Summary
Endothelium in embryonic hematopoietic tissues generates hematopoietic stem/progenitor cells; however, it is unknown how its unique potential is specified. We show that transcription factor Scl/Tal1 is essential for both establishing the hematopoietic transcriptional program in hemogenic endothelium and preventing its misspecification to a cardiomyogenic fate. Scl−/− embryos activated a cardiac transcriptional program in yolk sac endothelium, leading to the emergence of CD31+Pdgfrα+ cardiogenic precursors that generated spontaneously beating cardiomyocytes. Ectopic cardiogenesis was also observed in Scl−/− hearts, where the disorganized endocardium precociously differentiated into cardiomyocytes. Induction of mosaic deletion of Scl in Sclfl/fl Rosa26Cre-ERT2 embryos revealed a cell-intrinsic, temporal requirement for Scl to prevent cardiomyogenesis from endothelium. Scl−/− endothelium also upregulated the expression of Wnt antagonists, which promoted rapid cardiomyocyte differentiation of ectopic cardiogenic cells. These results reveal unexpected plasticity in embryonic endothelium such that loss of a single master regulator can induce ectopic cardiomyogenesis from endothelial cells.
Introduction
The hematopoietic and cardiovascular systems are the first organ systems to develop due to their essential functions in supporting the embryo. Blood cells, the heart and the vasculature are generated from lateral plate mesoderm (Fehling et al., 2003; Kattman et al., 2006) in a highly coordinated process that ultimately facilitates the circulation of blood cells in the endothelium-lined blood vessels. The high mortality from blood and cardiovascular diseases warrants studies to define the mechanisms that facilitate the establishment and regeneration of these cell types. However, the roadmap for generating stem/progenitor cells for the different mesodermal lineages is lacking, limiting our ability to derive these cells for clinical use.
It has now been established that the embryonic endothelium is not only a conduit for blood, but also serves as a source of hematopoietic stem/progenitor cells (HS/PCs) (Bertrand et al., 2010; Boisset et al., 2010; Chen et al., 2009; Eilken et al., 2009; Kissa and Herbomel, 2010; Lancrin et al., 2009; Zovein et al., 2008). This unique “hemogenic endothelium” is found during a defined developmental window in intra- and extra-embryonic tissues that generate HS/PCs, including the para-aortic splanchnopleura/aorta-gonad-mesonephros (pSP/AGM) region, the yolk sac and the placenta (Chen et al., 2009; Rhodes et al., 2008; Zovein et al., 2008). However, the mechanisms that govern hemogenic competence in the endothelium are poorly understood.
Fate choice toward cardiovascular and hematopoietic lineages begins when gastrulating cells invaginate through the primitive streak; timing and localization of ingression dictates which morphogens nascent mesodermal cells encounter while migrating to their destination (Kinder et al., 2001). Morphogens that regulate mesodermal fate choice include Wnt, Fgf, TGFβ and BMP (Nostro et al., 2008; Simoes et al., 2011). Combinations of these morphogens and their relative levels promote the patterning of the mesoderm to different cardiovascular and hematopoietic fates (Kattman et al., 2006; Kattman et al., 2011; Simoes et al., 2011).
The bHLH transcription factor Scl/Tal1 is a candidate transcription factor responsible for specifying hemogenic endothelium from mesoderm (Lancrin et al., 2009). Scl−/− embryos die by E 9.5 of mouse development due to absence of blood and poorly remodeled vasculature (Shivdasani et al., 1995; Visvader et al., 1998). The requirement for Scl to specify the hematopoietic fate is limited to a narrow window; conditional deletion of Scl using the hemato-endothelial specific Tie2/Tek Cre strain or the interferon-inducible Mx Cre strain do not disrupt the generation or maintenance of HS/PC pool, although erythropoiesis and megakaryopoiesis are compromised (Mikkola et al., 2003b; Schlaeger et al., 2005). Recent studies revealed that Scl becomes dispensable for HS/PC maintenance due to functional redundancy with a related bHLH factor Lyl1 (Souroullas et al., 2009).
Despite the critical role for Scl in the establishment of the hematopoietic system, little is known about how it generates HS/PCs. ChIP-sequencing in embryonic HS/PCs and erythroid cells revealed widespread binding of Scl and other hematopoietic transcription across thousands of genes, many of which regulate HSC development or maintenance (Kassouf et al., 2010; Wilson et al., 2010). However, these genome-wide binding studies have provided little information about the genes that depend on each of these factors for their expression.
Here we show that Scl establishes hemogenic endothelium by activating transcription factors required for HS/PC emergence and self-renewal. Moreover, our studies revealed an unexpected repressive role for Scl, as loss of Scl resulted in the generation of ectopic cardiomyocytes in yolk sac vasculature and the endocardium. These results uncover remarkable developmental plasticity in embryonic vasculature and identify Scl as a key determinant of endothelial fate choice.
Results
Scl establishes hemogenic competence in endothelium
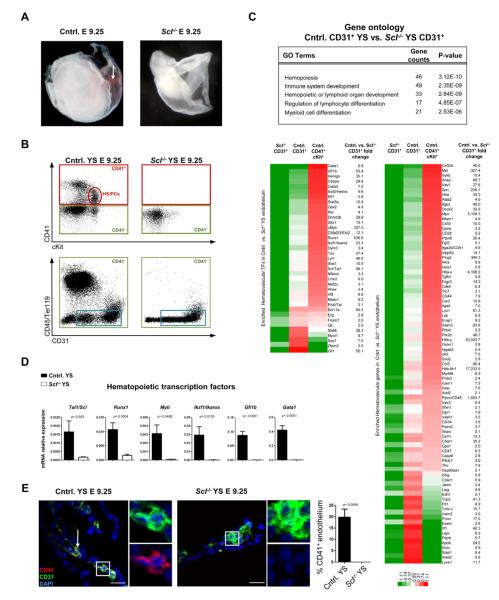
Embryos devoid of Scl lack embryonic red cells and HS/PCs in all hematopoietic tissues (Figure 1A, B, Figure S1A), providing an ideal in vivo model to study the establishment of the hematopoietic system. To determine which hematopoietic genes are activated in the endothelium in the yolk sac by Scl, we performed microarray analysis on Scl−/− and control CD31+ cells that were depleted of hematopoietic cells and compared them to CD41+cKit+ HS/PCs from control yolk sacs. Gene ontology (GO) enrichment analysis of the 1295 differentially expressed genes (Table S1a) downregulated in Scl−/− endothelium (>1.5 fold, p<0.05) revealed a robust Scl dependent hematopoietic transcriptional program in yolk sac endothelium. These genes included transcription factors regulating HSC development (Runx1, cMyb, Fli1), self-renewal (Etv6/Tel, Izkf1/Ikaros, Erg, Gfi1) and differentiation (Gata1 and Gfi1b), as well as surface proteins expressed on hematopoietic cells (Itga2b/CD41, a marker of nascent HS/PCs (Bertrand et al., 2005; Mikkola et al., 2003a) and the pan-hematopoietic marker Ptprc/CD45) (Figure 1C). qPCR on yolk sacs verified Scl dependence of key hematopoietic transcription factors (Figure 1D). Immunofluorescence (IF) validated robust co-expression of CD41 in CD31+ endothelium of control but not Scl−/− yolk sacs (Figure 1E). These data indicate that Scl activates a network of hematopoietic transcription factors necessary for hemogenic competence in the endothelium.
Figure 1. Scl establishes hemogenic endothelium in the yolk sac.
(A) Analysis of E 9.25 Scl−/− embryos documented lack of primitive red cells (arrow) and (B) CD41+cKit+ HS/PCs. (B) Sorting strategy used to isolate endothelial cells from control and Scl−/− yolk sacs (YS). Cells lacking hematopoietic markers CD41 (green boxes), CD45 and Ter119 but expressing CD31+ (blue boxes) were subjected to gene expression analysis. (C) Heat maps represent the relative expression and fold change of hematopoietic transcription factors (TFs) and other proteins significantly downregulated (>1.5 fold, p<0.05) in Scl−/− vs. control yolk sac endothelium; expression in control CD41+cKit+ HS/PCs is shown for comparison. Selected GO (gene ontology) categories derived from differentially expressed genes are listed. (D) qPCR for hematopoietic TFs on E 9.25 unfractionated yolk sacs. Mean ± SEM of 4 biological replicates normalized to HPRT is shown. (E) IF for Scl dependent HS/PC marker CD41 (n=3); insets are higher magnification, single channel + DAPI images of boxed areas. Scale bar represents 25 μm.
Loss of Scl provokes ectopic cardiogenesis in hemogenic tissues
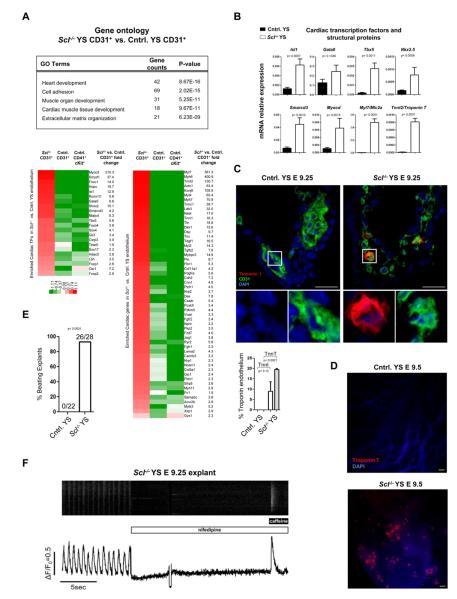
To evaluate whether Scl has a repressive function in the endothelium in hemogenic tissues, GO enrichment analysis was performed for the 965 genes upregulated in CD31+ cells in Scl−/− yolk sacs. Unexpectedly, several highly significant GO categories were related to cardiogenesis and heart function (Figure 2A). The upregulated genes included key cardiac transcription factors such as Isl1, Myocd, Smarcd3 and Tbx5, as well as several cardiomyocyte-specific structural proteins including Tnnt2, Myl7, Actc1, Myh6, Myh7 and Ryr2 (Figure 2A, Table S1b). qPCR analysis confirmed ectopic expression of cardiac genes in Scl−/− yolk sacs (Figure 2B), while FACS documented a population of cells expressing Troponin T (data not shown). IF revealed frequent co-expression of Troponin I (9.0 ± 4.5%) or T (19.5 ± 0.5%) in CD31+ cells in Scl−/− yolk sacs (Figure 2C), and whole mount staining identified clusters of ectopic cardiomyocytes scattered throughout the yolk sac (Figure 2D). Strikingly, explant cultures of Scl−/− yolk sacs produced colonies of synchronously beating cardiomyocytes (Movie S1A and B, Figure 2E). Formation of beating colonies from Scl−/− yolk sacs was highly reproducible (26/28), and in some cases, individual beating cells were observed after as few as 4 hours in culture, indicating that ectopic cardiomyocytes had matured in situ (Movie S1C). Cardiomyocytes from Scl−/− yolk sacs evidenced spontaneous calcium transients, which could be inhibited by the calcium channel blocker nifedipine. Moreover, ectopic cardiomyocytes were capable of storing Ca2+ within the sarcoplasmic reticulum, which was susceptible to caffeine-induced release (Figure 2F). These results demonstrate that Scl is critical for suppressing cardiomyogenesis in the yolk sac.
Figure 2. Lack of Scl induces cardiomyogenesis in yolk sac vasculature.
(A) Selected GO (gene ontology) categories encompassing genes upregulated in Scl−/− yolk sac endothelium are shown. Heat maps represent the relative expression and fold change of known cardiac transcription factors (TFs) and other proteins significantly upregulated (>1.5 fold, p<0.05) in Scl−/− vs. control yolk sac CD31+ cells. CD41+cKit+ HS/PCs are shown for comparison. (B) qPCR for cardiac TFs and cardiomyocyte structural genes on E 9.25 unfractionated yolk sacs. Mean ± SEM of 4 biological replicates normalized to HPRT is shown. (C) IF for Troponin I (n=2; Troponin T n=3) and CD31 on yolk sac sections; insets are higher magnification, single channel + DAPI images of boxed areas. Scale bar denotes 25 μm. (D) Whole mount IF for Troponin T on yolk sacs. Scale bar denotes 50 μm. (E) Frequency of yolk sacs generating beating colonies in explant culture (see Movie S1). (F) Scl−/− yolk sac explants were loaded with Fluo4-AM and spontaneous calcium transients were measured using confocal microscopy. Nifedipine (10μM) and caffeine (10mM) were added at the indicated times.
Scl activates hematopoiesis and represses cardiogenesis in placental vasculature
To investigate if Scl also modulates endothelial fate in other hematopoietic tissues, we performed microarray analysis on CD31+ cells in the placenta, another site that can generate multipotent HS/PCs (Rhodes et al., 2008; Zeigler et al., 2006). These results verified that also in placental endothelium, Scl activates many of the same hematovascular transcription factors as in the yolk sac, with the addition of Sox17 and Bmi1 that regulate self-renewal of fetal and adult HSCs (Kim et al., 2007; Park et al., 2003) (Table S2a). No significant expression of cardiac structural proteins was detected in Scl−/− placental endothelium; however, there was expression of transcription factors essential for cardiac progenitor cell function including Isl1 and Smyd1, as well as cardiac morphogenesis genes including Has2, Dkk1 and Vcan (Figure S1B and C, Table S2b). These results indicate that the misspecification of prospective hemogenic endothelium to the cardiac fate is not limited to the yolk sac.
Loss of Scl leads to the emergence of CD31+Pdgfrα+ cardiogenic cells from prospective hemogenic endothelium
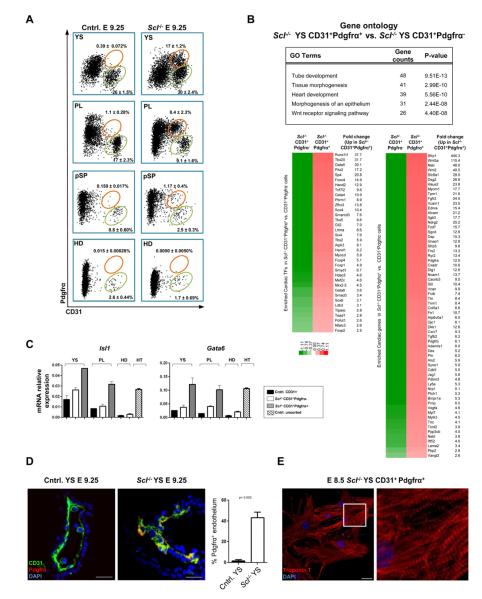
To identify the cardiogenic cells in the vasculature, we screened the microarray data for surface proteins upregulated in Scl−/− endothelium in both the yolk sac and the placenta. Pdgfrα, a marker of cardiogenic mesoderm (Kattman et al., 2011), was significantly upregulated in Scl−/− endothelium in both tissues (Figure 2A, Figure S1C). FACS analysis of E 9.25 conceptuses identified a sub-population of CD31+ cells co-expressing Pdgfrα in all hematopoietic tissues in Scl−/− embryos (Figure 3A). By E 8.5, CD31+Pdgfrα+ cells had already formed in the Scl−/− yolk sac, but were just emerging in the placenta (Figure S2A). CD31+Pdgfrα+ cells were not found in hematopoietic tissues in controls (Figure 3A), even at an earlier developmental stage (Figure S2A).
Figure 3. Scl represses the development of CD31+Pdgfrα+ cardiac cells in hemogenic tissues.
(A) FACS on cells depleted for CD41, CD45 and Ter119 showing CD31+Pdgfrα+ cells (orange gates) and CD31+ endothelial cells (green gates) in embryonic tissues (YS, yolk sac; PL, placenta; pSP, para-aortic splanchnopleura, HD, head). Mean ± SEM of at least 3 biological replicates is shown. (B) Gene expression analysis of CD31+Pdgfrα−/+ cells from Scl−/− yolk sacs. Differentially expressed (>1.5 fold, p<0.05) cardiac transcription factors (TFs) and other proteins enriched in CD31+Pdgfrα+ cells with their fold changes, as well as selected GO (gene ontology) categories, are shown. (C) qPCR for cardiac TFs on indicated populations sorted from YS, PL and HD; unfractionated heart (HT) was used as a positive control. Mean ± SD of technical replicates from pooled samples normalized to actin is shown. (D) IF for CD31 and Pdgfrα on yolk sac sections (n=3). Scale bar represents 25 μm.(See Movie S2). (E) CD31+Pdgfrα+ cells were sorted from E 8.5 Scl−/− yolk sacs, cultured and stained for Troponin T. Scale bar denotes 25 μm.
Microarray analysis comparing CD31+Pdgfrα− endothelial cells and CD31+Pdgfrα+ putative cardiogenic cells from Scl−/− yolk sacs revealed upregulation of cardiac transcription factors and structural proteins in CD31+Pdgfrα+ cells (Figure 3B, Table S3a), while the expression of many vascular genes (Kdr/Flk1, Esam1, Cdh5/Ve-cadherin etc.) was downregulated (Table S3b and Figure S2B, C). qPCR for Isl1 and Gata6 on sorted cells from the yolk sac and the placenta confirmed the upregulation of cardiac transcription factors in CD31+Pdgfrα+ cells (Figure 3C). The head endothelium in Scl−/− embryos neither generated CD31+Pdgfrα+ cells (Figure 3A) nor expressed cardiac transcription factors (Figure 3C). These data verify cardiac identity of CD31+Pdgfrα+ cells and suggest that the ectopic cardiogenesis in Scl−/− embryos is restricted to a specific type of endothelium.
Interestingly, comparison of gene expression in CD31+Pdgfrα− endothelial cells between Scl−/− and control yolk sacs revealed that the cardiogenic transcriptional program was initiated in endothelial cells (Figure S2D, Table S3c). Furthermore, IF for CD31 and Pdgfrα+ in Scl−/− yolk sacs documented integration of CD31+Pdgfrα+ cardiogenic cells within the vasculature (Figure 3D). To functionally assess the developmental potential of CD31+Pdgfrα− endothelium and CD31+Pdgfrα+ cells in Scl−/− embryos, these populations were sorted and plated in cardiac differentiation conditions. Both populations from Scl−/− yolk sac had the potential to generate cardiomyocytes, as evidenced by Troponin T expression, presence of organized sarcomeres and spontaneous beating (Figure 3E, S2E and Movies S2A, B and C). Altogether, these data indicate that, in the absence of Scl, endothelium in the yolk sac becomes misspecified to cardiac fate.
The requirement for Scl to repress cardiogenesis in hemogenic tissues is cell autonomous and temporally defined
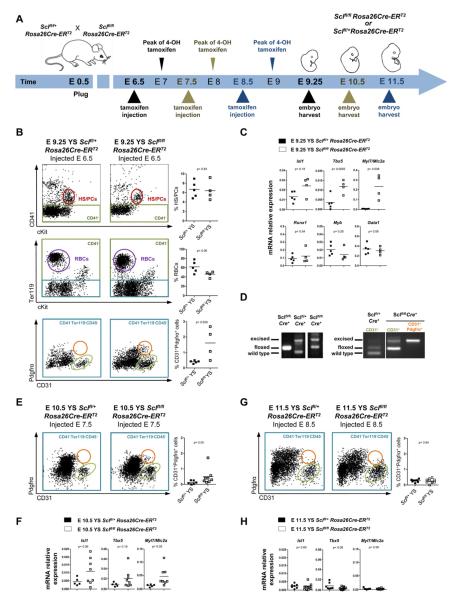
As the misspecified endothelium expressed both cell-intrinsic (e.g. transcription factors) and secreted factors (e.g. Wnt inhibitors Dkk1, Sfrp1, Sfrp5 and Frzb) that promote cardiac differentiation we asked if Scl acts cell autonomously to prevent cardiogenesis in hematopoietic tissues, and crossed tamoxifen-inducible Rosa26Cre-ERT2 mice with Sclfl/fl mice (Figure 4A). Injection of tamoxifen results in a peak of the active drug 12 hours later, which facilitates nuclear localization of Cre (Zovein et al., 2008). Injection of tamoxifen at E 6.5 did not result in a significant difference in the hematopoietic compartment; however, CD31+Pdgfrα+ cardiogenic cells appeared in a majority of Rosa26Cre-ERT2 Sclfl/fl yolk sacs (Figure 4B) and placentas (data not shown). Co-existence of hematopoiesis and ectopic cardiomyogenesis in Rosa26Cre-ERT2 Sclfl/fl yolk sacs was verified by qPCR (Figure 4C). Although the deletion at the Scl locus in Rosa26Cre-ERT2 Sclfl/fl yolk sacs was only partial, PCR for sorted CD31+Pdgfrα+ cardiogenic cells demonstrated full excision of both targeted alleles (Figure 4D). These data verify a cell-intrinsic requirement for Scl in repressing ectopic cardiogenesis,
Figure 4. Scl has a cell autonomous, temporally defined role in preventing ectopic cardiogenesis in hemogenic tissues.
(A) Tamoxifen-inducible Rosa26Cre-ERT2 mouse strain was used to delete Scl at desired time points. (B, E, G) FACS for CD41+cKit+ HS/PCs (red ovals) and Ter119+ RBCs (red blood cells; purple circles) in Rosa26Cre-ERT2 Sclfl/+ and Rosa26Cre-ERT2 Sclfl/fl yolk sacs of pregnant dams injected at E 6.5 (B), E 7.5 (E) and F 8.5 (G). Analysis for CD31+ cells (green ovals) and CD31+Pdgfrα+ cells (orange circles) is shown. Graphs show frequencies of HS/PCs, RBCs and CD31+Pdgfrα+ cells in individual embryos. (C, F, H) qPCR for cardiac and/or hematopoietic genes on yolk sacs following injection at E 6.5 (C), E 7.5 (F) and (H) E 8.5. Graphs show mRNA expression level normalized to HPRT in individual embryos. (D) PCR of embryonic head DNA (left) isolated from embryos injected at E 6.5 and harvested at E 9.5. PCR from CD31+Pdgfrα− vs. CD31+Pdgfrα+ cells sorted from Rosa26Cre-ERT2 Sclfl/fl yolk sacs (right).
To investigate whether the requirement for Scl to repress the cardiomyogenesis continues after mesodermal fates have diverged, injection was initiated at E 7.5 and the embryos harvested at E 10.5. Interestingly, many Rosa26Cre-ERT2 Sclfl/fl yolk sacs still generated CD31+Pdgfrα+ cells (Figure 4E) and expressed cardiac transcription factors and structural proteins (Figure 4F). CD31+Pdgfrα+ cells were also observed in 2/8 placentas (data not shown). However, when the deletion was induced a day later, at E 8.5, cardiac conversion was no longer observed despite comparable excision efficiency (Figures 4G and 4H, data not shown). These data suggest that Scl is required for the repression of the cardiac fate in hemogenic tissues for a limited developmental window.
Scl inhibits cardiomyocyte differentiation in the endocardium
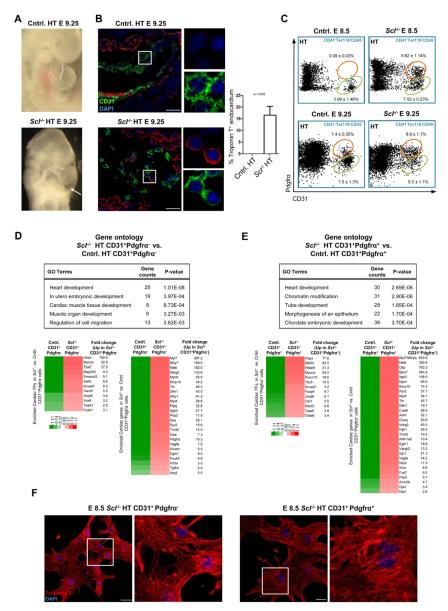
As the loss of Scl induced ectopic cardiogenesis from endothelium in hematopoietic tissues, we next examined Scl−/− hearts for evidence of endothelial misspecification. The heartbeat was initiated in Scl−/− hearts despite structural defects and pericardial effusion (Figure 5A and data not shown). Scl−/− outflow tracts and right ventricles were hypomorphic compared to the left ventricle and the atrial and ventricular septa were poorly formed. Moreover, the heart chambers were filled with poorly organized endocardium (Figures 5A and 5B). Strikingly, IF for CD31 and Troponin T revealed that a substantial population (16.7 ± 3.8%) of endocardial cells co-expressed cardiomyocyte-specific proteins (Figure 5B), demonstrating misspecification of Scl−/− endocardium to cardiomyocytic fate.
Figure 5. Scl−/− endocardium is misspecified to cardiomyogenic fate.
(A) Scl−/− hearts evidenced malformed outflow tract (dotted lines) and pericardial effusion (arrow). (B) IF for CD31 and Troponin T on heart sections (n=3). Scale bar represents 25 μm. (C) FACS for CD31+ endocardial cells (green ovals) and CD31+Pdgfrα+ cells (orange circles) in the heart at E 8.5 and E 9.25. Mean ± SEM of at least 3 biological replicates is shown. (D) Gene expression analysis of Scl−/− and control CD31+Pdgfrα− endocardial cells. Differentially expressed (>1.5 fold, p<0.05) cardiac transcription factors (TFs) and other proteins and their fold changes, as well as selected GO (gene ontology) categories enriched in Scl−/− CD31+Pdgfrα− cells, are shown. (E) Gene expression analysis of Scl−/− and control CD31+Pdgfrα+ cells from the heart. Differentially expressed (>1.5 fold, p<0.05) cardiac TFs and other proteins and their fold changes, as well as selected GO categories enriched in Scl−/− CD31+Pdgfrα+ cells, are shown. (F) IF for Troponin T on cells derived from E 8.5 Scl−/− CD31+Pdgfrα−/+ cells that were sorted and cultured (see Movie S3). Scale bar denotes 25 μm.
Notably, CD31+Pdgfrα+ cells were found in both Scl−/− and control hearts, although they emerged earlier (E 8.5) and were more frequent (E 9.25) in Scl−/− hearts (Figure 5C). Microarray analysis of CD31+Pdgfrα− and CD31+Pdgfrα+ cells from control and Scl−/− hearts revealed upregulation of the cardiomyogenic transcriptional program in both Scl−/− endocardium and CD31+Pdgfrα+ cells (Figure 5D and 5E; Table S4). Strikingly, both the endocardium and CD31+Pdgfrα+ cells from Scl−/− hearts generated beating cardiomyocytes that expressed Troponin T and displayed organized sarcomeres (Movies S3A, S3B and S3C; Figure 5F). These data indicate that Scl suppresses cardiomyocyte differentiation of endocardium and CD31+Pdgfrα+ cells in the heart.
The requirement for Scl to suppress cardiomyocyte fate extends perinatally in the heart
We next assessed whether the timing when Scl is required to suppress cardiomyocyte differentiation in the heart correlates with that in hemogenic tissues (Figure S3A). When induction was initiated at E 6.5 and embryos harvested at E 9.5, the expression of cardiac structural proteins (e.g. Myl7) was significantly increased in both CD31+Pdgfrα− endocardial cells and CD31+Pdgfrα+ cells in Rosa26Cre-ERT2 Sclfl/fl embryos (Figure S3B). Upregulation of Myl7 was also observed in CD31+Pdgfrα+ cells in Rosa26Cre-ERT2 Sclfl/fl hearts upon later induction at E 8.5 or 16.5 (harvest at E 11.5 and 19.5, respectively), after cardiogenic potential in the yolk sac had subsided (Figures S3C and S3D). Remarkably, in some cases, sorted CD31+Pdgfrα+ cells from Rosa26Cre-ERT2 Sclfl/fl perinatal hearts (E 19.5) generated spontaneously beating Troponin T+ cardiomyocytes (Figure S3D and Movie S4). These data indicate that the requirement for Scl to repress the cardiomyocyte fate continues perinatally in CD31+Pdgfrα+ cells in the heart (Figure S3E).
CD31+Pdgfrα+ putative cushion mesenchymal cells emerge from Ve-cadherin+Isl1+ precursors during normal development
As the requirement to repress cardiomyocyte differentiation in the later heart became confined to CD31+Pdgfrα+ cells, we sought to define their identity and origin. CD31+Pdgfrα+ cells in control hearts appeared first by E 9.25, their frequency peaked at around E 10.5 and then declined, although a small population persisted until early postnatal life (Figure S4A). Interestingly, CD31+Pdgfrα+ cells in control hearts at E 9.5 were localized specifically to the outflow tract (OFT) and the atrioventricular canal (AVC) (Figure S4B), in contrast to Scl−/− hearts where CD31+Pdgfrα+ cells were found throughout the heart (data not shown). Microarray analysis showed that the CD31+Pdgfrα+ cells in normal hearts were enriched for cardiac transcription factors Smarcd3, Hand1, Gata4 and Gata6 (Figure S4C, Table S4e and f), and expressed several transcription factors associated with EMT (epithelial-mesenchymal transition) including Twist1, Snai1, Twist2 and Zeb2. Furthermore, several genes involved in endocardial cushion morphogenesis, including Erbb3, Sox9, Msx1, Tgfb2 and Vcan, were upregulated in CD31+Pdgfrα+ cells. Based on the timing when CD31+Pdgfrα+ cells were found in normal hearts, their localization in the OFT and AVC, and gene expression we concluded that this population contains for precursors of cardiac cushion mesenchymal cells.
We next investigated whether the CD31+Pdgfrα+ cells in the heart have similar lineage relationship with endothelial cells as HS/PCs in hemogenic tissues, and performed lineage tracing using the endothelial Ve-cadherin Cre strain bred to the Rosa26-YFP reporter strain (Figure S4D) (Alva et al., 2006; Stadtfeld and Graf, 2005). In the heart, both CD31+Pdgfrα− endothelial cells as well as CD31+Pdgfrα+ cushion mesenchymal cells evidenced equally efficient marking by the Ve-cadherin Cre (Figure S4D), although the latter had downregulated endothelial genes (Figure S4E). The efficiency of lineage tracing in placental and yolk sac endothelium and circulating HS/PCs was similar to that observed in the heart (data not shown). These results further strengthen the hypothesis that CD31+Pdgfrα+ cells represent cushion mesenchymal cells, which are derived from endocardium (de Lange et al., 2004). Furthermore, lineage tracing using Isl1 Cre (Cai et al., 2003) – a transcription factor regulating multipotent cardiac progenitors (Moretti et al., 2006) - revealed that a large fraction of the endocardium and CD31+Pdgfrα+ cells originate from Isl1+ precursors (Figure S4F). Moreover, analysis of Isl1−/− hearts revealed that the establishment of CD31+Pdgfrα+ cells was Isl1 dependent (Figure S4G). These data suggest that CD31+Pdgfrα+ cells in the wild type heart represent cushion mesenchymal cells and are distinct from the ectopic cardiomyogenic CD31+Pdgfrα+ cells in Scl−/− hearts.
Runx1 and Isl1 do not have a critical effect on cardiac misspecification of endothelium
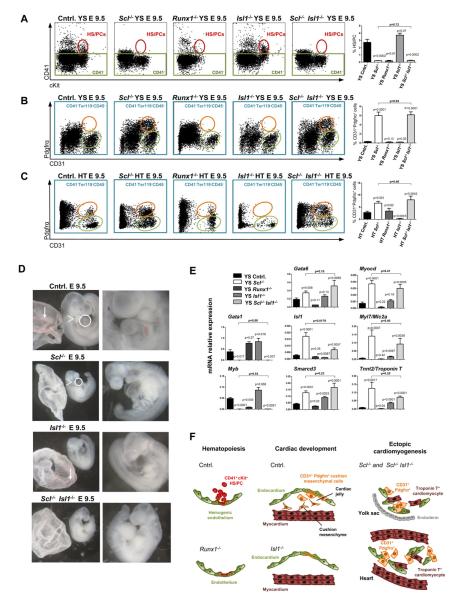
To understand the mechanisms downstream of Scl that modulate hematopoietic versus cardiac development in the endothelium, we asked if the emergence of CD41+cKit+ HS/PCs or CD31+Pdgfrα+ cardiogenic cells was dependent on known HS/PC or cardiac progenitor transcription factors that Scl regulates. Analysis of Runx1−/− embryos verified that the emergence of HS/PCs from hemogenic endothelium was Runx1 dependent (Figure 6A). However, in contrast to Scl−/− embryos, no CD31+Pdgfrα+ cells were detected in hematopoietic tissues in Runx1−/− embryos, nor was this population expanded in the heart (Figures 6B and 6C). Lack of ectopic cardiac differentiation in Runx1−/− yolk sacs was further confirmed by qPCR (Figure 6E). These results imply that, despite its pivotal role in the transition of hemogenic endothelium to HS/PCs, Runx1 is not required to repress cardiac conversion of endothelium (Figure 6F).
Figure 6. Runx1 and Isl1 are not major regulators of the ectopic cardiomyogenic cells.
FACS was performed on yolk sac (A and B) and hearts (C) isolated from control, Scl−/−, Runx1−/−, Isl1−/− and Scl−/− Isl1−/− embryos. All p values shown are with respect to control unless otherwise indicated. (A) CD41+cKit+ HS/PCs (red ovals) are shown in yolk sac and CD31+Pdgfrα+ cells (orange ovals) are shown in yolk sac (B) and heart (C). (D) Scl−/− Isl1−/− embryos lacked blood in the yolk sac (arrows, left panels), pharyngeal arches (arrowheads, left panels) and outflow tract (circles, left panels). (E) qPCR for hematopoietic and cardiac transcription factors and cardiomyocyte structural genes on E 9.5 unfractionated yolk sacs. Mean ± SEM of at least 2 (Runx1−/− = 2; all others ≥5) biological replicates normalized to HPRT is shown. (F) Runx1 and Isl1 are required for HS/PC generation and cardiac cushion development respectively; however, neither regulates ectopic cardiogenesis.
As Isl1−/− embryos showed a drastic reduction of CD31+Pdgfrα+ cells in the heart (Figure S4G), we asked whether generation of cardiogenic CD31+Pdgfrα+ cells in Scl−/− embryos was also dependent on Isl1, and generated Sc1−/− Isl1−/− embryos. Morphologically, Sc1−/− Isl1−/− embryos evidenced a compound phenotype with absence of blood, smaller embryonic size and lack of pharyngeal arches and cardiac outflow tract (Figure 6D). Interestingly, CD31+Pdgfrα+ cells were generated in the yolk sac and the heart of Sc1−/− Isl1−/− embryos, and cardiac transcription factors and structural proteins, including Gata6, Smarcd3, Myocd, Tnnt2 and Myl7, were ectopically expressed (Figure 6E). These data indicate that cardiac conversion in Sc1−/−endothelium is independent of Isl1 (Figure 6F).
Cardiomyocyte differentiation from endothelium is promoted by Wnt antagonism
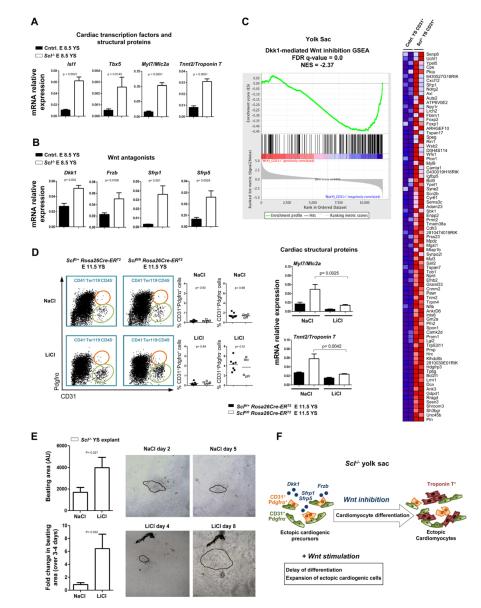
We next sought to define the mechanisms that promote the rapid cardiomyocyte differentiation in the endothelium of Sc1−/− yolk sacs and hearts. Notably, expression of cardiac transcription factors Isl1 and Tbx5 as well as structural proteins Myl7 and Tnnt2 were induced in Scl−/− yolk sacs by E 8.5, concurrent with the appearance of CD31+Pdgfrα+ cells (Figure 7A, S2A). Likewise, expression of several canonical Wnt antagonists including Dkk1, Frzb, Sfrp1 and Sfrp5 that promote terminal cardiomyocyte differentiation (David et al., 2008; Jaspard et al., 2000; Schneider and Mercola, 2001) was also observed at E 8.5 (Figure 7B). We therefore asked whether Scl−/− endothelial cells respond to Wnt inhibition. Indeed, GSEA (Gene Set Enrichment Analysis) comparing Scl−/− yolk sacs with expression data from differentiating mouse embryoid bodies treated with the Wnt inhibitor Dkk1 (Lindsley et al., 2008) revealed a highly significant correlation of gene expression, indicating that Scl−/− endothelial cells are responding to canonical Wnt inhibition (Figure 7C). Similar correlations were also observed in the heart and the placenta (Figures S5A, S5B).
Figure 7. Scl−/− endothelium expresses and responds to Wnt antagonists that promote cardiomyocyte differentiation.
(A) qPCR for cardiac transcription factors and cardiomyocyte structural genes and (B) Wnt antagonists on E 8.5 unfractionated yolk sacs. Mean ± SEM of 4 biological replicates normalized to HPRT is shown. (C) GSEA of expression data from E 9.25 Scl−/− and control yolk sac CD31+ cells compared to embryoid bodies treated with the Wnt antagonist Dkk1. The heat map depicts genes that are part of the core enrichment set. The False Discovery Rate (FDR) q-value and Normalized Enrichment Score (NES) are shown. (D) Pregnant dams were injected with tamoxifen on E 6.5 and treated daily with either NaCl (control) or LiCl (Wnt agonist). FACS analysis showing the frequency of CD31+Pdgfrα− and CD31+Pdgfrα+ cells after NaCl or LiCl treatment and qPCR for Myl7 and Tnnt2. (E) Scl−/− yolk sacs were cultured with either NaCl or LiCl; videos of beating clusters were taken and the area measured in arbitrary units (AU; n ≥27). Explants were imaged at two time points during the culture period (n ≥3); beating areas are outlined. (F) Schematic depicting the acceleration of differentiation of ectopic cardiogenic cells by Wnt inhibitors derived from Scl−/− endothelium; this could be attenuated by the Wnt agonist LiCl.
As Wnt inhibitors are known to promote differentiation of cardiac cells, we asked if the ectopic cardiogenic cells in Scl−/− tissues respond to Wnt agonists to preserve a more undifferentiated state. Pregnant dams carrying Rosa26Cre-ERT2 Sclfl/fl embryos were first injected with tamoxifen at E 6.5 to induce cardiogenic conversion and subsequently stimulated by the Wnt agonist LiCl, or control NaCl, after which yolk sacs were assayed for cardiomyocyte gene expression at E 11.5. Interestingly, the expression of cardiac structural proteins Myl7 and Tnnt2 was significantly reduced upon Wnt stimulation (Figure 7D). Moreover, when Scl−/− yolk sacs were plated in explant culture and supplemented either with LiCl or NaCl throughout the 9-10 day culture, the colonies containing beating cardiomyocytes were larger and their size continued to increase over several days upon Wnt stimulation (Figure 7E). These results indicate that in vivo, Scl−/− ectopic cardiogenic precursors experience Wnt antagonism that promotes cardiomyocyte differentiation; however, at least a subset of these cells retain the capacity to respond to Wnt stimulation (Figure 7F).
Discussion
Our work has uncovered unexpected cardiogenic potential in prospective hemogenic endothelium in blood-forming tissues and endocardium in the heart: upon loss of a single transcription factor, Scl, endothelium in these tissues gives rise to cardiogenic precursors that differentiate into spontaneously beating cardiomyocytes. The temporal window when Scl is required to repress cardiomyogenesis in hematopoietic tissues closely parallels the timing of HS/PC specification in hemogenic endothelium, while in the heart, the repressive function of Scl continues perinatally in CD31+Pdgfrα+ putative cushion mesenchymal cells. Previous studies had suggested that Scl has mesoderm patterning activity during early development, as overexpression of Scl in mesoderm during ES cell differentiation promoted hematopoiesis at the expense of cardiac and paraxial mesoderm (Ismailoglu et al., 2008). Likewise, modulation of Scl and Etsrp (zebrafish homologue for Etv2/ER71) expression altered the size of the heart field (Schoenebeck et al., 2007), and loss of Etsrp/Etv2 induced cardiomyocyte gene expression in endocardial progenitors (Palencia-Desai et al., 2011; Rasmussen et al., 2011). These studies delineate a critical function for Scl and its upstream regulator Etsrp/Etv2 in promoting hematovascular fates over cardiac potential during mesoderm specification. Nevertheless, our finding that loss of Scl in the endothelium in hemogenic tissues and the heart after mesodermal tissues have diverged can give rise to fully functional, beating cardiomyocytes reveals unforeseen developmental plasticity in embryonic endothelium and identifies Scl as a master regulator of endothelial fate choice.
Developmental fate decisions require precise orchestration of both cell intrinsic and microenvironmental regulatory mechanisms. Modulation of Wnt signaling has pivotal effects at multiple stages of cardiac mesoderm and progenitor cell development: Wnt antagonists induce cardiac specification from mesoderm and promote cardiomyocyte differentiation (Schneider and Mercola, 2001; Ueno et al., 2007) while activation of canonical Wnt signaling expands cardiac progenitors and maintain them in an undifferentiated state (Qyang et al., 2007). Our study suggests that ectopic cardiomyogenic cells progress through a Wnt responsive precursor state, the differentiation of which is promoted by Wnt antagonists. Nevertheless, our finding that cardiac conversion was cell-intrinsic to loss of Scl, and could not be induced solely by secretion of Wnt inhibitors by neighboring Scl−/− cells, is in an agreement with the notion that morphogen-induced mesoderm patterning requires relatively uncommitted cells. Recent studies have documented conversion of other mesodermal tissues to cardiomyocytes in vitro and in vivo by reprogramming with a combination of cardiac transcription factors (Gata4, Tbx5, Smarcd3 and/or Mef2c) (Ieda et al., 2010; Qian et al., 2012; Takeuchi and Bruneau, 2009); the same factors were also induced in Scl−/− yolk sac endothelial cells. Our data document that an altered cell-intrinsic regulatory program caused by loss Scl is sufficient to override the microenvironmental cues that dictate mesodermal fate.
Based on the localization of the CD31+Pdgfrα+ in OFT and AVC in normal hearts, their gene expression (cardiac transcription factors and genes related to EMT) and endothelial origin, we proposed that these cells represent cushion mesenchymal cells, which give rise to the septa and valves (Yamagishi et al., 2009). Thus, our data not only identifies novel markers to enrich putative cushion mesenchymal cells, but also uncovers a requirement for Scl to suppress cardiomyocyte differentiation in this endothelial-derived precursor. Moreover, analysis of Isl1−/− embryos revealed a novel role for Isl1 in CD31+Pdgfrα+ cushion mesenchymal cells. As Isl1 is required for the development of both the multipotent cardiac progenitors and CD31+Pdgfrα+ cushion mesenchymal cells, it was surprising that the ectopic CD31+Pdgfrα+ cells in Scl−/− yolk sacs or hearts were not dependent on Isl1 for their generation. These data indicate that, despite a similar surface phenotype, the CD31+Pdgfrα+ cells in control hearts and Scl−/− tissues represent different cells with distinct developmental potential, localization and regulation. Our data suggests that loss of Scl triggers a rapid differentiation program in the endothelium/endocardium that makes ectopic cardiogenesis independent of Isl1.
This work has revealed a broader than anticipated potential for embryonic endothelium as a prospective source of multiple mesodermal stem/progenitor cells, introducing the concept of “cardiogenic endothelium”. Our data revealed that endothelium with cardiomyogenic potential emerges in Scl−/− embryos in both hemogenic tissues as well as the heart, but not in non-hemogenic head endothelium, implying specificity of latent cardiogenic potential to a unique subset of endothelium. Furthermore, our finding about the distinct temporal windows when cardiogenic fate can be induced in hemogenic endothelium, endocardium or CD31+Pdgfrα+ putative cushion mesenchymal cells opens up important future questions to define the mechanisms that close the window of opportunity for cardiomyocyte development in different tissues. These data also raise the question to what extent endothelial-derived cardiac cells can contribute to normal heart development or regeneration, and how modulation of Scl expression could be exploited to improve the differentiation of pluripotent stem cells or partially committed precursors for therapeutic applications. These results call for future studies to examine the prospect of harnessing the latent cardiogenic potential in the vasculature for use in regenerative medicine, and to investigate whether similar developmental plasticity exists in other major cell fate decisions in the developing embryo.
Experimental procedures
Mouse models
Details about mouse models and genotyping can be found in Extended Experimental Procedures. Mice were maintained according to the guidelines of the UCLA Animal Research Committee.
FACS analysis and isolation of HS/PCs, endothelial and cardiac cells
Cells from embryonic tissues were isolated as described (Rhodes et al., 2008). For details about FACS, see Extended Experimental Procedures.
Differential gene expression analysis
RNA was isolated using RNeasy Micro kit (Qiagen), amplified using the NuGEN Pico kit and hybridized on Affymetrix Mouse Genome 430 2.0 Array GeneChip microarrays (GEO accession number GSE27445). For details how differentially expressed genes were identified, analyzed and verified, see Extended Experimental Procedures.
Analysis of tissue sections and whole mounts by confocal microscopy
Fixed frozen sections were prepared and stained as described (Rhodes et al., 2008) with the same antibodies used in FACS, Troponin I (1:400, Santa Cruz) or cardiac Troponin T (1:400, Sigma). Whole mount yolk sacs were prepared as described (Ahnfelt-Ronne et al., 2007) and stained for Troponin T. Confocal images were obtained on a Zeiss LSM 510 equipped with 405 nm, 488 nm, 543 nm and 633 nm lasers. Images were processed with ImageJ software (NIH).
Cardiac culture assays
Yolk sacs were mechanically dissociated by pipetting or enzymatically dissociated, sorted and plated on OP9 stromal cells in cardiac medium (α-MEM medium with 20% FBS, 5 ng/ml murine VEGF, 50 ng/ml human BMP4 and 30 ng/ml human bFGF (Peprotech)) or directly on Matrigel and fibronectin coated chamber slides (BD Biosciences) with or without 1μM XAV939 (Wnt antagonist; Sigma), 2 mM LiCl (Wnt agonist) or 2 mM NaCl (control). Videos were taken with a Nikon Diaphot 300 microscope and XY Clone software (Hamilton Thorne Biosciences). Analysis of beating area was performed using VirtualDub software (v1.9.11) and ImageJ (NIH).
Measurement of calcium transients
Yolk sacs were cultured on petri dishes (MatTek Corporation) coated with Matrigel in cardiac medium for 9-10 days. Before imaging, explants were loaded with the Ca2+-sensitive dye Fluo-4AM (Molecular Probes) in the presence of Pluronic F-127 (Molecular Probes). Spontaneous transients were imaged on a Zeiss LSM 5 Pascal confocal microscope equipped with a 488 nm laser.
In vivo manipulation of regulation
Scl deletion in Rosa26Cre-ERT2 Sclfl/fl embryos was induced by Tamoxifen (Sigma Aldrich T5648) or (Z)-4-hydroxytamoxifen (Sigma Aldrich H7094). Tamoxifen was suspended at 100 mg/mL in ethanol and mixed with sesame oil at a final concentration of 10 mg/mL (10% ethanol). Pregnant females were injected IP with 125 μL tamoxifen at the indicated days. Injections of 250 mg/kg NaCl and LiCl (Wnt agonist) were performed as described (Tian et al., 2010).
Statistical Analysis
For statistical analysis, Student’s unpaired two-tailed t-test was used for all comparisons.
Supplementary Material
Highlights.
Scl establishes hemogenic endothelium and prevents its conversion to cardiac fate
Scl inhibits precocious cardiomyocyte differentiation of endocardium in the heart
The requirement for Scl to repress cardiomyogenesis is cell-intrinsic and temporally defined
Ectopic cardiomyogenesis in Scl deficient tissues is promoted by Wnt antagonism
Acknowledgments
The authors thank Dr. Joshua Bloomekatz and Dr. Deborah Yelon for critical reading of this manuscript and sharing unpublished data and Guillaume Hagen for preparing the movies. The authors thank the BSCRC Flow Cytometry Core for FACS sorting. This work was funded by the CIRM New Faculty Award and NIH/RO1 (HL097766-01) grants to H.K.A.M. B.V.H. was supported by the Ruth L. Kirschstein NRSA GM07185 and NIH/NHLBI T32 HL69766. A.M-H was supported by successive fellowships from EMBO and HFSP. R.F. was funded in part by CIRM grant to S.K.K. C.J.B was funded by Rubicon grant 825.10.016 from the Netherlands Organization for Scientific Research.T.O. was supported by the EU through the European Social Fund (Mobilitas grant No. MJD284). S.H.O., an Investigator of the HHMI, is supported in part by a Center of Excellence in Molecular Hematology award from the NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: B.V.H. and A.M-H., project design, experimental work, data analysis and interpretation, manuscript preparation; R.S., R.F. and T.O., bioinformatic data analysis and interpretation, manuscript editing; H.N., C.J.B., J.S. experimental work, data analysis and interpretation, manuscript editing; Y.W., S.H. and J.Z., experimental work; X.L., J-N. C. and S.H.O., data interpretation, manuscript editing; M.P., bioinformatic method design, manuscript editing; S.K.K., S.M.E. and A.N., project design, data interpretation, manuscript editing; H.K.A.M., project design, data analysis and interpretation, manuscript preparation.
References
- Ahnfelt-Ronne J, Jorgensen MC, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An improved method for three-dimensional reconstruction of protein expression patterns in intact mouse and chicken embryos and organs. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2007;55:925–930. doi: 10.1369/jhc.7A7226.2007. [DOI] [PubMed] [Google Scholar]
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Giroux S, Golub R, Klaine M, Jalil A, Boucontet L, Godin I, Cumano A. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc Natl Acad Sci U S A. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nature cell biology. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, de Gier-de Vries C, Schneider MD, Webb S, van den Hoff MJ, Christoffels VM. Lineage and morphogenetic analysis of the cardiac valves. Circulation research. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailoglu I, Yeamans G, Daley GQ, Perlingeiro RC, Kyba M. Mesodermal patterning activity of SCL. Experimental hematology. 2008;36:1593–1603. doi: 10.1016/j.exphem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Jaspard B, Couffinhal T, Dufourcq P, Moreau C, Duplaa C. Expression pattern of mouse sFRP-1 and mWnt-8 gene during heart morphogenesis. Mechanisms of development. 2000;90:263–267. doi: 10.1016/s0925-4773(99)00236-1. [DOI] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Developmental cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell stem cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Loebel DA, Tam PP. Allocation and early differentiation of cardiovascular progenitors in the mouse embryo. Trends in cardiovascular medicine. 2001;11:177–184. doi: 10.1016/s1050-1738(01)00091-3. [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell stem cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003a;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003b;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell stem cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S, Kohli V, Kang J, Chi NC, Black BL, Sumanas S. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development. 2011;138:4721–4732. doi: 10.1242/dev.064998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell stem cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, et al. ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011;138:4801–4812. doi: 10.1242/dev.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HK. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell stem cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger TM, Mikkola HK, Gekas C, Helgadottir HB, Orkin SH. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105:3871–3874. doi: 10.1182/blood-2004-11-4467. [DOI] [PubMed] [Google Scholar]
- Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes & development. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Developmental cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- Simoes FC, Peterkin T, Patient R. Fgf differentially controls cross-antagonism between cardiac and haemangioblast regulators. Development. 2011;138:3235–3245. doi: 10.1242/dev.059634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell stem cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, et al. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Developmental cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes & development. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell stem cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Yamagishi T, Ando K, Nakamura H. Roles of TGFbeta and BMP during valvulo-septal endocardial cushion formation. Anatomical science international. 2009;84:77–87. doi: 10.1007/s12565-009-0027-0. [DOI] [PubMed] [Google Scholar]
- Zeigler BM, Sugiyama D, Chen M, Guo Y, Downs KM, Speck NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell stem cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.