Abstract
Neoplastic populations with stem cell potential have been most recently identified in human cutaneous melanoma, and initially characterized for their phenotypic profile. Being melanoma stem cells (MSC) the most desirable target of therapeutic intervention, we asked whether they express the epigenetically-regulated Cancer Testis Antigens (CTA) on which melanoma immunotherapy is increasingly focusing. Reverse transcription-PCR analyses identified the presence of the large majority of investigated CTA (i.e., MAGE, GAGE, NY-ESO and SSX families) in different MSC populations. MSC expressed MAGE-A proteins as detected by western blot; noteworthy, the distribution of MAGE-A proteins was highly homogeneous within given MSC populations as shown by confocal immunofluorescence. Promoter methylation studies unveiled a homogeneously-demethylated MAGE-A3 promoter that paired MAGE-A3 expression in MSC. Altogether these findings demonstrate that MSC can be efficiently targeted by CTA-directed immunotherapeutic approaches, and suggest that epigenetic patterns most likely drive the expression of CTA in MSC as previously shown for melanoma cells.
Keywords: cancer stem cells, melanoma, immunotherapy, DNA methylation, cancer testis antigens
Introduction
Cancer stem cells (CSC), considered to be tumor-sustaining cells, have been identified in several human malignancies including breast and brain tumors, and leukemias (Bonnet and Dick, 1997;Al-Hajj et al, 2003;Singh et al, 2004). CSC share biological properties with stem cells, being able to self-renew, differentiate into distinct progenies, and drive continuous tumor growth (Reya et al, 2001). Most recently, a multipotent stem cell-like neoplastic population, that propagates under appropriate culture conditions as non-adherent spheroids, has been isolated also from human melanomas both in vivo and in vitro (Fang et al, 2005). Based on their self-renewing, multi-lineage differentiation and highly tumorigenic properties, these “putative” melanoma stem cells (MSC) have been suggested to represent the ultimate target for the most effective treatment of melanoma patients (Fang et al, 2005). However, the initial phenotypic characterization of MSC has revealed that they differentially express melanoma markers (i.e., GD2, CSPG, MCAM and p70NGFR) as compared to their adherently growing, more differentiated, and less tumorigenic counterpart (Fang et al, 2005).
Targeting of MSC may indeed represent a novel and eventually more effective clinical approach for human melanoma; however, their specific antigenic profile may hamper the clinical outcome of immunotherapeutic strategies for these patients. In this context, we presently do not know whether and to what extent MSC express specific tumor-associated antigens (TAA) currently utilized as therapeutic targets in the clinical setting. This aspect is crucial for the ultimate effectiveness of treatment, and broadly applies to any TAA presently utilized in the melanoma clinic. The lack of the appropriate therapeutic targets on MSC might in fact result in the long-term resistance to treatment, sustained by the outgrowth of TAA-negative melanoma cell clones. Indeed, the emergence of TAA-negative cells able to evade immune control has been reported in melanoma patients undergoing immunologic treatments (Jager et al, 1997). Although immune escape of neoplastic cells represents a more general drawback in cancer immunotherapy, it is even more relevant in human melanoma since novel and most promising immunotherapeutic strategies are frequently undergoing clinical testing firstly in melanoma patients (Parmiani et al, 2003). Furthermore, the profound involvement of the immune system and of its therapeutic manipulation in contributing to the control of melanoma progression is a long-standing notion (Parmiani et al, 2003).
Cancer Testis Antigens (CTA), encompassing large families of methylation-regulated and strictly related TAA, first identified in human melanoma (Sigalotti et al, 2002;Zendman et al, 2003), represent the most promising immunotherapeutic targets currently utilized in melanoma patients. CTA are suitable targets due to their constitutive cellular and humoral immunogenicity, and to their expression being shared by neoplasms of different histotypes and restricted to tumor tissues, with the exception of testis and placenta (Zendman et al, 2003). Based on these notions, we asked the question whether MCS do, and to what extent, express CTA and if epigenetic patterns are involved in their expression by MSC.
Materials and Methods
Melanoma cell cultures
Cultures of NA and A melanoma cells were established as previously described (Fang et al, 2005).
Reverse Transcription-PCR Analysis
Total RNA extraction and reverse transcription (RT)-PCR reactions were done as previously described (Coral et al, 1999). The oligonucleotide primer sequences and gene-specific PCR amplification programs used are reported in the Table 1. The integrity of each RNA and oligodeoxythymidylic acid-synthesized cDNA sample was confirmed by the amplification of the β-actin housekeeping gene (Coral et al, 1999). Ten µl of each RT-PCR sample were run on a 2% agarose gel and visualized by ethidium bromide staining. The level of expression of each gene was scored accordingly to the intensity of the specific RT-PCR product, which was obtained by densitometric analysis of ethidium bromide-stained agarose gels using a Gel Doc 2000 documentation system and the QuantityOne densitometric analysis software (Bio-Rad, Milan, Italy). Samples were scored -, no RT-PCR product detectable; +, expression level ≤ 10% to that of the appropriate reference cell line; and ++, expression level > 10% to that of the appropriate reference cell line.
Table 1.
Primers and thermal protocols utilized for the RT-PCR evaluation of CTA and differentiation antigens expression in NA and A human melanoma cells.
| RT-PCR primers | ||||
|---|---|---|---|---|
| Gene | Sequence | Amplicon length |
Thermal cycle | |
| MAGE-A1 | Sense | CGGCCGAAGGAACCTGACCCAG | 421 bp | 30 cycles of 94°C 1 min, 72°C 3 min |
| Antisense | GCTGGAACCCTCACTGGGTTGCC | |||
| MAGE-A2 | Sense | AAGTAGGACCCGAGGCACTG | 230 bp | 30 cycles of 94°C 1 min, 67°C 2 min, 72°C 2min |
| Antisense | GAAGAGGAAGAAGCGGTCTG | |||
| MAGE-A3 | Sense | TGGAGGACCAGAGGCCCCC | 725 bp | 30 cycles of 94°C 1 min, 72°C 4 min |
| Antisense | GGACGATTATCAGGAGGCCTGC | |||
| MAGE-A4 | Sense | GAGCAGACAGGCCAACCG | 446 bp | 30 cycles of 94°C 1 min, 68°C 2 min, 72°C 2 min |
| Antisense | AAGGACTCTGCGTCAGGC | |||
| MAGE-A6 | Sense | TGGAGGACCAGAGGCCCCC | 727 bp | 30 cycles of 94°C 1 min, 71°C 2 min, 72° 3 min |
| Antisense | CAGGATGATTATCAGGAAGCCTGT | |||
| MAGE-A10 | Sense | AGCAGCCAAAAGGAGGAGAGTC | 409 bp | 30 cycles of 95°C 1 min, 65°C 2 min, 72°C 2 min |
| Antisense | TGACCTCCTCAGGGGTGCAGTA | |||
| GAGE 1–2 | Sense | GACCAAGACGCTACGTAG | 201 bp | 30 cycles of 94°C 1min, 55°C 2 min, 72° 3 min |
| Antisense | CCATCAGGACCATCTTCA | |||
| GAGE 1–6 | Sense | GCGGCCCGAGCAGTTCA | 239 bp | 30 cycles of 94°C 1 min, 56°C 2 min, 72° 3 min |
| Antisense | CCATCAGGACCATCTTCA | |||
| NY-ESO-1 | Sense | CACACAGGATCCATGGATGCTGCAGATGCGG | 379 bp | 35 cycles of 94°C 1 min, 59°C 1 min, 72°C 1 min |
| Antisense | CACACAAAGCTTGGCTTAGCGCCTCTGCCCTG | |||
| LAGE-1 | Sense | GCAGGATGGAAGGTGCCC | 399bp, 628bp | 30 cycles of 95°C 30 s, 62°C 1 min, 72 °C 2 min |
| Antisense | CTGGCCACTCGTGCTGGGA | |||
| SSX 1–5 | Sense | ACGGATCCCGTGCCATGAACGGAGACGAC | 663 bp | 35 cycles of 95°C 1 min, 67°C 1 min, 72° 2 min |
| Antisense | TTGTCGACAGCCATGCCCATGTTCGTGA | |||
| PRAME | Sense | CTGTACTCATTTCCAGAGCCAGA | 562 bp | 30 cycles of 94°C 1 min, 63°C 2 min, 72°C 3 min |
| Antisense | TATTGAGAGGGGTTTCCAAGGGGTT | |||
| Tyrosinase | Sense | TTGGCAGATTGTCTGTAGCC | 284 bp | 36 cycles of 94°C 30 s, 60°C 1 min, 72°C 1 min |
| Antisense | AGGCATTGTGCATGCTGCTT | |||
| MART-1 | Sense | CTGACCCTACAAGATGCCAAGAG | 602 bp | 24 cycles of 94°C 1 min, 60°C 1 min, 72°C 1 min |
| Antisense | ATCATGCATTGCAACATTTATTGATGGAG | |||
| GP100 | Sense | TATTGAAAGTGCCGAGATCC | 361 bp | 30 cycles of 94°C 1 min, 60°C 30 s, 72°C 1 min |
| Antisense | TGCAAGGACCACAGCCATC | |||
| β-actin | Sense | GGC ATC GTG ATG GAC TCC G | 615 bp | 21 cycles of 94°C 1 min, 68°C 2 min, 72°C 2 min |
| Antisense | GCT GGA AGG TGG ACA GCG A | |||
Real-time quantitative RT-PCR analysis
Real-time quantitative RT-PCR analyses were performed as described (Calabro et al, 2005). Briefly, total RNA was digested with RNAse free DNAse (Roche Diagnostics, Milan, Italy) to remove contaminating genomic DNA. Synthesis of cDNA was performed on 1 µg total RNA using MMLV reverse transcriptase (Invitrogen, Milan, Italy) and random hexamer primers (Promega, Milan, Italy), following manufacturers’ instructions. TaqMan quantitative PCR reactions were performed on 20 ng retrotranscribed total RNA in a final volume of 25 µl 1× TaqMan Universal Master Mix (Applyed Biosystems, Milan, Italy). TaqMan primers/probe sets were as follows: β-actin forward, CGAGCGCGGCTACAGCTT; β-actin reverse, CCTTAATGTCACGCACGATT; β-actin probe, FAM-ACCACCACGGCCGAGCGG-BHQ1; MAGE-A3 forward, TGTCGTCGGAAATTGGCAGTAT, MAGE-A3 reverse, CAAAGACCAGCTGCAAGGAACT; MAGE-A3 probe, FAM-TCTTTCCTGTGATCTTC-MGB. Real-time measurement of fluorescent signals was performed utilizing the ABI PRISM 7000 Sequence Detection System (Applyed Biosystems) and the copy number of MAGE-A3 and of the reference gene β-actin was established in each sample by extrapolation of a standard curve. The number of MAGE-A3 cDNA molecules in each sample was then normalized to the number of cDNA molecules of β-actin.
SDS-PAGE and Western blotting analysis
SDS-PAGE and Western blotting were performed as described (Maio et al, 1991). Cell lysates were size-fractionated by one dimensional SDS-PAGE on 10% polyacrylamide slab gels under reducing conditions and were electroblotted onto Hybond-C nitrocellulose membranes (GE Healthcare, Milan, Italy). Membranes were than incubated with 1 µg/ml anti-MAGE-A monoclonal antibody (mAb) clone 6C1 (Invitrogen), and blots were developed by the enhanced chemiluminescence technique, using the ECL kit (GE Healthcare).
Immunofluorescence staining
WM3130 A cells were seeded on 4 well glass chamber slides 24 hours prior to staining. WM3130 NA cells were pelleted, washed with HBSS and staining was performed in a 96 well plate. All cells were fixed in 4% paraformaldehyde for 20 min and blocked for 1 h in 1:50 fold dilution of goat serum in 1× PBS containing 0.1% Triton-X 100. Following blocking, cells were incubated for 1 hour with 10 µg/ml anti-MAGE-A mAb clone 6C1 (Invitrogen). As a control, cells were incubated with mouse IgG. After washing, cells were incubated for 1 h with 1:600 fold dilution of goat anti-mouse Alexafluor 488 secondary antibody (Invitrogen), and nuclei were stained with 1:1000 fold dilution of Hoechst 03323. Cells were than washed and mounted with Fluoromount G. All photographs were taken at 20× magnification.
Sodium Bisulfite Genomic DNA Modification and DNA Sequencing
The bisulfite reaction, converting all unmethylated but not methylated cytosines to uracil, was carried out as previously described (Rein et al, 1997). For the analysis of MAGE-A3 promoter methylation status, bisulfite-modified genomic DNA was PCR amplified using 50 pmol. each of sense (5’-GGT AGA ATT TAG TTT TAT TTT TGT T-3’) and antisense (5’-AAC CTA AAA ATC TTC CCC TAC-3’) primer in a total reaction mixture of 50 µl containing 1× PCR buffer, 1.5 mM MgCl2 and 1.25 U Taq DNA polymerase (Takara Bio Inc., Otsu, Japan). PCR thermal amplification protocol consisted of an initial denaturation step of 5 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, and 1 min at 72°C. Amplification products were directly cloned into the pCR2.1 plasmid vector using the TOPO TA kit (Invitrogen). Plasmid DNA from individual colonies was extracted by the NucleoSpin Plasmid kit (Macherey-Nagel, Düren, Germany) and sequenced by the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Results represent ten independent DNA clones.
Results and Discussion
Non adherent (NA) WM115 MSC, derived from the human melanoma cell line WM115, were previously described and functionally compared to their adherent (A) counterpart (Fang et al, 2005). Additional NA and A pairs were generated from the human melanoma cell lines WM1972, WM1976 and WM3130 utilizing identical experimental procedures (Fang et al, 2005). All pairs showed the biologic characteristics previously described for the WM115-derived pair (data not shown).
RT-PCR was utilized to investigate and compare the expression of a large panel of CTA in NA and A pairs from WM115, WM1972, WM1976 and WM3130 melanoma cells. Among investigated CTA, those that are currently utilized as therapeutic targets in melanoma and in other solid malignancies (Parmiani et al, 2003) were included. A striking overlap in the expression of the large majority of investigated CTA was identified between matched NA and A cells (Table 2). With the exception of SSX 1–5 for the WM1976 pair, investigated CTA were more frequently detectable in NA as compared to A cells (Table 2). Noteworthy, also the melanocytic differentiation antigens tyrosinase, MART-1 and gp100 were expressed in both NA and A cells in all pairs analyzed (Table 2).
Table 2.
RT-PCR analysis of CTA and differentiation antigens expression in NA and A human melanoma cells*.
| WM115 |
WM1972 |
WM1976 |
WM3130 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| NA | A | NA | A | NA | A | NA | A | ||
| CTA | MAGE-A1 | ++† | ++ | + | ++ | + | + | ++ | ++ |
| MAGE-A2 | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| MAGE-A3 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| MAGE-A4 | ++ | ++ | ++ | − | − | − | ++ | ++ | |
| MAGE-A6 | ++ | ++ | ++ | ++ | ++ | ++ | + | + | |
| MAGE-A10 | − | − | ++ | − | + | + | ++ | + | |
| GAGE 1–2 | ++ | ++ | ++ | + | + | − | ++ | ++ | |
| GAGE 1–6 | ++ | ++ | ++ | + | ++ | + | ++ | ++ | |
| NY-ESO-1 | ++ | ++ | ++ | ++ | + | + | ++ | − | |
| LAGE-1 | + | + | ++ | + | + | + | − | − | |
| SSX 1–5 | ++ | + | ++ | ++ | − | ++ | + | − | |
| PRAME | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| differentiation antigens | Tyrosinase | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| MART-1 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| Gp100 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
Total RNA was extracted from NA and A human melanoma cells and subjected to RT-PCR using gene-specific primers. RNA integrity and cDNA quality have been confirmed by amplification of the house-keeping gene β-actin.
Intensity of RT-PCR products: -, not detectable; +, weak; ++, strong.
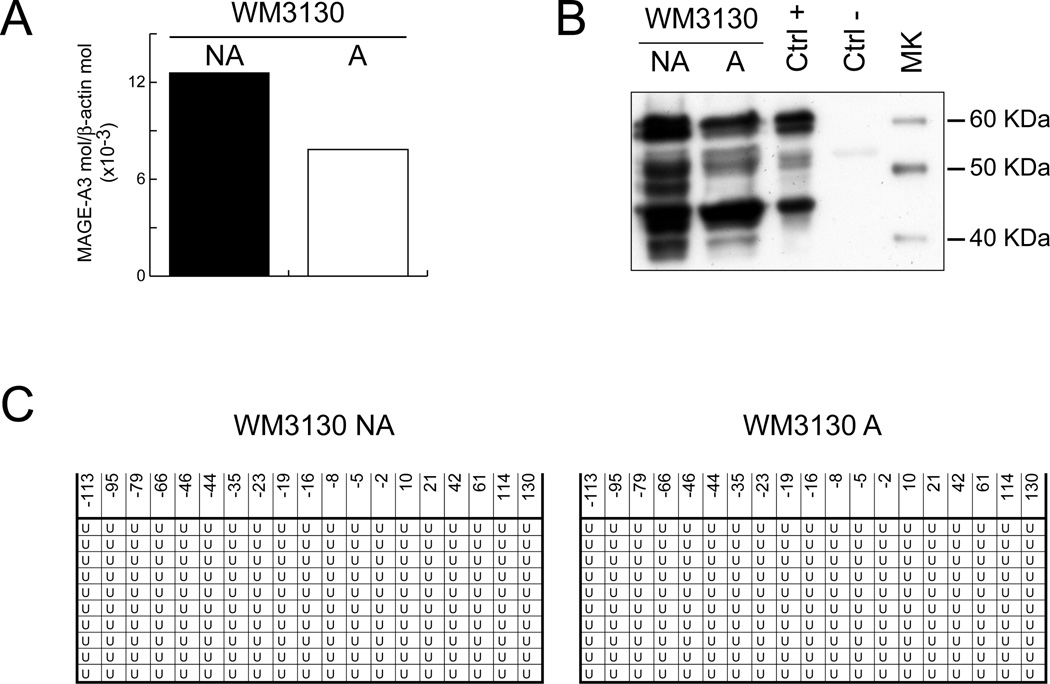
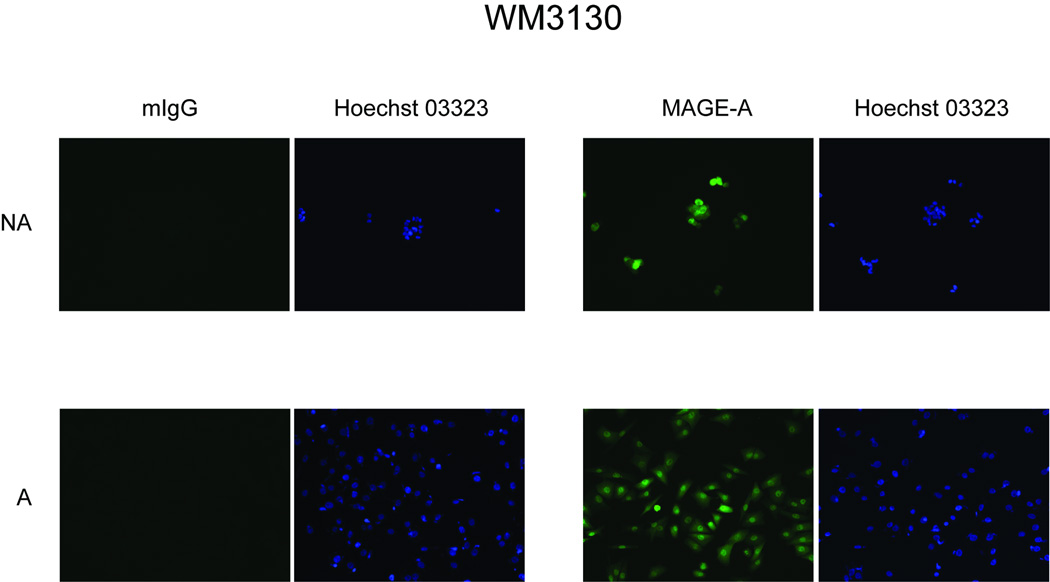
Among CTA analyzed, MAGE-A3 undoubtly represents the prototypic therapeutic CTA for cancer immunotherapy, owing to its frequent use as molecular target in ongoing clinical trials (Parmiani et al, 2003). Therefore, its molecular levels of expression were further characterized in selected NA and A pairs. Quantitative RT-PCR assays identified comparable levels of MAGE-A3 expression between NA (1.26×10−2) and A (7.84×10−3) cells derived from WM3130 melanoma cells (Fig. 1A). Furthermore, an identical pattern of immunoreactivity, compatible with the known apparent molecular weights of the MAGE-A proteins (45 to 75 KDa), was observed by Western blot analysis of NA and A cell lysates from WM3130 cells (Fig. 1B). Additionally, confocal immunofluorescence analyses demonstrated a highly homogeneous distribution of MAGE-A proteins within NA and A WM3130 cells populations (Fig 2). Consistent with the well-established role of DNA methylation in regulating presence and levels of CTA expression in human melanoma (Sigalotti et al, 2002;De Smet et al, 1996;Sigalotti et al, 2004), sequencing of bisulfite-modified genomic DNA demonstrated that MAGE-A3 promoter was fully unmethylated also in NA and A WM3130 cells (Fig. 1C).
Figure 1. MAGE-A3 expression and methylation status of its promoter in NA and A WM3130 melanoma cells.
(A) Total RNA was extracted from NA and A WM3130 melanoma cells, subjected to RT and utilized in real-time quantitative PCR with MAGE-A3- and β-actin-specific TaqMan primer/probe sets. MAGE-A3 expression was normalized to the expression of the housekeeping gene β-actin and data are reported as MAGE-A3 molecules/β-actin molecules. (B) Cell lysates from NA and A WM3130 melanoma cells, from Mel 275 melanoma cells employed as positive control (Ctrl +) and from peripheral blood mononuclear cells used as negative control (Ctrl -) were size-fractionated by a 10% one-dimensional SDS-PAGE under reducing conditions and blotted onto Hybond-C super-transfer nitrocellulose membranes. Membranes were incubated with 1 µg/ml anti-MAGE-A mAb clone 6C1 and were developed by the enhanced chemiluminescence technique. (C) Genomic DNA extracted from NA and A WM3130 melanoma cells was subjected to modification with sodium bisulfite that converts all the un-methylated cytosines to uracil leaving unaltered the methylated ones. MAGE-A3 promoter was PCR amplified from bisulfite-converted genomic DNA and cloned into the pCR2.1 plasmid. Ten individual colonies from each of NA and A WM3130 cells were sequenced and examined for methylation in the CpG dinucleotides contained in the amplified region. U represents unmethylated CpG dinucleotides. The position of CpG dinucleotides is reported with respect to the transcription start site (+1).
Figure 2. Immunofluorescence analysis of MAGE-A antigens expression in NA and A WM3130 melanoma cells.
NA and A WM3130 cells were fixed in 4% paraformaldehyde, blocked in 1:50 fold dilution of goat serum in 1× PBS/0.1% Triton-X 100, and incubated with the anti-MAGE-A mAb clone 6C1 or with mouse IgG (mIgG), as a control. After washing, cells were incubated with goat anti-mouse Alexafluor 488 secondary antibody, and nuclei were stained with Hoechst 03323. Photographs were taken at 20× magnification.
Besides providing further insights on the phenotypic profile of human MSC and on their potential susceptibility to epigenetic modeling, the consistent expression of therapeutic CTA in MSC herein reported bears relevant practical implications from the clinical viewpoint; among these: i, the presence and homogeneous distribution of CTA in MSC allows, in principle, for their therapeutic targeting in the course of CTA-based immunotherapy; ii, the concomitant expression of several CTA in MSC provides multiple molecules to be targeted by multivalent CTA-based immunotherapeutic strategies and possibly avoids the emergence of CTA-negative clones in the course of treatment; iii, the routine assessment of the biological eligibility of melanoma patients to CTA-based immunotherapeutic strategies can be done on whole melanoma lesions since they mirror the pattern of CTA expression of their stem cell population.
CSC are currently being proposed as the ultimate target population for therapeutic interventions. However, studies on their phenotypic and biomolecular characteristics, as well as on their susceptibility to conventional and experimental treatments, are still in their infancy. Along this line, ongoing studies on CSC most likely deal with highly enriched CSC rather than with pure CSC populations. Nevertheless, our first time identification in MSC of a homogeneous CTA expression, which is also inherited by their cellular progenies, contributes to further pave the way to novel immunotherapeutic strategies that will comprehensively target also MSC within the tumor mass. Whether these findings and their clinical implications will apply to other solid and hemopoietic human malignancies remains to be fully investigated.
Acknowledgments
Grant support:
Contract grant sponsor (M.M. and PG.N): Associazione Italiana per la Ricerca sul Cancro.
Contract grant sponsor (M.M. and PG.N): Lega Italiana per la Lotta contro i Tumori
Contract grant sponsor (M.H.): US National Institutes of Health; Contract grant number: CA 076674.
Contract grant sponsor (M.H.): US National Institutes of Health; Contract grant number: CA 25874.
Literature Cited
- Al-Hajj M, Wicha MS, ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Calabro L, Fonsatti E, Altomonte M, Pezzani L, Colizzi F, Nanni P, Gattei V, Sigalotti L, Maio M. Methylation-regulated expression of cancer testis antigens in primary effusion lymphoma: immunotherapeutic implications. J Cell Physiol. 2005;202:474–477. doi: 10.1002/jcp.20133. [DOI] [PubMed] [Google Scholar]
- Coral S, Sigalotti L, Gasparollo A, Cattarossi I, Visintin A, Cattelan A, Altomonte M, Maio M. Prolonged upregulation of the expression of HLA class I antigens and costimulatory molecules on melanoma cells treated with 5-aza-2'-deoxycytidine (5-AZA-CdR) J Immunother. 1999;22:16–24. doi: 10.1097/00002371-199901000-00003. [DOI] [PubMed] [Google Scholar]
- De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Jager E, Ringhoffer M, Altmannsberger M, Arand M, Karbach J, Jager D, Oesch F, Knuth A. Immunoselection in vivo: independent loss of MHC class I and melanocyte differentiation antigen expression in metastatic melanoma. Int J Cancer. 1997;71:142–147. doi: 10.1002/(sici)1097-0215(19970410)71:2<142::aid-ijc3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Maio M, Altomonte M, Tatake R, Zeff RA, Ferrone S. Reduction in susceptibility to natural killer cell-mediated lysis of human FO-1 melanoma cells after induction of HLA class I antigen expression by transfection with B2m gene. J Clin Invest. 1991;88:282–289. doi: 10.1172/JCI115289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmiani G, Castelli C, Rivoltini L, Casati C, Tully GA, Novellino L, Patuzzo A, Tosi D, Anichini A, Santinami M. Immunotherapy of melanoma. Semin Cancer Biol. 2003;13:391–400. doi: 10.1016/j.semcancer.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Rein T, Zorbas H, DePamphilis ML. Active mammalian replication origins are associated with a high-density cluster of mCpG dinucleotides. Mol Cell Biol. 1997;17:416–426. doi: 10.1128/mcb.17.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Sigalotti L, Coral S, Nardi G, Spessotto A, Cortini E, Cattarossi I, Colizzi F, Altomonte M, Maio M. Promoter methylation controls the expression of MAGE2, 3 and 4 genes in human cutaneous melanoma. J Immunother. 2002;25:16–26. doi: 10.1097/00002371-200201000-00002. [DOI] [PubMed] [Google Scholar]
- Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res. 2004;64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Zendman AJ, Ruiter DJ, van Muijen GN. Cancer/testis-associated genes: identification, expression profile, and putative function. J Cell Physiol. 2003;194:272–288. doi: 10.1002/jcp.10215. [DOI] [PubMed] [Google Scholar]