Abstract
A recent ion mobility – mass spectrometry (IM–MS) study of the nonapeptide bradykinin (BK, amino acid sequence Arg1–Pro2–Pro3–Gly4–Phe5–Ser6–Pro7–Phe8–Arg9) found evidence for 10 populations of conformations that depend upon the solution composition [J. Am. Chem. Soc. 2011, 133, 13810]. Here, the role of the three proline residues (Pro2, Pro3, and Pro7) in establishing these conformations is investigated using a series of seven analogue peptides in which combinations of alanine residues are substituted for prolines. IM–MS distributions of the analogue peptides, when compared to the distribution for bradykinin, indicate the multiple structures are associated with different combinations of cis and trans forms of the three proline residues. These data are used to assign the structures to different peptide populations that are observed under various solution conditions. The assignments also show the connectivity between structures when collisional activation is used to convert one state into another.
Introduction
Bradykinin (BK), a nine residue peptide (Arg1–Pro2–Pro3–Gly4–Phe5–Ser6–Pro7– Phe8–Arg9), is associated with blood pressure regulation and vasodilation, pain response, and inflammation.1,2 Since the 1949 discovery of its physiological effects,3 many investigators have attempted to determine the structure of BK. Nuclear magnetic resonance (NMR),4–7 circular dichroism,6–8 and molecular dynamics (MD)4–11 studies of the free peptide yield a partial characterization of the structure –identification of a β-turn involving the Ser6–Pro7–Phe8–Arg9 residues. No definitive structural information about the N-terminal portion of the peptide exists. Instead, this region is described as a highly flexible12 random coil7,13 that is believed to be unstructured in solution.14 Recently we found evidence for as many as 10 populations of different structural forms of free BK, that vary depending on the solution composition;15 this plurality of states is consistent with the inability to characterize the N-terminal region of the peptide. In 2008, Lopez et al. used NMR and MD techniques to show that BK binds to its B2 receptor in a relatively open geometry in which all three proline residues are in the trans conformation.14
That only a single BK conformer is complexed with its receptor raises a number of interesting questions. For example: what elements of structure exist in other forms of the free peptide? Can the inactive BK states convert into conformers that are capable of binding to the receptor? Or, do these other conformers represent dead ends that remain inactive? One can imagine that the many structures may be specific to other receptors, or even suggest the existence of other receptors that are yet to be discovered. Complete characterization of BK conformations in different environments is important in understanding its biological role, and may aid in the design of more effective receptor agonist and antagonist analogues.
This study focuses on understanding the origin of BK conformers that exist in the absence of BK receptors. After considering several factors that may influence conformer preferences of the unbound peptide, we conclude that the Pro2, Pro3, and Pro7 residues are key in establishing multiple conformations of the free peptide. Specifically, combinations of cis or trans forms of these three residues are responsible for some of the populations observed experimentally. Although cis and trans configurations of proline have been studied for many years (using numerous experimental approaches), the most convincing data about preferred configurations is derived from theory as well as statistical surveys of X-ray structures of non-redundant chains from the Protein Data Bank.16 Generally prolines in amino acid chains are found in the trans configuration (∼95% of the time).17 The present study integrates amino acid substitution chemistry and ion mobility – mass spectrometry (IM–MS) analysis for understanding the conformer preferences of biologically relevant peptides. The data provides insight into the critical role of cis and trans proline configurations in the populations of structures that are observed in solution. We find that the BK conformers present in solution have a high preponderance to incorporate cis Pro configurations. A discussion of variations in populations with different solution composition and the ability of states to interconvert is provided.
Experimental
Peptides and peptide modification
Table 1 provides a list of all of the peptides that were used in this study. A detailed description of how each was obtained is provided below. The BK peptide was purchased as a lyophilized acetate salt from Sigma-Aldrich (≥98%, St. Louis, MO, USA), and used without further purification. Amino-terminally acetylated BK was produced according to the protocol of Abello et al.18 N-hydroxysuccinimide (NHS) acetate (100 mM in DMSO) was added at a final concentration of 5 mM to 10 μM bradykinin in 100 mM sodium phosphate buffer (pH 8) and placed in a boiling water bath for 60 minutes. The sample (99% purity) was desalted with an Oasis HLB cartridge (Waters; Milford, MA, USA) prior to electrospray ionization (ESI).
Table 1.
Collision cross sections of [M+3H]3+ BK and analogue peptide conformations.
|
|
||||||
|---|---|---|---|---|---|---|
| Ωa (Å2) of Conformationb | ||||||
|
| ||||||
| Peptide | Sequence | % Purity | A | B | C | other |
| Bradykinin | RPPGFSPFR | ≥98 | 269 | 285 | 305 | 323,336,350 |
| BK-methyl | RPPGFSPFR-CH3 | 89 | 280 | 290 | 310 | 303 |
| Acetyl-BK | CH3CO-RPPGFSPFR | 99 | - | - | - | - |
| Pro2→ Ala BK | RAPGFSPFR | >99 | - | - | 304 | - |
| Pro3→Ala BK | RPAGFSPFR | 98 | - | 285 | 303 | 293 |
| Pro7→ Ala BK | RPPGFSAFR | 95 | - | 290 | - | - |
| Pro2,3→Ala BK | RAAGFSPFR | 97 | - | - | 304 | - |
| Pro2,7→ Ala BK | RAPGFSAFR | >99 | - | - | - | 292 |
| Pro3,7→Ala BK | RPAGFSAFR | >99 | - | 284 | - | - |
| Pro2,3,7→Ala BK | RAAGFSAFR | 95 | - | - | - | 293 |
Collision cross sections (Ω) of Pro→Ala analogue peptides are size-parameter shifted as described in the Experimental section
Conformation assignments A, B, and C are defined from ref. 46
Carboxy-terminal methylation of BK was carried out according to Ma et al.19 To 0.85 μg BK (lyophilized powder) was added 200 μL of methanolic HCL and incubated at 37°C for 2 hours. The solution was then dried in a Speed-Vac (Labconco; Kansas City, MO, USA) (89% purity), and resuspended for ESI.
Peptide synthesis
BK analogue peptides were synthesized on an Apex 396 peptide synthesizer (AAPPTec, Louisville, KY) by a standard Fmoc solid-phase peptide synthesis protocol. C-terminal Arg(Pbf) Wang-type polystyrene resin and Fmoc side-chain protected amino acids were purchased from Midwest Biotech (Fishers, IN, USA). Nα deprotections were carried out with 20% piperidine in dimethylformamide (DMF); double couplings were performed with 1,3-diisopropylcarbodiimide (DIC) / 6-chloro-1-hydroxibenzotriazol (6-Cl-HOBt). Peptides were cleaved from the resin with a solution of trifluoroacetic acid (TFA):triisopropylsilane (TIS):CH3OH (15:1:1), followed by precipitation into diethyl ether. Peptides were lyophilized, and purified by semi-preparative-scale reversed-phase liquid chromatography. Table 1 includes a list of all analogue sequences that are analyzed below as well as their estimated purities based on MS analysis. In general, sample purities range from ∼95 to >99% (i.e. HPLC grade).
ESI solutions
The various peptides were dissolved in 49:49:2CH3OH:H2O:CH3COOH to create ∼10 μM solutions. We note that a previous study15 utilized a wide range of ESI solvent compositions (0:100 to 100:0 CH3OH:H2O and 0:100 to 90:10 dioxane:H2O), which will be included in the discussion below.
Instrumentation
Ion mobility spectrometry techniques,20,21 instrumentation,22–25 and theory26–28 have been reviewed previously. A brief description is provided here. IM–MS measurements were carried out on a home-built ion mobility/time-of-flight mass spectrometer (Figure S1, Supporting Information). Solutions were electrosprayed from a NanoMate chip-based nano-ESI autosampler (Advion Biosciences, Inc., Ithaca, NY, USA). Ions were trapped in a Smith-geometry ion funnel, and pulsed into a ∼1.8-meter drift tube for mobility separation. Ion mobility measurements were performed under low-field conditions which employed 3.00 Torr He buffer gas and a drift field (E) of 10 V·cm-1. Mobility-separated ions were then focused into the source region and orthogonally accelerated into a two-stage reflectron-geometry time-of-flight mass spectrometer. Mass spectra were recorded every 50 μs resulting in a nested drift time (tD) and mass-to-charge (m/z) measurement.29
The instrument can also be used for IM–IM–MS experiments; this is accomplished by applying an electrostatic gate at the entrance of an ion funnel in the middle of the drift tube as described previously.30 This gate utilizes a delay pulse synchronized with the source pulse to select mobility-separated ions after ∼0.7 meters of drift length. A voltage can be applied at the conclusion of the ion funnel to collisionally activate the selected ions to form a new population of ions, which then separate through the remaining meter of drift tube.
Determination of experimental collision cross sections
The present study focuses on the +3 charge state. Therefore it is possible to convert drift times into values of collision cross section (Ω) and useful to compare mobility distributions directly on a cross section scale. This is a straightforward conversion as given by the following equation:31
where kb is Boltzmann's constant, T is temperature (300 K for these studies), mI is the mass of the ion, mB is the mass of the buffer gas (He), L is the length of the drift region, P is the pressure, and N is the number density of the gas at STP.
Correction of cross section distributions for Ala-substituted sequences by use of intrinsic size parameters
For direct comparison of BK analogue peptides (in which Ala residues are substituted for Pro residues) with BK, the size difference between Ala and Pro must be taken into consideration. For example, the mass of bradykinin is 1059.56 Da, and the mass of the three single-substituted Pro→Ala analogues is 1033.55 Da. The differences in the size of the Pro and Ala residues can be accounted for by using the intrinsic amino acid size parameters published by Valentine et al.32 and amino acid residue collision cross sections by Srebalus-Barnes et al.33 Assuming no other changes in the conformation, a single Ala substitution will shift the cross section by the difference in the intrinsic size parameters for Pro and Ala (i.e., 19.82–17.35 ≈ 2.5 Å2). Similarly, a two-residue substitution would shift the cross section scale by 4.9 Å2, and a substitution of Ala at all three Pro sites leads to a shift of 7.4 Å2 . As shown below, these shifts are very reproducible and when accounted for provide us with a means of understanding which types of structures are insensitive to Ala substitution (in which case the Pro residue is unimportant in establishing a given conformation) and which substitutions dramatically influence the distribution of structures that is observed.
Results and discussion
Background regarding the BK structure and charge configuration in the gas phase
One of the first issues that arises in relation to the structure of BK involves understanding the influence of the protonation state on conformation. Specifically, where are the protons located? The structure of the [BK+H]+ and [BK+2H]2+ ions in the gas phase have been studied extensively by a number of methods, including: isotopic H/D exchange;34–37 ion/ion reactions;38 ion dissociation techniques;39,40 cold ion spectroscopy;41 and IM–MS techniques.42–46 Williams and his coworkers39 used blackbody infrared radiative dissociation to study [BK+H]+ and its methyl ester ([BK-methyl+H]+). The idea is that modification of the basic or acidic groups restricts where the charges can reside. Based on their findings, they proposed that the [BK+H]+ ion exists as a salt-bridge structure. Results from H/D exchange34,37 and IM–MS42,43,47 studies are in agreement with the salt-bridge assignment for both [BK+H]+ and [BK+2H]2+.
The structure of gaseous [BK+3H]3+ ions has also been the subject of numerous studies;15,36,44,46,47 This ion has received considerable attention because multiple features are resolved in the IM–MS distribution and the intensities of these features vary substantially depending composition of the analyte solution, and instrumental conditions employed.15 On the basis of theoretical studies, Siu and coworkers proposed a non-salt bridged charge configuration for [BK+3H]3+ in which the two guanidine groups and the carbonyl oxygen of Phe5 are protonated;48 however, experimental support for this configuration is limited.
Experimental investigation of [BK+3H]3+ charge-site configuration
Because of the extensive work characterizing the charge-site configuration of BK in the gas phase, we begin by addressing this first for [BK+3H]3+. Our initial impression was that different charge configurations might be responsible for the multiple peaks that are observed in the IM–MS distributions.15,46 We proceed by modifying the ends of BK to restrict possible charge configurations –an approach that is analogous to that used by Williams and coworkers.
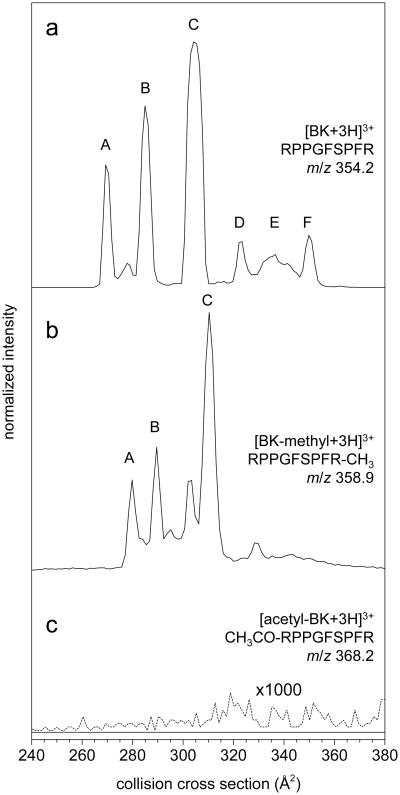
Figure 1 shows the ion mobility distributions that are recorded for the [BK+3H]3+, [BK-methyl+3H]3+, and [acetyl-BK+3H]3+ ions. A summation of the cross sections that are recorded for the different ions is provided in Table 1. Here, we focus on the three main features, previously assigned conformations A, B, and C.46 We focus on these peaks because they are the largest in the observed IM distributions (under the solution conditions employed) and we have previously studied the interconversion of these states in detail by ion selection and activation techniques.46 We note that the arguments that are presented below can be extended to the more elongated D through G states (reported previously15). The interested reader can evaluate these differences for states D, E, and F independently from the data we present below.
Figure 1.
Collision cross section distributions of (a) [BK+3H]3+, (b) [BK-methyl+3H]3+, and (c) [acetyl-BK+3H]3+. The three most abundant features, labeled A, B, and C, appear analogous between BK and the BK-methyl. No [M+3H]3+ ions were observed for acetylated BK, as shown by the 1000× zoom (dashed line) of the baseline.
Four configurations of charge sites resulting in a net charge of +3 have been considered previously.48 Each of these four configurations assigns protons to both highly basic guanido groups of arginine (accounting for two of the charges). With this in mind, the four charge-site assignments are completed as follows: (1) addition of a third proton at the amino terminus; (2) protonation of the backbone carbonyl oxygen of Phe5; (3) protonation of backbone carbonyl oxygen of Gly4; or (4) protonation of the amino terminus and a backbone carbonyl oxygen together with a deprotonation of the carboxyl terminus, resulting in a salt-bridged structure.
It is possible to test which of these configurations is present by analyzing the set of modified BKs described above. IM–MS analysis of the BK-methyl was performed to address the possibility of a salt-bridged structure (charge-site configuration 4). Methylation blocks the carboxyl terminus from deprotonating, hence preventing formation of a salt bridge between the C-terminus and a protonation site. The presence of unmodified BK ([M+2H]2+ at m/z 530.8 and [M+3H]3+ at m/z 354.2) and methyl-ester form ([BK-methyl +2H]2+ at m/z 537.8 and [BK-methyl+3H]3+ at m/z 358.9) are observed in the mass spectrum (not shown). If one (or any) of the peaks in the unmodified [BK+3H]3+ IM–MS distribution are due to the salt-bridge structure, they would not appear in the BK-methyl distribution. The three main features in the IM–MS distribution of the [BK-methyl+3H]3+ are similar in appearance to peaks A, B, and C observed for unmodified [BK+3H]3+. The addition of the methyl ester increases the cross sections by ∼5 to 10 Å2. The [BK-methyl+3H]3+ distribution also shows a partially resolved leading shoulder on peak C, termed C′, which was also observed previously for [BK+3H]3+.15 Overall, the [BK-methyl+3H]3+ distribution is remarkably similar to that for [BK+3H]3+. Thus, unlike the singly and doubly charged forms of BK, the results reported here for the methyl-ester form rule out conformers originating from salt-bridged ions for triply charged [BK+3H]3+ (configuration 4).
In order to investigate the three remaining charge-site configurations, i.e. where the third proton is located on the N-terminus (configuration 1), the Phe5 backbone carbonyl oxygen (configuration 2), or the Gly4 backbone carbonyl oxygen (configuration 3), acetyl-BK was synthesized and analyzed. Acetylation of BK blocks the N-terminus from being charged. Thus, if protonation at this site is required then one would expect to see no ion signal. If protonation at the amino terminus is optional, then [M+3H]3+ ions from charge-site assignments 2 and 3 should be observed. There is no evidence for [acetyl-BK+3H]3+ ions at m/z 368.2. It is important to note that [acetyl-BK+2H]2+ is observed at m/z 551.8 confirming acetyl-BK was indeed produced (Figure S2). These results require that the amino terminus be the site of the third proton.
The combination of results from the N- and C-terminally blocked BK peptides indicates only a single charge-site assignment exists –configuration 1, in which the two arginine side chains are protonated and the third proton resides on the amino terminus. The multiple peaks that are observed in Figure 1a must be imposed by some other structural factor.
Some other structural factor –cis/trans isomers of proline
Having ruled out the possibility of multiple charge assignments, we next consider whether different combinations of cis/trans Pro configurations (residues 2, 3, and 7 of BK) give rise to the multiple peaks observed by IM–MS. The substitution of Ala for a specific Pro residue is denoted here as, Pron→Ala, where n indicates the position of the substituted residue. A complete list of all of the Ala analogue peptides is provided in Table 1.
It is interesting to consider the implications of substituting Ala for Pro. Because trans configurations are intrinsically more stable, nearly all peptide bonds adopt this configuration (i.e., one in which the dihedral angle of the backbone Cα1–C′1–N2–Cα2 atoms is ∼180°); this configuration also reduces steric hindrance of the sidechains.49,50 For Xxx–nonPro bonds (where Xxx is any amino acid), the barrier between trans to cis rotation is approximately 20 kcal·mol-1 and the trans isomer is ∼2.5 kcal·mol-1 lower in energy.51 In the case of Xxx–Pro, however, the energy barrier is significantly lower (∼13 kcal·mol-1) and the energy difference is only ∼0.5 kcal·mol-1 between cis and trans forms of the peptide bond.50 We note that other substitutions could be used (e.g. D-Pro, sarcosine, pipecolic acid); however these substitutions, like Pro, can also readily isomerize to form both cis and trans populations. Here, we have selected Ala substitution because Xxx–Ala peptide bonds almost always exist as the trans isomer.
Single Pro→Ala substitution results
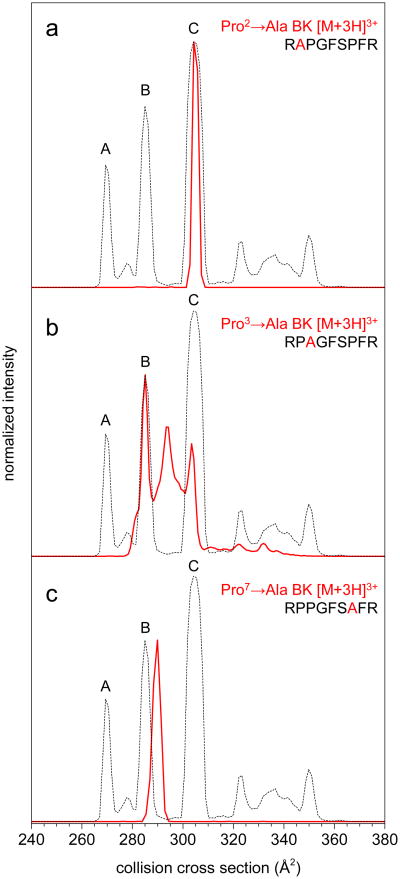
Figure 2 shows the cross section distributions (after correction for differences in the intrinsic sizes of Pro and Ala) for the three single-substituted analogues Pro2→Ala, Pro3→Ala, and Pro7→Ala. For ease of comparison each distribution is plotted on top of that recorded for the native sequence [BK+3H]3+ ion. The distribution for the Pro2→Ala analogue shows a single peak at 304 Å2 –the same position as the C conformer recorded for the native BK sequence. All other features that were observed for the native BK sequence are not observed for the Pro2→Ala analogue. Clearly the substitution of Ala at Pro2 has a major influence on the distribution of peptide structures. The result that conformations A and B (present for the native [BK+3H]3+ distribution) are not observed is revealing. These two conformers must require Pro2. Conformation C, however, persists in the analogue peptide when Ala takes the place of Pro2.
Figure 2.
Collision cross section distributions for [M+3H]3+ ions of the three single Pro→Ala substituted BK analogue peptides (red traces) overlaid with the distribution of [BK+3H]3+ (dashed trace). For direct comparison of the analogues to BK, the cross section scales for the three analogue peptides were shifted according to amino acid size parameters from ref. 33 to account for the Pro→Ala substitution; see Experimental section for details. Intensities were normalized to peak maxima of BK for ease of visualization.
The observation of only the C conformer in the Pro2→Ala analogue can also be used to assign the cis/trans configurations of the Arg1–Pro2 peptide bond. Remembering the energetic arguments presented above, the Arg1–Ala2 peptide bond in the Pro2→Ala analogue is almost certainly fixed in the trans configuration; whereas, Arg1–Pro2 in BK can be in either cis or trans. Because Arg–Ala has such a strong preference for trans, there should be no change in collision cross section if Pro2 is trans in native BK. Therefore, the agreement between the analogue and native BK sequence for peak C requires that for this conformer, the Arg1–Pro2 peptide bond in BK has a trans configuration. Because conformers A and B are not populated in the Pro2→Ala analogue we assign the Arg1–Pro2 peptide bond as the cis isomer form in BK conformers A and B.
This analysis is easily extended to the Pro3→Ala analogue. Figure 2 shows that substitution of Ala at Pro3 results in peaks that are in the same position as conformers B (285 Å2 for both the native and the Pro3→Ala analogue) and C (305 Å2 for the native peptide and 303 Å2 for the Pro3→Ala analogue). No features in the distribution for the Pro3→Ala analogue match the peak for native BK conformer A. The differences between the Pro2–Pro3 and Pro2–Ala3 require that the Pro2–Pro3 peptide bond for conformer A of the native BK sequence must exist in a cis configuration. Likewise, the Pro2–Pro3 peptide bond in conformers B and C must have a trans configuration.
Figure 2 also shows a third peak in the distribution for the Pro3→Ala analogue that falls between the peaks for conformers B and C. In order to investigate this new feature we carried out multidimensional IM–IM–MS experiments in which each of the three features in the distribution were selected and collisionally activated (Figure S3), as described previously.46,30 The high- and low-mobility peaks were found to convert into one another in the mid-drift tube ion activation region. Interestingly, it was not possible to select the middle peak; instead, selection (with no activation) resulted in a spectrum with two peaks on either side of the selection pulse that matched the high- and low-mobility peaks. This indicates the feature in the middle is not a third, stable conformation of Pro3→Ala BK; instead, the metastable peak is an indication of interconversion of conformers B and C between one another on the millisecond timescale of the IM separation. The results of these IM–IM–MS experiments suggest Pro3 plays a role in stabilizing conformers B and C thereby preventing low-barrier interconversion in native BK.
Finally, Figure 2 also shows the IM–MS distribution obtained for the Pro7→Ala analogue. A single major feature of slightly lower mobility than BK conformation B is observed. The size-parameter shifted cross section of [Pro7→Ala BK+3H]3+ is 290 Å2, which is approximately 2% difference from BK conformer B. When selected and activated in by IM–IM–MS, the Pro7→Ala peak shifts from 290 Å2 to 286.7 Å2, and overlaps with BK conformer B (Figure S4). These results suggest Ser6–Pro7 is in the cis configuration in BK conformations A and C, and the trans isomer is favored in B.
The results obtained from the single-substituted BK analogues are summarized in Table 2. None of the single-substituted analogues form peaks that overlap with BK conformation A, which indicates all three prolines must be present to establish this BK structure. This suggests BK conformer A is cis-Pro2, cis-Pro3, and cis-Pro7. Combining information from all three of the single Pro to Ala analogues also yields the assignments of cis-Pro2, trans-Pro3, trans-Pro7 for BK conformer B, and trans-Pro2, trans-Pro3, cis-Pro7 for BK conformer C.
Table 2.
Isomer forms of Pro in BK structures A, B, and C.a
| BK Conformera | Pro2 | Pro3 | Pro7 |
|---|---|---|---|
| A | cis | cis | cis |
| B | cis | trans | trans |
| C | trans | trans | cis |
Conformations A, B, and C are [BK+3H]3+ collision cross sections defined from ref. 46
Double Pro→Ala substitutions for verification of assignments
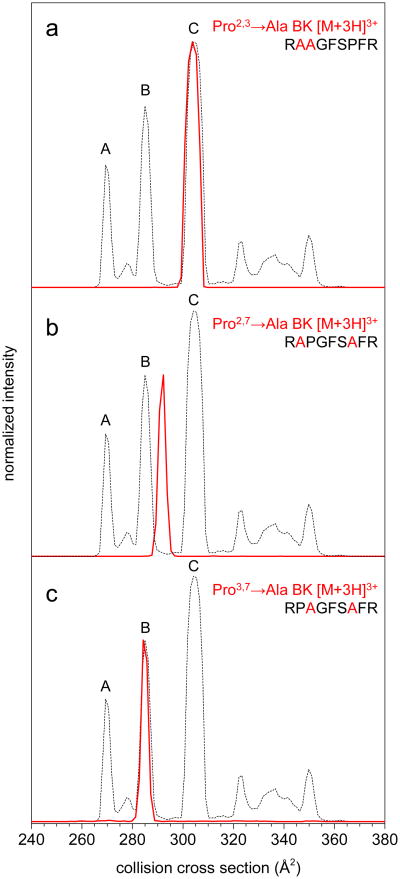
For the cis/trans analysis up to this point, cis Xxx–Pro is assigned in the cases where peaks are eliminated from the [BK+3H]3+ distribution, and trans Xxx–Pro is assigned when the Ala-substituted peak matches an original BK conformer. To further test the assignments in Table 2, three double-substituted Pro→Ala analogues were analyzed. Figure 3 shows the [M+3H]3+ size-parameter shifted cross section distributions of these analogues compared against that of [BK+3H]3+. The assignments derived from the data shown above for the single-substituted study suggest that fixing trans-Arg1–Ala2 and trans-Ala2–Ala3 would eliminate conformations A and B, and only conformer C could form with Pro7 in the cis peptide bond configuration (trans-Pro2, trans-Pro3, cis-Pro7 assignment for conformer C). The distribution for Pro2,3→Ala BK shows a single peak centered at 304 Å2 that matches well with BK conformer C, thus confirming the prediction and the following configurations of Pro residues: trans-Pro2; trans-Pro3; and, cis-Pro7.
Figure 3.
Comparison of collision cross section distributions of the three [M+3H]3+ double Pro→Ala substituted peptide analogues (red traces) with BK (dashed trace). Amino acid size parameters from ref. 33 were used to calibrate cross section scales of the analogues to BK. Intensities were normalized to peak maxima of BK for ease of visualization.
Pro2,7→Ala BK is a somewhat different case, in that fixing residues two and seven as trans does not match any of the cis/trans combinations in Table 2. Therefore, one would predict Pro2,7→Ala BK would not align with any of the three major BK peaks A, B, and C. As shown in Figure 3, the size-parameter shifted Ω for [Pro2,7→Ala BK+3H]3+ is ∼292 Å2, which is more than 2% larger than BK conformer B. It is important to note that when selected and activated by multidimensional IM, this peak does not change position (data not shown). Due to the relatively large difference in Ω between the analogue and BK conformers, and that the activated peak does not shift, it can be concluded that the peak for Pro2,7→Ala BK is not analogous to any of the BK structures. This is consistent with what would be expected on the basis of the single substitution results, as the two possible Pro2,7→Ala BK combinations trans-Pro2, cis-Pro3, trans-Pro7 and trans-Pro2, trans-Pro3, trans-Pro7 do not match any of the three cis/trans combinations in Table 2.
The case in which the second two prolines are fixed in trans configurations matches the cis-Pro2, trans-Pro3, trans-Pro7 assignment for conformer B in Table 2. The cross section distribution for Pro3,7→Ala BK in Figure 3 clearly shows a single feature overlapping with BK conformation B, thus confirming this assignment. In summary, all three double-substituted Pro→Ala BK analogues yield cis/trans configuration assignments that agree with the predictions from the data for the single-substituted analogues.
Triple-substituted (all trans) BK analogue
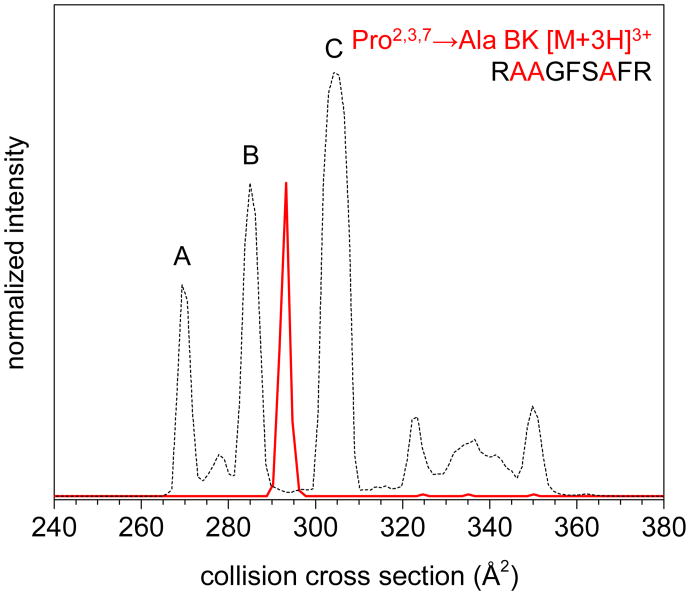
Finally, Figure 4 shows the IM–MS distribution for Pro2,3,7→Ala BK, where all three Pro residues are substituted for Ala. A single peak at Ω = 293 Å2 (size-parameter shifted) is observed. While this peak lies close to that of BK conformer B, its cross section is more than 2.5% larger. Additionally, the peak does not shift when selected and activated by the IM–IM–MS approach described above. As with Pro2,7→Ala BK, we conclude that the triple-substituted Pro→Ala sequence is not analogous to BK, and instead represents a new (all trans) conformation. This is also consistent with the cis/trans configuration assignments that were made from evaluation of the single- and double-substitution distributions.
Figure 4.
Cross section distribution of Pro2,3,7→Ala BK [M+3H]3+ (RAAGFSAFR) (red) and [BK+3H]3+ (dashed). Amino acid size parameters from ref. 33 were used to calibrate cross section scales of the analogues to BK (see text for details). Intensities were normalized to peak maxima of BK to aid in comparison of the two distributions.
Rationalizing transitions between states with cis/trans configuration assignments
It is interesting to consider the structural transitions that are observed with these assignments in mind. In previous work we have shown that it is possible to select and activate conformers A, B, and C and convert them into one another.46 The present cis/trans Pro assignments require in every case that two prolines isomerize from cis to trans or from trans to cis during this activation phenomenon. This is somewhat surprising, as one would expect the simplest transition would involve a single isomerization event. It is possible that these results indicate a concerted two-proline transition in this system. Alternatively, it could suggest our assignments are incomplete (only four of the eight possible Pro configurations for BK have been addressed). For example, conformer B might also be consistent with the cis-Pro2, trans-Pro3, cis-Pro7 configuration (on the basis of the slight mobility mismatch discussed for the Pro7→Ala distribution). In this scenario, the A to B transition requires a single cis to trans Pro3 isomerization, followed by a single cis to trans isomerization of Pro2 for the B to C transition.
Implications for solution-phase populations
From the statistically derived propensity of proline to prefer the trans isomer17 we would initially predict that a system with three proline residues has a 0.05, 0.0025, and 0.000125 probability of containing one, two, or three cis peptide bonds, respectively. Our current study allows us to test these predictions. Moreover, from our earlier paper15 we presented the dependence of IM peak intensities on the solution composition. Thus, the predictions can be evaluated for a range of solutions. Using the above assignments it is possible to obtain the populations of different cis/trans configuration forms across the range of different solution compositions that were evaluated.
Briefly, we find that BK in methanol exists as predominantly the cis-Pro2, trans-Pro3, trans-Pro7 isomer (39% of the total [BK+3H]3+ distribution); in aqueous environments, the trans-Pro2, trans-Pro3, cis-Pro7 form exists in highest abundance (43%). In a 90:10 dioxane:water solution (in which previous NMR studies have been conducted),5 approximately equal abundances of conformers B and C are observed (23 and 27%, respectively). Across all solution compositions studied,15 ∼10% of the total [BK+3H]3+ distribution exists in the cis-Pro2, cis-Pro3, cis-Pro7 isomer form; this is significantly higher than the 0.0125% predicted from statistical analysis of the Protein Data Bank. We speculate the various studies4,5 that describe the Ser6–Pro7–Phe8–Arg9 β-turn (where Ser6–Pro7 is trans) are in agreement with the cis-Pro2, trans-Pro3, trans-Pro7 isomer assignment (conformation B) in the present report.
Summary and conclusions
IM–MS distributions observed for single-, double-, and triple-substituted Pro→Ala substitutions in BK illustrate the important role of all three Pro residues in the origin of multiple BK conformations. All three Pro residues are essential for the formation of BK conformer A, which suggests A is the cis-Pro2, cis-Pro3, cis-Pro7isomer of the peptide. The requirement that all three Pro residues exist in this specific configuration, helps explain why conformation A only makes up 2% of the total gas-phase [BK+3H]3+ distribution in our previous studies.46 The results of the present study were also used to assign conformer C as the trans-Pro2, trans-Pro3, cis-Pro7 isomer form. The well-defined Ser6–Pro7–Phe8–Arg9 β-turn observed by NMR4,5 possesses a trans-Ser6–Pro7 peptide bond; this is in agreement with our assignment of the cis-Pro2, trans-Pro3, trans-Pro7 configuration for conformer B.
It is interesting to note that all previous efforts to computationally model this peptide ion have been directed at all-trans BK. This might explain why several different low-energy structures for BK have been reported in the literature.48,52 The present findings suggest both cis- and trans-Xxx–Pro should be evaluated in simulations of proline-containing polypeptides, which could help refine molecular dynamics approaches for small peptide systems.
Finally, we note that the abundance of cis Pro configurations in these studies, compared to the expectations of a preponderance of trans peptide bonds (derived statistically from protein crystal structures) was initially surprising to us. Overall, this finding suggests that it will be of interest to determine the proline configurations for other peptides. Moreover, the present work introduces a complexity into modeling proline-containing peptides. Unless the BK system is simply unusual, it may not be safe to assume that trans-Pro dominates the structures for smaller peptide systems.
Supplementary Material
Acknowledgments
The authors thank David Smiley and the DiMarchi Laboratory at Indiana University for assistance with the peptide synthesis used in this work. N. P. and D. C. gratefully acknowledge partial funding for instrumentation development from the NIH (RC1GM090797-02) and from the Indiana University METAcyte initiative that is funded by a grant from the Lilly Endowment. L. C. and D. R. acknowledge support for this research by The Robert A. Welch Foundation (A-1176).
Footnotes
Notes: The authors declare no competing financial interest
Supporting Information: Additional figures are included in a Supporting Information section. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hall JM, Morton IKM. The Pharmacology and Immunopharmacology of Kinin Receptors. In: Farmer SG, editor. The Handbook of Immunopharmacology: The Kinin System. San Diego, CA: Academic Press; 1997. pp. 1–43. [Google Scholar]
- 2.Bathon JM, Proud D. Annu Rev Pharmacol Toxicol. 1991;31:129–62. doi: 10.1146/annurev.pa.31.040191.001021. [DOI] [PubMed] [Google Scholar]
- 3.Rocha M, Silva E, Beraldo WT, Rosenfeld G. Am J Physiol. 1949;156:261–273. doi: 10.1152/ajplegacy.1949.156.2.261. [DOI] [PubMed] [Google Scholar]
- 4.Lee SC, Rusell AF, Laidig WD. Int J Peptide Protein Res. 1990;35:367–377. [PubMed] [Google Scholar]
- 5.Young JK, Hicks RP. Biopolymers. 1994;34:611–623. doi: 10.1002/bip.360340504. [DOI] [PubMed] [Google Scholar]
- 6.Cann JR, Liu X, Stewart JM, Gera L, Kotovych G. Biopolymers. 1994;34:869–878. doi: 10.1002/bip.360340706. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee C, Mukhopadhyay C. Biochem Biophys Res Commun. 2004;315:866–871. doi: 10.1016/j.bbrc.2004.01.134. [DOI] [PubMed] [Google Scholar]
- 8.Cann JR, Vatter A, Vovrek RJ, Stewart JM. Peptides. 1986;7:1121–1130. doi: 10.1016/0196-9781(86)90142-7. [DOI] [PubMed] [Google Scholar]
- 9.Nikiforovich GV. Int J Peptide Protein Res. 1994;44:513–531. doi: 10.1111/j.1399-3011.1994.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 10.Salvino JM, Seoane PR, Dolle RE. J Comput Chem. 1993;14:438–444. [Google Scholar]
- 11.Manna M, Mukhopadhyay C. Langmuir. 2011;27:3713–3722. doi: 10.1021/la104046z. [DOI] [PubMed] [Google Scholar]
- 12.Denys L, Bothner-By AA, Fisher G. Biochemistry. 1982;21:6531–6536. doi: 10.1021/bi00268a032. [DOI] [PubMed] [Google Scholar]
- 13.Ottleben H, Haasemann M, Ramachandran R, Gorlach M, Muller-Esterl W, Brown LR. Eur J Biochem. 1997;244:471–478. doi: 10.1111/j.1432-1033.1997.00471.x. [DOI] [PubMed] [Google Scholar]
- 14.Lopez JJ, Shukla AK, Reinhart C, Schwalbe H, Michel H, Glaubitz C. Angew Chem Int Ed. 2008;47:1668–1671. doi: 10.1002/anie.200704282. [DOI] [PubMed] [Google Scholar]
- 15.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. J Am Chem Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong H, Carlson HA. J Chem Theory Comput. 2006;2:342–353. doi: 10.1021/ct050182t. [DOI] [PubMed] [Google Scholar]
- 17.Vitagliano L, Berisio R, Mastrangelo A, Mazzarella L, Zagari A. Protein Sci. 2001;10:2627–2632. doi: 10.1110/ps.ps.26601a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abello N, Kerstjens HA, Postma DS, Bischoff R. J Proteome Res. 2007;6:4770–4776. doi: 10.1021/pr070154e. [DOI] [PubMed] [Google Scholar]
- 19.Ma M, Kutz-Naber KK, Li L. Anal Chem. 2007;79:673–681. doi: 10.1021/ac061536r. [DOI] [PubMed] [Google Scholar]
- 20.Hoaglund-Hyzer CS, Li J, Clemmer DE. Anal Chem. 2000;72:2737–2740. doi: 10.1021/ac0000170. [DOI] [PubMed] [Google Scholar]
- 21.Fenn LS, Mclean JA. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 22.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Int J Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 23.Tang K, Shvartsburg AA, Lee HN, Prior DC, Buschbach MA, Li FM, Tolmachev AV, Anderson GA, Smith RD. Anal Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu Rev Anal Chem. 2008;1(10):1–10. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 26.Shvartsburg AA, Jarrold MF. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 27.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem. 1996;100:16082–86. [Google Scholar]
- 28.Wyttenbach T, von Helden G, Batka JJ, Carlat D, Bowers MT. J Am Chem Soc. 1997;8:275–82. [Google Scholar]
- 29.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 30.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth H, Smith R, Clemmer DE. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 31.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Wiley; New York: 1988. [Google Scholar]
- 32.Valentine SJ, Counterman AE, Clemmer DE. J Am Soc Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 33.Srebalus-Barnes CA, Clemmer DE. J Phys Chem A. 2003;107:10566–10579. [Google Scholar]
- 34.Freitas MA, Hendrickson CL, Emmett MR, Marshall AG. J Am Soc Mass Spectrom. 1998;9:1012–1019. doi: 10.1016/S1044-0305(97)00287-0. [DOI] [PubMed] [Google Scholar]
- 35.Green MK, Lebrilla CB. Int J Mass Spectrom Ion Proc. 1998;175:15–26. [Google Scholar]
- 36.Schaaff TG, Stephenson JL, McLuckey SA. J Am Soc Mass Spectrom. 2000;11:167–171. doi: 10.1016/S1044-0305(99)00137-3. [DOI] [PubMed] [Google Scholar]
- 37.Mao D, Douglas DJ. J Am Soc Mass Spectrom. 2003;14:85–94. doi: 10.1016/S1044-0305(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 38.Schaaff TG, Stephenson JL, McLuckey SA. J Am Chem Soc. 1999;121:8907–8919. [Google Scholar]
- 39.Schnier PD, Price WD, Jockusch RA, Williams ER. J Am Chem Soc. 1996;118:7178–7189. doi: 10.1021/ja9609157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butcher DJ, Asano KG, Goeringer DE, McLuckey SA. J Phys Chem A. 1999;103:8664–8671. [Google Scholar]
- 41.Papadopoulos G, Svendsen A, Boyarkin OV, Rizzo TR. J Am Soc Mass Spectrom. 2012;23:1173–1181. doi: 10.1007/s13361-012-0384-0. [DOI] [PubMed] [Google Scholar]
- 42.Wyttenbach T, von Helden G, Bowers MT. J Am Chem Soc. 1996;118:8355–8364. [Google Scholar]
- 43.Wyttenbach T, Kemper PR, Bowers MT. Int J Mass Spectrom. 2001;212:13–23. [Google Scholar]
- 44.Guo Y, Wang J, Javahery G, Thomson BA, Siu KWM. Anal Chem. 2005;77:266–275. doi: 10.1021/ac048974n. [DOI] [PubMed] [Google Scholar]
- 45.Shvartsburg AA, Danielson WF, Smith RD. Anal Chem. 2010;82:2456–2462. doi: 10.1021/ac902852a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierson NA, Valentine SJ, Clemmer DE. J Phys Chem B. 2010;114:7777–7783. doi: 10.1021/jp102478k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Counterman AE, Valentine SJ, Srebalus CA, Henderson SC, Hoaglund CS, Clemmer DE. J Am Soc Mass Spectrom. 1998;9:743–759. doi: 10.1016/S1044-0305(98)00052-X. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez CF, Orlova G, Guo Y, Li X, Siu CK, Hopkinson AC, Siu KWM. J Phys Chem B. 2006;110:7528–7537. doi: 10.1021/jp046015r. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Gao J. J Am Chem Soc. 1988;110:4212–4216. [Google Scholar]
- 50.Stewart DE, Sarkar A, Wampler JE. J Mol Biol. 1990;214:253–260. doi: 10.1016/0022-2836(90)90159-J. [DOI] [PubMed] [Google Scholar]
- 51.Ramachandran GN, Sasisekaran V. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 52.Calvo F, Chirot F, Albrieux F, Lemoine J, Tsybin YO, Pernot P, Dugourd P. J Am Soc Mass Spectrom. 2012;23:1279–1288. doi: 10.1007/s13361-012-0391-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.