Abstract
The Schlafen (SLFN) family of proteins includes several mouse and human members. There is emerging evidence that members of this family of proteins are involved in important functions, such as the control of cell proliferation, induction of immune responses, and the regulation of viral replication. These proteins span across all species with great diversity, with 10 murine and 5 human isoforms. Recent work has established that mouse and human SLFN proteins are regulated by interferons (IFNs). Several Slfn genes were shown to be induced as classical interferon-stimulated genes, and emerging evidence suggests that these proteins play important roles in the growth inhibitory and antineoplastic effects of IFNs. In the current review, the known properties of mouse and human SLFNs are reviewed, and the implications of their emerging functions are discussed.
Introduction
The family of Schlafen (Slfn) genes (from the word schlafen, which in German means sleeping) includes several mouse and human member genes that have been implicated in the regulation of important biological functions in mammals. Originally, 4 Slfns (mouse Slfn1, Slfn2, Slfn3, and Slfn4) were identified in the mouse by the group of Steven Hedrick at the University of California at San Diego (Schwarz and others 1998). At that time, it was shown that these genes are differentially expressed during T-cell activation and thymocyte maturation (Schwarz and others 1998). Slfn 1, 2, and 4 were found to be expressed primarily in the thymus, spleen, and lymph nodes, while Slfn3 was detected primarily in the testis, with low expression in the thymus and heart (Schwarz and others 1998). Subsequent studies revealed that this multigene family is comprised of 3 groups, which can be classified among, other things, based on the size of the encoded proteins (Geserick and others 2004; Brady and others 2005; Bell and others 2006; Sohn and others 2007; Neumann and others 2008). Group I SLFNs includes proteins whose molecular masses range from 37 to 42 kDa; group II includes SLFN members with molecular masses between 58 and 68 kDa, while the molecular masses of group III members range from 100 to 104 kDa (Geserick and others 2004; Neumann and others 2008).
All SLFNs contain a Slfn box, a domain that is not found in other proteins and whose function is not well defined at this point (Geserick and others 2004; Neumann and others 2008). This motif lies near an AAA domain in SLFNs (Geserick and others 2004; Brady and others 2005; Neumann and others 2008). Although one could speculate that the AAA domain in SLFNs may function similarly to other AAA domains involved in GTP/ATP binding (Brady and others 2005; Neumann and others 2008), this remains to be directly studied. Another SLFN-specific domain selectively expressed in groups II and III, called the SWADL domain, is defined by the sequence Ser-Trp-Ala-Asp-Leu (Geserick and others 2004; Neumann and others 2008). A C-terminal extension exists only in group III SLFNs and is characterized by a motif that is homologous to the superfamily I of RNA helicases (Geserick and others 2004). Altogether, there is evidence that SFLN protein members have domains that are involved in RNA metabolism or RNA structure-modeling activity (Geserick and others 2004). Furthermore, these extensions contain a nuclear localization signal (RKRRR), further indicating that group III Slfns are involved in nuclear mechanisms (Neumann and others 2008). While the function of most Slfns is not yet known, several studies have implicated some members of this family of proteins in specific cellular functions, as discussed below.
Mouse Slfns and Their Functions
There are 10 known or predicted mouse Slfn genes (Slfn1, 1L, 2, 3, 4, 5, 8, 9, 10, and 14) (Ferguson and others 2005; Neumann and others 2008; Bustos and others 2009). These genes are distributed among the 3 mouse Slfn groups. Group I consists of Slfn1 and Slfn2, which are the shortest Slfns (37–42 kDa), while group II includes Slfn3 and Slfn4, whose sizes range between 58 and 68 kDa (Bustos and others 2009). Finally, group III includes the large Slfns (Slfn5, Slfn8, Slfn9, Slfn10, and Slfn14), which have a molecular mass of ∼100 kDa, because they contain the C-terminal extension (Neumann and others 2008; Bustos and others 2009). From the 10 mouse Slfns, only 3 have been studied extensively: Slfn1, Slfn2, and Slfn3 (Sohn and others 2007; Lee and others 2008; Katsoulidis and others 2009; Patel and others 2009a; Brady and others 2005; Walsh and others 2012; Patel and others 2009b; Berger and others 2010; Condamine and others 2010; Horton and Powell 2010; Yuan and others 2010; Ahmadi and Veinotte 2011; Oh and others 2011). Slfn2 has been implicated in a variety of functions, such as contributing to an immune response, to differentiation, and to cell growth. In particular, one Slfn2 mutation (elektra mutation) renders mice more susceptible to both bacterial and viral infections (Berger and others 2010). This Slfn2-associated immune defect likely reflects the fact that T cells expressing this mutation (Slfn2-I135N) undergo apoptosis via the intrinsic pathway, in response to activation or expansion stimuli (Berger and others 2010). Other studies have shown that Slfn2 is induced by RANKL, a member of the tumor necrosis factor family (Lee and others 2008). Knockdown of mouse Slfn2 using siRNAs was shown to decrease the activation of c-Jun and expression of NFATc1, resulting in decreased osteoclastogenesis (Lee and others 2008). Based on this study, it appears that Slfn2 can function as a potent inducer of osteoclastogenesis (Lee and others 2008). Notably, a mechanism for transcriptional induction of the Slfn2 gene in response to LPS has been described, involving an NF-kB- and AP-1-dependent mechanism (Sohn and others 2007).
Beyond these functions, there is evidence that Slfn2 is also involved in the control of malignant cell proliferation (Katsoulidis and others 2009). It has been shown that engagement of the mouse Type I interferon (IFN) receptor results in upregulation of Slfn2 in a Stat- and p38-MAPK-dependent manner, ultimately resulting in suppression of cyclin D1 expression and inhibition of malignant cell proliferation (Katsoulidis and others 2009). Furthermore, Slfn2 knockdown in that context was shown to lead to an increase in anchorage-independent growth (Katsoulidis and others 2009). Thus, it appears that depending on the inducing stimulus, more than one transactivation mechanism can regulate Slfn2 expression, ultimately resulting in suppression of cellular proliferation. As in the case of Slfn2, Slfn1 also appears to control cell growth by regulating the expression of cyclin D1 (Brady and others 2005). Slfn1 has also been implicated in the regulation of endothelial progenitor cell proliferation, functioning downstream of transient receptor potential canonical-1 (Kuang and others 2012).
Group II Slfns include 2 members, Slfn3 and Slfn4. Slfn3 also appears to be upregulated during T-cell activation. There is evidence that Slfn3 is expressed in CD4(+) CD25(+) circulating T cells, and that Slfn3 mRNA expression is downregulated upon activation and proliferation in CD4(+) CD25(+) Tregs (Condamine and others 2010). These findings suggest a possible role for this Slfn family member in regulation of the immune response by controlling T-cell activation and differentiation. In the intestinal mucosa, Slfn3 plays an important role in regulating intestinal differentiation (Patel and others 2009b; Yuan and others 2010) and is involved in intestinal development and maturation (Walsh and others 2012). As with group I Slfns, Slfn3 exhibits antiproliferative properties, as evidenced by the fact that ectopic expression of Slfn3 in human colon cancer cells significantly decreased proliferation as well as the expression of proliferating cell nuclear antigen, a protein expressed during the S phase of the cell cycle and a marker of proliferation (Patel and others 2009a). Furthermore, Slfn3 expression increases the expression of p27, a negative cell cycle regulator that inhibits the expression of CDK2, further confirming that Slfn3 functions as a negative regulator or cell growth (Patel and others 2009b). Importantly, there is evidence that ectopic Slfn3 expression inhibits multiple characteristics of cancer stem cells—enriched colon cancer cells—and appears to sensitize them to reverse FOLFOX chemotherapy resistance (Oh and others 2011).
Slfn4 is a not well-characterized member of the group II Slfns. Indeed, only a single study exists to date that has investigated the function and properties of Slfn4 (van Zuylen and others 2011). Slfn4 expression was found to be induced by Toll-like Receptor agonists in mouse bone marrow-derived macrophages (van Zuylen and others 2011). Interestingly, Slfn4 mRNA expression in IFNAR-1−/− macrophages was severely decreased, suggesting autocrine IFN-β-dependent expression (van Zuylen and others 2011). This finding is consistent with another study that demonstrated induction of Slfn4 mRNA expression by exogenous Type I IFN-treatment of NIH 3T3 fibroblasts (Katsoulidis and others 2009). Slfn4 mRNA expression is downregulated during differentiation (van Zuylen and others 2011), while overexpression of Slfn4 in transgenic mice resulted in decreased monocyte numbers and inflammatory macrophages in the peritoneal cavity of these mice (van Zuylen and others 2011). Notably, hematopoietic activity was increased in the livers and spleens of these mice (van Zuylen and others 2011), consistent with a unique regulatory role of Slfn4 in the control of myelopoiesis.
While group I and II Slfns are localized in the cytoplasm, group III Slfns contain a nuclear localization signal and are primarily localized to the nucleus (Neumann and others 2008). Even though very little is known about the function of this mouse Slfn group, there has been some suggestion based on colocalization studies that at least one member of the group, Slfn 9, colocalizes with the active/phosphorylated form of RNA polymerase II and with the SC-35 protein (Neumann and others 2008), which is a component of the splicing apparatus, suggesting a possible role for this protein in transcription and/or RNA splicing. There is evidence implicating group III mouse Slfns in hematopoietic cell differentiation (Geserick and others 2004), but among the helicase-containing Slfns, only Slfn8 affects the progression of the cell cycle of stimulated T cells, an observation that appears to be cell-type specific (Geserick and others 2004).
Human SLFNs and Their Functions
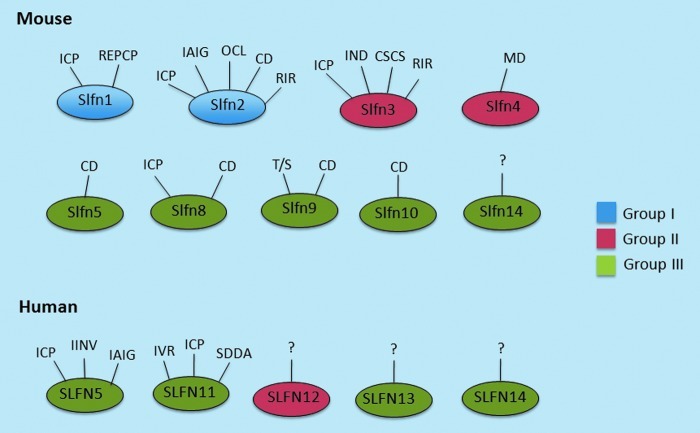
There are 5 human SLFN isoforms: SLFN5, SLFN11, SLFN12, SLFN13, and SLFN14 (Bustos and others 2009; Katsoulidis and others 2010; Li and others 2012; Razzak 2012; Barretina and others 2012; Zoppoli and others 2012). Unlike the mouse Slfns, there is only one cytoplasmic human SLFN, SLFN12, which lacks the helicase domain and belongs in the group II category (Fig. 1). The remaining human SLFNs harbor a helicase domain as well as a nuclear localization signal, placing them in group III (Fig. 1). As related to the group III human SLFNs, studies have uncovered important functions for 2 of them, SLFN5 and SLFN11. Human SLFN5 has been shown to exhibit negative regulatory effects on anchorage-independent malignant melanoma cell growth and invasion of malignant melanoma cells in collagen (Katsoulidis and others 2010). Interestingly, when the expression of SLFN5 was compared in normal melanocytes and malignant melanoma cell lines, there was suppression of SLFN5 mRNA expression in the malignant melanoma lines, suggesting selection against SLFN5 during emergence of the malignant phenotype (Katsoulidis and others 2010).
FIG. 1.
Summary of documented and/or proposed biological effects of Slfns in different systems. SFLN, Schlafen; CD, cell differentiation; CSCS, cancer stem cell sensitization; ICP, inhibition of cell proliferation; IND, induction of intestinal differentiation; IAIG, inhibition of anchorage-independent growth; IINV, inhibition of malignant cell invasion; MD, myeloid differentiation; RIR, regulation of the immune response; IVR, inhibition of viral replication; REPCP, regulation of endothelial progenitor cell proliferation; SDDA, sensitization to DNA-damaging agents; T/S, transcription/splicing. Color images available online at www.liebertpub.com/jir
There have been recent studies investigating the functional relevance of human SLFN11 (Barretina and others 2012; Li and others 2012; Razzak 2012; Zoppoli and others 2012). One study recently published in Nature demonstrated that SLFN11 specifically blocks the production of retroviruses such as HIV-1 (Li and others 2012). The mechanism by which this inhibition occurs was described, and appears to involve blocking expression of viral proteins in a codon usage-defined manner (Li and others 2012). Thus, human SLFN11 appears to play a key role in the control of HIV infection in humans, and it is possible that its selective targeting may lead to the development of new antiviral drugs.
Beyond important antiviral properties, other recent studies have demonstrated that SLFN11 has an important role in sensitizing malignant cells to topoisomerase inhibitors (Barretina and others 2012; Zoppoli and others 2012), as well as alkylating agents and other DNA-damaging agents (Zoppoli and others 2012). These findings raise the possibility of a strategy to enhance the antineoplastic effects of certain agents by developing drugs that promote SLFN11 expression. It will also be important to examine the potential involvement of other human SLFNs in the generation of responses to a variety of anticancer agents, as this may lead to the development of a novel antineoplastic agents and combination treatments.
Regulation of SLFN Proteins by IFNs
IFNs play key and essential roles in innate immunity and in immune surveillance against cancers (Stark and others 1998; Borden and others 2007). Extensive work over the years has shown that Type I IFNs bind to specific cell surface receptors and activate receptor-associated Jak kinases that engage the Stat pathways (Darnell 1997; Platanias and Fish 1999; Aaronson and Horvath 2002; Kaur and others 2005; Platanias 2005; Stark and Darnell 2012), as well as other signaling pathways downstream of Jaks, including the p38 Map kinase pathway, which is also required for optimal transcriptional activation of interferon-stimulated genes (ISGs; reviewed in Platanias and Fish 1999; Platanias 2003, 2005). There is also recent evidence that activation of the Mnk pathway downstream of Mek/Erk is important for mRNA translation of ISGs and the generation of IFN responses (Joshi and others 2009, 2011).
Recent studies established that mouse Slfns (Katsoulidis and others 2009) and human SLFNs (Katsoulidis and others 2010) are inducible during engagement of the Type I IFN receptor. Treatment of mouse cells with IFNα strongly induced mRNA expression for several Slfn proteins, including Slfn1, Slfn2, Slfn5, and Slfn8 (Katsoulidis and others 2009). Using different knockout mouse embryonic fibroblast cell lines for different signaling intermediates in IFN-signaling pathways, it was shown that Stat1 was required for induction of all IFN-inducible mouse Slfn genes, while Stat3 was required for all inducible Slfns, except Slfn5 (Katsoulidis and others 2009). Similarly, the p38 MAPK was also required, except in the cases of Slfn5 and Slfn8, which were p38 MAPK independent (Katsoulidis and others 2009). Thus, the patterns of induction of mouse Slfn genes appear to be similar to the ones seen in the case of classic ISGs. Similarly, there is evidence that human SLFNs are induced in response to engagement of the human Type I IFN receptor (Katsoulidis and others 2009, 2010). Remarkably, mRNA expression for all human SLFNs studied, including SLFN5, SLFN11, SLFN12, and SLFN13, was induced in normal melanocytes, while only SLFN5 was inducible in melanoma cell lines (Katsoulidis and others 2010). The divergent patterns of mRNA expression for different human SLFNs may reflect differences in gene transcription, and the functional relevance of such differences remains to be determined in future studies. It should also be noted that a recent study demonstrated that, as in the case of ISG15, mRNA translation of SLFN5 appears to be regulated by the effector of the mTOR/S6K pathway, PDCD4 (Kroczynska and others 2012). This suggests that mRNA translation/protein expression of SLFNs may depend on the same upstream signaling pathways that may regulate mRNA translation of classic ISGs, suggesting a similar mechanism of overall regulation (Fig. 2). Viewed altogether, there is accumulating evidence that mouse and human SLFNs have important roles in the generation of IFN-inducible responses.
FIG. 2.
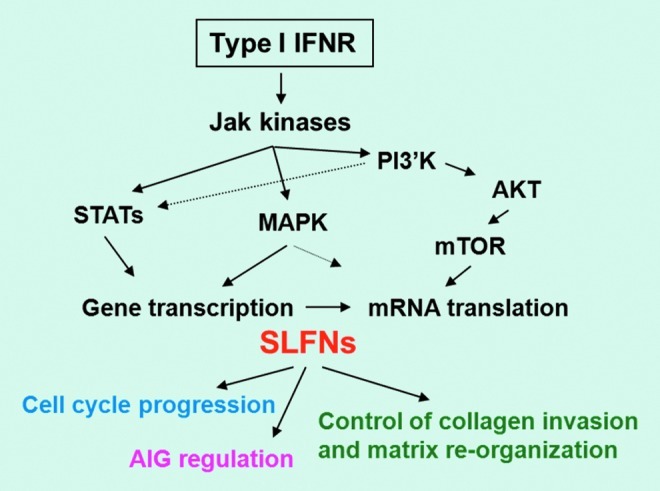
Proposed model for the regulation of SLFN functions by Type I interferons (IFNs) in malignant cells. PI 3′K, phosphatidylinositol 3′ kinase; mTOR, mammalian target of rapamycin; AIG, anchorage-independent growth; MAPK, mitogen-activated protein kinases; STAT, signal transducer and activator of transcription. Color images available online at www.liebertpub.com/jir
Acknowledgment
We thank Elisabeth Stecki for expert secretarial assistance.
Author Disclosure Statement
No competing financial interests exist.
References
- Aaronson DS. Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296(5573):1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- Ahmadi S. Veinotte LL. Effect of Schlafen 2 on natural killer and T cell development from common T/natural killer progenitors. Pak J Biol Sci. 2011;14(22):1002–1010. doi: 10.3923/pjbs.2011.1002.1010. [DOI] [PubMed] [Google Scholar]
- Barretina J. Caponigro G. Stransky N. Venkatesan K. Margolin AA. Kim S. Wilson CJ. Lehár J. Kryukov GV. Sonkin D. Reddy A. Liu M. Murray L. Berger MF. Monahan JE. Morais P. Meltzer J. Korejwa A. Jané-Valbuena J. Mapa FA. Thibault J. Bric-Furlong E. Raman P. Shipway A. Engels IH. Cheng J. Yu GK. Yu J. Aspesi P., Jr. de Silva M. Jagtap K. Jones MD. Wang L. Hatton C. Palescandolo E. Gupta S. Mahan S. Sougnez C. Onofrio RC. Liefeld T. MacConaill L. Winckler W. Reich M. Li N. Mesirov JP. Gabriel SB. Getz G. Ardlie K. Chan V. Myer VE. Weber BL. Porter J. Warmuth M. Finan P. Harris JL. Meyerson M. Golub TR. Morrissey MP. Sellers WR. Schlegel R. Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TA. de la Casa-Esperon E. Doherty HE. Ideraabdullah F. Kim K. Wang Y. Lange LA. Wilhemsen K. Lange EM. Sapienza C. de Villena FP. The paternal gene of the DDK syndrome maps to the Schlafen gene cluster on mouse chromosome 11. Genetics. 2006;172(1):411–423. doi: 10.1534/genetics.105.047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. Krebs P. Crozat K. Li X. Croker BA. Siggs OM. Popkin D. Du X. Lawson BR. Theofilopoulos AN. Xia Y. Khovananth K. Moresco EM. Satoh T. Takeuchi O. Akira S. Beutler B. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol. 2010;11(4):335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC. Sen GC. Uze G. Silverman RH. Ransohoff RM. Foster GR. Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G. Boggan L. Bowie A. O'Neill LA. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem. 2005;280(35):30723–30734. doi: 10.1074/jbc.M500435200. [DOI] [PubMed] [Google Scholar]
- Bustos O. Naik S. Ayers G. Casola C. Perez-Lamigueiro MA. Chippindale PT. Pritham EJ. de la Casa-Esperon E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene. 2009;447(1):1–11. doi: 10.1016/j.gene.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine T. Le Luduec JB. Chiffoleau E. Beriou G. Louvet C. Heslan M. Tilly G. Cuturi MC. Characterization of Schlafen-3 expression in effector and regulatory T cells. J Leukoc Biol. 2010;87(3):451–456. doi: 10.1189/jlb.0609410. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr Stats and gene regulation. Science. 1997;277(5332):1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Ferguson DA. Chiang JT. Richardson JA. Graff J. eXPRESSION: an in silico tool to predict patterns of gene expression. Gene Expr Patterns. 2005;5(5):619–628. doi: 10.1016/j.modgep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Geserick P. Kaiser F. Klemm U. Kaufmann SH. Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16(10):1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- Horton MR. Powell JD. Quieting T cells with Slfn2. Nat Immunol. 2010;11(4):281–282. doi: 10.1038/ni0410-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S. Kaur S. Redig AJ. Goldsborough K. David K. Ueda T. Watanabe-Fukunaga R. Baker DP. Fish EN. Fukunaga R. Platanias LC. Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc Natl Acad Sci U S A. 2009;106(29):12097–12102. doi: 10.1073/pnas.0900562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S. Sharma B. Kaur S. Majchrzak B. Ueda T. Fukunaga R. Verma AK. Fish EN. Platanias LC. Essential role for Mnk kinases in type II interferon (IFNgamma) signaling and its suppressive effects on normal hematopoiesis. J Biol Chem. 2011;286(8):6017–6026. doi: 10.1074/jbc.M110.197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsoulidis E. Carayol N. Woodard J. Konieczna I. Majchrzak-Kita B. Jordan A. Sassano A. Eklund EA. Fish EN. Platanias LC. Role of Schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J Biol Chem. 2009;284(37):25051–25064. doi: 10.1074/jbc.M109.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsoulidis E. Mavrommatis E. Woodard J. Shields MA. Sassano A. Carayol N. Sawicki KT. Munshi HG. Platanias LC. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285(51):40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S. Uddin S. Platanias LC. The PI3′ kinase pathway in interferon signaling. J Interferon Cytokine Res. 2005;25(12):780–787. doi: 10.1089/jir.2005.25.780. [DOI] [PubMed] [Google Scholar]
- Kroczynska B. Sharma B. Eklund EA. Fish EN. Platanias LC. Regulatory effects of programmed cell death 4 (PDCD4) protein in interferon (IFN)-stimulated gene expression and generation of type I IFN responses. Mol Cell Biol. 2012;32(14):2809–2822. doi: 10.1128/MCB.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang CY. Yu Y. Wang K. Qian DH. Den MY. Huang L. Knockdown of transient receptor potential canonical-1 reduces the proliferation and migration of endothelial progenitor cells. Stem Cells Dev. 2012;21(3):487–496. doi: 10.1089/scd.2011.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK. Choi HK. Yoo HJ. Shin J. Lee SY. RANKL-induced schlafen2 is a positive regulator of osteoclastogenesis. Cell Signal. 2008;20(12):2302–2308. doi: 10.1016/j.cellsig.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Li M. Kao E. Gao X. Sandig H. Limmer K. Pavon-Eternod M. Jones TE. Landry S. Pan T. Weitzman MD. David M. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B. Zhao L. Murphy K. Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370(1):62–66. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- Oh PS. Patel VB. Sanders MA. Kanwar SS. Yu Y. Nautiyal J. Patel BB. Majumdar AP. Schlafen-3 decreases cancer stem cell marker expression and autocrine/juxtacrine signaling in FOLFOX-resistant colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2011;301(2):G347–G355. doi: 10.1152/ajpgi.00403.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BB. Yu Y. Du J. Rishi AK. Sarkar FH. Tarca AL. Wali A. Majumdar AP. Schlafen 3, a novel gene, regulates colonic mucosal growth during aging. Am J Physiol Gastrointest Liver Physiol. 2009a;296(4):G955–G962. doi: 10.1152/ajpgi.90726.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VB. Yu Y. Das JK. Patel BB. Majumdar AP. Schlafen-3: a novel regulator of intestinal differentiation. Biochem Biophys Res Commun. 2009b;388(4):752–756. doi: 10.1016/j.bbrc.2009.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. The p38 mitogen-activated protein kinase pathway and its role in interferon signaling. Pharmacol Ther. 2003;98(2):129–142. doi: 10.1016/s0163-7258(03)00016-0. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27(11):1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- Razzak M. Genetics: Schlafen 11 naturally blocks HIV. Nat Rev Urol. 2012;9(11):605. doi: 10.1038/nrurol.2012.188. [DOI] [PubMed] [Google Scholar]
- Schwarz DA. Katayama CD. Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9(5):657–668. doi: 10.1016/s1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- Sohn WJ. Kim D. Lee KW. Kim MS. Kwon S. Lee Y. Kim DS. Kwon HJ. Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol. 2007;44(13):3273–3282. doi: 10.1016/j.molimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Stark GR. Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR. Kerr IM. Williams BR. Silverman RH. Screiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- van Zuylen WJ. Garceau V. Idris A. Schroder K. Irvine KM. Lattin JE. Ovchinnikov DA. Perkins AC. Cook AD. Hamilton JA. Hertzog PJ. Stacey KJ. Kellie S. Hume DA. Sweet MJ. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS One. 2011;6(1):e15723. doi: 10.1371/journal.pone.0015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MF. Hermann R. Sun K. Basson MD. Schlafen 3 changes during rat intestinal maturation. Am J Surg. 2012;204(5):598–601. doi: 10.1016/j.amjsurg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. Yu Y. Sanders MA. Majumdar AP. Basson MD. Schlafen 3 induction by cyclic strain regulates intestinal epithelial differentiation. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G994–G1003. doi: 10.1152/ajpgi.00517.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppoli G. Regairaz M. Leo E. Reinhold WC. Varma S. Ballestrero A. Doroshow JH. Pommier Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109(37):15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]