. Dewell S. Chung CW. Chandwani R. Marazzi I. Wilson P. Coste H. White J. Kirilovsky J. Rice CM. Lora JM. Prinjha RK. Lee K. Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nisoli E. Tonello C. Cardile A. Cozzi V. Bracale R. Tedesco L. Falcone S. Valerio A. Cantoni O. Clementi E. Moncada S. Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 101.Nyunoya T. Monick MM. Klingelhutz A. Yarovinsky TO. Cagley JR. Hunninghake GW. Cigarette smoke induces cellular senescence. Am J Respir Cell Mol Biol. 2006;35:681–688. doi: 10.1165/rcmb.2006-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nyunoya T. Monick MM. Klingelhutz AL. Glaser H. Cagley JR. Brown CO. Matsumoto E. Aykin-Burns N. Spitz DR. Oshima J. Hunninghake GW. Cigarette smoke induces cellular senescence via Werner's syndrome protein down-regulation. Am J Respir Crit Care Med. 2009;179:279–287. doi: 10.1164/rccm.200802-320OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogiwara H. Ui A. Otsuka A. Satoh H. Yokomi I. Nakajima S. Yasui A. Yokota J. Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- 104.Ota H. Akishita M. Eto M. Iijima K. Kaneki M. Ouchi Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. J Mol Cell Cardiol. 2007;43:571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 105.Ota H. Eto M. Kano MR. Ogawa S. Iijima K. Akishita M. Ouchi Y. Cilostazol inhibits oxidative stress-induced premature senescence via upregulation of Sirt1 in human endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:1634–1639. doi: 10.1161/ATVBAHA.108.164368. [DOI] [PubMed] [Google Scholar]
- 106.Pastukh VM. Zhang L. Ruchko MV. Gorodnya O. Bardwell GC. Tuder RM. Gillespie MN. Oxidative DNA damage in lung tissue from patients with COPD is clustered in functionally significant sequences. Int J Chron Obstruct Pulmon Dis. 2011;6:209–217. doi: 10.2147/COPD.S15922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Philibert RA. Sears RA. Powers LS. Nash E. Bair T. Gerke AK. Hassan I. Thomas CP. Gross TJ. Monick MM. Coordinated DNA methylation and gene expression changes in smoker alveolar macrophages: specific effects on VEGF receptor 1 expression. J Leukoc Biol. 2012;92:621–631. doi: 10.1189/jlb.1211632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pinamonti S. Leis M. Barbieri A. Leoni D. Muzzoli M. Sostero S. Chicca MC. Carrieri A. Ravenna F. Fabbri LM. Ciaccia A. Detection of xanthine oxidase activity products by EPR and HPLC in bronchoalveolar lavage fluid from patients with chronic obstructive pulmonary disease. Free Radic Biol Med. 1998;25:771–779. doi: 10.1016/s0891-5849(98)00128-2. [DOI] [PubMed] [Google Scholar]
- 109.Potente M. Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 110.Potente M. Dimmeler S. NO targets SIRT1: a novel signaling network in endothelial senescence. Arterioscler Thromb Vasc Biol. 2008;28:1577–1579. doi: 10.1161/ATVBAHA.108.173682. [DOI] [PubMed] [Google Scholar]
- 111.Qiu W. Baccarelli A. Carey VJ. Boutaoui N. Bacherman H. Klanderman B. Rennard S. Agusti A. Anderson W. Lomas DA. DeMeo DL. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med. 2012;185:373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Racey LA. Byvoet P. Histone acetyltransferase in chromatin. Evidence for in vitro enzymatic transfer of acetate from acetyl-coenzyme A to histones. Exp Cell Res. 1971;64:366–370. doi: 10.1016/0014-4827(71)90089-9. [DOI] [PubMed] [Google Scholar]
- 113.Rahman I. The role of oxidative stress in the pathogenesis of COPD: implications for therapy. Treat Respir Med. 2005;4:175–200. doi: 10.2165/00151829-200504030-00003. [DOI] [PubMed] [Google Scholar]
- 114.Rahman I. Adcock IM. Oxidative stress and redox regulation of lung inflammation in COPD. Eur Respir J. 2006;28:219–242. doi: 10.1183/09031936.06.00053805. [DOI] [PubMed] [Google Scholar]
- 115.Rahman I. Gilmour PS. Jimenez LA. MacNee W. Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol Cell Biochem. 2002:234–235. 239–248. [PubMed] [Google Scholar]
- 116.Rahman I. Kinnula VL. Gorbunova V. Yao H. SIRT1 as a therapeutic target in inflammaging of the pulmonary disease. Prev Med. 2012;54(Suppl):S20–S28. doi: 10.1016/j.ypmed.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rahman I. MacNee W. Oxidant/antioxidant imbalance in smokers and chronic obstructive pulmonary disease. Thorax. 1996;51:348–350. doi: 10.1136/thx.51.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rahman I. MacNee W. Role of transcription factors in inflammatory lung diseases. Thorax. 1998;53:601–612. doi: 10.1136/thx.53.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rahman I. Marwick J. Kirkham P. Redox modulation of chromatin remodeling: impact on histone acetylation and deacetylation, NF-kappaB and pro-inflammatory gene expression. Biochem Pharmacol. 2004;68:1255–1267. doi: 10.1016/j.bcp.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 120.Rahman I. van Schadewijk AA. Crowther AJ. Hiemstra PS. Stolk J. MacNee W. De Boer WI. 4-Hydroxy-2-nonenal, a specific lipid peroxidation product, is elevated in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;166:490–495. doi: 10.1164/rccm.2110101. [DOI] [PubMed] [Google Scholar]
- 121.Rajendrasozhan S. Yang SR. Caito S. Rahman I. Nucleocytoplasmic shuttling and post-translational modifications of sirtuin in response to cigarette smoke lead to increased acetylation of NF-kappaB and FOXO3. Am J Respir Crit Care Med. 2008;177:A266. [Google Scholar]
- 122.Rajendrasozhan S. Yang SR. Kinnula VL. Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rajendrasozhan S. Yao H. Rahman I. Current perspectives on role of chromatin modifications and deacetylases in lung inflammation in COPD. COPD. 2009;6:291–297. doi: 10.1080/15412550903049132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Repine JE. Bast A. Lankhorst I. Oxidative stress in chronic obstructive pulmonary disease. Oxidative Stress Study Group. Am J Respir Crit Care Med. 1997;156:341–357. doi: 10.1164/ajrccm.156.2.9611013. [DOI] [PubMed] [Google Scholar]
- 125.Rodier F. Coppe JP. Patil CK. Hoeijmakers WAM. Munoz DP. Raza SR. Freund A. Campeau E. Davalos AR. Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodier F. Munoz DP. Teachenor R. Chu V. Le O. Bhaumik D. Coppe JP. Campeau E. Beausejour CM. Kim SH. Davalos AR. Campisi J. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Roth SY. Denu JM. Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 128.Saccani S. Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sarg B. Koutzamani E. Helliger W. Rundquist I. Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 130.Sato T. Seyama K. Sato Y. Mori H. Souma S. Akiyoshi T. Kodama Y. Mori T. Goto S. Takahashi K. Fukuchi Y. Maruyama N. Ishigami A. Senescence marker protein-30 protects mice lungs from oxidative stress, aging, and smoking. Am J Respir Crit Care Med. 2006;174:530–537. doi: 10.1164/rccm.200511-1816OC. [DOI] [PubMed] [Google Scholar]
- 131.Seligson DB. Horvath S. McBrian MA. Mah V. Yu H. Tze S. Wang Q. Chia D. Goodglick L. Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sengupta N. Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 133.Shi Y. Lan F. Matson C. Mulligan P. Whetstine JR. Cole PA. Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 134.Siafakas NM. Tzortzaki EG. Sourvinos G. Bouros D. Tzanakis N. Kafatos A. Spandidos D. Microsatellite DNA instability in COPD. Chest. 1999;116:47–51. doi: 10.1378/chest.116.1.47. [DOI] [PubMed] [Google Scholar]
- 135.Song JY. Lim JW. Kim H. Morio T. Kim KH. Oxidative stress induces nuclear loss of DNA repair proteins Ku70 and Ku80 and apoptosis in pancreatic acinar AR42J cells. J Biol Chem. 2003;278:36676–36687. doi: 10.1074/jbc.M303692200. [DOI] [PubMed] [Google Scholar]
- 136.Sood A. Petersen H. Blanchette CM. Meek P. Picchi MA. Belinsky SA. Tesfaigzi Y. Wood smoke exposure and gene promoter methylation are associated with increased risk for COPD in smokers. Am J Respir Crit Care Med. 2010;182:1098–1104. doi: 10.1164/rccm.201002-0222OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suga T. Kurabayashi M. Sando Y. Ohyama Y. Maeno T. Maeno Y. Aizawa H. Matsumura Y. Kuwaki T. Kuro OM. Nabeshima Y. Nagai R. Disruption of the klotho gene causes pulmonary emphysema in mice. Defect in maintenance of pulmonary integrity during postnatal life. Am J Respir Cell Mol Biol. 2000;22:26–33. doi: 10.1165/ajrcmb.22.1.3554. [DOI] [PubMed] [Google Scholar]
- 138.Sundar IK. Chung S. Hwang JW. Lapek JD., Jr. Bulger M. Friedman AE. Yao H. Davie JR. Rahman I. Mitogen- and stress-activated kinase 1 (MSK1) regulates cigarette smoke-induced histone modifications on NF-kappaB-dependent genes. PLoS One. 2012;7:e31378. doi: 10.1371/journal.pone.0031378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sundar IK. Mullapudi N. Yao H. Spivack SD. Rahman I. Lung cancer and its association with chronic obstructive pulmonary disease: update on nexus of epigenetics. Curr Opin Pulm Med. 2011;17:279–285. doi: 10.1097/MCP.0b013e3283477533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sundar IK. Rahman I. Vitamin d and susceptibility of chronic lung diseases: role of epigenetics. Front Pharmacol. 2011;2:50. doi: 10.3389/fphar.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Suzuki M. Wada H. Yoshino M. Tian L. Shigematsu H. Suzuki H. Alaa M. Tamura H. Fujiwara T. Nagato K. Motohashi S. Moriya Y. Hoshino H. Yoshida S. Shibuya K. Hiroshima K. Nakatani Y. Yoshino I. Molecular characterization of chronic obstructive pulmonary disease-related non-small cell lung cancer through aberrant methylation and alterations of EGFR signaling. Ann Surg Oncol. 2010;17:878–888. doi: 10.1245/s10434-009-0739-3. [DOI] [PubMed] [Google Scholar]
- 142.Suzuki MM. Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 143.Szulakowski P. Crowther AJ. Jimenez LA. Donaldson K. Mayer R. Leonard TB. MacNee W. Drost EM. The effect of smoking on the transcriptional regulation of lung inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174:41–50. doi: 10.1164/rccm.200505-725OC. [DOI] [PubMed] [Google Scholar]
- 144.Tan S. One HAT size fits all? Nat Struct Biol. 2001;8:8–10. doi: 10.1038/83098. [DOI] [PubMed] [Google Scholar]
- 145.Taplick J. Kurtev V. Kroboth K. Posch M. Lechner T. Seiser C. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J Mol Biol. 2001;308:27–38. doi: 10.1006/jmbi.2001.4569. [DOI] [PubMed] [Google Scholar]
- 146.Taplick J. Kurtev V. Lagger G. Seiser C. Histone H4 acetylation during interleukin-2 stimulation of mouse T cells. FEBS Lett. 1998;436:349–352. doi: 10.1016/s0014-5793(98)01164-8. [DOI] [PubMed] [Google Scholar]
- 147.Thannickal VJ. Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 148.Thomson S. Mahadevan LC. Clayton AL. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol. 1999;10:205–214. doi: 10.1006/scdb.1999.0302. [DOI] [PubMed] [Google Scholar]
- 149.Tian X. Fang J. Current perspectives on histone demethylases. Acta Biochim Biophys Sin (Shanghai) 2007;39:81–88. doi: 10.1111/j.1745-7270.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- 150.Tjeertes JV. Miller KM. Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.To Y. Ito K. Kizawa Y. Failla M. Ito M. Kusama T. Elliott WM. Hogg JC. Adcock IM. Barnes PJ. Targeting phosphoinositide-3-kinase-delta with theophylline reverses corticosteroid insensitivity in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:897–904. doi: 10.1164/rccm.200906-0937OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tomita K. Barnes PJ. Adcock IM. The effect of oxidative stress on histone acetylation and IL-8 release. Biochem Biophys Res Commun. 2003;301:572–577. doi: 10.1016/s0006-291x(02)03029-2. [DOI] [PubMed] [Google Scholar]
- 153.Torchia J. Glass C. Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 154.Tsou JA. Hagen JA. Carpenter CL. Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21:5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 155.Tsuji T. Aoshiba K. Nagai A. Cigarette smoke induces senescence in alveolar epithelial cells. Am J Respir Cell Mol Biol. 2004;31:643–649. doi: 10.1165/rcmb.2003-0290OC. [DOI] [PubMed] [Google Scholar]
- 156.Tsuji T. Aoshiba K. Nagai A. Alveolar cell senescence in patients with pulmonary emphysema. Am J Respir Crit Care Med. 2006;174:886–893. doi: 10.1164/rccm.200509-1374OC. [DOI] [PubMed] [Google Scholar]
- 157.Tsuji T. Aoshiba K. Nagai A. Alveolar cell senescence exacerbates pulmonary inflammation in patients with chronic obstructive pulmonary disease. Respiration. 2010;80:59–70. doi: 10.1159/000268287. [DOI] [PubMed] [Google Scholar]
- 158.Van Gool F. Galli M. Gueydan C. Kruys V. Prevot PP. Bedalov A. Mostoslavsky R. Alt FW. De Smedt T. Leo O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med. 2009;15:206–210. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vaquero A. Scher M. Erdjument-Bromage H. Tempst P. Serrano L. Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 160.Vaquero A. Scher M. Lee D. Erdjument-Bromage H. Tempst P. Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 161.Vaute O. Nicolas E. Vandel L. Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vaziri H. Dessain SK. Ng Eaton E. Imai SI. Frye RA. Pandita TK. Guarente L. Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 163.Vempati RK. Jayani RS. Notani D. Sengupta A. Galande S. Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wada H. Kagoshima M. Ito K. Barnes PJ. Adcock IM. 5-Azacytidine suppresses RNA polymerase II recruitment to the SLPI gene. Biochem Biophys Res Commun. 2005;331:93–99. doi: 10.1016/j.bbrc.2005.03.138. [DOI] [PubMed] [Google Scholar]
- 165.Wagner M. Brosch G. Zwerschke W. Seto E. Loidl P. Jansen-Durr P. Histone deacetylases in replicative senescence: evidence for a senescence-specific form of HDAC-2. FEBS Lett. 2001;499:101–106. doi: 10.1016/s0014-5793(01)02524-8. [DOI] [PubMed] [Google Scholar]
- 166.Wang H. Cao R. Xia L. Erdjument-Bromage H. Borchers C. Tempst P. Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 167.Wang J. Jacob NK. Ladner KJ. Beg A. Perko JD. Tanner SM. Liyanarachchi S. Fishel R. Guttridge DC. RelA/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep. 2009;10:1272–1278. doi: 10.1038/embor.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Z. Zang C. Rosenfeld JA. Schones DE. Barski A. Cuddapah S. Cui K. Roh TY. Peng W. Zhang MQ. Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wilting RH. Yanover E. Heideman MR. Jacobs H. Horner J. van der Torre J. DePinho RA. Dannenberg JH. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wolffe AP. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 171.Workman JL. Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 172.Wright JL. Churg A. Short-term exposure to cigarette smoke induces endothelial dysfunction in small intrapulmonary arteries: analysis using guinea pig precision cut lung slices. J Appl Physiol. 2008;104:1462–1469. doi: 10.1152/japplphysiol.00520.2007. [DOI] [PubMed] [Google Scholar]
- 173.Wu ZH. Shi Y. Tibbetts RS. Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 174.Wyatt TA. Heires AJ. Sanderson SD. Floreani AA. Protein kinase C activation is required for cigarette smoke-enhanced C5a-mediated release of interleukin-8 in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1999;21:283–288. doi: 10.1165/ajrcmb.21.2.3636. [DOI] [PubMed] [Google Scholar]
- 175.Xu WS. Parmigiani RB. Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 176.Yang J. Jubb AM. Pike L. Buffa FM. Turley H. Baban D. Leek R. Gatter KC. Ragoussis J. Harris AL. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70:6456–6466. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang L. Mei Q. Zielinska-Kwiatkowska A. Matsui Y. Blackburn ML. Benedetti D. Krumm AA. Taborsky GJ., Jr. Chansky HA. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem J. 2003;369:651–657. doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang SR. Chida AS. Bauter MR. Shafiq N. Seweryniak K. Maggirwar SB. Kilty I. Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 179.Yang SR. Valvo S. Yao H. Kode A. Rajendrasozhan S. Edirisinghe I. Caito S. Adenuga D. Henry R. Fromm G. Maggirwar S. Li JD. Bulger M. Rahman I. IKK alpha causes chromatin modification on pro-inflammatory genes by cigarette smoke in mouse lung. Am J Respir Cell Mol Biol. 2008;38:689–698. doi: 10.1165/rcmb.2007-0379OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang SR. Wright J. Bauter M. Seweryniak K. Kode A. Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 181.Yang SR. Yao H. Rajendrasozhan S. Chung S. Edirisinghe I. Valvo S. Fromm G. McCabe MJ., Jr. Sime PJ. Phipps RP. Li JD. Bulger M. Rahman I. RelB is differentially regulated by IkappaB Kinase-alpha in B cells and mouse lung by cigarette smoke. Am J Respir Cell Mol Biol. 2009;40:147–158. doi: 10.1165/rcmb.2008-0207OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang T. Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8:E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yao H. Arunachalam G. Hwang JW. Chung S. Sundar IK. Kinnula VL. Crapo JD. Rahman I. Extracellular superoxide dismutase protects against pulmonary emphysema by attenuating oxidative fragmentation of ECM. Proc Natl Acad Sci U S A. 2010;107:15571–15576. doi: 10.1073/pnas.1007625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yao H. Chung S. Hwang JW. Rajendrasozhan S. Sundar IK. Dean DA. McBurney MW. Guarente L. Gu W. Ronty M. Kinnula VL. Rahman I. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest. 2012;122:2032–2045. doi: 10.1172/JCI60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yao H. Edirisinghe I. Yang SR. Rajendrasozhan S. Kode A. Caito S. Adenuga D. Rahman I. Genetic ablation of NADPH oxidase enhances susceptibility to cigarette smoke-induced lung inflammation and emphysema in mice. Am J Pathol. 2008;172:1222–1237. doi: 10.2353/ajpath.2008.070765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yao H. Hwang JW. Moscat J. Diaz-Meco MT. Leitges M. Kishore N. Li X. Rahman I. Protein kinase C zeta mediates cigarette smoke/aldehyde- and lipopolysaccharide-induced lung inflammation and histone modifications. J Biol Chem. 2010;285:5405–5416. doi: 10.1074/jbc.M109.041418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yao H. Rahman I. Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol Appl Pharmacol. 2011;254:72–85. doi: 10.1016/j.taap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yao H. Rahman I. Perspectives on translational and therapeutic aspects of SIRT1 in inflammaging and senescence. Biochem Pharmacol. 2012 doi: 10.1016/j.bcp.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.You H. Mak TW. Crosstalk between p53 and FOXO transcription factors. Cell Cycle. 2005;4:37–38. doi: 10.4161/cc.4.1.1401. [DOI] [PubMed] [Google Scholar]
- 190.Yuan J. Pu M. Zhang Z. Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang X. Yang Z. Khan SI. Horton JR. Tamaru H. Selker EU. Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang Y. Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 193.Zhou R. Han L. Li G. Tong T. Senescence delay and repression of p16INK4a by Lsh via recruitment of histone deacetylases in human diploid fibroblasts. Nucleic Acids Res. 2009;37:5183–5196. doi: 10.1093/nar/gkp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou X. Sun H. Chen H. Zavadil J. Kluz T. Arita A. Costa M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70:4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]