Abstract
Discovered as antiviral cytokines, interferons (IFNs) are now also recognized for their capacity to inhibit the growth of malignant cells via activation of programmed cell death, better known as apoptosis. In this review, we will cover recent advances made in this field, as it pertains to the various proposed mechanisms of IFN-induced apoptosis and the characterization of IFN-responsive genes not previously known to have apoptotic function. Also mentioned here is a description of the activation and crosstalk of survival signaling pathways as a mode of IFN resistance that remains a persistent clinical adversary to overcome and the future of IFNs as antitumor agents.
The Interferons
Interferons (IFNs) are naturally secreted glycoproteins produced by almost every cell type as a mechanism of host defense in response to microbial attack (Rees 1990) and tumor cells (Young 1971). They were coined “interferons”, because they protected cells by interfering with viral infection (Nagano and others 1954; Lindenmann and Isaacs 1957). Ensuing studies revealed that they had additional biological functions, including immunomodulatory, antiangiogenic, antiproliferative, and apoptotic activities. IFNs are a family of cytokines classified into 3 different groups. Type I IFNs consist of IFN-α (comprised of 13 subtypes), IFN-β, IFN-κ, IFN-δ, IFN-ε, IFN-τ, IFN-ω, and IFN-ξ. Type II IFN contains one member, IFN-γ. Type III IFNs consist of 3 members: IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28A) (Kontsek and others 2003; Vilcek 2003). Each type of IFN interacts with its specific cell surface receptor complex. Nearly every cell expresses receptors for type I and type II IFNs, whereas receptors for type III IFNs are cell type-restricted (Lasfar and others 2011).
Elucidation of how IFNs transmit signals inside cells to exert their biological effects is credited to the joint efforts of Stark and Darnell (2012), who uncovered the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway. Extracellular binding of IFN to its receptor causes changes in the receptor structure, allowing JAKs associated with the intracellular portion of the receptor subunits to transphosphorylate one another and become activated. Activated JAKs then tyrosine-phosphorylate the receptor and STAT proteins. As dimers, activated STATs localize from the cytoplasm to the nucleus and bind to IFN-responsive DNA elements named ISRE and GAS to initiate transcription of IFN-stimulated genes (ISGs).
IFN-Induced Apoptosis of Malignant Cells
All 3 types of IFNs can induce apoptosis of tumor cells (Chawla-Sarkar and others 2001; Steen and Gamero 2010). Tumor cells exposed to IFNs show the classical features of apoptosis such as cell shrinkage, membrane blebbing, chromatin condensation, and DNA fragmentation (Lokshin and others 1995). It is important to remark that not all tumor cells, irrespective of their origin, are susceptible to the apoptotic effects of IFNs (Trubiani and others 1994; Rodríguez-Villanueva and McDonnell 1995). This suggests the existence of unique transcriptional signatures present in tumor cells susceptible to IFN-induced apoptosis that are distinctive from tumor cells that are resistant to or growth inhibited by IFNs.
Endothelial cells (ECs) present within the tumor microenvironment can contribute indirectly to the apoptotic effects of IFNs. Apoptosis of ECs by IFNs causes restriction of blood flow within the tumor vasculature, leading to shrinkage of tumors (Zhang and others 2010). This indirect antiangiogenic effect of IFNs is a highly desirable outcome against tumors that rely on the tumor vasculature to disseminate to other organs. Contributing factors to this effect are CXC chemokines, particularly CXCL10 (also an ISG), which promotes its antitumor effects via a decrease in microvessel density as observed in melanoma tumor xenografts (Feldman and others 2006).
Another unique cell population that has attracted recent attention is cancer stem cells (CSCs). They represent a small fraction of cells in a tumor. CSCs are reported to be one of the causes of recurrence of various cancers that acquire resistance to anticancer drugs and/or radiation therapy (Hemmings 2010). Recently, a side population with stem cell features isolated from primary ovarian cancer samples was shown to be drastically reduced by IFN-α exposure (Moserle and others 2008). This observation was reproduced in vivo, in which IFN-α treatment caused regression of established tumors in mice that contained a large fraction of this side population. In a different study, IFN-α robustly induced apoptosis of paclitaxel (PTX)-resistant KF28TX ovarian cancer cells, while a marginal effect was seen in the parental PTX-sensitive KF28 cells. This marked difference in IFN response was attributed to KF28TX being enriched in a resistant CSC side population (Kobayashi and others 2011). These observations illustrate that IFN therapy may be used to eradicate CSCs.
Mechanisms of IFN-Induced Apoptosis
Despite extensive studies, it remains unclear how the fate of a tumor cell is shaped by IFNs, that is, to choose between cell death and growth arrest. What is clear is that the JAK/STAT signaling pathway remains fundamental in initiating the apoptotic signals of IFNs. For instance, in the human fibrosarcoma cell line 2fTGH, in which IFN-α induces growth arrest, apoptosis was induced only when IFN-α was used in combination with vanadate, a protein tyrosine phosphatase inhibitor (Gamero and Larner 2001). In addition, the expression of catalytically active JAK1 and TYK2 tyrosine kinases, STAT1, and STAT2 were all essential for the induction of apoptosis by IFN-α. In the case of STAT2, this molecule contains a motif in the Src-homology-2 (SH2) domain composed of 4 amino acid residues consisting of proline, tyrosine, threonine, and lysine found conserved in STAT1 and STAT3 that modulate the apoptotic effects of IFN-α (Scarzello and others 2007). Mutation of tyrosine to phenylalanine prolonged the activation of STAT1/STAT2 heterodimers and induction of apoptosis in the IFN-apoptosis-resistant 2fTGH cells. In contrast, mutation of proline to leucine antagonized these effects by preventing dimerization of activated STAT1/STAT2 (Gamero and others 2004). Alternate signaling pathways also contribute to IFN-induced apoptosis. The apoptotic effects of IFN-α can be facilitated by activation of phosphoinositide-3-kinase (PI3K) and mammalian target of rapamycin, as their pharmacological inhibition moderately inhibited apoptosis of leukemic Daudi cells (Thyrell and others 2004). STAT1 was found to be essential, but its action was independent of PI3K, thus showing cooperation of 2 signaling pathways to trigger apoptosis (Arulampalam and others 2011). Contradictory studies, however, show that suppressing the activation of the PI3K/AKT survival signaling pathway by IFN-β treatment correlated with apoptosis of neuroblastoma cells (Dedoni and others 2010) and colorectal cancer cells (Lei and others 2005). In the 2 glioblastoma cell lines (T98G and U87MG), the combination of IFN-γ and all-trans-retinoic acid reduced cell viability and promoted cell death. While in T98G cells, induction of apoptosis was due to suppression of the PI3K/AKT pathway, apoptosis of U87MG occurred independently of the tumor suppressor PTEN (the negative regulator of the AKT pathway) and linked to decreased activity of the NF-κB survival pathway (Zhang and others 2007). A later study revealed that these 2 lines produced different amounts of the proapoptotic protein TRAIL in response to IFN treatment (Sgorbissa and others 2011).
Downstream events of the PI3K/AKT signaling pathway have been explored. IFN-α causes the activation of the ERK1/2 and JNK pathways culminating in apoptosis via the mitochondrial-dependent death pathway and involving the activation of proapoptotic Bcl-2 family members Bax and Bak. Blockade of these 2 signaling pathways prevented apoptosis without affecting activation of the upstream JAK/STAT pathway, confirming that activation of the ERK1/2 and JNK pathways are downstream events (Panaretakis and others 2008). Moreover, other studies highlight the role of activated Bid (a proapoptotic member of the Bcl-2 family) as a necessary component in IFN-induced cell death via TRAIL induction in ovarian cancer and glioblastoma (Tsuno and others 2012). In pancreatic β-cells, treatment with tumor necrosis factor (TNF)-α and IFN-γ induced apoptosis via activation of the proapoptotic protein Bim (another Bcl-2 family member) that occurred in a STAT1-dependent manner (Barthson and others 2011). In contrast, downregulation of the antiapoptotic Bcl-xL protein by TNF-α and IFN-γ sensitized colon carcinoma cells to TRAIL-mediated apoptosis (Liu and others 2011).
The NF-κB survival pathway also modulates the apoptotic effects of IFNs. One study in particular showed that type I IFN signaling in lymphoblastoid cells activated NF-κB signaling via NIK and TRAF2 and promoted cell survival, in a manner independent of PI3K/AKT-activated NF-κB signaling (Yang and others 2005). NF-κB activity is also linked to activation of calcium signaling, which reciprocally antagonizes the apoptotic effects of type I IFNs in T cells (Yue and others 2012). Defective calcium signaling caused either by a loss of the calcium channel Orai1 or by overexpression of dominant-negative Orai1, which blocked calcium entry, sensitized T cells to die after exposure to type I IFNs. Restoration of Orai1 protected T cells from undergoing apoptosis with type I IFNs. Furthermore, the transcriptional responses to type I IFNs were also negatively regulated by Orai1 via NF-κB, as pharmacological inhibition of NF-κB or addition of calcium chelators reversed this effect. Although the importance of the JAK/STAT signaling pathway cannot be understated, it is clear that additional signaling pathways are involved in the regulation of IFN-induced apoptosis.
IFN-Inducible Genes with Apoptotic and Survival Roles
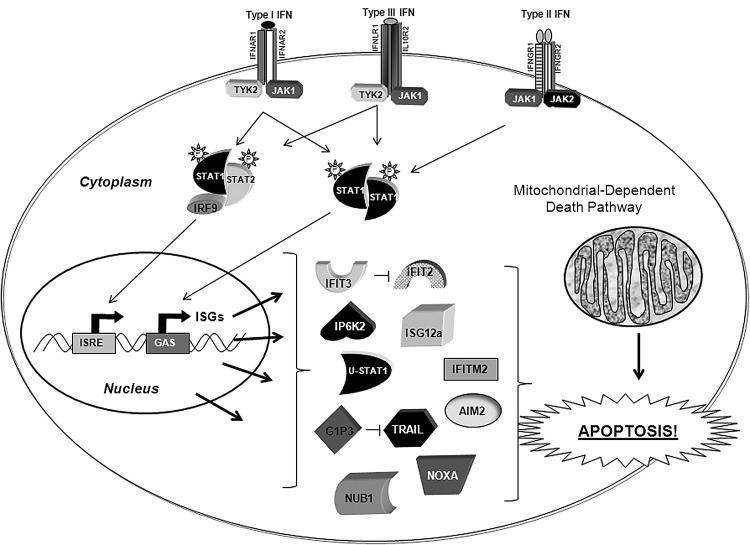
The multiple biological effects of IFNs are mediated through the action of ∼2,000 IFN-stimulated gene (ISG) products (Samarajiwa and others 2009). While in the last decade, a number of ISGs implicated in the antiviral and immune response have been characterized, there is still limited information on the identity of ISGs with apoptotic function. Below, we review ISGs recently characterized for having either proapoptotic or survival functions (see Fig. 1). A list containing most of these genes is presented in Tables 1 and 2. A more detailed description of the vast majority of previously identified apoptotic ISGs can be found in published reviews (Clemens 2003; Pokrovskaja and others 2005).
FIG. 1.
Interferon-stimulated genes (ISGs) involved in IFN-induced apoptosis and IFN resistance. Unphosphorylated (U)-STAT1, G1P3, and IFIT3 are the genes with antiapoptotic/survival function. G1P3 inhibits TRAIL-mediated cell death, and IFIT3 interacts with IFIT2 and disables its proapoptotic activity. AIM1, IP6K2, IFIT2, IFITM2, ISG12a, NUB1, NOXA, and TRAIL are the genes with apoptotic function. Note that some of these proteins are present in more than one cellular compartment. Please refer to Tables 1 and 2 for their correct subcellular localization. Many of these apoptotic gene products promote cell death via activation of the intrinsic mitochondrial-dependent death pathway.
Table 1.
Apoptotic Interferon-Stimulated Gene
| Gene | Localization | References |
|---|---|---|
| AIM2 | Cytoplasmic and nuclear | Chen and others 2006; Luan and others 2008; Ludlow and others 2008; Gariglio and others 2011; Fridman and others 2008. |
| IFITM2 (1-8D) | Cytoplasm and plasma membrane | Daniel-Carmi and others 2009. |
| IP6K2 (Inositol hexakisphosphate-2) (RID-2) | Nuclear and cytoplasmic (Rat), cytoplasmic (Human) | Nalaskowski and others 2003; Shames and Minna 2008; Morrison and others 2001, 2002; Koldobskiy and others 2010. |
| ISG12a (IFI27, p27) | Nuclear membrane and mitochondrial | Martensen and others 2001; Cheriyath and others 2011; Rosebeck and others 2008. |
| ISG54 (IFIT2) | Cytoplasmic | Stawowcyk and others 2011; Fensteri and others 2011. |
| NOXA | Cytoplasmic and mitochondrial | Sun and Leaman 2005; Knowlton and others 2012. |
| NUBI | Predominantly nuclear | Hosono and others 2010. |
Table 2.
Antiapoptotic/Survival Interferon-Stimulated Gene
| Gene | Localization | References |
|---|---|---|
| IF16 (6–16, G1P3) | Nuclear and mitochondrial | Gugliesi and others 2010; Cheriyath and others 2007; Gariglio and others 2011; Tahara and others 2005. |
| ISG60 (IFIT3) | Cytoplasmic | Stawowczyk and others 2011. |
| STAT1 (unphosphorylated) | Cytosolic and nuclear | Luszczek and others 2010; Khodarev and others 2007. |
FAM14: This family of genes encodes highly conserved small hydrophobic proteins with 46 genes in 25 organisms. In humans, the most well-known FAM14 members include FAM14D (ISG12a), FAM14A (ISG12b), FAM14B (ISG12c), and FAM14C (G1P3) (Cheriyath and others 2011). The distribution of human (h) ISG12a and mouse ISG12b1 to the mitochondria suggests their role in mitochondrial function. Indeed, transient expression of ISG12a resulted in decreased cell viability and enhanced sensitivity to DNA-damage-induced apoptosis (Rosebeck and Leaman 2008). ISG12a expression enhanced etoposide-induced cytochrome c release, Bax activation, and loss of mitochondrial membrane potential, all pointing to the activation of the intrinsic mitochondrial-dependent death pathway. Bcl-2 overexpression or treatment with a pan-caspase inhibitor abrogated the proapoptotic effects of hISG12a. siRNA-mediated silencing of ISG12a also prevented sensitization of cells to etoposide-induced apoptosis by either ectopic ISG12a or IFN pretreatment. Together, these findings support ISG12 as a driver of IFN-induced apoptosis.
Unlike FAM14D, FAM14C (G1P3) is a survival factor displaying antiapoptotic activity. Studies conducted in the G1P3-deficient gastric cancer cell TMK1 showed apoptosis sensitivity to 5-fluorouracil (5-FU), cycloheximide, and serum starvation, while ectopic expression of G1P3 conferred protection against apoptosis (Tahara and others 2005). In human myeloma cells, IFN-α2b treatment was found to stabilize mitochondria and inhibit caspase-3 activation and TRAIL-mediated apoptosis, whereas RNAi-mediated silencing of G1P3 restored IFN-induced apoptosis (Cheriyath and others 2007). In addition, ectopically expressed G1P3 localized to the mitochondria and inhibited the apoptotic activity of TRAIL, indicating that IFN-induced G1P3 was acting as a survival factor inhibiting apoptosis. It is worth noting that increased G1P3 expression has been observed in breast (Shen and others 2005), gastric, head and neck, and hepatocellular cancers (Cheriyath and others 2011). Furthermore, several cancer therapies also induce the expression of G1P3, including PTX in ovarian cancer and ionizing radiation (IR) in the breast, prostate, and gliomas, supporting the notion that G1P3 is a stress-induced protein, and its antiapoptotic activity might play a role in the development of therapeutic resistance (Bani and others 2004; Tsai and others 2007).
IFIT2 (ISG54): This gene was one of the first ISGs to be discovered (Larner and others 1986). Surprisingly, its biological role remained unknown until very recently when it was identified as a gene possessing apoptotic (Stawowczyk and others 2011) and antiviral function (Fensterl and others 2012). IFIT2 encodes a 54-kDa protein that is closely related to 2 other ISGs, IFIT1 (ISG56) and IFIT3 (ISG60), which are also binding partners of IFIT2. Expression of IFIT2 alone without IFN stimulation is able to induce apoptosis via activation of the mitochondrial-dependent and p53-independent apoptotic pathway (Stawowczyk and others 2011). Expression of green fluorescent protein (GFP)-tagged ISG54 in HeLa cells increased the number of GFP-positive apoptotic cells, and IFIT2 knockdown protected cells from IFN-α-induced apoptosis. Activation of apoptosis was dependent on the presence of 2 members of the Bcl2 family of proapoptotic proteins, Bax and Bak (Stawowczyk and others 2011). It remains to be evaluated if IFIT2 expression is modulated in response to cancer therapies, and if tumors that acquire resistance to different therapeutic modalities have defective IFIT2 expression.
IP6K2: Inositol hexakisphosphate kinase-2, or RID-2, is a cytoplasmic kinase that directs the catalysis of inositol hexakisphosphate (InsP6) to diphosphoinositol pentakisphosphate (InsP7/PP-InsP5). It was identified as an IFN-regulated gene with proapoptotic activity using an antisense knockout approach in an ovarian cancer cell line. Its expression is regulated post-transcriptionally by IFN-β (Morrison and others 2001). Further studies revealed that overexpression of IP6K2 in ovarian cancer cells sensitized them to apoptosis by IFN-β through the extrinsic death receptor pathway (Morrison and others 2002). The proapoptotic function of IP6K2 is negatively regulated by the heat shock protein HSP90 in a number of human cancer cell lines. IP6K2 binding to HSP90 disables its catalytic activity (Chakraborty and others 2008). In another study, direct interactions between IP6K2 and p53 were found to be required in p53-mediated apoptosis of colorectal cancer cells netting a reduction in the expression of growth-arrest genes (Koldobskiy and others 2010).
HIN-200 family: Members of this family are strongly induced by IFNs in a number of cell types (Lengyel 2008). IFI16 is one member of this family that is not only activated by IFNs, but also by RNA viruses (Choubey and others 2010). The presence of a PYRIN-PAAD-DAPIN (PYD) domain in IFI16 suggests interactions with transcription factors involved in apoptosis via caspase activation (Gariglio and others 2011). The proapoptotic effects of IFI16 were demonstrated when silencing of IFI16 in EC increased resistance to apoptosis by IFN-β priming and double-stranded RNA treatment. In contrast, IFI16 overexpression in EC resulted in the activation of caspase-2 and caspase-3, causing massive EC apoptosis (Gugliesi and others 2010). AIM2 is another member of the HIN-200 family with apoptotic activity. Forced expression of AIM2 in breast cancer cells resulted in the accumulation of cells in the apoptotic sub-G1 population, suggesting that AIM2-expressing cells underwent apoptosis. In line with this proapoptotic activity, AIM2 was shown to negatively regulate the TNF-α/NF-κB antiapoptotic survival pathway (Chen and others 2006). Members of this family also display antiproliferative (Luan and others 2008), differentiation (Ludlow and others 2008), and senescence effects (Fridman and Tainsky 2008).
NUB1: NEDD8 ultimate buster I is an IFN-inducible gene encoding a 69-kDa nuclear protein. NUB1 interacts with NEDD8 and promotes the recruitment of components of the NEDD8 conjugation system to the proteasome for their degradation (Kamitani and others 2001). Induction of NUB1 by IFN-α is detected in renal cell carcinoma cell lines and associated with cell growth inhibition (Hosono and others 2010). In this study, the IFN-α-resistant renal cell carcinoma cell line A498 was used for further studies, because NUB1 is not induced by IFN-α. Knockdown of NUB1 in IFN-α-sensitive 4TUHR cells augmented cell proliferation with IFN-α stimulation, while in contrast, overexpression of NUB1 in A498 cells led to an increase in cell cycle regulator proteins cyclin E and p27 and apoptosis. Future studies will reveal whether IFN-induced NUBI expression in other types of cancer may be linked to an apoptotic response.
STAT1: Tyrosine phosphorylation is required for this transcription factor to be activated, localize to the nucleus, and initiate gene transcription. However, this view has recently changed as unphosphorylated STAT1 (U-STAT1) can drive the expression of a small group of ISGs with immunomodulatory and survival functions. U-STAT1 can prolong the expression of ISGs initially stimulated by IFN-activated STAT1 (Cheon and Stark 2009; Cheon and others 2011). Earlier studies suggested that U-STAT1 may be implicated in tumor resistance to cancer therapies. High STAT1 levels were detected in radiation-resistant tumors (Khodarev and others 2004), and sensitivity to radiation was rescued by STAT1 silencing (Khodarev and others 2007). Recent studies now suggest that high U-STAT1 expression may be driving drug resistance in small-cell lung cancer cells (Luszczek and others 2010). These studies indicate that IFNs not only may be responsible for causing short-term expression of cytostatic and apoptotic proteins, via phosphorylated-STAT1, but also may promote resistance to DNA-damaging agents to facilitate tumor survival, via U-STATs.
Resistance to IFN-Induced Apoptosis
One formidable obstacle to overcome in the clinic is the treatment of patients whose tumors have acquired resistance to IFN immunotherapy. Different mechanisms of resistance have been proposed that, among others, implicate several members of the STAT family of transcription factors. Tumor–stromal interactions can promote the selection and amplification of tumor cells with a prometastatic aggressive phenotype. For example, in vivo passage of tumor cells with IR treatment generated tumor cells with constitutive high levels of STAT1 relative to parental tumor cells that rendered them resistant to IR, IFN-α, and IFN-γ treatment (Khodarev and others 2004; Khodarev and others 2007). Reducing the STAT1 levels was sufficient to reverse resistance. In a different study, analysis of murine B16 melanoma clones established in the lungs of animals showed variable levels of activation in the IFN/STAT1 signaling pathway (Khodarev and others 2009). Tumor cells with high IFN/STAT1 transcriptional profile showed resistance to IFN-γ, IR, and doxorubicin compared to low expressors of the IFN/STAT1 pathway. These 2 studies clearly denote that STAT1 plays dual roles in cancer, and the expression level of STAT1 may be a key determinant between resistance and susceptibility to therapy modalities.
Recently, the importance of STAT2 deficiency in disabling IFN-induced apoptosis was described in 2 different apoptosis-sensitive human cell lines, Daudi and H123 Jurkat. Apoptosis resistance arose when cells were maintained under the continuous selective pressure of IFN treatment, which resulted in a loss of STAT2 (Du and others 2009; Romero-Weaver and others 2010). In the B-lymphoblast Daudi cell line lacking STAT2, it was also observed that the loss of STAT2 led to diminished apoptosis induced by the chemotherapeutic drugs doxorubicin, staurosporine, and campothecin. The same effects, however, were not seen in the STAT2-deficient H123 Jurkat cell line (our unpublished observations). Notably, this discrepancy in response to DNA-damaging agents may be dependent on the tumor cell type. Nevertheless, the exact protective molecular mechanism driven by the absence of STAT2 remains unknown.
IFNs are known to activate the proto-oncogene STAT3, and activation of the NF-κB survival pathway by IFNs requires STAT3, thus implicating both in cellular resistance to IFN-induced apoptosis (Yang and others 2000, 2005). Disabling IFN-induced activation of NF-κB in Daudi cells via expression of the super-repressor IκBα, a natural inhibitor of NF-κB, made these cells susceptible to IFN-induced apoptosis (Yang and others 2000). This finding strongly suggests that activation of the NF-κB pathway by IFNs via STAT3 promotes cell survival by counteracting the lethal effects of IFNs. In the case of STAT5, this transcription factor also regulates the expression of prosurvival NF-κB activators Bcl10, Traf2, and Traf5, which were found to be weakly regulated by STAT3 in lymphoid cells (Nagy and others 2006). STAT5 can be found expressed at high levels in IFN-α-resistant melanoma cell lines and in melanoma biopsies. Sensitivity to IFN-α can be acquired by silencing STAT5. Reciprocally, overexpression of STAT5 in IFN-α-sensitive melanoma cells can abrogate the antigrowth effects of IFN-α (Wellbrock and others 2005).
Other proposed contributors of resistance to IFNs include micro-RNAs (miRs). They are small noncoding RNAs that suppress mRNA translation. mIR-21 was previously reported to promote metastasis and support the survival and proliferation of tumor cells. In fact, miR-21 is found overexpressed in a number of cancers, including glioblastoma, prostate, and breast (Löffler and others 2007; Folini and others 2010). Recently, mIR-21 was shown to be induced by IFN-α in a STAT3-dependent manner in various cancer cell lines. In particular, knockdown of miR-21 sensitized prostate cancer cells to IFN-induced apoptosis, whereas expression of miR-21 in apoptosis-sensitive tumor cells conferred resistance (Yang and others 2010). Thus, miR-21 partakes in a negative feedback loop to inhibit IFN-induced apoptosis. Furthermore, a recent study identified a population of hepatocellular carcinoma (HCC) cells displaying resistance to IFN that contained increased levels of miR-146a (Tomokuni and others 2011). In other studies, overexpression of miR Let-7a induced resistance in cell lines, otherwise susceptible to the apoptotic effects of IFN-γ, PTX, and doxorubicin (Tsang and Kwok 2008). The apoptotic mediator caspase-3 was identified to be the target of Let-7a.
Combination Therapies to Potentiate the Apoptotic Effects of IFNs
Beginning in the 1980s, multiple clinical trials around the world were designed to test the antitumor effects of IFN-α in a variety of malignancies that ranged from hairy cell leukemia and chronic myelogenous leukemia (CML) to melanoma and other types of solid tumors. IFNs purified from leukocytes were initially used, but were later replaced by recombinant forms of the cytokine (Nagata and others 1980). The early and promising results of these clinical trials prompted the Food and Drug Administration to approve the use of IFN-α as an anticancer agent in the mid-1980s. Although IFN immunotherapy is effective in a small subset of patients with cancer when used as a single agent, this form of therapy is also associated with high toxicity, causing treatment to be discontinued (Ascierto and others 2012). Several clinical studies are now exploring the use of IFN in combination with other treatment modalities to improve the benefits of IFN therapy (Garbe and others 2011; Picozzi and others 2011). Already in progress is the use of IFN-α supplemented with the antiviral prodrug ribavirin, an inhibitor of RNA metabolism. This combination therapy is being used to prevent recurrence and occurrence of HCC in difficult-to-treat patients with high titers of hepatitis C virus (Kudo 2011). In addition, patients with HCC show increased responsiveness to IFN therapy when combined with the pyrimidine analog 5-FU to block DNA replication. Synergistic effects are due to enhanced activation of the JAK/STAT pathway as well as caspase activation (Koike and others 2006).
In the treatment of other types of cancer such as renal cell carcinoma, the combinatory effects of IFN-α with a number of chemotherapeutic drugs have shown greater potential over chemotherapy alone (Amato 2005). Addition of the proteasome inhibitor bortezomib to IFN therapy appears to be promising in the treatment of myeloma (Chim 2010) and lymphoma (Abou-Merhi and others 2007). Other chemotherapeutic agents such as the alkylating agent temozolomide have shown very positive results when combined with IFN regimens. When tested in glioma cell lines, temozolomide produced greater apoptotic gene expression patterns when combined with IFN-β in a synergistic manner (Yoshino and others 2011).
Finally, small-molecule inhibitors tailored to disrupt specific signaling pathways have also been advocated in recent years as novel therapeutic approaches to be used in combination with IFN therapy. For example, treatment of a patient with CML with IFN-α and tyrosine kinase inhibitor imatinib was effective in overcoming a previously developed imatinib resistance (Itonaga and others 2012). Furthermore, targeted delivery of chemotherapeutics, particularly in the form of fusion proteins, is a promising concept in treating a number of malignancies that are already showing positive results (Schrama and others 2006). Another tactic being pursued is to target a therapeutic agent to the tumor site using the specificity of an antibody that recognizes a tumor antigen to decrease any toxicity associated with nonspecific antitumor agents. This has been accomplished with IFN-α via fusion to antiCD20 antibodies, and so far it shows potent antitumor activity against B-cell lymphoma (Xuan and others 2010). A similar approach was performed earlier by fusing IFN-α to anti-HER2/neu antibody to target murine HER2/neu tumors. This strategy prolonged the in vivo half-life of IFN-α and when tested in vivo, the antitumor effects were enhanced when compared with recombinant IFN-α alone (Huang and others 2007).
Closing Remarks
Despite advances made in the past decades, the underlying molecular mechanisms of how IFNs influence tumor cells to choose the apoptotic path remain poorly characterized. However, some important achievements that must be recognized include the characterization of additional ISGs with apoptotic and survival function. Another important observation was the elucidation of signaling pathways that crosstalk with the JAK/STAT pathway to promote IFN-induced apoptosis. Most notable are the role of STATs in the survival and apoptotic activities of IFNs and the suppressive roles miRs play in disabling apoptosis of cancer cells. While several signaling mechanisms of IFN-induced apoptosis and resistance have been proposed, other challenges remain. This includes the identification of unique molecules that disable or alternatively, and when induced will promote death of tumor cells that are normally growth inhibited by IFNs. Nevertheless, progress made in understanding how IFNs promote cell death represents a leap forward and provides ideal opportunities to design novel strategies for overcoming resistance and enhancing the therapeutic effects of IFNs.
Acknowledgment
This work was supported by a grant from the National Cancer Institute (RO1CA140499 to AMG).
Author Disclosure Statement
No competing financial interest in connection with this article to report.
References
- Abou-Merhi R. Khoriaty R. Arnoult D. El Hajj H. Dbouk H. Munier S. El-Sabban ME. Hermine O. Gessain A. de Thé H. Mahieux R. Bazarbachi A. PS-341 or a combination of arsenic trioxide and interferon-alpha inhibit growth and induce caspase-dependent apoptosis in KSHV/HHV-8-infected primary effusion lymphoma cells. Leukemia. 2007;21(8):1792–1801. doi: 10.1038/sj.leu.2404797. [DOI] [PubMed] [Google Scholar]
- Amato RJ. Renal cell carcinoma: review of novel single-agent therapeutics and combination regimens. Ann Oncol. 2005;16(1):7–15. doi: 10.1093/annonc/mdi002. [DOI] [PubMed] [Google Scholar]
- Arulampalam V. Kolosenko I. Hjortsberg L. Björklund AC. Grandér D. Tamm KP. Activation of STAT1 is required for interferon-alpha-mediated cell death. Exp Cell Res. 2011;317(1):9–19. doi: 10.1016/j.yexcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Ascierto PA. Gogas HJ. Grob JJ. Algarra SM. Mohr P. Hansson J. Hauschild A. Adjuvant interferon alfa in malignant melanoma: An interdisciplinary and multinational expert review. Crit Rev Oncol Hematol. 2012 doi: 10.1016/J.critrevonc.2012.07.004. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bani MR. Nicoletti MI. Alkharouf NW. Ghilardi C. Petersen D. Erba E. Sausville EA. Liu ET. Giavazzi R. Gene expression correlating with response to paclitaxel in ovarian carcinoma xenografts. Mol Cancer Ther. 2004;3(2):111–121. [PubMed] [Google Scholar]
- Barthson J. Germano CM. Moore F. Maida A. Drucker DJ. Marchetti P. Gysemans C. Mathieu C. Nuñez G. Jurisicova A. Eizirik DL. Gurzov EN. Cytokines tumor necrosis factor-alpha and interferon-gamma induce pancreatic beta-cell apoptosis through STAT1-mediated Bim protein activation. J Biol Chem. 2011;286(45):39632–39643. doi: 10.1074/jbc.M111.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A. Koldobskiy MA. Sixt KM. Juluri KR. Mustafa AK. Snowman AM. van Rossum DB. Patterson RL. Snyder SH. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2008;105(4):1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla-Sarkar M. Leaman DW. Borden EC. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res. 2001;7(6):1821–1831. [PubMed] [Google Scholar]
- Chen IF. Ou-Yang F. Hung JY. Liu JC. Wang H. Wang SC. Hou MF. Hortobagyi GN. Hung MC. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol Cancer Ther. 2006;5(1):1–7. doi: 10.1158/1535-7163.MCT-05-0310. [DOI] [PubMed] [Google Scholar]
- Cheon H. Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106(23):9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H. Yang J. Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31(1):33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyath V. Glaser KB. Waring JF. Baz R. Hussein MA. Borden EC. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest. 2007;117(10):3107–3117. doi: 10.1172/JCI31122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyath V. Leaman DW. Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31(1):173–181. doi: 10.1089/jir.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS. Updated survivals and prognostic factor analysis in myeloma treated by a staged approach use of bortezomib/thalidomide/dexamethasone in transplant eligible patients. J Transl Med. 2010;8:124. doi: 10.1186/1479-5876-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Duan X. Dickerson E. Ponomareva L. Panchanathan R. Shen H. Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30(6):371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens MJ. Interferons and apoptosis. J Interferon Cytokine Res. 2003;23(6):277–292. doi: 10.1089/107999003766628124. [DOI] [PubMed] [Google Scholar]
- Daniel-Carmi V. Makovitzki-Avraham E. Reuven EM. Goldstein I. Zilkha N. Rotter V. Tzehoval E. Eisenbach L. The human 1-8D gene (IFITM2) is a novel p53 independent pro-apoptotic gene. Int J Cancer. 2009;125(12):2810–2819. doi: 10.1002/ijc.24669. [DOI] [PubMed] [Google Scholar]
- Dedoni S. Olianas MC. Onali P. Interferon-beta induces apoptosis in human SH-SY5Y neuroblastoma cells through activation of JAK-STAT signaling and down-regulation of PI3K/Akt pathway. J Neurochem. 2010;115(6):1421–1433. doi: 10.1111/j.1471-4159.2010.07046.x. [DOI] [PubMed] [Google Scholar]
- Du Z. Fan M. Kim JG. Eckerle D. Lothstein L. Wei L. Pfeffer LM. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J Biol Chem. 2009;284(41):27808–27815. doi: 10.1074/jbc.M109.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ED. Weinreich DM. Carroll NM. Burness ML. Feldman AL. Turner E. Xu H. Alexander HR., Jr Interferon gamma-inducible protein 10 selectively inhibits proliferation and induces apoptosis in endothelial cells. Ann Surg Oncol. 2006;13(1):125–133. doi: 10.1245/ASO.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Fensterl V. Wetzel JL. Ramachandran S. Ogino T. Stohlman SA. Bergmann CC. Diamond MS. Virgin HW. Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8(5):e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folini M. Gandellini P. Longoni N. Profumo V. Callari M. Pennati M. Colecchia M. Supino R. Veneroni S. Salvioni R. Valdagni R. Daidone MG. Zaffaroni N. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman AL. Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27(46):5975–5987. doi: 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamero AM. Larner AC. Vanadate facilitates interferon alpha-mediated apoptosis that is dependent on the Jak/Stat pathway. J Biol Chem. 2001;276(17):13547–13553. doi: 10.1074/jbc.M007948200. [DOI] [PubMed] [Google Scholar]
- Gamero AM. Sakamoto S. Montenegro J. Larner AC. Identification of a novel conserved motif in the STAT family that is required for tyrosine phosphorylation. J Biol Chem. 2004;279(13):12379–12385. doi: 10.1074/jbc.M310787200. [DOI] [PubMed] [Google Scholar]
- Garbe C. Eigentler TK. Keilholz U. Hauschild A. Kirkwood JM. Systemic review of medical treatment in melanoma: current status and future prospects. Oncologist. 2011;16(1):5–24. doi: 10.1634/theoncologist.2010-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio M. Mondini M. De Andrea M. Landolfo S. The multifaceted interferon-inducible p200 family proteins: from cell biology to human pathology. J Interferon Cytokine Res. 2011;31(1):159–172. doi: 10.1089/jir.2010.0106. [DOI] [PubMed] [Google Scholar]
- Gugliesi F. De Andrea M. Mondini M. Cappello P. Giovarelli M. Shoenfeld Y. Meroni P. Gariglio M. Landolfo S. The proapoptotic activity of the Interferon-inducible gene IFI16 provides new insights into its etiopathogenetic role in autoimmunity. J Autoimmun. 2010;35(2):114–123. doi: 10.1016/j.jaut.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Hemmings C. The elaboration of a critical framework for understanding cancer: the cancer stem cell hypothesis. Pathology. 2010;42(2):105–112. doi: 10.3109/00313020903488773. [DOI] [PubMed] [Google Scholar]
- Hosono T. Tanaka T. Tanji K. Nakatani T. Kamitani T. NUB1, and interferon-inducible protein, mediates antiproliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br J Cancer. 2010;102(5):873–882. doi: 10.1038/sj.bjc.6605574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH. Chintalacharuvu KR. Morrison SL. Targeting IFN-alpha to B cell lymphoma by a tumor-specific antibody elicits potent antitumor activities. J Immunol. 2007;179(10):6881–6888. doi: 10.4049/jimmunol.179.10.6881. [DOI] [PubMed] [Google Scholar]
- Itonaga H. Tsushima H. Hata T. Matsuo E. Imanishi D. Imaizumi Y. Kawaguchi Y. Fukushima T. Doi Y. Mori S. Kamihira S. Tomonaga M. Miyazaki Y. Successful treatment of a chronic-phase T-315I-mutated chronic myelogenous leukemia patient with a combination of imatinib and interferon-alfa. Int J Hematol. 2012;95(2):209–213. doi: 10.1007/s12185-012-1005-1. [DOI] [PubMed] [Google Scholar]
- Kamitani K. Kito K. Fukuda-Kamitani T. Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276(49):46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- Khodarev NN. Beckett M. Labay E. Darga T. Roizman B. Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci U S A. 2004;101(6):1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev NN. Minn AJ. Efimova EV. Darga TE. Labay E. Beckett M. Mauceri HJ. Roizman B. Weichselbaum RR. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67(19):9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- Khodarev NN. Roach P. Pitroda SP. Golden DW. Bhayani M. Shao MY. Darga TE. Beveridge MG. Sood RF. Sutton HG. Beckett MA. Mauceri HJ. Posner MC. Weichselbaum RR. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4(6):e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton JJ. Dermody TS. Holm GH. Apoptosis induced by mammalian reovirus is beta interferon (IFN) independent and enhanced by IFN regulatory factor 3- and NF-kappaB-dependent expression of Noxa. J Virol. 2012;86(3):1650–1660. doi: 10.1128/JVI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y. Seino K. Hosonuma S. Ohara T. Itamochi H. Isonishi S. Kita T. Wada H. Kojo S. Kiguchi K. Side population is increased in paclitaxel-resistant ovarian cancer cell lines regardless of resistance to cisplatin. Gynecol Oncol. 2011;121(2):390–394. doi: 10.1016/j.ygyno.2010.12.366. [DOI] [PubMed] [Google Scholar]
- Koike K. Takaki A. Tatsukawa M. Suzuki M. Shiraha H. Iwasaki Y. Sakaguchi K. Shiratori Y. Combination of 5-FU and IFNalpha enhances IFN signaling pathway and caspase-8 activity, resulting in marked apoptosis in hepatoma cell lines. Int J Oncol. 2006;29(5):1253–1261. [PubMed] [Google Scholar]
- Koldobskiy MA. Chakraborty A. Werner JK., Jr Snowman AM. Juluri KR. Vandiver MS. Kim S. Heletz S. Snyder SH. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2010;107(49):20947–20951. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontsek P. Karayianni-Vasconcelos G. Kontseková E. The human interferon system: characterization and classification after discovery of novel members. Acta Virol. 2003;47(4):201–215. [PubMed] [Google Scholar]
- Kudo M. Adjuvant therapy after curative treatment for hepatocellular carcinoma. Oncology. 2011;81(Suppl 1):50–55. doi: 10.1159/000333259. [DOI] [PubMed] [Google Scholar]
- Larner AC. Chaudhuri A. Darnell JE., Jr Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J Biol Chem. 1986;261(1):453–459. [PubMed] [Google Scholar]
- Lasfar A. Abushahba W. Balan M. Cohen-Solal KA. Interferon lambda: a new sword in cancer immunotherapy. Clin Dev Immunol. 2011;2011:349575. doi: 10.1155/2011/349575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H. Furlong PJ. Ra JH. Mullins D. Cantor R. Fraker DL. Spitz FR. AKT activation and response to interferon-beta in human cancer cells. Cancer Biol Ther. 2005;4(7):709–715. doi: 10.4161/cbt.4.7.1767. [DOI] [PubMed] [Google Scholar]
- Lengyel P. From RNase L to the multitalented p200 family proteins: an exploration of the modes of interferon action. J Interferon Cytokine Res. 2008;28(5):273–281. doi: 10.1089/jir.2008.3993.HP. [DOI] [PubMed] [Google Scholar]
- Lindenmann J. Isaacs A. [Research on viral interference] Schweiz Z Pathol Bakteriol. 1957;20(5):640–646. [PubMed] [Google Scholar]
- Liu F. Hu X. Zimmerman M. Waller JL. Wu P. Hayes-Jordan A. Lev D. Liu K. TNFalpha cooperates with IFN-gamma to repress Bcl-xL expression to sensitize metastatic colon carcinoma cells to TRAIL-mediated apoptosis. PLoS One. 2011;6(1):e16241. doi: 10.1371/journal.pone.0016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler D. Brocke-Heidrich K. Pfeifer G. Stocsits C. Hackermüller J. Kretzschmar AK. Burger R. Gramatzki M. Blumert C. Bauer K. Cvijic H. Ullmann AK. Stadler PF. Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Lokshin A. Mayotte JE. Levitt ML. Mechanism of interferon beta-induced squamous differentiation and programmed cell death in human non-small-cell lung cancer cell lines. J Natl Cancer Inst. 1995;87(3):206–212. doi: 10.1093/jnci/87.3.206. [DOI] [PubMed] [Google Scholar]
- Luan Y. Lengyel P. Liu CJ. p204, a p200 family protein, as a multifunctional regulator of cell proliferation and differentiation. Cytokine Growth Factor Rev. 2008;19(5–6):357–369. doi: 10.1016/j.cytogfr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE. Purton LE. Klarmann K. Gough DJ. Hii LL. Trapani JA. Keller JR. Clarke CJ. Johnstone RW. The role of p202 in regulating hematopoietic cell proliferation and differentiation. J Interferon Cytokine Res. 2008;28(1):5–11. doi: 10.1089/jir.2007.0070. [DOI] [PubMed] [Google Scholar]
- Luszczek W. Cheriyath V. Mekhail TM. Borden EC. Combinations of DNA methyltransferase and histone deacetylase inhibitors induce DNA damage in small cell lung cancer cells: correlation of resistance with IFN-stimulated gene expression. Mol Cancer Ther. 2010;9(8):2309–2321. doi: 10.1158/1535-7163.MCT-10-0309. [DOI] [PubMed] [Google Scholar]
- Martensen PM. Søgaard TM. Gjermandsen IM. Buttenschøn HN. Rossing AB. Bonnevie-Nielsen V. Rosada C. Simonsen JL. Justesen J. The interferon alpha induced protein ISG12 is localized to the nuclear membrane. Eur J Biochem. 2001;268(22):5947–5954. doi: 10.1046/j.0014-2956.2001.02545.x. [DOI] [PubMed] [Google Scholar]
- Morrison BH. Bauer JA. Hu J. Grane RW. Ozdemir AM. Chawla-Sarkar M. Gong B. Almasan A. Kalvakolanu DV. Lindner DJ. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21(12):1882–1889. doi: 10.1038/sj/onc/1205265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BH. Bauer JA. Kalvakolanu DV. Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276(27):24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moserle L. Indraccolo S. Ghisi M. Frasson C. Fortunato E. Canevari S. Miotti S. Tosello V. Zamarchi R. Corradin A. Minuzzo S. Rossi E. Basso G. Amadori A. The side population of ovarian cancer cells is a primary target of IFN-alpha antitumor effects. Cancer Res. 2008;68(14):5658–5668. doi: 10.1158/0008-5472.CAN-07-6341. [DOI] [PubMed] [Google Scholar]
- Nagano Y. Kojima Y. Sawai Y. [Immunity and interference in vaccinia; inhibition of skin infection by inactivated virus] C R Seances Soc Biol Fil. 1954;148(7–8):750–752. [PubMed] [Google Scholar]
- Nagata S. Taira H. Hall A. Johnsrud L. Streuli M. Ecsödi J. Boll W. Cantell K. Weissmann C. Synthesis in E. coli of a polypeptide with human leukocyte interferon activity. Nature. 1980;284(5754):316–320. doi: 10.1038/284316a0. [DOI] [PubMed] [Google Scholar]
- Nagy ZS. Rui H. Stepkowski SM. Karras J. Kirken RA. A preferential role for STAT5, not constitutively active STAT3, in promoting survival of a human lymphoid tumor. J Immunol. 2006;177(8):5032–5040. doi: 10.4049/jimmunol.177.8.5032. [DOI] [PubMed] [Google Scholar]
- Nalaskowski MM. Bertsch U. Fanick W. Stockebrand MC. Schmale H. Mayr GW. Rat inositol 1,4,5-trisphosphate 3-kinase C is enzymatically specialized for basal cellular inositol trisphosphate phosphorylation and shuttles actively between nucleus and cytoplasm. J Biol Chem. 2003;278(22):19765–19776. doi: 10.1074/jbc.M211059200. [DOI] [PubMed] [Google Scholar]
- Panaretakis T. Hjortsberg L. Tamm KP. Björklund AC. Joseph B. Grandér D. Interferon alpha induces nucleus-independent apoptosis by activating extracellular signal-regulated kinase 1/2 and c-Jun NH2-terminal kinase downstream of phosphatidylinositol 3-kinase and mammalian target of rapamycin. Mol Biol Cell. 2008;19(1):41–50. doi: 10.1091/mbc.E07-04-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi VJ. Abrams RA. Decker PA. Traverso W. O'Reilly EM. Greeno E. Martin RC. Wilfong LS. Rothenberg ML. Posner MC. Pisters PW. Multicenter phase II trial of adjuvant therapy for resected pancreatic cancer using cisplatin, 5-fluorouracil, and interferon-alfa-2b-based chemoradiation: ACOSOG Trial Z05031. Ann Oncol. 2011;22(2):348–354. doi: 10.1093/annonc/mdq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaja K. Panaretakis T. Grandér D. Alternative signaling pathways regulating type I interferon-induced apoptosis. J Interferon Cytokine Res. 2005;25(12):799–810. doi: 10.1089/jir.2005.25.799. [DOI] [PubMed] [Google Scholar]
- Rees RC. MHC restricted and non-restricted killer lymphocytes. Blood Rev. 1990;4(3):204–210. doi: 10.1016/0268-960x(90)90049-x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villanueva J. McDonnell TJ. Induction of apoptotic cell death in non-melanoma skin cancer by interferon-alpha. Int J Cancer. 1995;61(1):110–114. doi: 10.1002/ijc.2910610119. [DOI] [PubMed] [Google Scholar]
- Romero-Weaver AL. Wang HW. Steen HC. Scarzello AJ. Hall VL. Sheikh F. Donnelly RP. Gamero AM. Resistance to IFN-alpha-induced apoptosis is linked to a loss of STAT2. Mol Cancer Res. 2010;8(1):80–92. doi: 10.1158/1541-7786.MCR-08-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebeck S. Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13(4):562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- Samarajiwa SA. Forster S. Auchettl K. Hertzog PJ. INTERFEROME: the database for interferon regulated genes. Nucleic Acids Res. 2009;37:D852–D857. doi: 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarzello AJ. Romero-Weaver AL. Maher SG. Veenstra TD. Zhou M. Qin A. Donnelly RP. Sheikh F. Gamero AM. A Mutation in the SH2 domain of STAT2 prolongs tyrosine phosphorylation of STAT1 and promotes type I IFN-induced apoptosis. Mol Biol Cell. 2007;18(7):2455–2462. doi: 10.1091/mbc.E06-09-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrama D. Reisfeld RA. Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5(2):147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- Sgorbissa A. Tomasella A. Potu H. Manini I. Brancolini C. Type I IFNs signaling and apoptosis resistance in glioblastoma cells. Apoptosis. 2011;16(12):1229–1244. doi: 10.1007/s10495-011-0639-4. [DOI] [PubMed] [Google Scholar]
- Shames DS. Minna JD. IP6K2 is a client for HSP90 and a target for cancer therapeutics development. Proc Natl Acad Sci U S A. 2008;105(5):1389–1390. doi: 10.1073/pnas.0711993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D. He J. Chang HR. In silico identification of breast cancer genes by combined multiple high throughput analyses. Int J Mol Med. 2005;15(2):205–212. [PubMed] [Google Scholar]
- Stark GR. Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawowczyk M. Van Scoy S. Kumar KP. Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286(9):7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen HC. Gamero AM. Interferon-lambda as a potential therapeutic agent in cancer treatment. J Interferon Cytokine Res. 2010;30(8):597–602. doi: 10.1089/jir.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. Leaman DW. Involvement of Noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J Biol Chem. 2005;280(16):15561–15568. doi: 10.1074/jbc.M412630200. [DOI] [PubMed] [Google Scholar]
- Tahara E., Jr Tahara H. Kanno M. Naka K. Takeda Y. Matsuzaki T. Yamazaki R. Ishihara H. Yasui W. Barrett JC. Ide T. Tahara E. G1P3, an interferon inducible gene 6–16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol Immunother. 2005;54(8):729–740. doi: 10.1007/s00262-004-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyrell L. Hjortsberg L. Arulampalam V. Panaretakis T. Uhles S. Dagnell M. Zhivotovsky B. Leibiger I. Grandér D. Pokrovskaja K. Interferon alpha-induced apoptosis in tumor cells is mediated through the phosphoinositide 3-kinase/mammalian target of rapamycin signaling pathway. J Biol Chem. 2004;279(23):24152–24162. doi: 10.1074/jbc.M312219200. [DOI] [PubMed] [Google Scholar]
- Tomokuni A. Eguchi H. Tomimaru Y. Wada H. Kawamoto K. Kobayashi S. Marubashi S. Tanemura M. Nagano H. Mori M. Doki Y. miR-146a suppresses the sensitivity to interferon-alpha in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414(4):675–680. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- Trubiani O. Bosco D. Di Primio R. Interferon-gamma (IFN-gamma) induces programmed cell death in differentiated human leukemic B cell lines. Exp Cell Res. 1994;215(1):23–27. doi: 10.1006/excr.1994.1309. [DOI] [PubMed] [Google Scholar]
- Tsai MH. Cook JA. Chandramouli GV. DeGraff W. Yan H. Zhao S. Coleman CN. Mitchell JB. Chuang EY. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67(8):3845–3852. doi: 10.1158/0008-5472.CAN-06-4250. [DOI] [PubMed] [Google Scholar]
- Tsang WP. Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13(10):1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- Tsuno T. Mejido J. Zhao T. Phillips T. Myers TG. Bekisz J. Zoon KC. BID is a critical factor controlling cell viability regulated by IFN-alpha. J Immunother. 2012;35(1):23–31. doi: 10.1097/CJI.0b013e3182372dcf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek J. Novel interferons. Nat Immunol. 2003;4(1):8–9. doi: 10.1038/ni0103-8. [DOI] [PubMed] [Google Scholar]
- Wellbrock C. Weisser C. Hassel JC. Fischer P. Becker J. Vetter CS. Behrmann I. Kortylewski M. Heinrich PC. Schartl M. STAT5 contributes to interferon resistance of melanoma cells. Curr Biol. 2005;15(18):1629–1639. doi: 10.1016/j.cub.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Xuan C. Steward KK. Timmerman JM. Morrison SL. Targeted delivery of interferon-alpha via fusion to anti-CD20 results in potent antitumor activity against B-cell lymphoma. Blood. 2010;115(14):2864–2871. doi: 10.1182/blood-2009-10-250555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer LM. Interferon induces NF-kappa B-inducing kinase/tumor necrosis factor receptor-associated factor-dependent NF-kappa B activation to promote cell survival. J Biol Chem. 2005;280(36):31530–31536. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer SR. Basu L. Kim JG. Pfeffer LM. IFNalpha/beta promotes cell survival by activating NF-kappa B. Proc Natl Acad Sci U S A. 2000;97(25):13631–13636. doi: 10.1073/pnas.250477397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Yue J. Fan M. Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70(20):8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino A. Tashiro S. Ogino A. Yachi K. Ohta T. Fukushima T. Watanabe T. Katayama Y. Okamoto Y. Sano E. Tsumoto K. Gene expression profiles predicting the response to IFN-beta and a combination of temozolomide and IFN-beta in malignant gliomas. Int J Oncol. 2011;39(3):529–542. doi: 10.3892/ijo.2011.1068. [DOI] [PubMed] [Google Scholar]
- Young CW. Interferon induction in cancer: with some observations on the clinical aspects of poly 1:c. Med Clin North Am. 1971;55(3):721–728. doi: 10.1016/s0025-7125(16)32513-5. [DOI] [PubMed] [Google Scholar]
- Yue C. Soboloff J. Gamero AM. Control of type I interferon-induced cell death by Orai1-mediated calcium entry in T cells. J Biol Chem. 2012;287(5):3207–3216. doi: 10.1074/jbc.M111.269068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. Banik NL. Ray SK. Combination of all-trans retinoic acid and interferon-gamma suppressed PI3K/Akt survival pathway in glioblastoma T98G cells whereas NF-kappaB survival signaling in glioblastoma U87MG cells for induction of apoptosis. Neurochem Res. 2007;32(12):2194–2202. doi: 10.1007/s11064-007-9417-7. [DOI] [PubMed] [Google Scholar]
- Zhang T. Sun HC. Zhou HY. Luo JT. Zhang BL. Wang P. Wang L. Qin LX. Ren N. Ye SL. Li Q. Tang ZY. Interferon alpha inhibits hepatocellular carcinoma growth through inducing apoptosis and interfering with adhesion of tumor endothelial cells. Cancer Lett. 2010;290(2):204–210. doi: 10.1016/j.canlet.2009.09.009. [DOI] [PubMed] [Google Scholar]