Abstract
To compensate for the low sensitivity of magnetic resonance imaging (MRI), nanoparticles have been developed to deliver high payloads of contrast agents to sites of disease. Here, we report the development of supramolecular MRI contrast agents using the plant viral nanoparticle tobacco mosaic virus (TMV). Rod-shaped TMV nanoparticles measuring 300×18 nm were loaded with up to 3,500 or 2,000 chelated paramagnetic gadolinium (III) ions selectively at the interior (iGd-TMV) or exterior (eGd-TMV) surface, respectively. Spatial control is achieved through targeting either tyrosine or carboxylic acid side chains on the solvent exposed exterior or interior TMV surface. The ionic T1 relaxivity per Gd ion (at 60 MHz) increases from 4.9 mM−1s−1 for free Gd(DOTA) to 18.4 mM−1s−1 for eGd-TMV and 10.7 mM−1s−1 for iGd-TMV. This equates to T1 values of ~ 30,000 mM−1s−1 and ~ 35,000 mM−1s−1 per eGd-TMV and iGd-TMV nanoparticle. Further, we show that interior-labeled TMV rods can undergo thermal transition to form 170 nm-sized spherical nanoparticles containing ~ 25,000 Gd chelates and a per particle relaxivity of almost 400,000 mM−1s−1 (15.2 mM−1s−1 per Gd). This work lays the foundation for the use of TMV as a contrast agent for MRI.
Keywords: viral nanoparticle, tobacco mosaic virus, magnetic resonance imaging, contrast agent
1. Introduction
Magnetic resonance imaging (MRI) is an emerging technology used for detection of disease and for follow-up diagnosis after surgery or treatment. While MRI shows great potential because of its high spatial resolution, deep soft tissue contrast, and use of non-ionizing radiation, its low sensitivity remains a drawback. To overcome this shortcoming, paramagnetic contrast agents, such as Magnevist (an FDA approved chelated gadolinium reagent), can be used to enhance the detection sensitivity of MRI[1]. The conjugation of contrast agents to a macromolecular platform further enhances imaging sensitivity[1,2]. Reduced molecular tumbling rates of gadolinium ions after conjugation to such nanoparticles results in increased longitudinal relaxivities[3]. Furthermore, multivalent display results in increased local concentration, both events are contributing to increased sensitivity. Various nanoparticle systems have been explored as supramolecular contrast agents; these include dendrimers[4,5], liposomes[6], perfluorocarbons[7], silica[8], as well as protein cages and virus-based nanoparticles, also termed viral nanoparticles (VNPs)[9–16].
VNPs, specifically plant viruses and bacteriophages, have received tremendous attention in recent years. They have been developed as research tools and platforms for materials science as well as for potential nanomedical applications[17,18]. The propensity to self-assemble around a cargo (the genome) and to deliver this cargo to specific cells and tissues, make viruses ideal candidates for site-specific delivery of therapeutics and/or contrast agents[19,20]. Indeed, several VNP-based technologies are in clinical testing for gene delivery and oncolytic virotherapy[21–23]. VNPs are attractive materials because of their high degree of symmetry, polyvalency, monodispersity, and their genetic or chemical programmability. Several VNP structures have been solved to atomic resolution; which allows tailoring with a high degree of spatial control. Using chemoselective bioconjugation reactions[24,25], VNPs can be modified with imaging contrast agents, therapeutic moieties, and/or targeting ligands such as peptides or antibodies[20]. For example, preclinical imaging of prostate tumors has been demonstrated using cowpea mosaic virus (CPMV) modified with prostate cancer-specific peptide ligands (bombesin) and near infrared imaging dyes[26]. Moving toward translational research, several research groups have engineering VNPs with paramagnetic MRI contrast agents[9–12,14–16]. Similar to other nanoparticles, increased relaxivities are achieved based on reduced tumbling rate of the contrast agent[27]. For example, bacteriophage MS2, a 27 nm sphere, was loaded with ~180 chelated Gd molecules using a TOPO ligand and was able to achieve ionic relaxivities of up to 41.2 mM−1s−1 per Gd ion and 7,416 mM−1s−1 per nanoparticle[9]. In comparison, Magnevist has a relaxivity 5.2 mM−1s−1 [12].
To date, research and development of VNP-based MRI contrast agents has focused on spherical platforms, however, this may not be optimal. A growing body of data suggests advantageous pharmacokinetic and disease targeting properties of rod-shaped, elongated materials[28–34]. In agreement, recent data from our laboratory indicate increased tumor homing of filamentous potato virus X (PVX) compared to cowpea mosaic virus (CPMV)[35]. The use of rod-shaped nanoparticles or VNPs as MRI contrast agents, however, remains unexplored. To address this gap, we turned toward the development and evaluation of tobacco mosaic virus (TMV) as a scaffold for multivalent display of paramagnetic MRI contrast agents. TMV is a rod-shaped VNP measuring 300×18 nm with a solvent-accessible 4 nm-wide interior channel. What makes TMV particularly interesting is the recent discovery that TMV can undergo thermal transition to form RNA-free spherical nanoparticles (SNPs)[36]. The size of SNPs can be tightly tuned through adjustment of TMV concentration with sizes ranging from 100–300 nm (0.1–1.0 mg/ml) to 300–800 nm (1–10 mg/ml). The opportunity to synthesize supramolecular MR contrast agents of varying shapes but identical building blocks (i.e. TMV coat proteins) may provide a unique tool to gain further insights into the structure-function relationship of nanomaterials. Here we start this research by describing the synthesis and materials properties of Gd-loaded TMV rods and spheres.
Each TMV nanorod is formed from 2130 copies of an identical coat protein that is helically arranged around a single strand RNA[37]. The rigid structure of TMV displays 2.2 times more coat proteins per cubic nanometer than its spherical (~30 nm diameter) VNP counterparts, thus allowing more efficient loading of cargos (contrast agents, therapeutics, and or targeting ligands). Engineering TMV particles has yielded a variety of materials for tissue engineering scaffolds[38–40], vaccine development[41–44], and a wide array of electronic materials[19,45,46]. Its application as drug delivery vehicle and contrast agent, however, is novel.
Here, we report the formulation of a novel class of MRI contrast agents based on TMV nanorods and spheres. We show that contrast agent-loaded rod-shaped TMV can undergo thermal transition to form a spherical contrast agent. Conjugation of rod-shaped TMV with DOTA-Gd at either the exterior surface or interior channel was achieved using a combination of amide coupling, diazonium chemistry, and Cu(I)-catalyzed azide alkyne cycloaddition reactions. Particle modification and stability is confirmed with MALDI-TOF mass spectroscopy (MS), inductively coupled plasma optical emission spectrometry (ICP-OES), denaturing gel electrophoresis (SDS-PAGE), size exclusion chromatography (SEC), and transmission and scanning electron microscopy (TEM and SEM). Thermal re-shaping was then applied to generate high-relaxivity nanospheres. TMV rods with relaxivities up to ~ 35,000 mM−1s−1 and TMV SNPs with relaxivities of close to 400,000 mM−1s−1 were generated (measured at 60 MHz); these formulations display the highest relaxivities reported to date using VNP scaffolds. Finally, MR phantoms of varying concentrations were imaged using a pre-clinical 7.0T and a clinical 1.5T MRI.
2. Experimental
TMV propagation
TMV was propagated in N. benthamiana plants. TMV was extracted in yields of 4.5 mg of virus per gram infected leaf material using established extraction methods[47]. Virus concentration in plant extracts was determined by UV-Vis absorbance (ε260 nm = 3.0 mg−1 mL cm−1), and virus integrity was determined by size exclusion chromatography (SEC), and transmission and scanning electron microscopy (TEM and SEM) imaging (see below).
TMV bioconjugation
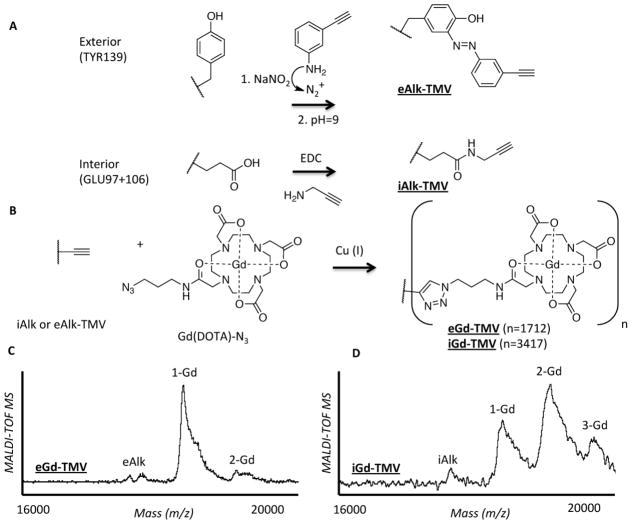
To decorate the exterior TMV surface, the phenol ring of tyrosine underwent an electrophilic substitution (pH=9, 30 min.) with the diazonium salt generated from 3-ethynylaniline (25 molar equivalents (eq)) to incorporate a terminal alkyne. The resulting nanorods are designated eAlk-TMV[40]. Similarly, a terminal alkyne was incorporated onto the interior channel of TMV by targeting glutamic acid residues, designated iAlk-TMV[48]. This was achieved by mixing propargyl amine (25 eq) with EDC (ethyldimethylpropylcarbodiimide, 45 eq) and HOBt (n-hydroxybenzotriazole, 45 eq) for 24 hours. The HOBt is used to suppress EDC side product formation. Following sucrose gradient ultracentrifugation purification, the structural integrity of the particles was confirmed with TEM and SEC and the labeling efficiency was confirmed with MALDI-TOF MS (shown in Supporting Information). Efficient conjugation of Gd(DOTA) azide to terminal alkyne labeled TMV (eAlk- and iAlk-TMV) is accomplished via a copper-catalyzed azide-alkyne cycloaddition[40,48] (CuAAC) to form exterior or interior Gd conjugated TMV, designated eGd-TMV and iGd-TMV, respectively (Figure 2). Initially, GdCl3 was incubated with commercially available (azido-mono amide-1,4,7,10-tetraazacyclododecane-N,N′,N″,N‴-tetraacetic acid; Macrocyclics) DOTA azide while maintaining a pH ~6–7 (adjusted periodically with NaOH) over a six day-period to produce Gd(DOTA) azide. For both interior and exterior conjugation, the same protocol was used. Briefly, alkyne-labeled TMV (2 mg/ml) in 0.1 M potassium phosphate buffer pH 7.0 was mixed with Gd(DOTA) azide (5 eq to CP), aminoguanidine (2 mM), ascorbic acid (2 mM), and copper sulfate (1 mM) for 15 minutes. The reaction mix was purified using a 10–40% sucrose gradient and ultracentrifugation (gradient images in Supporting Information), and analyzed by TEM, SEM, SEC, and MALDI-TOF MS (see below).
Figure 2.
(A) Schematic illustration of the bioconjugation reactions used to incorporate terminal alkynes to the interior and exterior of TMV. (B) Schematic illustration of the CuAAC reaction to label TMV particles with Gd(DOTA). MALDI-TOF MS of (C) eGd-TMV and (D) iGd-TMV. In the MALDI-TOF MS, peaks labeled with eAlk and iAlk refer to the alkyne labeled proteins, 1-Gd, 2-Gd, and 3-Gd refer to coat proteins labeled with one, two, and three Gd(DOTA).
Thermal transition to SNPs
The standard protocol for thermal transition of native TMV rods to SNPs is heating of the sample at 0.1 mg/mL for 10 seconds at 96°C with a Peltier thermal cycler. Alternatively, for iGd-TMV, PEG 8 kDa (0.5% w/v) was added to the reaction mix and incubation time was increased to 15 seconds (see discussion in the main text).
MALDI-MS analysis
For MALDI-MS analysis, native and modified TMV (were denatured using guanidine hydrochloride (6 μL, 6 M) to the sample at 10–20 μg in 24μL 0.1 M potassium phosphate buffer and mixing for 5 min at room temperature. Denatured proteins were spotted on MTP 384 massive target plate using Zip-TipsμC18 (Millipore). MALDI-MS analysis was performed using a Bruker Ultra-Flex I TOF/TOF mass spectrometer.
Size exclusion chromatography (SEC)
All labeled particles were analyzed by SEC using a Superose6 column on the ÄKTA Explorer chromatography system (GE Healthcare). Samples (100 μg/100 μL) were analyzed at a flow rate of 0.5 mL/min using 0.1 M potassium phosphate buffer (pH 7.0).
Transmission electron microscopy (TEM)
Drops of TMV rods or SNPs in DI water were placed on copper TEM grids (5 μL, 0.1 mg/mL), allowed to adsorb for 5 minutes, washed with DI water, and negatively stained with 2% (w/v) uranyl acetate for 2 minute. Samples were examined using a Zeiss Libra 200FE transmission electron microscope operated at 200 kV.
Gel electrophoresis
Denaturing gel electrophoresis was used to analyze protein subunits, specifically proteins were analyzed on denaturing 4–12% NuPAGE gels (Invitrogen) using 1x MOPS running buffer (Invitrogen) and 10 μg of sample. After separation, the gel was photographed using an AlphaImager (Biosciences) imaging system after staining with Coomassie Blue. ImageJ software (rsbweb.nih.gov/ij/, Supporting Information) was used for band analysis and to determine the protein concentration per SNP.
Scanning electron microscopy (SEM)
Samples were dried onto glass cover slips and then mounted on the surface of an aluminium pin stub with use of double-sided adhesive carbon discs (Agar Scientific). The stubs were then sputter-coated with gold in a high-resolution sputter coater (Agar Scientific, Ltd.) and transferred to a Hitachi 4500 scanning electron microscope.
ICP-OES measurements
The Gd per VNP ratio was determined using an ICP-OES (Perkin-Elmer ICP-OES 3300 DV) located in the Geology Department at Kent State University.
Relaxivity measurements
The ionic relaxivity of the engineered VNPs was tested using a pre-clinical 7.0T (300 MHz) MRI (Bruker BioSpec 70/30USR), a clinical 1.5T (64 MHz) MRI (Siemens Espree), and a Bruker Minispec mq60 relaxometer (60 MHz). A standard inversion recovery sequence protocol was used to determine the T1 values on each of the instruments.
3. Results and Discussion
3.1 Spatially-controlled loading of MR contrast agents to the exterior and interior surface of TMV
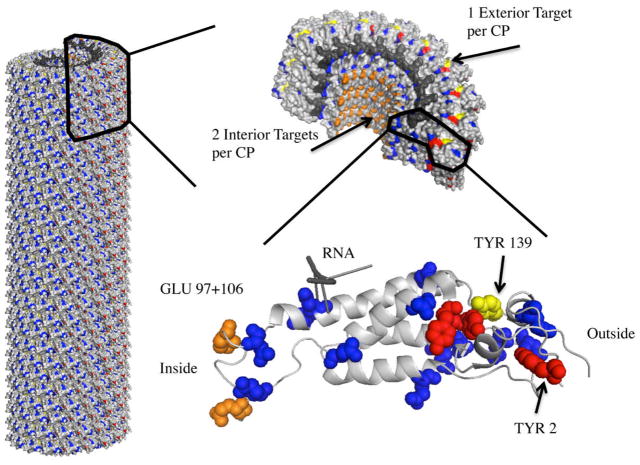
TMV was propagated in N. benthamiana plants. Pure TMV nanorods were extracted in yields of 4.5 mg of virus per gram infected leaf material using established extraction methods[47]. The exterior and interior surfaces of TMV’s hollow rod can be efficiently functionalized using previously established bioconjugation protocols[24,25,40,48]. Figure 1 shows the high-resolution crystal structure of TMV highlighting carboxylic acids (orange and blue) and tyrosine (yellow and red) side chains (PDB ID 2TMV). Previous studies have indicated that exterior tyrosine 139 residues (Figure 1, yellow) and interior glutamic acid 97 and 106 residues (Figure 1, orange) can be modified and functionalized using diazonium coupling or carbodiimide-based conjugation reactions[40,48].
Figure 1.
A PyMol image of tobacco mosaic virus highlighting the interior glutamic acids, GLU97 and GLU106 (orange), exterior tyrosine, TYR139 (yellow), for bioconjugation. Additional glutamic and aspartic acid residues (blue) and tyrosine residues (red) are highlighted for reference.
In this study, we explored both, the exterior and interior surface, for labeling with chelated gadolinium compounds. Using the interior surface provides the advantage that the exterior surface remains available for further tailoring with tissue-specific ligands. On the other hand, attaching the contrast agent to the exterior surface provides an opportunity to load the interior with drugs. To decorate the exterior TMV surface, tyrosine residues were targeted with the diazonium salt generated from 3-ethynylaniline to yield eAlk-TMV[40]. Similarly, a terminal alkyne was incorporated onto the interior channel of TMV by targeting glutamic acid residues, designated iAlk-TMV[48]. Following sucrose gradient ultracentrifugation purification, the structural integrity of the particles was confirmed with TEM and SEC and the labeling efficiency was confirmed with MALDI-TOF MS (shown in Supporting Information).
Efficient conjugation of Gd(DOTA) azide to terminal alkyne labeled TMV (eAlk- and iAlk-TMV) was accomplished via a copper-catalyzed azide-alkyne cycloaddition[40,48] (CuAAC) to form exterior or interior Gd conjugated TMV, designated eGd-TMV and iGd-TMV, respectively (Figure 2). eGd-TMV and iGd-TMV formulations were purified using a 10–40% sucrose gradient and ultracentrifugation (gradient images in Supporting Information). This reaction gave an overall yield of 50–60% i/eGd-TMV, as confirmed by UV-Vis absorption to measure the TMV concentration (Abs260nm=3 for 1 mg/ml) and SDS-PAGE. The structural integrity of the modified TMV particles was confirmed with sucrose gradients (matching light scattering region to native TMV), SEC, and TEM (see Supporting Information). A representative TEM image of iGd-TMV is shown in Figure 3B. The successful incorporation of Gd-DOTA-azide onto the interior or exterior surfaces of TMV was confirmed using MALDI-TOF MS (Figure 2C and 2D) and ICP-OES (see Table 1).
Figure 3.
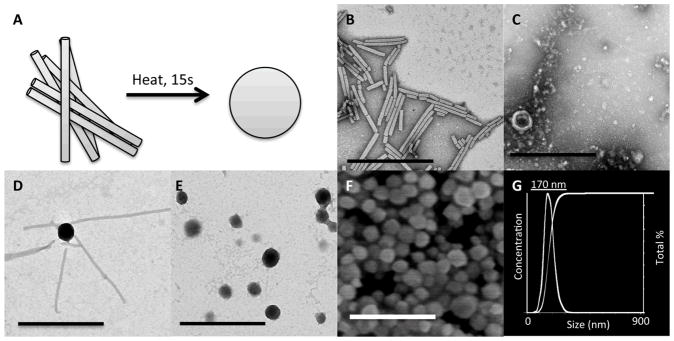
(A) Schematic illustration of the thermal transition from rod-shaped TMV to spherical nanoparticles. Representative TEM images of (B) iGd-TMV, (C) eGd-TMV to SNP, (D) iGd-TMV to SNP, 10 seconds, and (E) iGd-TMV to SNP 15 seconds. (F) SEM image of iGd-SNPs with (G) the corresponding DLS (Nanosight size analyzer). Scale bars = 500 nm.
Table 1.
Longitudinal relaxivity values for Gd-TMV particles.
| Relaxivity per Gd (and per particle) mM−1s−1 | ||||
|---|---|---|---|---|
| VNP | Gd/VNP | 60 MHz | 64 MHz | 300 MHz |
| eGd-TMV | 1,712 | 18.4 (31,501) | 15.7 (26,896) | 6.7 (11,402) |
| iGd-TMV | 3,417 | 10.7 (36,562) | 11.0 (37,519) | 4.7 (15,932) |
| iGd-SNP | 25,815 | 15.2 (392,388) | 13.2 (340,758) | 3.7 (95,515) |
| Gd(DOTA) | 1 | 4.9 | 4.9 | 4.9 |
For eGd-TMV, the mass spectrum (MS) shown in Figure 2C displays peaks attributed to eAlk-TMV/wt-TMV coat proteins (CP) (eAlk, 17713 m/z), CPs with one (1-Gd, 18339 m/z) and two (2-Gd, 19094 m/z) Gd(DOTA) molecules attached. Similarly, the MS of iGd-TMV shown in Figure 2D displays peaks attributed to iAlk-TMV/wt-TMV CPs (iAlk, 17618 m/z), CPs with one (1-Gd, 18359 m/z), two (2-Gd, 19044 m/z), and three (3-Gd, 19671 m/z) Gd(DOTA) molecules attached. The differences in mass values obtained are attributed to the DOTA molecule only, indicating that the chelated Gd ions did not remain chelated to DOTA after ionization. It should be noted that the presence of Gd was confirmed using ICP-OES (see below).
Interestingly, MALDI-TOF MS characterizations of both the exterior and interior labeling indicate one additional amino acid modification per CP than previously reported[40,48,49]. For exterior conjugation, the MS indicates a majority of the CPs that make up eGd-TMV have one Gd(DOTA), likely attached to TYR139, a small amount of CPs that are un-labeled, and a small amount of CPs that contain two Gd(DOTA) molecules per CP. Of the four tyrosine residues contained in the TMV coat protein, TYR139, TYR2, TYR70, and TYR72 (see Figure 1), only Tyr139 has proven to be the primary reactive tyrosine[49]. Based on the crystal structure it appears that TYR2 is solvent-exposed (more than TYR70 and TYR72), and we think that TYR2 is the potential second attachment site (see Figure 1). For interior labeling, glutamic acids GLU97 and GLU106 have been proven to undergo bioconjugation[48,49] while the third modification site remains unclear. The TMV coat protein includes several aspartic and glutamic acids that could serve as potential attachment sites (highlighted in blue in Figure 1).
Quantitative labeling of TMV’s interior and exterior surface with Gd was confirmed using ICP-OES. Data indicate that the exterior (eGd-TMV) was loaded with 1,712 Gd per particle and the interior (iGd-TMV) was loaded with 3,417 Gd per particle. The ICP-OES results are in agreement with the MALDI-TOF MS results. Overall, these results are exciting because previous studies have not indicated labeling of a second tyrosine or third carboxylic acid[40,49].
3.2 Thermal transition of contrast agent loaded TMV rods into SNPs
Inspired by the capability of TMV to form uniform SNPs (Figure 3A)[36], we explored the thermal transition of chemically modified TMV particles (Figure 3). Initially, we tested our exterior modified TMV particles (eGd-TMV). Here, we found that no SNPs were formed after heating for 10 seconds at 96°C (Figure 3C). We expanded those conditions to extended incubation times (up to 30 seconds) and addition of additives such as PEG, urea, guanidinium chloride, triton X-100 and at high and low ionic strengths and found that no SNPs were formed. Finally, we attempted to form SNPs with eAlk-TMV, thinking the DOTA group was too large and blocking the assembly. Again the SNPs did not form and only broken protein aggregates were found. This may suggest that TYR139 might play a role in the formation and stability of SNPs.
Subsequently, we tested the thermal transition of interior modified TMV (iGd-TMV) to SNPs. Using the standard protocol, i.e. heating for 10 seconds at 96°C with a Peltier thermal cycler, formation of SNPs was not noticeable. While a lack of rod-shaped particles indicated that TMV was denatured, only irregular protein aggregates were observed. Similarly, a variety of additives were used to either increase or decrease TMVs stability (listed above). We found that addition of PEG 8 kDa (0.5% w/v) to the reaction mix improved the stability of SNPs and decrease non-specific protein aggregation. After heating for 10 seconds, we found more regular SNP formation, however, the transition was incomplete, meaning that many rod-shaped TMV particles were still detectable in the sample (Figure 3D). The rods appeared to be feeding into the SNPs indicating an end-in feeding/melting mechanism. Next, we increased the incubation time from 10 to 15 seconds and found that all of the rods were transitioned to SNPs, as seen with TEM (Figure 3E) and SEM (Figure 3F). A longer heating time is required to fully transition iGd-TMV into iGd-SNPs compared to native TMV. We propose that the requirement of additional incubation time is because the interior modified TMV are more stable than native TMV. More stable interior modified TMV particles were confirmed using a differential scanning calorimeter (Supporting Information). Native TMV was found to fall apart at 65°C, whereas iAlk-TMV remained stable until a temperature of 80°C was reached. Modification of glutamic acids 97 and 106 has been shown to decrease the electrostatic repulsion between CPs[37,50,51], therefore leading to stronger attraction between CPs.
Analysis of the SNP size distribution was done using TEM (Figure 3E), SEM (Figure 3F) and dynamic light scattering (DLS) with a Nanosight size analyzer (Figure 3G) and a standard DLS instrument (Brookhaven). We found that there is some variability from experiment to experiment with the size of the SNP batches varying between 150 nm to 200 nm. Within a particular batch, however, there is a narrow size distribution. The SNP batch utilized for the described studies, measured a hydrodynamic radius of 170 ± 41 nm in diameter as determined by Nanosight (Figure 3G) and DLS (not shown). This is in agreement with TEM and SEM measurements that indicated a SNP size of 152 nm ± 58 nm (the smaller size is explained that in SEM and TEM dried samples are measured whereas Nanosight and DLS record the hydrodynamic radii).
The Gd loading per SNP was determined using a combination of ICP-OES for Gd concentration and SDS-PAGE for protein concentration. Here, when we assume that the coat proteins form densely packed SNPs upon thermal transition, a 170 nm-sized SNP would contain ~75,400 coat proteins (35.4 times the number of coat protein found in a single TMV rod). The protein concentration was estimated using SDS-PAGE protein gel electrophoresis followed by Coomassie staining and band analysis using ImageJ software (rsbweb.nih.gov/ij/, Supporting Information). Based on the SDS-PAGE and size analysis, we estimated that a 1 mg/ml solution of SNPs contained 4.73×1011 particles/mL, or a molar concentration of 7.05×10−10M. ICP-OES analysis of the same 1 mg/ml solution of SNPs contained 2.85ppm Gd, or 1.82×10−5M, yielding SNPs with 25,815 Gd per SNP (see Table 1).
3.3 Ionic relaxivity of Gd-loaded TMV rods and SNPs
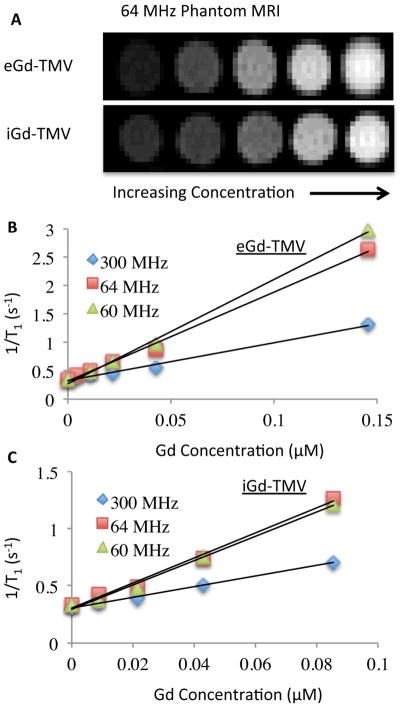
The ionic relaxivity of the engineered VNPs was tested using a pre-clinical 7.0T (300 MHz) MRI (Bruker BioSpec 70/30USR), a clinical 1.5T (64 MHz) MRI (Siemens Espree), and a Bruker Minispec mq60 relaxometer (60 MHz). A standard inversion recovery sequence protocol was used to determine the T1 values on each of the instruments. Shown in Figure 4A is the inversion recovery image (T1 = 2000 ms) of iGd-TMV and eGd-TMV phantoms taken on the clinical MRI (64 MHz, 1.5T). The concentrations increase from left to right with the phantom on far left being water. In order to determine the ionic relaxivities of Gd, 1/T1 (units = 1/seconds) was plotted against the concentration of Gd (in μM) for each formulation and field strength; the slope of each correlation line is the ionic relaxivity (r1, see Figure 4B+C). The relaxivity of the entire particle was computed by multiplying the ionic relaxivity by the number Gd ions per particle determined by ICP-OES. Similar analysis was performed on the Gd-SNPs (data are summarized in Table 1). Next, ionic relaxivity values were determined using a pre-clinical MRI and relaxometer with a similar inversion recovery sequence (see Table 1). As expected, the ionic relaxivity increases at lower field strengths[52].
Figure 4.
(A) Phantom images of tubes containing eGd-TMV, iGd-TMV and Gd(DOTA) with the corresponding Gd concentrations in μM; measured using a clinical 1.5T MRI. Plot of 1/T1 versus Gd concentration (mM) for eGd-TMV (B) and iGd-TMV (C) taken from three MR sources. The slopes of the plots correspond to the ionic relaxivity. Data were collected at varying field strengths (300 MHz, 64 MHz, and 60 MHz).
As with other macromolecular carriers, the relaxivity of DOTA chelated Gd ions was found to increase after conjugation to TMV, compared to free Gd(DOTA) in solution. The increase in ionic relaxivity is greater for exterior labeling of TMV compared to the interior labeling, 18.4 mM−1s−1 and 10.7 mM−1s−1, respectively. This is primarily attributed to the difference in molecular attachment site. Exterior modification is carried out targeting tyrosine side chains and interior loading is accomplished through modification of glutamic acids. The ring structure of the tyrosine side chain induces rigidity, whereas the alkyl chain in the glutamic acids is comparatively flexible. The more rigid the attachment site, the higher the enhancement in relaxivity. This is consistent with previous reports that showed that amino acid stiffness lowers the tumbling rate thus increasing relaxivity[9,10]. For example, exterior lysine residues of MS2 were labeled with bis(HOPO) ligands to chelate Gd, they exhibited an ionic T1 relaxivity of 23.2 mM−1s−1, while interior tyrosine residues labeled with the same bis(HOPO) ligand demonstrated an ionic T1 relaxivity of 31.0 mM−1s−1 (at 60 MHz)[10].
Additionally, after transition of iGd-TMV to SNPs, the ionic relaxivity increases from 10.7 mM−1s−1 to 15.2 mM−1s−1 at 60 MHz. One potential explanation for the relaxivity increase is because transition to SNPs provides a more rigid macromolecule, therefore further reducing the molecular tumbling rate (tR)[53]. Additionally, because the ionic relaxivity at high magnetic field strength (7.0T) remains low, we can assume that the diffusional correlation times (tD) and water proton residency times (tm) remain unaffected[54–57]. Here, it is important to note the drastic difference in per nanoparticle relaxivity between TMV rods and spheres. Based on ICP results, the iGd-TMV particles have 3,417 Gd atoms per rod, which gives a per particle relaxivity of 36,562 mM−1s−1, while the Gd-SNPs contain 25,815 Gd atoms per sphere giving a per particle relaxivity of 392,388 mM−1s−1.
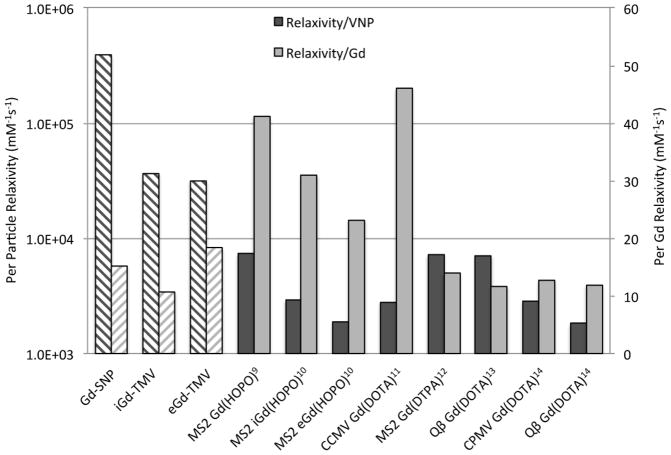
Based on their biocompatibility, monodispersity and ability to undergo multiple rounds of site-selective chemical and/or genetic modification, several icosahedral VNPs have previously been utilized as scaffolds for the presentation of MRI contrast agents, these include cowpea chlorotic mottle virus (CCMV) [11,16], cowpea mosaic virus (CPMV) [14], and bacteriophages MS2[9,10,12,15] and Qβ[13,14]. In all cases enhancements of ionic T1 relaxivities above FDA-approved Magnevist (data are summarized in Figure 5). The Gd-TMV particles generated in this study show similar T1 relaxivity enhancements to VNPs decorated with Gd chelated with DOTA or DTPA ligands, see Figure 5. While the ionic relaxivity of TMV is comparable to other magnetic VNPs, the rod-shaped Gd loaded TMV particles have a four times higher per particle relaxivity (of more than 30,000 mM−1s−1) compared to icosahedral VNPs (see Figure 5). These results are exciting and expected because while the size of TMV is bigger (volume = 7.6×104 nm3 of TMV vs 1.4×104 nm3 for a 30 nm-sized icosahedron), the relaxivity per volume ratio (R1/V) is similar. The R1/V for eGd-TMV and iGd-TMV is 0.41 and 0.48, respectively, while the R1/V for the spherical VNPs ranges from 0.13[14] to 0.52[9] (Full table in Supporting Information). To date the highest per Gd relaxivities were reported by the Francis Lab, who utilized a special HOPO ligand[9,53]. The combination of the HOPO ligand and TMV scaffold is expected to yield a powerful contrast agent (future studies).
Figure 5.
Graph comparing the nanoparticle relaxivities (left axis) and ionic relaxivities (right axis) of TMV particles formed in this paper (pattern) against other Gd-VNPs (solid) at 60 MHz (refs 9, 10 and 13) or 64 MHz (refs 11, 12, and 14).
Finally, the enhancement in ionic relaxivity per Gd is maintained after thermal transition to SNPs. Phantom MRI tests indicate that the relaxivity is even further enhanced. The per particle relaxivity of the SNPs (4×105 mM−1s−1) bridges the gap between contemporary VNPs (T1 relaxivity near 104 mM−1s−1) and dendrimers[4], silica nanoparticles[54] and perfluorocarbons[55], which have per particle T1 relaxivities in the 106 mM−1s−1 range.
4. Conclusion
In conclusion, we report the development of a novel plant viral-based nanoparticle platform for potential applications in MRI. Covalent attachment of chelated gadolinium ions to the supramolecular carrier leads to enhanced ionic relaxivity of the Gd ions based on reduced tumbling rates. Multivalent display leads to relaxivity per nanoparticle four times higher than over VNP contrast agents. Furthermore, the transition of rod-shaped TMV to SNPs improved the ionic T1 relaxivity per Gd; while further increasing the loading of Gd per particle yielding a protein based MRI contrast agent with a T1 relaxivity of 400,000 mM−1s−1. We hypothesize that the conformational changes from TMV rod into SNP provides a more rigid scaffold, which may further reduce molecular tumbling and therefore increase the ionic T1 relaxivity per Gd further. The Gd-loaded TMV rods and spheres reach T1 relaxivities comparable to state-of-the-art dendrimers. This work lays the foundation for the application of Gd-modified TMV nanoparticles for MR imaging.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIBIB grant P30 EB011317 (to NFS). Lauren Randolph, Brian So, and Paul Chariou (BME, CWRU) are thanked for assistance with TMV virus growth and purification. We would also like to thank Dr. Lan Lu with her generous assistance in calculating the relaxivities.
Footnotes
Supporting Information. Additional TEM, MALDI-TOF MS, DLS, sucrose gradient, SEC and SDS-PAGE with lane analysis characterization.
References
- 1.Yan GP, Robinson L, Hogg P. Magnetic resonance imaging contrast agents: Overview and perspectives. Radiography. 2007;13:e5–e19. [Google Scholar]
- 2.Caravan P. Strategies for increasing the sensitivity of gadolinium based MRI contrast agents. Chemical Society Reviews. 2006;35:512. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]
- 3.Waters EA, Wickline SA. Contrast agents for MRI. Basic Research in Cardiology. 2008;103:114–121. doi: 10.1007/s00395-008-0711-6. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z, Thorek DLJ, Tsourkas A. Gadolinium-Conjugated Dendrimer Nanoclusters as a Tumor-Targeted T1 Magnetic Resonance Imaging Contrast Agent. Angewandte Chemie International Edition. 2009;9999 doi: 10.1002/anie.200905133. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floyd WC, et al. Conjugation Effects of Various Linkers on Gd(III) MRI Contrast Agents with Dendrimers: Optimizing the Hydroxypyridinonate (HOPO) Ligands with Nontoxic, Degradable Esteramide (EA) Dendrimers for High Relaxivity. Journal of the American Chemical Society. 2011;133:2390–2393. doi: 10.1021/ja110582e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamaly N, et al. Bimodal Paramagnetic and Fluorescent Liposomes for Cellular and Tumor Magnetic Resonance Imaging. Bioconjugate Chemistry. 2008;19:118–129. doi: 10.1021/bc7001715. [DOI] [PubMed] [Google Scholar]
- 7.Neubauer AM, et al. Gadolinium-modulated 19F signals from perfluorocarbon nanoparticles as a new strategy for molecular imaging. Magnetic Resonance in Medicine. 2008;60:1066–1072. doi: 10.1002/mrm.21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan AK, Klemm PJ, Raymond KN, Landry CC. Silica microparticles as a solid support for gadolinium phosphonate magnetic resonance imaging contrast agents. Journal of the American Chemical Society. 2012;134:8046–8049. doi: 10.1021/ja302183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garimella PD, Datta A, Romanini DW, Raymond KN, Francis MB. Multivalent, High-Relaxivity MRI Contrast Agents Using Rigid Cysteine-Reactive Gadolinium Complexes. Journal of the American Chemical Society. 2011;133:14704–14709. doi: 10.1021/ja204516p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooker JM, Datta A, Botta M, Raymond KN, Francis MB. Magnetic Resonance Contrast Agents from Viral Capsid Shells: A Comparison of Exterior and Interior Cargo Strategies. Nano Letters. 2007;7:2207–2210. doi: 10.1021/nl070512c. [DOI] [PubMed] [Google Scholar]
- 11.Liepold L, et al. Viral capsids as MRI contrast agents. Magn Reson Med. 2007;58:871–879. doi: 10.1002/mrm.21307. [DOI] [PubMed] [Google Scholar]
- 12.Anderson EA, et al. Viral Nanoparticles Donning a Paramagnetic Coat: Conjugation of MRI Contrast Agents to the MS2 Capsid. Nano Letters. 2006;6:1160–1164. doi: 10.1021/nl060378g. [DOI] [PubMed] [Google Scholar]
- 13.Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, Finn MG. Functional virus-based polymer-protein nanoparticles by atom transfer radical polymerization. Journal of the American Chemical Society. 2011;133:9242–9245. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasuhn DE, Yeh RM, Obenaus A, Manchester M, Finn MG. Viral MRI contrast agents: coordination of Gd by native virions and attachment of Gd complexes by azide?alkyne cycloaddition. Chemical Communications. 2007:1269. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 15.Datta A, et al. High Relaxivity Gadolinium Hydroxypyridonate Viral Capsid Conjugates: Nanosized MRI Contrast Agents 1. Journal of the American Chemical Society. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 16.Allen M, et al. Paramagnetic viral nanoparticles as potential high-relaxivity magnetic resonance contrast agents. Magn Reson Med. 2005;54:807–812. doi: 10.1002/mrm.20614. [DOI] [PubMed] [Google Scholar]
- 17.Lee SY, Lim JS, Harris MT. Synthesis and application of virus-based hybrid nanomaterials. Biotechnol Bioeng. 2012;109:16–30. doi: 10.1002/bit.23328. [DOI] [PubMed] [Google Scholar]
- 18.Steinmetz NF. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine: Nanotechnology, Biology, and Medicine. 2010;6:634–641. doi: 10.1016/j.nano.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witus LS, Francis MB. Using synthetically modified proteins to make new materials. Accounts of Chemical Research. 2011;44:774–783. doi: 10.1021/ar2001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yildiz I, Shukla S, Steinmetz NF. Applications of viral nanoparticles in medicine. Current Opinion in Biotechnology. 2011;22:901–908. doi: 10.1016/j.copbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chitale R. Merck hopes to extend gardasil vaccine to men. J Natl Cancer Inst. 2009;101:222–223. doi: 10.1093/jnci/djp014. [DOI] [PubMed] [Google Scholar]
- 22.Liu TC, Galanis E, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 23.Shirakawa T. Clinical trial design for adenoviral gene therapy products. Drug News Perspect. 2009;22:140–145. doi: 10.1358/dnp.2009.22.3.1354090. [DOI] [PubMed] [Google Scholar]
- 24.Hermanson GT. Bioconjugation Techniques. Academic Press; 2008. [Google Scholar]
- 25.Pokorski JK, Steinmetz NF. The Art of Engineering Viral Nanoparticles. Molecular Pharmaceutics. 2011;8:29–43. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz NF, et al. Intravital imaging of human prostate cancer using viral nanoparticles targeted to gastrin-releasing Peptide receptors. Small. 2011;7:1664–1672. doi: 10.1002/smll.201000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Zhong X, Wang L, Yang L, Mao H. Improving the magnetic resonance imaging contrast and detection methods with engineered magnetic nanoparticles. Theranostics. 2012;2:86–102. doi: 10.7150/thno.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai S, Vijayan K, Cheng D, Lima EM, Discher DE. Micelles of Different Morphologies—Advantages of Worm-like Filomicelles of PEO-PCL in Paclitaxel Delivery. Pharm Res. 2007;24:2099–2109. doi: 10.1007/s11095-007-9335-z. [DOI] [PubMed] [Google Scholar]
- 29.Chauhan VP, et al. Fluorescent nanorods and nanospheres for real-time in vivo probing of nanoparticle shape-dependent tumor penetration. Angew Chem Int Ed Engl. 2011;50:11417–11420. doi: 10.1002/anie.201104449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christian DA, et al. Flexible filaments for in vivo imaging and delivery: persistent circulation of filomicelles opens the dosage window for sustained tumor shrinkage. Mol Pharm. 2009;6:1343–1352. doi: 10.1021/mp900022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decuzzi P, et al. Size and shape effects in the biodistribution of intravascularly injected particles. Journal of Controlled Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Geng Y, et al. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentile F, et al. The effect of shape on the margination dynamics of non-neutrally buoyant particles in two-dimensional shear flows. J Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Lee SY, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 35.Shukla S, et al. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Molecular pharmaceutics. 2012 doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atabekov J, Nikitin N, Arkhipenko M, Chirkov S, Karpova O. Thermal transition of native tobacco mosaic virus and RNA-free viral proteins into spherical nanoparticles. Journal of General Virology. 2011;92:453–456. doi: 10.1099/vir.0.024356-0. [DOI] [PubMed] [Google Scholar]
- 37.Klug A. The tobacco mosaic virus particle: structure and assembly. Philos Trans R Soc Lond B Biol Sci. 1999;354:531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee LA, et al. Mutant Plant Viruses with Cell Binding Motifs Provide Differential Adhesion Strengths and Morphologies. Biomacromolecules. 2012;13:422–431. doi: 10.1021/bm2014558. [DOI] [PubMed] [Google Scholar]
- 39.Kaur G, et al. Regulation of osteogenic differentiation of rat bone marrow stromal cells on 2D nanorod substrates. Biomaterials. 2010;31:1732–1741. doi: 10.1016/j.biomaterials.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruckman MA, et al. Surface Modification of Tobacco Mosaic Virus with “Click” Chemistry. ChemBioChem. 2008;9:519–523. doi: 10.1002/cbic.200700559. [DOI] [PubMed] [Google Scholar]
- 41.Staczek J, Bendahmane M, Gilleland LB, Beachy RN, Gilleland HE., Jr Immunization with a chimeric tobacco mosaic virus containing an epitope of outer membrane protein F of Pseudomonas aeruginosa provides protection against challenge with P. aeruginosa. Vaccine. 2000;18:2266–2274. doi: 10.1016/s0264-410x(99)00571-x. [DOI] [PubMed] [Google Scholar]
- 42.Koo M, et al. Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proc Natl Acad Sci U S A. 1999;96:7774–7779. doi: 10.1073/pnas.96.14.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, et al. A modified TMV-based vector facilitates the expression of longer foreign epitopes in tobacco. Vaccine. 2006;24:109–115. doi: 10.1016/j.vaccine.2005.09.060. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, et al. Expression of foot-and-mouth disease virus epitopes in tobacco by a tobacco mosaic virus-based vector. Vaccine. 2003;21:4390–4398. doi: 10.1016/s0264-410x(03)00428-6. [DOI] [PubMed] [Google Scholar]
- 45.Miller RA, et al. Impact of Assembly State on the Defect Tolerance of TMV-Based Light Harvesting Arrays. Journal of the American Chemical Society. 2010;132:6068–6074. doi: 10.1021/ja909566z. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, et al. Virus-enabled silicon anode for lithium-ion batteries. ACS Nano. 2010;4:5366–5372. doi: 10.1021/nn100963j. [DOI] [PubMed] [Google Scholar]
- 47.Niu Z, et al. Assembly of Tobacco Mosaic Virus into Fibrous and Macroscopic Bundled Arrays Mediated by Surface Aniline Polymerization. Langmuir. 2007;23:6719–6724. doi: 10.1021/la070096b. [DOI] [PubMed] [Google Scholar]
- 48.Wu LY, et al. Electrospinning fabrication, structural and mechanical characterization of rod-like virus-based composite nanofibers. Journal of Materials Chemistry. 2011;21:8550–8557. [Google Scholar]
- 49.Schlick TL, Ding Z, Kovacs EW, Francis MB. Dual-Surface Modification of the Tobacco Mosaic Virus. Journal of the American Chemical Society. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- 50.Culver JN, Dawson WO, Plonk K, Stubbs G. Site-directed mutagenesis confirms the involvement of carboxylate groups in the disassembly of tobacco mosaic virus. Virology. 1995;206:724–730. doi: 10.1016/s0042-6822(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 51.Lu B, Stubbs G, Culver JN. Carboxylate interactions involved in the disassembly of tobacco mosaic tobamovirus. Virology. 1996;225:11–20. doi: 10.1006/viro.1996.0570. [DOI] [PubMed] [Google Scholar]
- 52.Caravan P, Farrar CT, Frullano L, Uppal R. Influence of molecular parameters and increasing magnetic field strength on relaxivity of gadolinium- and manganese-based T1 contrast agents. Contrast Media & Molecular Imaging. 2009;4:89–100. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raymond KN, Pierre VrC. Next Generation, High Relaxivity Gadolinium MRI Agents †. Bioconjugate Chemistry. 2005;16:3–8. doi: 10.1021/bc049817y. [DOI] [PubMed] [Google Scholar]
- 54.Rieter WJW, et al. Hybrid silica nanoparticles for multimodal imaging. Angewandte Chemie International Edition. 2007;46:3680–3682. doi: 10.1002/anie.200604738. [DOI] [PubMed] [Google Scholar]
- 55.Morawski AM, et al. Targeted nanoparticles for quantitative imaging of sparse molecular epitopes with MRI. Magnetic Resonance in Medicine. 2004;51:480–486. doi: 10.1002/mrm.20010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.